Abstract
Vertebrate Tpr and its yeast homologs Mlp1/Mlp2, long coiled-coil proteins of nuclear pore inner basket filaments, are involved in mRNA export, telomere organization, spindle pole assembly, and unspliced RNA retention. We identified Arabidopsis thaliana NUCLEAR PORE ANCHOR (NUA) encoding a 237-kD protein with similarity to Tpr. NUA is located at the inner surface of the nuclear envelope in interphase and in the vicinity of the spindle in prometaphase. Four T-DNA insertion lines were characterized, which comprise an allelic series of increasing severity for several correlating phenotypes, such as early flowering under short days and long days, increased abundance of SUMO conjugates, altered expression of several flowering regulators, and nuclear accumulation of poly(A)+ RNA. nua mutants phenocopy mutants of EARLY IN SHORT DAYS4 (ESD4), an Arabidopsis SUMO protease concentrated at the nuclear periphery. nua esd4 double mutants resemble nua and esd4 single mutants, suggesting that the two proteins act in the same pathway or complex, supported by yeast two-hybrid interaction. Our data indicate that NUA is a component of nuclear pore-associated steps of sumoylation and mRNA export in plants and that defects in these processes affect the signaling events of flowering time regulation and additional developmental processes.
INTRODUCTION
The nuclear pore complex (NPC) is a large multiprotein complex that is the sole gateway of macromolecular trafficking between the cytoplasm and the nucleus. The mammalian and yeast NPC consists of multiple copies of the ∼30 different nucleoporins (Nups). Together, they form a channel-like structure of eightfold symmetry that has been roughly divided into three elements: a nuclear basket, a central pore, and cytoplasmic fibrils. While a small number of Nups are anchored to the nuclear envelope membrane, others form a protein scaffold or line the central pore cylinder with FG-repeat-containing hydrophobic domains. Nuclear import and export receptors traffic through the pore bound to their cargos, and the Ran cycle provides spatial information on the directionality of the transport (reviewed in Tran and Wente, 2006).
Recently, several reports have demonstrated that Nups are involved in functions beyond being building blocks of the NPC. Some Nups are highly dynamic and appear in locations away from the pore (Griffis et al., 2002; Rabut et al., 2004). Several Nups have mitotic functions, for example, involvement in kinetochore assembly (reviewed in Chan et al., 2005). Possibly the most exciting new function of Nups is their ability to dock specific enzymatic activities to the NPC, thereby providing spatial regulation for the respective activities. An example that has been known for several years is the docking of the mammalian Ran GTPase activating protein RanGAP1 to the outer surface of the NPC by the nucleoporin RanBP2, where it hydrolyzes the RanGTP bound to export complexes (Matunis et al., 1998). Mammalian RanGAP1 requires sumoylation to bind to RanBP2, and subsequently it was found that RanBP2 itself is a SUMO E3 ligase (Matunis et al., 1998; Pichler et al., 2002).
At the nuclear side of the pore, the nuclear basket is also involved in regulating SUMO modification. The Nups Mlp1/Mlp2 in yeast (Saccharomyces cerevisiae) and Nup153 in mammals dock a SUMO protease to the NPC (Ulp1 in yeast and SENP2 in mammals) (Zhang et al., 2002; Panse et al., 2003). Mlp1 is also a docking site for heterogeneous nuclear ribonucleoproteins (hnRNPs) (Green et al., 2003), and mammalian hnRNPs have been shown to be sumoylated (Vassileva and Matunis, 2004). It has been proposed that Mlps act as a quality control checkpoint for mRNA export (Galy et al., 2004).
At the outer pore surface, another nucleoporin acts as an anchor/activator of a step in mRNA export. The nucleoporin Gle1 binds the DEAD box helicase Dbp5 and together with the soluble inositol polyphosphate InsP6 activates the ATPase activity of Dbp5. This leads to a highly localized activation of mRNA remodeling, likely involved in a release step of mRNA export (Alcazar-Roman et al., 2006; Weirich et al., 2006). Together, these examples support the emerging picture that precise spatial regulation is crucial for a number of nuclear functions and emphasize that this spatial regulation can be provided by anchoring enzymatic activities to nuclear pore proteins.
While the components of the animal and yeast NPC have been identified in proteomic studies (Rout et al., 2000; Cronshaw et al., 2002), the plant NPC is still largely a black box. Recently, putative plant Nups have been identified by genetic screens for the seemingly unrelated signaling pathways involved in plant-microbe interactions, cold tolerance, and the action of the plant hormone auxin (Zhang and Li, 2005; Dong et al., 2006; Kanamori et al., 2006; Parry et al., 2006). The identified proteins have convincing sequence similarity to Nup96, Nup160, and Nup133, which are all components of the Nup107-160 complex, suggesting its conservation during evolution. The Nup107-160 complex is believed to provide a core scaffolding function for NPC assembly in mammals (Harel et al., 2003; Walther et al., 2003). Partial in vivo depletion of this complex from HeLa cells via RNA interference or immunodepletion from in vitro Xenopus laevis nuclear assembly reactions leads to defects in the assembly of the NPC, which suggests their pivotal roles (Harel et al., 2003; Walther et al., 2003). The various phenotypes associated with the mutations of putative plant Nups underline that many cellular and developmental processes involve the communication between the nucleus and the cytoplasm via the NPC.
A nuclear rim-associated activity with developmental functions is desumoylation mediated by the SUMO protease EARLY IN SHORT DAYS4 (ESD4). esd4 mutants flower extremely early and have pleiotropic alterations in shoot development (Reeves et al., 2002). The early flowering phenotype is consistent with reduced expression of the flowering repressor FLOWERING LOCUS C (FLC). In line with the in vitro SUMO protease activity of ESD4, esd4 mutants accumulate more SUMO conjugates and have less free SUMO than wild-type plants (Murtas et al., 2003). These data suggest a connection in Arabidopsis thaliana between SUMO homeostasis and flowering-time regulation.
Here, we show that mutants of an Arabidopsis protein, NUCLEAR PORE ANCHOR (NUA), with similarity to the inner nuclear basket proteins Tpr (for Translocated Promoter Region), Mlp1/Mlp2 (for Myosin-like proteins 1 and 2), and Megator flower extremely early and have several phenotypic characteristics in common with esd4 mutants. NUA is located at the inner nuclear envelope, and nua esd4 double mutants resemble nua and esd4 single mutants, indicating that the two proteins might act in a shared pathway or complex, supported by their interaction in yeast two-hybrid assays. nua mutant alleles show an increase in SUMO conjugates and reduction of free SUMO, an increase in nuclear poly(A)+ RNA, and altered expression of several genes involved in flowering-time regulation. We propose that NUA acts as a docking site at the inner nuclear pore for activities required for desumoylation and mRNA export and that disruption of this docking affects the expression of key regulators of plant development.
