Abstract
E3 ubiquitin ligases (E3s) target proteins for degradation by the 26S proteasome. In SKP1/CDC53/F-box protein–type E3s, substrate specificity is conferred by the interchangeable F-box protein subunit. The vast majority of the 694 F-box proteins encoded by the Arabidopsis thaliana genome remain to be understood. We characterize the VIER F-BOX PROTEINE (VFB; German for FOUR F-BOX PROTEINS) genes from Arabidopsis that belong to subfamily C of the Arabidopsis F-box protein superfamily. This subfamily also includes the F-box proteins TRANSPORT INHIBITOR RESPONSE1 (TIR1)/AUXIN SIGNALING F-BOX (AFB) proteins and EIN3 BINDING F-BOX proteins, which regulate auxin and ethylene responses, respectively. We show that loss of VFB function causes delayed plant growth and reduced lateral root formation. We find that the expression of a number of auxin-responsive genes and the activity of DR5:β-glucuronidase, a reporter for auxin reponse, are reduced in the vfb mutants. This finding correlates with an increase in the abundance of an AUXIN/INDOLE-3-ACETIC ACID repressor. However, we also find that auxin responses are not affected in the vfb mutants and that a representative VFB family member, VFB2, cannot functionally complement the tir1-1 mutant. We therefore exclude the possibility that VFBs are functional orthologs of TIR1/AFB proteins.
INTRODUCTION
Eukaryotes use the ubiquitin proteasome system for the targeted proteolysis of regulatory proteins, such as cell cycle regulators and transcription factors (Hershko and Ciechanover, 1998; Schwechheimer and Schwager, 2004). Protein degradation by the 26S proteasome is generally preceded by the polyubiquitylation of degradation targets. Polyubiquitylation is the result of the consecutive activities of an E1 ubiquitin activating enzyme (E1), an E2 ubiquitin conjugating enzyme (E2), and an E3 ubiquitin ligase (E3) (Hershko and Ciechanover, 1998). E3s confer specificity to the system in that they specifically recognize the degradation targets and bring them into the vicinity of E2 enzymes (Deshaies, 1999; Cardozo and Pagano, 2004; Schwechheimer and Villalobos, 2004). Different types of E3 ligases have been described in eukaryotes. The so-called Cullin-RING E3s include the SCF (for SKP1/CDC53/F-Box protein), VCB (for Von-Hippel-Lindau, Elongin C, Elongin B), BTB/POZ (for Bric-a-Brac, Tramtrack and Broad Complex/Pox Virus, and Zinc-Finger), and DCX (for Damaged DNA Binding Protein1, Cullin 4A, X-box) complexes (Deshaies, 1999; Kamura et al., 1999; Higa et al., 2003; Xu et al., 2003; Cardozo and Pagano, 2004; Wertz et al., 2004). Common to these Cullin-RING E3s are the RING BOX1 (RBX1) and Cullin subunits (Cullin1 in SCF, Cullin2 or Cullin 5 in VCB, Cullin 3 in BTB/POZ, and Cullin 4 in DCX). For each Cullin-RING E3, substrate specificity is brought about by the association of the Cullin-RBX1 core complex with specific substrate receptor proteins, such as the F-box proteins of the SCF complexes.
Cullins are modified by the ubiquitin-related protein NEDD8 (neddylation), and neddylation is essential for E3 function (Hori et al., 1999). Although some experiments indicate that neddylation may control the assembly of E3 complexes or the association of E3 ligases with E2 ubiquitin conjugating emzymes, it needs to be said that the precise role of neddylation for E3 function is at present not understood (Kawakami et al., 2001; Bornstein et al., 2006; Wu et al., 2006). NEDD8 is deconjugated from Cullins (deneddylation) through the activity of COP9 signalosome (CSN) subunit 5 (CSN5) (Schwechheimer et al., 2001; Cope et al., 2002; Chiba and Tanaka, 2004; Dohmann et al., 2005). Multiple studies from a range of organisms have shown that CSN interacts with Cullin-RING E3s and that proper E3 function is dependent on CSN and more specifically on CSN5-mediated deneddylation (Lyapina et al., 2001; Schwechheimer et al., 2001; Schwechheimer, 2004). Some recent studies suggest that deneddylation is required for E3 subunit stability or E3 complex assembly (Hetfeld et al., 2005; Wee et al., 2005; Wu et al., 2005; Bornstein et al., 2006). Furthermore, it has been proposed that CSN is part of a proteasome-related complex (Peng et al., 2001, 2003). Regardless of its precise molecular mode of action, it can be said that CSN acts as a global regulator of Cullin-RING E3 activities and that loss of CSN function in higher eukaryotes causes severe growth defects (Freilich et al., 1999; Lykke-Andersen et al., 2003; Schwechheimer, 2004; Tomoda et al., 2004; Dohmann et al., 2005). Since it is expected that hundreds of Cullin-RING E3 functions are impaired in csn mutants, their severe phenotype may be the result of the combined defects caused by the impaired function of individual E3 activities.
F-box proteins are the interchangeable substrate receptor subunits of SCF-type E3s (Deshaies, 1999; Skowyra et al., 1999; Cardozo and Pagano, 2004). The F-box domain, which is in most cases located at the protein's N terminus, mediates the interactions with the adaptor protein of the Suppressor of Kinetochore1 (SKP1) protein family (Cardozo and Pagano, 2004). In addition, most F-box proteins contain recognizable protein–protein interaction domains, such as Leu-rich repeats (LRRs), WD40 repeats, or kelch repeats that are thought to recognize degradation substrates (Gagne et al., 2002; Jin et al., 2004). The Arabidopsis thaliana genome is predicted to encode 694 F-box proteins, suggesting that protein degradation via SCF complexes is a predominant control mechanism in plants (Gagne et al., 2002). To date, the biological function of only ∼20 F-box proteins has been elucidated, and these F-box proteins have been implicated in a wide range of physiological processes, such as cell cycle control, circadian rhythms, floral and development, as well as different phytohormone responses (Schwechheimer and Villalobos, 2004).
The C subfamily is the best characterized family of Arabidopsis F-box proteins. Most members of this protein family, which comprises 140 family members, contain LRRs as protein–protein interaction domains (Gagne et al., 2002). An unexpectedly large number of C subfamily F-box proteins regulate plant hormone responses: TRANSPORT INHIBITOR RESPONSE1 (TIR1) and its three homologs, designated AUXIN SIGNALING F-BOX PROTEIN1 (AFB1) to AFB3, mediate the degradation of AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) repressors in response to auxin (Ruegger et al., 1998; Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005); CORONATINE INSENSITIVE1 (COI1) controls the degradation of an as yet unknown protein in response to jasmonates (Xie et al., 1998); and EIN3 BINDING F-BOX1 (EBF1) and EBF2 regulate ethylene signaling via degradation of the transcription factor ETHYLENE INSENSITIVE3 (EIN3) (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). Interestingly, this subfamily also includes SKP2;1 and SKP2;2, the putative Arabidopsis orthologs of the mammalian SKP2 protein, which promotes the degradation of E2F transcription factors during the cell cycle (Marti et al., 1999; del Pozo et al., 2002a). Hence, hormone response regulatory F-box proteins may be derived from the evolutionarily conserved cell cycle regulatory SKP2s (Calderon-Villalobos et al., 2007).
The vast majority of the 140 F-box proteins that belong to the C subfamily remain to be characterized. We were interested in an F-box protein family, which we designated VIER F-BOX PROTEINE (VFB; German for FOUR F-BOX PROTEINS). We found that plants defective in all four VFB genes are delayed in general growth and are defective in lateral root formation. Transcript profiling experiments revealed that a number of auxin-responsive protein encoding genes are repressed in the vfb mutants, a finding that correlates with the apparent accumulation of the unstable AUXIN RESISTANT3 (AXR3)/IAA17 repressor. However, our further experiments also indicate that auxin-dependent growth responses are normal and that AXR3/IAA17 turnover is unaltered in vfb mutants. Since VFB2, a representative VFB family member, cannot functionally replace TIR1, we hypothesize that the VFB and TIR1/ABF F-box proteins act in distinct signaling pathways.
RESULTS
Characterization of the VFB F-Box Protein Family
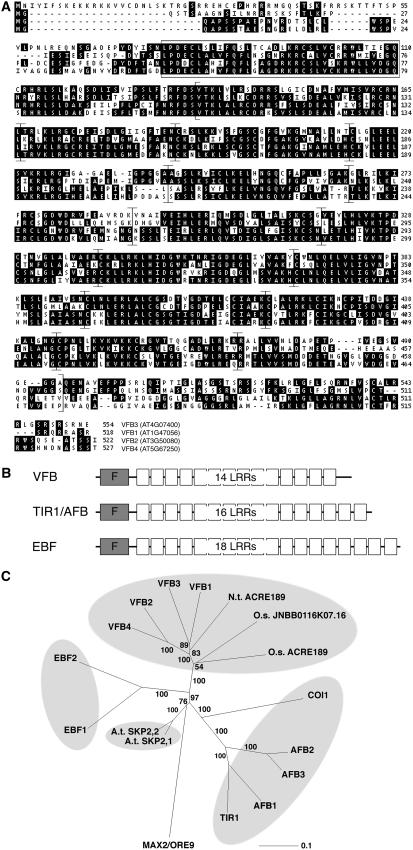
The four F-box proteins AT1G47056, AT3G50080, AT4G07400, and AT5G67250 share significant sequence similarity (56 to 69% identity and 63 to 80% similarity), and together they form a distinct F-box protein family within the C subfamily of the Arabidopsis F-box protein superfamily (Figures 1A and 1C) (Gagne et al., 2002). We named this protein family VFB and designated its individual members VFB1 (AT1G47056), VFB2 (AT3G50080), VFB3 (AT4G07400), and VFB4 (AT5G67250). The F-box protein C subfamily also includes the cell cycle regulatory SKP2 proteins, the TIR1/AFB auxin receptor F-box proteins, and the ethylene response F-box proteins EBF1 and EBF2 (Figure 1C) (Xie et al., 1998; del Pozo et al., 2002a; Guo and Ecker, 2003; Potuschak et al., 2003; Dharmasiri et al., 2005a; Kepinski and Leyser, 2005). Common features of these F-box proteins are the presence of an N-terminal F-box domain and a series of LRRs, which are predicted to interact with the respective degradation substrates (Figure 1B). We failed to identify nonplant proteins with a clear homology to the Arabidopsis VFB proteins. However, we identified a putative tobacco (Nicotiana tabacum) VFB homolog designated AVR9/CF-9 RAPIDLY ELICITED PROTEIN 189 (ACRE189), whose expression is induced following flagellin treatment, and two putative rice (Oryza sativa) VFB homologs ACRE189 and JNBb0002J11.1 (Figure 1C) (Navarro et al., 2004). Since we were unable to assign these proteins unequivocally to a specific Arabidopsis VFB protein, we assume that the Arabidopsis VFBs and their putative orthologs from tobacco and rice have a common ancestor.
