Abstract
Gibberellic acid (GA) promotes seed germination, elongation growth, and flowering time in plants. GA responses are repressed by DELLA proteins, which contain an N-terminal DELLA domain essential for GA-dependent proteasomal degradation of DELLA repressors. Mutations of or within the DELLA domain of DELLA repressors have been described for species including Arabidopsis thaliana, wheat (Triticum aestivum), maize (Zea mays), and barley (Hordeum vulgare), and we show that these mutations confer GA insensitivity when introduced into the Arabidopsis GA INSENSITIVE (GAI) DELLA repressor. We also demonstrate that Arabidopsis mutants lacking the three GA INSENSITIVE DWARF1 (GID1) GA receptor genes are GA insensitive with respect to GA-promoted growth responses, GA-promoted DELLA repressor degradation, and GA-regulated gene expression. Our genetic interaction studies indicate that GAI and its close homolog REPRESSOR OF ga1-3 are the major growth repressors in a GA receptor mutant background. We further demonstrate that the GA insensitivity of the GAI DELLA domain mutants is explained in all cases by the inability of the mutant proteins to interact with the GID1A GA receptor. Since we found that the GAI DELLA domain alone can mediate GA-dependent GID1A interactions, we propose that the DELLA domain functions as a receiver domain for activated GA receptors.
INTRODUCTION
The phytohormone gibberellic acid (GA) promotes important processes of plant growth and development, such as seed germination, elongation growth, and flowering time (Richards et al., 2001). The GA signaling pathway is controlled by the DELLA repressors, which are characterized by their N-terminal DELLA domain (Pysh et al., 1999). The Arabidopsis thaliana genome encodes five highly homologous DELLA protein repressors, including GA INSENSITIVE (GAI) and REPRESSOR OF ga1-3 (RGA) (Peng et al., 1997; Silverstone et al., 1998; Richards et al., 2001). While GAI and RGA have overlapping functions as repressors of elongation growth, RGA-LIKE1 (RGL1) and RGL2 play a predominant role in controlling germination and floral development, respectively (Dill and Sun, 2001; King et al., 2001; Lee et al., 2002; Cheng et al., 2004; Tyler et al., 2004). The DELLA repressors are inactivated in response to GA by ubiquitin proteasome–dependent protein degradation (Silverstone et al., 2001; Fu et al., 2002; McGinnis et al., 2003; Sasaki et al., 2003). A 17–amino acid deletion in the conserved DELLA domain, which is the mutation present in the dominant Arabidopsis gai-1 mutant, renders mutant gai and rga proteins insensitive to GA induced proteolysis, and plants expressing these mutant DELLA repressors are GA-insensitive, dark-green, late-flowering dwarfs (Peng and Harberd, 1997; Dill and Sun, 2001; Silverstone et al., 2001; Fleck and Harberd, 2002; Itoh et al., 2002; Dill et al., 2004). Interestingly, mutations in and of the DELLA domain were also identified in dwarfing alleles of the DELLA repressors Reduced height1 (Rht1) from wheat (Triticum aestivum), dwarf8 (d8) from maize (Zea mays), and Slender1 (Sln1) from barley (Hordeum vulgare), and these mutations were hypothesized to be the molecular cause for the GA insensitivity of the respective alleles (Gale and Marshall, 1976; Peng et al., 1999b; Chandler et al., 2002). In line with this hypothesis, it was demonstrated in the case of the barley Sln1D allele that the SLN1 protein produced by this mutant is partially impaired in GA-dependent SLN1 degradation (Gubler et al., 2002). Conversely, the understanding of the molecular mechanism underlying the dwarfing phenotypes of the wheat Rht1 alleles is not understood but of particular importance since their use in breeding permitted to generate the lodging-resistant high-yield wheat varieties of the so-called green revolution (Gale and Marshall, 1973, 1976; Peng et al., 1999b).
In Arabidopsis, the GA-dependent degradation of GAI and RGA is promoted by the F-box protein SLEEPY1 (SLY1), which functions as the degradation substrate receptor subunit of the E3 ubiquitin ligase SCFSLY1. sly1 mutants fail to degrade GAI and RGA, and the sly1 mutant phenotype is suppressed by gai and rga loss-of-function alleles. SLY1 interacts in the yeast two-hybrid system with GAI and RGA, and the gai-1 gain-of-function phenotype is suppressed by sly1 gain-of-function alleles with increased affinity for the DELLA repressors (Peng et al., 1999a; Dill et al., 2004; Fu et al., 2004; Tyler et al., 2004). The SLY1 DELLA protein interaction also occurs when the DELLA domain is deleted. Thus, the possibility that the DELLA domain serves as an interaction domain for SLY1 has been excluded.
The identification of the GA INSENSITIVE DWARF1 (GID1) proteins as soluble GA receptors in rice (Oryza sativa) and Arabidopsis was a major breakthrough in the understanding of GA signaling (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). In rice and Arabidopsis, the analysis of GID1 proteins revealed that these GA receptors interact in a GA-dependent manner with the DELLA proteins from the respective species (Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006). A recent study shows that loss of the three Arabidopsis GID1 receptors results in GA insensitivity and that the N-terminal DELLA and VHYNP domains of the DELLA protein RGA are required for GID1 interactions in Arabidopsis (Griffiths et al., 2006).
As introduced above, several DELLA domain mutations have been described that result in GA-insensitive growth in different plant species. In most cases, the consequences of these mutations on DELLA protein behavior had not been tested at the molecular level, and how these mutations affect GA signaling remained to be addressed. In this article, we characterize plants expressing gai variants with DELLA domain mutations that had previously been identified in DELLA repressors from maize, wheat, and barley. In all cases examined, these mutations result in GA-insensitive plant growth and a stabilization of the mutant gai proteins. Consistent with a recently published report, we also found that all three Arabidopsis GID1 genes participate in GA responses, and we extend this analysis by showing that the growth repression of the GA receptor mutants is largely caused by GAI and RGA. Finally, we show that the GAI DELLA domain is required and sufficient for interactions with the Arabidopsis GA receptor protein GID1A. We therefore conclude that the DELLA domain serves as a receiver domain for activated GID1 GA receptors.