RESULTS
Identification of NUA, an Arabidopsis Protein Similar to Mammalian Tpr, Drosophila Megator, and Yeast Mlp1/Mlp2
NUA was identified in a targeted phenotypic screen of 36 T-DNA insertion mutants in Arabidopsis genes coding for long coiled-coil proteins that might play a structural-organizational role in the nucleus or the endomembrane system. The genes were selected by the following criteria from the ARABI-COIL Arabidopsis coiled-coil protein database (Rose et al., 2004). The proteins should be at least 500 amino acids long, with a coiled-coil coverage of at least 50% and either a nuclear localization signal (NLS) or at least one predicted transmembrane domain. For all selected open reading frames, T-DNA insertion lines generated by the Salk Institute Genomic Analysis Laboratory (SIGnAL; Alonso et al., 2003) were acquired from the ABRC (Ohio State University), and segregating populations were screened for visible phenotypes to identify proteins with an experimentally approachable biological role. nua-1 was identified as an extreme early-flowering mutant that was stunted in growth and had phyllotaxy defects in the inflorescence.
The translated open reading frame of NUA has significant sequence similarity to mammalian Tpr, an inner nuclear pore-associated long coiled-coil protein. Figure 1A shows the coiled-coil domains in NUA in comparison to human Tpr, Drosophila melanogaster Megator, and yeast Mlp1 and Mlp2. The size of the predicted protein, the length of the coiled-coil domain, the presence of a non-coiled-coil C-terminal tail, and the distribution of predicted NLS are very similar in NUA and the known Tpr-like proteins (Kuznetsov et al., 2002). Using the full-length NUA protein in WU-BLAST against the plant Gene Indices, homologous partial protein sequences with a significant identity and similarity (>35 and 50%, respectively) can be recovered from various plant species, including rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), and potato (Solanum tuberosum). Hence, NUA is evolutionarily conserved both among and within different kingdoms.
Figure 1.
Characterization of NUA T-DNA Insertion Lines.
(A) The coiled-coil domain prediction of NUA (At1g79280) using COILS, and comparison with human Tpr, Drosophila Megator, and yeast Mlp1 and Mlp2. Black bars show predicted coiled-coil domains. The dimerization and NPC association domain of Tpr are also depicted. Bar = 200 amino acids (aa).
(B) Position of T-DNA insertions and PCR primers. The confirmed insertion sites in nua-1, nua-2, nua-3, and nua-4 are indicated by vertical arrows above and below the schematic exon-intron structure of the NUA gene. Exons are depicted as gray bars and introns as black lines. Horizontal arrows indicate the positions of PCR primers, and brackets indicate the positions of the RT-PCR products shown in (D). The area surrounding the nua-2 insertion site is shown enlarged, and the structure of the ∼200 and ∼300-bp fragments derived from RT-PCR with NUA-D primers in nua-2 is indicated. ex, exon; int, intron.
(C) Immunoprecipitation of NUA from Arabidopsis wild-type and nua-1, nua-2, and nua-3 seedlings. Arrowheads depict NUA bands, and asterisks mark unspecific IgG bands also seen without plant extract (data not shown). Molecular weights are indicated on the left.
(D) RT-PCR products derived from primer pairs indicated on the right (as shown in [B]) and plant tissues as indicated on the top. Approximate sizes of the fragments amplified with the primer pair for NUA-D in nua-2 are indicated on the left. Two exposures are shown for the fragments amplified with NUA-D primers. The primer combinations in each reaction were as follows: NUA-A, F1 + LP115409; NUA-B, F1 + SALK_057101LP; NUA-C, RP057101.1 + R1(1599); NUA-D, RP069922 + LP069922; NUA-E, CS821281FP + R2(3798); NUA-F, RP079795 + R3(4993). All primer sequences are listed in Supplemental Table 2 online. Tub, tubulin 2 (At5g62690).
Based on the existing EST and microarray data (Zimmermann et al., 2004), NUA appears to be expressed ubiquitously in all tissues and during all developmental stages. This was also confirmed by RT-PCR with different tissues, including root, stem, flower, silique, and cauline leaf (see Supplemental Figure 1A online).
Characterization of NUA T-DNA Insertion Lines
Three additional T-DNA insertion alleles of NUA were identified (Figure 1B). Flanking sequences of the T-DNA insertion sites were amplified by PCR and sequenced. The original allele nua-1 (SALK_057101) has an insertion at nucleotide +1855 (with +1 being the A of the ATG) (within intron 8), the nua-2 (SALK_069922) insertion causes a 45-bp deletion between nucleotides +5657 and +5902, and nua-3 (SAIL_505_H11) has an insertion at nucleotide +7531 (within exon 29). The insertion site of nua-4 (WiscDsLox297300_17E) was mapped to nucleotide +2145 (within exon 9).
The NUA cDNA was cloned by RT-PCR (see Methods), and the partial cDNA fragment encoding amino acids 518 to 1248 (see Supplemental Figure 4 online) was used to express His-tagged protein in Escherichia coli and generate an antiserum (anti-NUA) that detects a protein of an extrapolated molecular mass of 220 kD. Figure 1C shows that after immunoprecipitation no 220-kD protein was detected in nua-1 (note that the short N-terminal fragment potentially expressed in nua-1 would not be detected by anti-NUA). A weak band of approximately full-length size was detected in nua-2 and a band corresponding to a truncated protein of ∼100 kD in nua-3 (Figure 1C). NUA protein was also absent in nua-4 (see Supplemental Figure 3C online).
Analysis of NUA transcripts by RT-PCR showed that the locus is transcribed both upstream and downstream of the T-DNA insertion in nua-1, nua-3, and nua-4 but that no transcript was detected across the insertion (Figure 1D; see Supplemental Figure 3B online). By contrast, a small amount of full-length transcript in addition to truncated transcripts was detected across the nua-2 insertion (Figure 1D). The truncated bands were sequenced and indicated that alternative splicing leads to in-frame deletions of 135 bp (exon 22) or 135 + 99 bp (exons 22 and 23) (Figure 1B). All PCR products were derived from specific cDNA templates since they were not present in the control reactions in which reverse-transcriptase was omitted (data not shown) and since all primer pairs span introns. We speculate that either mRNA is synthesized through the T-DNA insertion (which is confirmed in the nua-2 mutant) or that a transcript reads out of the T-DNA into the NUA gene. In any case, no protein in the NUA open reading frame is made downstream of the T-DNA insertions of nua-1 and nua-4 because it would be detectable with our antibody. In summary, we conclude that nua-1 and nua-4 are likely null mutations. In nua-2, a small amount of full-length protein and/or almost-full-length proteins with short internal deletions are present. In nua-3, a partial protein of ∼100 kD is made.