Figure 1.
The VFB Proteins Are LRR-Containing F-Box Proteins of the C Subfamily of the Arabidopsis F-Box Protein Superfamily.
(A) Pretty box representation of a Clustal alignment of the VFB proteins VFB1 (AT1G47056), VFB2 (AT3G50080), VFB3 (AT4G07400), and VFB4 (AT5G67250). The F-box domain is boxed, and the individual LRRs are flanked by brackets and were identified as described by Gagne et al. (2004).
(B) Schematic view of the VFB F-box proteins in comparison with the auxin receptor TIR1/AFB F-box proteins and the ethylene response EBF F-box proteins. F, F-box domain.
(C) Unrooted phylogenetic tree of the Arabidopsis VFB proteins, two apparent homologs from rice annotated as ACRE189 and JNBb0002J11.1, one apparent homolog from tobacco annotated as ACRE189, and several related and previously characterized F-box proteins from Arabidopsis. At, Arabidopsis; O.s., O. sativa; N.t., N. tabacum. Bootstrap values are indicated at the nodes. Please refer to the text for other abbreviations. Accession numbers are listed in Methods.
VFB2 Interacts with the SCF Complex Adaptor Protein ASK2 and Localizes to the Cytoplasm
F-box proteins interact via their F-box domain with the evolutionarily conserved SKP1 adaptor protein, which in turn binds to the Cullin1 subunit of SCF complexes (Deshaies, 1999; Zheng et al., 2002; Risseeuw et al., 2003). To show that the VFB proteins have retained the ability to bind SKP1 proteins, we used VFB2 as a representative family member to examine its interaction with ARABIDOPSIS SKP1-2 (ASK2), a predominant member of the Arabidopsis SKP1 protein family, using the yeast two-hybrid system (Risseeuw et al., 2003; Zhao et al., 2003). Our analysis revealed an interaction between VFB2 and ASK2 but not between VFB2 and the two other SCF complex components RBX1 and CUL1 (Figures 2A and 2B) (del Pozo and Estelle, 1999; Schwechheimer et al., 2002; Risseeuw et al., 2003). As expected, a VFB2 deletion variant lacking the F-box domain (VFB2ΔF-box) failed to interact with ASK2 in the yeast assay (Figures 2A and 2B). Due to the high sequence conservation between the individual VFB family members, we assume that these findings can be extended to VFB1, VFB3, and VFB4 and suggest that these proteins act as F-box protein subunits of SCFVFB complexes.
Figure 2.
Interactions and Localization of the VFB F-Box Proteins.
(A) A yeast two-hybrid interaction study between VFB2 and the SCF complex components ASK2, RBX1, and CUL1 reveals an F-box domain–dependent interaction between VFB2 and ASK2. ASK2, RBX1, and CUL1 full-length proteins were expressed from pJG4-5. VFB2 and VFB2ΔF-box, a VFB2 variant lacking the protein's N terminus, including the F-box domain, were expressed from pEG202. Blue staining corresponds to LacZ reporter activity and indicates protein–protein interaction.
(B) Protein expression of the respective fusion proteins in yeast was confirmed using fusion protein–specific antibodies, anti-B42 (left panel) and anti-LexA (right panels). Fusion proteins of the expected molecular mass were identified, namely pJG4-6 B42 activation domain (12 kD), CUL1 (98 kD), RBX1 (25 kD), ASK2 (30 kD), pEG202 LexA DNA binding domain (26 kD), pEG202VFB2 (84 kD), and pEG202VFB2ΔF-box (76 kD).
(C) Confocal microscopy images of Arabidopsis protoplasts transiently transformed with fluorescent protein fusions of VFB proteins. From left to right: confocal image of GFP or YFP fusion protein, confocal image of Hoechst 33342–stained nuclei, overlay of GFP/YFP and Hoechst 33342 images, and Nomarski image.
To examine the subcellular localization of the VFB proteins, we generated 35S:VFB2:GREEN FLUORESCENT PROTEIN (GFP), 35S:VFB3:GFP, 35S:VFB4:GFP, and 35S:YFP:VFB2 for the 35S promoter of Cauliflower mosaic virus-driven expression of VFB2:GFP, VFB3:GFP, VFB4:GFP, and YELLOW FLUORESCENT PROTEIN (YFP):VFB2 fusion proteins, respectively. While we were unable to detect a VFB3:GFP-derived fluorescent signal with 35S:VFB3:GFP, we found that 35S:VFB2:GFP, 35S:VFB4:GFP, and 35S:YFP:VFB2 localize to the cytoplasm when expressed in a transient manner in Arabidopsis protoplasts (Figure 2C). Similarly, a VFB2 promoter (2-kb fragment)–driven VFB2:GFP fusion protein also localized predominantly to the cytoplasm (data not shown). Taken together, this suggests that the VFB F-box proteins act in the cytoplasm. However, these results have to be treated with appropriate caution since we could not confirm the functionality of the fusion constructs (e.g., by mutant complementation).
VFB Gene Expression
To examine the expression pattern of the four VFB genes, we fused 1-kb VFB gene promoter fragments to the β-glucuronidase (GUS) reporter. At least 10 transgenic lines were generated and analyzed for each construct. While the promoter:GUS constructs for VFB1, VFB2, and VFB4 showed strong GUS staining in many plant tissues, including the vascular system, leaves, flowers, and roots, the VFB3:GUS construct stained only weakly in any tissue examined (Figures 3B to 3G). VFB2 and VFB4 also seem to be expressed in the cotyledons of bent cotyledon stage embryos but not at earlier stages of embryo development (Figure 3A; data not shown). In most cases, the tissue- and organ-specific expression pattern of all four genes, as detected by promoter:GUS fusions, reflects well the organ-specific expression detected by RT-PCR (Figure 3H). Most importantly, this analysis also confirmed the weak expression of the VFB3 gene as indicated by the GUS experiments.
Figure 3.
Expression Analysis Predicts Distinct but Also Overlapping Expression Patterns for the VFB Genes.
(A) to (G) Transgenic plants expressing promoter:GUS fusions for each of the four VFB genes were examined in bent cotyledon stage embryos (A), flowers (B), leaves (C), roots (D), and root tips (E). Noteworthy is the strong expression of VFB1 in stomata (F) and of VFB4 in lateral roots (G). Representative staining patterns are shown.
(H) RT-PCR analysis of VFB gene expression in flowers (F), stems (ST), roots (R), leaves (L), and seedlings (SD). 35 PCR cycles were needed to obtain significant gene amplification. ACTIN served as a normalization control of the experiment.
Since our analysis had shown that the two most closely related VFB proteins, VFB2 and VFB4, have very similar expression patterns, we also analyzed publicly available microarray data for VFB expression (Zimmermann et al., 2004). In a set of 1021 experiments with the Affymetrix ATH1 GeneChip, we found VFB2 and VFB4 to be coexpressed in the vast majority of tissue types and experimental conditions (see Supplemental Figure 1A online). Also in agreement with our own expression analysis, VFB3 is only very weakly expressed in microarray analyses (see Supplemental Figure 1B and 1C online). The fourth member of the VFB gene family, VFB1, is not represented on the Affymetrix ATH1 GeneChip. Taken together, our expression analyses show that VFB1, VFB2, and VFB4 are actively transcribed members of the VFB gene family, that VFB2 and VFB4 expression is coregulated, and that VFB3 is the least strongly expressed member of this gene family.
vfb1 vfb2 vfb3 Triple Mutants Show No Apparent Growth Defects
To study the role of the VFB genes in plant growth and development, we characterized homozygous T-DNA insertion mutants available for VFB1 (SALK_128933; vfb1-1), VFB2 (SALK_047600; vfb2-1), VFB3 (SALK_054809; vfb3-1), and VFB4 (GABI_414F09; vfb4-1) (Figure 4A). The absence of the respective full-length transcripts was confirmed in vfb1-1, vfb2-1, and vfb3-1 mutant alleles, which carry insertions in the only exon of the respective gene (Figure 4B). By contrast,VFB4 expression appeared to be unaltered in the vfb4-1 allele, which carries a T-DNA insertion 26 bp upstream of the ATG start codon. We thus concluded that vfb4-1 is not a VFB4 knockdown or knockout allele (Figure 4B). We failed to detect apparent phenotypes in the vfb1-1, vfb2-1, and vfb3-1 mutants and, assuming that the genes may have redundant functions, went on to generate double and triple mutant combinations (Figures 4C and 4D). However, neither of these mutants revealed any apparent defects compared with the wild type (e.g., when challenged with any of the major phytohormones or with salt stress) (Figure 4D; see Supplemental Figure 2 online).
