Abstract
Analyses of Arabidopsis thaliana defense response to the damping-off oomycete pathogen Pythium irregulare show that resistance to P. irregulare requires a multicomponent defense strategy. Penetration represents a first layer, as indicated by the susceptibility of pen2 mutants, followed by recognition, likely mediated by ERECTA receptor-like kinases. Subsequent signaling of inducible defenses is predominantly mediated by jasmonic acid (JA), with insensitive coi1 mutants showing extreme susceptibility. In contrast with the generally accepted roles of ethylene and salicylic acid cooperating with or antagonizing, respectively, JA in the activation of defenses against necrotrophs, both are required to prevent disease progression, although much less so than JA. Meta-analysis of transcriptome profiles confirmed the predominant role of JA in activation of P. irregulare–induced defenses and uncovered abscisic acid (ABA) as an important regulator of defense gene expression. Analysis of cis-regulatory sequences also revealed an unexpected overrepresentation of ABA response elements in promoters of P. irregulare–responsive genes. Subsequent infections of ABA-related and callose-deficient mutants confirmed the importance of ABA in defense, acting partly through an undescribed mechanism. The results support a model for ABA affecting JA biosynthesis in the activation of defenses against this oomycete.
INTRODUCTION
The success of plants in colonizing so many different environments where they have to cope with a plethora of biotic and abiotic challenges indicates that evolution has provided them with efficient defense mechanisms. Plants possess both preformed and inducible layers of defense to resist pathogen invasion. Constitutive physical and chemical barriers prevent the establishment of most plant–pathogen interactions. However, should the pathogen overcome these constitutive defenses, its recognition leads to the induction of a multitude of defenses through the genetic reprogramming of the cell.
Our understanding of the complex mechanisms by which plants first detect, and then defend against, different microbial pathogens has advanced considerably over the last few decades. Central to this progress has been the identification and characterization of plant disease resistance genes that facilitate pathogen strain–specific recognition and the identification of signal transduction pathways that link pathogen recognition with a targeted response (Nimchuk et al., 2003; Glazebrook, 2005). Three phytohormones, salicylic acid (SA), jasmonic acid (JA), and ethylene (ET), have been shown to play a major role in the regulation of these signal transduction pathways. This regulation is not achieved through the isolated activation of each single hormonal pathway but rather through a complex regulatory network that connects the different pathways enabling each to assist or antagonize the others as required to fine-tune the defense response to the individual pathogen. Thus, it is generally accepted (although may be oversimplified) that SA plays a major role in activation of defenses against biotrophic pathogens, whereas JA and ET are more usually associated with defense against necrotrophic pathogen attack. Additionally, SA and JA/ET defense pathways are mutually antagonistic (Thomma et al., 2001; Kunkel and Brooks, 2002; Turner et al., 2002; Rojo et al., 2003; Glazebrook, 2005; Lorenzo and Solano, 2005; van Loon et al., 2006).
Although mechanistic explanations of this antagonistic and cooperative crosstalk are scarce, several examples suggest its regulation through the differential modulation of transcription factor (and cofactors) activity by the different hormones. Thus, antagonism between JA and SA pathways requires the activation of proteins such as NPR1 and WRKY70 that activate expression of SA-responsive genes while repressing JA-responsive genes (Spoel et al., 2003; Li et al., 2004). Conversely, cooperation of JA and ET in the activation of defenses against necrotrophs can be explained by their concerted activation of Ethylene Response Factor1 (ERF1), which induces defense gene expression and plant resistance (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003). However, in contrast with the case of pathogen infection, ET and JA antagonize one another in the activation of responses to wounding. The fine-tune regulation of this antagonism depends on the balance of activation by both hormones of ERF1 and MYC2, another transcription factor differentially regulating two branches of the JA signaling pathway (Lorenzo et al., 2004).
The current understanding of plant defense responses described above has been achieved through the study of a limited number of models, which may be constraining our view of the plant–pathogen interaction and the true capacity of plants to defend against pathogens. In fact, random sequencing approaches of microbial populations from seawater samples have recently demonstrated that we still only know a relatively small percentage of gene functions existing in nature (Venter et al., 2004). Applied to the plant–pathogen field, discovery of new plant defense mechanisms should be favored by broadening the range of pathogens and plant species under study. With this aim, we have characterized the interaction between Pythium irregulare and Arabidopsis thaliana.
Pythium is commonly regarded as a soil-borne vascular pathogen. It is particularly virulent in seedlings, although it can infect mature aboveground tissue in several plant species, including Arabidopsis (van der Plaats-Niterink, 1981; Martin, 1995). Species within this genus range from virulent to opportunistic pathogens, producing seedling damping-off and root and stem rots that may cause economically important losses in crops and ornamental plants (Miller and Sauve, 1975; Farr et al., 1989). P. irregulare has been reported to produce both lytic enzymes (Deacon, 1979) and phytotoxins (Brandenburg, 1950) that would degrade plant tissue, thus behaving as a necrotrophic pathogen. Being an oomycete, and thus phylogenetically distant from the most extensively studied species of fungi (Kamoun et al., 1999), the supposed necrotrophic lifestyle, and the fact that JA and ET are required for effective defenses against different species from this genus also differentiate Pythium from other oomycetes (Knoester et al., 1998; Staswick et al., 1998; Vijayan et al., 1998).
To gain a deeper insight of the Arabidopsis–P. irregulare host–pathogen interaction, it is imperative to understand both the physical infection process and the molecular consequences. To that end, we have (1) characterized the infection process and (2) studied the plant molecular defense pathways involved in resistance through genetic and genomic analysis. In addition to the comprehensive characterization of the interaction, this combined analysis has identified ABA as a signal required for plant resistance to P. irregulare and other necrotrophic pathogens.
RESULTS
Characterization of P. irregulare Colonization of Arabidopsis Tissue
Appresoria were observed during early stages of infection of both roots and leaves of mature plants (Figure 1A; see Supplemental Figure 1A online). Following penetration of the first host cell, hyphal ramification gave rise to multidigitate haustoria-like structures (Figure 1B; see Supplemental Figure 1B online). The individual lobes of the haustoria-like structures continued to lengthen until an opposing cell wall was encountered, whereupon constricted hyphae attempted penetration of the adjoining host cell wall (Figure 1C; see Supplemental Figure 1C online). Further ramification of hyphal tissue occurred frequently, giving rise to a dense network that penetrated all host tissue types. This colonization eventually lead to tissue collapse and ultimately wet rot. Fungal growth was predominantly but not exclusively intracellular (Figure 1C; see Supplemental Figure 1C online).
Figure 1.
Microscopy Analysis of Arabidopsis Infection by P. irregulare.
(A) Appresorial (ap) formation on leaf epidermis. Penetration pegs (pp) may also be observed (Trypan blue stain). Bar = 30 μm.
(B) Multidigitate haustoria-like structure (hl) formed upon invasion of the epidermal cell by runner hyphae (rh). The cell wall (cw) is also fluorescent due to callose deposition (Analine blue stain). Bar = 25 μm.
(C) Constricted hyphae (hy) penetration through the root cell wall (cw) (Trypan blue stain). Bar = 5 μm.
(D) Localized plant cell death upon direct infection of P. irregulare via runner hyphae (rh). Bar = 25 μm.
(E) Trailing necrosis (tn) of infected mesophyll cells. Bar = 25 μm.
Hyphal swellings were occasionally produced where fungal growth was prolific and allowed to progress for 48 h (see Supplemental Figure 1D online). Oogonium and oospores were only observed in culture media (see Supplemental Figure 1E online), presumably because in planta infections were rarely allowed to progress beyond 24 h, this being the optimal time for discernment of differential resistance between defense mutants.
Trypan blue stain indicated that only infected cells in physical contact with the hyphae died (Figure 1D), suggesting that this oomycete does not produce lytic enzymes or phytotoxins during infection. Consistent with this, culture filtrates were not able to reproduce disease symptoms in Arabidopsis (data not shown). Thus, despite reports that P. irregulare produces both lytic enzymes (Deacon, 1979) and phytotoxins (Brandenburg, 1950), no evidence of extracellular toxin production was seen during this pathogen–host interaction.
Additionally, trailing necrosis occurred during infection of susceptible tissue, where hyphae were observed to extend ahead of dead cells (i.e., those retaining the Trypan blue stain) (Figure 1E) similar to that witnessed for Phytophthora infestans during infection of partially resistant plants (Kamoun et al., 1999). If toxins or lytic enzymes were being produced and dissipated in functionally important quantities within the plant tissue, cell death might be expected to extend ahead of hyphal growth, similar to that of Botrytis cinerea (Clark and Lorbeer, 1976; Govrin and Levine, 2000).
Characterization of Plant Defense Response
Results described above showed that P. irregulare infection of Arabidopsis has neither typical necrotrophic nor biotrophic characteristics. Additionally, oomycetes are phylogenetically distinct from many of the more extensively studied fungal pathogens (Kamoun et al., 1999). Thus, to understand how the plant counteracts hyphal invasion, we used mutants affected in hormonal and nonhormonal defense pathways to dissect the involvement of known plant defense mechanisms.
