Abstract
A tetraploidy left Arabidopsis thaliana with 6358 pairs of homoeologs that, when aligned, generated 14,944 intragenomic conserved noncoding sequences (CNSs). Our previous work assembled these phylogenetic footprints into a database. We show that known transcription factor (TF) binding motifs, including the G-box, are overrepresented in these CNSs. A total of 254 genes spanning long lengths of CNS-rich chromosomes (Bigfoot) dominate this database. Therefore, we made subdatabases: one containing Bigfoot genes and the other containing genes with three to five CNSs (Smallfoot). Bigfoot genes are generally TFs that respond to signals, with their modal CNS positioned 3.1 kb 5′ from the ATG. Smallfoot genes encode components of signal transduction machinery, the cytoskeleton, or involve transcription. We queried each subdatabase with each possible 7-nucleotide sequence. Among hundreds of hits, most were purified from CNSs, and almost all of those significantly enriched in CNSs had no experimental history. The 7-mers in CNSs are not 5′- to 3′-oriented in Bigfoot genes but are often oriented in Smallfoot genes. CNSs with one G-box tend to have two G-boxes. CNSs were shared with the homoeolog only and with no other gene, suggesting that binding site turnover impedes detection. Bigfoot genes may function in adaptation to environmental change.
INTRODUCTION
Functional DNA sequence changes at a lower rate over evolutionary time than sequence without function. Exon sequence tends to be conserved, whereas functionless sequence is randomized by substitution, lost by conversion, or deleted entirely. Therefore, if two genes or chromosomal regions have diverged from a common ancestor, be they in different species (orthologs) or duplications within the same genome (paralogs), those few noncoding regions that retain a high degree of sequence similarity provide a measure of noncoding DNA function, where function is inferred from conservation (Hardison, 2000, 2003). In flowering plants, the alignment algorithm BLAST-2-sequences (Tatusova and Madden, 1999) has been used successfully to detect conserved noncoding sequences (CNSs) in comparisons of maize (Zea mays) and rice (Oryza sativa) orthologous genes (Kaplinsky et al., 2002; Inada et al., 2003) and to find intragenomic CNSs in Arabidopsis thaliana (Thomas et al., 2007) by comparing the syntenic duplicates (syntenic paralogs and homoeologs) retained following its most recent tetraploidy (called the α-event). The two genomes from the α-event (Simillion et al., 2002; Bowers et al., 2003; Maere et al., 2005) diverged to approximately the same extent as have maize-rice and man-mouse (Kaplinsky et al., 2002); all of these have diverged sufficiently such that sequence conservations are not due to neutral carryover. Table 1 defines important terms we use: CNS, αCNS, gene space, and phylogenetic footprint. It should be understood that function does not assure that noncoding sequence will be conserved. Some sequences evolve quickly; others defy detection. CNSs are a subset of functional noncoding sequences.
Table 1.
Definitions Involving CNSs and Their Identification
| Plant CNS: A pairwise bl2seq (Tatusova and Madden, 1999) hit between the nonprotein-coding sequences near usefully diverged, orthologous genes. These sequences are at least 15-bp long with an e-value equal to or more significant than a 15/15 exact nucleotide match, and without complexity filtration (Kaplinsky et al., 2002; Inada et al., 2003). BLAST results for Arabidopsis are displayed and may be researched in a custom viewer (http://synteny.cnr.berkeley.edu/AtCNS). “Useful” levels of divergence are not so small that conservation occurs by carryover from the ancestor, without selection, but not so great that detection is impeded, as will be further defined in the text. |
| Plant αCNS: As above, but the chromosomal regions are homoeologous (syntenous and paralogous) remnants of a usefully diverged tetraploidy event (α) in the lineage (Thomas et al., 2006). Subfunctionalization is expected of homoeologous pairs, but not orthologous pairs. |
| Gene space: Gene space is computed after CNSs have been identified for a paired region, and each CNS has been sorted to gene. The segment of genome between the most 5′ (upstream) and most 3′ CNS or untranslated region plus ∼500 bp on each side (depending on neighboring features; Thomas et al., 2007). Within a gene space are exons, UTRs, CNSs, known motifs, positions where specific TFs reside, and any feature that is fixed at a chromosomal locus. |
| Phylogenetic footprint: The most inclusive term for conserved sequence, whether two or multiple sequences, and without stipulations as to the extent of divergence. A CNS is a type of phylogenetic footprint. |
Haberer et al. (2004) demonstrated that intragenomic (α) phylogenetic footprints in proximal promoters of Arabidopsis syntenic gene pairs could be found with an anchored alignment tool (Brudno et al., 2004), anchoring on the 5′ATG. However, these workers did not explore sequence outside of the 500-bp proximal promoter. Guo and Moose (2003) used LAGAN (Brudno et al., 2003) with a Vista display (Mayor et al., 2000) to explore CNSs within the grasses, especially between maize and rice, concluding that transcription factor binding sites are within some CNSs and that CNSs were sometimes shared among different grasses.
This report explores the functions of 14,944 intragenomic CNSs (αCNSs) retained from a set of 3179 gene space pairs retained from the α-event in the Arabidopsis lineage (Thomas et al., 2006, 2007). These data can be accessed by a custom Web application called the Arabidopsis Bl2seq Viewer available at http://synteny.cnr.berkeley.edu/AtCNS. According to Thomas et al. (2007), these αCNSs exist in gene spaces with a modal frequency of zero and a mean of 1.7. The median αCNS length is 25 bp, and these sites are distributed in all regions of a gene with a general preference to be 5′ of a gene. The ratio of CNSs within the gene space, going from 5′ to 3′, 5′:5′UTR:intron:3′UTR:3′ is 3.0:2.0:1.2: 1.0: 1.2, giving a 5′/3′ bias of 2.3. Thomas et al. (2007) also showed that genes with certain functions, especially transcription factors responding early to external stimuli, as estimated by their gene ontology (GO) annotation, tended to associate with different numbers of CNSs; this functional category correlation with CNS richness validated the notion that CNSs are functional. Finally, Thomas et al. (2007) concluded that these CNSs are not simple sequences and not likely to either encode or bind small RNAs. Thus, the likely general function of an αCNS must be to bind protein or, perhaps, carbohydrates.
CNSs have been extensively studied, especially in mammals. However, using the typical mammalian definition of CNS (e.g., ≥70% identity; ≥100 bp in length [Loots et al., 2000]), comparably diverged plant genes would have no CNSs. Putative CNS function includes matrix attachment regions (Avramova et al., 1998; Glazko et al., 2003), single and multiple transcription factor (TF) binding sites (Hardison, 2000; Loots et al., 2000; Levy et al., 2001; Dubchak and Frazer, 2003; Guo and Moose, 2003; Hardison, 2003; Thomas et al., 2003; Loots and Ovcharenko, 2004; Bejerano et al., 2005; Siepel et al., 2005), chromosome-level regulatory regions (Loots et al., 2000), DNase I hypersensitive sites (Gottgens et al., 2001), and enhancers of vertebrate animal genes (such as sonic hedgehog; Goode et al., 2005). These latter highly conserved enhancers have been shown to mark bivalent states of chromatin associated with downregulation of genes in stem cells (Bernstein et al., 2006). In plants, one intronic CNS has been genetically analyzed and functions normally to prevent ectopic expression of the homeobox gene kn1 (Inada et al., 2003).
Given the evidence for TF binding sites in CNSs, we expect that some of our CNSs will carry binding motifs for known plant TFs. Because not all TF binding motifs are known, perhaps some short sequence motifs will be enriched in CNSs compared with nonconserved noncoding sequences and will later be found to bind TFs. A typical animal promoter might carry as many as 6 to 15 clusters of TF binding sites, with each TF binding site being between 5 and 12 bp, and each cluster binding four to eight different TF proteins (Wray, 2003). Although evidence indicates that plant proximal 5′ regions carry such cis-modules (Vandepoele et al., 2006), it is not yet clear how to apply animal results to plant genes, especially plant genes with very long 5′ or 3′ gene spaces. The PLACE cis-acting site database (Higo et al., 1999) lists 458 experimentally derived plant motifs (August, 2006). The median PLACE site is 8 bp long with some degree of sequence degeneracy. While finding these sites in a plant genome is trivial, proving a site is functional is a daunting task. Algorithms analyzing DNA sequence only rarely find function (Tompa et al., 2005) without the addition of expert knowledge such as coexpression or phylogenetic relatedness (Prakash et al., 2004; Van Hellemont et al., 2005). Using phylogenetic relatedness, ∼1400 enhancers are deeply (>300 million years) conserved in vertebrates (Woolfe et al., 2005).
One cis-acting binding motif is known to be vastly overrepresented in the plant genome, a microsatallite, and thus was truly expected to be enriched in CNSs: GAGA. GAGA (CTCT) motifs are known to bind particular proteins, and these are known to interact with chromatin remodeling complexes to alter animal gene expression (Lehman, 2004; Meister et al., 2004; Kooiker et al., 2005). Simple repeated sequences are also known to mark regions of the human X chromosome that avoid silencing (McNeil et al., 2006). To not exclude GAGA or any similar regulatory sequence, we included simple sequences in our CNS list. In so doing, we accepted the increased noise level expected when vastly overrepresented sequences locate in α-syntenous gene space by chance alone. As it turned out, few αCNSs are simple sequence repeats (Thomas et al., 2007).
Genes retained after either a local or whole-genome duplication are a biased subset of ancestral gene content (Freeling and Thomas, 2006). Therefore, the αCNSs may be a biased sample of functional sites simply because the genes available for CNS analysis are themselves biased. In the Arabidopsis lineage, genes in the large molecular function GO categories “transcription factor activity” and “protein kinase,” along with other genes whose products interact in complexes, tend to be retained following tetraploidies at frequencies nearly double expectations (Blanc and Wolfe, 2004; Seoighe and Gehring, 2004; Maere et al., 2005). So far, the combined data of gene retention following either local or whole-genome duplications in eukaryotes fits with the predictions of the gene balance hypothesis (Freeling and Thomas, 2006), but vertebrate tetraploidy data are more difficult to acquire. The gene balance hypothesis (Veitia, 2002; Birchler et al., 2003; Papp et al., 2003; Birchler et al., 2005) predicts that genes whose products function in subunit–subunit interactions or participate at the top of regulatory cascades will tend to be more susceptible to gene dosage change and therefore will be overretained following tetraploidy. For this reason, connected genes tend to be retained as pairs following tetraploidy. Since CNSs can only be detected in α-gene pairs, our CNS database is obviously biased toward connected genes.
RESULTS
Gene Retainability and CNS Richness Are Weakly or Not Correlated
Our CNS database is necessarily confined to the analysis of those 25% of genes that have α-pairs. To extrapolate from our data on functional noncoding sequence to the entire genome, we needed to access the relationship, if any, between retention post-tetraploidy and CNS richness. When genes are sorted by GO category and each was compared for both retention and CNS richness, no correlation was found (data not shown; see Supplemental Table 1 online). Because GO categories have overlapping gene contents, we attempted the same correlation among genes in different, nonoverlapping TF families (DATF families in Supplemental Table 1 online; Guo et al., 2005 at http://datf.cbi.pku.edu.cn/, July, 2005). There was no obvious correlation (graph not shown; see data in Supplemental Table 1 online). Had every gene in Arabidopsis been retained following the tetraploidy, and thus been available for analysis, we would have found approximately fourfold the number of αCNSs.
