Abstract
The silencing phenotype in Arabidopsis thaliana lines with an inverted repeat transgene under the control of a phloem-specific promoter was manifested in regions around veins due to a mobile signal of silencing. Genetic analysis implicates RNA-DEPENDENT RNA POLYMERASE2 (RDR2) and an RNA polymerase IVa subunit gene (NRPD1a) in the signaling mechanism. We also identified an SNF2 domain–containing protein (CLASSY1) that acts together with RDR2 and NRPD1a in the spread of transgene silencing and in the production of endogenous 24-nucleotide short interfering RNAs (siRNAs). Cytochemical analysis indicates that CLASSY1 may act in the nucleus with NRPD1a and RDR2 in the upstream part of RNA silencing pathways that generate a double-stranded RNA substrate for Dicer-like (DCL) nucleases. DCL3 and ARGONAUTE4 act in a downstream part of the pathway, leading to endogenous 24-nucleotide siRNA production, but are not required for intercellular signaling. From genetic analysis, we conclude that another downstream part of the pathway associated with intercellular signaling requires DCL4 and at least one other protein required for 21-nucleotide trans-acting siRNAs. We interpret the effect of polymerase IVa and trans-acting siRNA pathway mutations in terms of a modular property of RNA silencing pathways.
INTRODUCTION
The non-cell-autonomous nature of RNA silencing in plants is evident in transgenic and virus-infected plants. Transgenic silencing, for example, may initiate spontaneously or be induced in localized regions and subsequently spread throughout the plant (Palauqui et al., 1997; Voinnet and Baulcombe, 1997; Voinnet et al., 1998). Similarly, virus-induced silencing has effects beyond the infected tissue (Ruiz et al., 1998; Schwarz et al., 2002; Szittya et al., 2002). Local movement of the signal is through plasmodesmata (Voinnet et al., 1998), and systemic spread is through the phloem (Ryabov et al., 2004).
The spread of a silencing phenotype is always highly nucleotide sequence–specific, indicating that the signal includes an RNA, probably the double-stranded (ds) or short interfering (si) RNA generated in RNA silencing (Hamilton and Baulcombe, 1999; Mallory et al., 2001; Hamilton et al., 2002; Himber et al., 2003; Mallory et al., 2003; Dunoyer et al., 2005). The Dicer-like (DCL) RNaseIII family enzymes cleave dsRNA into siRNAs, and in one experimental system, the spread of silencing was lost in a DCL4 mutant, consistent with the idea that siRNA production is required for signal production (Dunoyer et al., 2005). This is an attractive idea because siRNAs, being 21 to 24 nucleotides long, are small enough to pass through plasmodesmata and because phloem sap samples contain siRNA-like species (Yoo et al., 2004). However, longer transcripts of both sense and antisense orientations are also present in phloem sap, and it remains possible that long silencing RNAs are also mobile (Ruiz-Medrano et al., 1999; Haywood et al., 2005).
Cell-to-cell movement through plasmodesmata and long-distance translocation of silencing through the phloem are sometimes considered separate mechanisms, because they can be inhibited differentially by cadmium treatment, by viral proteins, or by mutation of plant genes (Ueki and Citovsky, 2001; Himber et al., 2003; Schwach et al., 2005). One interpretation of these differences invokes distinctions between the transport mechanisms through plasmodesmata that connect nonvascular cells or that mediate movement of the signal either into or out of the phloem. However, the spread of RNA silencing might also be affected by dilution as the silencing signal moves away from its source. Signal moving through plasmodesmata in cells that are adjacent to the primary source would be abundant and could have a significant direct effect. However, as the signal moves longer distances through the phloem, it would be progressively diluted. In this situation, an amplification step involving the production of secondary siRNAs would be required for strong silencing. Thus, the differences between short- and long-distance signaling could be related to the requirement for amplification through secondary siRNA production as well as the different mechanisms associated with movement between cells and through the phloem (Himber et al., 2003).
There are several precedents for secondary siRNA production mechanisms that could provide the amplification of silencing when the signal is diluted. In plants, the mechanism is different from that in Caenorhabditis elegans, although, in both organisms, an RNA-dependent RNA polymerase (RDR) is involved (Axtell et al., 2006; Ruby et al., 2006; Pak and Fire, 2007; Sijen et al., 2007). In plants, an RNA becomes a template for RDR if it is targeted by one or two primary siRNAs or by a related type of silencing RNA known as microRNA (miRNA) (Axtell et al., 2006). The regions of this RNA adjacent to the target sites are then converted into dsRNA by the RDR, and a DCL cleaves this dsRNA into the secondary siRNAs (Axtell et al., 2006). A requirement for RDR6 in systemic silencing is supporting evidence for the involvement of amplification in the long-distance spread of silencing (Himber et al., 2003; Schwach et al., 2005). In some instances, there is evidence that the signal amplification mechanism involves a relay process in which cells receiving the signal become a secondary source of signal. Eventually, the source of primary signal is no longer required and silencing persists even after its removal from the plant (Palauqui and Vaucheret, 1998; Voinnet et al., 1998).
There are multiple RDR genes in Arabidopsis thaliana (Yu et al., 2003), and to date, only RDR6 has been associated with silencing signals. It is involved in an amplification mechanism associated with long-distance movement through the vascular system (Schwach et al., 2005) but not with short-distance spread (Himber et al., 2003). RDR2 has been implicated in a nuclear process (Herr et al., 2005; Kanno et al., 2005b; Onodera et al., 2005; Pontier et al., 2005; Li et al., 2006; Pontes et al., 2006), but until the work described here, it had not been associated with spreading. The siRNAs in this RDR2-mediated nuclear mechanism are 24 nucleotides long (Xie et al., 2004), and the end result is RNA-directed DNA methylation and histone modification at targeted chromatin loci (Huettel et al., 2006). Genetic analysis identified proteins in this pathway, including NRPD1a and NRPD2a, that are putative subunits of RNA polymerase (Pol) IVa. Pol IVa uses either DNA or RNA templates to generate single-stranded RNA templates for dsRNA production by an RDR, RDR2 (Herr et al., 2005; Kanno et al., 2005b; Onodera et al., 2005; Pontier et al., 2005). The dsRNA is then cleaved by DCL3 to produce the 24-nucleotide siRNAs that are recruited by an ARGONAUTE4 (AGO4) effector protein (Zilberman et al., 2003; Xie et al., 2004; Qi et al., 2005).
Here, we describe an experimental system for the analysis of the non-cell-autonomous or spreading process in RNA silencing. Like a similar system described elsewhere (Himber et al., 2003; Dunoyer et al., 2005), it involves a silencing transgene that is expressed specifically in the phloem companion cells, so that a silencing phenotype in cells away from the vascular system requires the spread of a silencing signal. It is likely that the mechanism of spreading in our plants is the same as in the previous work, because the spreading phenotype in both systems is insensitive to RDR6 loss of function but sensitive to DCL4 mutations (Himber et al., 2003; Dunoyer et al., 2005). However, our findings extend the previous analysis by showing how the upstream proteins in the Pol IVa silencing pathway, including an SNF2 domain–containing protein (CLASSY1 [CLSY1]), are required for spreading. Downstream proteins in this pathway, DCL3 and AGO4, are not required for spreading because their loss of function results in enhanced spreading of the silencing signal. These results reveal an unexpected link between the RNA silencing mechanisms associated with spreading and chromatin silencing. They also illustrate how an upstream mechanism for dsRNA production may feed into multiple downstream mechanisms for siRNA production involving, in our system, either DCL3 or DCL4. This modular behavior is likely to be a general feature of RNA silencing pathways.
RESULTS
Mutagenesis of a Silencing Signal Reporter Line in Arabidopsis
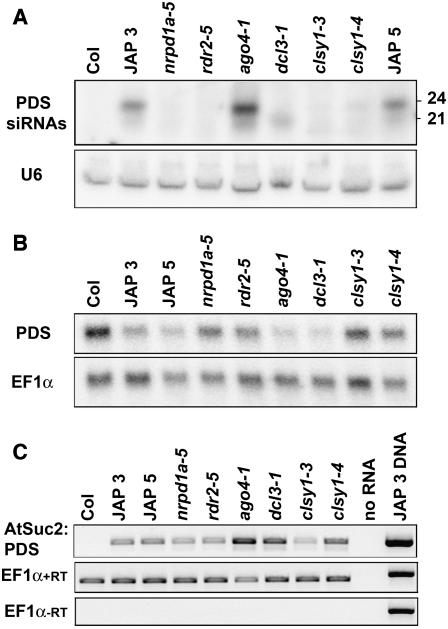
To study the spread of RNA silencing, we generated a transgenic plant line in which the initiation and reporting of silencing are in separate tissues. A PHYTOENE DESATURASE (PDS) inverted repeat was expressed in these plants under the control of the phloem-specific Suc2 promoter (Figure 1A). Movement of the silencing signal out of the phloem led to silencing in the surrounding mesophyll cells, which was manifested as photobleaching (Figure 1B) in wild-type plants (PSuc2:PDS-labeled JAP [for Jawohl:AtSuc2:PDS] lines 3 and 5). Both 24- and 21-nucleotide siRNAs were produced in the PSuc2:PDS plants, indicating that the transgene may activate two or more silencing pathways (Figure 1C). The insertion locus of the JAP 3 line contains an inverted repeat of two truncated or rearranged copies of the T-DNA, while the insertion locus of the JAP 5 line remains uncharacterized (see Supplemental Figure 1 online). It is likely that both loci mediate the same silencing mechanisms, because the silencing phenotypes described below are indistinguishable.