RESULTS
DELLA Domain Mutations Impair GA-Promoted Protein Degradation and Plant Growth
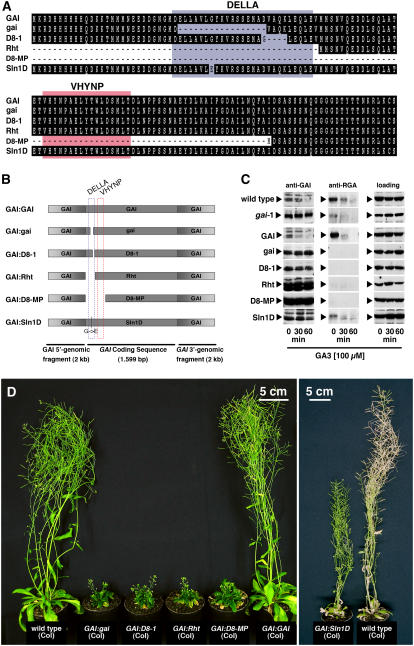
The dominant GA-insensitive Arabidopsis gai-1 mutant expresses a mutant gai protein with a 17–amino acid deletion of the highly conserved N-terminal DELLA domain (Peng et al., 1997). Following the identification of the DELLA domain deletion in Arabidopsis gai-1, DELLA domain mutations were also identified in dwarfing alleles of DELLA repressor genes from several crop species, including wheat, maize, and barley (Gale and Marshall, 1976; Peng et al., 1999b; Chandler et al., 2002). Based on the apparent importance of the DELLA domain for GA-induced DELLA protein degradation, it was hypothesized (but with the exception of the barley protein never shown) that these mutations lead to a stabilization of the respective DELLA proteins (Dill et al., 2001; Gubler et al., 2002). Interestingly, there is no obvious correlation between the extent of growth repression conferred by the different DELLA mutations and their expected severity. For example, while a 5–amino acid deletion in the DELLA domain seems to be responsible for extreme dwarfism in D8-1 mutant maize, a predicted large truncation of the protein's N terminus, including the DELLA domain and the neighboring VHYNP domain, suppresses plant growth only moderately in maize D8-Mp1 mutants (see Supplemental Figure 1A online) (Harberd and Freeling, 1989; Winkler and Freeling, 1994; Peng et al., 1999b). This gives rise to the hypotheses that either the nature of the DELLA domain mutations or the specific genetic backgrounds determine the severity conferred by these mutations.
We wanted to study the effect of the different DELLA domain mutations in a homogenous genetic background. To this end, we generated transgenic Arabidopsis plants that contain genomic fragments for the expression of wild-type Arabidopsis GAI or GAI variants carrying DELLA domain mutations reported for the dwarfing alleles from Arabidopsis gai-1, maize D8-1 and D8-Mp1, wheat Rht-B1b and Rht-D1b, and barley Sln1D (Figures 1A and 1B) (Peng et al., 1997, 1999b; Chandler et al., 2002; Gubler et al., 2002). For each construct, at least 10 transgenic lines were generated, and eight lines were analyzed at the biochemical and physiological level. While endogenous wild-type GAI and wild-type GAI expressed from the GAI:GAI transgene is efficiently degraded in response to GA, we found that the mutant gai proteins are fully stabilized in lines expressing variants with a partial or full deletion of the DELLA domain (GAI:gai and gai-1, GAI:D8-1, GAI:Rht, and GAI:D8-MP) (Figure 1C). In agreement with the previously reported observation that the SLN1D protein from barley is still partially GA sensitive, GAI:Sln1D plants, producing a GAI variant with a single amino acid substitution in the DELLA domain, express a partially stabilized gai mutant protein (Figure 1C) (Gubler et al., 2002).
Figure 1.
Physiologically Relevant DELLA Domain Mutations Stabilize GAI.
(A) Clustal alignment of the N-terminal 140–amino acid residues of Arabidopsis GAI and gai mutant variants that were designed based on the mutations identified in Arabidopsis gai-1, maize D8-1 and D8-MP, wheat Rht-B1b/D1b (Rht), and barley Sln1D alleles (see Supplemental Figure 1A online for a detailed alignment).
(B) Schematic representation of the GAI-derived transgenes carrying the mutations shown in (A). The mutations in the GAI coding sequence, but not the flanking 5′- and 3′-GAI genomic regions, are drawn to scale.
(C) Immunoblots with specific anti-GAI and anti-RGA antibodies (see Supplemental Figure 2 online) using 45 μg of protein extract from GA-treated inflorescences of wild-type, gai-1 mutant, and transgenic plants. A cross-reacting band serves as a loading control.
(D) Wild-type and transgenic 7.5-week-old (left panel) and 8.5-week-old (right panel) Arabidopsis plants.
In all cases, the degree of GAI stabilization correlates well with the level of growth suppression in the transgenic lines. All transgenic plants expressing fully stabilized GAI variants are GA-insensitive, dark-green, late-flowering dwarfs that are phenotypically indistinguishable from the Arabidopsis gai-1 mutant (Figure 1D; see Supplemental Figures 1B and 3 online). Again in agreement with the previously reported observation that the Sln1D allele from barley is still partially GA sensitive, we observed a partial reduction of elongation growth and a delay in the onset of flowering in GAI:Sln1D lines that was significantly less severe than that observed in lines expressing fully stabilized mutant gai proteins (Figure 1D).
When we examined the consequences of GAI stabilization on RGA protein accumulation, we noticed that RGA protein levels are strongly reduced in all lines expressing stabilized GAI variants (Figure 1C). Since GA-insensitive mutants, such as gai-1, were reported to contain increased levels of GA (Peng and Harberd, 1997), we reasoned that increased GA-promoted RGA degradation may be responsible for this effect. Indeed, when we treated plants expressing stabilized GAI variants with the GA biosynthesis inhibitor paclobutrazol (PAC), RGA protein levels increased, suggesting that GA levels regulate DELLA repressor abundance, at least in part, by promoting their degradation (see Supplemental Figure 2B online). In summary, we conclude that partial or full deletions of the DELLA domain, as previously reported for dwarfing alleles of several crop species, cause (when introduced into Arabidopsis GAI) GA insensitivity with respect to GA-promoted protein degradation and GA-promoted plant growth. Hence, the differences in the severity of dwarfing mutations, such as the D8-1 and D8-Mp mutations from maize, may be attributable to differences in the genetic background of these alleles.
The Three Arabidopsis GID1 Genes Participate in GA Responses
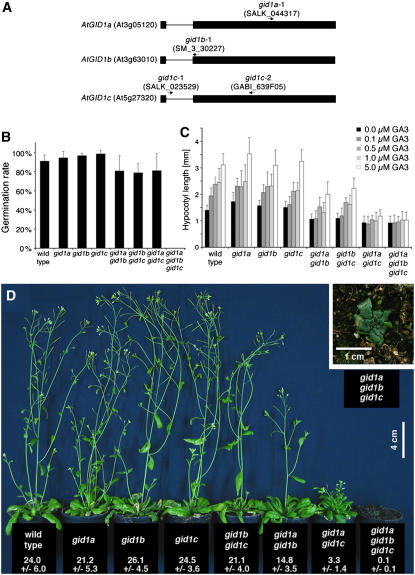
The biological role of the three apparent Arabidopsis homologs (GID1A, AT3G05120; GID1b, AT3G63010; and GID1c, At5G27320) of the rice GA receptor GID1 was recently determined, and it was found that the three GID1 genes have redundant functions in mediating GA responses (Griffiths et al., 2006). We also analyzed GA responses in T-DNA insertion mutants for each of the three Arabidopsis GID1 genes (Figure 2A). For our analysis, we selected three mutant alleles with in-gene in-exon T-DNA insertions, namely, gid1a-1, gid1b-1, and gid1c-2. Our gene and allele nomenclature is identical to the one used in the previous publications of these genes (Griffiths et al., 2006; Nakajima et al., 2006), and with the exception of the gid1c-2 allele used in our study, identical mutant alleles were used for physiological analysis and genetic experiments. We found that while single mutants of each of the three GID1 genes do not have obvious defects in GA-controlled growth responses, such as germination, GA-induced hypocotyl elongation, elongation growth, or flowering time, gid1 double and triple mutants are partially (double mutants) or fully (triple mutants) impaired in these responses (Figures 2B to 2D). Therefore, our gid1 triple mutants display a complete suppression of GA responses and are phenotypically indistinguishable from severe GA biosynthesis mutants, such as ga1-3, in that they fail to germinate and, after manual removal of the seed coat, develop into dark-green severely dwarfed plants with a maximum rosette diameter of 1 cm (Figures 2B to 2D). In contrast with ga1-3, which is the most severe GA biosynthesis mutant described in the literature (Koornneef and van der Veen, 1980), and in contrast with the gid1 triple mutant described in a recent publication (Griffiths et al., 2006), our gid1 triple mutants never flower even in long-day conditions (8 h dark/16 h light), continuous light conditions, or when treated with GA3 (see Supplemental Figure 4 online). This difference in phenotype severity may be attributable to the fact that we used the allele gid1c-2 to generate the gid1a-1 gid1b-2 gid1c-2 triple mutant, whereas the above-mentioned report (Griffiths et al., 2006) made use of the allele gid1c-1. The T-DNA insertion of the gid1c-2 mutation (GABI_639F05) resides in the second exon of the GID1c gene, and this mutation may affect gene function more severely than the T-DNA insertion in gid1c-1 (SALK_023529), which is located in the GID1c intron (Figure 2A). Taken together, based on our genetic analyses and the biochemical analyses conducted by others (Griffiths et al., 2006; Nakajima et al., 2006), we conclude that the three Arabidopsis GID1 proteins have redundant functions as GA receptors and that gid1 triple mutants are insensitive to GA.