GAGA Motifs and PLACE TF Binding Motifs in αCNSs
GAGA
On the basis of published work (see Introduction), we expected that CNSs would preferentially contain GAGA repeats. We expected significantly more GAGA sequences in CNS sequence than in an equal amount of control noncoding sequence (see Methods); this we call “enrichment.” For GA6 with less than two mismatches (≤10/12), CNS enrichment was 0.8, meaning that there were actually more GA6-type sequences in control noncoding sequence than in CNS: GA6 = 0.6; GA5 = 0.9; GA4 = 1.16; and [GA] GA[GA]AG[GA][GA]A (Kooiker et al., 2005) = 1.0. We expected a GAGA CNS enrichment proportion to be significantly >1. This expectation was not met.
PLACE cis-Acting Binding Motifs
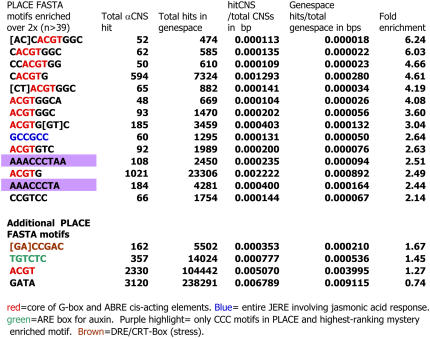
Data on CNS function suggests that some CNSs might be clusters of known TF binding sites (see Introduction). The PLACE Angiosperm cis-acting sequence database (Higo et al., 1999) had accumulated 431 such TF binding motifs on August 1, 2005. Each motif and its reverse complement, in IUPAC (Prosite-type) format, were found in both αCNS sequences and the control noncoding sequences within the same gene spaces. By requiring that at least 50 CNSs were hit by any motif reduced the list to 136 TF binding motifs. Enrichments ranged from 6.2-fold to 0.46-fold with a median at 0.9-fold. Fourteen motifs gave CNS enrichments more than twofold. Table 2 shows these data ranked descending for enrichment in CNSs; the top 14 and a few additional PLACE motifs are included here. The top-ranked eight motifs and 10 of the top 14 carry a G-box: CACGTG (Williams et al., 1992; Menkens et al., 1995; Gao et al., 2004). The specific PLACE sequence CACGTGGC, the most CNS-enriched PLACE motif (6.2-fold), is part of a Type I-AA_G-ABRE element, CCACGTGGC, know to operate in genes responsive to both abscisic acid (ABA) and external stress (Choi et al., 2000). TF binding sites for jasmonic acid response (pathogenic stress; Brown et al., 2003) and three additional known boxes (color-coded in Figure 1 and cited at PLACE) were enriched more than twofold in our αCNSs. Boxes DRE-CRT and ARE (thought to confer response to auxin) are enriched to a lesser extent. We conclude that the most CNS-enriched motif, the G-box, lies in a noncoding gene space near particular gene pairs. The 268 CNSs with a perfect G-box are noted in the CNS list (see Supplemental Table 2 online). The G-box CNSs are usually positioned 5′ of the ATG (229 are 5′, eight are in intron, 29 are 3′, and one is near a gene we called ourselves, designated -oa) at a mean distance of 1588 bp and a range from 40 to 8958 bps upstream of the A of the 5′ATG. Note that this distance is well 5′ of that considered to be the normal promoter region of a gene.
Table 2.
DATF Families and Counts in July, 2005, Showing Nonrandom Distribution of Both CNS Richness and Frequency of Genes That Are Bigfoot; Bigfoot Genes Are Possible among Retained Pairs Only
| TF Families | Total Genes | Genes Retained | Retention Frequency | Average No. CNS/Gene | Average Length Total CNS/Gene | Fraction Bigfoot | Bigfoot Frequency |
|---|---|---|---|---|---|---|---|
| TF:LUG | 1 | 0 | 0.00 | ||||
| TF:NZZ | 1 | 0 | 0.00 | ||||
| TF:SAP | 1 | 0 | 0.00 | ||||
| TF:CCAAT-DR1 | 2 | 2 | 1.00 | 2.00 | 75.00 | 0 of 2 | |
| TF:ULT | 2 | 2 | 1.00 | 3.00 | 66.00 | 0 of 2 | |
| TF:S1Fa-like | 2 | 0 | 0.00 | ||||
| TF:VOZ | 2 | 0 | 0.00 | ||||
| TF:GIF | 3 | 2 | 0.67 | 0.00 | 0.00 | 0 of 2 | |
| TF:HRT-like | 3 | 2 | 0.67 | 0.00 | |||
| TF:MBF1 | 3 | 2 | 0.67 | 0.00 | 0.00 | 0 of 2 | |
| TF:PBF-2-like (Whirly) | 3 | 2 | 0.67 | 1.00 | 21.00 | 0 of 2 | |
| TF:LFY | 3 | 0 | 0.00 | ||||
| TF:PcG | 4 | 2 | 0.50 | 0.00 | 0.00 | 0 of 2 | |
| TF:C2C2-YABBY | 5 | 0 | 0.00 | ||||
| TF:FHA | 5 | 0 | 0.00 | ||||
| TF:EIL | 6 | 2 | 0.33 | 0.00 | 0.00 | ||
| TF:LIM | 6 | 4 | 0.67 | 0.50 | 10.50 | ||
| TF:BES1 | 6 | 2 | 0.33 | 5.00 | 140.00 | 0 of 2 | |
| TF:ALFIN | 7 | 6 | 0.86 | 1.00 | 19.33 | 0 of 6 | |
| TF:CAMTA | 7 | 6 | 0.86 | 1.00 | 32.67 | 0 of 6 | |
| TF:ARID | 7 | 4 | 0.57 | 3.00 | 132.50 | 0 0f 2 | |
| TF:E2F/DP | 8 | 2 | 0.25 | 0.00 | 0.00 | ||
| TF:CPP | 8 | 4 | 0.50 | 8.00 | 239.50 | 0 of 4 | |
| TF:PLATZ | 9 | 4 | 0.44 | 5.00 | 140.00 | 0 of 4 | |
| TF:HMG | 10 | 4 | 0.40 | 0.00 | 0.00 | 0 of 4 | |
| TF:CCAAT-HAP2 | 10 | 6 | 0.60 | 1.67 | 38.67 | 2 of 6 | 0.33 |
| TF:TAZ | 10 | 6 | 0.60 | 3.33 | 117.67 | 0 of 6 | 0.00 |
| TF:GRF | 10 | 4 | 0.40 | 6.00 | 286.50 | 0 of 4 | |
| TF:SRS | 10 | 6 | 0.60 | 11.00 | 514.67 | 2 of 6 | 0.33 |
| TF:TUB | 11 | 2 | 0.18 | 2.00 | 37.00 | 0 of 2 | |
| TF:CCAAT-HAP3 | 11 | 2 | 0.18 | 7.00 | 131.00 | 0 of 2 | |
| TF:PHD | 12 | 2 | 0.17 | 3.00 | 68.00 | 0 of 2 | |
| TF:JUMONJI | 13 | 2 | 0.15 | 1.00 | 28.00 | 0 of 2 | |
| TF:CCAAT-HAP5 | 13 | 4 | 0.31 | 2.00 | 73.00 | 0 of4 | |
| TF:GARP-ARR-B | 13 | 6 | 0.46 | 2.67 | 100.33 | 2 of 6 | 0.33 |
| TF:ABI3/VP1 | 13 | 6 | 0.46 | 13.67 | 528.33 | 4 of 6 | 0.67 |
| TF:Nin-like | 14 | 8 | 0.57 | 3.00 | 82.25 | 0 f 8 | 0.00 |
| TF:ZIM | 15 | 10 | 0.67 | 1.40 | 28.00 | 0 of 10 | 0.00 |
| TF:ZF-HD | 15 | 4 | 0.27 | 13.50 | 450.00 | 4 of 4 | 1.00 |
| TF:GeBP | 16 | 4 | 0.25 | 0.00 | 0.00 | 0 of 4 | |
| TF:SBP | 17 | 6 | 0.35 | 2.67 | 101.67 | 2 0f 6 | 0.33 |
| TF:ARF | 23 | 2 | 0.09 | 6.00 | 211.00 | 2 of 2 | 1.00 |
| TF:HSF | 24 | 4 | 0.17 | 2.50 | 79.50 | 0 of 4 | |
| TF:TCP | 24 | 8 | 0.33 | 5.13 | 179.00 | 2 of 8 | 0.25 |
| TF:Trihelix | 29 | 12 | 0.41 | 2.33 | 68.67 | 2 of 12 | 0.17 |
| TF:C2C2-GATA | 29 | 16 | 0.55 | 5.38 | 210.75 | 4 of 16 | 0.25 |
| TF:AUX/IAA | 29 | 16 | 0.55 | 5.63 | 211.75 | 2 of 16 | 0.13 |
| TF:C2C2-co-like | 31 | 16 | 0.52 | 3.00 | 118.25 | 0 of 16 | 0.00 |
| TF:GRAS | 33 | 15 | 0.45 | 3.27 | 85.67 | 6 of 15 | 0.40 |
| TF:C3H | 35 | 16 | 0.46 | 2.06 | 48.13 | 2 of 16 | 0.13 |
| TF:C2C2-DOF | 36 | 16 | 0.44 | 6.19 | 213.88 | 6 of 16 | 0.38 |
| TF:B3 | 39 | 4 | 0.10 | 6.50 | 116.00 | 0 of 4 | |
| TF:AS2 | 42 | 8 | 0.19 | 4.25 | 118.75 | 2 of 8 | 0.25 |
| TF:GARP-G2-like | 43 | 16 | 0.37 | 7.00 | 239.13 | 2 of 16 | 0.13 |
| TF:WRKY | 73 | 24 | 0.33 | 3.75 | 109.00 | 4 of 24 | 0.17 |
| TF:bZIP | 75 | 30 | 0.40 | 3.87 | 122.40 | 2 of 16 | 0.13 |
| TF:HB | 94 | 44 | 0.47 | 9.64 | 333.95 | 24 of 44 | 0.55 |
| TF:MADS | 108 | 20 | 0.19 | 4.38 | 138.57 | 0 of 20 | 0.00 |
| TF:NAC | 116 | 40 | 0.34 | 5.05 | 185.00 | 8 of 40 | 0.20 |
| TF:C2H2 | 131 | 44 | 0.34 | 5.82 | 225.91 | 8 of 44 | 0.18 |
| TF:AP2/EREBP | 146 | 75 | 0.51 | 5.16 | 158.67 | 28 of 75 | 0.37 |
| TF:bHLH | 168 | 68 | 0.40 | 5.75 | 188.32 | 14 of 68 | 0.21 |
| TF:MYB | 207 | 100 | 0.48 | 6.00 | 213.90 | 28 of 100 | 0.28 |
| Totals | 1852 | 724 | 0.39 |
Figure 1.
The Fasta Motifs from the PLACE Database That Are Most Enriched in CNSs Compared with Adjacent Noncoding NonCNS Gene Space.
The color codes are explained within the figure. Citations for each motif are at PLACE (http://www.dna.affrc.go.jp/PLACE/).