Contribution of the JA, ET, and SA Pathways
Consistent with previous analyses of JA-deficient or -insensitive mutants (Staswick et al., 1998; Vijayan et al., 1998) and the described role of JA in the activation of responses against oomycetes (Coego et al., 2005), the JA-insensitive coi1-1 mutants were highly susceptible to P. irregulare (Figure 2A), indicating that JA is a major signal for activation of defenses against this oomycete.
Figure 2.
P. irregulare Infection of Arabidopsis Defense Mutants.
(A) Infection of JA/ET/SA-related mutants. Mean disease area (MDA) per infected leaf (mm2) 24 h after inoculation with a mycelial plug. Asterisks indicate significant difference (P < 0.05) from Col-0 using the Student's t test.
(B) Endogenous JA and SA levels (ng/g fresh weight [FW]) in Arabidopsis wild-type (Col-0) and JA/ET/SA-related mutant plants (coi1-16, ein2-5, and sid2-1) after infection with P. irregulare for 0, 6, and 12 h (h after infection [HAI]). Seedlings were grown on Johnson's media plates for 7 d prior to infection. Values are means ± se of three independent replicate experiments.
(C) Infection of nonhormonal mutants. Asterisks indicate significant difference (P < 0.05) from Col-0 (or Landsberg in the case of Ler) using the Student's t test.
Consistent with results in transgenic tobacco (Nicotiana tabacum) plants (Geraats et al., 2002), ET-insensitive ein2-5 mutants showed a significantly increased susceptibility to the oomycete, indicating that ET is required to achieve fully active plant defenses. However, the weaker effect of ein2 compared with that of coi1 within this study indicates that ET is not as critical to the P. irregulare defense response as it is for other necrotrophs.
Interestingly, SA-related mutants (npr1 and sid2-1) showed a susceptibility similar to ET-insensitive mutants, indicating that SA, instead of antagonizing JA-dependent defenses, also contributes to overall resistance (Figure 2A).
To further understand this apparent cooperation between JA and SA, the levels of both hormones were measured in wild-type and JA/SA/ET-related mutants after P. irregulare infection. Supporting their role in defense against the oomycete, levels of both hormones increased rapidly in wild-type plants after infection (Figure 2B). The increase in JA levels was comparable in wild-type and sid2-1 plants and lower in coi1-16 mutants, indicating that SA does not prevent JA biosynthesis. In contrast with the assumed cooperation between ET and JA, but consistent with previous reports of ET-mediated repression of JA-regulated gene expression (Rojo et al., 2003; Lorenzo et al., 2004), JA levels were dramatically increased in ein2-5 mutants than in wild-type plants. In contrast with wild-type plants, an increase in SA levels was not observed in the biosynthetic mutant sid2-1. Furthermore, and consistent with the negative regulation of SA biosynthesis by JA/ET, the levels of SA in coi1-16 and ein2-5 mutants increased over wild-type levels.
These results support a role for both defense pathways (JA/ET and SA) in the resistance against P. irregulare.
Involvement of Other Nonhormonal Defense Pathways
We analyzed the susceptibility of nonhormonal defense-related mutants, including nonhost-related mutants (pen1 and pen2; Collins et al., 2003; Assaad et al., 2004), mutants affected in responses to biotrophs (edr1, pad4, and eds1; Frye and Innes, 1998; Falk et al., 1999; Jirage et al., 1999), and the erecta mutation (essential for resistance to some necrotrophs; Godiard et al., 2003; Llorente et al., 2005). Among them, only pen2 and erecta (in Columbia-0 [Col-0] and Landsberg erecta [Ler] backgrounds, respectively) showed a significantly increased susceptibility to P. irregulare colonization (Figure 2C). This suggests that plant resistance to the initial establishment and progression of disease by P. irregulare depends on avoidance of penetration mediated by PEN2 and, very likely, other primary defense layers, since pen2 mutants are not fully susceptible to the oomycete. Additionally, signaling of defense activation may start from ERECTA (and other partially redundant proteins).
Transcriptomic Profiling of Arabidopsis Infection by P. irregulare
To further characterize the plant molecular response to this oomycete, whole-genome transcriptional profiles of infected wild-type, coi1-1, ein2-5, and sid2-1 plants were obtained using two-color long-oligonucleotide microarrays (see Methods).
Seven transcriptome comparisons were directly made in the two-color chips, four within-genotype comparisons between infected versus noninfected seedlings from each genotype (Col-0, coi1-1, ein2-5, and sid2-1), and three counter genotype comparisons between infected wild-type seedlings and each of the infected hormone-related mutants (Col-0 infected versus coi1-1 infected, Col-0 infected versus ein2-5 infected, and Col-0 infected versus sid2-1 infected). A cluster view of all genes differentially expressed in at least one of the seven comparisons can be found in Supplemental Figure 2 online. The complete list of genes can be found in Supplemental Table 1 online.
The combination of these two types of comparison (within genotype and counter genotype) allowed for the differentiation of two gene classes of interest: (1) JA/ET/SA-dependent genes that are differentially expressed following P. irregulare infection in Col-0 and are also dependent on at least one of the three hormonal pathways being studied (129 genes) and (2) JA/ET/SA-independent genes that are differentially expressed following P. irregulare infection in wild-type plants but appear independent of the three hormonal pathways being studied (1385 genes).
In addition to these two groups, 217 genes appear to be dependent upon at least one of the three hormonal pathways tested but are not differentially expressed in the wild type following P. irregulare infection. Most of these genes, being constitutively over/underexpressed in hormonal mutants, are of limited interest to this pathogen defense study and will not be considered further.
In addition to the statistical methods described elsewhere in this article, validation of microarray data was achieved by RNA gel blot analysis. As shown in Supplemental Figure 3 online, expression of all randomly chosen genes correlated well with the microarray data.
Gene Ontology (GO) analysis of differentially expressed genes using FatiGO (http://fatigo.bioinfo.cipf.es/) further supported the quality of the microarray data. FatiGO analysis of JA/ET/SA-dependent gene data showed that most of these genes belong to defense-related GO categories (Figure 3A). Among them, JA/ET-dependent defenses dominate (Figure 3A). Metabolism of amino acid derivatives, aromatic compounds, and phenylpropanoids were also among the categories containing many of these genes. Interestingly, many genes within this group (JA/ET/SA-dependent genes) belong to two categories related to ABA (response to ABA stimulus and response to water deprivation), suggesting a role for ABA in the plant response to P. irregulare (Figure 3A).
Figure 3.
GO Categorization of Arabidopsis Genes Differentially Expressed after Inoculation with P. irregulare using FatiGo.
(A) 129 JA/ET/SA-dependent genes categorized at biological process level 5.
(B) 296 JA/ET/SA-independent genes categorized at biological process level 3.
(C) 149 JA/ET/SA-independent genes, previously identified as having a cellular physiological or metabolic function (first two categories in [B]), categorized at biological process level 9.
Percentages relate to total number of genes with an ontology at each level. Only categories >5% of total number of genes shown.
Analysis of JA/ET/SA-independent genes showed that most of these genes belong to two major categories: cellular physiological process and metabolism (Figure 3B). Significantly, a deeper analysis of the genes within these two categories (using more detailed GO levels) showed that the majority of the genes belong to metabolic processes involved in the biosynthesis of defensive secondary metabolites, such as lignin, indol-glucosinolates (Trp and glucose derivatives), flavonoids, and nicotinamide (Figure 3C).
JA/ET/SA-Dependent Genes Induced by P. irregulare
The susceptibility of hormone-insensitive mutants (especially coi1) makes this group of genes particularly interesting. A total of 129 genes were identified as being dependent on both P. irregulare infection and at least one of the three hormonal defense pathways analyzed (JA, ET, and SA). Consistent with the lower susceptibility of ein2 and sid2 mutants compared with coi1, hierarchical clustering shows that the contribution of ET and SA to the activation of gene expression is much lower than that of JA (Figure 4). Thus, expression of more than half (70 of 129) of the JA/ET/SA-dependent genes induced in wild-type plants depends on COI1.
Figure 4.
Expression of JA/ET/SA-Dependent Genes.
Hierarchical cluster analysis of 129 genes differentially expressed in wild-type (Col-0) Arabidopsis and at least one of the hormone-deficient mutants (sid2-1, ein2-5, and coi1-1) compared with wild-type plants following P. irregulare infection (P). Genes (rows) and experiments (columns) were clustered with The Institute for Genomic Research (TIGR) multi-experiment viewer using Euclidean distance and complete linkage. Gene subclusters of interest that are discussed in the text are indicated by labeled vertical bars to the right of the image.