Figure 1.
A Transgenic Experimental System for the Analysis of Silencing Signal Spreading.
(A) T-DNA of the PSuc2:PDS construct. LB and RB indicate the left and right T-DNA borders, respectively. Arrows indicate the directions of transcription from the promoters and transcriptional start sites. pNOS, pANOS, and pA35S indicate the nopaline synthase promoter and terminator and the cauliflower mosaic virus 35S terminator, respectively. The intron is At WRKY33 intron 1. Pat indicates the phosphinothricin-N-acetyltransferase gene, which gives resistance to dl-phosphinothricin. The pJawohl 8 backbone surrounding the T-DNA is not shown.
(B) Phenotypes of the two PSuc2:PDS lines (JAP 3 and JAP 5) used in this study compared with the parental Columbia-0 (Col-0) line. Also shown are the phenotypes of the PSuc2:PDS (JAP 3) line in the ago4-1, dcl3-1, nrpd1a-1, nrpd2a-1, and rdr2-2 backgrounds. All plants except for wild-type Col-0 are homozygous for the PSuc2:PDS construct. Plants were grown in controlled-environment rooms with an 8-h photoperiod.
(C) Detection of PDS siRNAs in Col-0 and the PSuc2:PDS lines (JAP 3 and JAP 5). Low molecular weight RNA gel blot analysis was performed on 50 μg of total nucleic acids isolated from the aerial portions of 1-month-old plants. The probe was a 32P-labeled PDS transcript.
We investigated the role of the 24-nucleotide siRNA pathway in the PSuc2:PDS phenotype by introgressing mutant alleles of previously characterized RNA silencing genes. As described previously with a similar Suc2 promoter–driven silencing transgene (Dunoyer et al., 2005), the rdr6(sde1-1) allele had no effect (L.M. Smith, unpublished data) and dcl4-2 caused reduced spreading (Figure 6B). Of the other tested mutations, nrpd1a-1, nrpd2a-1, and rdr2-2 resulted in almost complete loss of the photobleaching phenotype. Surprisingly, dcl3-1 and ago4-1 caused enhanced silencing, manifested as photobleaching across most of the leaf lamina (Figure 1B). There was no effect of drd1-6 and nrpd1b(drd3-1) on the PDS silencing phenotype in plants in which the presence of the homozygous mutation and the transgene was detected by PCR (L.M. Smith, unpublished data). It is likely, therefore, that the Pol IVb pathway is not involved in the spread of RNA silencing in these plants (Kanno et al., 2005b).
Figure 6.
Phenotype Analysis of Double Mutants.
(A) Photobleaching phenotypes of double mutants of the 24-nucleotide pathway. Parental lines are also shown as indicated. Plants were grown in controlled-environment rooms with an 8-h photoperiod. All plants except for wild-type Col-0 are homozygous for the PSuc2:PDS construct.
(B) dcl3-1 dcl4-2 double and single mutants exhibiting intermediate photobleaching compared with the two single mutants. All plants are hemizygous for the PSuc2:PDS construct.
A screen of ethyl methanesulfonate–mutagenized PSuc2:PDS plant lines (JAM lines, for Jawohl:AtSuc2:PDS mutant) revealed plants with both reduced and enhanced silencing. Among the reduced silencing plants, there were three new nrpd1a mutants (nrpd1a-5 to nrpd1a-7) and two new rdr2 mutants (rdr2-4 and rdr2-5). There was also a new dcl3 mutant with an enhanced silencing phenotype (dcl3-2). The molecular description of these mutations is included in Supplemental Table 1 online.
We also recovered two reduced PDS silencing ethyl methanesulfonate mutants that had a late-flowering phenotype that cosegregated with reduced PDS silencing. These plants carried mutations at fpa and fca and are described elsewhere (I. Baurle, C. Dean, L.M. Smith, and D.C. Baulcombe, unpublished data). Additionally there were reduced PDS silencing mutants that did not carry alleles of fpa, fca, nrpd1a, nrpd2a, or rdr2. The mutants discussed here that are at unidentified loci are in JAM lines 14, 23, 32, 51, and 55. There were also mutations at the CLSY1 locus that are described in detail below. As a first step in characterizing these mutants, we assayed their levels of endogenous siRNAs, trans-acting siRNAs (tasiRNAs), and miRNAs.
We predicted that loss of function in genes acting together with NRPD1a, NRPD2a, and RDR2 would be associated with a reduction in 24-nucleotide siRNAs from both repetitive (At SN1 and siRNA 1003) and/or low-copy-number (clusters 2 and 55) loci (Figure 2; see Supplemental Figure 2 online for full figure). Five lines (nrpd1a-5, nrpd1a-6, nrpd1a-7, clsy1-4, and rdr2-5) had reduced levels of all endogenous 24-nucleotide siRNAs tested (Figure 2; see Supplemental Figure 2 online). Eight other JAM lines (clsy1-1, clsy1-2, clsy1-3, clsy1-5, clsy1-6, rdr2-4, JAM 51, and JAM 55) had partial reductions in 24-nucleotide siRNA 1003 and siRNA At SN1 (Figure 2; see Supplemental Figure 2 online). These plants had wild-type levels of 24-nucleotide siRNAs from the cluster 2 and 55 loci. Therefore, with 13 of 20 mutant lines showing a reduction in 24-nucleotide siRNAs, these results reinforce an association of the 24-nucleotide siRNA pathway with the spread of RNA silencing. We also tested for mutations in silencing pathways affecting tasiRNAs (Allen et al., 2005) or miRNAs (Han et al., 2004; Kurihara and Watanabe, 2004; Xie et al., 2004). Three mutants (JAM 14, 23, and 32; see Supplemental Figure 2 online) had lower tasiRNA levels (siRNA1511/255) than did wild-type plants, but there were no mutants with altered levels of miRNAs miR159 or miR167. The spread of RNA silencing, therefore, involves elements of both 24-nucleotide siRNA and tasiRNA silencing pathways but not the miR159 and miR167 pathways.
Figure 2.
Detection of Endogenous siRNAs and miRNAs in the Spread of Silencing Mutants.
Low molecular weight RNA gel blot analysis was performed on 20 μg of total nucleic acids isolated from young inflorescence tissue. Each plant line is indicated above the lane. The probes were hybridized to the same blot that was stripped and reprobed. Analysis is shown here for two clsy1 mutants, the two new rdr2 alleles, a representative nrpd1a mutant, and JAM 14 as a representative of the mutants in the tasiRNA synthesis pathway; full analysis of all mutant lines is given in Supplemental Figure 2 online. siRNA1511 is derived from TAS2, and siRNA255 is derived from the three TAS1 loci.
A Novel SNF2 Domain Protein in RNA Silencing
Six mutations affecting 24-nucleotide siRNAs (clsy1-1 through clsy1-6) mapped to the At3g42670 gene that encodes an SNF2 domain–containing protein (Figures 3A and 3B). We now refer to this gene as CLSY1 based on an amino acid motif at positions 555 to 560 of the predicted protein sequence (Figure 3C). The 108-nucleotide 3′ untranslated region (UTR) of the CLSY1 mRNA overlaps the 3′ UTR of At3g42660 and has been described previously (available through The Arabidopsis Information Resource at www.arabidopsis.org/ [Salanoubat et al., 2000] and at www.plantgdb.org/AtGDB/ [Zhu et al., 2003]). The 5′ UTR of CLSY1 is complex, because we detected multiple CLSY1 cDNAs by RT-PCR (5′ rapid amplification of cDNA ends [RACE]) that extend up to 559 nucleotides before the presumed translational start site (−559; Figure 3D; see Supplemental Tables 2 and 3 online). Of the 41 cDNAs sequenced, 35 represented transcriptional start points on the 5′ side of the presumed initiation codon with differentially spliced introns and short open reading frames in the upstream region.
Figure 3.
Structures of the Gene and the Encoded Protein for the SNF2 Domain–Containing Protein CLSY1.