Figure 2.
Loss of Arabidopsis GID1 GA Receptor Function Results in GA Insensitivity.
(A) Schematic representation of the genomic organization of the three Arabidopsis GID1 orthologs GID1A to GID1C. Exons are shown as black boxes and introns as lines. The positions of T-DNA insertions and the names of the gid1 mutant alleles are indicated by arrows.
(B) Seed germination rate after 4 d as evaluated by root emergence of the gid1a-1, gid1b-1, and gid1c-2 alleles and their double and triple mutant combinations (n ≥ 100). In the case of the gid1 triple mutant, germination rate was calculated based on the germination rate of a homozygous gid1a gid1b double mutant segregating for the gid1c mutation.
(C) Hypocotyl elongation in response to GA measured from 5-d-old seedlings grown on GA3-containing media as indicated. The gid1 triple mutant seedlings used in this experiment were manually removed from the seed coat (n ≥ 10).
(D) Phenotype of 4-week-old gid1 mutants as indicated in the panel. The average and sd of the height of 4-week-old gid1 mutants is indicated below the genotypes (n ≥ 8).
gid1 Mutants Are GA Insensitive with Respect to GA-Promoted DELLA Protein Degradation and GA-Controlled Transcriptional Responses
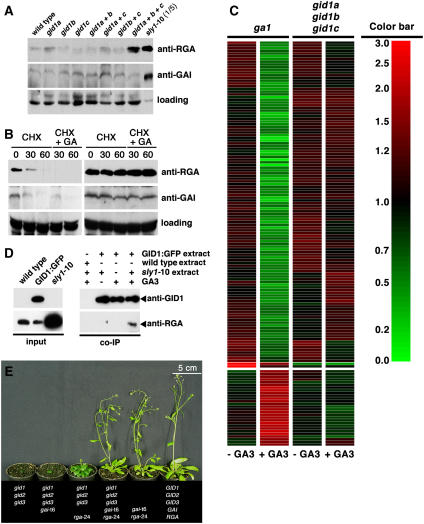
To examine whether the GA insensitivity observed in the gid1 triple mutant correlates with DELLA repressor stabilization, we tested RGA and GAI abundance in the gid1 mutants (Figure 3A). Interestingly, neither the gid1 single mutants nor the gid1 double mutants showed the increase in DELLA protein levels expected for mutants impaired in GA perception. Although minor differences in RGA and GAI abundance may be observed in the comparison of individual gid1 single and double mutants in the experiment shown in Figure 3A, these differences were generally not reproducible and cannot be considered significant. However, we observed a significant and reproducible accumulation of RGA but not of GAI in the gid1 triple mutant (Figure 3A). Furthermore, following GA treatments that cause complete degradation of these proteins in the wild type, RGA and GAI were found to be fully stabilized in the gid1 triple mutant (Figure 3B). We thus conclude that the gid1 triple mutants are GA insensitive with respect to DELLA protein degradation and that GAI and RGA accumulate to different levels in the gid1 receptor mutants.
Figure 3.
GAI and RGA Are Major Growth Repressors in gid1 Triple Mutants.
(A) Abundance of RGA and GAI in the gid1 mutants. Immunoblots with specific anti-RGA and anti-GAI antibodies (see Supplemental Figure 2 online) using 45 μg of total protein extract from the wild type and gid1 mutants as indicated and 9 μg from sly1-10 mutants. A cross-reacting band serves as loading control.
(B) RGA and GAI are stabilized in gid1 triple mutants. Immunoblot with anti-RGA and anti-GAI antibodies using 45 μg of protein extract from wild-type and gid1 triple mutant plants after cycloheximide (CHX; 50 μM) and GA3 (100 μM) treatments as indicated. A cross-reacting band serves as loading control.
(C) gid1 triple mutants are GA insensitive with respect to GA-regulated gene expression. Clustered gene tree of the 148 GA-regulated genes in the ga1 mutant (two columns on the left) and the gid1 triple mutants (two columns on the right) in the absence and presence of GA as indicated. Coloring of the gene tree is according to the color bar on the right. The complete list of GA-regulated genes and raw expression data are provided in Supplemental Table 1 online.
(D) The GID1:GFP fusion protein coimmunoprecipitates with RGA in a GA-dependent manner. Immunoblots with anti-GID1 and anti-RGA antibodies using 45 μg of total protein extract as input control (left panel) and after coimmunoprecipitation (co-IP; right panel).
(E) The rga-24 and gai-t6 mutations suppress the gid1 triple mutant phenotype (6-week-old plants are shown). All plants are homozygous for the Columbia ER allele. At least four quadruple and quintuple mutant plants with identical phenotypes were identified. The rga-24 gai-t6 double mutants with the Columbia ER allele segregating from the cross are indistinguishable from the Columbia wild type.
Since we could show that gid1 triple mutants are GA insensitive at the physiological and biochemical level, we next examined whether their GA insensitivity also extends to GA-regulated transcription. To this end, we dissected ga1 and gid1 triple mutant seedlings from seeds, allowed the dissected seedlings to grow for 1 week on growth media, and subjected the phenotypically identical mutant seedlings for 1 h to 100 μM GA3 or a mock control treatment (three biological replicate samples for each mutant and each treatment). With the exception of the GA1 and GID1 genes mutated in the respective loss-of-function mutants, gene expression analyses with the Arabidopsis ATH1 gene chip identified only a single gene that is differentially expressed between mock-treated ga1 and mock-treated gid1a-c (data not shown). Thus, ga1 and gid1 mutants are not only identical at the phenotypic level but also at the level of gene expression. In turn, the analysis of the GA-treated ga1 mutant resulted in the identification of 120 GA-repressed and 28 GA-induced genes (Figure 3C; see Supplemental Table 1 online). In line with previous gene expression studies, this analysis identified GAI as being induced in response to GA and GID1A, GID1b, SLY1, and several genes encoding proteins required for GA biosynthesis as being repressed in response to GA (see Supplemental Table 1 online) (Cao et al., 2006; Griffiths et al., 2006). Importantly, the expression of the 148 genes regulated by GA in ga1 was not affected by the GA treatment in the gid1 triple mutants (Figure 3C; see Supplemental Table 1 online). Furthermore, the direct comparison of the expression profiles of mock-treated and GA-treated gid1 triple mutants did not lead to the identification of any GA-regulated genes in the gid1 triple mutant. Thus, our findings suggest that all GA-regulated transcriptional responses are mediated by the GID1 GA receptors in Arabidopsis, at least at the seedling stage.