Figure 4.
vfb1, vfb2, and vfb3 Mutants Do Not Have Apparent Phenotypes.
(A) The VFB genes are composed of a single exon (black bar). The T-DNA insertion lines characterized in this study are indicated.
(B) RT-PCR (30 cycles) of VFB gene expression in the vfb1-1, vfb2-1, and vfb3-1 mutants (top three panels) reveals that the full-length transcripts are missing from the respective mutants. The T-DNA insertion upstream of the VFB4 open reading frame in GABI_414F09 does not affect VFB4 expression (fourth panel from the top).
(C) RT-PCR (30 cycles) for VFB gene expression in the vfb1-1 vfb2-1 vfb3-1 triple mutants.
(D) Single and triple vfb mutants (5-week-old plants) do not show apparent phenotypes when grown under standard growth conditions.
Suppression of VFB4 by RNA Interference Causes Growth Defects
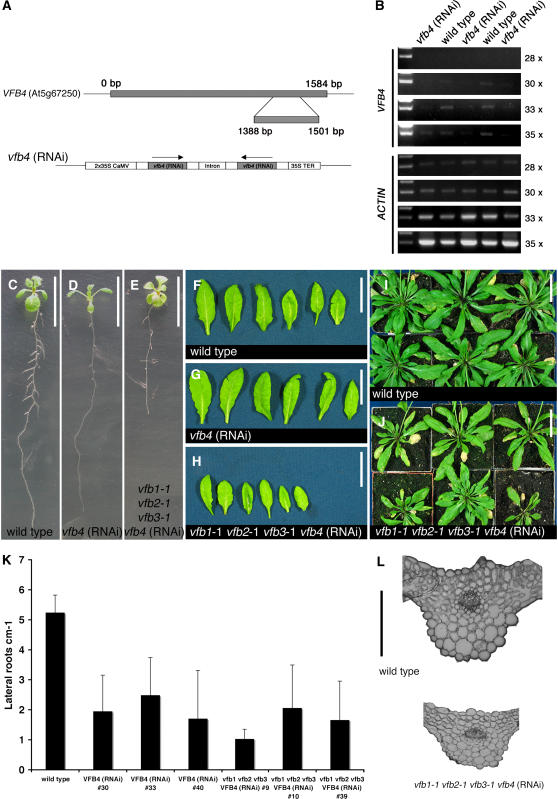
Since the mutant combinations generated so far did not include a mutant of the VFB4 gene, and since we knew that VFB4 is a strongly expressed family member, we used RNA interference (RNAi) to suppress VFB4 function. To this end, a 114-bp fragment of the VFB4 3′-end, the region with the highest sequence divergence between all four VFB genes, was selected to generate the vfb4 (RNAi) construct (Figure 5A). The vfb4 (RNAi) transgene was introduced into the wild type and the vfb1-1 vfb2-1 vfb3-1 triple mutant, and 20 transgenic lines were generated in each background. By RT-PCR we showed a reduction of VFB4 gene expression in vfb4 (RNAi) plants (Figure 5B). The analysis of the progeny of the vfb4 (RNAi) and vfb1-1 vfb2-1 vfb3-1 vfb4 (RNAi) transgenic plants revealed defects in lateral root formation in the seedlings and, in the case of the vfb1-1 vfb2-1 vfb3-1 vfb4 (RNAi) plants, also a delay in root elongation and a general delay in plant growth as illustrated, for example, by their reduced rosette size (∼50%) during early stages of vegetative growth (Figures 5C to 5K). Cross sections of leaves indicate that the growth defects observed in the vfb1-1 vfb2-1 vfb3-1 vfb4 (RNAi) mutants are due to a reduction in cell size and not due to a reduction of cell cycle activity (Figure 5L). Taken together, we showed that the VFB4 gene together with the other three family members is required for normal plant growth.
Figure 5.
RNAi Suppression of VFB4 Reveals a Role for the VFB Gene Family in Development.
(A) Schematic representation of the VFB4 gene (top panel) and the vfb4 (RNAi) construct (bottom panel) that was selected for suppression of VFB4 expression. Magnified is the 114-bp region between base pair 1388 and base pair 1501 of the VFB4 open reading frame, which shows the highest sequence divergence between all four VFB family members.
(B) RT-PCR (28, 30, 33, and 35 cycles) of three vfb4 (RNAi) lines and two wild-type controls indicates that VFB4 gene expression is reduced in selected vfb4 (RNAi) lines.
(C) to (E) A comparative analysis of 10-d-old light-grown seedlings reveals differences in root growth and lateral root formation between the wild type (C), the wild type transformed with the vfb4 (RNAi) construct (D), and vfb1-1 vfb2-1 vfb3-1 mutants containing vfb4 (RNAi) (E). Bars = 5 mm.
(F) to (J) vfb4 RNAi suppression in the vfb1-1 vfb2-1 vfb3-1 triple mutant ([H] and [J]) but not in the wild type ([G] and [I]) causes growth delay as indicated by the plants' reduced rosette size. Shown are representative leaves ([F] to [H]) and rosettes ([I] and [J]) of 3-week-old plants. Bars = 5 mm in (F) to (H) and 2.5 cm in (I) and (J).
(L) Average and sd of lateral roots formed per centimeter in 10-d-old Arabidopsis seedlings. n ≥ 20.
(M) A cross section from rosette leaves indicates that growth differences in the vfb1-1 vfb2-1 vfb3-1 vfb4 (RNAi) mutants are due to reduced cell size and not due to reduced cell number. Bar = 3 mm.
Gene Expression Profiling of the vfb Mutants
To gain insight into the molecular mechanisms that underlie the phenotype observed in the vfb1-1 vfb2-1 vfb3-1 vfb4-1 (RNAi) mutants (vfb mutants), we performed gene expression profiling using Affymetrix ATH1 GeneChips of 10-d-old vfb mutant and wild-type seedlings. Using Bioconductor analysis tools, we identified 213 genes that are induced and 220 genes that are repressed in the vfb mutants compared with the wild type (odds of differential expression ≥95% and fold induction ≥1.5; see Supplemental Tables 1 and 2 online). While the gene products of the misregulated genes have a wide variety of biochemical functions, we noted that two gene families are overrepresented within the group of repressed genes, namely, auxin response and signaling genes (21 genes) and genes encoding cell wall metabolic enzymes (24 genes) (Table 1). The group of repressed auxin signaling genes includes the AUX/IAA repressors IAA1 (AXR5), IAA2, IAA3 (SHORT HYPOCOTYL2 [SHY2]), IAA5, IAA6, IAA19 (MASSUGU2 [MSG2]), and IAA29 as well as 10 SMALL AUXIN UP RNAs (SAURs) (Gil et al., 1994; Leyser et al., 1996; Tian and Reed, 1999; Marchant et al., 2002; Tatematsu et al., 2004; Yang et al., 2004; Overvoorde et al., 2005). The AUX/IAA genes encode unstable repressors that control the expression of auxin-induced genes in the absence of auxin (Reed, 2001; Tiwari et al., 2004; Overvoorde et al., 2005). The expression of all AUX/IAAs and at least six of the 10 SAUR genes identified in our study are auxin induced according to publicly available expression data, while the expression of cell wall metabolic enzyme genes is not under auxin control (Table 1) (Reed, 2001; Tiwari et al., 2004; Zimmermann et al., 2004). Since this finding suggested that the VFB genes act upstream of auxin response, we introduced the DR5:GUS reporter into the vfb mutant background (Ulmasov et al., 1997b). The DR5:GUS construct contains multiple binding sites for AUXIN RESPONSE FACTOR transcriptional activators whose activities are repressed by the AUX/IAA repressors (Ulmasov et al., 1997a). In agreement with previous reports, we detected DR5:GUS staining in the leaf margins, at sites of lateral root initiation, and in the root tips of wild-type seedlings. Interestingly, the staining at these sites was reduced in the vfb mutant background, suggesting—in agreement with our microaray data—that auxin-responsive gene expression is impaired in the vfb mutant background (Figures 6A to 6F).
Table 1.