(A) Intron–exon structure of the coding region showing the mutations identified in the PSuc2:PDS screen.
(B) Diagrammatic representation of the predicted 1256–amino acid CLSY1 protein indicating proposed structural domains and the effect of four mutations on the protein-coding region. The acidic domain is a region of 18 amino acids of which 15 are Glu or Asp. SNF2 is the area defined by pfam00176, and helicase C (HelicC) is the region defined by cd00079, as identified by National Center for Biotechnology Information conserved domains search.
(C) Protein sequence of CLSY1 indicating the motifs discussed in the text. The nuclear localization signal and CLASSY sequence are indicated in boldface type, while the SNF2 and helicase C domains are underlined.
(D) Structure of the CLSY1 5′ UTR. The graph indicates the relative transcriptional start site of 41 cDNAs sequenced relative to the translational start site (0). cDNAs indicated by black dots have an intron (−452 to −46 nucleotides) of the length indicated by the black line above the graph spliced out of the RNA. The one cDNA with an intron from −461 to −46 nucleotides is indicated by a dark gray dot, with a dark gray line above representing the intronic region. cDNAs with a shorter intron (−343 to −46 nucleotides; dashed line) are represented by striped dots, while cDNAs with no introns removed are indicated by white dots. Upstream open reading frames (uORFs) coded within the 5′ UTR are shown by representative lines below the graph. See Supplemental Tables 2 and 3 online for further details.
It is possible that the true 5′ end of CLSY1 mRNA is farther upstream of the region shown in Figure 3D. However, we do not favor this possibility, because RT-PCR and RNA gel blot analysis showed no evidence for transcripts from this upstream region. Additionally, we complemented the PDS silencing phenotype and the 24-nucleotide siRNA phenotypes of clsy1-3 and clsy1-4 with a transgene comprising 1.52 kb in the promoter regions and the presumed open reading frame (see Supplemental Figure 3 online). Therefore, it is likely that CLSY1 is one of a minority of Arabidopsis genes (Rogozin et al., 2001; Kawaguchi and Bailey-Serres, 2005) with upstream open reading frames and in which translational regulation is achieved by processes such as ribosomal shunting or an internal ribosomal entry site.
CLSY1 is in the same plant-specific subfamily of SNF2 domain–containing proteins as DRD1, an SNF2 domain–containing protein implicated in DNA methylation and transcriptional gene silencing, along with Pol IVb (NRBD1b and NRPD2a) (Kanno et al., 2004, 2005a, 2005b). All six proteins of the DRD1/CLSY1 subfamily share divergence at 17 amino acids that are otherwise conserved (but not invariant) in 70% of the Arabidopsis SNF2 domain–containing proteins (see Supplemental Figure 4A online). The subfamily is arranged into three pairs of similar proteins (see Supplemental Figure 4B online). These homologs include a protein (At2g21450) that is 59% similar to DRD1, At5g20420, which is 81% similar to CLSY1 (and hereafter referred to as CLSY2), and two proteins (At3g24340 and At1g05480) that are 35 to 46% similar to DRD1 and CLSY1 (see Supplemental Figure 4B online). The SNF2 domain of these six proteins in this subfamily group is most closely related to the RAD54 subfamily of eukaryotic SNF2 domain–containing proteins. However, based on the helicase C domain, the DRD1/CLSY1 subfamily is most closely related to the RAD16 subfamily of eukaryotic SNF2 proteins (see Supplemental Figure 4C online).
No animal proteins have significant sequence similarity to CLSY1 outside of the SNF2 and helicase domains. However, there are close plant homologs with similarity over the full length of the protein sequence from Olimarabidopsis pumila (90.5% amino acid sequence identity and 93.6% sequence similarity), Medicago truncatula (46.8% identity and 58.3% similarity), and Oryza sativa (29.7% identity and 43.5% similarity; see Supplemental Figure 4D online for alignments). Outside of the SNF2 and helicase domains, these four homologs are most similar in the first 216 amino acids of CLSY1 at the N-terminal domain, although there are no obvious motifs or predicted structures in this domain to hint at function.
Other clues to CLSY1 function include a predicted nuclear localization signal of KKRKK at residues 364 to 368 of CLSY1 and features in the SNF2 and helicase C domains that can be inferred by homology modeling. The Protein Homology/analogY Recognition Engine (http://www.sbg.bio.ic.ac.uk/phyre/html/index.html [Bates et al., 2001]) predicted structural similarity of residues 630 to 1256 to the zebrafish Rad54 protein (Thoma et al., 2005). A homology model of residues 630 to 1256 was developed and then optimized via energy minimization protocols using a molecular graphics and modeling package (http://www.accelrys.com/) (Figure 4A). The template structure is fundamentally composed of six domains: a 52-residue N-terminal domain, a 75-residue C-terminal domain, two RecA-like domains found in helicases, which can be referred to as NTRD and CTRD (for N- and C-terminal RecA-like domains, respectively), and two α-helical domains, HD1 and HD2, that are inserted within each RecA-like domain. Compared with the template structure, the CLSY1 model has not only all six domains in place but also has four substantial insertions that occur primarily in two domains. Two of the insertions occur in the NTRD and two in the HD2 domains of the protein. It is surprising, however, that in the three-dimensional model, the insertions occur in or around the putative DNA binding region of the protein. Basic amino acid residues of CLSY1 predicted to bind the acidic DNA backbone are Arg-770, Lys-776, Lys-778, Lys-787, Lys-995, Lys-999, Lys-1029, Lys-1048, Arg-1086, Arg-1097, and Lys-1099 (Figure 4B). More than 50% of these residues (i.e., Arg-770, Lys-776, Lys-778, Lys-787, Lys-995, and Lys-999) originate from the insertions. Hence, these insertions are likely to affect the manner in which nucleic acid is bound by CLSY1 and may play important role(s) in defining the protein function.
Figure 4.
Model of the C-Terminal Section of CLSY1 Based on Similarity with Rad54.
(A) The crystal structure of Rad54 (Thoma et al., 2005) is superimposed on the predicted structure for the C-terminal domain of CLSY1 as a SwissPDB image. Rad54 is shown in yellow (coils), orange (α-helices), and blue (β-sheets), while CLSY1 is indicated in white (coils), red (α-helices), and green (β-sheets). Variant loops in the DNA binding region are indicated by arrows, with the loop specified by the blue arrow specific to CLSY1 and CLSY2.
(B) The predicted structure for CLSY1 is shown with DNA in the putative binding cleft. The side chains of some basic residues that may contribute to DNA binding are shown in a stick model.
Structural modeling also predicts similarity of Rad54 and CLSY1 in motifs responsible for the binding and hydrolysis of ATP, which is required for chromatin remodeling activity (Eisen et al., 1995). In the template crystal structure, the position of the γ-phosphate of ATP is occupied by sulfate rather than a phosphate ion. Residues predicted to form hydrogen bonds with the γ-phosphate of ATP by assessing hydrogen bonds formed by the given sulfate ion are Gly-180, Gly-182, Lys-183, Arg-600, and Arg-603 (Thoma et al., 2005). When we used Swiss PDB viewer to determine hydrogen bonds by the given sulfate ion of the template molecule under the conditions of no hydrogens on display and angle criterion set to zero, some additional hydrogen bonds were computed. These bonds can be summarized as Leu-181 (backbone N: one H-bond), Lys-183 (backbone N and side chain: two H-bonds), Thr-184 (backbone N and side chain: two H-bonds), Cys-572 (backbone N: one H-bond), Gln-596 (side chain: one H-bond), Arg-600 (side chain: one H-bond), and Arg-603 (side chain: three H-bonds). The corresponding residues in the CLSY1 model are Ala-716, Lys-718, Thr-719, Glu-1142, Gln-1166, Arg-1170, and Arg-1173. By comparison, these residues showed that CLSY1 had a potential ATP binding site that was ∼71% identical to that of the template molecule Rad54. When sulfate was modeled at the corresponding site of CLSY1 and hydrogen bonds were computed, identical numbers of hydrogen bonds between the sulfate ion and the corresponding residues of CLSY1 were produced. Hence, CLSY1 may bind and hydrolyze ATP in a mechanism that is analogous to that of Rad54.