Bigfoot and Smallfoot Genes
The Problem Caused by a Few Hundred Very CNS-Rich Genes
Thomas et al. (2007) showed that some genes are particularly CNS rich, and these genes tend to fall into GO categories that are TF or MIR, each with a mean of approximately five CNSs/gene. The most CNS-enriched genes (10 to 20 CNSs/gene space) populate GO categories characterized by “response to…” plant hormones, internal or exogenous stress, light, desiccation, jasmonic acid, and so forth. This previous work engendered the hypothesis that CNS richness characterizes, in particular, genes that respond first to signals of all sorts: the first responder hypothesis. However, the average paired gene in Arabidopsis has 1.7 CNSs, and the modal gene has 0. These more CNS average genes tend to encode enzymes or structural proteins. The result is that a few CNS-rich genes dominate our CNS database with their many and potentially special CNSs. To address this problem, we created two subdatabases, one from the longest gene spaces in Arabidopsis (Bigfoot genes) and the other from genes with a modest number, three to five, of CNSs/gene (called Smallfoot genes). Table 3, row 1, quantifies the genes and sequences that define these two subdatabases, and Table 3 compares them. Twenty-four genes were shared.
Table 3.
Comparison of Features of Bigfoot and Smallfoot Genes, Where Bigfoot Genes Have >4 bp of Syntenous, CNS-Rich Space 3′+5′ of the CDS, Smallfoot Genes Have Three to Five CNSs, and 24 Genes Are Both Bigfoot and Smallfoot
| Feature | Bigfoot Genes | Smallfoot Genes |
|---|---|---|
| Number of genes (CNSs; total bp CNS; total bp control noncoding) | 252 genes (2897; 459,548 bp; 26,141,400 bp) | 1197 genes (7532; 252,934 bp; 5,426,926 bp) |
| GO categories | Narrow spectrum, 78% TFs; many respond to environmental or hormonal signals (Table 4) | Broad spectrum. Signal transduction, then transcription, metabolism, and cytoskeleton (Table 5) |
| Does the direction of transcription influence enrichment/purification of a random 7-mer? | None significantly preferred complement versus reverse complement, and only one pair had a nominal P < 0.001 (see Supplemental Table 3 online) | Many 7-mers were polar. Of the eight significantly enriched, two were significantly polar and four were polar at P nominal <0.001. The next most significantly enriched were 13% polar (see Supplemental Table 4 online) |
| How important is purifying selection in determining the overall sequence of gene space? | Very. The difference between slope 0.45 and the expected 1.0 of the hits CNS versus hits control of the trend line of Figure 3A implies massive purification of CNSs. | Very. The difference between the slope 0.51 and 1.0 on the similar plot as discussed for Bigfoot genes attests to importance of purification (data not shown). |
| Which 7-mer sequences tend to be purified from CNSs (removed)? | Most purified are long runs of A or T, and purified are almost any sequence with more than three As or Ts in a row. In general, [A+T] is purified. More research is needed. | Similar to Bigfoot genes. A general definition of CNS should include sequences purified and sequences under positive selection. |
| Which 7-mers are enriched? Are they TF binding sites? | The most significant carry CACGTG (G-box binds bZIP or HLH TFs), plus a few known TF binding sites, but most are unknown (see Supplemental Table 3 online). | One of the eight significant 7-mers could bind a MYC TF (see Supplemental Table 4 online). As with Bigfoot genes, significant and “worth interest” 7-mers are mostly unknown (e.g., 5′-CTTCTTC). |
Bigfoot Genes
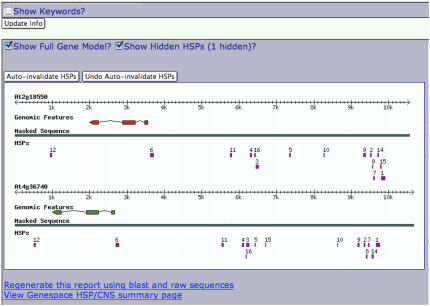
Figure 2 shows a screenshot of a Bigfoot gene pair in the Arabidopsis Bl2seq Viewer (Thomas et al., 2006, 2007). Note that it is transcribed from right to left. More often than not, the 126 pairs of Bigfoot genes exist in a region of chromosome devoid of other genes, with CNSs populating the void (Figure 1). Comparisons of vertebrate genomes identified gene deserts that are adjacent to TF and developmental genes and are full of deeply conserved noncoding sequences (Ovcharenko et al., 2005); Bigfoot gene space seems to be a plant convergence on the gene desert phenomenon. The 252 Bigfoot genes are noted in Supplemental Table 1 online. Using GOStat (see Methods), there were 140 to 142 genes each in four overrepresented (P = 0.00) GO categories: 0005488, binding; 0003677, DNA binding; 0003700, TF activity; and 0003676, nucleic acid binding. Other significantly overrepresented GO categories (P < 3 × 10−8) were several with the phrase “regulation of…” and GO:0008755 hormone-mediated signaling. Bigfoot genes are predominantly (∼66%) TF genes. Other very broad categories of Bigfoot gene pairs are 16 unknown protein pairs, six protein kinase pairs, 18 enzyme pairs, one RNA binding pair and, interestingly, two DVL gene pairs (A12N061 and A08N202). DVL genes (genes that were not in The Arabidopsis Information Resource [TAIR]; May, 2005), are a group of genes encoding small polypeptides that, when overexpressed, confer developmental phenotypes (Wen et al., 2004). The GOstat results for Bigfoot genes (Table 4) expand upon the “first responder” list of GO terms that are most CNS rich of all α-pairs in Arabidopsis (Thomas et al., 2007). It was not a surprise that particularly long gene spaces (Bigfoot) tended to be CNS rich.
Figure 2.
Partial Viewer Screen Shot of Frozen Gene Space of an HD-ZIP TF Homoeologous Gene Pair, an Exemplary Bigfoot Gene Space.
Orientation is −/−; transcription is right to left. The space is enclosed by CNS1 at the 5′ end and CNS12 at the 3′ end. CNS10 is colored blue because it is oriented backward (−/+) from exon orientation. The solid colored exons of the models (GenBank from TAIR, The Institute for Genomic Research [TIGR] version 5) have no untranslated region annotations, suggesting that cDNA evidence was lacking. CNS15 is colored red, denoting that it was invalidated during the proofing stages of CNS database construction (Thomas et al., 2007).
Table 4.
256 Bigfoot Genes Are Enriched for Particular GO Categories (Ranked by Significance)
| Rank | GO | Gross Function | GO Description | Count of 256 | Total Count | GOStat P Value |
|---|---|---|---|---|---|---|
| 1 | GO:0006355 | Transcription | Regulation of transcription, DNA dependent | 54 | 1114 | 0 |
| 2 | GO:0006350 | Transcription | Transcription | 96 | 1965 | 0 |
| 3 | GO:0045449 | Transcription | Regulation of transcription | 96 | 1845 | 0 |
| 4 | GO:0006139 | Transcription | Nucleoside, -tide, base NA metabolism | 96 | 3144 | 0 |
| 5 | GO:0019222 | Regulation of… | Regulation of metabolism | 97 | 1915 | 0 |
| 6 | GO:0003677 | Transcription | DNA binding | 104 | 2792 | 0 |
| 7 | GO:0003700 | Transcription | TF activity | 104 | 2060 | 0 |
| 8 | GO:0050791 | Regulation of… | Regulation of physiological processes | 97 | 2118 | 0 |
| 9 | GO:0019219 | Regulation of… | Nucleic acid regulation of | 96 | 1859 | 0 |
| 10 | GO:0050794 | Regulation of… | Regulation of cellular processes | 96 | 2079 | 0 |
| 11 | GO:0031323 | Regulation of… | Regulation of physiological process | 96 | 1896 | 0 |
| 12 | GO:0050789 | Regulation of… | Regulation of biological process | 97 | 2331 | 0 |
| 13 | GO:0003676 | Transcription | NA binding | 106 | 3956 | 0 |
| 14 | GO:0051244 | Regulation of… | Regulation of cellular physiological process | 96 | 2075 | 0 |
| 15 | GO:0006351 | Transcription | Transcription, DNA-dependent | 54 | 1158 | 4.52E-86 |
| 16 | GO:0005634 | Nucleus | Nucleus | 78 | 2536 | 2.24E-75 |
| 17 | GO:0044238 | Metabolism | Primary metbolism | 113 | 8953 | 1.42E-28 |
| 18 | GO:0044237 | Metabolism | Cellular metabolism | 116 | 9761 | 3.61E-26 |
| 19 | GO:0008152 | Metabolism | Metabolism | 121 | 11086 | 4.75E-23 |
| 20 | GO:0043231 | Organelle | Intracellular membrane-bound organelle | 114 | 10654 | 3.07E-20 |
| 21 | GO:0043227 | Organelle | Membrane-bound organelle | 114 | 10655 | 3.07E-20 |
| 22 | GO:0050875 | Organelle | Cellular physiological process | 125 | 12540 | 1.67E-19 |
| 23 | GO:0043229 | Organelle | Intracellular organelle | 115 | 11089 | 4.68E-19 |
| 24 | GO:0043226 | Organelle | Organelle | 115 | 11090 | 4.68E-19 |
| 25 | GO:0009987 | Cell | Cellular process | 126 | 13047 | 3.02E-18 |
| 26 | GO:0005622 | Cell | Intracellular | 119 | 11949 | 3.92E-18 |
| 27 | GO:0042221 | Response to… | Response to chemical stimulus | 25 | 1142 | 5.34E-14 |
| 28 | GO:0009719 | Response to… | Response to endogenous stimulus | 24 | 947 | 1.34E-09 |
| 29 | GO:0009723 | Response to… | Response to ethylene stimulus | 11 | 138 | 1.38E-09 |
| 30 | GO:0009725 | Response to… | Response to hormone stimulus | 20 | 655 | 1.95E-09 |
| 31 | GO:0009628 | Response to… | Response to abiotic stimulus | 25 | 1627 | 9.37E-08 |
| 32 | GO:0005623 | Cell | Cell | 130 | 18237 | 1.01E-06 |
| 33 | GO:0009873 | Response to… | Ethylene-mediated signaling pathway | 6 | 51 | 1.88E-06 |
| 34 | GO:0009753 | Response to… | Response to jasmonic acid stimulus | 8 | 124 | 2.02E-06 |
| 35 | GO:0009861 | Response to… | Response to wounding stress | 8 | 154 | 1.02E-05 |
| 36 | GO:0009651 | Response to… | Response to salt stress | 7 | 112 | 1.33E-05 |
| 37 | GO:0000160 | Signal transduction | Two-component signal transduction (phosphorylation) | 6 | 74 | 1.56E-05 |
| 38 | GO:0009751 | Response to… | Response to salyclic acid stimulus | 7 | 121 | 2.11E-05 |
| 39 | GO:0006970 | Response to… | Response to osmotic stress | 7 | 134 | 3.95E-05 |
| 40 | GO:0009611 | Response to… | Response to wounding | 8 | 210 | 8.55E-05 |
| 41 | GO:0009737 | Response to… | Response to ABA stimulus | 7 | 177 | 0.00023 |
| 42 | GO:0009733 | Response to… | Response to auxin stimulus | 8 | 261 | 0.00040 |
| 43 | GO:0009814 | Response to… | Response to pathogen | 8 | 264 | 0.00040 |
We investigated whether or not Bigfoot genes occur at random in the phylogenetic trees of gene families. To answer this question, we looked more closely at the 56 TF families recognized by DATF (Table 2). Twenty-four of 44 α-paired homeobox (HB) genes are Bigfoot (55%), but 0 of 20 MADS box α-paired genes are Bigfoot. The range of percentage of Bigfoot among all TF families is greater yet (even though some families have so few α-paired genes that they cannot be studied at this time). Breaking the 24 HB Bigfoot genes into subfamilies, the HD-ZIPI and HD-ZIPII subfamilies are almost all Bigfoot (90%), while those genes in the sister HD-ZIPIII and -IV subfamilies are more like the average TF. The Bigfoot notation measures a CNS property that is positively correlated with CNS richness but is a unique feature of gene space. As with CNS richness, Bigfoot genes characterize some gene lineages but not others.