NUA T-DNA Insertion Alleles Flower Early in Long Days and Short Days and Have Pleiotropic Developmental Defects
All four lines flowered early in long days (Figure 2A, Table 1). nua-1 and nua-4 were the most extreme alleles and bolted with 4 to 5 rosette leaves in long days, while nua-2 and nua-3 bolted with 8 to 10 and 6 to 8 leaves, respectively (wild-type Columbia: 10 to 12 leaves). nua-1, nua-2, and nua-3 also flowered early in short days (Table 1), with nua-1 again being the most severe, nua-3 being moderate, and nua-2 being a mild allele (nua-4 was not tested). In addition to early flowering, nua-1, nua-4, and nua-3 also showed stunted inflorescences (Figure 2B; see Supplemental Figure 3A online) and smaller, narrower rosette leaves (Figure 2A), while adult nua-2 plants were indistinguishable from the wild type.
Figure 2.
Phenotypic Characteristics of nua Mutant Alleles.
(A) Seedlings at 25 d after germination, grown under long-day conditions.
(B) Plants at 34 d after germination, grown under long-day conditions.
(C) Phyllotaxy defects of nua-1.
(D) Silique phenotype of nua-1.
(E) Flower phenotype of nua-1.
Table 1.
Flowering Time of Wild-Type Columbia and nua Mutants, Given as Number of Rosette Leaves at Time of Bolting
| Columbia | nua-1 | nua-2 | nua-3 | nua-4 | |
|---|---|---|---|---|---|
| Long day | 12.2 ± 1.0 (45 plants) | 4.0 ± 0.0 (53 plants) | 8.0 ± 0.9 (16 plants) | 6.1 ± 0.6 (16 plants) | 4.2 ± 0.4 (53 plants) |
| Short day | 50.6 ± 2.0 (11 plants) | 7.4 ± 0.5 (53 plants) | 45.6 ± 1.8 (10 plants) | 33.6 ± 1.2 (6 plants) | ND |
The ± indicates sd. Long day: 16 h light/8 h dark, 75 to 125 μmol s−1 m−2. Short day: 8 h light/16 h dark, 85 to 95 μmol s−1 m−2. ND, not determined.
nua-1 and nua-4 showed several additional developmental alterations not found in nua-2 and nua-3 (Figures 2C to 2E; see Supplemental Figure 3A online). The inflorescence of the mutants showed some abnormalities with a reduced number of flower buds on the top of the main inflorescence (Figure 2C). Siliques were found at unexpected positions; two or three siliques were positioned at one node or at the top of the main inflorescence. In addition, indeterminate shoots were found in one node together with a silique and a cauline leaf. The size of siliques of the mutants was shorter and more stunted compared with the wild type (Figure 2D), with fewer developed seeds per silique. Furthermore, the majority of flowers did not set seeds. The stamens of the mutants were shorter and the size of petals was slightly smaller than the wild type (Figure 2E).
Together, we conclude that loss of NUA leads to severe developmental defects and that the isolated T-DNA insertions comprise an allelic series of increasing severity in the order nua-2, nua-3, and nua-1/nua-4.
Subcellular Localization of NUA
We used immunofluorescence with anti-NUA to characterize the subcellular localization of NUA in root tip cells of Arabidopsis seedlings. In interphase cells, NUA is clearly located at the nuclear envelope (Figure 3). During prometaphase, at the onset of spindle formation, the antibody decorated a structure in the vicinity of the spindle, which did not directly colocalize with tubulin (Figures 3A to 3D). During metaphase, this signal was less obvious (Figures 3E to 3H). During cytokinesis, NUA reappeared at the nuclear envelope (Figures 3I to 3L). Supplemental Figure 2 online shows that the NE signal is absent in nua-1, indicating that anti-NUA specifically decorates NUA at the nuclear envelope. To determine if NUA is associated with the inner nuclear envelope, we performed double labeling in plants expressing a green fluorescent protein (GFP) fusion of WIP1, an Arabidopsis outer nuclear envelope/NPC-associated protein (X.M. Xu and I. Meier, unpublished data). Figure 4 shows that the NUA signal could be clearly resolved inside the GFP-WIP1 signal, indicating that NUA is indeed associated with the inner surface of the nuclear envelope.
Figure 3.
NUA Is Located at the Nuclear Envelope during Interphase and in the Vicinity of the Spindle during Prometaphase.
Immunofluorescence images of root tip cell files in interphase ([A] to [D] and [I] to [L]), prometaphase ([A] to [D]), metaphase ([E] to [H]), and late cytokinesis ([I] to [L]). Green, anti-NUA; red in (D), (H), and (L) and magenta in (B), (C), (F), (G), (J), and (K), anti-tubulin; blue in (D), (H), and (L), 4′,6-diamidino-2-phenylindole. (A), (E), and (I) show the green channel only, and (B), (F), and (J) show the red channel only (false colored in magenta). (C), (G), and (K) show the red and green channels, and (D), (H), and (L) show the red, green, and blue channels. The arrowhead in (A) indicates the interphase nuclear envelope, and the arrow in (A) indicates the spindle-like structure in prometaphase. Bars = 10 μm in (D), (H), and (L).
Figure 4.
NUA Is Localized at the Inner Side of the Nuclear Envelope, Shown by Colocalization with the Outer Nuclear Envelope/NPC-Localized Protein GFP-WIP1.
NUA and GFP-WIP1 were detected by rabbit anti-NUA antibody (A) and mouse anti-GFP antibody (B), respectively, with appropriate secondary antibodies. The overlay image is shown in (C) (green, GFP-WIP1; magenta, NUA). The dashed box in (C) was enlarged, and the green and magenta fluorescence profiles were analyzed in (D). Bar = 5 μm.
nua-1 esd4-2 Double Mutant Analysis
The observed pleiotropic phenotype of nua-1 and nua-4 is reminiscent of the esd4-1 and esd4-2 mutations (Reeves et al., 2002; Murtas et al., 2003). To investigate their genetic interaction, homozygote nua-1 plants were crossed with esd4-2 (SALK_032317) plants, resulting in a wild-type phenotype for all seedlings in the F1 generation. Double mutants in the F2 generation were indistinguishable from nua-1 and esd4-2 in terms of flowering time under long-day conditions (Figures 5A and 5B). The only difference observed was that nua-1 esd4-2 had even shorter stamens and reduced fertility than the single mutants (Figure 5C; data not shown). These data suggest that nua-1 and esd4-2 might act in a shared pathway or complex that affects flowering time as well as vegetative and inflorescence development but that they act additively in stamen development. This notion is supported by the interaction of NUA and ESD4 in yeast two-hybrid assays, indicating that ESD4 binds specifically to the N-terminal 533 amino acids of NUA (see Supplemental Figure 4 online).