Auxin Signaling and Response Genes and Cell Wall Metabolic Genes Are Repressed in vfb1-1 vfb2-1 vfb3-1 vfb4 (RNAi) Mutants (vfb Mutants) Compared with the Wild Type
| Fold Repression | Description | Protein Function | Adjusted P Value | Raw Data (Min to Max) Wild Type | Raw Data (Min to Max) Wild Type |
|---|---|---|---|---|---|
| Auxin signaling | |||||
| 10.43 | AT4G32280* | Auxin-responsive AUX/IAA family protein 29 (lAA29) | 2.62e-06 | 94.76 (54.55 to 130.2) | 8.632 (7.184 to 10.23) |
| 3.27 | AT3G15540* | Auxin-responsive AUX/IAA family protein 19 (lAA19) | 3.08e-05 | 109.2 (103.3 to 115.9) | 33.6 (27.75 to 37.8) |
| 2.97 | AT1G04240* | Auxin-responsive AUX/IAA family protein 3 (IAA3) | 2.1e-05 | 198.6 (171.9 to 243.3) | 66.2 (63.8 to 68.37) |
| 2.44 | AT3G23030* | Auxin-responsive AUX/IAA family protein 2 (IAA2) | 3.01e-05 | 1109 (1015 to 1264) | 454 (414.2 to 490.5) |
| 2.25 | AT1G52830* | Auxin-responsive AUX/IAA family protein 6 (IAA6) | 8.35e-05 | 21.39 (20.44 to 21.96) | 9.579 (7.938 to 10.45) |
| 2.23 | AT4G14560* | Auxin-responsive AUX/IAA family protein 1 (IAA1) | 1.67e-05 | 73.78 (62.95 to 83.6) | 32.93 (31.08 to 34.74) |
| 2.08 | AT1G15580* | Auxin-responsive AUX/IAA family protein 5 (IAA5) | 2.43e-05 | 16.79 (13.13 to 20.2) | 7.967 (7.582 to 8.677) |
| Auxin response | |||||
| 4.35 | AT5G18060* | Auxin-responsive protein (SAUR) | 1.59213e-04 | 150.7 (122.9 to 205.8) | 33.61 (32 to 34.59) |
| 3.26 | AT4G34770* | Auxin-responsive protein (SAUR) | 1.65e-06 | 65.48 (56.96 to 77.05) | 20.46 (15.62 to 26.81) |
| 2.71 | AT3G03840 | Auxin-responsive protein (SAUR) | 5.75141e-04 | 37.43 (32.37 to 43.88) | 14.02 (10.18 to 17.41) |
| 2.63 | AT1G29430* | Auxin-responsive protein (SAUR) | 4.65316e-04 | 212.1 (156.8 to 292) | 78.34 (70.26 to 88.68) |
| 2.36 | AT1G29500* | Auxin-responsive protein (SAUR) | 5.23e-05 | 165.1 (146.7 to 201) | 69.71 (59.58 to 76.61) |
| 2.32 | AT1G29440* | Auxin-responsive protein (SAUR) | 2.71175e-04 | 91.05 (63.75 to 118.8) | 38.24 (34.9 to 43.61) |
| 2.25 | AT2G21200 | Auxin-responsive protein (SAUR) | 1.59213e-04 | 16.64 (15.88 to 17.89) | 7.394 (7.133 to 7.703) |
| 2.06 | AT4G34760* | Auxin-responsive protein (SAUR) | 2.3e-05 | 335.1 (310.8 to 352.2) | 162.4 (151 to 170.3) |
| 1.75 | AT5G20820 | Auxin-responsive protein (SAUR) | 2.55523e-04 | 16.87 (14.48 to 21) | 9.542 (8.918 to 10.47) |
| 1.67 | AT5G54510 | Auxin-responsive protein (GH3) | 2.02999e-04 | 476.4 (421.8 to 506) | 288.8 (216.6 to 341.1) |
| 1.63 | AT4G36110 | Auxin-responsive protein (SAUR) | 1.11912e-04 | 16.84 (15.15 to 19.09) | 10.5 (7.632 to 12.42) |
| Auxin transport | |||||
| 1.90 | AT2G01420 | Auxin transport protein (PIN4) | 4.77277e-04 | 304.1 (292.4 to 318.9) | 160.4 (151 to 176.4) |
| 1.85 | AT1G70940 | Auxin transport protein (PIN3) | 3.90964e-04 | 269 (247.5 to 305.5) | 145.4 (129.1 to 164.9) |
| 1.80 | AT1G23080 | Auxin transport protein (PIN7) | 8.19e-05 | 189.8 (169.3 to 202.2) | 107.5 (87.13 to 141.9) |
| Cell wall metabolism | |||||
| 2.83 | AT1G03870 | Fasciclin-like arabinogalactan protein (FLA9) | 4.92e-05 | 1494 (1433 to 1615) | 536 (415.9 to 641.6) |
| 2.59 | AT1G65310 | Xyloglucan:xyloglycosyl transferase | 9.63e-06 | 59.11 (52.88 to 67.11) | 22.85 (19.78 to 26.07) |
| 2.57 | AT5G65390 | Arabinogalactan protein (AGP7) | 1.96e-05 | 280.2 (253.2 to 305.8) | 110 (94.63 to 131.5) |
| 2.52 | AT1G02640 | Glycosyl hydrolase family 3 protein | 7.34e-06 | 269.6 (250.4 to 289.7) | 109.4 (78.45 to 127.9) |
| 2.44 | AT5G48900 | Pectate lyase family protein | 4.83028e-04 | 281.9 (244.2 to 301.3) | 115,4 (102,7 to 125,4) |
| 2.32 | AT4G25260 | Invertase/pectin methylesterase inhibitor family protein | 6.57e-05 | 146.7 (133.9 to 168.7) | 63.11 (56.31 to 67.26) |
| 2.32 | AT2G06850 | Xyloglucan:xyloglycosyl transferase | 1.06e-05 | 4020 (3945 to 4058) | 1743 (1588 to 1986) |
| 2.11 | AT1G11545 | Xyloglucan:xyloglycosyl transferase | 9.27e-06 | 137.3 (116 to 157.7) | 64.62 (61.89 to 69.93) |
| 2.03 | AT3G62660 | Glycosyl transferase family 8 protein | 1.48006e-04 | 183.1 (170.3 to 198.1) | 92.14 (71.46 to 119.9) |
| 2.00 | AT3G54920 | Pectate lyase | 1.51e-05 | 458.3 (446.6 to 471.7) | 231.1 (197.5 to 269.7) |
| 1.98 | AT5G10430 | Arabinogalactan protein (AGP4) | 3.45e-05 | 68.06 (60.68 to 76.66) | 35.09 (27.94 to 45.71) |
| 1.89 | AT1G24170 | Glycosyl transferase family 8 protein | 1.92478e-04 | 380.6 (354.5 to 393.9) | 203.1 (164.4 to 224.9) |
| 1.80 | AT5G62340 | Invertase/pectin methylesterase inhibitor family protein | 6.2108e-04 | 24.8 (21.47 to 27.55) | 14.03 (10.31 to 17.62) |
| 1.72 | AT3G49220 | Pectinesterase family protein | 2.0067e-04 | 535.5 (501.7 to 596) | 310.7 (296.8 to 329.1) |
| 1.68 | AT1G62770 | Invertase/pectin methylesterase inhibitor family protein | 5.33077e-04 | 30.39 (26.87 to 32.93) | 18.23 (15.45 to 21.33) |
| 1.67 | AT1G10550 | Xyloglucan:xyloglycosyl transferase | 2.21e-05 | 19.01 (15.29 to 25.81) | 11.08 (9.709 to 12.15) |
| 1.67 | AT1G80280 | Hydrolase, α/β-fold family protein | 1.89723e-04 | 107.1 (72.49 to 139.9) | 62.3 (52.53 to 69.17) |
| 1.64 | AT2G28080 | Glycosyltransferase family protein | 5.22086e-04 | 189.7 (166.9 to 217.3) | 115.2 (114.8 to 115.5) |
| 1.61 | AT3G62110 | Glycoside hydrolase family 28 protein | 2.17015e-04 | 237.3 (217.5 to 261.2) | 148.4 (125.3 to 168.3) |
| 1.59 | AT1G52070 | Jacalin lectin family protein | 4.88818e-04 | 35.68 (32.97 to 39.08) | 22.74 (17.85 to 28.14) |
| 1.58 | AT3G23730 | Xyloglucan:xyloglycosyl transferase | 5.87e-06 | 143.1 (118.4 to 167.3) | 90.02 (82.75 to 102.7) |
| 1.57 | AT3G55500 | Expansin (EXP16) | 3.47e-05 | 21.73 (16.9 to 29.44) | 13.68 (10.95 to 16.72) |
| 1.56 | AT4G00080 | Invertase/pectin methylesterase inhibitor family protein | 6.16975e-04 | 21.52 (18.43 to 23.52) | 13.84 (11.83 to 15.88) |
| 1.52 | AT4G12730 | Fasciclin-like arabinogalactan protein (FLA2) | 1.7138e-04 | 279.3 (242.8 to 339.3) | 181.2 (176.1 to 185.9) |
The asterisk indicates genes that are induced by auxin.
Figure 6.
Auxin Responses Are Impaired in the vfb1-1 vfb2-1 vfb3-1 vfb4 (RNAi) Mutants.
(A) to (F) The activity of the DR5:GUS was compared between the wild type and vfb1-1 vfb2-1 vfb3-1 vfb4 (RNAi) mutants. A general reduction of GUS reporter activity was observed in leaf margins, lateral root initials, and root tips in the vfb1-1 vfb2-1 vfb3-1 vfb4 (RNAi) mutants ([B], [D], and [F]) compared with the wild type ([A], [C], and [E]). Bars = 1 mm.
(G) Auxin sensitivity assay of vfb mutants compared with the wild type and the auxin-insensitive tir1-1 mutant. Average and sd of absolute root elongation after 5 d on dexamethasone-containing medium without auxin: wild type, 2.47 ± 0.75 cm; tir1-1, 2.54 ± 0.48 cm; vfb2-1, 2.45 ± 0.28 cm; vfb1-1 vfb2-1 vfb3-1, 2.02 ± 0.77 cm; vfb1-1 vfb2-1 vfb3-1 vfb4 (RNAi), 1.73 ± 0.33 cm; vfb1-1 vfb2-1 vfb3-1 vfb4 (RNAi), 1.54 ± 0.69 cm. n = 10.
(H) The AUX/IAA fusion protein AXR3:GUS accumulates in the vfb mutants compared with the wild type. Bar = 0.5 mm.
(I) AXR3:GUS turnover is unaltered in the vfb mutants compared with the wild type. Two representative roots are shown for each time point.
(J) Average and sd of lateral root initiation of 1-cm root segments on 2,4-D–containing media as described by Ruegger et al. (1998). n = 10.
(K) Average and sd of relative root elongation in response to auxin in the wild type, tir1-1, and tir1-1 transformed with a dexamethasone-inducible TIR1-MYC transgene. Absolute root elongation after 5 d on dexamethasone-containing medium without auxin: wild type, 3.29 ± 0.27 cm; tir1-1, 2.76 ± 0.78 cm; tir1-1 (TIR1-MYC), 2.08 ± 0.79 cm. n = 10.