The extreme susceptibility of coi1 to P. irregulare indicates that the genes more effectively contributing to the overall resistance are those differentially expressed in the Col-0 versus COI profile (COI1-dependent genes). A complete list of these genes can be found in Supplemental Table 2 online. Among them, clusters 1 and 5 in Figure 4 exemplify genes coregulated by ET and JA (dependent on both ein2 and coi1), and cluster 3 includes genes specifically regulated by JA (COI1-dependent). Cluster 1 includes five PDF genes that, in addition to being among the genes most widely used as markers of JA/ET-dependent defenses, have been shown to confer resistance to fungal pathogens (Gao et al., 2000). Cluster 5 includes genes also involved in defense, such as Hevein-like, b-CHI, GST, and PR1, as well as unknown proteins (putatively new defense-related proteins) and transcription factors (two MYB-related proteins) that might play a role in their regulation. Most of the JA-specific genes (cluster 3) are JA biosynthetic genes (Lipoxygenase, OPR3, and AOC-like), known JA-regulated genes (CORI, VSP1, VSP2, MBP, and TAT), and transcription factors (NAM/NAC and ERF family members).
In addition to the ET/JA cooperation (clusters 1 and 5), examples of other types of hormonal crosstalk can be found within this clustering analysis. For instance, positive and negative interactions between SA and ET are shown in clusters 2, 4, and 7.
Interestingly, although the negative interaction between ET/JA and SA has been widely documented, this antagonism does not appear to be relevant in the case of P. irregulare (only a few examples can be found within clusters 1, 5, and 6), and even positive interactions between SA and JA can be found within cluster 3 where several genes whose expression depends on COI1 are also dependent on SID2. This result, together with the increase in the levels of hormones and the increased susceptibility of the corresponding mutants, further supports at the molecular level the assertion that in this case (P. irregulare–Arabidopsis interaction), SA does not exert a negative effect on JA/ET-dependent defenses but rather cooperates with them. In spite of the generally assumed antagonistic interaction between SA- and JA/ET-dependent defenses, examples of cooperation have also been reported (Berrocal-Lobo et al., 2002; Devadas et al., 2002; O'Donnell et al., 2003; Berrocal-Lobo and Molina, 2004).
Meta-Analysis of JA/ET/SA-Dependent Genes
To gather additional information on the regulation of these genes by other hormones and pathogens, meta-analysis was performed by clustering JA/ET/SA-dependent gene data obtained in this work together with available expression data for these genes following hormonal and pathogen treatments in the GENEVESTIGATOR database (www.genevestigator.ethz.ch/; Zimmermann et al., 2004). Of the nine hormonal treatment experiments available (ABA, 1-aminocyclopropane-1-carboxylic acid, brassinolide, ET, gibberellic acid, indole-3-acetic acid, methyl jasmonate [MeJA], SA, and Zeatin), MeJA clustered most closely to the wild-type response to P. irregulare infection, confirming that JA is the major signal activating responses to this oomycete (Figure 5; see Supplemental Table 3 online). In accordance with this, the COI1-dependent profile is, in general, the opposite to the MeJA profile, with only minor differences likely due to the availability of only one time point of the MeJA treatment, indicating that most (if not all) of the JA-activated responses to P. irregulare are mediated by COI1.
Figure 5.
Meta-Analysis of JA/ET/SA-Dependent Gene Data.
Cluster of JA/ET/SA-dependent genes with available expression data from hormonal and pathogen treatments within the GENEVESTIGATOR database (expression data available for only 107 of the 129 genes; Zimmermann et al., 2004). Genes (rows) and experiments (columns) were clustered with the TIGR multi-experiment viewer using Euclidean distance and complete linkage. Gene subclusters of interest that are discussed in the text are indicated by labeled vertical bars. Overrepresented cis-elements within each gene cluster were identified by TAIR motif analysis, Botany Beowulf Cluster Promomer, and Gibbs motif sampler. Statistical significance (P value from binomial distribution) is shown in parenthesis where possible.
Interestingly, besides JA, the second signal more closely related to the pattern of gene activation by P. irregulare in wild-type plants is ABA. More than one-third (39 of 119) of the genes upregulated by P. irregulare are also upregulated by the hormone, suggesting that it may be an important signal in the activation of defenses against this oomycete. Furthermore, approximately half of these genes (19 of 39 ABA-regulated genes) are also regulated by JA, suggesting that either both signals cooperate or one hormone precedes the other in the activation of this set of genes.
In addition to the ABA and JA signals, P. irregulare's infection profile also clusters with profiles of responses to herbivory (Pieris rapae) and a necrotrophic fungal pathogen (B. cinerea), indicating that the hormone-dependent response to P. irregulare is similar to that of necrotrophic pathogens and chewing insects.
By contrast, profiles from two biotrophic fungal pathogens (Erysiphe orontii and Erysiphe cichoracearum) cluster together, distant from P. irregulare, indicating that plants regulate the expression of most of the genes in this set (JA/ET/SA-dependent induced by P. irregulare) in a different way (in many cases the opposite) depending on the lifestyle of the pathogen. This result fully agrees with the general view that defenses against biotrophs and necrotrophs are essentially antagonistic.
Identification of cis-Regulatory Elements
To gain further insight into the transcriptional regulation of JA/ET/SA-dependent responses to P. irregulare, we analyzed the presence of putative cis-regulatory elements and trans-acting factors responsible for the transcriptional regulation of the genes in each gene cluster by searching for statistically overrepresented sequences in their promoters. Using motif analysis (The Arabidopsis Information Resource [TAIR]), Promomer (http://bbc.botany.utoronto.ca), and motif sampler (Thijs et al., 2002), several statistically significant candidate cis-regulatory elements were found for each cluster (only known boxes are shown; Figure 5). Consistent with the regulation by ET and JA of genes in cluster 1a (see Supplemental Table 3 online), the GCC-box was the predominant element found in this cluster. This element is specifically recognized by members of the ERF family of transcription factors, some of which are well-known defense regulators (Lorenzo et al., 2003; Gutterson and Reuber, 2004). Also consistent with the regulation of clusters 2 and 4 by JA, the G-box and related elements (T/G-box) were the most significantly overrepresented sequences. These elements are regulated by basic domain/leucine zipper or MYC transcription factors, some of which are also involved in defense (Siberil et al., 2001; Boter et al., 2004; Lorenzo et al., 2004; Nickstadt et al., 2004). Interestingly, the ABA response element (ABRE) is overrepresented in clusters 2 and 4 where most of the ABA-regulated genes concentrate. In addition to ABRE, another cis-element related to dehydration was identified, the dehydration-responsive element. The presence of the ABRE and dehydration-responsive elements further supports an important role for ABA and dehydration in the regulation of responses to P. irregulare. Additional boxes with likely functional importance in the regulation of these genes are an H-box– and a P-box–related element (both MYB-related binding sites) and a W-box (Figure 5). Candidate transcription factors regulating these cis-elements can be identified within the genes induced by the oomycete.
JA/ET/SA-Independent Genes Induced by P. irregulare
It is noteworthy that the vast majority of differentially expressed genes (1385) are independent of the three hormones studied. GO analysis showed that the majority of the genes with a GO description are related to the metabolism of defensive compounds (Figure 3B), indicating that many of these inducible defenses may be activated by other, so far unknown, processes/signals.
To further understand the role of these genes in plant defense and to identify new signals/pathways involved in resistance to P. irregulare, meta-analysis was performed by clustering JA/ET/SA-independent gene data obtained in this work together with available expression data from the GENEVESTIGATOR database. A full view of the cluster including all genes can be found in Supplemental Figure 4 online. Figure 6 shows a clustered tree view of the experiments, and Figure 7 shows a full cluster view of the most related profiles to P. irregulare expression patterns. To facilitate the analysis, only genes with a false discovery rate <1% and log ratio >1.5 are shown.
Figure 6.
Meta-Analysis of JA/ET/SA-Independent Gene Data.
Tree view of Arabidopsis–P. irregulare infection (P) experiments with GENEVESTIGATOR (stress response) experiments when cluster analysis included only JA/ET/SA-independent genes. Thus, the four within-genotype comparisons [Col-0(P)-Col-0, ein2(P)-ein2, coi1(P)-coi1, and sid2(P)-sid2] show very similar gene expression patterns and cluster together. Data were clustered with the TIGR multi-experiment viewer using Euclidean distance and complete linkage.
Figure 7.
Meta-Analysis of JA/ET/SA-Independent Gene Data.
Cluster view of 296 JA/ET/SA-independent genes (selected with a false discovery rate <1% and log ratio >1.5) with available expression data from the most closely related experiments within the GENEVESTIGATOR database (Zimmermann et al., 2004). Genes (rows) and experiments (columns) were clustered with the TIGR multi-experiment viewer using the Pearsons uncentered distance and complete linkage. Gene subclusters of interest that are discussed in the text are indicated by labeled vertical bars. Overrepresented (black lettering) and underrepresented (green lettering) cis-elements within each gene cluster were identified by TAIR motif analysis, Botany Beowulf Cluster Promomer, and Gibbs motif sampler. WBS, WRKY binding site; ARE, anthocyanin regulatory element. Statistical significance (z-score significance [WRKY binding site and ARE] and P value from binomial distribution [all others]) is shown in parenthesis.