GID1 Receptors and the DELLA Repressors Interact at the Biochemical and Genetic Levels
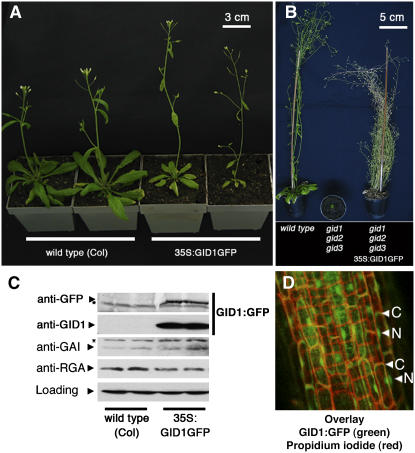
Next, we generated a transgenic line expressing a fusion protein of GID1A and green fluorescent protein (GFP) under control of the 35S promoter of Cauliflower mosaic virus (35S:GID1:GFP). Overexpression of GID1:GFP in the wild-type background resulted in a slight reduction of flowering time and correlated with a slight reduction in RGA protein levels when compared with wild-type plants (Figures 4A and 4C). Furthermore, the 35S:GID:GFP transgene was able to complement the germination, elongation, and flowering time defects of the gid1 triple mutant, indicating that the fusion protein retained functionality (Figure 4B). As described for the rice GID1 protein, the GFP-tagged Arabidopsis GID1A protein localizes to the nucleus, the presumed site of action of the DELLA repressors, and to the cytoplasm. Furthermore, the localization of the GID1A receptor was not altered by GA treatments or by treatments of the GA biosynthesis inhibitor PAC (Figure 4D; data not shown) (Silverstone et al., 2001; Fleck and Harberd, 2002; Ueguchi-Tanaka et al., 2005).
Figure 4.
Transgenic 35S:GID1GFP plants overexpress a functional GID1:GFP fusion protein.
(A) The 35S:GID1GFP transgenic plants flower earlier and elongate faster than wild-type plants. Two representative three-week-old wild-type and 35S:GID1GFP lines are shown.
(B) The 35S:GID1GFP transgene complements the gid1 triple mutant phenotype. 35S:GID1GFP was introgressed into a segregating gid1 mutant, and complementing lines were identified by genotyping for the three gid1 mutant alleles and the 35S:GID1GFP transgene in the F2 generation.
(C) Immunoblots with 45 μg of protein extract indicate that the overexpression of GID1:GFP correlates with a slight reduction of RGA but not of GAI protein levels. Asterisks indicate cross-reacting bands.
(D) GID1:GFP localizes to the nucleus (N) and the cytoplasm (C). Overlay image of confocal images of GID1:GFP (green) as detected in roots of 35S:GID1GFP transgenic plants and propidium iodide (red), which was used to outline the cellular boundaries.
To gain evidence for the in vivo interaction between GID1A and the DELLA repressors, we immunoprecipitated GID1:GFP with an anti-GFP affinity matrix. Using protein extracts of sly1-10 mutant seedlings, which accumulate the DELLA repressors, we were able to coimmunoprecipitate RGA in a GA-dependent manner (Figure 3D). Our attempts to coimmunoprecipitate GID1:GFP and GAI in a similar experiment were not successful, most likely due to the comparatively lower affinity of the anti-GAI antibody. Nevertheless, this experiment indicates that GID1A can interact with DELLA proteins in vivo.
In rice, the introduction of a SLENDER RICE loss-of-function mutant completely suppresses the gid1 mutant phenotype. In a similar experiment, we introduced GAI and RGA loss-of-function alleles (gai-t6 and rga-24) into the Arabidopsis gid1 triple mutant background. In the resulting quintuple mutant, we observed a dramatic suppression of almost all aspects of the gid1 mutant phenotype, including the defects in germination, elongation growth, and flowering time (Figure 3E). Quadruple mutants lacking only GAI or RGA suppressed the gid1 triple mutant phenotype to a comparatively minor extent, but the suppression by rga-24 was significantly stronger than that by gai-t6 (Figure 3E). Since the gai-t6 and rga-24 mutations are in a different Arabidopsis ecotype (Landsberg erecta) than the gid1 mutations (Columbia), we were concerned with the effect of the genetic background on the mutants' phenotypes. Since the complexity of the genetic experiment and the severity of the mutant phenotypes prevented us from performing repeated backcrosses, we restricted our analysis to quadruple and quintuple mutants that were genotyped as being homozygous wild type for the Columbia allele of the ERECTA (ER) gene. ER is mutated in Landsberg erecta, and this mutation is at least in part responsible for the differences in elongation growth between the Columbia and Landsberg erecta ecotypes (Torii et al., 1996). A minimum of four plants was identified for each quadruple and quintuple mutant in the Columbia ER background, and all plants belonging to the same group were phenotypically indistinguishable. Taken together, our genetic and biochemical data strongly support the notion that GID1 proteins interact with DELLA proteins in a GA-dependent manner in vivo and that the DELLA repressors GAI and RGA are the major growth repressors in the absence of GA signaling in an Arabidopsis GA receptor mutant background.
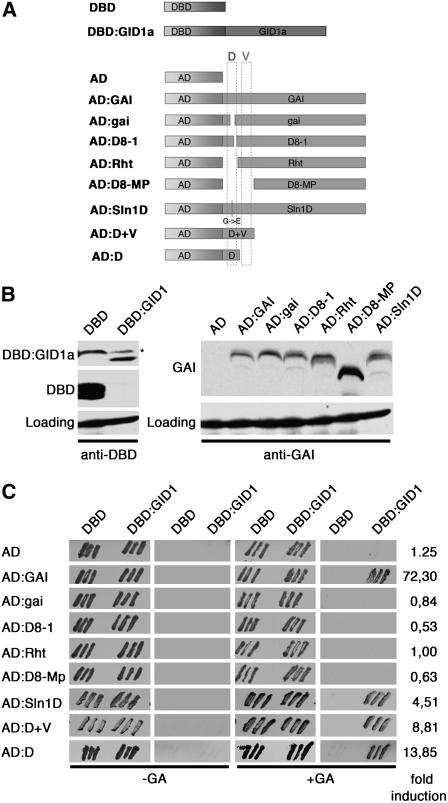
The GAI DELLA Domain Mediates GID1A GA Receptor Interactions
Since mutations of the GAI DELLA domain and mutations of the GID1 GA receptors confer GA-insensitive growth (Figures 1 and 2), and since DELLA proteins and GID1 proteins interact in vivo (Figure 3), we reasoned that the GAI DELLA domain itself may mediate the interactions with the GID1A GA receptor. Following up on experiments made by others, we used the yeast two-hybrid system to demonstrate that GAI can interact with GID1A in a GA-dependent manner in yeast (Nakajima et al., 2006). We then tested the gai mutant variants in this interaction assay and found that all gai mutant variants with a partial or full deletion of the DELLA domain fail to interact with GID1A (Figure 5). This finding is in agreement with recent reports that the DELLA domain of RGA is required for GID1A interactions (Griffiths et al., 2006). Consistent with our observation that plants expressing GAI:Sln1D are partially impaired in GA responses, we detected an ∼10-fold reduction of the interaction between the Sln1D mutant variant and GID1A over a range of GA concentrations (Figure 5C; data not shown). In a complementary experiment, we observed that the GAI DELLA domain alone (amino acids 1 to 73) is sufficient to mediate GA-dependent interactions with GID1A and that the presence of the adjacent VHYNP domain (amino acids 1 to 115) does not contribute to the strength of this interaction. Thus, our results support the notion that the DELLA domain serves as a receiver domain for the GA receptor following GA binding and that the loss of this interaction is the molecular cause for the GA insensitivity observed in the previously reported dwarfing alleles of DELLA repressor genes from a range of species.