Figure 5.
The nua-1 esd4-2 Double Mutant Resembles nua-1 and esd4-2 in Flowering Time and Stunted Growth Characteristics, whereas Additive Effects Exist for Stamen Length.
(A) Wild-type, nua-1, esd4-2, and nua-1 esd4-2 seedlings at 21 d after germination, gown under long-day conditions.
(B) Plants after 34 d in long-day conditions.
(C) Close-up for open flowers. Arrows indicate top of stamens.
Effects of nua Mutants on Sumoylation and Flowering Gene Expression
ESD4 is a SUMO protease associated with the nuclear envelope (Reeves et al., 2002; Murtas et al., 2003). Mutations in ESD4 lead to an increase in SUMO conjugates and a decrease in free SUMO (Murtas et al., 2003). To test if mutations in NUA have a similar molecular phenotype, we investigated the SUMO conjugation pattern in nua-1, nua-2, nua-3, and nua-4, compared with the T-DNA insertion allele esd4-2 and wild-type Columbia plants (Figure 6A; see Supplemental Figure 3D online). In esd4-2, the level of high molecular weight SUMO conjugates was increased, while the free SUMO level was reduced, as described previously (Murtas et al., 2003). We found that nua-1 and nua-4 phenocopy esd4-2 and that nua-3 leads to an intermediate and nua-2 to the least increase of SUMO conjugates. The level of free SUMO was reciprocally altered in all alleles.
Figure 6.
nua Mutant Alleles Lead to Increasing Accumulation of SUMO Conjugates and Altered Expression of Genes Involved in Different Flowering Pathways.
(A) Protein extracts from 10-d-old seedlings were probed with an anti-SUMO1 antibody. Free SUMO is reduced (arrowhead), and the amount of SUMO conjugates increased (bracket) in nua mutants, like previously shown for esd4 mutants. The effect increases in severity in the order of nua-2, nua-3, and nua-1. The asterisk indicates the putative SUMO dimer.
(B) RT-PCR of tubulin 2 (Tub), FLC, MAF4, FT, SOC1, LFY, MYB33, and MYB65 mRNAs in Arabidopsis wild type, nua-1, nua-2, nua-3, nua-4, and esd4-2.
ESD4 mutations have been shown to decrease the mRNA level of the floral repressor FLC and increase mRNA levels of the floral activators FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) (Reeves et al., 2002). To test whether the nua mutants would also phenocopy these effects, we tested mRNA abundance of FLC, FT, and SOC1 by semiquantitative RT-PCR. Indeed, as shown in Figure 6B, FLC mRNA level is strongly reduced in nua-1 and nua-4, comparable to esd4-2, and is somewhat reduced in nua-2 and nua-3, with nua-2 showing the weakest effect. Consistently, FT and SOC1 mRNA levels were found inversely affected.
Since nua-1 and nua-4 flower significantly earlier than the flc-3 null mutant (four to five compared with 10 to 11 rosette leaves in long-day conditions; see Supplemental Table 1 online) (Michaels and Amasino, 1999, 2001), additional factors involved in flowering-time regulation are likely affected in nua mutants. Additionally, the ESD4 mutation was also suggested to promote flowering both through and independently of FLC (Reeves et al., 2002). To probe into additional effects of NUA and ESD4 mutations on the flowering pathways, we tested mRNA levels of several additional floral repressors and floral integrators. MAF4, one of the FLC paralogs also implied in floral repression (Ratcliffe et al., 2003), was decreased in nua-1, nua-3, nua-4, and esd4-2 (Figure 6B). Furthermore, the phytohormone gibberellin is known to promote floral transition strongly under short-day conditions via activating the floral integrator LEAFY (LFY) by GAMYB transcription factors (Achard et al., 2004 and references therein). When the expression levels of LFY, MYB33, and MYB65 were analyzed, elevation in nua-1 and nua-4 mutants was apparent (Figure 6B), again supporting the notion that several flowering pathways were affected in the nua mutants. We noted that LFY, MYB33, and MYB65 expression levels were not significantly altered in esd4-2, supporting an overlapping, but not identical, effect of the two gene knockouts on flowering regulator expression.
GFP-ESD4 Localization Is Not Altered in nua Mutants
If NUA acted as a nuclear pore anchor for ESD4, as predicted for the interaction between Ulp1 and Mlp1 in yeast, depletion of NUA should lead to a release of ESD4 from the nuclear periphery. We tested this model by investigating the localization of GFP-ESD4 in wild-type Columbia and the nua mutant alleles. Figure 7A shows that GFP-ESD4 has a nuclear location with enrichment at the nuclear envelope in root cell files, consistent with previous reports for ESD4-GFP (Murtas et al., 2003). Figures 7B to 7D and Supplemental Figure 3E online show that no significant changes in this pattern were seen in nua-1, nua-2, nua-3, and nua-4, suggesting that the increase in SUMO conjugates in nua mutants is unlikely to be based on delocalization of ESD4.
Figure 7.
The Concentration of ESD4 on the Nuclear Periphery Is Not Abolished in nua Mutants.
GFP fluorescence was observed at the nuclear periphery, in root cell files of Arabidopsis seedlings transformed with GFP-ESD4. Bars = 10 μm.
(A) Wild-type background.
(B) nua-1 background.
(C) nua-2 background.
(D) nua-3 background.
nua Mutants Accumulate Nuclear Poly(A)+ RNA
Both mammalian Tpr and yeast Mlp1/Mlp2 have been implicated in affecting mRNA export. While poly(A)+ RNA accumulates in nuclei immunodepleted of Tpr (Shibata et al., 2002) and in cells that overexpress nuclear Tpr fragments (Bangs et al., 1998), double mutants of Mlp1 and Mlp2 show no nuclear mRNA accumulation (Kosova et al., 2000). However, they are affected in the nuclear retention of mis-spliced RNAs (Galy et al., 2004), and Mlp1 binds hnRNPs (Green et al., 2003). Using in situ hybridization with an oligo(dT) probe, we tested if the nua mutant alleles accumulate more nuclear poly(A)+ RNA compared with the wild type. Figure 8 and Supplemental Figure 3F online show that, indeed, all four mutant alleles show higher nuclear signals than the wild type. Significantly, the amount of nuclear signal detected correlated well with the severity of the mutant alleles in terms of flowering time, SUMO conjugate abundance, flowering regulator expression, and developmental defects. In all assays employed, nua-1 and nua-4 were the most severe alleles, followed by nua-3 as intermediate and nua-2 as the mildest allele.