Consistent with the JA/ET/SA-independent nature of the regulation of these genes, the four within-genotype profiles cluster tightly together, showing that once JA/ET/SA-dependent genes are eliminated, the response of all four genotypes is similar, further supporting the quality of the data (Figures 6 and 7).
Similar to JA/ET/SA-dependent genes, P. irregulare response profiles clustered with the necrotroph B. cinerea and the only other oomycete (P. infestans) profile included within the analysis and show little similarity to the profiles of responses to biotrophs. This result is consistent with our previous conclusions from the genetic analysis and further supports the plant's necrotrophic-like response to this oomycete (Figure 6).
In addition to B. cinerea and P. infestans, several profiles of responses to ABA and abiotic stresses, such as those induced by ozone and osmotic and salt treatments, clustered within this group (Figures 6 and 7). In fact, most of the genes up- or downregulated by P. irregulare infection are also regulated by these stresses (Figure 7). For instance, clusters 1 and 3 (a, b, c, and d) in Figure 7, representing >90% of the genes analyzed, are commonly regulated by most of the stresses in the cluster.
Interestingly, the only hormone profile clustering close to P. irregulare (and B. cinerea and P. infestans) profiles is ABA treatment. In fact, when analyzed in more detail, ABA was found to be the main signal regulating the expression of many of the P. irregulare–responsive genes, particularly those included within clusters 3b and 3d, which represent ∼30% of the genes in this analysis. These data support the previous observation using JA/ET/SA-dependent genes that ABA may play a role in the activation of effective defenses against P. irregulare.
While the vast majority of gene clusters are nonspecific for P. irregulare infection, clusters specifically regulated by this oomycete can be identified (cluster 2, Figure 7; see Supplemental Table 4 online). The specificity of the regulation of these genes by P. irregulare suggests that they may have a defined role in the determination of particular responses to this oomycete.
As with JA/ET/SA-dependent genes, a search for sequences statistically overrepresented within promoters of JA/ET/SA-independent genes identified several candidate cis-regulatory elements (Figure 7). Among them, two are associated with ABA-regulated responses, ABRE and RY (an element recognized by B3 transcription factors, including ABI3, that regulate seed-specific promoters; Vicente-Carbajosa and Carbonero, 2005). In addition, WRKY binding sites seem also to be particularly relevant since they appeared overrepresented in several clusters (2, 3a, and 3c). Candidate transcription factors recognizing these elements have been found within each cluster, and the corresponding KO mutants are being analyzed for susceptibility/resistance to P. irregulare.
ABA Is Required for Plant Defense
To further discriminate if, as suggested above, ABA may be required for overall plant resistance, ABA hormone levels were measured in wild-type and JA/ET/SA/ABA-related mutants after P. irregulare infection, and ABA-deficient mutants were tested for their susceptibility to P. irregulare. As shown in Figure 8A, ABA levels increased rapidly in the wild type and all mutants tested (except in aba2-12) after infection, further supporting a role for this hormone in activation of defenses. Moreover, all three ABA mutants tested, either impaired in ABA biosynthesis (aao3-2 and aba2-12) or insensitive to ABA (abi4), showed an increased susceptibility to P. irregulare compared with the wild-type background, indicating that ABA is a positive signal involved in the activation of effective defenses against this pathogen (Figure 8B).
Figure 8.
Role of ABA and Callose in Resistance to P. irregulare.
(A) Endogenous ABA levels (ng/g fresh weight) in Arabidopsis wild-type (Col-0) and JA/ET/SA/ABA-related mutant plants (coi1-16, ein2-5, sid2-1, and aba2-12) after infection with P. irregulare for 0, 6, and 12 h. Seedlings were grown on Johnson's media plates for 7 d prior to infection. Values are means ± se of three independent replicate experiments.
(B) Susceptibility of Arabidopsis ABA-related mutants (biosynthesis, aba2-12 and aao3-2; insensitive, abi4-1) and a callose-deficient mutant (pmr4) to P. irregulare compared with the wild type (Col-0). Mean disease area (mm2) per infected leaf 24 h after infection. Asterisks indicate significant difference (P < 0.05) from Col-0 using the Student's t test.
(C) B. cinerea (open bars) and A. brassicicola (closed bars) infection of Arabidopsis ABA-related mutants (biosynthesis, aba2-12 and aao3-2; insensitive, abi4-1) compared with the wild type (Col-0). Mean disease area (mm2) per infected leaf either 4 (B. cinerea) or 10 (A. brassicicola) d after infection. Asterisks indicate significant difference (P < 0.05) from Col-0 using the Student's t test.
(D) to (G) Callose deposition in Arabidopsis following infection with P. irregulare.
(D) Localized callose deposition (apposition [app]) around site of pathogen contact. Plant cell wall (cw) and P. irregulare hyphae (hy) are also indicated. Bar = 5 μm.
(E) Normal callose deposition in wild-type (Col-0) Arabidopsis following P. irregulare infection (haustoria-like infection structure [hl]) encompassed the entire plant cell wall (cw). Bar = 30 μm.
(F) Callose deposition in the Arabidopsis ABA mutant aao3-2 following P. irregulare infection (haustoria-like infection structure [hl]) encompassed the entire plant cell wall (cw). Bar = 15 μm.
(G) Spotted callose deposition (sp) within the Arabidopsis callose mutant (pmr4) cell wall (cw) following infection with P. irregulare and production of haustoria-like infection structure (hl). Bar = 30 μm.
Two additional necrotrophs (Alternaria brassicicola and to B. cinerea) have been tested to further understand the breadth of ABA's role in pathogen resistance. As shown in Figure 8C, all three ABA-related mutants tested were more susceptible to A. brassicicola but (surprisingly) more resistant to B. cinerea.
These results demonstrate that although the role of ABA in pathogen resistance is not restricted to P. irregulare, ABA is not a positive signal for plant defense against all necrotrophs. Thus, other properties of the pathogenic infection may be determinant of the role of ABA in each plant–pathogen interaction, rather than the pathogen's assigned lifestyle.
ABA has been previously proposed to play a role in priming of callose biosynthesis after pathogen recognition, which suggests a putative mechanism explaining the role of ABA in defense activation (Ton and Mauch-Mani, 2004). Callose (β-1,3-glucan) is normally associated with cell wall appositions (papillae) and has been suggested to be a fortifying agent deposited rapidly after pathogen recognition to inhibit pathogen penetration of the cell (Aist, 1976; Jacobs et al., 2003; Ton and Mauch-Mani, 2004). To ascertain the role of callose formation in the case of P. irregulare, we studied the deposition of this compound and its effect on resistance after inoculation with the oomycete of wild-type plants and callose-deficient mutants (pmr4).
Aniline blue staining for 1→3-β-d-glucan (callose) suggested that the deposition of this compound occurred in recently infected wild-type cells. This was infrequently only concentrated in appositions or papilla around the area of penetration (Figure 8D; see Supplemental Figure 1F online), more normally being seen to encompass the entire cell wall and possibly the walls of adjoining cells (Figure 8E). By contrast, pmr4 mutants, impaired in callose biosynthesis, deposited callose only in specks around the cell wall (Figure 8G). Moreover, pmr4 mutants showed an enhanced susceptibility to the oomycete compared with wild-type plants (Figure 8B), thus indicating that callose deposition plays a role in defense against P. irregulare.
Taken altogether, these results suggest that ABA may exert its role, at least in part, through priming of callose production (Ton and Mauch-Mani, 2004). Nevertheless, the susceptibility of pmr4 mutants was lower than that of ABA-deficient mutants, suggesting that priming of callose is not the only defense mechanism regulated by ABA. Moreover, aniline blue staining demonstrated that whereas no biosynthesis of callose was observed in pmr4 after infection with P. irregulare (Figure 8G), the level of callose production in ABA-deficient mutants was indistinguishable from that of the wild type (Figure 8F). Therefore, in addition to its role in callose production, ABA-dependent resistance has to be exerted through a callose-independent mechanism.
To further understand this mechanism, microarray profiles of ABA biosynthesis mutants (aba2-12) were compared with those of wild-type plants following the infection of both with P. irregulare. This identified 38 ABA-dependent genes among those regulated by the pathogen in wild-type plants (Figure 9; see Supplemental Table 5 online). Meta-analysis showed that within this ABA-dependent group, two major classes of genes could be identified: ABA-regulated genes and JA-regulated genes.
Figure 9.
Meta-Analysis of ABA-Dependent Gene Data.
Cluster view of 38 aba2-12–dependent genes also induced/repressed by P. irregulare in wild-type (Col-0) plants, with available expression data from JA- and ABA-treated experiments within the GENEVESTIGATOR database (Zimmermann et al., 2004). Genes (rows) were clustered with the TIGR multi-experiment viewer using the Pearsons uncentered distance and complete linkage. Gene subclusters of interest that are discussed in the text are indicated by labeled vertical bars.
These results reinforce the previous conclusion that ABA is an important signal in the activation of plant defenses through the transcriptional reprogramming of the cell. Moreover, the deficiency in the activation of JA-induced genes in aba2-12 indicates that ABA either precedes or cooperates with JA in the activation of this set of defense genes.