(L) Average and sd of root elongation in response to auxin in the wild type, tir1-1, and tir1-1 containing a dexamethasone-inducible VFB2 transgene pTA:VFB2. n = 10. Absolute root elongation after 3 d on dexamethasone-containing medium without auxin: wild type, 1.29 ± 0.23 cm; tir1-1, 1.23 ± 0.18 cm; tir1-1 (pTA:VFB2#1), 1.19 ± 0.32 cm; tir1-1 (pTA:VFB2#22), 1.26 ± 0.23 cm.
(M) VFB2 expression as quantified by RT-PCR (27 PCR cycles) in the wild type and tir1-1 containing a dexamethasone (Dex)–inducible VFB2 transgene 4 h after dexamethasone (30 μM) induction.
Auxin response via proteasomal degradation of AUX/IAA proteins is controlled by the auxin receptor F-box protein TIR1 and its three AFB homologs (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005). Similar to the vfb mutants, tir1 mutants have fewer lateral roots than the wild type and fail to efficiently induce the transcription of auxin-induced genes (Ruegger et al., 1998). In addition, tir1 mutants are auxin insensitive when grown on auxin-containing medium. To examine whether the vfb mutants also share this feature of the tir1 mutant phenotype, we tested auxin sensitivity in vfb mutants. When we treated seedling roots with concentrations of the synthetic auxin 2,4-D that lead to growth arrest in the wild type but not in the tir1-1 mutant allele, we detected no differences between the wild type and the vfb mutants (Figure 6G). Next, we analyzed the stability of the AUX/IAA repressor AXR3/IAA17 by introducing the HS:AXR3NT-GUS construct, which allows expression of an AXR3:GUS fusion protein under control of a heat shock promoter, into the vfb mutants (Gray et al., 2001). Although we detected stronger AXR3:GUS signals in the vfb mutants compared with the wild type, we found that the turnover of the fusion protein is not significantly altered in the mutants when we followed protein abundance after heat shock treatment (Figures 6H and 6I). In line with these experiments, which suggest that auxin responses are not significantly impaired in vfb mutants, we also found that auxin induces lateral root formation in the vfb mutants to the same extent as it does in the wild type (Figure 6J). Finally, we examined whether VFB2 as a representative VFB family member can functionally replace TIR1 by introducing a dexamethasone-inducible construct for the expression of VFB2 into the tir1-1 mutant background. A similar experimental approach had previously been used to show that the inducible expression of wild-type TIR1 rescues the tir1-1 mutant phenotype (Figure 6K) (Ruegger et al., 1998; Gray et al., 1999). Since our experiments indicate that VFB2 expression cannot rescue the auxin-resistant phenotype of tir1-1 following induction with dexamethasone, we conclude that VFB2 cannot functionally replace TIR1 (Figures 6L and 6M). In summary, these experiments suggest that the loss of VFB function results in the morphological and molecular defects that are similar to the defects in mutants with loss of auxin signaling but that the VFB proteins, or at least VFB2, do not have the same function as TIR1. Thus, the effect of the VFB proteins on the auxin signaling pathway may be indirect.
A Partial Overlap between the Gene Expression Defects of vfb Mutants and Mutants of the CSN
CSN interacts with SCF-type E3 ubiquitin ligases and is required for proper E3 activity (Schwechheimer et al., 2001). Arabidopsis csn mutants have severe phenotypes that ultimately lead to growth arrest at the early seedling stage (Kwok et al., 1996; Schwechheimer, 2004; Dohmann et al., 2005). Since Arabidopsis may contain close to 1000 different Cullin-RING E3s, the phenotype of the csn mutants may be the combination of the growth defects conferred by malfunction of the individual E3-dependent pathways. Since CSN may be viewed as a global regulator of E3 activities, genes that are misexpressed in vfb mutants as identified above are expected to represent, at least in part, a fraction of the genes that are misexpressed in csn mutants (Schwechheimer et al., 2001, 2002; Dohmann et al., 2005). We tested this hypothesis by comparing the gene expression changes between vfb mutants and csn4 mutants that are deficient in CSN function since they carry a CSN-destabilizing mutation in CSN4. In the comparison of 10-d-old vfb mutants and 7-d-old csn mutants and their respective wild-type controls, we identified 61 genes that are induced and 120 genes that are repressed in both mutants when applying the same criteria as described above for the vfb gene expression profiling experiment (see Supplemental Tables 3 and 4 online). The list of genes whose expression is misregulated in both mutants includes many as yet uncharacterized signaling components and transcriptional regulators. Most importantly, however, this list includes all seven AUX/IAA genes, eight SAUR genes, and 12 genes encoding cell wall metabolic enzymes. Therefore, these gene families may represent common downstream targets of SCFVFB complexes and CSN.
DISCUSSION
The Four VFB Proteins Belong to the C Subfamily of the Arabidopsis F-Box Protein Superfamily
In this study, we characterized the four VFB F-box proteins from Arabidopsis, which form a distinct protein family within the C subfamily of the Arabidopsis F-box protein superfamily. The VFB proteins and the other bona fide members of this subfamily contain LRR domains for degradation substrate interaction. While the human genome encodes only 21 LRR-containing F-box proteins, plants have undergone a dramatic expansion of this F-box protein family (Jin et al., 2004). The most exhaustive survey of this protein family predicts 202 LRR-containing F-box proteins (Gagne et al., 2002). Plants seemingly have recruited F-box proteins of the C subfamily to regulate phytohormone responses, axillary branching, and senescence (Xie et al., 1998; Woo et al., 2001; Stirnberg et al., 2002; Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004; Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005). In addition, our study suggests that the VFB proteins are plant growth regulators with an important role in controlling lateral root formation and plant growth in general. Interestingly, the LRR F-box protein family also includes the predicted Arabidopsis orthologs of the human cell cycle regulatory F-box protein SKP2. SKP2 is required for the degradation of the cell cycle transcriptional regulator E2F and the cell cycle inhibitors p27Kip1 and p21Cip1 in humans (Marti et al., 1999; Sutterluty et al., 1999; Ungermannova et al., 2005). While the Arabidopsis genome does not contain apparent orthologs of p27Kip1 and p21Cip1, the E2F regulators and their degradation via SKP2 proteins are conserved between humans and plants (del Pozo et al., 2002a). Therefore, the At SKP2 F-box proteins may represent an evolutionary link to the human F-box protein family. The close evolutionary relationship between the cell cycle regulatory At SKP2 proteins and the other family members of the C subfamily invites the hypothesis that these plant-specific F-box proteins also may control plant growth by affecting cell cycle activity. While this hypothesis may potentially hold true for some members of this F-box protein family, it is very unlikely for others. The proteins that are targeted for degradation by the VFBs remain to be identified, and revealing their identity using molecular, genetic, or biochemical approaches will be pivotal to gain an insight into how and as part of which pathway these F-box proteins regulate plant development.
Auxin Responses and the VFB F-Box Proteins
Mutants deficient in VFB function have a reduced number of lateral roots. Lateral root formation requires a functional auxin pathway, and numerous auxin signaling mutants with reduced lateral root formation have been identified. These include mutants that express stabilized versions of the AUX/IAA regulators IAA3/SHY2 and IAA19/MSG2 and loss-of-function mutants of the proteins required for AUX/IAA degradation, including the TIR1/AFB F-box proteins, the proteins required for the neddylation of the SCF Cullin subunit, and the proteins required for Cullin deneddylation, notably CSN (Leyser et al., 1993; Tian and Reed, 1999; Gray et al., 2001; Schwechheimer et al., 2001; del Pozo et al., 2002b; Fukaki et al., 2002; Tian et al., 2003; Zenser et al., 2003; Tatematsu et al., 2004; Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005). In the absence of auxin, AUX/IAA proteins repress their own expression and the expression of other auxin-induced genes. In the presence of auxin, AUX/IAA repressors are degraded, allowing for the expression of auxin-induced genes. Due to the stabilization of the AUX/IAA proteins in the various mutants mentioned above, auxin-induced gene expression is impaired (Tian et al., 2002; Overvoorde et al., 2005). We observed a similar repression of AUX/IAA and auxin-induced gene expression in the vfb mutants. We therefore tested the hypothesis that VFB F-box proteins are also required for auxin reponses. However, our physiological experiments indicate that auxin-induced growth responses and the turnover of the AUX/IAA protein AXR3/IAA17 are normal in vfb mutants. Our further experiments indicate that the VFB proteins cannot functionally replace TIR1 and that the VFB proteins when fused to GFP or YFP localize to the cytoplasm. Identical protein fusions for TIR1 had shown that these proteins accumulate in the nucleus (Dharmasiri et al., 2005a, 2005b). Although their different subcellular localization does not exclude the possibility that both proteins target the same substrates for degradation, it can be considered unlikely that both proteins control the same components within the cell. We therefore suggest that the VFB proteins affect the expression of auxin-induced genes but function through a pathway that is distinct from that of TIR1/AFB protein-regulated protein degradation.
Our microarray analysis also revealed that a number of cell wall metabolic genes are misregulated in the vfb mutants. Interestingly, a recent gene expression analysis of the axr3-1/iaa17-1 gain-of-function mutant, which expresses a stabilized axr3/iaa17 mutant protein, also showed the repression of a broad set of cell wall metabolic enzyme genes (Overvoorde et al., 2005). Since the analysis of microarray data indicates that the expression of these cell wall metabolic enzyme genes is not induced by auxin, it may be that this gene expression pattern is a molecular consequence of an interupted developmental program that is common to both types of mutants, such as the failure to efficiently induce lateral root formation.