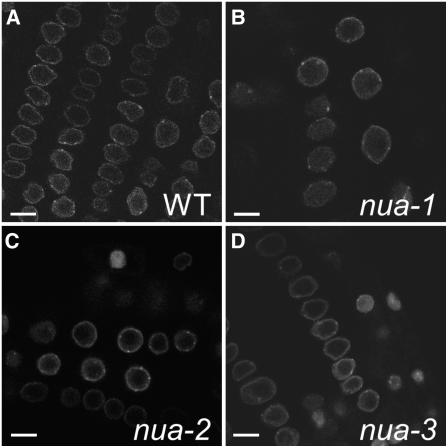
Figure 8.
Poly(A)+ RNA Accumulates in Nuclei of nua Mutants.
Whole-mount in situ hybridization of 10-d-old seedling petioles with fluoresceine-labeled oligo(dT) probe shows increasing severity of nuclear poly(A)+ mRNA retention in nua-2, nua-3, and nua-1. Insets show single nuclei at a higher magnification. All images were taken at identical gain settings. The experiment was repeated twice. Bars = 20 μm.
DISCUSSION
Mammalian Tpr (Park et al., 1986) and its closest structural relatives in Drosophila (Zimowska et al., 1997) and yeast (Strambio-de-Castillia et al., 1999) are large coiled-coil proteins located on the nucleoplasmic side of the NPC (Cordes et al., 1997; Zimowska et al., 1997; Bangs et al., 1998; Strambio-de-Castillia et al., 1999). While Mlp1/Mlp2 deletion mutants in yeast are viable, knockout mutants of Megator are embryonic lethal (Qi et al., 2004). Megator depletion via RNAi leads to a reduction of cells going through mitosis (Qi et al., 2004). To our knowledge, no Tpr knockout mutants have so far been reported in vertebrates. We show here that a likely knockout of the single Arabidopsis Tpr/Megator/Mlp1/Mlp2 homolog NUA is viable. Loss of NUA leads to complex developmental phenotypes, including early flowering, stunted growth, defects in stamen and silique development, and changes in phyllotaxy. Our data indicate that loss of NUA affects a number of regulatory pathways, likely by affecting the expression level of key regulators, as shown here for the example of flowering-time regulators.
Several immuno-electron microscopy studies have suggested that Tpr is located at the nuclear basket of the NPC (Cordes et al., 1997; Frosst et al., 2002; Krull et al., 2004). Nevertheless, on the light microscopy level, a continuous nuclear envelope staining was seen in several recent studies of human Tpr (Hase et al., 2001; Hase and Cordes, 2003; Krull et al., 2004). Similarly, in our system, we do not detect a punctate pattern for NUA. This does not contradict nuclear pore localization but simply indicates that a higher level of resolution (such as immunogold labeling) is required to investigate the precise ultrastructural location of NUA.
We have identified an allelic series of four T-DNA insertion alleles that have increasing severity of several correlating whole-plant and molecular phenotypes. In the order of nua-2, nua-3, and nua-1/nua-4, we have observed an increase in severity of early flowering, accumulation of sumoylated proteins, nuclear accumulation of poly(A)+ RNA, and altered expression of several flowering-time regulators. This suggests that these events are connected by the activity of NUA. Molecular characterization of the NUA insertion alleles indicates that nua-1 and nua-4 are likely null alleles, that nua-2 is a knockdown allele, and that nua-3 expresses an ∼100-kD N-terminal fragment of NUA. nua-3 acts as a functional knockdown of intermediate severity, indicating that part of the NUA activity involved in SUMO homeostasis, RNA metabolism, and flowering-time regulation resides in the N-terminal fragment expressed in nua-3.
Yeast Mlp1 and Mlp2 function in anchoring the SUMO protease Ulp1 to the NPC (Zhao et al., 2004). Mlp1/Mlp2 deletion mutants in yeast exhibit a clonal lethality phenotype caused by an increase of extrachromosomal 2-μm circle DNA and show an enhanced sensitivity to DNA-damaging drugs (Galy et al., 2000; Kosova et al., 2000; Zhao et al., 2004). This phenotype is consistent with SUMO pathway mutants and can be suppressed by overexpression of Ulp1, whereas deletion of Ulp1 or delocalization of Ulp1 through deletion of its targeting domain mimics Mlp1/Mlp2 deletion mutants (Zhao et al., 2004). These data indicate that Mlp1/Mlp2 are required for Ulp1 function, most likely through its anchoring at the NPC. By contrast, the mammalian SUMO protease SENP2 is anchored to the inner nuclear pore by binding Nup153, the same nucleoporin that is involved in anchoring Tpr to the nuclear basket (Hang and Dasso, 2002; Zhang et al., 2002).
Our data show that in Arabidopsis, NUA and ESD4 are both associated with the nuclear periphery, and the accumulation of sumoylated proteins in nua mutants indicates that NUA is involved in desumoylation. The close similarity of the whole-plant and molecular phenotypes of nua and esd4 knockout mutants, together with their ability to interact in yeast two-hybrid assays, are consistent with a model in which the two proteins interact at the nuclear periphery, thereby affecting mRNA accumulation, desumoylation, and alterations in gene expression. If the interaction of NUA and ESD4 were analogous to Mlp1/Mlp2 and Ulp1, we would expect that ESD4 lost nuclear rim localization in a nua null mutant. However, our data show that GFP-ESD4 is still targeted to the nuclear rim in nua-1 and nua-4, suggesting that the desumoylation defect observed in nua mutants is not caused by the delocalization of ESD4. In addition, the interaction between NUA and ESD4 in yeast two-hybrid assays might be indirect or additional proteins might tether ESD4 to the NPC. Further experiments are needed to test these hypotheses. Our current working model for the functional interaction between NUA and ESD4 is therefore that ESD4 and possibly other Arabidopsis SUMO proteases and NUA have to be in close proximity at the NPC for wild-type-level desumoylation to occur. This could, for example, be envisioned if sumoylated proteins bind to NUA, thereby being presented to localized SUMO protease activity.
Our finding that nua mutants accumulate nuclear poly(A)+ RNA indicates a function of NUA in mRNA export and/or nuclear mRNA turnover. There are several lines of evidence for a role of Tpr-like proteins in mRNA export. Overexpression of mammalian Tpr and its depletion through the injection of antibodies leads to an accumulation of poly(A)+ RNA in the nucleus, partially in enlarged clusters containing splicing factors (Bangs et al., 1998; Shibata et al., 2002). Overexpression of Mlp1 in yeast causes nuclear accumulation of poly(A)+ RNA, while no nuclear poly(A)+ RNA accumulation was seen in double deletion mutants of Mlp1/Mlp2 (Kosova et al., 2000). The C-terminal domain of yeast Mlp1 interacts with hnRNPs via Nab2, and a model has been proposed for Mlp1 to act as a checkpoint for RNA export (Green et al., 2003). Mlp1 is responsible for the nuclear retention of unspliced mRNAs, and deletion of Mlp1 leads to a leakage of intron-containing RNAs into the cytoplasm (Galy et al., 2004). Our data demonstrate an effect on mRNA export by a likely null mutant of a Tpr-like protein and contrast the yeast data that show no effect in an Mlp1/Mlp2 null mutant. This indicates that while homologs of Tpr in different kingdoms all appear to function in an aspect of mRNA export, their precise role probably differs.