The Relationship of the Pol IVa siRNA Pathway and the Signaling of Silencing
To further explore the relationship of the Pol IV siRNA pathway and silencing signaling, we characterized the PDS siRNAs in the wild type (JAP 3 and JAP 5) and mutant lines of Arabidopsis by RNA gel blot analysis. Both the 21- and 24-nucleotide siRNAs were reduced in the clsy1-3, clsy1-4, nrpd1a-5, and rdr2-5 mutant plants (Figure 5A), suggesting that the mutated proteins are required for either primary or secondary PDS siRNA production. By contrast, in ago4-1, the siRNAs of both size classes were more abundant than in wild-type plants, consistent with the increased photobleaching phenotype in this line. The dcl3-1 mutant plants also exhibited increased photobleaching and had higher levels of 21-nucleotide siRNAs than did the wild-type plants (JAP 3) but not the 24-nucleotide siRNAs. This result suggests that the 24-nucleotide siRNAs but not the 21-nucleotide siRNAs are made by DCL3 and that the 24-nucleotide siRNAs are not required for movement of the silencing signal, consistent with previous findings (Himber et al., 2003; Dunoyer et al., 2005).
Figure 5.
PDS and PSuc2:PDS RNAs in Wild-Type and Mutant Plants.
(A) Detection of PDS siRNAs in mutant backgrounds. Low molecular weight RNA gel blot analysis was performed on 50 μg of total nucleic acids isolated from the aerial portions of 1-month-old plants. The PDS probe was prepared by T7 transcription from a T7:PDS template amplified from genomic DNA using primers T7 PDS 5′ and PDS 5862r. The U6 signal is included as a loading control.
(B) PDS mRNA levels in 24-nucleotide siRNA pathway mutants. A total of 1.75 μg of poly(A) RNA from 1-month-old plants was analyzed by high molecular weight RNA gel blot. A randomly primed probe from a PDS PCR product (amplified using primers PDS left and PDS right) was used to detect PDS transcript. EF1α was probed as a loading control.
(C) RT-PCRs prepared with the Qiagen one-step RT-PCR kit on total RNA from 1-month-old seedlings. The bottom panel shows a negative control of EF1α RT-PCRs in which the reverse transcription step was omitted from the protocol. Thirty-four cycles of amplification were used for the transgene, and 26 cycles of amplification were used for EF1α.
In keeping with the photobleaching phenotypes, the PDS mRNA levels were inversely correlated with effects of the mutations on siRNA (Figure 5B). Thus, in the reduced silencing mutants nrpd1a, rdr2, and clsy1, PDS mRNA levels were higher than in the JAP parent. Conversely, in the enhanced-photobleaching mutants ago4-1 and dcl3-1, the PDS mRNA levels were substantially lower than in the JAP parent.
To explain the enhanced silencing mutant phenotypes, we propose that the PSuc2:PDS transgene is normally self-silenced through a mechanism that is dependent on AGO4 and DCL3. Loss of function in either protein releases the self-silencing mechanism, and the increased accumulation of the PDS dsRNA generates the enhanced silencing phenotype. To test this hypothesis, we assayed the PSuc2:PDS transgene transcript by RT-PCR with primers specific for the transgene rather than the endogenous PDS RNA (Figure 5C). Consistent with our hypothesis, PSuc2:PDS transgene transcripts were substantially more abundant in ago4-1 and dcl3-1 than in the wild-type parent. Conversely, in nrpd1a-5, rdr2-5, and clsy1-3, the reduced-spreading phenotype correlates with low levels of transgene transcripts. We consider that this reduction is seen because the RT-PCR detected a mixture of primary Pol II–derived transcripts from the transgene and secondary RNAs generated by Pol IVa or RDR2. These secondary RNAs would be absent in nrpd1a-5, rdr2-5, and clsy1-3. The anomalous increase in transgene RNA in clsy1-4 is discussed below.
To investigate the relationship between self-silencing and the NRPD1a/RDR2/CLSY1 pathway required for the spread of PDS silencing from the phloem, we generated double mutant plants. According to our hypothesis, the ago4-1 allele in these double mutants would suppress the PSuc2:PDS self-silencing pathway, whereas rdr2-2 and clsy1-3 would suppress the pathway required for the spread of silencing. We expected that if RDR2 and CLSY1 are essential for spreading, the ago4 rdr2 and ago4 clsy1 mutants would exhibit loss or reduction of the ago4-1–enhanced PDS silencing phenotype.
However, that was not the result (Figure 6A). Double mutants exhibited the same enhanced silencing as in the single mutant ago4-1, and we conclude that RDR2 and CLSY1 are only required for the spread of silencing if AGO4 and DCL3 are present. The simplest interpretation of these results is that spreading is dependent on dsRNA from the PSuc2:PDS transgene. In a self-silencing background involving AGO4 and DCL3, the dsRNA production required for spreading would be generated through the combined action of Pol IV and RDR2. It may be that this requirement is related to the compound nature of the transgene inserts (shown in Supplemental Figure 1 online for JAP 3). In the absence of the self-silencing mechanism, dsRNA produced by direct transcription of the transgene would support the spreading mechanism.
The limited-spreading phenotype in plants with functional AGO4 and DCL3 is suppressed by dcl4-2. Therefore, to determine whether the enhanced-spreading mechanism is through the same mechanism, we generated dcl3-1 dcl4-2 double mutants. If the enhanced-spreading signal is long dsRNA in the double mutants, for example, it would be unaffected by dcl4. However, as shown in Figure 6B, the PSuc2:PDS lines with a dcl3-1 dcl4-2 genotype exhibited suppression of the full enhanced silencing phenotype relative to the dcl3-1 single mutant. Thus, the spreading phenotype in the presence or absence of DCL3 function is influenced by DCL4. The failure to achieve complete suppression of silencing in the double mutant is likely to reflect the functional redundancy of DCL proteins: DCL2 and DCL1 would be produced in the dcl3-1 dcl4-2 genotype plants and could substitute for the lack of DCL4 (Gasciolli et al., 2005).
CLSY1 Mutations Disrupt the Subcellular Localization of the Pol IVa Pathway Proteins RDR2 and Pol IVa
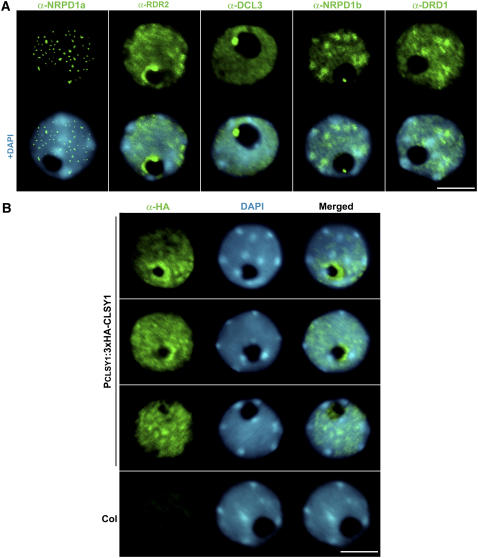
The Pol IVa silencing pathway had been defined as NRPD1a/NRPD2a>RDR2>DCL3>AGO4>Pol IVb/DRD1 based on epistatic effects of mutations in the upstream pathway components on the subcellular localization of the downstream proteins (Pontes et al., 2006). Therefore, to place CLSY1 in this pathway, we analyzed the effect of CLSY1 mutation on the localization of the NRPD1a Pol IVa subunit, RDR2, and DRD1. In addition, we analyzed the subcellular localization of CLSY1.
We localized CLSY1 using an antibody to the N-terminal 3× hemagglutinin (HA) epitope tag in the CLSY1 transgenic complementation lines. Unlike NRPD1a, which is located in punctate foci in the nucleoplasm and is absent from the nucleolus (which is apparent as the portion of the nucleus that is not stained by the fluorescent DNA binding dye 4′,6-diamidino-2-phenylindole [DAPI]), CLSY1 was localized in a distinctive ring around the inner periphery of the nucleolus in addition to being dispersed throughout the nucleoplasm (Figures 7A and 7B). The localization pattern of CLSY1 is similar to the localization pattern of RDR2, which also forms a ring or crescent along the inner perimeter of the nucleolus. However, unlike CLSY1, RDR2 is also localized to a distinctive nucleolar dot (Figure 7A) in which DCL3, AGO4, and siRNAs were previously shown to localize, suggesting that the dot corresponds to an siRNA processing center (Li et al., 2006; Pontes et al., 2006).
Figure 7.
Subcellular Localization of CLSY1 Compared with 24-Nucleotide siRNA Pathway Proteins.
(A) Immunolocalization of NRPD1a, RDR2, DCL3, NRPD1b, and DRD1 in nuclei of wild-type cells (green fluorescence). Counterstaining of nuclei with DAPI (blue fluorescence) reveals the nucleolus as a black hole. Neither NRPD1a nor DRD1 localizes within the nucleolus, but RDR2, DCL3, and NRPD1b all localize to a nucleolar dot at the edge of the nucleolus, corresponding to the apparent 24-nucleotide siRNA processing center, in addition to nucleoplasmic localization. RDR2 also forms a characteristic crescent (as shown) or a complete ring around the inner perimeter of the nucleolus. Bar = 5 μm.