To discriminate between these two possibilities, JA hormone levels as well as a JA precursor (12-oxo-phytodienoic acid) were measured in wild-type plants and aba2-12 mutants after P. irregulare infection. As shown in Figure 10, the increase in JA (or its precursor) following infection (12 h after infection) is much lower in aba2-12 mutants than in wild-type plants. This indicates that ABA synthesis is required for JA production and the activation of plant defenses against P. irregulare.
Figure 10.
JA and JA-Precursor Levels in Wild-Type and ABA Mutant Plants.
Endogenous JA and 12-oxo-phytodienoic acid (OPDA) (ng/g fresh weight) levels in Arabidopsis wild-type (Col-0) and ABA mutant (aba2-12) plants following infection with P. irregulare for 0, 6, and 12 h. Seedlings were grown on Johnson's media plates for 7 d prior to infection. Values are means ± se of three independent replicate experiments.
DISCUSSION
The study of Arabidopsis–P. irregulare host–pathogen interaction reported here highlights the importance of broadening our understanding of the interaction of diverse pathogens with this important model species.
Microscopy analysis has shown that P. irregulare infection of Arabidopsis has neither purely typical necrotrophic nor biotrophic characteristics. The infection process starts with the production of appressoria and haustoria-like structures. Compatible host–pathogen interactions show further hyphal ingression to be primarily intracellular, moving more rapidly through the vasculature and invading all tissues. Although no lytic enzymes or toxins have been detected, this progressive invasion ultimately provokes plant cell death.
Analysis of defense-related mutants has shown that Arabidopsis resistance to the establishment and progression of disease by P. irregulare depends initially on avoidance of penetration, mediated by PEN2 and, very likely, other primary defense layers, since pen2 mutants are not fully susceptible to the pathogen. These results are in line with previous reports demonstrating that PEN2 (encoding a glucosyl hydrolase) controls the ingress of a broader range of pathogens, including both biotrophs and necrotrophs, than other penetration proteins (Lipka et al., 2005; Nurnberger and Lipka, 2005).
Recognition and signaling of defense activation may start from ERECTA (and other partially redundant proteins, since erecta mutants are not fully susceptible). Due to its structural similarity to receptor-like kinases, ERECTA has been suggested to recognize a pathogen-associated molecular pattern (Godiard et al., 2003; Llorente et al., 2005). The susceptibility of erecta mutants suggests the likely importance of this receptor-like kinase in the recognition of P. irregulare.
Role of JA, ET, and SA in Defense against P. irregulare
While P. irregulare has biotrophic-like infection structures, the genetic and genomic characterization of defense-related hormone-signaling mutants has shown that the plant's response to P. irregulare is more similar to responses against necrotrophic pathogens. Thus, expression profiles in response to P. irregulare are closely related to those of B. cinerea and different from those of biotrophic pathogens such as Erisyphe spp.
In accordance with this and as shown by the extreme susceptibility of the JA-insensitive coi1 mutants, the main signal activating defenses is JA, the role of which in necrotrophic pathogen defense has been widely documented (Rojo et al., 2003; Reymond et al., 2004; Lorenzo and Solano, 2005). Additionally, although of lesser importance than JA, ET and SA play significant roles in disease resistance against P. irregulare. Cooperation of SA with JA/ET-dependent defenses has been previously reported (Berrocal-Lobo et al., 2002; Devadas et al., 2002; O'Donnell et al., 2003; Berrocal-Lobo and Molina, 2004). However, these results contrast with the generally assumed roles of SA and JA/ET in the activation of two independent and antagonistic defense pathways and highlight the flaws inherent within oversimplified models. Pathogen lifestyles are not often readily attributable to purely biotrophic or necrotrophic classes. Thus, in our view, depending on the particular characteristics of each pathogen, the complex regulatory network involving JA, ET, and SA will establish the required interactions to fine-tune the appropriate defenses. Thus, cooperation or antagonism may be regulated (and has been selected through evolution) to adapt to the specific pathogen.
Transcriptomic analysis and meta-analysis confirmed these conclusions showing that JA regulates more than half the JA/ET/SA-dependent genes, whereas the contribution of ET and SA is more modest.
It is noteworthy, however, that profiles of responses to hemibiotrophic bacteria (both virulent and avirulent strains of Pseudomonas syringae pv maculicola ES4326) are very closely related to the P. irregulare–wild type profile, being particularly evident in the case of genes regulated by JA or ABA (clusters 2a and 4 in Figure 5). Interestingly, this particular P. syringae strain produces a JA analog (coronatine) as a mechanism of pathogenicity (Hendrickson et al., 2000). The activation of JA-dependent responses by coronatine is supposed to counteract SA-dependent defenses facilitating bacterial infection (Cui et al., 2005). Coronatine also promotes stomatal reopening, favoring bacterial entry into the plant (Melotto et al., 2006). Thus, although P. irregulare and P. syringae have different lifestyles and promote very different defense responses, they are coincident in this particular set of genes likely because of the coronatine mimicry of JA. By analogy, the fact that ABA regulates a set of these genes (i.e., cluster 2a) suggests that the coronatine mimicry of JA may not be the only mechanism inhibiting SA-dependent defenses by P. syringae and that ABA might also play a role. In fact, this is consistent with previous reports showing that ABA-deficient Arabidopsis mutants have higher levels of salicylate-induced genes and are more resistant to P. syringae, whereas ABA treatment promotes the opposite behavior (Mohr and Cahill, 2003; Thaler and Bostock, 2004). The SA-dependent inhibition of these genes is evident from their upregulated expression in sid2 mutants infected with P. irregulare (see clusters 2a and 3 in Figure 5). In line with our results, this ABA/SA antagonism could be explained by an indirect effect based on the ABA induction of JA biosynthesis.
JA-, ET-, and SA-Independent Defenses
Transcriptome analysis indicated that the majority of genes differentially expressed following P. irregulare infection were independent of the three defense hormones, JA, ET, and SA, but more related to secondary metabolism (i.e., biosynthesis of defensive compounds), highlighting the importance of these metabolites in defense. The striking overlap of expression patterns between JA/ET/SA-independent genes and profiles from abiotic stress (osmotic, salt, and ozone) may suggest that plant cells use similar mechanisms (the same type of genes) to cope with different stress situations or that some of these stresses are part of the infection process. In fact, among abiotic stresses, ozone exposure, which promotes plant cell death, is the closest profile to P. irregulare, suggesting that many of the common genes in this analysis maybe associated with the cell damage accompanying the infection. In addition, Pythium has been shown to be a vascular pathogen promoting clogging of the vasculature and wilting of the plant (Martin, 1995; Kamoun, 2003). Thus, it should not be surprising that osmotic/salt-related stress profiles and ABA profiles are closely related to P. irregulare infection profiles. A similar hypothesis has been proposed in the case of wounding, where dehydration is a major component of the response (Reymond et al., 2000, 2004). The fact that B. cinerea and P. infestans profiles are also included within this experimental cluster suggests that the abiotic and cell death components of the stress induced after pathogen infection is not exclusive to P. irregulare but rather common to other pathogens.
Role of ABA in Pathogen Defense
The role of ABA in pathogen defense is, so far, poorly understood and even controversial. Whereas several reports have shown an inverse correlation between ABA levels and resistance to pathogens with different lifestyles in several plant species, others have suggested a positive role of this hormone in activation of defense gene expression and pathogen resistance (Mauch-Mani and Mauch, 2005). In the case of bacterial leaf pathogens, ABA plays an important role in the activation of stomatal closure that, as part of the innate immune system, represents a barrier against bacterial infection (Melotto et al., 2006). Thus, ABA-deficient mutants are more susceptible to P. syringae infection. However, it has also been shown that (1) ABA increases susceptibility by counteracting SA-dependent defenses, and (2) ABA-dependent priming of callose biosynthesis promotes enhanced resistance to some pathogens (Ton and Mauch-Mani, 2004). Since callose has been shown to have a detrimental effect on SA-dependent defenses (Nishimura et al., 2003), both issues (1 and 2) support that ABA should be expected to have a negative effect on resistance against biotrophs. However, the effect of ABA on resistance to necrotrophs remains unclear.
Interestingly, meta-analysis of transcriptomic data showed that ABA upregulated approximately one-third of the plant genes induced by P. irregulare, suggesting an important role of this hormone in defense activation. This hypothesis contrasts with previous analysis showing that induction of some ET/JA-regulated defense genes, such as PDF1.2, HEL, and b-CHI, is prevented by ABA (Anderson et al., 2004). However, the genomic view in our work shows that the ABA repression described by Anderson et al. (2004) affects only a reduced group of ET/JA-regulated genes, whereas the major effect of ABA is the opposite, activating many ABA-specific and ABA/JA-related defense genes.