A Specific Subset of Genes Is Misregulated in vfb and csn Mutants
CSN is an evolutionarily conserved regulator of Cullin-RING E3s. Arabidopsis csn mutants are characterized by their constitutive photomorphogenic phenotype, which includes loss of skotomorphogenic growth, chloroplast differentiation, and the expression of light-induced genes in dark-grown seedlings (Chamovitz et al., 1996; Kwok et al., 1996). The seedling-lethal csn mutants also fail to develop a proper root and lateral roots. The severe pleiotropic csn mutant phenotype may be the result of the malfunction of the hundreds of Cullin-RING E3s that can be formed in Arabidopsis (Schwechheimer and Villalobos, 2004). In Arabidopsis, SCFTIR1, SCFCOI1, and SCFUFO have already been shown to interact physically with CSN (Schwechheimer et al., 2001; Feng et al., 2003; Wang et al., 2003). This, taken with numerous reports from other eukaryotes, strongly suggests that all Cullin-RING E3 functions are CSN dependent (Schwechheimer, 2004). We have examined the overlap between the misexpressed genes in the vfb mutants and the misexpressed genes in the csn mutants. We found that 181 genes are misexpressed in the vfb and csn mutants. Strikingly, the group of repressed genes includes a number of genes whose expression is normally controlled by auxin. Therefore, this analysis identified reduced auxin response as a common denominator of both types of mutants, and malfunction of the proposed SCFVFB complex may account at least in part for the repression of auxin-induced genes in the csn mutants.
METHODS
Biological Material and T-DNA Insertion Mutants
All experiments were performed using Arabidopsis thaliana ecotype Columbia. The T-DNA insertion mutants SALK_128933 (vfb1-1), SALK_047599 (see below), SALK_047600 (vfb2-1), and SALK_054809 (vfb3-1) were identified in the SIGNAL database and obtained from the Nottingham Arabidopsis Stock Centre (Alonso et al., 2003). The T-DNA insertion line GABI_414F09 (vfb4-1) was identified and obtained from the GABI-Kat collection (Rosso et al., 2003). T-DNA insertions in the SALK lines were verified using the primer LbB1 5′-GCGTGGACCGCTTGCTGCAACT-3′ in combination with the primers 5′-ATCGATAGGTAAGCATCACGCTAACGAAT-3′ (SALK_128933; vfb1-1), 5′-ATGGGCCAAGCTCCGTCGTCTCCGGCGGAACCAAACGTA-3′ (SALK_047600; vfb2-1), and 5′-GCCGGCTACTGGCTTCGACTTGATTCTGA-3′ (SALK_054809; vfb3-1). We were unable to confirm the predicted T-DNA insertion of SALK_047599 in VFB2. The T-DNA insertion of GABI_414F09 (vfb4-1) was confirmed and analyzed using the primers GABI T-DNA 5′-CCCATTTGGACGTGAATGTAGACAC-3′ and the two gene-specific primers 5′-ATATTGACCATCATACTCATTGC-3′ and 5′-AGTAGAACTACTAGCATTATCATTGTGAGACCA-3′. Double and triple vfb mutant combinations were generated by crosses of the respective single mutants.
The reporter lines DR5:GUS (T. Guilfoyle, Columbia, MO) and HS:AXR3NT-GUS (S. Kepinski and O. Leyser, York, UK) were crossed into a vfb1-1 vfb2-1 vfb3-1 vfb4 (RNAi) mutant line (Gray et al., 2001). Plants homozygous for the vfb mutations and vfb4 (RNAi) were identified and analyzed in the F3 generation. For heat shock induction, seedlings were induced for 1 h at 37°C and then incubated for 1 h at room temperature and stained for GUS expression at the times indicated.
Phylogenetic Analysis
The protein sequence alignment of the VFB proteins (Figure 1A) was performed using the Clustal algorithm (gap penalty 10; gap length penalty 10) of the Lasergene DNAStar suite (Higgins et al., 1992). LRRs were predicted based on alignments presented by Gagne et al. (2004). The unrooted phylogenetic tree of plant F-box proteins (Figure 1B) was generated with the ClustalW algorithm (http://www.ebi.ac.uk/clustalw/) using default settings (see Supplemental Figure 3 online; Thompson et al., 1994). An unrooted dendrogram was inferred with PAUP* version 4.0b10 (Swofford, 2002) using the BIONJ variant (Gascuel, 1997) of neighbor joining (Saitou and Nei, 1987). Genetic distances were calculated as mean numbers of amino acid differences, ignoring sites with gaps in pairwise comparisons. Branch support was inferred from 1000 rounds of nonparametric bootstrapping (Felsenstein, 1985).
Cloning Procedures
For yeast two-hybrid analysis, full-length VFB2 and a deletion variant lacking the F-box domain (VFB2ΔF-box) were amplified using the primers 5′-CTCGAGTCTGAGATTCTCCCTTTCTTACCATGTATC-3′ and 5′-CTCGAGATGGGCAAGCTCCGTCGTCTCCGGCGGAA-3′ in combination with 5′-CTCGAGTTTTGCTCAATGGAACTAGTAGCTTCACT-3′. The fragments were introduced as Xho1 fragments into pEG202. The ASK2, RBX1, and CUL1 prey constructs were described previously (Schwechheimer et al., 2001; Risseeuw et al., 2003). Two-hybrid interactions were performed as described by Schwechheimer and Deng (2002). The expression of the respective fusion proteins was verified using antibodies directed against the B42 domain of fusion proteins expressed from pJG4-5 (anti-B42, Q19; Santa Cruz Biotechnology) and the LexA domain of fusion proteins expressed from pEG202 (anti-LexA; Invitrogen).
To generate a VFB2:GFP fusion, the VFB2 open reading frame was amplified by PCR with the primers 5′-attB1-TCATGGGCCAAGCTCCGTCGTCTCGGGCG-3′ and 5′-attB2-CAATGGAACTAGTAGCTTCACTTTG-3′. The fragment was introduced into pDONR201 using Gateway technology (Invitrogen), sequence verified, and then transferred into the Gateway-compatible vector 35-S-(GW)-GFP (J. Parker, Cologne, Germany) to obtain 35S:VFB2:GFP. 35S:VFB3:GFP and 35S:VFB4:GFP were cloned in an identical manner using 5′-attB1-GAATATTTACATTTTTAG-3′ and 5′-attB2-TTCATTCCGGGACCTGCTTCG-3′ or 5′-attB1-GGCCAAGCGCCGTCGTCTACG-3′ and 5′-attB2-AGTAGAACTACTAGCATTATCA-3′. To generate the YFP:VFB2 fusion, the VFB2 gene was amplified with the oligonucleotides 5′-attB1-TCATGCGCCAAGCTCCGTCGTCTCCG-3′ and 5′-attB2-CTCAAATGGAACTAG TAGCTTCACTTTG-3′. As above, the fragment was introduced into pDONR201, sequence verified, and then transferred into the Gateway-compatible vector pExtag-YFP-GW (L. Noel and J. Parker, Cologne, Germany) to obtain 35S:YFP:VFB2. All Gateway vector manipulations were performed using Gateway reagents following the manufacturer's instructions. VFB2:VFB2:GFP was obtained by replacing the 35S promoter in VFB2:GFP by a 1-kb VFB2 promoter fragment amplified by PCR using the primers 5′-GATATCAATAAAAAGTTCTTCAATAACTTT-3′ and 5′-CTCGAGTGTTGTGTGGTTGAGTATTGGGTAAGGT-3′. The subcellular localization of the fusion proteins was examined in transiently transformed Arabidopsis protoplasts using a Leica TCS confocal microscope. Nuclei were stained using Hoechst 33342 as described by Meadows and Potrykus (1981).
To suppress VFB4 gene expression by RNAi, a 114-bp gene-specific fragment was amplified using the primers 5′-attB1-TCAGGTTGAAACCGTTGTGGAGG-3′ and 5′-attB2-CACCTAATCATCGCTAATCTAC-3′. The fragment was inserted into pDONR201 using Gateway technology, sequence verified, and then transferred to the destination vector pJawohl17 (I. Somssich, Cologne, Germany) to generate vfb4 (RNAi). Using Agrobacterium tumefaciens–mediated transformation, the construct was introduced into the wild type and vfb1-1 vfb2-1 vfb3-1 triple mutant plants. At least 20 transgenic plants were generated for each construct.
For tir1-1 complementation analysis, a construct for the dexamethasone-inducible expression of VFB2 was obtained by inserting the VFB2 open reading frame as an XhoI fragment into pTA7002, and the resulting pTA:VFB2 construct was transformed into the tir1-1 mutant background (Aoyama and Chua, 1997). VFB2 gene expression was induced with 30 μM dexamethasone for 4 h for RT-PCR analysis and throughout the experiment for auxin sensitivity assays.
VFB Gene Expression Analyses
To generate VFB promoter:GUS constructs, DNA fragments corresponding to an ∼1000-bp fragment upstream of the genes' ATG start codons were amplified using the primer combinations 5′-GAATTCTCCTATCGATCCAATTAACTGGTT-3′ and 5′-CCATGGACGAAAGTCTC CGGTGGTCGTTCGT-3′ (VFB1), 5′-GAATTCCTTCCGATAGAGTGTAACTTGGTC-3′ and 5′-CCATGGTTGTGTGGTGAGTATTGGGTAAGG-3′ (VFB2), 5′-GAATTCAGAAGCTAGTCGATGCTCTACAGAGCCGGA-3′ and 5′-CCATGGTTTTCTTTACAA GTTTTAATATAC-3′ (VFB3), and 5′-GAATTCGTGGCACTTTCTTTTCTAAGTGTT-3′ and 5′-CCATGGCCAACCGAATGGTTTCGATTTCCT-3′ (VFB4). The fragments were introduced into pCR2.1 (Invitrogen), sequence verified, and then cloned into the EcoRI and NcoI sites of pCAMBIA1391Z to generate the constructs VFB1:GUS through VFB4:GUS. The constructs were transformed into Arabidopsis wild-type plants, and at least 10 transgenic lines were analyzed for GUS expression. GUS staining was performed overnight at 37°C.