(B) Localization of CLSY1 by virtue of its HA epitope tag in clsy1 PCLSY1:3xHA-CLSY1 lines. Nuclei were counterstained with DAPI (blue). Typical localization patterns in independently transformed lines in the clsy1-3 and clsy1-4 mutant backgrounds are shown. No signal is detected in nontransgenic Col-0, as expected. Bar = 5 μm.
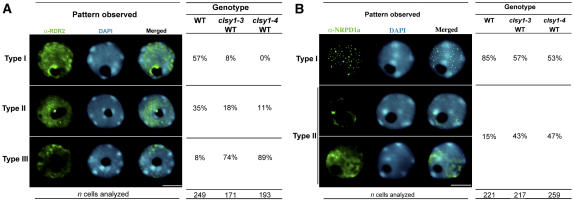
Overall, CLSY1 displayed a localization pattern that suggests a possible role at a step intermediate between NRPD1a and RDR2. Consistent with this hypothesis, RDR2 localization was severely disrupted in clsy1 mutants (Figure 8A; see Supplemental Figure 5A online). In both clsy1-3 and clsy1-4 mutants, the RDR2 signal was reduced and the nucleolar dot signal was absent in ∼90% of the nuclei examined. Therefore, it is likely that CLSY1 acts upstream of RDR2 in the silencing pathway and is required for the proper localization of RDR2 in the nucleolus.
Figure 8.
Effect of the clsy1 Alleles on RDR2 and NRPD1a Localization.
Immunolocalization using antibodies raised against the native RDR2 (A) and NRPD1a (B) is shown in green. Representative nuclei corresponding to RDR2 or NRPD1a interphase distribution patterns are presented. The frequency of nuclei observed in Arabidopsis Col-0 and in clsy1-3 and clsy1-4 mutants is shown (see also Supplemental Table 4 online).
(A) Disruption of RDR2 localization. Localization patterns are classified as follows: type I, nucleoplasmic foci, nucleolar dot, and nucleolar perimeter ring; type II, nucleoplasmic foci and nucleolar dot, no nucleolar perimeter ring; type III, nucleoplasmic foci only (no nucleolar dot or ring). Bar = 5 μm.
(B) clsy1 mutant alleles have subtle effects on NRPD1a localization. The interphase nuclei were sorted according to the localization patterns observed as follows: type I, large number of small foci in the nucleoplasm and near the chromocenter periphery; type II, one to three large foci or diffuse labeling with no detectable foci. Bar = 5 μm.
NRPD1a localization was also affected to some extent in clsy1 mutants, although to a lesser degree than RDR2 (Figure 8B; see Supplemental Figure 5B online). In wild-type cells (Col-0, JAP 3, and JAP 5), NRPD1a was typically localized to numerous punctate foci in the nucleoplasm (79 to 85% frequency). However, NRPD1a was also found (15 to 21% frequency) in a diffuse labeling pattern in which no more than one to three disorganized foci are apparent in wild-type nuclei (see Supplemental Table 4 online). In clsy1-3 and clsy1-4 mutants, the frequency of nuclei displaying the punctate NRPD1a pattern typical of wild-type cells was reduced by ∼20 to 25% (to a frequency of 53 to 65%), and a larger proportion of the nuclei (35 to 47%) displayed the more diffuse NRPD1a localization pattern. Therefore, clsy1 mutations have a subtle but detectable influence on NRPD1a localization in addition to having dramatic effects on RDR2 localization. It is possible that the effects of clsy1 mutations on NRPD1a may be indirect, due to disruption of RDR2 and downstream steps of the pathway that are part of a positive feedback loop for maintaining Pol IVa-dependent silencing. Alternatively, a more plausible scenario is that CLSY1 acts upstream of RDR2 and downstream of, or in partnership with, Pol IVa.
We also investigated the subcellular localization of two other proteins (NRPD1b and DRD1). These homologs of NRPD1a and CLSY1 have been implicated in a Pol IV–related pathway of RNA silencing (Kanno et al., 2005a, 2005b), but corresponding loss-of-function mutations had no effect on the PSuc2:PDS silencing phenotype, as described above. The localization of DRD1 was unaffected in either the clsy1-3 or the clsy1-4 background, as expected (see Supplemental Figure 6 online), although localization of NRPD1b to the nucleolar dot was compromised (see Supplemental Figure 7 online), most likely due to the disruption of RDR2 localization in an upstream step of the pathway (Li et al., 2006; Pontes et al., 2006).
DISCUSSION
Silencing Pathways and the Silencing Signal
Genetic analysis, as presented here, identified proteins affecting the spread of the silencing phenotype. Based on cytological evidence, our data support the sequence Pol IVa > CLSY1 > RDR2 or Pol IVa/CLSY1 > RDR2 (with Pol IVa and CLSY1 acting in partnership in the latter scenario) in the upstream part of a pathway, leading to the spread of PDS silencing (Figures 7 and 8). It is likely that this upstream part of the pathway generates dsRNA for cleavage by DCL proteins. The downstream part of this pathway is discussed below.
An alternative upstream mechanism for dsRNA production could be by direct Pol II–mediated transcription of the self-complementary regions in the PSuc2:PDS transgene RNA, as is likely with transgene (Kanno et al., 2005b) and endogenous inverted repeat loci (Zhang et al., 2007). It is unlikely that these directly transcribed self-complementary transcripts are required for the spreading phenotype in plants with functional AGO4 and DCL3, because there is a loss of spreading in nrpd1a, rdr2, and clsy1 genotype plants (Figure 1B): the proteins encoded by these mutant loci would not be required if the dsRNA is produced by direct transcription. However, if DCL3 or AGO4 is not fully functional, the PDS dsRNA production does not require the Pol IVa > RDR2 mechanism. Thus, in plants with dcl3-1 or ago4-1 mutation, there is an enhanced silencing phenotype that, in the ago4-1genotype at least, is insensitive to rdr2 or clsy1 (Figure 6A).
To explain the different rdr2 and clsy1 effects in plants with either wild-type or mutant forms of AGO4/DCL3, we invoke a self-silencing mechanism. The directly transcribed dsRNA from PSuc2:PDS would initiate this self-silencing mechanism through a mechanism that is dependent on DCL3 and AGO4. After the onset of self-silencing, the production of PDS dsRNA to mediate the spread of PDS silencing would be dependent on CLSY1, Pol IVa, and RDR2. In the absence of the self-silencing mechanism, dsRNA production by direct transcription would be enhanced (Figure 5C) and the synthesis of PDS dsRNA to mediate spreading would be independent of CLSY1, Pol IVa, and RDR2. A summary model of this interpretation is shown in Figures 9A and 9B.
Figure 9.
Model for RNA Silencing in the PSuc2:PDS Line.
(A) In wild-type plants, we propose that two parallel pathways of PDS silencing and PSuc2:PDS self-silencing operate through amplification by Pol IVa/CLSY1/RDR2 and siRNA production by DCL4. By contrast, the PSuc2:PDS self-silencing pathway is proposed to function through Pol II, DCL3, and AGO4.
(B) In the absence of DCL3 or AGO4, transcription of the inverted repeat by Pol II and Pol IVa may lead to the production of excess siRNAs through DCL4 and increased PDS silencing.
Our interpretation of the downstream part of the PDS RNA silencing pathways is based on the reduced silencing phenotype of dcl4-2 mutants (Figure 6B). These phenotypes indicate that DCL4 plays a role downstream of the PSuc2:PDS foldback RNA in dcl3-1 plants (Figure 6B) and are consistent with DCL4 acting downstream of CLSY1, Pol IVa, and RDR2 if the proposed transgene self-silencing pathway operates (Figure 9). It is likely that DCL4 generates the 21-nucleotide siRNAs that are associated with enhanced silencing in dcl3-1 (Figure 5A). Other proteins acting in this downstream part of the pathway may have been mutated in PSuc2:PDS plants that, like dcl4-2, are defective in tasiRNA production (see Supplemental Figure 2 online); therefore, it is likely that the downstream part of the pathway leading to the spread of PDS silencing out of the veins is similar to the downstream part of the pathway leading to tasiRNA production.
Unfortunately, these genetic analyses cannot identify the stage in the pathway in which the silencing signal is produced. The ambiguity exists because genetic analysis does not generate information about the cells in which the various pathway genes are expressed. A plausible scenario from the results presented here, in agreement with previous suggestions (Dunoyer et al., 2005), is that the signal includes the siRNAs derived by DCL4-mediated processing of dsRNA. However, there are other equally plausible interpretations. It could be, for example, that DCL4 is only required in cells that receive the signal and that an earlier precursor RNA is, or is part of, the informational molecule that moves between cells. Alternatively, it could be that all of the genes defined by our analysis act downstream of signal production. They could all play a role in cells that receive a silencing signal that is generated through an as yet undefined mechanism.