The indication that ABA is a signal required for necrotrophic pathogen resistance was substantiated by the increase in ABA levels after infection and when analysis of ABA-deficient or -insensitive mutants showed them to be more sensitive to P. irregulare and A. brassicicola than wild-type plants. In accordance with these results, several groups have reported mutants (or transgenic plants) with altered resistance to both pathogens and ABA-dependent abiotic stresses, further supporting our hypothesis (Mengiste et al., 2003; Chini et al., 2004). Moreover, treatment with ABA protects Arabidopsis plants against A. brassicicola and Plectosphaerella cucumerina (Ton and Mauch-Mani, 2004), further indicating that ABA is necessary and sufficient to enhance defense responses against several necrotrophic pathogens.
However, ABA is not a positive signal for plant defense against all necrotrophs since it has a negative effect on plant resistance against B. cinerea and Fusarium oxysporum (Audenaert et al., 2002; Anderson et al., 2004; Abuqamar et al., 2006; this work). Thus, other properties of the pathogenic infection, rather than the pathogen lifestyle, have to be determinant of the role of ABA in each plant-pathogen interaction.
The increased susceptibility of pmr4 mutants, impaired in callose biosynthesis, suggests that ABA may exert its role, at least in part, through priming of callose production (Ton and Mauch-Mani, 2004). Nevertheless, ABA-deficient mutants do not show a significant defect in callose production compared with wild-type plants in response to P. irregulare infection and still show higher susceptibility to this oomycete than pmr4 mutants. These results strongly suggest that priming of callose is not the only defense mechanism regulated by ABA. In support of this hypothesis, microarray data and measurements of ABA levels in aba2-12 biosynthetic mutants demonstrated that ABA is required for JA biosynthesis and JA-dependent defense gene expression after infection with P. irregulare. Thus, similar to the proposed wound response mechanism in solanaceous plants (Hildmann et al., 1992; Pena-Cortes et al., 1995; Leon et al., 2001), ABA affects JA biosynthesis, suggesting that it precedes JA in the activation of defenses against this oomycete.
In summary, our results indicate that ABA is a component of the signaling network activating plant defenses necessary for resistance against some (but not all) necrotrophic pathogens. ABA enhances defenses through at least two independent mechanisms: callose priming and regulation of defense gene expression through activation of JA biosynthesis. The differential role of the hormone within different plant–pathogen interactions suggests that ABA levels may be key to the fine-tuning of plant defenses against particular pathogens, a modulation system previously suggested for ET (Pierik et al., 2006).
METHODS
Genetic Backgrounds of Material
Pythium irregulare was identified by microscopy and the genotype confirmed by sequencing of the internal transcribed spacer (ITS) region using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (Kageyama et al., 1997).
With the exception of Ler, all mutants were in a Col-0 background.
Seed Sterilization and Growth
Seeds were surface sterilized with 75% bleach prior to plating on sterile Johnson's growth medium as described by Lorenzo et al. (2003). Homozygous coi1 was selected using 50 μM JA amended Johnson's media. In vitro growth of seedlings was at 21°C with a photoperiod of 16 h light/8 h dark. While seedlings for soil inoculation were transferred at 7 d old, those for transcriptome analysis were transferred to three concentric rings with diameters measuring 55, 69, and 90 mm on Murashige and Skoog media at 4 d old. In vitro growth conditions remained as previously described, but soil-transferred seedlings were moved to a phytochamber with a 12-h-light/12-h-dark photoperiod, 21°C, and 70% RH. To encourage a fuller rosette, ABA mutants were grown in a phytochamber with 8 h light/16 h dark, 21°C, and 70% RH.
P. irregulare Growth Conditions
P. irregulare was cultured on potato dextrose agar (Oxoid CM0139) at 21°C and plugs taken from the growing edge stored underwater at 4°C. Fresh cultures from these stocks were used for inoculation of soil-grown plants and transcriptome seedlings. All plugs for inoculation were taken from the growing edge of a fresh P. irregulare colony. Periodically (approximately every 6 months), fresh 4°C stocks were established using infected leaf tissue to grow an aseptic colony.
Disease Inoculation and Assessment of Soil-Grown Plants
Four weeks after being transferred to soil, plants were inoculated. A single 1.5-mm-diameter agar plug, taken from a fresh-growing colony edge, was placed centrally on the upper surface of each of two lower rosette leaves per plant. This was then covered with 50 to 100 μL of sterilized distilled water. Clear plastic lids covered the inoculated plants to maintain 100% humidity, and infections were allowed to progress for either 24 or 48 h. Leaves were collected and immediately either stained with Trypan blue (Koch and Slusarenko, 1990) or Analine blue (Dietrich et al., 1994). Leaves were mounted in glycerol solutions and Leica DMR and Leica MZFLIII microscopes with fluorescence capabilities used for the examination of infected tissue.
Two diameters, perpendicular to each other, were measured for each disease lesion and a mean obtained. The production of runner hyphae and concomitant establishment of distinct lesions per single inoculum necessitated the conversion of lesion radii to area, thereby enabling total disease areas per leaf to be calculated. Thus, disease area per leaf was calculated as the sum of all disease lesion areas:
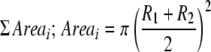 |
All infections of Arabidopsis thaliana mutant genotypes were repeated at least three times with similar results. Between six and nine plants of each genotype were infected per replicate experiment.
Hormone Analysis
Seeds were plated on Johnson's media in a grid formation so that each was 1 cm from the next. Seed sterilization and growth conditions were as described above. After 7 d, plates were inoculated with P. irregulare mycelium taken from the growing edge of a fresh colony. Multiple points of inoculation that were equidistant from each seedling ensured seedlings were infected at the same time. Tissue was collected 0, 6, and 12 h after infection (0, 18, and 24 h after inoculation, respectively), immediately weighed, and frozen until hormone quantification. Pooled whole-plant samples (n = 3) containing 100 mg of tissue were homogenized, derivatized, vapor phase extracted, and analyzed by isobutane chemical-ionization gas chromatography–mass spectrometry as previously described (Schmelz et al., 2004). The initial extraction solution was modified to contain a 62:100:1 ratio of H20:1-propanol:concentrated HCl.
Microarray Hybridization and Analysis
Seedling Inoculation and Tissue Collection
Three days after seedling transfer, MS plates were centrally inoculated with a 4.5-mm P. irregulare–containing agar plug. From the central point, growth of the pathogen took 24 h to reach the first ring of seedlings and 48 h to reach the outer ring. Thus, plants growing on the inner ring had been in contact with P. irregulare for ∼24 h and the middle ring for ∼10 h before the outer ring plants were infected. Whole seedlings from transcriptome plates were harvested and immediately frozen when P. irregulare hyphae infected the outer ring of plants.
Four Arabidopsis genotypes were included in the initial transcriptome analysis, comprising the wild type (Col-0) and three mutants (coi1-1, ein2-5, and sid2-1). The four genotypes were represented in each independent plate, divided into four equal sectors each containing 18 seedlings equally divided between the three rings. Nine replicate experiments were done on different days, each with an identical, but noninoculated, control plate. Additional transcriptome analyses of aba2-12 mutant plants were done identically to the initial analysis although with only one of the four plate sectors occupied.
RNA Gel Blot Analysis
RNA was extracted from the nine individual experiments and then pooled, in equal proportions, into three groups of three. RNAwiz was used to extract the RNA from frozen tissue according to the manufacturer's instructions (Ambion). Following purification with RNeasy (Qiagen), RNAs were subjected to electrophoresis on 1.5% formaldehyde/agarose gels and blotted to Hybond N+ membranes (Amersham). All probes were labeled with 50 μCi of [α-32P]dCTP. Blots were exposed for 12 to 24 h on a PhosphorImager screen (Molecular Dynamics).
RNA Quantification and Quality Determination
RNA was quantified with a NanoDrop ND-100 spectrophotometer (NanoDrop Technologies). RNA quality was assessed with a 2100 Bioanalyzer from Agilent Technologies.
RNA Amplification and Labeling
RNA was amplified with the MessageAmp aRNA amplification kit from Ambion following the instruction manual. To allow later labeling with Cy fluorophores, aminoallyl UTP (Ambion) was added to the mix of the T7 RNA polymerase-driven aRNA amplification reaction. The amount and quality of aRNA obtained was assessed as before. The aminoallyl-labeled aRNA (10 μg) was incubated in 1 M Na2CO3 with 8 nmol of dye monofunctional NHS ester (Cy3/Cy5) RPN 5661 (Amersham Biosciences) at room temperature in the dark for 1 h. Then, 35 μL of 0.1 M sodium acetate, pH 5.2, was added and incubated for a further 5 min in the dark. The Cy-labeled aRNA was purified with the Megaclear kit from Ambion and measured with the Nanodrop ND-100 spectrophotometer.
Hybridization
Three biological replicates were independently hybridized for each transcriptomic comparison.
Microarray slides were composed of synthetic 70-mer oligonucleotides from the Operon Arabidopsis Genome Oligo Set Version 1.0 (Qiagen) spotted on aminosilane-coated slides (Telechem) by the University of Arizona. Slides were rehydrated and UV cross-linked according to the supplier's website (http://ag.arizona.edu/microarray/methods.html). The slides were then washed twice for 2 min in 0,1% SDS and in ethanol for 30 s. Arrays were drained with a 2000-rpm spin for 2 min. Slides were prehybridized in 6× SSC, 0.5% SDS (w/v), and 1% BSA (w/v) at 42°C for 1 h, followed by five rinses with milliQ water. Excess water was drained with a 2000-rpm spin for 2 min.