RT-PCR
RT-PCR was performed based on a previously published protocol (Frohman et al., 1988). Total RNA was extracted from 100 mg of seedling (7 d old) or plant material using the Qiagen RNeasy kit. Five micrograms of RNA was used for reverse transcription with SuperScript II reverse transcriptase (Invitrogen) primed with the oligo(dT) primer 5′-GACTCGAGTCGACATCGA(17T)-3′. One microliter of the reverse transcription reaction was used for PCR to analyze gene expression of the individual VFB genes using the following primer combinations: 5′-GACGACCACCGGAGACTTTCGTGGATGGGCCA-3′ and 5′-ATCGATAGTAAGCATCACGCTAACGAAT-3′ (VFB1), 5′-ATGGGCCAAGCTCCGTCGTCTCCGGCGGAACCAAACGTA-3′ and 5′-AATGGAACTAGTAGCTTCACTTTGA-3′ (VFB2), 5′-GGGTGGCATTCTTGCCTATTTCTCCACAT-3′ and 5′-GCCGGCTACTGGCTTCGACTTGATTCTGA-3′ (VFB3), and 5′-ATGGGCCAAGCGCCGTCGTCTACGGCG-3′ and 5′-AGTAGAACTACTAGCATTATCATTGTGAGACCA-3′ (VFB4). ACTIN (At3g18780) expression was examined as a control using the primers 5′-ATTCAGATGCCCAGAAGTCTTGTTC-3′ and 5′-GCAAGTGCTGTGATTTCTTTGCTCA-3′. PCR was performed for 28, 30, 33, and 35 cycles to assure that the amplification was within the linear range of the reaction.
Physiological Assays
For auxin sensitivity assays, 5-d-old Arabidopsis seedlings were transferred to 2,4-D–containing medium. Root elongation after transfer to auxin-containing medium was measured after another 3 to 5 d and calculated relative to the untreated control samples. The average and sd of 10 seedlings per experimental condition were determined and plotted. Lateral root initiation experiments were performed as described by Ruegger et al. (1998).
Microarray Analysis
Three biological replicate samples were harvested from 10-d-old light-grown wild-type and vfb1-1 vfb2-1 vfb3-1 vfb4-1 (RNAi) mutant seedlings. RNA for microarray analysis was extracted from each replicate using the RNeasy kit (Qiagen), and complementary RNA was prepared from 6 μg of total RNA from each replicate as described in the Affymetrix Expression Analysis Technical Manual using the One-Cycle Target Labeling and Control Reagents (Affymetrix). In brief, double-stranded cDNA was synthesized and biotin-labeled target cRNA was prepared by cDNA in vitro transcription in the presence of biotinylated UTP and CTP. After purification, cRNA was fragmented and used to hybridize the Arabidopsis ATH1 GeneChip array (Affymetrix). Hybridization, washing, staining, scanning, and data collection were performed for each replicate sample independently in an Affymetrix GeneChip Fluidics Station 450 and GeneArray scanner.
Microarray analyses were performed in the R statistical programming environment using the Bioconductor modules Limma and Affy (Ihaka and Gentleman, 1996; Irizarry et al., 2002; Gentleman et al., 2004; Smyth, 2004). Raw expression values from each probe set were read from the original CEL files through the Affy module, and then the Robust Multichip Average algorithm was applied to the data set for background adjustment, quantile normalization, and summarization using median polish (Irizarry et al., 2003). The Limma module was then used to fit a linear model to the data for all pairwise comparisons of mutant versus control microarrays, and the empirical Bayes method was used to reduce the genewise sample variance (Smyth, 2004). The P values from the hypothesis tests were adjusted for multiple testing with the Benjamini and Hochberg method to control for false positives (Benjamini and Hochberg, 1995). In addition, the empirical Bayes approach automatically adjusts raw P values for multiple testing and generates a B-statistic that may also be used for ranking differentially expressed genes (Smyth, 2004). An identical experiment followed by identical data analysis was conducted with 7-d-old light-grown csn4 (SALK_043720) mutant seedlings. The microarray data were submitted to the Gene Expression Omnibus and are available under the accession numbers GSM88040 to GSM88042 (wild type of vfb mutant experiment), GSM88046 to GSM88048 (vfb mutant), GSM88055 to GSM88057 (wild type of csn4 experiment), and GSM88049 to GSM88051 (csn4 mutant).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers and Arabidopsis Genome Initiative locus identifiers: VFB1 (AT1G47056), VFB2 (AT3G50080), VFB3 (AT4G07400), VFB4 (AT5G67250), Oryza sativa ACRE189 (XP_450015.1), O. sativa JNBb0002J11.1 (XP_473092.1), Nicotiana tabacum ACRE189 (AAP03878.1), EBF1 (AT2G25490), EBF2 (AT5G25350), TIR1 (AT3G62980), AFB1 (AT4G03190), AFB2 (AT3G26810), AFB3 (AT1G12820), COI1 (AT2G39940), MAX2/ORE9 (AT2G42620), and CSN4 (AT5G42970). The microarray data have accession numbers GSM3863 (vfb mutant experiment) and GSM3865 (csn4 mutant experiment).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. VFB Gene Expression Analysis of Public Microarray Data Reveals Coregulation of VFB2 and VFB4 Expression.
Supplemental Figure 2. Growth Response Assays Reveal No Differences between the Wild-Type and vfb1-1 vfb2-1 vfb3-1 Mutant Seedlings.
Supplemental Figure 3. ClustalW Alignment of the F-Box Proteins Shown in Figure 1C.
Supplemental Table 1. List of Genes Induced in the vfb Mutants Compared with the Wild Type.
Supplemental Table 2. List of Genes Repressed in the vfb Mutants Compared with the Wild Type.
Supplemental Table 3. List of Genes Induced in the vfb Mutants and in the csn4 Mutant Seedlings Compared with the Wild Type.
Supplemental Table 4. List of Genes Repressed in the vfb Mutants and in the csn4 Mutant Seedlings Compared with the Wild Type.
Supplementary Material
Acknowledgments
We thank our colleagues for helpful comments on the manuscript. We acknowledge Markus Schmid, Mitch Levesque, and Detlef Weigel (Max Planck Institute for Developmental Biology, Tübingen, Germany) for providing access to their microarray analysis facility and for their help with chip hybridization and data analysis and Michael Weiss (Tübingen University) for the bootstrap analysis. We also thank the Nottingham Arabidopsis Stock Centre and GABI-Kat for providing T-DNA insertion lines. This research was supported by grants from the Arabidopsis Functional Genomics Network (SCHW 741/4) and from the Sonderforschungsbereich 446 (SFB446) of the Deutsche Forschungsgemeinschaft.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Claus Schwechheimer (claus.schwechheimer@zmbp.uni-tuebingen.de).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 633–657. [DOI] [PubMed] [Google Scholar]
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11 605–612. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser B 57 289–300. [Google Scholar]
- Bornstein, G., Ganoth, D., and Hershko, A. (2006). Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc. Natl. Acad. Sci. USA 103 11515–11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Villalobos, L.I., Nill, C., Marrocco, K., Kretsch, T., and Schwechheimer, C. (2007). The evolutionarily conserved Arabidopsis thaliana F-box protein AtFBP7 is required for efficient translation during temperature stress. Gene 392 106–116. [DOI] [PubMed] [Google Scholar]
- Cardozo, T., and Pagano, M. (2004). The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5 739–751. [DOI] [PubMed] [Google Scholar]
- Chamovitz, D.A., Wei, N., Osterlund, M.T., von Arnim, A.G., Staub, J.M., Matsui, M., and Deng, X.-W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86 115–121. [DOI] [PubMed] [Google Scholar]
- Chiba, T., and Tanaka, K. (2004). Cullin-based ubiquitin ligase and its control by NEDD8-conjugating system. Curr. Protein Pept. Sci. 5 177–184. [DOI] [PubMed] [Google Scholar]
- Cope, G.A., Suh, G.S.B., Aravind, L., Schwarz, S.E., Zipursky, S.L., Koonin, E.V., and Deshaies, R.J. (2002). Role for predicted metalloprotease motif of JAB1/Csn5 in cleavage of NEDD8 from CUL1. Science 298 608–611. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., Boniotti, M.B., and Gutierrez, C. (2002. a). Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 2002 3057–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., Dharmasiri, S., Hellmann, H., Walker, L., Gray, W.M., and Estelle, M. (2002. b). AXR1-ECC1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin reponse. Plant Cell 14 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.L., and Estelle, M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and Cullin/RING H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15 435–467. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005. a). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., Weijers, D., Lechner, E., Yamada, M., Hobbie, L., Ehrismann, J., Jürgens, G., and Estelle, M. (2005. b). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9 109–119. [DOI] [PubMed] [Google Scholar]
- Dohmann, E.M., Kuhnle, C., and Schwechheimer, C. (2005). Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 17 1967–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., Ma, L., Wang, X., Xie, D., Dinesh-Kumar, S.P., Wei, N., and Deng, X.-W. (2003). The COP9 signalosome interacts physically with SCFCOI1 and modulates jasmonate responses. Plant Cell 15 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution Int. J. Org. Evolution 39 783–791. [DOI] [PubMed] [Google Scholar]
- Freilich, S., Oron, E., Kapp, Y., Nevo-Caspi, Y., Orgad, S., Segal, D., and Chamovitz, D.A. (1999). The COP9 signalosome is essential for development of Drosophila melanogaster. Curr. Biol. 9 1187–1190. [DOI] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29 153–168. [DOI] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S.H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Smalle, J., Gingerich, D.J., Walker, J.M., Yoo, S.-D., Yanagisawa, S., and Vierstra, R. (2004). Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA 101 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel, O. (1997). BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14 685–695. [DOI] [PubMed] [Google Scholar]
- Gentleman, R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil, P., Liu, Y.-C., Orbovic, V., Verkamp, E., Poff, K.L., and Green, P.J. (1994). Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol. 104 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13 1678–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414 271–276. [DOI] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115 667–677. [DOI] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67 425–479. [DOI] [PubMed] [Google Scholar]
- Hetfeld, B.K., Helfrich, A., Kapelari, B., Scheel, H., Hofmann, K., Guterman, A., Glickman, M., Schade, R., Kloetzel, P.M., and Dubiel, W. (2005). The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr. Biol. 15 1217–1221. [DOI] [PubMed] [Google Scholar]
- Higa, L.A.A., Mihaylov, I.S., Banks, D.P., Zhneg, J., and Zhang, H. (2003). Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5 1008–1015. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., Bleasby, A.J., and Fuchs, R. (1992). CLUSTAL V: Improved software for multiple sequence alignment. Comput. Appl. Biosci. 8 189–191. [DOI] [PubMed] [Google Scholar]
- Hori, T., Osaka, F., Chiba, T., Miyamoto, C., Okabayashi, K., Shimbara, N., Kato, S., and Tanaka, K. (1999). Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18 6829–6834. [DOI] [PubMed] [Google Scholar]
- Ihaka, R., and Gentleman, R. (1996). Language of data analysis and graphics. J. Comput. Graph. Stat. 5 299–314. [Google Scholar]
- Irizarry, R., Gautier, L., and Cope, L. (2002). An R Package for Analyses of Affymetrix Oligonucleotide Arrays. (New York: Springer).