Figure 5.
The DELLA Domain Is Required and Sufficient for the Interaction with the GID1A Receptor.
(A) Schematic representation of the constructs used for the yeast two-hybrid interaction study. The positions of the DELLA (D) and the VHYNP (V) domains are indicated.
(B) Immunoblots with anti-GAI (AD constructs; anti-GAI antibodies) and anti-GAL4 (DBD constructs; anti-GAL4 [DBD] RK5C1; Santa Cruz Biotechnology) antibodies demonstrating the expression of the respective fusion proteins in yeast. The asterisk indicates a cross-reacting band. The expression levels of the AD:D and AD:D+V constructs in yeast could not be examined because the anti-GAI peptide antibody does not recognize the GAI N terminus.
(C) Result of the yeast two-hybrid interaction study demonstrating that the DELLA domain is required and sufficient for the GA-dependent GID1A interactions. The first and third columns show yeast transformants (growth control); the second and fourth columns show the result of a qualitative LacZ filter lift assay conducted with yeast grown for 2 d in the absence (left) and presence (right) of 100 μM GA3 (Duchefa). Indicated on the right are the fold inductions of LacZ enzyme activity of DBD:GID1 AD:construct combinations grown on GA3 over untreated samples as determined using the Galacton-Star luminescent reagent. In agreement with the qualitative assays, the DBD AD:construct combinations (negative controls) did not result in differential LacZ activity in the absence and presence of GA (data not shown).
DISCUSSION
We were interested in determining the molecular function of the DELLA domain of the DELLA repressor proteins. To this end, we introduced previously identified DELLA domain mutations from Arabidopsis, maize, wheat, and barley into Arabidopsis GAI (Figure 1A). Genetic evidence suggested that these mutations are responsible for GA-insensitive growth in the respective alleles (Peng et al., 1999b; Chandler et al., 2002). Transgenic Arabidopsis plants expressing GAI variants with a deletion of or in the DELLA domain were phenotypically indistinguishable GA-insensitive dwarfed plants that express stabilized gai proteins (Figure 1; see Supplemental Figure 1B online). The single amino acid exchange mutation designed based on the barley Sln1D mutant allele showed an intermediate phenotype with respect to plant growth and mutant gai protein stabilization, and these data are entirely consistent with a previous study on the SLN1D protein from barley (Gubler et al., 2002). In turn, our interaction studies with the Arabidopsis GID1A GA receptor and the GAI DELLA repressor mutant proteins indicate that the loss (or reduction in the case of Sln1D) of GA responsiveness can be explained by the loss (or reduction) of GA-dependent GID1A binding (Figure 5). Since we also found that the DELLA domain alone is sufficient to mediate GA-dependent GAI interactions in the yeast two-hybrid system, we propose that the DELLA domain functions as the receiver domain for the activated GID1A receptor. Thus, loss of GA receptor interaction may be molecular cause for GA-insensitive growth of the respective dwarfing alleles from wheat, maize, and barley. At the same time, we of course realize that the ultimate proof for this hypothesis needs to be furnished in the respective crop species, as already done in the case of barley Sln1D (Gubler et al., 2002). In the case of the wheat Rht1 alleles, another but related mechanistic explanation to the one mentioned above may be valid. Based on the nature of the Rht1 mutations, it may be possible that these alleles express an N-terminally truncated GAI ortholog, as used in our study, as well as a peptide containing only the protein's N terminus, including the DELLA domain (Peng et al., 1999b). It cannot be excluded that the expression of this DELLA domain-only peptide titrates out GID1 interaction partners and thereby causes indirectly the stabilization of functional full-length DELLA repressors.
We characterized mutants lacking the three genes encoding GID1 GA receptor function in Arabidopsis. To a large extent, our phenotypic analysis is consistent with a report on the function of these genes that was published recently by others (Griffiths et al., 2006). In addition to the phenotypic analysis provided in this other report, we could show that gid1 double mutants are impaired to the same extent in germination, while the gid1 triple mutants are unable to germinate (Figure 2B). By contrast, we found that GA-induced hypocotyl elongation and plant height of adult plants are significantly more affected in gid1a gid1c mutants than in double mutant combinations containing gid1b (Figures 2C and 2D). Analyses of the gid1 double mutants at the adult stage further indicate that gid1a mutations make a stronger contribution to the adult phenotype than the two other genes. These observations are also consistent with the observations published recently (Griffiths et al., 2006). By contrast, however, we noted that our gid1 triple mutant never flowered and therefore has a more severe phenotype than the previously described gid1 triple mutant. We attribute this difference to the different gid1c alleles that were used in the two studies. We made use of the gid1c-2 allele, which may be stronger than the gid1c-1 allele used in the other study. gid1c-2 has an insertion in the large exon of the GID1c gene, while the insertion in gid1c-1 is in the gene's intron and could potentially be removed by splicing (Figure 2A).
Our analyses also extend the understanding of the interaction between the GID1 proteins and the DELLA repressors (Figure 3). While we show that GAI and RGA are stabilized in the gid1 triple mutant and that both proteins act as repressors in the gid1 triple mutant background (Figures 3B and 3E), we also show that GA-regulated transcription is fully impaired in the gid1 triple mutants (Figure 3C). In this experiment, we had envisioned three different scenarios: First, GA-regulated transcription may be fully repressed in the gid1 triple mutant, indicating that all transcriptional GA responses are mediated by the three GID1 GA receptors. Second, GA-regulated transcription may be globally reduced but not fully repressed, a finding indicative of the existence of other proteins with a general role in GA perception and signal transduction. (An alternative explanation for this scenario could have been that the gid1 triple mutants are not null alleles.) Third, GA-regulated gene expression of a subset of GA-regulated genes can still occur, a finding indicative for the existence of other receptor proteins that control a subset of GA responses. Our gene expression analysis strongly supports the first scenario, suggesting that all transcriptional GA responses, at least at the growth stage examined, are mediated by the GID1 proteins.