SUMO modification has been discussed as a possible mechanism to control nucleocytoplasmic transport of proteins (for review, see Pichler and Melchior, 2002). Recently, hnRNPs, RNA helicases, and other proteins involved in RNA metabolism were identified as substrates for SUMO modification in mammals (Li et al., 2004; Vassileva and Matunis, 2004; Vertegaal et al., 2004). This suggests a previously unrecognized link between the SUMO pathway and mRNA metabolism. Our data, showing that nua mutants are affected both in SUMO homeostasis and nuclear RNA accumulation, indicate that such a link also exists in plants.
Loss of NUA leads to a number of developmental defects, the most striking one being extreme early flowering under both long-day and short-day conditions. Early flowering is consistent with the reduction of FLC expression and a concomitant increase of expression of FT and SOC1, a reduction of MAF4 expression, and an increase of MYB33, MYB65, and LFY. Additional untested factors involved in flowering-time regulation might also be affected in nua mutants. If the observed effect on mRNA in nua mutants also included a perturbation of microRNA (miRNA) export or turnover, it is conceivable that miRNA targets are misexpressed in nua mutants. Interestingly, both flowering-time regulation and anther development include miRNA-regulated steps (Achard et al., 2004; Millar and Gubler, 2005). In the future, it will be important to determine if miRNA export is indeed affected in nua mutants and how the observed defects in SUMO and mRNA homeostasis are molecularly connected.
METHODS
Plant Material and Growth Conditions
For T-DNA insertion mutants nua-1 (SALK_057101), nua-2 (SALK_069922), nua-3 (SAIL_505_H11), nua-4 (WiscDsLox297300_17E), and esd4-2 (SALK_032317), T3 or T4 bulk seeds were acquired from the ABRC. The nua-1 esd4-2 double mutant was identified in the F2 generation from crosses between nua-1 and esd4-2. Arabidopsis thaliana wild-type and T-DNA lines were grown on soil under standard long-day conditions (16 h light/8 h dark) or short-day conditions (8 h light/16 h dark) or on Murashige and Skoog plates under constant light.
Flowering-Time Measurements
Plants were grown on soil in short-day or long-day conditions. Flowering time was measured by counting the total number of rosette leaves at the time of bolting. Data shown are the mean value and sd of 11 to 53 samples per line.
PCR-Based Genotyping of T-DNA Insertion Lines
Genomic DNA was extracted as described (Krysan et al., 1999). Primers were designed using SIGnAL iSect tools (http://signal.salk.edu/tdnaprimers.html). All primer sequences are summarized in Supplemental Table 2 online. The exact T-DNA insertion sites were determined by sequencing the PCR products derived from primer combinations of gene-specific primers plus T-DNA-specific primers. In nua-1 and nua-3, primer combinations LP057101.1/LBa1 and CS821281RP/pCSA110-LB were used, respectively. In nua-2, primer combinations RP069922/LBa1 and LP069922/New-RB-primer were used. In nua-4, the insertion site was mapped to nucleotide +2145 (within exon 9) using primer combinations CS850695FP/p745-primer and CS850695RP/p745-primer.
Cloning of NUA Full-Length cDNA and Plasmid Construction
Due to the large size of the predicted NUA coding sequence (> 6 kb), the NUA cDNA was cloned in four fragments by RT-PCR using the ThermoScript RT-PCR system (Invitrogen). Partial fragments were cloned into the pENTR/D-TOPO vector and confirmed by sequencing. The full-length cDNA was assembled using the unique AatII, ScaI, and XmaI restriction enzyme sites and moved into the Gateway destination vectors pDEST17, pDEST22, and pDEST32 (Invitrogen). To validate the assembled >6-kb cDNA, full-length NUA transcript was reverse-transcribed with gene-specific primers. After amplification using the BD Advantage 2 PCR enzyme system (BD Biosciences Clontech), the 6282-bp cDNA was fully sequenced and found identical to the assembled cDNA (see Supplemental Figure 1 online). The ESD4 cDNA (Reeves et al., 2002) in the pENTR/SD/D-TOPO vector was received from the ABRC. The gene was sequenced for confirmation and moved into the GFP vector pK7WGF2 (Karimi et al., 2002).
Arabidopsis Transformation
Arabidopsis Columbia wild type, nua-1, nua-2, and nua-3 were transformed by floral dipping (Clough and Bent, 1998) with Agrobacterium tumefaciens strain ABI harboring plasmid pK7WGF2-ESD4. Primary transformants were selected for kanamycin resistance.
RT-PCR
Total RNA was extracted with the RNeasy plant mini kit (Qiagen) from 10-d-old seedlings (a stage at which none of the mutants had bolted yet) grown on Murashige and Skoog plates under long-day conditions. After digestion with DNase I (amplification grade; Invitrogen), cDNA was synthesized with oligo(dT) primers from ∼3 μg total RNA using the Thermoscript RT-PCR system (Invitrogen). cDNA templates were PCR amplified with gene-specific primers (see Supplemental Table 2 online) for quantification of mRNA levels of flowering time integrator genes. Twenty-eight cycles were used for tubulin 2, 35 cycles for FLC, 30 cycles for FT, 26 cycles for SOC1, 35 cycles for LFY, and 33 cycles for MYB33 and MYB65.
Sequence Analysis
Coiled-coil domains were predicted using the Protean software from the Lasergene package (DNASTAR) and the algorithm MultiCoil (Wolf et al., 1997).
Protein Expression, Purification, and Antibody Production
The anti-NUA antibody (OSU156) was generated against a partial recombinant protein (amino acids 518 to 1248). The N-terminal 6xHis-tagged protein was purified from Escherichia coli BL21-AI using Ni-NTA resin according to the QIAexpressionist manual (Qiagen) and preparative SDS-PAGE. The rabbit antiserum was generated by Cocalico Biologicals.