- Irizarry, R.A., Bolstad, B.M., Collin, F., Cope, L.M., Hobbs, B., and Speed, T.P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J., Cardozo, T., Lovering, R.C., Elledge, S.J., Pagano, M., and Harper, J.W. (2004). Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18 2573–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura, T., Koepp, D.M., Conrad, M.N., Skowyra, D., Moreland, R.J., Iliopoulos, O., Lane, W.S., Kaelin, W.G.J., Elledge, S.J., Conaway, R.C., Harper, J.W., and Conaway, J.W. (1999). RBX1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284 657–661. [DOI] [PubMed] [Google Scholar]
- Kawakami, T., Chiba, T., Suzuki, T., Iwai, K., Yamanaka, H., Minato, N., Suzuki, H., Shimbara, N., Hidaka, Y., Osaka, F., Omata, M., and Tanaka, K. (2001). NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Piekos, B., Miséra, S., and Deng, X.-W. (1996). A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 110 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, H.M., Lincoln, C.A., Timpte, C., Lammer, D., Turner, J., and Estelle, M. (1993). Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364 161–164. [DOI] [PubMed] [Google Scholar]
- Leyser, H.M.O., Pickett, F.B., Dharmasiri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10 403–413. [DOI] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Zhou, C., Wolf, D.A., Wei, N., Shevchenko, A., and Deshaies, R.J. (2001). COP9 signalosome promotes cleavage of NEDD8-CUL1 conjugates. Science 292 1382–1385. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen, K., Schaefer, L., Menon, S., Deng, X.-W., Miller, J.B., and Wei, N. (2003). Disruption of the COP9 signalosome CSN2 subunit in mice causes deficient cell proliferation, accumulationn of p53 and cyclin E, and early embryonic death. Mol. Cell. Biol. 23 6790–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant, A., Bhalerao, R., Casimiro, I., Eklof, J., Casero, P.J., Bennett, M., and Sandberg, G. (2002). AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, A., Wirbelauer, C., Scheffner, M., and Krek, W. (1999). Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1 14–19. [DOI] [PubMed] [Google Scholar]
- Meadows, M.G., and Potrykus, I. (1981). Hoechst 33258 as a vital stain for plant cell protoplasts. Plant Cell Rep. 1 77–79. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Zipfel, C., Rowland, O., Keller, I., Robatzek, S., Boller, T., and Jones, J.D. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde, P.J., et al. (2005). Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17 3282–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Z., Shen, Y., Feng, S., Wang, X., Chittetei, B.N., Vierstra, R.D., and Deng, X.W. (2003). Evidence for a physical association of the COP9 signalosome, the proteasome, and specific SCF E3 ligases in vivo. Curr. Biol. 13 R504–R505. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Staub, J.M., Serino, G., Kwok, S.F., Kurepa, J., Bruce, B., Vierstra, R.D., Wei, N., and Deng, X.-W. (2001). The cellular level of PR500, a protein complex related to the 19S regulatory particle of the proteasome, is regulated in response to stresses in plants. Mol. Biol. Cell 12 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak, T., Lechner, E., Parmentier, Y., Yanagisawa, S., Grava, S., Koncz, C., and Genschik, P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115 679–689. [DOI] [PubMed] [Google Scholar]
- Reed, J. (2001). Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 6 420–425. [DOI] [PubMed] [Google Scholar]
- Risseeuw, E.P., Daskalchuk, T.E., Banks, T.W., Liu, E., Cotelesage, J., Hellmann, H., Estelle, M., Somers, D.E., and Crosby, W.L. (2003). Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 34 753–767. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y.-F., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C. (2004). The COP9 signalosome (CSN): An evolutionarily conserved proteolysis regulator in eukaryotic development. Biochim. Biophys. Acta 1695 45–54. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., and Deng, X.W. (2002). Studying protein-protein interactions with the yeast two-hybrid system. In Molecular Plant Biology, P.M. Gilmartin and C. Bowler, eds (Oxford, UK: Oxford University Press), pp. 173–198.
- Schwechheimer, C., and Schwager, K. (2004). Regulated proteolysis and plant development. Plant Cell Rep. 23 353–364. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.-W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292 1379–1382. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., and Deng, X.-W. (2002). Multiple ubiquitin-ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C., and Villalobos, L.I. (2004). Cullin-containing E3 ubiquitin ligases in plant development. Curr. Opin. Plant Biol. 7 677–686. [DOI] [PubMed] [Google Scholar]
- Skowyra, D., Koepp, D.M., Kamura, T., Conrad, M.N., Conaway, R.C., Conaway, J.W., Elledge, S.J., and Harper, J.W. (1999). Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science 284 662–665. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3 3. [DOI] [PubMed] [Google Scholar]
- Stirnberg, P., van de Sande, K., and Leyser, H.M.O. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129 1131–1141. [DOI] [PubMed] [Google Scholar]
- Sutterluty, H., Chatelain, E., Marti, A., Wirbelauer, C., Senften, M., Muller, U., and Krek, W. (1999). p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1 207–214. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2002). PAUP*. Phylogenetic Analysis Using Parsimony. (Sunderland, MA: Sinauer Associates).
- Tatematsu, K., Kumagai, S., Muto, H., Sato, A., Watahiki, M.K., Harper, R.M., Liscum, E., and Yamamoto, K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Q., Nagpal, P., and Reed, J.W. (2003). Regulation of Arabidopsis SHY2/IAA3 protein turnover. Plant J. 36 643–651. [DOI] [PubMed] [Google Scholar]
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126 711–721. [DOI] [PubMed] [Google Scholar]
- Tian, Q., Uhlir, N.J., and Reed, J.W. (2002). Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda, K., Yoneda-Kato, N., Fukumoto, A., Yamanaka, S., and Kato, J.-y. (2004). Multiple functions of Jab1 are required for early embryonic development and growth potential in mice. J. Biol. Chem. 279 43013–43018. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997. a). ARF1, a transcription factor that binds to auxin response elements. Science 276 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997. b). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermannova, D., Gao, Y., and Liu, X. (2005). Ubiquitination of p27Kip1 requires physical interaction with cyclin E and probable phosphate recognition by SKP2. J. Biol. Chem. 280 30301–30309. [DOI] [PubMed] [Google Scholar]
- Wang, X., Feng, S., Nakayama, N., Crosby, W.L., Irish, V.F., Deng, X.W., and Wei, N. (2003). The COP9 signalosome interacts with SCFUFO and participates in Arabidopsis flower development. Plant Cell 15 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee, S., Geyer, R.K., Toda, T., and Wolf, D.A. (2005). CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat. Cell Biol. 7 387–391. [DOI] [PubMed] [Google Scholar]
- Wertz, I.E., O'Rourke, K.M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R.J., and Dixit, V.M. (2004). Human De-Etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 303 1371–1374. [DOI] [PubMed] [Google Scholar]
- Woo, H.R., Chung, K.M., Park, J.-H., Oh, S.A., Ahn, T., Hong, S.H., Jang, S.K., and Nam, H.G. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.T., Chan, Y.R., and Chien, C.T. (2006). Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol. 16 362–369. [DOI] [PubMed] [Google Scholar]
- Wu, J.T., Lin, H.C., Hu, Y.C., and Chien, C.T. (2005). Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat. Cell Biol. 7 1014–1020. [DOI] [PubMed] [Google Scholar]
- Xie, D.-X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, L., Wei, Y., Reboul, J., Vaglio, P., Shin, T.-H., Vidal, M., Elledge, S.J., and Harper, J.W. (2003). BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing Cul-3. Nature 425 316–321. [DOI] [PubMed] [Google Scholar]
- Yang, X., Lee, S., So, J.H., Dharmasiri, S., Dharmasiri, N., Ge, L., Jensen, C., Hangarter, R., Hobbie, L., and Estelle, M. (2004). The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1). Plant J. 40 772–782. [DOI] [PubMed] [Google Scholar]
- Zenser, N., Dreher, K.A., Edwards, S.R., and Callis, J. (2003). Acceleration of Aux/IAA proteolysis is specific for auxin and independent of AXR1. Plant J. 35 285–294. [DOI] [PubMed] [Google Scholar]
- Zhao, D., Ni, W., Feng, B., Han, T., Petrasek, M.G., and Ma, H. (2003). Members of the Arabidopsis-SKP1-like gene family exhibit a variety of expression patterns and may play diverse roles in Arabidopsis. Plant Physiol. 133 203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, N., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F-boxSkp2 SCF ubiquitin ligase complex. Nature 416 703–709. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.