Abstract
In yeast and mammals, the AAA ATPase Vps4p/SKD1 (for Vacuolar protein sorting 4/SUPPRESSOR OF K+ TRANSPORT GROWTH DEFECT1) is required for the endosomal sorting of secretory and endocytic cargo. We identified a VPS4/SKD1 homolog in Arabidopsis thaliana, which localizes to the cytoplasm and to multivesicular endosomes. In addition, green fluorescent protein–SKD1 colocalizes on multivesicular bodies with fluorescent fusion protein endosomal Rab GTPases, such as ARA6/RabF1, RHA1/RabF2a, and ARA7/RabF2b, and with the endocytic marker FM4-64. The expression of SKD1E232Q, an ATPase-deficient version of SKD1, induces alterations in the endosomal system of tobacco (Nicotiana tabacum) Bright Yellow 2 cells and ultimately leads to cell death. The inducible expression of SKD1E232Q in Arabidopsis resulted in enlarged endosomes with a reduced number of internal vesicles. In a yeast two-hybrid screen using Arabidopsis SKD1 as bait, we isolated a putative homolog of mammalian LYST-INTERACTING PROTEIN5 (LIP5)/SKD1 BINDING PROTEIN1 and yeast Vta1p (for Vps twenty associated 1 protein). Arabidopsis LIP5 acts as a positive regulator of SKD1 by increasing fourfold to fivefold its in vitro ATPase activity. We isolated a knockout homozygous Arabidopsis mutant line with a T-DNA insertion in LIP5. lip5 plants are viable and show no phenotypic alterations under normal growth conditions, suggesting that basal SKD1 ATPase activity is sufficient for plant development and growth.
INTRODUCTION
Endosomes are dynamic compartments with a variable biochemical composition, structure, and function (Gruenberg and Stenmark, 2004; Miaczynska et al., 2004). In animal cells, sorting endosomes recycle membrane proteins back to the plasma membrane or trans-Golgi network (TGN), sequester membrane proteins into internal vesicles for degradation, or retain proteins in their limiting membrane that, upon fusion with vacuoles, are delivered to the vacuolar membrane (Gruenberg and Stenmark, 2004; Russell et al., 2006). In addition, endosomes play important roles during trafficking of newly synthesized proteins from the Golgi to the vacuole, and for this reason they are often called prevacuolar compartments.
An important element of the degradation pathway in endosomes involves the formation of internal vesicles from invaginations of limiting membrane domains that contain membrane proteins targeted for degradation. These endosomes are therefore called multivesicular endosomes or multivesicular bodies (MVBs). When MVBs fuse with vacuoles/lysosomes, the internal vesicles are degraded in the vacuolar lumen by hydrolases (Reggiori and Pelham, 2001; Katzmann et al., 2002; Gruenberg and Stenmark, 2004).
The endosomal invagination process is unique because, unlike most characterized vesiculation processes, the vesiculating membrane buds away from the cytoplasm. This invagination process requires the concentration of membrane proteins into specific membrane domains and the initiation of budding into the endosomal lumen. Approximately 18 class E vacuolar protein sorting (VPS) proteins in yeast and 27 in mammals have been found to be involved in the endosomal invagination process (Babst, 2005). Ubiquitination of membrane-bound receptors is now well established as a signal for sorting into internal vesicles of MVBs (Hicke and Dunn, 2003; Gruenberg and Stenmark, 2004). When the ubiquitinated receptors reach the sorting endosome, flat clathrin coats highly enriched in hepatocyte growth factor–regulated Tyr kinase substrate form on the endosomal membranes (Raiborg et al., 2002; Murk et al., 2003). Three multisubunit complexes called ESCRT-I, -II, and -III (endosomal sorting complexes required for transport) are also required for protein sorting into MVB internal vesicles in yeast and mammalian cells (Katzmann et al., 2002, 2003; Bowers et al., 2004; Babst, 2005; Hurley and Emr, 2006). Once the three ESCRTs are assembled onto the endosomal membrane, an AAA (for ATPases associated with various cellular activities) ATPase called Vps4p in yeast and SUPPRESSOR OF K+ TRANSPORT GROWTH DEFECT1 (SKD1) in mouse is required for endosomal membrane invagination, presumably by releasing the ESCRTs from the membrane. The binding of Vps4p/SKD1 to the membrane is regulated by its ATPase cycle, being membrane-associated in its ATP-bound form and cytoplasmic in its ADP-bound form.