While our study shows that the GAI DELLA domain alone (amino acids 1 to 73) is sufficient to mediate interactions with the GID1A receptor, the recently published study on the interaction between GID1 proteins and RGA suggests that the RGA DELLA domain (RGA amino acids 1 to 70) alone is not sufficient to mediate this interaction (Griffiths et al., 2006). In addition to the fact that these two studies use different vector systems, we would like to note that our GAI DELLA domain fragment (GAI amino acids 1 to 73) corresponds based on sequence homologies to the first 89 amino acids of RGA, due to the extended N terminus of RGA. Thus, our fragment covers a considerably longer part of the GAI N terminus downstream of what we define as the DELLA domain than the RGA DELLA domain fragment used in the other study (Figure 1A; see Supplemental Figure 1A online). The longer GAI DELLA domain fragment used in our study may provide additional flexibility to the DELLA domain and thereby permit interactions with the GA receptor. It may also be that the functional DELLA domain is longer than what is generally defined as the DELLA domain (Figure 1A; see Supplemental Figure 1A online) since, in the absence of structural protein data, it is difficult to delimit the boundaries of the DELLA domain solely based on the degree of sequence conservation.
In our experiments, we also detected an effect of GAI stabilization on the abundance of RGA. All lines expressing stabilized mutant gai proteins had strongly reduced RGA levels. GA biosynthesis is under negative feedback control of GA, and this feedback control mechanism is known to be impaired in GA signaling mutants that fail to degrade GAI or RGA, such as gai-1 (Peng and Harberd, 1997). We therefore reasoned that an increase in GA levels promotes the accelerated degradation of RGA in plants expressing stabilized gai variants. We could substantiate this hypothesis by our finding that RGA levels increase in plants expressing stabilized gai when GA biosynthesis is inhibited with the GA biosynthesis inhibitor PAC. Thus, although we did not examine other levels of control, such as gene transcription, the reduction of RGA levels may at least in part be explained by increases in GA levels after GAI stabilization. Similar observations were made in previous studies where, for example, GAI protein levels were found to be increased in the absence of RGA in ga1-3 rga-24 and sly1-10 rga-24 loss-of-function mutants compared with the ga1-3 or sly1-10 single mutants (Dill et al., 2004). It thus appears that the abundance of DELLA repressors is under global control and that a mechanism is in place that controls DELLA protein levels. While one element of this control mechanism is certainly SLY1-mediated GA-dependent DELLA protein turnover, the precise mode of regulation remains to be established. A differential accumulation of the DELLA repressors is also apparent in several of our mutant analyses. For example, RGA and GAI accumulate and are stabilized in the sly1-10 mutant, which lacks the F-box protein that promotes RGA and GAI degradation. In turn, only RGA but not GAI accumulates in the gid1 triple mutant, but neither protein responds to GA in the GA receptor mutant background (Figures 3A and 3B). This difference may be explained by a differential GA-independent turnover of GAI by SLY1. This hypothesis may be supported by the observation that SLY1 interacts significantly better with GAI than with RGA in the yeast two-hybrid system, notably in the absence of GA (Dill et al., 2004). Such a hypothetical GA-independent turnover may explain the differential accumulation of the two repressors in the gid1 triple mutant. Although we have not been able to resolve such a GAI turnover in our experiments, we cannot exclude its existence and consider it a suitable mechanism to explain the differences in basal DELLA protein levels.
Independent of the molecular mechanism that underlies the differential accumulation of GAI and RGA in specific mutants, it also needs to be said that there is a striking discrepancy in the severity of the phenotypes of the (weak) sly1-10 and the (strong) gid1 triple mutant, especially in relation to the (strong and weak) accumulation of the DELLA repressors (e.g., Figure 3A). Thus, there is no good correlation between DELLA protein accumulation, the ability to degrade the repressors, and the severity of a phenotype. It is therefore very likely that, besides GA-dependent protein degradation, alternative molecular mechanisms, such as posttranslational modifications, regulate DELLA repressor activity. Several reports point to a role of phosphorylation in controlling DELLA protein function, but the regulatory role of this modification with respect to DELLA repressor activity is unclear (Sasaki et al., 2003; Gomi et al., 2004; Itoh et al., 2005). Glycosylation by the glycosyl transferase SPINDLY is a second modification that may be implicated in DELLA repressor activity, or more specifically in repressor activation (Silverstone et al., 2007). The identification of the proteins that exert regulatory functions on the DELLA repressors and the identification of the site, nature, and function of DELLA protein modifications will further increase our understanding of the GA signaling pathway in the future.
METHODS
Plant Material
All gid1 alleles are from Arabidopsis thaliana (ecotype Columbia) and were obtained from the Nottingham Arabidopsis Stock Centre and the GABI-KAT facility (Max-Planck Institute for Plant Breeding Research) (Alonso et al., 2003; Rosso et al., 2003). Homozygous mutants were identified by PCR-based genotyping: gid1a-1 was genotyped using 5′-attB1-CGGATCCTGGCTGCGAGCGATGAAGTTAATC-3′ and 5′-attB2-CTGCAGTTAACATTCCGCGTTTACAAACGCCG-3′ to test for the wild-type gene and 5′-attB2-CTGCAGTTAACATTCCGCGTTTACAAACGCCG-3′ and LBb1 to test for the T-DNA insertion. gid1b-1 was genotyped using 5′-ATGGCTGGTGGTAACGAAGTCAACC-3′ and 5′-CTAAGGAGTAAGAAGCACAGGACTTGAC-3′ to test for the wild-type gene and 5′-CTAAGGAG TAAGAAGCACAGGACTTGAC-3′ together with 5′-CTTATTTCAGTAAGAGTGTGGGGTTTTGG-3′ to test for the insertion. gid1c-2 was genotyped using 5′-ACCAGCTGATGCTGGCACTTCACCAAGT-3′ and 5′-GGCATTCTGCGTTT ACAAATGCAGCTATCT-3′ to test for the wild-type gene and 5′-CAGACAGTGGTTCCTCTCAATACA-3′ together with 5′-CCCATTTGGACGTGAATGTAGACAC-3′ to test for the insertion. rga-24 was genotyped using 5′-GGTGATTTTCACGGTGGTTG-3′ and 5′-TCGCTTAGTAGTTAGTACTC-3′ to amplify the wild-type gene fragment and 5′-CATAGACCATAGTATTCGTGA-3′ and 5′-TCGCTTAGTAGTTAGTACTC-3′ to amplify the mutant allele. gai-t6 was genotyped using 5′-CTAGATCCGACATTGAAGGA-3′ and 5′-AGCATCAAGATCAGCTAAAC-3′ to amplify the wild-type gene and 5′-CTAGATCCGACATTGAAGGA-3′ and 5′-TCGGGTACGGGATTTTCGCAT-3′ to amplify the gai-t6 insertion allele. The alleles gid1a-1, gid1b-1, and gid1c-2 were used to generate mutant combinations. The genotypes of important mutant combinations, such as the gid1a gid1b gid1c rga-24 gai-t6 mutants, were confirmed three times using independent genomic DNA preparations.