Immunoprecipitation and Sumoylation Assays
For immunoprecipitation, all steps were performed at 4°C. One milliliter of tissue powder from 12-d-old seedlings was suspended in 1 mL of buffer containing 50 mM Tris-Cl, 150 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 3 mM DTT, 1 mM PMSF, and protease inhibitor cocktail (Sigma-Aldrich). After centrifugation at 16,000g, the supernatant was incubated with anti-NUA (1:500 dilution) bound to protein A-sepharose for 3 h. The immunoprecipitates were resuspended in 50 μL 3× SDS-PAGE loading buffer and subjected to SDS-PAGE. For the analysis of sumoylation profiles, total protein from 2-week-old seedlings was extracted as described (Thompson et al., 2005). Approximately 100 μg of protein was separated on a two-layer SDS-PAGE gel to resolve both the free SUMO and high molecular weight conjugates (8% top half and 15% bottom half). After SDS-PAGE, proteins were then transferred to a polyvinylidene difluoride membrane (Bio-Rad), probed with anti-NUA (1:2000) or anti-AtSUMO1 (Abcam), and subsequently probed with peroxidase-conjugated anti-rabbit secondary antibody (GE Healthcare; 1:15,000). For detection, the Supersignal West Pico Chemiluminescent Substrate for the HRP system (Pierce) was used.
Immunolocalization
Whole-mount immunolocalization in Arabidopsis root tip cells was performed as described (Friml et al., 2003) using anti-NUA (1:100), monoclonal anti-α-tubulin (1:100, DM1A; Sigma-Aldrich), monoclonal anti-NPC (1:250, QE5, recognizing Nup214, Nup153, and p62 in mammals; Covance), monoclonal anti-GFP (2.5 μg/mL; Molecular Probes), primary antibodies, and appropriate secondary antibodies conjugated to Alexa Fluor 488 or 568 (Invitrogen). DNA was counterstained by 4′,6-diamidino-2-phenylindole (Sigma-Aldrich). The images in Figure 3 were collected from a Leica TCS SP2 AOBS confocal laser scanning microscope equipped with four lasers (red helium neon 633 nm, green helium neon 543 nm, argon 458/476/488/496/514 nm, and argon UV). The images in Figure 4 and Supplemental Figure 2 online were collected on a PCM 2000/Nikon Eclipse 600 confocal laser scanning microscope as described (Rose and Meier, 2001).
In Situ Hybridization
The poly(A)+ RNA in situ hybridization was conducted essentially as described (Gong et al., 2005) with minor modifications. Briefly, samples were taken from equivalent portions of young leaves at similar developmental stages (2-week-old, four true leaves) from Columbia wild type or nua mutants and were fixed in glass vials by adding 8 to 10 mL of fixation cocktail containing a mixture of 50% fixation buffer (120 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, 2.7 mM KCl, 0.1% Tween 20, 80 mM EGTA, 5% formaldehyde, and 10% DMSO) and 50% heptane to completely immerse the sample. Samples were shaken gently for 30 min at room temperature. After dehydration twice for 5 min each in absolute methanol and three times for 5 min each in absolute ethanol, the samples were incubated for 30 min in 1:1 (v/v) ethanol:xylene and then washed twice for 5 min each with absolute ethanol, twice for 5 min each with absolute methanol, and once for 5 min with 1:1 (v/v) methanol:fixation buffer without formaldehyde. The samples were post-fixed in fixation buffer for 30 min at room temperature. After fixation, the samples were rinsed twice with fixation buffer without formaldehyde and once with 1 mL of perfect Hyb Plus hybridization buffer (Sigma-Aldrich; H-7033). To each glass vial, 1 mL of hybridization buffer was added, and samples were prehybridized for 1 h at 50°C. After prehybridization, 10 pmol 45-mer oligo(dT) labeled with one molecule of fluoresceine at the 5′-end (synthesized by MWG Company) was used for hybridization at 50°C in darkness for at least 8 h. After hybridization, the samples were washed once for 60 min in 2× SSC (0.3 M NaCl and 0.03 M sodium citrate) and 0.1% SDS at 50°C and once for 20 min in 0.2× SSC and 0.1% SDS at 50°C in darkness. Confocal images were acquired as described (Rose and Meier, 2001).
Yeast Two-Hybrid Assays
Yeast strain PJ69-4A was used. Competent cell preparation and yeast transformation was performed as described (Dohmen et al., 1991).
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EF426860.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Expression Pattern of NUA and the Validation of the Full-Length NUA cDNA.
Supplemental Figure 2. Immunofluorescence in Wild-Type and nua-1 Root Tip Cells.
Supplemental Figure 3. Characterization of the nua-4 Mutant.
Supplemental Figure 4. NUA Self-Interacts and Interacts with ESD4 in Yeast Two-Hybrid Assays.
Supplemental Table 1. Flowering Time of Wild Type Columbia, nua Mutants, and flc-3 Null Mutant, Given as Number of Rosette Leaves at the Time of Bolting.
Supplemental Table 2. Primer Sequences Used in This Study.
Supplementary Material
Acknowledgments
We thank SIGnAL and the ABRC for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, Richard Amasino for the flc-3 line, Biao Ding for the use of his confocal microscope, Jelena Brkljacic for fruitful discussions, David E. Somers for critical reading of the manuscript, and Chao (Sylvia) He and Kyle Kennedy for technical assistance. Financial support by the National Science Foundation is greatly acknowledged.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Iris Meier (meier.56@osu.edu).
Online version contains Web-only data.