Vps4p dimerizes in vitro and assembles into large complexes (10 to 12 subunits) upon ATP binding (Babst et al., 1998). The N-terminal domain of VPS4p/SKD1 interacts with Vps20 (Yeo et al., 2003), one of the ESCRT III components, and with CHARGED MULTIVESICULAR BODY PROTEIN1 (CHMP1)/Did2p (Scott et al., 2005a; Nickerson et al., 2006; Vajjhala et al., 2006), whereas the C-terminal domain binds the adaptor proteins mammalian LYST-INTERACTING PROTEIN5/SKD1 BINDING PROTEIN1 (LIP5/SBP1) (Scott et al., 2005b) and yeast Vta1p (for Vps twenty associated 1 protein) (Yeo et al., 2003; Azmi et al., 2006). In addition, other class E VPS proteins, such as Vps2p, Bro1, and Snf7/CHMP4, have been also shown to interact with Vps4p/SKD1 (Bowers et al., 2004; Fujita et al., 2004; Lin et al., 2005). These results indicate that Vps4p/SKD1 is a crucial player during MVB formation. When VPS4 expression is suppressed or a dominant-negative (ATPase-deficient) form of Vps4p/SKD1 is overexpressed, aberrant endosomes called class E compartments arise. However, these endosomal alterations do not compromise cell viability in either yeast or mammalian cells (Babst et al., 1998; Nara et al., 2002; Lin et al., 2005). Vps4p/SKD1 proteins appear to be conserved across eukaryotes, and some organisms have more than one VPS4 paralog. For example, humans and other mammals express two closely related Vps4 proteins, VPS4A and VPS4B/SKD1, whereas budding yeast has a single Vps4 protein that is ∼60% identical to both human proteins.
As in animal and yeast cells, the endosomal/prevacuolar system in plants plays important roles in cellular functions. However, while there are undoubtedly many similarities between the endosomal systems of all eukaryotic cells, the endosomal/prevacuolar system of plants appears to have some unique properties. For example, plants are thought to have specialized mechanisms that allow individual cells to maintain a diversity of trafficking pathways (Surpin and Raikhel, 2004) and more than one type of vacuole (Paris et al., 1996; Robinson and Hinz, 1999; Otegui et al., 2005; Robinson et al., 2005). In addition, whereas mammalian cells have distinct early endosomes that received endocytosed membranes and recycle plasma membrane proteins, in plants, the TGN or a TGN subdomain appears to act as an early compartment in the endocytic pathway (Dettmer et al., 2006; Lam et al., 2007). Recent studies have also shown the central role of endocytosis and the endosomal compartment in key plant processes, such as embryo differentiation (Geldner, 2003), gravitropism (Silady et al., 2004), cell type specification (Shen et al., 2003), stomatal movement (Shope et al., 2003), cytokinesis (Dhonukshe et al., 2005; Spitzer et al., 2006), cell wall remodeling (Herman and Lamb, 1991; Baluska, 2002), and the regulation of auxin transport (Geldner et al., 2001; Grebe et al., 2003; Muday et al., 2003; Paciorek et al., 2005; Samaj et al., 2005; Jaillais et al., 2006; Sieburth et al., 2006). Plant endosomes are also important compartments in the trafficking of soluble vacuolar proteins from the Golgi to the vacuole (Jürgens, 2004; Kotzer et al., 2004; Surpin and Raikhel, 2004; Tse et al., 2004; Otegui et al., 2006).
Although it has become clear in recent years that endosomal compartments play important and complex roles during plant growth and development, the molecular machinery that regulates their functions is only partially known. The retromer complex that mediates the recycling of receptors for vacuolar proteins from endosomes back to the TGN in yeast and mammalian cells seems to be conserved in plants (Jaillais et al., 2006; Oliviusson et al., 2006; Shimada et al., 2006). Likewise, the class E VPS genes responsible for the degradation/invagination pathway in animal and yeast cells seem to have the corresponding homologs in plants, at least based on sequence similarity analyses (Mullen et al., 2006; Spitzer et al., 2006; Winter and Hauser, 2006). However, only a few plant class E VPS genes have been studied to date: a VPS4/SKD1 homolog in the ice plant Mesembryanthemum crystallinum (Jou et al., 2004; Jou et al., 2006), two putative CHMP1 homologs, Sal1 in Zea mays (Shen et al., 2003) and CHMP1 in Nicotiana benthamiana (Yang et al., 2003), and ELCH, an Arabidopsis homolog of Vps23p/TSG101, which is an ESCRTI subunit in yeast and mammals (Spitzer et al., 2006). However, the current knowledge on the specific functions of these genes in plant endosomal trafficking is very limited.