T-DNA Constructs for Plant Transformation
To generate GAI:GAI, GAI:gai, etc., the open reading frames of GAI and gai mutant variants were obtained by overlap extension PCR and introduced into the vector pGREEN0179 containing 2-kb GAI upstream and downstream sequences (Hellens et al., 2000). All constructs were designed and cloned in an identical manner. In detail, the 2-kb GAI promoter fragment was amplified by PCR from ecotype Columbia genomic DNA using 5′-CTCGAGTATTACTTCTTTAGAAAAAATAATGTTTGG-3′ and 5′-GAATTCATTGGCGATGGATCCCATGGTTGGTTTTTTTTCAG-3′ and inserted as an XhoI-BamHI fragment into pGREEN0179. Subsequently, the 2-kb GAI downstream sequence was amplified using 5′-GGATCCATCGCCAATGAATTCTAGATGGTGGCTCAATGAATTGATC-3′ and 5′-GAGCTCCGTGGCGGAAGTACCGCTGAAAG-3′ and inserted as a BamHI-SacI fragment into the above construct. Into the resulting BamHI and EcoRI restriction sites, full-length GAI and gai were ligated after amplification from wild-type and gai-1 mutant genomic DNA. PCR fragments containing gene-specific deletions were then generated and used to replace part of the GAI wild-type sequence: For GAI:Rht1, a PCR product was obtained with 5′-GGATCCGTTATGATGTCTAATGTTCAAGAA-3′ and 5′-CTGCAGTCACCCAGGTCGAAGCGCAAGAG-3′. For GAI:Sln1D, an overlap PCR product was inserted that had been obtained with the primer pairs 5′-GGATCCAAGAGAGATCATCATCATC-3′ and 5′-CATTTCGGATGACCTAACCTTGTACTCAAGAACAGCTAGAAGCTCATCCAT-3′ as well as 5′-ATGGATGAGCTTCTAGCTGTTCTTGAGTACAAGGTTAGGTCATCCGAATG-3′ and 5′-CTGCAGTCACCCAGGTCGAAGCGCAAGAG-3′. For GAI(D8-1), an overlap PCR product was obtained with the primer pairs 5′-GGATCCAAGAGAGATCATCATCATC-3′ and 5′-CTACATAACTTCAAGCTGCTCGAGCCCAGCCATTTCGGATGACCTAACCTTGTAAC-3′ as well as 5′-GGTTACAAGGTTAGGTCATCCGAAATGGCTGGGCTGGAGCAGCTTGAAGTTATGATG-3′ and 5′-CTGCAGTCACCCAGGTCGAAGCGCAAGAG-3′ for GAI(D8-1). At least 10 transgenic lines were generated in the Columbia ecotype, and eight lines were analyzed at the phenotypic level.
35S:GID1GFP was generated by inserting the GID1A open reading frame amplified by RT-PCR with the primers 5′-attB1-ATGGCTGCGAGCGATGAAGTTAATCT-3′ and 5′-attB2-CACATTCCGCGTTTACAAACGCCGAAATC-3′ into 35S-GW-GFP (a gift from the Max Planck Institute for Plant Breeding Research) via pDONR201 using the Gateway system. At least 10 transgenic lines were generated in the Columbia ecotype, and eight lines were analyzed at the phenotypic level. The 35S:GID1GFP transgene was introduced into the gid1 triple mutant background. Complementing 35S:GID1GFP gid1 triple mutant plants were identified in the F2 generation by genotyping as described above.
Immunoblots and Immunoprecipitation
Specific anti-GAI and anti-RGA peptide antibodies were raised in rabbits and affinity-purified against the specific peptides GGDTYTTNKRLKC (amino acids 127 to 139 of GAI) and KRDHHQFQGRLSNHGC (amino acids 2 to 16 of RGA; Eurogentec). Both peptide sequences are specific for the respective DELLA proteins. Immunoblots were performed using 45 μg of total plant protein extract prepared in extraction buffer (50 mM Tris-HCl, 150 NaCl, 0.5% Triton X-100, 10 μM MG132, 0.1 μM PMSF, and Sigma-Aldrich protease inhibitor cocktail, pH 7.5). For immunoprecipitation of GID1:GFP, 400 μg of plant extract from the wild type or a 35S:GID1GFP-expressing line was preincubated with 20 μL of anti-GFP agarose (Vector Laboratories) in a binding buffer (50 mM Tris-HCl, 150 NaCl, 10 μM MG132, 0.1 μM PMSF, and Sigma-Aldrich protease inhibitor cocktail, pH 7.5). Following three washes in wash buffer (50 mM Tris-HCl and 150 NaCl), GID1:GFP fusion protein bound to the matrix was mixed with 200 μg of protein extract from 7-d-old sly1-10 mutant seedlings in the presence and absence of 100 μM GA3 and incubated for 3 h in binding buffer. Following three washes in wash buffer, the resin was resuspended in loading buffer, and equal volumes were loaded for immunoblot analysis. GID1:GFP was detected with an anti-GFP antibody (Invitrogen) or an anti-GID1 peptide antibody raised in rabbits and affinity-purified against the GID1A-specific peptide FGGNERTESEKSLDG (Eurogentec).
Microarray Analysis
For gene expression profiling, nongerminating ga1 and gid1 triple mutant seedlings were dissected and grown on solid growth medium for 1 week in continuous light. For each mutant, three replicate samples (biological replicates) were subjected to a mock spray treatment with water (uninduced control), and three replicate samples were subjected to a spray treatment with 100 μM GA3 (Duchefa). After 1 h, the plant material was frozen in liquid nitrogen, and RNA was extracted from each replicate using the RNeasy kit (Qiagen). Complementary RNA was prepared from 1 μg of total RNA from each replicate as described using the MessageAmp II Biotin-Enhanced Signal Round aRNA amplification kit (Ambion). In brief, double-stranded artificial DNA was synthesized and biotin-labeled target artificial RNA was prepared by artificial DNA in vitro transcription in the presence of biotinylated UTP and CTP. After purification, artificial RNA was fragmented and used to hybridize the Arabidopsis ATH1 GeneChip array (Affymetrix). Hybridization, washing, staining, scanning, and data collection were performed for each replicate sample independently in an Affymetrix GeneChip Fluidics Station 450 and GeneArray Scanner. The microarray computational analysis was performed on CEL data files and analyzed using the robust multiarray average GC method (gcRMA) of the GeneSpring software (version 7.2; Agilent Technologies). Means of the three replicate values were analyzed for each data set. Data sets with expression levels below 50 were excluded from comparative analysis (noise level of expression cutoff). Genes were considered as induced or repressed if their mean expression level deviated >1.75 in a comparison of two samples. Statistical significance of gene expression was tested using a one-way analysis of variance test combined with a Benjamini and Hochberg false discovery rate multiple correction algorithm (GeneSpring version 7.2) with an adjusted P value < 0.05 set as cutoff. The gene tree was generated with the 148 genes identified as being GA regulated using the Pearson correlation and average linkage algorithms of the GeneSpring software. The microarray data were submitted to the Gene Expression Omnibus and are available under the accession numbers GSM177119 to GSM177121 (ga1 mutant experiment), GSM177122 to GSM177124 (ga1 mutant + GA3), GSM177125 to GSM177127 (gid1a-c mutant experiment), and GSM177128 to GSM177130 (gid1a-c mutant experiment + GA3).
Microscopy
The intracellular distribution of GID1:GFP was examined in roots of 35S:GIDGFP plants using a Leica TCS SP2 confocal microscope. Propidium iodide was used to outline the cell boundaries.