Smallfoot Genes
We then subjected a list of 1197 Smallfoot genes to the same analyses described for Bigfoot genes. Table 5 shows the GOStat results: 45 terms were enriched at P < 0.001, and 20 of them involve signal transduction; the six highest ranked terms identify protein kinase genes, including GO:004713, protein Tyr kinase activity (rank 1, P = 9E-12), and GO:004674. GO:0045449, regulation of transcription (rank 8, P = 1.9E-9), and GO:003700, TF activity (rank 24, P = E-7), are also represented. Lower-ranked terms include cytoskeleton and RNA helicase. Note that no term involving signal transduction was significantly enriched in Bigfoot genes (Table 4), so Bigfoot and Smallfoot genes function quite differently.
Table 5.
1197 Smallfoot Genes Are Enriched for Particular GO Categories (Ranked by Significance)
| Rank | GO | Gross Function | GO Description | Count of 1197 | Total Count | GOStat P Value |
|---|---|---|---|---|---|---|
| 1 | GO:0004713 | Signal transduction | Protein Tyr kinase activity | 46 | 650 | 9.09E-12 |
| 2 | GO:0005524 | Signal transduction | ATP binding | 84 | 1630 | 8.22E-11 |
| 3 | GO:0004674 | Signal transduction | Protein Ser-Thr kinase activity | 56 | 927 | 1.39E-10 |
| 4 | GO:0030554 | Signal transduction | A nucleotide binding | 84 | 1663 | 1.80E-10 |
| 5 | GO:0017076 | Signal transduction | Purine nucleotide binding | 91 | 1909 | 1.06E-09 |
| 6 | GO:0006468 | Signal transduction | Protein–amino acid phosphorylation | 60 | 1071 | 1.06E-09 |
| 7 | GO:0045449 | Transcription | Regulation of transcription | 88 | 1845 | 1.77E-09 |
| 8 | GO:0019219 | Transcription | Nucleic acid metbolism | 88 | 1859 | 2.62E-09 |
| 9 | GO:0000166 | Signal transduction | Nucleotide binding | 99 | 2186 | 2.93E-09 |
| 10 | GO:0051244 | Regulation of… | Regulation of physiological process | 95 | 2075 | 3.27E-09 |
| 11 | GO:0050794 | Regulation of… | Regulation of cellular process | 95 | 2079 | 3.27E-09 |
| 12 | GO:0016773 | Signal transduction | P-transferase activated alcohol acceptor | 64 | 1210 | 3.27E-09 |
| 13 | GO:0016310 | Signal transduction | Phosphorylation | 62 | 1166 | 4.67E-09 |
| 14 | GO:0004672 | Signal transduction | Protein kinase activity | 57 | 1038 | 4.95E-09 |
| 15 | GO:0031323 | Regulation of… | Regulation of cellular metabolism | 88 | 1896 | 5.34E-09 |
| 16 | GO:0050791 | Regulation of … | Regulation of physiological process | 95 | 2118 | 8.65E-09 |
| 17 | GO:0019222 | Regulation of … | Regulation of metabolism | 88 | 1915 | 9.13E-09 |
| 18 | GO:0006796 | Signal transduction | Phosphate metabolism | 63 | 1229 | 1.84E-08 |
| 19 | GO:0006793 | Signal transduction | Phosphorus metabolism | 63 | 1230 | 1.84E-08 |
| 20 | GO:0006350 | Transcription | Transcription | 88 | 1965 | 4.08E-08 |
| 21 | GO:0043283 | Enzyme | Biopolymer metabolism | 113 | 2742 | 5.32E-08 |
| 22 | GO:0006464 | Signal transduction | Protein modification | 72 | 1513 | 6.06E-08 |
| 23 | GO:0050789 | Regulation of… | Regulation of biological processes | 99 | 2331 | 9.50E-08 |
| 24 | GO:0003700 | Transcription | TF activity | 90 | 2060 | 1.01E-07 |
| 25 | GO:0043412 | Signal transduction | Biopolymer modification | 73 | 1637 | 1.39E-06 |
| 26 | GO:0016772 | Signal transduction | Transferring P-containing groups | 81 | 1896 | 2.27E-06 |
| 27 | GO:0006139 | Signal transduction | Nucleobase, -side, -tide, nucleic acid metabolism | 120 | 3144 | 2.41E-06 |
| 28 | GO:0005634 | Nucleus | Nucleus | 101 | 2536 | 2.92E-06 |
| 29 | GO:0016301 | Signal transduction | Kinase activity | 72 | 1657 | 5.52E-06 |
| 30 | GO:0007169 | Signal transduction | Transmembrane receptor protein Tyr kinase signaling pathway | 16 | 137 | 6.17E-06 |
| 31 | GO:0007167 | Signal transduction | Enzyme-linked receptor protein signaling pathway | 16 | 137 | 6.17E-06 |
| 32 | GO:0003677 | Transcription | DNA binding | 106 | 2792 | 1.92E-05 |
| 33 | GO:0005515 | Regulation of… | Protein binding | 82 | 2027 | 2.35E-05 |
| 34 | GO:0044238 | Metabolism | Primary metabolism | 275 | 8953 | 4.34E-05 |
| 35 | GO:0007010 | Cytoskeleton | Cytoskeletal organization and biogenesis | 17 | 191 | 0.00010 |
| 36 | GO:0015630 | Cytoskeleton | Microtubule cytoskeleton | 13 | 125 | 0.00024 |
| 37 | GO:0030163 | Enzyme | Protein catabolism | 19 | 294 | 0.00027 |
| 38 | GO:0007166 | Signal transduction | Cell surface receptor-linked signal transduction | 16 | 187 | 0.00028 |
| 39 | GO:0007018 | Cytoskeleton | Microtubule-based movement | 9 | 63 | 0.00041 |
| 40 | GO:0003676 | Transcription | Nucleic acid binding | 134 | 3956 | 0.00043 |
| 41 | GO:0046910 | Enzyme | Pectinesterase inhibitor activity | 9 | 64 | 0.00045 |
| 42 | GO:0007017 | Cytoskeleton | Microtubule-based process | 11 | 101 | 0.00064 |
| 43 | GO:0003724 | Enzyme | RNA helicase activity | 5 | 17 | 0.00075 |
| 44 | GO:0005875 | Cytoskeleton | Microtubule-associated complex | 9 | 69 | 0.00076 |
| 45 | GO:0043285 | Enzyme | Biopolymer catabolism | 19 | 315 | 0.00100 |
Enriched and Purified Motifs in Bigfoot αCNSs
Bigfoot Genes
As presented previously, most Bigfoot genes encode TFs, most are CNS rich, and all use an exceptional amount of chromosomes for conserved function (Figure 2, Table 2). Even so, Bigfoot CNSs comprise only 4.3% of the average Bigfoot gene space. The Bigfoot subdatabase contains 2538 αCNS sequences (see Supplemental Table 2 online) whose total sequence lengths sum to 91,970 bps of nonoverlapping, aligned (5′-3′) αCNS sequence. These CNSs are distributed around Bigfoot genes with a greater overall 5′:3′ bias (3.1:1) than the similar statistic for the average gene (1.8:1). The frequency and distribution of these CNSs will, of course, influence the frequency and distribution of motifs located within CNSs; that's why these CNS distribution data are reviewed here.
Random 7-bp Motif Enrichment and Purification
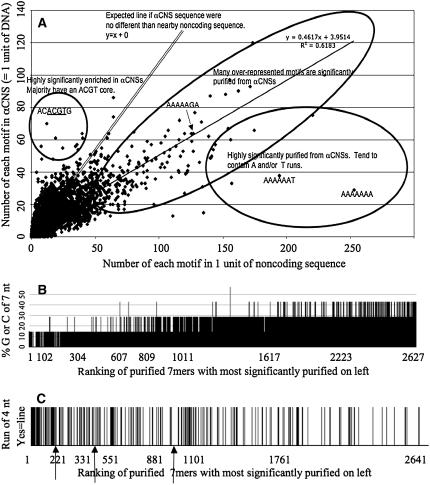
Supplemental Table 3 online lists all 16,384 (47) possible 7-bp DNA sequence motifs and data on their properties, hit numbers, and hit locations in the aligned Bigfoot subdatabase. Data on each motif in the database include number and frequency of nonoverlapping hits to αCNS (for both complement and reverse complement), hits to gene space control sequences, the ratio of αCNS hits to control hits (called “fold enrichment” if αCNSs are preferred and “fold purification” if noncoding nonCNS gene space is preferred), and a χ2 evaluation of significance (see Methods) for those 7-mers with hits totaling 10 or more; nominal P values are listed and evaluated in a special column based on Bonferroni corrections for multiple tests (see Methods). Figure 3A plots the number of occurrences of each of the 16,384 motifs plus its reverse complement in αCNSs (y axis) versus an equal amount of gene space control sequences (x axis; see Methods). Given normal distributions, and no difference between αCNS sequence and nearby noncoding sequence, a trend line in Figure 3A should emerge with a slope of 1.0; this slope, the null hypothesis, reflects a 1:1 motif hit ratio between CNS and control noncoding sequence. Unexpectedly, the slope of the trend line in Figure 3A is 0.46. Many of the 7-bp motifs that are most overrepresented in noncoding space (enclosed in the largest oval in Figure 3A) have been purified from αCNS sequences, so much so that the slope of the trend line is far below 1.0. Those motifs most overrepresented in noncoding sequence in general (high A+T sequence and especially in runs; Figures 3B and 3C) tend not to be in CNSs. There are 150 significantly purified 7-mers and 282 additional 7-mers that have low enough P values to be worth interest (see Methods). Supplemental Tables 3 and 4 online have these 7-mer rows color-coded in pink and blue, respectively.
Figure 3.
The Nature of Random 7-Mer Hits to Bigfoot Gene αCNSs.
(A) Plot is the number of each 7-mer motif in CNSs (y axis) versus number of hits in control noncoding DNA (x axis) where the expected hit ratio is 1:1 (primary data; see Supplemental Table 3 online). Each point is a particular 7-mer. The slope of the correlation line, 0.46 and not 1.0, and the volume of the points that define it (large oval) imply that many overrepresented 7-mers are removed (purified) from αCNSs (rightmost oval is the most purified). Some 7-mers are most enriched in αCNSs (circle at left). The 14 most significantly enriched 7-mers, in descending order of their significance, are as follows (all P < E-5): 5′-ACACGT, CACGTGT, CACGTGA, TCACGTG, CACGTGC, GCACGTG, CACGTGG, CCACGTG, ACGTGGC, GCCACGT, CATGTGA, TCACATG (the MYCATERD1 box at PLACE: dehydration stress [Tran et al., 2004]), GGACCAC, and GTGGTCC (not in PLACE).