Here, we characterize Arabidopsis thaliana SKD1, a homolog of Vps4p/SKD1. We show that Arabidopsis SKD1 localizes to the cytoplasm and to endosomes. The overexpression of SKD1E232Q, which shows no ATPase activity in vitro, affects multivesicular endosome architecture and function and leads to a lethal phenotype in plants. We have also identified Arabidopsis LIP5 as a strong positive regulator of SKD1 activity. Interestingly, a lip5 null mutant does not exhibit phenotypic alterations in normal growth conditions, suggesting that basal SKD1 ATPase activity is sufficient for plant development and growth.
RESULTS
Identification of SKD1 Homologs in Arabidopsis
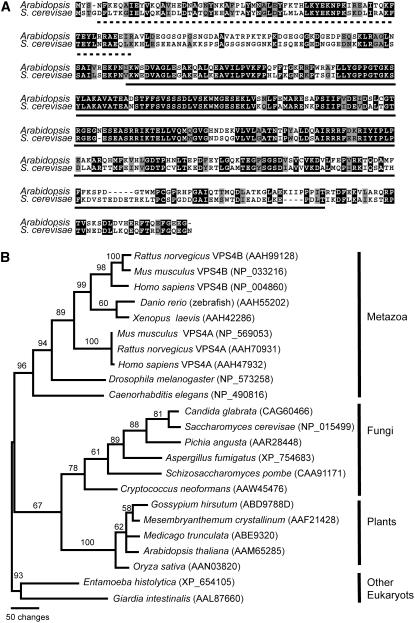
Based on a BLAST search, we identified only one putative VPS4/SKD1 homolog in the Arabidopsis genome, At2g27600 (SKD1). We isolated Arabidopsis SKD1 cDNA by RT-PCR of RNA extracted from leaves, roots, and flowers. The nucleotide sequence predicted a protein of 435 amino acid residues and ∼48.5 kD that is 54% identical and 67% similar to yeast Vps4p (Figure 1A). The analysis of the predicted protein sequence indicated that, like yeast Vps4p and mammalian SKD1 (Fujita et al., 2004; Scott et al., 2005a, 2005b; Takasu et al., 2005), At-SKD1 has a modular structure, with an N-terminal region containing a microtubule interacting and trafficking domain, a canonical AAA ATPase cassette, and a C-terminal domain (Figure 1A).
Figure 1.
Vps4p/SKD1 in Arabidopsis and Other Eukaryotes.
(A) Sequence alignment of the deduced amino acid sequences of Arabidopsis SKD1 (At2g27600) and Saccharomyces cerevisae Vps4p. The AAA ATPase domain is underlined.
(B) Phylogenetic analysis of SKD1/VPS4 sequences using PAUP*4.0b10. The bootstrap values are shown above each branch.
To understand the phylogenetic relationship of Vps4p/SKD1-related proteins in plants and other eukaryotes, we compared amino acid sequences from 23 phylogenetically diverse organisms. The resulting phylogram showed a well-supported clade (bootstrap value = 95%) containing all the multicellular organisms considered in this analysis (Figure 1B). Whereas fungi and plants seem to all contain only one copy of the gene, a gene duplication event in the vertebrate clade seems to account for the occurrence of two VPS4 copies in mammals, VPS4A and VPS4B/SKD1
Arabidopsis SKD1 Is Expressed in All Tissues and Localizes to the Cytoplasm and MVBs
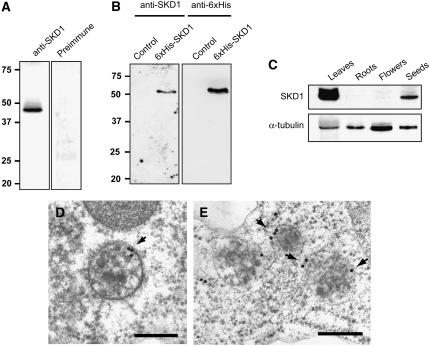
We raised peptide antibodies against At-SKD1. Since >170 proteins in Arabidopsis contain AAA ATPase domains (Winter and Hauser, 2006), we chose three peptides outside the AAA ATPase SKD1 cassette for raising antibodies. The serum containing polyclonal antibodies against the three peptides identified a band of ∼48 kD in Arabidopsis protein extracts (Figure 2A). To further characterize the affinity of the antisera, we expressed At-SKD1 in Escherichia coli fused to a 6xHis tag. The antibody recognized a band at ∼50 kD size only in the protein extracts from bacterial cells expressing 6xHis-SKD1 but not in the control (bacterial cells transformed with an empty plasmid; Figure 2B), suggesting the antibody does recognize the At-SKD1 protein.
Figure 2.
Distribution and Localization of SKD1 in Arabidopsis.
(A) Protein gel blot analysis of total protein extract from Arabidopsis plants with the anti-SKD1 peptide antibodies. The antibodies cross-reacted with a band of ∼48 kD. The preimmune serum did not recognize any band on the same extract.