To test these various mechanisms, it will be necessary to express different genes in the pathway in separate cells. In the context of long-distance movement, this separation can be achieved relatively easily by reciprocal grafting of mutant and wild-type stocks and scions. Data to be presented elsewhere (C. Brosnan and B. Carroll, personal communications) will describe this approach. For short-distance movement, an alternative method would use genetic mosaics. There would be spread of silencing into sectors that do not express upstream genes required for signal production, but not if the sector knocks out downstream genes in the pathway required for reception. Conversely, there will be spread of signal from sectors that lack functions required for signal reception, but not, of course, from sectors that do not produce the signal. In the absence of this type of information, we cannot conclude anything about the identity of the silencing signal.
Modules in Silencing Pathways
There are multiple RNA silencing pathways in plants (Baulcombe, 2004). Correspondingly, there are multigene families encoding silencing-related proteins and various types of short silencing RNAs, including miRNAs, tasiRNAs, natural antisense siRNAs, and the 24-nucleotide Pol IV pathway siRNAs. These pathways were originally described as separate parallel processes, but it is now increasingly apparent that they interact or operate sequentially. The processing of tasiRNA precursors, for example, is dependent on miRNAs (Axtell et al., 2006) and illustrates how pathways can operate sequentially.
The potential for pathways to interact is illustrated by the features of the PDS spreading pathway described here. The upstream part resembles the 24-nucleotide endogenous siRNA pathway involving CLSY1, Pol IVa, and RDR2. However, in the PSuc2:PDS system, this putative process for dsRNA production is uncoupled from the downstream part of the endogenous pathway involving DCL3 and AGO4. The upstream process is instead linked with DCL4 and perhaps other proteins that operate downstream of RDR6 in the tasiRNA pathway. Our interpretation of these findings is that the silencing pathways have upstream and downstream modules that can combine to form different pathways. The DCL4 module is associated with the spreading of PDS silencing, whereas the DCL3/AGO4 module would lead to the self-silencing of the PSuc2:PDS transgene (Figure 9).
Modularity is also apparent in the biogenesis of natural antisense siRNA pathways, in which the NRPD1a Pol IVa subunit is involved (Borsani et al., 2005). However, unlike the Pol IVa endogenous siRNA pathway, which involves RDR2 and DCL3, or the PDS spreading pathway described here, with RDR2 and DCL4, the natural antisense siRNA Pol IVa pathway involves RDR6, DCL1, and DCL2. The complexity in this natural antisense siRNA pathway may be the consequence of multiple modular pathways operating sequentially. Presumably, the properties of an RNA entering a silencing pathway, and features of the chromosomal domain in which that RNA is transcribed, are factors that influence the flow of RNA between the different modules.
Are there endogenous pathways leading to the spread of RNA silencing between cells? At present, we cannot answer that question; only viral and transgene silencing RNAs have been unequivocally associated with spreading. However, based on the requirement of DCL4 for the spread of transgene silencing (Figure 6B) (Dunoyer et al., 2005), it is a plausible speculation that DCL4-dependent tasiRNAs or miRNAs (Rajagopalan et al., 2006) may be mobile. If there are any DCL4- and Pol IVa–dependent siRNAs associated with spreading, like the transgene system described here, they may include DCL4-dependent 21/22-nucleotide species.
SNF2 Helicase Proteins in RNA Silencing
CLSY1 is the second SNF2 domain–containing protein to be associated with nuclear RNA silencing in plants. The other, DRD1, is a homolog of CLSY1 that was identified in a mutant screen of a Pol IVb–dependent silencing system (Kanno et al., 2004, 2005b). Pol IVa differs from Pol IVb in the largest subunit (NRPD1a versus NRPD1b); therefore, we propose that CLSY1 and DRD1 have analogous functions for Pol IVa and Pol IVb, respectively. Arabidopsis also encodes four other SNF2 helicase proteins that are highly similar to DRD1 and CLSY1 (Kanno et al., 2004) that may also have roles as other variant RNA silencing modules.
Cytochemistry indicates that CLSY1 localizes within a perinucleolar ring similar to RDR2 (Figure 7), and structural analysis indicates that it is likely to be an ATPase, like Rad54 (Figure 4). The differences between CLSY1 and Rad54 in the DNA binding domain do not rule out DNA binding activity of this protein. However, the likely insertions in the DNA binding site compared with Rad54 may make this protein capable of binding RNA or RNA–DNA duplex structures. CLSY1's nucleolar localization, where it might be required for RDR2 activity, and evidence that Pol IVa may be an RDR (Pontes et al., 2006) are consistent with a possible function for CLSY1 on RNA templates. By analogy with the role of SNF2 helicases on chromatin remodeling, it could be that CLSY1 remodels ribonucleoprotein structures and thereby influences the availability of the RNA to polymerases. The availability of an epitope-tagged version of CLSY1 (Figure 7; see Supplemental Figure 3 online) should allow us to investigate directly the physical interactions of CLSY1—with both protein and nucleic acid—that are required in RNA silencing.
In this article, we characterize two mutant alleles of CLSY1: clsy1-3 and clsy1-4. Both exhibited reduced spreading of PDS silencing but had unexpected phenotypic differences. The clsy1-4 allele, with a missense G592E mutation, exhibited extensive loss of the endogenous 24-nucleotide siRNAs (Figure 2), reduced cytological signal for NRPD1a (see Supplemental Figure 7B online), and, unexpectedly, a higher level of the PSuc2:PDS transgene transcript (Figure 5C). By contrast, the clsy1-3 allele, with a nonsense mutation at position Gln-475, had little or no effect on the endogenous 24-nucleotide siRNAs (Figure 2), a lesser effect on the cytological signal for NRPD1a (see Supplemental Figure 7B online), and a slightly reduced level, relative to the wild type, of the PSuc2:PDS transgene transcript. Formally, we cannot rule out the possibility that the clsy1-4 mutation is affected by a second mutation. However, our genetic data in mapping, from the phenotype of backcrossed plants in Supplemental Figure 3 online and in immunolocalization assays, require that any such second mutation must be closely linked to CLSY1. Therefore, it is likely that the phenotype difference between the clsy1-3 and clsy1-4 plants is due to the mutations at the CLSY1 locus. To explain these differences, we propose that clsy1-4 is a neomorphic mutation encoding a misfunctional protein. This neomorphic property must be weak, because the mutation is fully recessive in heterozygotes with a wild-type allele (L.M. Smith, unpublished data).
We envision that the misfunctional CLSY1-4 protein might allow Pol IVa-directed production of siRNA precursor RNAs but prevent the accessibility of these RNAs as substrates for RDR2-mediated amplification and subsequent processing into siRNAs (Figure 2). This misfunctionality could explain the hyperaccumulation of PSuc2:PDS RNA in the clsy1-4 plants (Figure 5C). These long RNAs would be a precursor of PDS siRNAs and, in a wild-type plant, would accumulate at a low level, because they would be targeted in a self-silencing process by the same PDS siRNAs. In a clsy1-3 plant with a loss of protein, these precursor RNAs would also accumulate at a low level (Figure 5C), because they would not be synthesized. The misfunctional CLSY1-4 protein might allow the biosynthesis of these RNAs, but because the corresponding siRNAs would not be produced (Figure 5A), the self-silencing process would not operate. It could also be envisioned that the hyperaccumulation of these siRNA precursors could interfere with the localization of proteins at or immediately downstream of CLSY1. Further exploration of these variant clsy1 phenotypes will allow us to understand the links between the Pol IVa pathway, the spread of silencing, and the modular nature of silencing pathways.
METHODS
DNA Constructs
The pJawohl 8 vector (I. Somssich, Max Planck Institute for Plant Breeding Research) was used as the backbone for the PSuc2:PDS construct. The Suc2 promoter was amplified from Arabidopsis thaliana using primers AtSuc2 left and AtSuc2 right (see Supplemental Table 5 online for all primer sequences) and cloned into pGEM-T Easy (Promega). The 35S promoter was excised using AscI and XhoI restriction sites and replaced with the Suc2 promoter. A cDNA fragment of PDS was cloned into pENTR4 (Invitrogen) with primers PDS left and PDS right containing NotI and SalI restriction sites. The PDS inverted repeat of the PSuc2:PDS construct was formed using an LR clonase (Invitrogen) reaction.
The PCLSY1:3xHA-CLSY1 complementation construct was assembled in pGreen-Gateway pGGW28 (N-terminal HA tag) (Herr et al., 2005). The CLSY1 promoter and 5′ UTR (−7 to −1520 bp) were amplified with primers At3g42670 −1520F +KpnI and At3g42670 −1R +XbaI and cloned into the XbaI and KpnI sites of pGGW28. The CLSY1 coding sequence was amplified from Arabidopsis DNA using primers At3g42670 1F and At3g42670 4178R + TGA and cloned into pENTR/DTOPO (Invitrogen). LR clonase was used to transfer the coding sequence into the pGGW28-promoter construct.