For the hybridization, equal amounts of dye of each aRNA labeled with either Cy3 or Cy5, ranging from 200 to 300 pmol, were mixed with 20 ug of poly(A) and 20 μg of yeast tRNA (Sigma-Aldrich) in a volume of 9 μL. To this volume, 1 μL of RNA fragmentation buffer was added (RNA fragmentation reagents; Ambion) and after 15 min at 70°C, 1 μL of stop solution. Formamide, 20× SSC, 50× Denhardt's, and 20% SDS were added to a final concentration of 50% formamide, 6× SSC, 5× Denhardt's, and 0.5% SDS. This mix was boiled for 3 min at 95°C and then added to the prehybridized slide. Hybridization took place overnight at 37°C in a hybridization chamber. Arrays were then washed for 5 min at 37°C in 0.5× SSC and 0.1% SDS, twice for 5 min at room temperature (21°C) with 0.5× SSC and 0.1% SDS, three times with 0.5× SSC at room temperature, and 5 min with 0.1× SSC. The slides were then drained with a 2000-rpm spin for 2 min. The slides were stored in darkness until they were scanned.
The scanning was done with a GenePix 400B scanner (Molecular Devices) at 10-μm resolution. The images were quantified with GenePix Pro 5.1.
Images from Cy3 and Cy5 channels were equilibrated and captured with a GenePix 4000B (Axon) and spots quantified using GenPix Pro 5.1 software (Axon). The data from each scanned slide were first scaled and normalized using the Lowess method, before being log-transformed. The mean of the three replicate log-ratio intensities and their standard deviations were generated.
Microarray Analysis
The expression data were normalized and statistically analyzed using the LIMMA package (Smyth and Speed, 2003). LIMMA is part of Bioconductor, an R language project (Ihaka and Gentleman, 1996). First, the data set was filtered based on the spot quality. A strategy of adaptive background correction was used that avoids exaggerated variability of log ratios for low-intensity spots. For local background correction, the normexp method in LIMMA to adjust the local median background was used. The resulting log ratios were print-tip loess normalized for each array (Smyth and Speed, 2003). To have similar distribution across arrays and to achieve consistency among arrays, log-ratio values were scaled using as scale estimator the median absolute value (Smyth and Speed, 2003).
Assessment of Differentially Expressed Genes
Linear model methods were used for determining differentially expressed genes. Each probe was tested for changes in expression over replicates using an empirical Bayes moderated t statistic (Smyth, 2004). To control the false discovery rate, P values were corrected using the method of Benjamani and Hochberg (1995) and Berrocal-Lobo and Molina (2004). The expected false discovery rate was controlled to be <5% (or 1% where specified). Genes were considered to be differentially expressed if the corrected P values were <0.05 (or <0.01 where specified). In addition, only genes with a fold change more than twofold were considered for further analysis. After removing repetition, 1731 differentially expressed genes (in at least one experiment) were merged. Data from Affymetrix microarrays were downloaded from the meta-analysis program of the GENEVESTIGATOR database. Previously, and just for clustering purposes, a scaling factor was applied to gain consistency between the two platforms. To assure a normal distribution of mean 0 and sd equal to 1, a z-score transformation was performed for each gene. First, the mean and sd were calculated for each row. Subsequently, each value (of the row) was transformed by subtracting the mean and dividing by the sd. All hierarchical clusters were calculated and drawn using the TIGR MeV (Saeed et al., 2003) software provided by TIGR.
Three biological replicates were made for each transcriptomic comparison.
Accession Number
Microarray data were deposited with MIAMEXPRESS under accession number E-TABM-258
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Infection of Arabidopsis Tissue by P. irregulare.
Supplemental Figure 2. Cluster of All Genes Significantly Induced or Repressed following Infection.
Supplemental Figure 3. Verification of Microarray Data.
Supplemental Figure 4. Meta-Analysis of JA/ET/SA-Independent Gene Data.
Supplemental Table 1. All Arabidopsis Genes Differentially Expressed in at Least One of the Seven Comparisons following P. irregulare Infection.
Supplemental Table 2. JA/ET/SA- and P. irregulare–Dependent Arabidopsis Genes Included in Figure 4.
Supplemental Table 3. Gene Data Included in Meta-Analysis of JA/ET/SA-Dependent Genes in Figure 5.
Supplemental Table 4. JA/ET/SA-Independent Genes Specifically Upregulated in Arabidopsis by P. irregulare Infection in Gene Cluster 2 in Figure 7.
Supplemental Table 5. ABA- and P. irregulare–Dependent Arabidopsis Genes Included in Figure 9.
Supplementary Material
Acknowledgments
We thank C. Castresana, J. Paz-Ares, and members of the R.S. lab for critical reading of the manuscript and stimulating discussions. We also thank P.L. Rodriguez-Egea who kindly provided seeds of ABA-related mutants. pmr4 mutants were kindly supplied by M. Nishimura and S. Sommerville. edr1 and pen mutants were kindly supplied by R. Innes and P. Schulze-Lefert, respectively. This work was financed by grants to R.S. from the Spanish Ministerio de Ciencia y Tecnología (BIO2001-0567, BIO2004-02502, and GEN2003-20218-C02-02) and from the Comunidad de Madrid (07G/0048/2000, 07B/0044/2002, and GR/SAL/0674/2004) and by European Union Grant HPRN-CT-2000-00093 to J.-J.S.-S. B.A.T.A. was supported by postdoctoral fellowships from the European Union (CRISP project HPRN-CT-2000-00093) and by the Spanish Ministerio e Educación y Ciencia (GEN2003-20218-C02-02).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Roberto Solano (rsolano@cnb.uam.es).
Online version contains Web-only data.
References
- Abuqamar, S., Chen, X., Dhawan, R., Bluhm, B., Salmeron, J., Lam, S., Dietrich, R.A., and Mengiste, T. (2006). Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J. 48 28–44. [DOI] [PubMed] [Google Scholar]
- Aist, J. (1976). Papillae and related wound plugs of plant cells. Annu. Rev. Phytopathol. 14 145–163. [Google Scholar]
- Anderson, J.P., Badruzsaufari, E., Schenk, P.M., Manners, J.M., Desmond, O.J., Ehlert, C., Maclean, D.J., Ebert, P.R., and Kazan, K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad, F.F., Qiu, J.L., Youngs, H., Ehrhardt, D., Zimmerli, L., Kalde, M., Wanner, G., Peck, S.C., Edwards, H., Ramonell, K., Somerville, C.R., and Thordal-Christensen, H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15 5118–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert, K., De Meyer, G.B., and Hofte, M.M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamani, Y., and Hochberg, Y. (1995). Controlling the false discovery rate. J. R. Stat. Soc. [Ser A] 57 289–300. [Google Scholar]
- Berrocal-Lobo, M., and Molina, A. (2004). Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 17 763–770. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29 23–32. [DOI] [PubMed] [Google Scholar]
- Boter, M., Ruiz-Rivero, O., Abdeen, A., and Prat, S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg, E. (1950). Uber die bildung von toxinen in der gattung Pythium und ihre wirkung auf die pflanzen. Nachrichtenbl. Deut. Pflanzenschutzdienst. 2 69–70. [Google Scholar]
- Chini, A., Grant, J.J., Seki, M., Shinozaki, K., and Loake, G.J. (2004). Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 38 810–822. [DOI] [PubMed] [Google Scholar]
- Clark, C.A., and Lorbeer, J.W. (1976). Comparative histopathology of Botrytis squamosa and B. cinerea on onion leaves. Phytopathology 66 1279–1289. [Google Scholar]
- Coego, A., Ramirez, V., Gil, M.J., Flors, V., Mauch-Mani, B., and Vera, P. (2005). An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell 17 2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, N.C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J.L., Huckelhoven, R., Stein, M., Freialdenhoven, A., Somerville, S.C., and Schulze-Lefert, P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425 973–977. [DOI] [PubMed] [Google Scholar]
- Cui, J., Bahrami, A.K., Pringle, E.G., Hernandez-Guzman, G., Bender, C.L., Pierce, N.E., and Ausubel, F.M. (2005). Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl. Acad. Sci. USA 102 1791–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon, J.W. (1979). Cellulose decomposition by Pythium and its relevance to substrate-groups of fungi. Trans. Br. Mycol. Soc. 72 469–477. [Google Scholar]
- Devadas, S.K., Enyedi, A., and Raina, R. (2002). The Arabidopsis hrl1 mutation reveals novel overlapping roles for salicylic acid, jasmonic acid and ethylene signalling in cell death and defence against pathogens. Plant J. 30 467–480. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77 565–577. [DOI] [PubMed] [Google Scholar]
- Falk, A., Feys, B.J., Frost, L.N., Jones, J.D., Daniels, M.J., and Parker, J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr, D.F., Bills, G.F., Chamuris, G.P., and Rossman, A.Y. (1989). Fungi on Plants and Plant Products in the United States. (St. Paul, MN: American Phytopathological Society Press).