(B) and (C) These graphs illustrate the nucleotide content of purified 7-mers. The x axis is all purified 7-mers ranked from most significantly purified (lowest P value) on left to least significant on right, with arrows denoting the boundaries of the three nominal P value groups that are below P = 0.05 (95% confidence is all to the left of the rightmost arrow).
(B) Plots percentage of GC of 7-mer (y axis) versus significance of purification.
(C) Plots “yes or no” to the question, “Is there a run of four nucleotides in this 7-mer?” versus significance of purification, where a vertical line denotes “yes.” The three arrows denote, from left to right, nominal P values for significance of purification: P = 10−5, 10−3, and 0.05. Note that 7-mer purification is elevated with high percentage of AT and runs of the same nucleotide.
We found 14 motifs, seven pairs, that are significantly enriched (overrepresented, the opposite of purified) in Bigfoot αCNSs; those furthest from and above the trend line are circled in Figure 3. Most of these motifs have a core ACGT. Each point on Figure 3, each representing the sum of complement and reverse complement hits, was evaluated for significance of difference from an expected (via null hypothesis) 1:1 αCNS:control ratio (see Methods). All data points enclosed within circles or ovals in Figure 3A are significantly unexpected (see Supplemental Table 3 online).
Among those 7-mers enriched in αCNSs, 14 (including reverse complements) are enriched significantly. These 14 are listed in the legend of Figure 3. Ten of these 14 motifs carry a complete G-box, or are consistent with overlapping a complete G box, and all have the core ACGT. The additional significantly enriched 7-mer motifs were experimentally unprecedented: a CATG core and a CCAC/GTGG core. The fold enrichment for these 14 most significant 7-bp sequences varies from 3.21 (TCACATG and reverse complement: 61 hits) to 9.89 (CACGTGC and reverse complement: 40 hits). Among all 8-mers with hits in Bigfoot gene αCNSs >9, GCACGTGC was most enriched at 23-fold (n = 10). This maximum-enriched 8-mer is clearly related to the most enriched PLACE motif (stress and ABA responsiveness) discussed previously: CACGTGGC. Among all 8 bp, this PLACE motif also ranks among the most enriched in the Bigfoot αCNSs at 9.7-fold (n = 22).
There are 28 7-mers, those that hit Bigfoot CNSs in the “worth interest” category of significance; these usually share core motifs CATG, ACGT (G-box core), or CCAC/GTGG that are known to be associated with significantly enriched 7-mers and sometimes group into additional orderly patterns.
GAGA motifs are not significantly enriched. The GAGAGAG/AGAGAGA 7-mers (and reverse complements) were 2.1-fold enriched with an insignificant P value, one below our “worth interest” category. The most significantly enriched GAGA-like sequence is GGAGAAG and its reverse complement at 2.75-fold enrichment (nominal P = 0.003), with 30 hits in the Bigfoot αCNS; this 7-mer is also not “worth interest.”
Sequences That Are Missing (Purified) from the αCNSs of Bigfoot Genes
We use the term “purified” to be the opposite of “enriched” to connote the selective process that presumably removed unfit sequence from functional regions of gene space. A total of 150 7-bp sequences are significantly purified from BF αCNSs, and an additional 282 7-mers are purified at “worth interest” levels of significance. Many of the significantly purified 7-mers are highly abundant in noncoding space, which accounts for the 0.46 slope of the trend line in Figure 3A. Figures 3B and 3C plot all purified 7-mers that hit a Bigfoot CNS at least 10 times in the order of their purification significance, with the most significant on the left (arrows in Figure 3C at P nominal < 0.00001, P nominal < 0.001, and P nominal < 0.05). The x axes are percentage of GC of the 7-mer (Figure 3B) and 7-mers with runs of 4 nucleotides (Figure 3C). It can be seen from Figures 3B and 3C that significant purification involves runs of A and T and generally high percentage of AT, the very sequences that characterize the bulk of noncoding space.
Distribution of Significantly Enriched or Purified 7-bp Sequences within Bigfoot Gene Space and Strand Preference
The 5′:3′ ratio of the 2538 CNSs around Bigfoot genes, the genes used for this 7-mer analysis, is 3.1: 1, and the “worth interest” 7-mers 3.8:1 5′:3′. Those 14 most significantly enriched 7-mers exhibit a 5′:3′ ratio of 7.3:1. The mean 5′ αCNS hit position for the significantly enriched 7-mers is 3.15 kb upstream of the 5′ATG; the mean 3′ position is 1.9 kb downstream from the stop codon upstream. Data for the 28 “worth interest” 7-mers are similar. These average locations within Bigfoot gene space are well outside the transcription unit and outside the 0 to −500-bp proximal promoter that is generally studied experimentally.
None of the 7-mers to Bigfoot CNSs showed preference for orientation with regard to the direction of transcription; there were neither significant nor “worth interest” motifs (see Supplemental Table 3 online). This is in contrast with similar data for Smallfoot genes, as will be shown.
Smallfoot Gene Data
We prepared an aligned subdatabase of αCNSs with genes labeled as Smallfoot genes as described and subjected this gene list to the same analyses as was the Bigfoot gene list (Supplemental Table 5 online is the 7-mer to Smallfoot genes data sheet). Tables 4 and 5 show that the gene GO annotations of Bigfoot and Smallfoot genes are very different. In general, the 7-mers enriched were very different as well, as summarized in Table 3 and Supplemental Table 4 online. Those eight sequences (complement only) enriched significantly and 53 sequences enriched at a “worth interest” significance level were, with one possible exception (a MYC gene binding site; see Supplemental Table 4 online), without experimental precedent and not the same as were hit in the Bigfoot database. The most significantly enriched 7-mer is 5′CTTCTTC, and there are several more reasonably similar sequences ranking among the 61 most significant. There are several sequences that are 5′CACG-like, all of unknown function. In general, 7-mers that significantly hit Smallfoot CNSs are approximately five times more numerous but about half as enriched as are the comparable Bigfoot 7-mers (Table 3). If we had twice as many 7-mer hits, as would be the case with a twice-as-large database, there would be a manifold increase in the number of significantly enriched 7-mers.
For 7-mers to Bigfoot genes, it didn't matter whether the complement or reverse complement was used. Smallfoot gene CNSs hit significantly by 7-mers are often biased (Table 3; see Supplemental Tables 3 and 4 online) to one strand or the other. This directionality fits the compact nature of Smallfoot genes and tends to validate the 7-mer motif's functionality in relation to transcription or translation of Smallfoot genes. Twenty-five percent of significantly enriched and 5% of “worth interest”-enriched 7-mers are biased significantly (Bonferroni-corrected; see Methods). These percentages rise to 50 and 13%, respectively, if strand bias is judged at the P nominal < 0.001 level. Even purified sequences to Smallfoot genes show strand bias at approximately one-third the values of enriched 7-mers. Under each of these conditions, every 7-mer to Bigfoot gene CNS showed zero stand bias; Bigfoot CNSs provided an exceptionally useful control.
G-Boxes in CNSs Are Not Significantly Associated with Conserved Neighbor 7-Mers or Motifs, Except Other G-Boxes
The G-box, CACGTG, has been studied in detail for two decades. Some of these studies have found associated motifs that, with the G-box, comprise a cis-acting array. However, the G-boxes have been in core promoters (not as far upstream as our CNS G-boxes). Studies on ribulose-1-5-bisphosphate carboxylase small subunit genes, for example, have discovered a conserved modular array consisting of a G-box and an adjacent I-box (Arguello-Astorga and Herrera-Estralla, 1998), and a GCC motif (jasmonic acid, GCCGCC) was found near a G-box in pmt1a gene of tobacco (Nicotiana tabacum; Xu and Timko, 2004), and both were necessary for jasmonic acid induction. Weaker associations between G-boxes and adjacent sequences have been reported (Giuliano et al., 1988). The Arabidopsis G-box and an I-box–like motif (ATAATCCA) were associated with photosynthesis GO terms in Arabidopsis (Vanderpoele et al., 2006). None of these G-box–linked motifs or any PLACE motif was found significantly overrepresented in the same CNS as a G-box. The single exception is another G-box (see Supplemental Table 2 online). The frequency of finding an exact G-box in a CNS is 0.02 (297/14,944). Of the 136 G-box CNSs that were long enough to extend 20 bp on both sides, nine had at least one other exact G-box in this 46-bp sequence, for a frequency of 0.07. A crude expectation for two or more G-boxes in any one CNS by chance would be (0.02)2 = 0.0004. The observed 0.07 exceeds null hypothesis expectations by 175-fold. We also used the DIALIGN Web application (Morgenstern et al., 2006) on all 267 G-box CNSs and their reverse complements, anchored on the G-boxes and strict global positioning around all G-boxes, to search for statistically significant nucleotides or motifs conserved near G-boxes; no motifs were found other than the G-box itself. We compiled a table of all CNSs and all significantly enriched Bigfoot 7-mers and looked for any co-occurrence of 7-mer pairs to CNSs; we found nothing significant.
Evaluation of Shared CNSs
Some sorts of negative data are important. With trivial exceptions, there were zero Arabidopsis αCNSs, not even our most significant CNSs, detectable by bl2seq in other Arabidopsis gene space except the duplicate from the α-tetraploidy. Supplemental Table 5 online contains these data. We examined 23 Bigfoot gene pairs with an out-group gene from the same lineage, a gene categorized by Bowers et al. (2003) to have diverged from α-duplicate genes as part of the β-tetraploidy. (The β-tetraploidy, more ancient than the α-tetraploidy, occurred after the monocot-dicot split [Bowers et al., 2003], but it remains unclear whether or not the β-tetraploidy is in poplar [Tuskan et al., 2006].) Not one CNS was conserved between either α-gene and any β-out-group gene. These three-gene comparisons were at the tips of each of the 23 gene trees. The β-tetraploidy event has been estimated to have happened ∼100 million years ago (Bowers et al., 2003; Maere et al., 2005). Therefore, unlike the case in vertebrates, where some enhancer CNSs are conserved for >350 million years (see Introduction), the maximum level of CNS conservation in higher plants is relatively shallow. That does not mean that the function specified by these CNSs is not conserved; it means that we cannot detect such conservation at the sequence level.
Supplemental Table 5 online also conveys an exception to the above result: occasionally CNSs are duplicated, usually in tandem, within the gene space of a single gene.
DISCUSSION
Limitations of Intragenomic Footprints
We can only analyze 25% of the gene content of Arabidopsis, which is the gene content that was retained in pairs following α-tetraploidy. We found that those genes retained, and studied, are not expected to have more or fewer CNSs than the remaining 75% of the minimized genome would have had they all retained post-tetraploidy or if there were a brassicoid genome diverged to the same extent as are α-pairs. To our knowledge, no Arabidopsis relative useful for pairwise (CNS) research has been sequenced to date.