Transgenic Plant Lines
The PSuc2:PDS construct was transformed into Col-0 plants and the PCLSY1:3xHA-CLSY1 construct was transformed into clsy1-4 plants using the Agrobacterium tumefaciens floral dip method (Bechtold et al., 1993). Complementation of clsy1-3 with PCLSY1:3xHA-CLSY1 involved crossing the clsy1-4 transgenic lines to clsy1-3. Transformants were obtained by growth on F1 Arabidopsis mix and selection with BASTA (PSuc2:PDS) or growth on GM supplemented with hygromycin (35 mg/L; Sigma-Aldrich; PCLSY1:3xHA-CLSY1).
PSuc2:PDS seeds (JAP 3 and JAP 5 lines) were mutagenized with 1 mM ethyl methanesulfonate, and M1 plants were grown in a total of 195 pools of ∼50 plants. Putative RNA silencing mutants were selected based on the photobleaching phenotype in the M2 generation.
Genetic Mapping
Genomic DNA samples for mapping were prepared using the method described by Edwards et al. (1991) on 96-well format microtiter plates. A low-resolution chromosomal location was found for each mutation using 24 plants with a loss of PDS silencing and a series of 18 insertion/deletion markers that are polymorphic between Col-0 and Landsberg erecta. Supplemental Table 5 online gives the sequences of all PCR primers used as genetic markers. The loss of silencing phenotype in plants with clsy1 alleles was associated with an overrepresentation of Col-0 DNA at markers LID 32 and LID 24 between 7.6 and 17.35 Mb of chromosome 3. Using the Monsanto Arabidopsis Polymorphism Database (Jander et al., 2002), we generated additional insertions/deletions and cleaved amplified polymorphism (CAP) markers between 11.15 and 16.35 Mb. An additional 2000 plants were used for high-resolution genetic mapping, with no recombination events detected between markers CAP L58 (14.30 Mb) and CAP L74 (14.85 Mb). Of the 61 genes in this interval that do not code for retroelements, At3g42670, an SNF2 domain–containing protein, was selected as the best candidate and was located 4 kb from the CAP L22 and CAP L23 markers, at which no recombinant plants were found. Sequencing of At3g42670 DNA identified single point mutations in the JAM 33, JAM 35, JAM 36, JAM 44, JAM 47, and JAM 54 lines, as detailed in the main text.
Genotyping of clsy1 Alleles
Genotyping of clsy1 alleles was performed using 10-μL PCR as follows. The clsy1-3 mutation (JAM 54) was detected by amplification with primers JAM 54 CAP F and CAP R and restriction by AciI to give fragments of 36, 51, 142, and 323 bp (wild type) and 36, 193, and 323 bp (clsy1-3). The clsy1-4 mutation was detected by amplification of the region with primers JAM 44 CAP F and CAP R and restriction by TaqI to give fragments of 7, 39, and 411 bp (wild type) and 7, 39, 108, and 303 bp (clsy1-4). The clsy1-6 mutation was detected by amplification with primers At3g42670 2191 F and At3g42670 3529 R and digestion by XbaI to give fragments of 603 and 762 bp (wild type) and 275, 487, and 603 bp (clsy1-6). clsy1-1, clsy1-2, and clsy1-5 alleles were genotyped by DNA sequencing of At3g42670.
RNA Detection and Characterization
Poly(A) RNA for use in high molecular weight RNA gel blots was purified using the MicroPoly(A)Purist kit (Ambion). A total of 400 μg of total RNA from the aerial sections of 1-month-old plants was used as the starting material for poly(A) RNA purification, and two rounds of purification were performed. RNA was electrophoresed on a 1% agarose formaldehyde denaturing gel and then transferred to a Hybond N+ membrane (Amersham Biosciences). RNA was cross-linked to the membrane, and hybridizations were performed in modified Church buffer (0.5 M sodium phosphate buffer, pH 7.2, 7% SDS, and 10 mM EDTA). PDS and EF1α RNAs were detected using randomly primed DNA probes labeled with the Rediprime II kit (Amersham Biosciences) prepared from RT-PCRs of PDS (primers JAP for RPA and JAP 3′ end of transcript; see Supplemental Table 5 online for primer sequences) and EF1α (primers EF1α F and EF1α R). Probes were purified using MicroSpin G-25 Sephadex columns (Amersham Biosciences).
RT-PCR was performed with the Qiagen one-step RT-PCR kit according to the manufacturer's protocol. Primers used for detection of the PSuc2:PDS hairpin were PDS right and JAP 3′ end of transcript, while primers for the control amplification of EF1α were EF1α F and EF1α R, as given above.
RACE kits from three different suppliers were used for 5′ RACE of CLSY1. The Marathon cDNA amplification kit (Clontech Laboratories) was used with gene-specific primers (At3g42670 600R and At3g42670 500R) and the supplied adaptor primers (AP1 and AP2). The GeneRacer kit (Invitrogen) protocol was performed with gene-specific primers that were either generic or specific for the splice variants in the 5′ UTR of CLSY1. The third RACE kit used was the 5′/3′ RACE kit (Roche). First-strand synthesis was performed using a gene-specific primer, followed by PCR with a gene-specific primer and the supplied oligo(dT) anchor primer. Nested PCR was performed using splice variant–specific primers with the supplied PCR anchor primer. RACE PCR products were cloned into the pGEM-T Easy plasmid (Promega). Colonies containing insertions of the correct size were selected by colony PCR and sequenced. Results were analyzed using AlignX (Vector NTi) with manual adjustments as required.
Endogenous siRNAs were detected as described previously (Herr et al., 2005). The PDS siRNA probe was prepared by T7 transcription from a T7:PDS template amplified from genomic DNA using primers T7 PDS 5′ and PDS 5862r.
Bioinformatics
Homologs of CLSY1 were identified through the BLASTP algorithm at National Center for Biotechnology Information and through the Arabidopsis Transcription Factor Database. Alignments of the full-length SNF2 domain–containing proteins and helicase C and SNF2 domains from homologs were constructed using the AlignX (Vector NTi) program with manual correction as required. Phylogenetic trees of the Arabidopsis SNF2 domain–containing proteins were constructed using 1000 iterations of the neighbor-joining method and were bootstrapped. The putative nuclear localization signal was identified through the PredictNLS program (Cokol et al., 2000). A description of the method used for protein modeling is given in the text.
Protein Localization
Localization of the 3× HA–tagged CLSY1 protein, DCL3, RDR2, DRD1, NRPD1a, and NRPD1b was performed as detailed by Pontes et al. (2006). Briefly, interphase nuclei were isolated from leaves of 28-d-old plants, spread on glass slides, and fixed in 4% paraformaldehyde. Slides were incubated overnight at 4°C with antibodies raised against peptides corresponding to NRPD1a, RDR2, NRPD1b, or DRD1. Antibodies were diluted 1:400 in 1% BSA. The PCLSY1:3xHA-CLSY1 protein was detected using a mouse monoclonal anti-HA primary antibody (Sigma-Aldrich; 1:200 dilution). Antibody–antigen complexes were detected using goat anti-chicken secondary antibody conjugated to Alexa 488 (Molecular Probes; 1:400 dilution) or anti-mouse antibody conjugated to fluorescein isothiocyanate (Sigma-Aldrich; 1:400 dilution). Secondary antibody incubations were for 1 h at 37°C. DNA was counterstained with 1 μg/mL DAPI (Sigma-Aldrich) in Vectashield (Vector Laboratories). Images were obtained using a Nikon Eclipse E800i epifluorescence microscope equipped with a Photometrics Coolsnap ES Mono digital camera and were processed using Adobe Photoshop software (Adobe Systems).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: SUC2 (At1g22710), PDS (At4g14210), CLSY1 (At3g42670), NRPD1a (At1g63020), NRPD1b (At2g40030), NRPD2a (At3g23780), DRD1 (At2g16390), RDR2 (At4g11130), DCL3 (At3g43920), AGO4 (At2g27040), RDR6 (At3g49500), DCL4 (At5g20320), and CLSY2 (At5g20420).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Structure of the T-DNA Insertion Site in the JAP 3 Line.
Supplemental Figure 2. Detection of Endogenous siRNAs and miRNAs in the Spread of Silencing Mutants.
Supplemental Figure 3. Complementation of the clsy1-3 and clsy1-4 Mutants with a PCLSY1:3xHA-CLSY1 Transgene.
Supplemental Figure 4. Bioinformatic Comparison of CLSY1 with Other SNF2 Domain–Containing Proteins.
Supplemental Figure 5. Immunolocalization of RDR2 and NRPD1a.