- Frye, C.A., and Innes, R.W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, A.G., Hakimi, S.M., Mittanck, C.A., Wu, Y., Woerner, B.M., Stark, D.M., Shah, D.M., Liang, J., and Rommens, C.M. (2000). Fungal pathogen protection in potato by expression of a plant defensin peptide. Nat. Biotechnol. 18 1307–1310. [DOI] [PubMed] [Google Scholar]
- Geraats, B.P., Bakker, P.A., and van Loon, L.C. (2002). Ethylene insensitivity impairs resistance to soilborne pathogens in tobacco and Arabidopsis thaliana. Mol. Plant Microbe Interact. 15 1078–1085. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43 205–227. [DOI] [PubMed] [Google Scholar]
- Godiard, L., Sauviac, L., Torii, K.U., Grenon, O., Mangin, B., Grimsley, N.H., and Marco, Y. (2003). ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 36 353–365. [DOI] [PubMed] [Google Scholar]
- Govrin, E.M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10 751–757. [DOI] [PubMed] [Google Scholar]
- Gutterson, N., and Reuber, T.L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7 465–471. [DOI] [PubMed] [Google Scholar]
- Hendrickson, E.L., Guevera, P., Penaloza-Vazquez, A., Shao, J., Bender, C., and Ausubel, F.M. (2000). Virulence of the phytopathogen Pseudomonas syringae pv. maculicola is rpoN dependent. J. Bacteriol. 182 3498–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildmann, T., Ebneth, M., Pena-Cortes, H., Sanchez-Serrano, J.J., Willmitzer, L., and Prat, S. (1992). General roles of abscisic and jasmonic acids in gene activation as a result of mechanical wounding. Plant Cell 9 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka, R., and Gentleman, R. (1996). R: A language for data analysis and graphics. J. Comput. Graph. Statist. 5 299–314. [Google Scholar]
- Jacobs, A.K., Lipka, V., Burton, R.A., Panstruga, R., Strizhov, N., Schulze-Lefert, P., and Fincher, G.B. (2003). An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15 2503–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama, K., Ohyama, A., and Hyakumachi, M. (1997). Detection of Pythium ultimum using polymerase chain reaction with species-specific primers. Plant Dis. 81 1155–1160. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. (2003). Molecular genetics of pathogenic oomycetes. Eukaryot. Cell 2 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S., Huitema, E., and Vleeshouwers, V.G. (1999). Resistance to oomycetes: A general role for the hypersensitive response? Trends Plant Sci. 4 196–200. [DOI] [PubMed] [Google Scholar]
- Knoester, M., van Loon, L.C., van den Heuvel, J., Hennig, J., Bol, J.F., and Linthorst, H.J.M. (1998). Ethylene-insensitive tobacco lacks nonhost resistance against soil-borne fungi. Proc. Natl. Acad. Sci. USA 95 1933–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, E., and Slusarenko, A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, B.N., and Brooks, D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5 325–331. [DOI] [PubMed] [Google Scholar]
- Leon, J., Rojo, E., and Sanchez-Serrano, J.J. (2001). Wound signaling in plants. J. Exp. Bot. 52 1–9. [DOI] [PubMed] [Google Scholar]
- Li, J., Brader, G., and Palva, E.T. (2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka, V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183. [DOI] [PubMed] [Google Scholar]
- Llorente, F., Alonso-Blanco, C., Sanchez-Rodriguez, C., Jorda, L., and Molina, A. (2005). ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 43 165–180. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O., Chico, J.M., Sanchez-Serrano, J.J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O., Piqueras, R., Sanchez-Serrano, J.J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O., and Solano, R. (2005). Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 8 532–540. [DOI] [PubMed] [Google Scholar]
- Martin, F.N. (1995). Pythium. In Pathogenesis and Host Specificity in Plant Diseases: Histopathological, Biochemical, Genetic and Molecular Bases, Vol. 2, U.S. Singh, K. Kohmoto, and R.P. Singh, eds (Oxford, UK: Elsevier), pp. 17–36.
- Mauch-Mani, B., and Mauch, F. (2005). The role of abscisic acid in plant-pathogen interactions. Curr. Opin. Plant Biol. 8 409–414. [DOI] [PubMed] [Google Scholar]
- Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126 969–980. [DOI] [PubMed] [Google Scholar]
- Mengiste, T., Chen, X., Salmeron, J., and Dietrich, R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15 2551–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, H.M., and Sauve, R.J. (1975). Etiology and control of Pythium stem rot of geranium. Plant Dis. Reptr. 59 122–126. [Google Scholar]
- Mohr, P., and Cahill, D. (2003). Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct. Plant Biol. 30 461–469. [DOI] [PubMed] [Google Scholar]
- Nickstadt, A., Thomma, B.P.H.J., Feussner, I., Kangasjärvi, J., Zeier, J., Löffler, C., Scheel, D., and Berger, S. (2004). The jasmonate-insensitive mutant jin1 shows increased resistance to biotrophic as well as necrotrophic pathogens. Mol. Plant Pathol. 5 425–434. [DOI] [PubMed] [Google Scholar]
- Nimchuk, Z., Eulgem, T., Holt, B.F., and Dangl, J.L. (2003). Recognition and response in the plant immune system. Annu. Rev. Genet. 37 579–609. [DOI] [PubMed] [Google Scholar]
- Nishimura, M.T., Stein, M., Hou, B.H., Vogel, J.P., Edwards, H., and Somerville, S.C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301 969–972. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T., and Lipka, V. (2005). Non-host resistance in plants: New insights into an old phenomenon. Mol. Plant Pathol. 6 335–345. [DOI] [PubMed] [Google Scholar]
- O'Donnell, P.J., Schmelz, E., Block, A., Miersch, O., Wasternack, C., Jones, J.B., and Klee, H.J. (2003). Multiple hormones act sequentially to mediate a susceptible tomato pathogen defense response. Plant Physiol. 133 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Cortes, H., Fisahn, J., and Willmitzer, L. (1995). Signals involved in wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proc. Natl. Acad. Sci. USA 92 4106–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik, R., Tholen, D., Poorter, H., Visser, E.J., and Voesenek, L.A. (2006). The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci. 11 176–183. [DOI] [PubMed] [Google Scholar]
- Reymond, P., Bodenhausen, N., Van Poecke, R.M., Krishnamurthy, V., Dicke, M., and Farmer, E.E. (2004). A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16 3132–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P., Weber, H., Damond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, E., Solano, R., and Sanchez-Serrano, J.J. (2003). Interactions between signaling compounds involved in plant defense. J. Plant Growth Regul. 22 82–98. [Google Scholar]
- Saeed, A.I., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34 374–378. [DOI] [PubMed] [Google Scholar]
- Schmelz, E.A., Engelberth, J., Tumlinson, J.H., Block, A., and Alborn, H.T. (2004). The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 39 790–808. [DOI] [PubMed] [Google Scholar]
- Siberil, Y., Doireau, P., and Gantet, P. (2001). Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur. J. Biochem. 268 5655–5666. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3. [DOI] [PubMed]
- Smyth, G.K., and Speed, T. (2003). Normalization of cDNA microarray data. Methods 31 265–273. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Yuen, G.Y., and Lehman, C.C. (1998). Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 15 747–754. [DOI] [PubMed] [Google Scholar]
- Thaler, J.S., and Bostock, R.M. (2004). Interactions between abscisic-acid-mediated responses and plant resistance to pathogens and insects. Ecology 85 48–58. [Google Scholar]
- Thijs, G., Marchal, K., Lescot, M., Rombauts, S., De Moor, B., Rouze, P., and Moreau, Y. (2002). A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. 9 447–464. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P., Penninckx, I.A., Broekaert, W.F., and Cammue, B.P. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13 63–68. [DOI] [PubMed] [Google Scholar]
- Ton, J., and Mauch-Mani, B. (2004). Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38 119–130. [DOI] [PubMed] [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14(Suppl): S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Plaats-Niterink, A.J. (1981). Monograph of the genus Pythium. Stud. Mycol. 21 1–242. [Google Scholar]
- van Loon, L.C., Geraats, B.P., and Linthorst, H.J. (2006). Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11 184–191. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304 66–74. [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa, J., and Carbonero, P. (2005). Seed maturation: Developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 49 645–651. [DOI] [PubMed] [Google Scholar]
- Vijayan, P., Shockey, J., Levesque, C.A., Cook, R.J., and Browse, J. (1998). A role for jasmonate in pathogen defense of Arabidopsis. Proc. Natl. Acad. Sci. USA 95 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- When this work was in press, Hernández-Blanco et al. (2007) published results concerning the role of ABA in disease resistance that support results described in this article.
- Hernández-Blanco, C., et al. (2007). Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19 890–903. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.