References
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131 3357–3365. [DOI] [PubMed] [Google Scholar]
- Alcazar-Roman, A.R., Tran, E.J., Guo, S., and Wente, S.R. (2006). Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8 711–716. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Bangs, P., Burke, B., Powers, C., Craig, R., Purohit, A., and Doxsey, S. (1998). Functional analysis of Tpr: Identification of nuclear pore complex association and nuclear localization domains and a role in mRNA export. J. Cell Biol. 143 1801–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, G.K., Liu, S.T., and Yen, T.J. (2005). Kinetochore structure and function. Trends Cell Biol. 15 589–598. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cordes, V.C., Reidenbach, S., Rackwitz, H.R., and Franke, W.W. (1997). Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J. Cell Biol. 136 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw, J.M., Krutchinsky, A.N., Zhang, W., Chait, B.T., and Matunis, M.J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R.J., Strasser, A.W., Honer, C.B., and Hollenberg, C.P. (1991). An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7 691–692. [DOI] [PubMed] [Google Scholar]
- Dong, C.H., Hu, X., Tang, W., Zheng, X., Kim, Y.S., Lee, B.H., and Zhu, J.K. (2006). A putative Arabidopsis nucleoporin AtNUP160 is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 26 9533–9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J., Benkova, E., Mayer, U., Palme, K., and Muster, G. (2003). Automated whole mount localisation techniques for plant seedlings. Plant J. 34 115–124. [DOI] [PubMed] [Google Scholar]
- Frosst, P., Guan, T., Subauste, C., Hahn, K., and Gerace, L. (2002). Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 156 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy, V., Gadal, O., Fromont-Racine, M., Romano, A., Jacquier, A., and Nehrbass, U. (2004). Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116 63–73. [DOI] [PubMed] [Google Scholar]
- Galy, V., Olivo-Marin, J.C., Scherthan, H., Doye, V., Rascalou, N., and Nehrbass, U. (2000). Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403 108–112. [DOI] [PubMed] [Google Scholar]
- Gong, Z., Dong, C.H., Lee, H., Zhu, J., Xiong, L., Gong, D., Stevenson, B., and Zhu, J.K. (2005). A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D.M., Johnson, C.P., Hagan, H., and Corbett, A.H. (2003). The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc. Natl. Acad. Sci. USA 100 1010–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis, E.R., Altan, N., Lippincott-Schwartz, J., and Powers, M.A. (2002). Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell 13 1282–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang, J., and Dasso, M. (2002). Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277 19961–19966. [DOI] [PubMed] [Google Scholar]
- Harel, A., Orjalo, A.V., Vincent, T., Lachish-Zalait, A., Vasu, S., Shah, S., Zimmerman, E., Elbaum, M., and Forbes, D.J. (2003). Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11 853–864. [DOI] [PubMed] [Google Scholar]
- Hase, M.E., and Cordes, V.C. (2003). Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell 14 1923–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase, M.E., Kuznetsov, N.V., and Cordes, V.C. (2001). Amino acid substitutions of coiled-coil protein Tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Mol. Biol. Cell 12 2433–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori, N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kosova, B., Pante, N., Rollenhagen, C., Podtelejnikov, A., Mann, M., Aebi, U., and Hurt, E. (2000). Mlp2p, a component of nuclear pore attached intranuclear filaments, associates with nic96p. J. Biol. Chem. 275 343–350. [DOI] [PubMed] [Google Scholar]
- Krull, S., Thyberg, J., Bjorkroth, B., Rackwitz, H.R., and Cordes, V.C. (2004). Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell 15 4261–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov, N.V., Sandblad, L., Hase, M.E., Hunziker, A., Hergt, M., and Cordes, V.C. (2002). The evolutionarily conserved single-copy gene for murine Tpr encodes one prevalent isoform in somatic cells and lacks paralogs in higher eukaryotes. Chromosoma 111 236–255. [DOI] [PubMed] [Google Scholar]
- Li, T., Evdokimov, E., Shen, R.F., Chao, C.C., Tekle, E., Wang, T., Stadtman, E.R., Yang, D.C., and Chock, P.B. (2004). Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc. Natl. Acad. Sci. USA 101 8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M.J., Wu, J., and Blobel, G. (1998). SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., and Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtas, G., Reeves, P.H., Fu, Y.F., Bancroft, I., Dean, C., and Coupland, G. (2003). A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15 2308–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse, V.G., Kuster, B., Gerstberger, T., and Hurt, E. (2003). Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5 21–27. [DOI] [PubMed] [Google Scholar]
- Park, M., Dean, M., Cooper, C.S., Schmidt, M., O'Brien, S.J., Blair, D.G., and Vande Woude, G.F. (1986). Mechanism of met oncogene activation. Cell 45 895–904. [DOI] [PubMed] [Google Scholar]
- Parry, G., Ward, S., Cernac, A., Dharmasiri, S., and Estelle, M. (2006). The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18 1590–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler, A., Gast, A., Seeler, J.S., Dejean, A., and Melchior, F. (2002). The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108 109–120. [DOI] [PubMed] [Google Scholar]
- Pichler, A., and Melchior, F. (2002). Ubiquitin-related modifier SUMO1 and nucleocytoplasmic transport. Traffic 3 381–387. [DOI] [PubMed] [Google Scholar]
- Qi, H., Rath, U., Wang, D., Xu, Y.Z., Ding, Y., Zhang, W., Blacketer, M.J., Paddy, M.R., Girton, J., Johansen, J., and Johansen, K.M. (2004). Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol. Biol. Cell 15 4854–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut, G., Doye, V., and Ellenberg, J. (2004). Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6 1114–1121. [DOI] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Kumimoto, R.W., Wong, B.J., and Riechmann, J.L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, P.H., Murtas, G., Dash, S., and Coupland, G. (2002). early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129 5349–5361. [DOI] [PubMed] [Google Scholar]
- Rose, A., Manikantan, S., Schraegle, S.J., Maloy, M.A., Stahlberg, E.A., and Meier, I. (2004). Genome-wide identification of Arabidopsis coiled-coil proteins and establishment of the ARABI-COIL database. Plant Physiol. 134 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, A., and Meier, I. (2001). A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc. Natl. Acad. Sci. USA 98 15377–15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., Aitchison, J.D., Suprapto, A., Hjertaas, K., Zhao, Y., and Chait, B.T. (2000). The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 148 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, S., Matsuoka, Y., and Yoneda, Y. (2002). Nucleocytoplasmic transport of proteins and poly(A)+ RNA in reconstituted Tpr-less nuclei in living mammalian cells. Genes Cells 7 421–434. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia, C., Blobel, G., and Rout, M.P. (1999). Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144 839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, A.R., Doelling, J.H., Suttangkakul, A., and Vierstra, R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, E.J., and Wente, S.R. (2006). Dynamic nuclear pore complexes: Life on the edge. Cell 125 1041–1053. [DOI] [PubMed] [Google Scholar]
- Vassileva, M.T., and Matunis, M.J. (2004). SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol. Cell. Biol. 24 3623–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertegaal, A.C., Ogg, S.C., Jaffray, E., Rodriguez, M.S., Hay, R.T., Andersen, J.S., Mann, M., and Lamond, A.I. (2004). A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 279 33791–33798. [DOI] [PubMed] [Google Scholar]
- Walther, T.C., Alves, A., Pickersgill, H., Loiodice, I., Hetzer, M., Galy, V., Hulsmann, B.B., Kocher, T., Wilm, M., Allen, T., Mattaj, I.W., and Doye, V. (2003). The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113 195–206. [DOI] [PubMed] [Google Scholar]
- Weirich, C.S., Erzberger, J.P., Flick, J.S., Berger, J.M., Thorner, J., and Weis, K. (2006). Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 8 668–676. [DOI] [PubMed] [Google Scholar]
- Wolf, E., Kim, P.S., and Berger, B. (1997). MultiCoil: A program for predicting two- and three-stranded coiled coils. Protein Sci. 6 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and Li, X. (2005). A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17 1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Saitoh, H., and Matunis, M.J. (2002). Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22 6498–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Wu, C.Y., and Blobel, G. (2004). Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimowska, G., Aris, J.P., and Paddy, M.R. (1997). A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J. Cell Sci. 110 927–944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.