Yeast Two-Hybrid System
Yeast two-hybrid constructs were obtained by insertion of GAI and its mutant variants into the vector pGAD424(+2) as BamHI-PstI fragments excised from the pGREEN0179-based GAI constructs described above to generate AD:GAI, AD:gai, etc. (Roder et al., 1996). Constructs expressing only the DELLA domain (AD:D) and the DELLA and VHYNP domains (AD:D+V) were constructed by insertion of a PCR fragment obtained with the primers 5′-GGATCCAAGAGAGATCATCATCATC-3′ and 5′-CTGCAGTCAAGTCTCAGTAGCGAGTTGAGA-3′ to generate AD:D or 5′-GGATCCAAGAGAGATCATCATCATC-3′ and 5′-CTGCAGTCAAGCGAACTGATTGAGAATCGCGTC-3′ to generate AD:D+V. The GID1A open reading frame was obtained by RT-PCR from Arabidopsis ecotype Columbia mRNA and inserted in frame into the yeast two-hybrid vector pGBT9. Yeast two-hybrid interaction studies were performed in the yeast strains Y187 and Y190 using standard protocols (Schwechheimer and Deng, 2002). In brief, DBD and DBD:GID1A constructs were transformed into Y190 and mated with Y187 strains harboring AD, AD:GAI, AD:gai, etc. DBD and AD plasmids were selected on dropout medium lacking Leu and Trp. LacZ filter lift assays were performed with yeast clones grown on dropout medium lacking Leu and Trp in the absence and presence of 100 μM GA3. Quantitative LacZ assays were conducted in a Mithras LB940 luminometer (Berthold) using protein extracts prepared from clones grown in liquid dropout medium lacking Leu and Trp in the absence and presence of 100 μM GA3 using the Galacton-Star reagent (Tropix) as a luminescent substrate. Average relative light units/μg protein were determined from four replicates, and fold induction was calculated.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: ER (AT2G26330), GID1A (AT3G05120), GID1b (AT3G63010), GID1c (AT5G27320), GAI (AT1G14920), RGA (AT2G01570), and SLY1 (AT4G24210).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Arabidopsis GAI with Maize, Wheat, and Barley Orthologs, and Transgenic GAI:gai Plants Mimic the gai-1 Gain-of-Function Mutant Phenotype.
Supplemental Figure 2. Anti-GAI and Anti-RGA Antibodies Specifically Recognize the Respective DELLA Protein.
Supplemental Figure 3. Mutants Expressing Stabilized GAI Variants Are GA Insensitive.
Supplemental Figure 4. Growth of ga1 GA Biosynthesis Mutants but Not That of gid1 Triple Mutants Can Be Normalized by GA Treatment.
Supplemental Table 1. Genes That Are Repressed or Induced by 100 μM GA3 Treatment (1 h) in a ga1 Allele (SALK_109115, Columbia Ecotype) and Their Expression in the gid1a-c Triple Mutant.
Supplementary Material
Acknowledgments
We thank Gerd Jürgens and Erika Isono (Tübingen University) for critical comments on the manuscript. We also thank Nicholas Harberd (John Innes Centre, Norwich, UK) and Camille Steber (Washington State University, Pullman, WA) for sharing mutant seeds. This work is supported by grants from the Deutsche Forschungsgemeinschaft (Schw751/4 and Sch751/5) and the Sonderforschungsbereich SFB446 to C.S. and by a fellowship from the Landesgraduiertenförderung Baden-Württemberg to E.M.N.D.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Claus Schwechheimer (claus.schwechheimer@zmbp.uni-tuebingen.de).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Cao, D., Cheng, H., Wu, W., Soo, H.M., and Peng, J. (2006). Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, P.M., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of barley cv. Himalaya. Molecular and physiological characterization. Plant Physiol. 129 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D.E., Cao, D., Luo, D., Harberd, N.P., and Peng, J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055–1064. [DOI] [PubMed] [Google Scholar]
- Dill, A., Jung, H.-S., and Sun, T.-p. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., and Sun, T. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., Thomas, S.G., Hu, J., Steber, C.M., and Sun, T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck, B., and Harberd, N.P. (2002). Evidence that the Arabidopsis nuclear gibberellin signalling protein GAI is not destabilised by gibberellin. Plant J. 32 935–947. [DOI] [PubMed] [Google Scholar]
- Fu, X., Richards, D.E., Ait-Ali, T., Hynes, L.W., Ougham, H., Peng, J., and Harberd, N.P. (2002). Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X., Richards, D.E., Fleck, B., Xie, D., Burton, N., and Harberd, N.P. (2004). The Arabidopsis mutant sleepy1gar2–1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D., and Marshall, G.A. (1973). Insensitivity to gibberellins in dwarf wheats. Ann. Bot. (Lond.) 37 729–735. [Google Scholar]
- Gale, M.D., and Marshall, G.A. (1976). The chromosomal location of GAI 1 and RHT 1, genes for gibberellin insensitivity and semi-dwarfism, in a derivative of Norin 10 wheat. Heredity 37 283–289. [Google Scholar]
- Gomi, K., Sasaki, A., Itoh, H., Ueguchi-Tanaka, M., Ashikari, M., Kitano, H., and Matsuoka, M. (2004). GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 37 626–634. [DOI] [PubMed] [Google Scholar]
- Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.L., Powers, S.J., Gong, F., Phillips, A.L., Hedden, P., Sun, T.P., and Thomas, S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Chandler, P.M., White, R.G., Llewellyn, D.J., and Jacobsen, J.V. (2002). Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol. 129 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd, N., and Freeling, M. (1989). Genetics of dominant gibberellin-insensitive dwarfism in maize. Genetics 121 827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Sasaki, A., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Hasegawa, Y., Minami, E., Ashikari, M., and Matsuoka, M. (2005). Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol. 46 1392–1399. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.E., Moritz, T., and Harberd, N.P. (2001). Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., and van der Veen, J.H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 58 257–263. [DOI] [PubMed] [Google Scholar]
- Lee, S., Cheng, H., King, K.E., Wang, W., He, Y., Hussain, A., Lo, J., Harberd, N.P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis, K.M., Thomas, S.G., Soule, J.D., Strader, L.C., Zale, J.M., Sun, T.P., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46 880–889. [DOI] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., and Harberd, N.P. (1997). Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol. 113 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., et al. (1999. b). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Peng, J., Richards, D.E., Moritz, T., Cano-Delgado, A., and Harberd, N.P. (1999. a). Extragenic suppressors of the Arabidopsis gai mutation alter the dose–response relationship of diverse gibberellin responses. Plant Physiol. 119 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh, L.D., Wysocka-Diller, J.W., Camilleri, C., Bouchez, D., and Benfey, P.N. (1999). The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18 111–119. [DOI] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-Ali, T., and Harberd, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 67–88. [DOI] [PubMed] [Google Scholar]
- Roder, K.H., Wolf, S.S., and Schweizer, M. (1996). Refinement of vectors for use in the yeast two-hybrid system. Anal. Biochem. 241 260–262. [DOI] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., and Deng, X.W. (2002). Studying protein-protein interactions with the yeast two-hybrid system. In Molecular Plant Biology, P.M. Gilmartin and C. Bowler, eds (Oxford, UK: Oxford University Press), pp. 173–198.
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H.S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T.P. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Tseng, T.S., Swain, S.M., Dill, A., Jeong, S.Y., Olszewski, N.E., and Sun, T.P. (2007). Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 143 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, L., Thomas, S.G., Hu, J., Dill, A., Alonso, J.M., Ecker, J.R., and Sun, T.P. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.Y., Hsing, Y.I., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Winkler, R.G., and Freeling, M. (1994). Physiological genetics of the dominant gibberellin-nonresponsive maize dwarfs, Dwarf8 and Dwarf9. Planta 193 341–348. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.