Without an out-group branching off the Arabidopsis lineage before the α-tetraploidy, we cannot measure subfunctionalized or neofunctionalized (gain-of-function) sequences, although post-tetraploidy duplicate genes certainly diverge (Gu et al., 2002a, 2002b; Makova and Li, 2003; Raes and Van de Peer, 2003; Gu et al., 2004; Haberer et al., 2004; Li et al., 2005; Rastogi and Liberles, 2005; Roth et al., 2006). However, sometimes divergence is slower than expected (Koonin, 2005; Chapman et al., 2006), or divergence may vary by gene functional category (Ha et al., 2007). What we measure are sequences shared by homoeologs for the entirety of the time since tetraploidy. These 7470 noncoding sequence pairs in our αCNS database are likely to specify the same function today as they provided in the ancestral gene before tetraploidy. Cotton (Gossypium hirsutum), a candidate out-group, is simply too ancient to facilitate Arabidopsis–cotton CNS discovery (data not shown), and poplar is even more distant. Papaya (Carica papaya; in the family Caricacae, in the same order as Arabidopsis) could constitute a useful out-group when the sequence is finished (Lai et al., 2006). Lack of a properly positioned out-group or usefully diverged genomic sequences fundamentally limits this and all current work using the α-tetraploidy of Arabidopsis.
Results That Validate the Functionality of CNSs
Table 6 summarizes our major results. Three results imply CNS functionality independent of evolutionary conservation. (1) CNS richness varies with gene GO category (Thomas et al., 2007). Genes annotated GO “respond to…” are most CNS-rich, then TF genes in general, then signal transduction genes, then metabolic genes, and then, with zero CNSs, genes encoding ribosomal subunits and components of the mitochondria. Our GO term analyses of Bigfoot (Table 4) and Smallfoot (Table 5) genes substantiates this result. (2) The most significantly CNS-enriched motifs in all CNSs and in the Bigfoot CNS list are G-box motifs (Figures 2 and 3A). G-boxes are proven to bind known families of TFs (Menkens et al., 1995; Toledo-Ortiz et al., 2003), lending credibility to our database. (3) The significantly and worth interest CNS-enriched motifs in the Smallfoot but not (0%) Bigfoot CNSs are often aligned in relation to the direction of transcription. Twenty-five percent of the significantly enriched and 6% of those worth interest are polar, and these values increase to 50 and 13%, respectively, if we reduce the significance level to P nominal <0.001. Bigfoot 7-mers show zero polarity. The CNS enrichment significance of complement versus reverse complement is given in Supplemental Tables 3 and 4 online. CNS polarity, based on the direction of transcription, implies a function physically linked to the transcription process, which fits Smallfoot gene spaces with their CNSs being close to or within the transcriptional units. We could think of no alternative to function that could account for these three results, but experimental verification of αCNS function remains outstanding. Presumably, this function is to bind or affect the movement (e.g., boundary elements) of regulatory factors. Small RNAs binding gene transcripts are unlikely candidates for binding αCNS sequence (Thomas et al., 2007), although such binding is possible for 1.3% of the CNSs. We are left with the hypothesis that CNSs generally bind proteins or other macromolecules. CNSs of the sort that are aligned near Smallfoot gene exons behave like typical TF binding sites. On the other hand, CNSs far upstream of Bigfoot exons, even though they may carry a G-box, are more like enhancers.
Table 6.
Four Major Conclusions as Clues to CNS Functions
|
Many CNSs May Be Explained by Selection for or against Particular Sequences, but Almost All of These Sequence Motifs Were Unknown Previously
As seen from the slope of the regression line in Figure 3A (0.46 and not the expected 1.0) and in the description of the similar line comparing Bigfoot and Smallfoot genes in Table 3 (0.51 and not the expected 1.0), it is clear that CNS regions of noncoding gene space have been significantly purified of many particular 7-mer sequences. CNSs are defined largely by what sequences they do not contain. Apparently, when CNSs are interrupted by many sorts of sequences, function is impeded. Thus, a CNS must tend to be a module of function rather than a chance closeness of independent binding sequences. In other words, a CNS generally does not have spacer DNA that could be any sequence. We know that runs of five or six of any nucleotide and runs of four of A or T are particularly removed from CNS space. AAAAAAA in Smallfoot CNSs is purified at 0.13 in CNS versus in noncoding control space (P = 0).
With exceptions, Bigfoot and Smallfoot CNSs have had very similar sequences removed but are characterized by very different enriched 7-mers (see Supplemental Tables 3 and 4 online). Almost all of our significant or worth interest 7-mers have no experimental history. For example, there are nine 7-mers of the 50 most enriched in Smallfoot genes marked “High GC” (e.g., 5′-CGTGGCC, 5′-CGGCGCC, or 5′-GAGCCGT of Supplemental Table 4 online). The core CCGTCC may be similar to a few of these; this is called Box A (Logeman et al., 1995) in the PLACE database (Higo et al., 1999). Our results indicate that the linear sequence of conserved noncoding DNA sometimes explains function. We need much more CNS data (10-fold) in plants if we are to generate enough hits to fully evaluate linear DNA sequence as an explanation for CNS function. The sorghum sequence as a rice comparison should be perfectly suited to this aim (Inada et al., 2003). It remains possible that a conserved CNS sequence is but one of several or many that maintains identical functional conformations.
Genes with Zero CNSs
It is curious why the modal gene carries zero CNSs and is also expected to contain many TF binding sites. We think there is an explanation: the expected TF binding sites are either conserved as motifs but not as recognizable linear sequence or that there has been enough binding site turnover among redundant sequences in a cluster to obscure the conserved function, as has been demonstrated in Drosophila (Ludwig et al., 1998, 2005; Moses et al., 2004, 2006) and mammals (Frith et al., 2006). When Arabidopsis α-gene pairs are anchored on the 5′-A of their ATG and compared using a modified local alignment program, such as DIALAIGN (Brudno et al., 2003; used by Haberer et al., 2004), convincing footprints were detected in the 500-bp upstream region. These less significant intragenomic footprints include potentially the TF binding sites that are missing in our modal pair of homoeologs. Motif-finding algorithms coupled with phylogenetic footprinting between poplar and Arabidopsis have been successful in finding convincing arrays of TF binding sites in the promoters of dicot MADS box TF genes (De Bodt et al., 2006). Additionally, Vandepoele et al. (2006) found motifs in Arabidopsis genes using three input data sets: genome sequence of Arabidopsis, genome-wide expression data for Arabidopsis, and the complete genome of the related out-group dicot, poplar. Confining themselves to 500 to 1000 bp upstream of exon 1, these workers used alignments to find Arabidopsis-poplar orthologs and used a nonalignment-based MotifSampler (Thijs et al., 2001, 2002) to find overrepresented, conserved (functional) short proximal promoter cis-acting motifs among coregulated genes. There is adequate evidence that a TF binding site in the proximal promoters of Arabidopsis genes can sometimes be identified if the correct tools are used. Presumably, future research, perhaps using binding site reconstruction tools such as MONKEY (Moses et al., 2004, 2006), will find that CNSs, and especially the larger CNSs, contain more than one binding motif, or that anchoring on CNSs might permit finding conserved motifs adjacent to CNSs and far from exons. The fact remains that we did not find any motif associations within CNSs with the exception that G-boxes tend to occur multiple times in CNSs.
We suggest that plant researchers interested in cis-regulatory modules include all noncoding gene space as we define it (Table 1) and not just a bit of 5′ sequence.
CNS Function and Gene Expression
The most significant short sequence enriched in CNSs is the G-box; this is a clue as to the role(s) of CNSs in gene expression. However, because the historically documented G-box is the most significantly CNS-enriched TF binding motif does not mean that the G-box–containing CNSs should exhibit historical properties. It is possible that the G-box has many functions and those CNS-associated have not been studied. The placement of the average G-box is in a CNS 1.5 kb upstream of exon 1, and the GO annotation of the G-box–containing gene spaces did not include response to light signals. This far 5′ location is not far by Bigfoot gene standards with their mean CNS 3.1 kb 5′. Perhaps the G-boxes within CNSs are as yet unstudied enhancers. On the other hand, research on G-box–like motifs has linked them to several different functions. The ACGT core embedded in a number of related sequences is known to bind TGA-type bZIP TFs (Izawa et al., 1993), although this core is not always necessary for binding (de Pater et al., 1994). The phenotypic consequences of such binding can involve both positive and negative regulation of transcription in response to developmental cues, ABA, auxin, pathogenic stress, salicylic acid, and ethylene (Tucker et al., 2002). However, the GOStat results on our genes with G-box CNSs did not yield any significant term in the “response to…” genre except “response to ethylene.” Further research on these distant CNS G-boxes and their genes is needed.
All three of the original studies of CNSs in plants (Kaplinsky et al., 2002; Guo and Moose, 2003; Inada et al., 2003) noted that,compared with man-mouse, there were far fewer CNSs in plants and they were far shorter, so much so that the definition used by mammalian workers to define a CNS would find no CNSs in plants at all. This observation was predicated on the comparability of data from man-mouse and maize-rice, both with exons diverged to ∼85% nucleotide sequence identity over long high-scoring pairs from BLAST-2-sequences. Our work on αCNSs in Arabidopsis is consistent with the observation that plants have vastly fewer and shorter CNSs than mammals. Most compelling is the difference in distribution of CNSs along the grass or Arabidopsis chromosome versus the man-mouse chromosome. In plants, CNSs are almost always clustered near exons so that sorting them to gene was feasible; mammalian intergenic regions are often covered with CNSs, and raising bl2seq mismatch penalties does not help. Sorting them to gene without experimental data would generate ambiguities. Inada et al. (2003) suggested that CNSs largely keep stem cell/meristem genes from being ectopically expressed, where they might cause disease/cancer, and that many more genes use this regulatory mode in mammals compared with plants due to several differences in biology. This hypothesis was based on one case study and some arguments. Our results support this useful but untested hypothesis.
CNS Richness, Bigfoot Genes, and Evolvability
When faced with stress, such as must occur with environmental change, animals move and plants endure on penalty of extinction. Major trends in animal form and development might be seen primarily as adaptations involving organismal and cellular movement, with trends involving synapses becoming paramount over evolutionary time in the metazoan lineage. Perhaps the comparable major (most complex, perhaps) trends during higher plant evolution might be in those gene lineages that, when expressed as functional modules, modify abilities to endure. Over the last 500 million years of plant evolution, the maximums of adaptation to environmental extremes have probably gone up and not down over time; maximums of plant morphological complexity have certainly gone up (Freeling and Thomas, 2006). Table 4 details the many “response to…” GO categories that are significantly overrepresented in the Bigfoot database. We suggest that Bigfoot genes and other CNS-rich genes include those special endurance genes, genes we have lumped together in the general category “first responder” to environmental signals. As an illustration, the CNS-rich TF family HB, the even more CNS-rich subfamily, HD-ZIP (homeodomain-basic leucine zipper), and especially the Bigfoot HD-ZIPI class are known to be induced by environmental stress (Henriksson et al., 2005). The relationship between CNSs and signal response invites further research. Recent research from the Z.J. Chen laboratory (Ha et al., 2007) found that α-duplicates that were induced by environmental stress had more divergent expression patterns than α-duplicates expressed programmatically in the course of development.