Supplemental Figure 6. Immunolocalization of DRD1 in a clsy1 Mutant Background.
Supplemental Figure 7. Immunolocalization of NRPD1b in a clsy1 Mutant Background.
Supplemental Table 1. Isolation of New NRPD1a, RDR2, and DCL3 Alleles.
Supplemental Table 2. 5′ RACE of CLSY1.
Supplemental Table 3. Upstream Open Reading Frames in the 5′ UTR of CLSY1.
Supplemental Table 4. Immunolocalization of the PCLSY1:3xHA-CLSY1 Transgenic Line and 24-Nucleotide Pathway Proteins in the clsy1 Mutant Background.
Supplemental Table 5. Primer Sequences.
Supplementary Material
Acknowledgments
We acknowledge the contribution of Jagger Harvey in the mapping of nrpd1a-7 (JAM 42) and David Studholme for assistance with protein sequence alignments and database searches. The Gatsby Charitable Foundation and a European Union training network (Silencing in Different Organisms) provided funding for this work. Cytological work in the Pikaard laboratory was funded by National Institutes of Health Grant R01GM-077590 to C.S.P. and by Fellowship SFRH/BPD/17508/2004 from the Fundação Para a Ciência e Tecnologia, Portugal, to O.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: David C. Baulcombe (david.baulcombe@tsl.ac.uk).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., Jan, C., Rajagopalan, R., and Bartel, D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127 565–577. [DOI] [PubMed] [Google Scholar]
- Bates, P., Kelley, L., MacCallum, R., and Sternberg, M. (2001). Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins 45 39–46. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. III 316 1194–1199. [Google Scholar]
- Borsani, O., Zhu, J., Verslues, P.E., Sunkar, R., and Zhu, J.-K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol, M., Nair, R., and Rost, B. (2000). Finding nuclear localization signals. EMBO Rep. 1 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer, P., Himber, C., and Voinnet, O. (2005). DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 37 1356–1360. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, J.A., Sweder, K.S., and Hanawalt, P.C. (1995). Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 23 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli, V., Mallory, A.C., Bartel, D.P., and Vaucheret, H. (2005). Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15 1494–1500. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in post-transcriptional gene silencing in plants. Science 286 950–952. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., Voinnet, O., Chappell, L., and Baulcombe, D.C. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M.-H., Goud, S., Song, L., and Fedoroff, N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood, V., Yu, T.S., Huang, N.C., and Lucas, W.J. (2005). Phloem long-distance trafficking of gibberellic acid-insensitive RNA regulates leaf development. Plant J. 42 49–68. [DOI] [PubMed] [Google Scholar]
- Herr, A.J., Jensen, M.B., Dalmay, T., and Baulcombe, D. (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308 118–120. [DOI] [PubMed] [Google Scholar]
- Himber, C., Dunoyer, P., Moissiard, G., Ritzenthaler, C., and Voinnet, O. (2003). Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22 4523–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel, B., Kanno, T., Daxinger, L., Aufsatz, W., Matzke, A.J.M., and Matzke, M. (2006). Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 25 2828–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Rounsley, S.D., Bush, D.F., Levin, I.M., and Last, R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129 440–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno, T., Aufsatz, W., Jaligot, E., Mette, M.F., Matzke, M., and Matzke, A.J. (2005. a). A SNF2-like protein facilitates dynamic control of DNA methylation. EMBO Rep. 6 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno, T., Huettel, B., Mette, M.F., Aufsatz, W., Jaligot, E., Daxinger, L., Kreil, D.P., Matzke, M., and Matzke, A.J. (2005. b). Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 37 761–765. [DOI] [PubMed] [Google Scholar]
- Kanno, T., Mette, F., Kreil, D.P., Aufsatz, W., Matzke, A.J.M., and Matzke, M. (2004). Involvement of putative SNF2 chromatin remodelling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 14 801–805. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, R., and Bailey-Serres, J. (2005). mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 33 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C.F., Pontes, O., El-Shami, M., Henderson, I.R., Bernatavichute, Y.V., Chan, S.W.L., Lagrange, T., Pikaard, C., and Jacobsen, S.E. (2006). An ARGONAUTE4-containing nuclear processing center colocalized with cajal bodies in Arabidopsis thaliana. Cell 126 93–106. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., et al. (2001). HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Mlotshwa, S., Bowman, L.H., and Vance, V.B. (2003). The capacity of transgenic tobacco to send a systemic RNA silencing signal depends on the nature of the inducing transgene locus. Plant J. 35 82–92. [DOI] [PubMed] [Google Scholar]
- Onodera, Y., Haag, J.R., Ream, T., Nunes, P.C., Pontes, O., and Pikaard, C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120 613–622. [DOI] [PubMed] [Google Scholar]
- Pak, J., and Fire, A. (2007). Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315 241–244. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.-C., Elmayan, T., Pollien, J.-M., and Vaucheret, H. (1997). Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui, J.-C., and Vaucheret, H. (1998). Transgenes are dispensable for the RNA degradation step of cosuppression. Proc. Natl. Acad. Sci. USA 95 9675–9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes, O., Li, C.F., Nunes, P.C., Haag, J.R., Ream, T., Vitins, A., Jacobsen, S.E., and Pikaard, C.S. (2006). The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126 79–92. [DOI] [PubMed] [Google Scholar]
- Pontier, D., Yahubyan, G., Vega, D., Bulski, A., Saez-Vasquez, J., Hakimi, M.A., Lerbs-Mache, S., Colot, V., and Lagrange, T. (2005). Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19 2030–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y., Denli, A.M., and Hannon, G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19 421–428. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin, I., Kochetov, A., Kondrashov, F., Koonin, E., and Milanesi, L. (2001). Presence of ATG triplets in 5′ untranslated regions of eukaryotic cDNAs correlates with a ‘weak’ context of the start codon. Bioinformatics 17 890–900. [DOI] [PubMed] [Google Scholar]
- Ruby, J.G., Jan, C., Player, C., Axtell, M.J., Lee, W., Nusbaum, C., Ge, H., and Bartel, D.P. (2006). Large-scale sequencing reveals 21U-RNAs and additional micro-RNAs and endogenous siRNAs in C. elegans. Cell 127 1193–1207. [DOI] [PubMed] [Google Scholar]
- Ruiz, M.T., Voinnet, O., and Baulcombe, D.C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126 4405–4419. [DOI] [PubMed] [Google Scholar]
- Ryabov, E.V., van Wezel, R., Walsh, J., and Hong, Y. (2004). Cell-to-cell, but not long-distance, spread of RNA silencing that is induced in individual epidermal cells. J. Virol. 78 3149–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanoubat, M., et al. (2000). Sequence and analysis of chromosome 3 of the plant Arabidopsis thaliana. Nature 408 820–822. [DOI] [PubMed] [Google Scholar]
- Schwach, F., Vaistij, F.E., Jones, L., and Baulcombe, D.C. (2005). An RNA-dependent RNA-polymerase prevents meristem invasion by Potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138 1842–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Haley, B., and Zamore, P.D. (2002). Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10 537–548. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Steiner, F.A., Thijssen, K.L., and Plasterk, R.H.A. (2007). Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315 244–247. [DOI] [PubMed] [Google Scholar]
- Szittya, G., Silhavy, D., Dalmay, T., and Burgyan, J. (2002). Size-dependent cell-to-cell movement of defective interfering RNAs of Cymbidium ringspot virus. J. Gen. Virol. 83 1505–1510. [DOI] [PubMed] [Google Scholar]
- Thoma, N.H., Czyzewski, B.K., Alexeev, A.A., Mazin, A.V., Kowalczykowski, S.C., and Pavletich, N.P. (2005). Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat. Struct. Mol. Biol. 12 350–356. [DOI] [PubMed] [Google Scholar]
- Ueki, S., and Citovsky, V. (2001). RNA commutes to work: Regulation of plant gene expression by systemically transported RNA molecules. Bioessays 23 1087–1090. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., and Baulcombe, D.C. (1997). Systemic signalling in gene silencing. Nature 389 553. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA. Cell 95 177–187. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, B.-C., Kragler, F., Varkonyi-Gasic, E., Haywood, V., Archer-Evans, S., Lee, Y.M., Lough, T.J., and Lucas, W.J. (2004). A systemic small RNA signaling system in plants. Plant Cell 16 1979–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D., Fan, B., MacFarlane, S.A., and Chen, Z. (2003). Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant Microbe Interact 16 206–216. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Henderson, I.R., Lu, C., Green, P.J., and Jacobsen, S.E. (2007). Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. USA 104 4536–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Bilgin, M., and Snyder, M. (2003). Proteomics. Annu. Rev. Biochem. 72 783–812. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus specific siRNA accumulation and DNA and histone methylation. Science 299 716–719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.