Most Arabidopsis genes are positioned close to one another on the chromosome and have zero αCNSs. Bigfoot genes are not only exceptional in size, but, as first responders, function high in the regulatory cascade. If CNS richness implies a high level of gene regulation, then Bigfoot genes are, at the same time, high-level regulatory genes and also the most highly regulated (Inada et al., 2003; Thomas et al., 2007). This enigma suggests a systems sort of regulatory model. During the course of higher plant evolution, mutations (subfunctionalizations and neofunctionalizations) in particular genes, important genes, may help explain adaptations to changing environments. Bigfoot genes present exceptionally large targets for regulatory mutation. Since Bigfoot genes are exceptionally enriched in “response to…” genes (Table 4), the report that “response to…” homoeologs may diverge in expression particularly rapidly (Ha et al., 2007) supports these conclusions. All else being equal, it seems wise to look at alleles, derivatives and cooptions involving these large, CNS-rich, first responder genes to better understand the genetic basis of plant endurance.
METHODS
α-Pairs, Gene Spaces, and the αCNS Database
We used the Arabidopsis thaliana intragenic CNS database as by Thomas et al. (2007) and its supplemental data. When pairs, gene spaces, or CNSs gain information due to this study, that information is added to our data tables as Supplemental Tables 1 and 2 online.
Gene Categories and Statistics
Genes were categorized by GO term (per Thomas et al., 2007), as MIR genes, and those genes that were also TFs were subcategorized into DATF's 56 families (Database of Arabidopsis Transcription Factors, July, 2005; http://datf.cbi.pku.edu.cn/) (Guo et al., 2005). Except for MIR genes, genes encoding RNA were not counted in this study. When a gene in a category list had an uncalled, unannotated, or vaguely annotated homoeologous partner, we did not add the poorly annotated gene to the GO annotation lists. However, we did update the DATF lists, thereby increasing the number of genes above that on the original list (documented in Supplemental Table 1 online, columns B and C). Genes we called ourselves in the process of validating α-gene pairs were not used when GO terms were needed. Our analysis did not find new MIR genes except as additional duplicates in known gene spaces; MIR genes were from “RNA Families Database of Alignments and CMs” or Rfam (Thomas et al., 2007). Gene lists derived from the annotation keyword Bigfoot and Smallfoot and gene lists from motif enrichment analyses, like “contains a G-box CNS” (lists from Supplemental Table 1 online) were evaluated for GO category representation using GOStat (http://gostat.wehi.edu.au) (Beissbarth and Speed, 2004), using TAIR GO annotations, with a P value cutoff ≤0.001.
Analysis of CNS Sequence for Over- and Underrepresentation of Particular DNA cis-Acting Motifs or 7-Mer Sequences
We used the χ2 test to evaluate the significance of a PLACE (cis-acting binding sequence) motif or random 7-mer hit to CNSs versus hits to control noncoding gene space. We limited data to having 10 hits minimum to facilitate this test. The results are the nominal P values displayed in Supplemental Tables 3 and 4 online. In instances where many, usually thousands, of motifs/7-mers were assessed in the same experiment, nominal P values were multiplied by this repeat number, generating a Bonferroni-corrected P value. Values are considered significant if their corrected P value is <0.05 and color-coded pink in our spreadsheets. Based on observations to be presented, motifs/sequences giving values with P values just below Bonferroni significance (nominal P < 0.001) are identified as “worth interest,” but are not called “significant”; these are color-coded blue.
The 7-mers were chosen because they were the longest random sequences that gave us adequate numbers of hits to CNSs >9 and because Guo and Moose (2003) used them previously.
CNS sequences were from Thomas et al. (2007); we include these in Supplemental Table 2 online. The control nongenic sequences were prepared by extracting the total gene space from our manual annotations and then by partitioning sequence fragments into categories representing αCNSs, noncoding nonCNS controls, and exons. We also prepared CNS and control noncoding sequence fragments from a subdatabase composed only of gene spaces from 126 pairs of genes labeled as Bigfoot pairs and 1197 pairs called Smallfoot genes. CNS and control fragments were from the gene pairs actually used to prepare the database. There were always far more noncoding nonCNS control sequences than CNS sequences. Therefore, we normalized the hits to our control sequences so we could compare our αCNS hits to control hits, assuming a 1:1 correlation would be expected, or that there would be no difference in 7-mer content between αCNS and control sequence. When there were ≥10 CNS hits, significance of difference from the 1:1 expectation was estimated by χ2 and corrected for multiple tests (see above). Most ratios were not significantly different than 1.0. Those ratios >1.0 were marked as “O” in Supplemental Tables 3 and 4 online, and those ≤1 were marked as “U” whether or not the difference from 1.0 was significant or “worth interest.”
Evaluation of Our Results and Conclusions with TIGR Assembly Version 5 in Light of Version 6
After we froze our αCNS database, TAIR released a newer version of their annotations, TIGR assembly 6. Using the lists of changes available from the TAIR website (ftp://ftp.arabidopsis.org/Genes/TAIR6_genome_release/), we estimated the effect these changes might have on our data. Of the 6358 genes we analyzed in pairs, 551 genes were revised. Of these, 228 changed gene models, 195 changed protein structures only, and 353 new splice variants were added. No genes used in our analysis were deleted or had splice variants removed. No “new” exons are represented as αCNSs in our database, presumably because this would require misannotation of both homoeologs, not just one. The αCNS database prepared using version 5 gene annotation did not require correction in light of version 6 annotation (Thomas et al., 2007).
Websites Cited
Websites used in our study include the following: Public Arabidopsis Synteny Viewer 1.0 (http://synteny.cnr.berkeley.edu/AtCNS/), Arabidopsis Small RNA Project (http://asrp.cgrb.oregonstate.edu/), BLASTView (ensemble) set on Arabidopsis TIGR assembly (http://atensembl.arabidopsis.info/Multi/blastview?species=arabidopsis_thaliana), Database of Arabidopsis Transcription Factors (http://datf.cbi.pku.edu.cn/), GOstat application (http://gostat.wehi.edu.au), MultAlin: Multiple sequence alignment by Florence Corpet (http://prodes.toulouse.inra.fr/multalin/multalin.html), NCBI Blast: National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/), PLACE: Plant cis-acting sequence database (http://www.dna.affrc.go.jp/PLACE/), R project for statistical computing (http://www.r-project.org), and TAIR Arabidopsis assembly 6 download (ftp://ftp.arabidopsis.org/Genes/TAIR6_genome_release/).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Merged Gene and Pairs List, with Data Sorted by Gene or Pair.
Supplemental Table 2. Master αCNS List with Data Sorted by αCNS.
Supplemental Table 3. 7-Mer to Bigfoot Data Sheet, with Data Sorted by Random 7-Mer Sequence.
Supplemental Table 4. 7-Mer to Smallfoot Data Sheet.
Supplemental Table 5. Results on Shared αCNSs.
Supplementary Material
Acknowledgments
We thank Damon Lisch for discussions, our College of Natural Resources for partial subsidy of the Statistics and Bioinformatics Consulting Service, and especially the National Science Foundation (DBI-034937 to M.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Michael Freeling (freeling@nature.berkeley.edu).
Online version contains Web-only data.
References
- Arguello-Astorga, G., and Herrera-Estralla, L. (1998). Evolution of light-regulated plant promoters. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 525–555. [DOI] [PubMed] [Google Scholar]
- Avramova, Z., Tikhonov, A., Chen, M., and Bennetzen, J.L. (1998). Matrix attachment regions and structural colinearity in the genomes of two grass species. Nucleic Acids Res. 26 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth, T., and Speed, T. (2004). GOstat: Find statistically over-represented gene ontologies within groups of genes. Bioinformatics 1 1–2. [DOI] [PubMed] [Google Scholar]
- Bejerano, G., Siepel, A.C., Kent, W.J., and Haussler, D. (2005). Computational screening of conserved genomic DNA in search of functional noncoding elements. Nat. Methods 2 535–545. [DOI] [PubMed] [Google Scholar]
- Bernstein, B.E., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125 315–326. [DOI] [PubMed] [Google Scholar]
- Birchler, J.A., Auger, D.L., and Riddle, N.C. (2003). In search of the molecular basis of heterosis. Plant Cell 15 2236–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J.A., Riddle, N.C., Auger, D.L., and Veitia, R.A. (2005). Dosage balance in gene regulation: Biological implications. Trends Genet. 21 219–226. [DOI] [PubMed] [Google Scholar]
- Blanc, G., and Wolfe, K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J.E., Chapman, B.A., Rong, J., and Paterson, A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422 433–438. [DOI] [PubMed] [Google Scholar]
- Brown, R., Kazan, K., McGrath, K., Maclean, D.J., and Manners, J.M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene in Arabidopsis. Plant Physiol. 132 1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno, M., Do, C.B., Cooper, G.M., Kim, M.F., Davydov, E., Green, E.D., Sidow, A., and Batzoglou, S. (2003). LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno, M., Steinkamp, R., and Morgenstern, B. (2004). The CHAOS/DIALIGN WWW server for multiple alignment of genomic sequences. Nucleic Acids Res. 32 W41–W44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, B.A., Bowers, J.E., Feltus, F.A., and Paterson, A.H. (2006). Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc. Natl. Acad. Sci. USA 103 2730–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275 1723–1730. [DOI] [PubMed] [Google Scholar]
- De Bodt, S., Theissen, G., and Van de Peer, Y. (2006). Promoter analysis of MADS-box genes in eudicots through phylogenetic footprinting. Mol. Biol. Evol. 23 1293–1303. [DOI] [PubMed] [Google Scholar]
- de Pater, S., Katagiri, F., Kijne, J., and Chua, N.H. (1994). bZIP proteins bind to a palindromic sequence without an ACGT core located in a seed-specific element of the pea lectin promoter. Plant J. 6 133–140. [DOI] [PubMed] [Google Scholar]
- Dubchak, I., and Frazer, K. (2003). Multi-species sequence comparison: The next frontier in genome annotation. Genome Biol. 4 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling, M., and Thomas, B.C. (2006). Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16 805–814. [DOI] [PubMed] [Google Scholar]
- Frith, M.C., Ponjavic, J., Fredman, D., Kai, C., Kaweai, J., Carninci, P., Hayshizaki, Y., and Sandelin, A. (2006). Evolutionary turnover of mammalian transcription start sites. Genome Res. 16 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y., Li, J., Strickland, E., Hua, S., Zhao, H., Chen, Z., Qu, L., and Deng, X.W. (2004). An Arabidopsis promoter microarray and its initial usage in the identification of HY5 binding targets in vitro. Plant Mol. Biol. 54 683–699. [DOI] [PubMed] [Google Scholar]
- Giuliano, G., Pichersky, E., Malik, V.S., Timko, M.P., Scolnik, P.A., and Cashmore, A.R. (1988). An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 85 7089–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko, G.V., Koonin, E.V., Rogozin, I.B., and Shabalina, S.A. (2003). A significant fraction of conserved noncoding DNA in human and mouse consists of predicted matrix attachment regions. Trends Genet. 19 119–124. [DOI] [PubMed] [Google Scholar]
- Goode, D.K., Snell, P., Smith, S.F., Cooke, J.E., and Elgar, G. (2005). Highly conserved regulatory elements around the SHH gene may contribute to the maintenance of conserved synteny across human chromosome 7q36.3. Genomics 86 172–181. [DOI] [PubMed] [Google Scholar]
- Gottgens, B., Gilbert, J.G., Barton, L.M., Grafham, D., Rogers, J., Bentley, D.R., and Green, A.R. (2001). Long-range comparison of human and mouse SCL loci: Localized regions of sensitivity to restriction endonucleases correspond precisely with peaks of conserved noncoding sequences. Genome Res. 11 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z., Cavalcanti, A., Chen, F.C., Bouman, P., and Li, W.H. (2002. b). Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 19 256–262. [DOI] [PubMed] [Google Scholar]
- Gu, Z., Nicolae, D., Lu, H.H., and Li, W.H. (2002. a). Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 18 609–613. [DOI] [PubMed] [Google Scholar]
- Gu, Z., Rifkin, S.A., White, K.P., and Li, W.H. (2004). Duplicate genes increase gene expression diversity within and between species. Nat. Genet. 36 577–579. [DOI] [PubMed] [Google Scholar]
- Guo, A., He, K., lLu, D., Bai, S., Gu, X., Wei, L., and Luo, J. (2005). DATF: A database of Arabidopsis transcription factors. Bioinformatics 21 2568–2569. [DOI] [PubMed] [Google Scholar]
- Guo, H., and Moose, S.P. (2003). Conserved noncoding sequences among cultivated cereal genomes identify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell 15 1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, M., Li, W.H., and Chen, Z.J. (2007). External factors accelerate expression divergence between duplicate genes. Trends Genet. 23 162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer, G., Hindemitt, T., Meyers, B.C., and Mayer, K.F. (2004). Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol. 136 3009–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison, R.C. (2000). Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 16 369–372. [DOI] [PubMed] [Google Scholar]
- Hardison, R.C. (2003). Comparative genomics. PLoS Biol. 1 E58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson, E., Olsson, A.S., Johannesson, H., Johansson, H., Hanson, J., Engstrom, P., and Soderman, E. (2005). Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol. 139 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada, D.C., Bashir, A., Lee, C., Thomas, B.C., Ko, C., Goff, S.A., and Freeling, M. (2003). Conserved noncoding sequences in the grasses. Genome Res. 13 2030–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, T., Foster, R., and Chua, N.H. (1993). Plant bZIP protein DNA binding specificity. J. Mol. Biol. 230 1131–1144. [DOI] [PubMed] [Google Scholar]
- Kaplinsky, N.J., Braun, D.M., Penterman, J., Goff, S.A., and Freeling, M. (2002). Utility and distribution of conserved noncoding sequences in the grasses. Proc. Natl. Acad. Sci. USA 99 6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooiker, M., Airoldi, C.A., Losa, A., Manzotti, P.S., Finzi, L., Kater, M.M., and Colombo, L. (2005). BASIC PENTACYSTEINE1, a GA binding protein that induces conformational changes in the regulatory region of the homeotic Arabidopsis gene SEEDSTICK. Plant Cell 17 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E.V. (2005). Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39 309–338. [DOI] [PubMed] [Google Scholar]
- Lai, C., Yu, Q., Hou, S., Skelton, R., Jones, M., Lewis, K., Murry, J., Guan, M., Agbayani, R., Moore, P., Ming, R., and Presting, G. (2006). Analysis of papaya BAC end sequences reveals first insights into the organization of a fruit tree genome. Mol. Genet. Genomics 276 1–12. [DOI] [PubMed] [Google Scholar]
- Lehman, M. (2004). Anything else but GAGA: A nonhistone protein complex reshapes chromatin structure. Trends Genet. 20 15–22. [DOI] [PubMed] [Google Scholar]
- Levy, S., Hannenhalli, S., and Workman, C. (2001). Enrichment of regulatory signals in conserved noncoding sequence. Bioinformatics 17 871–877. [DOI] [PubMed] [Google Scholar]
- Li, W.H., Yang, J., and Gu, Z. (2005). Expression divergence between duplicate genes. Trends Genet. 21 1–6. [DOI] [PubMed] [Google Scholar]
- Logeman, E., Parniske, M., and Halbrook, K. (1995). Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc. Natl. Acad. Sci. USA 92 5905–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots, G.G., Locksley, R.M., Blankespoor, C.M., Wang, Z.E., Miller, W., Rubin, E.M., and Frazer, K.A. (2000). Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288 136–140. [DOI] [PubMed] [Google Scholar]
- Loots, G.G., and Ovcharenko, I. (2004). rVISTA 2.0: Evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 32 W217–W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, M.Z., Palsson, A., Alekseeva, E., Bergman, C.M., Nathan, J., and Kreitman, M. (2005). Functional evolution of a cis-regulatory module. PLoS Biol. 3 e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, M.Z., Patel, N.H., and Kreitman, M. (1998). Functional analysis of eve stripe 2 enhancer evolution in Drosophila: Rules governing conservation and change. Development 125 949–958. [DOI] [PubMed] [Google Scholar]
- Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., and Van de Peer, Y. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102 5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makova, K.D., and Li, W.-H. (2003). Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 13 1638–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, C., Brudno, M., Schwartz, J.R., Poliakov, A., Rubin, E.M., Frazer, K.A., Pachter, L.S., and Dubchak, I. (2000). VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16 1046–1047. [DOI] [PubMed] [Google Scholar]
- McNeil, J., Smith, K., Hall, L., and Lawrence, J. (2006). Word frequency analysis reveals enrichment of dinucleotide repeats on the human X chromosome and [GATA]n in the X escape region. Genome Res. 16 477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, R.J., Williams, L.A., Monfared, M.M., Gallagher, T.L., Kraft, E.A., Nelson, C.G., and Gasser, C.S. (2004). Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J. 37 426–438. [DOI] [PubMed] [Google Scholar]
- Menkens, A.E., Schindler, U., and Cashmore, A.R. (1995). The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20 506–510. [DOI] [PubMed] [Google Scholar]
- Morgenstern, B., Prohaska, S.J., Pohler, D., and Stadler, P.F. (2006). Multiple sequence alignment with user-defined anchor points. Algorithms Mol. Biol. 1 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses, A., Chiang, D., Pollrd, D., Iyer, V., and Eisen, M. (2004). MONKEY: Identifying conserved transcription factor binding sites in multiple alignments using a binding-specific evolutionary model. Genome Biol. 5 R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses, A.M., Pollard, D.A., Nix, D.A., Iyer, V.N., Li, X.Y., Biggin, M.D., and Eisen, M.B. (2006). Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput. Biol. 2 e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko, I., Loots, G.G., Nobrega, M.A., Hardison, R.C., Miller, W., and Stubbs, L. (2005). Evolution and functional classification of vertebrate gene deserts. Genome Res. 15 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp, B., Pal, C., and Hurst, L.D. (2003). Dosage sensitivity and the evolution of gene families in yeast. Nature 424 194–197. [DOI] [PubMed] [Google Scholar]
- Prakash, A., Blanchette, M., Sinha, S., and Tompa, M. (2004). Motif discovery in heterogeneous sequence data. Pac. Symp. Biocomput. 9 348–359. [DOI] [PubMed] [Google Scholar]
- Raes, J., and Van de Peer, Y. (2003). Gene duplication, the evolution of novel gene functions, and detecting functional divergence of duplicates in silico. Appl. Bioinformatics 2 91–101. [PubMed] [Google Scholar]
- Rastogi, S., and Liberles, D.A. (2005). Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Biol. 5 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, C., Rastogi, S., Arvestad, L., Dittmar, K., Light, S., Ekman, D., and Liberles, D.A. (2006). Evolution after gene duplication: Models, mechanisms, sequences, systems, and organisms. J. Exp. Zoolog. B Mol. Dev. Evol. 308 58–73. [DOI] [PubMed] [Google Scholar]
- Seoighe, C., and Gehring, C. (2004). Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 20 461–464. [DOI] [PubMed] [Google Scholar]
- Siepel, A., et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion, C., Vandepoele, K., Van Montagu, M.C., Zabeau, M., and Van de Peer, Y. (2002). The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 99 13627–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova, T.A., and Madden, T.L. (1999). BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174 247–250. [DOI] [PubMed] [Google Scholar]
- Thijs, G., Lescot, M., Marchal, K., Rombauts, S., De Moor, B., Rouze, P., and Moreau, Y. (2001). A higher order background model improves detection of regulatory elements by Gibbs sampling. Bioinformatics 17 1113–1122. [DOI] [PubMed] [Google Scholar]
- Thijs, G., Marchal, K., Lescot, M., Rombauts, S., De Moor, B., Rouze, P., and Moreau, Y. (2002). A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. 9 447–464. [DOI] [PubMed] [Google Scholar]
- Thomas, B.C., Pedersen, B., and Freeling, M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, B.C., Rapaka, L., Lyons, E., Pedersen, B., and Freeling, M. (2007). Intragenomic conserved noncoding sequences in Arabidopsis. Proc. Natl. Acad. Sci. USA 104 3348–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J.W., et al. (2003). Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424 788–793. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz, G., Huq, E., and Quail, P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa, M., Li, N., Bailey, T.L., Church, G.M., De Moor, B., Eskin, E., Favorov, A.V., Frith, M.C., Fu, Y., Kent, W.J., Makeev, V.J., Mironov, A.A., et al. (2005). Assessing computational tools for the discovery of transcription factor binding sites. Nat Biotechnol. 23 137–144. [DOI] [PubMed] [Google Scholar]
- Tran, L., Nakashima, K., Sakuma, Y., and Yamaguchi-Shinozaki, K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a draught-responsive cis-element in the early response to dehydration stress1 promoter. Plant Cell 16 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M.L., Whitelaw, C.A., Lyssenko, N.N., and Nath, P. (2002). Functional analysis of regulatory elements in the gene promoter for an abscission-specific cellulase from bean and isolation, expression, and binding affinity of three TGA-type basic leucine zipper transcription factors. Plant Physiol. 130 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan, G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K., Casneuf, T., and Van de Peer, Y. (2006). Identification of novel regulatory modules in dicot plants using expression data and comparative genomics. Genome Biol. 7 R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hellemont, R., Monsieurs, P., Thijs, G., de Moor, B., Van de Peer, Y., and Marchal, K. (2005). A novel approach to identifying regulatory motifs in distantly related genomes. Genome Biol. 6 R113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia, R.A. (2002). Exploring the etiology of haploinsufficiency. Bioessays 24 175–184. [DOI] [PubMed] [Google Scholar]
- Wen, J., Lease, K.A., and Walker, J.C. (2004). DVL, a novel class of small polypeptides: Overexpression alters Arabidopsis development. Plant J. 37 668–677. [DOI] [PubMed] [Google Scholar]
- Williams, M.E., Foster, R., and Chua, N.H. (1992). Sequences flanking the hexameric G-box core CACGTG affect the specificity of protein binding. Plant Cell 4 485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe, A., et al. (2005). Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, G.A. (2003). Transcriptional regulation and the evolution of development. Int. J. Dev. Biol. 47 675–684. [PubMed] [Google Scholar]
- Xu, B., and Timko, M. (2004). Methyl jasmonate induced expression of the tobacco putrescine N-methyltransferase genes requires both G-box and GCC-motif elements. Plant Mol. Biol. 55 743–761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.