(B) Protein gel blots of protein extracts from E. coli cells expressing 6xHis-tagged Arabidopsis SKD1 or control cells transformed with empty vector. The anti-SKD1 antibodies recognized a band ∼50 kD in the extract for 6xHis-SKD1–expressing cells but not in the control. A band of similar size was recognized by the anti-6xHis antibodies.
(C) Expression of SKD1 in different tissues. α-Tubulin was used as a loading control.
(D) and (E) Immunolocalization of SKD1 on plastic sections of Arabidopsis embryo cells. Labeling was detected on the cytoplasm and on discrete domains of the MVB-limiting membrane (arrows). Bars = 200 nm.
Immunoblot analysis of protein extracts from leaves, roots, flowers, and seeds indicates that SKD1 is expressed in all of these tissues but at different levels. The highest SKD1 protein level was detected in leaves, whereas the lowest level was in roots (Figure 2C).
Once the specificity of the antibodies was tested in immunoblots, we performed immunolabeling experiments on plastic sections of high-pressure frozen/freeze-substituted developing Arabidopsis embryos. The antibodies labeled only the cytoplasm and ∼60% of the MVBs analyzed (n = 50). Interestingly, the gold labeling seems to concentrate on discrete domains of the MVB limiting membrane (Figures 2D and 2E, arrows).
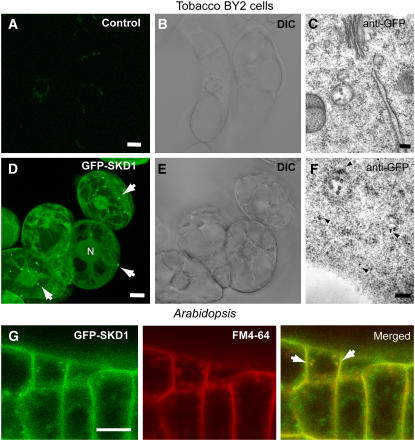
GFP-SKD1 Localizes to the Cytoplasm and to Multivesicular Endosomes in Tobacco BY2 Cells and in Arabidopsis
After confirming the localization of native SKD1 in cytoplasm and MVBs by immunogold labeling, we expressed At-SKD1 fused to green fluorescent protein (GFP) under the control of the 35S promoter of Cauliflower mosaic virus (CaMV35S) in tobacco (Nicotiana tabacum) Bright Yellow 2 (BY2) cells to analyze the dynamics of SKD1-positive compartments (Figures 3A to 3F). The fluorescence signal from GFP-SKD1 was localized to the cytoplasm and to small punctate structures distributed randomly throughout the cell (Figure 3D, arrows). In contrast with the localization of the SKD1 native protein revealed by immunolabeling, GFP fluorescent signal was also observed in the nucleus of the GFP-SKD1 BY2 cells (Figure 3). The nuclear GFP signal is likely due to the partial degradation of GFP-SKD1 as shown in immunoblots of total protein extracts (see Supplemental Figure 1 online). Gold immunolabeling studies on high-pressure frozen/freeze-substituted GFP-SKD1 BY2 cell lines showed that the fusion protein does localize to the cytoplasm and MVBs (Figure 3F), like the native SKD1 protein in nontransformed Arabidopsis cells (Figures 2D and 2E). We then obtained Arabidopsis plants stably expressing GFP-SKD1 under the control of the CaMV35S promoter and confirmed that the fusion protein shows the same localization pattern seen in tobacco BY2 cells (Figure 3G). Partial degradation of the GFP-SKD1 fusion protein was also observed in Arabidopsis (see Supplemental Figure 1 online). These transgenic plants grow normally, suggesting that the overexpression of GFP-SKD1 does not cause deleterious effects in plant cells.
Figure 3.
Expression of GFP-SKD1.
(A) to (F) Expression of GFP-AtSKD1 in tobacco BY2 cells. DIC, differential interference contrast.
(A) to (C) Untransformed control cells.
(D) to (F) Cells expressing Pro35S:GFP-AtSKD1. N, nucleus.
(D) Projection of 70 confocal slices. The GFP signal localizes to the cytoplasm and small structures (arrows).
(F) Immunogold labeling with anti-GFP on high-pressure frozen/freeze-substituted transgenic cells. Arrowheads indicate anti-GFP gold labeling on cytoplasm and MVBs.
(G) Transgenic Pro35S:GFP-SKD1 Arabidopsis roots incubated in FM4-64. Ten minutes after incubation, most of the GFP-SKD1–positive compartments were loaded with FM4-64 (arrows).
Bars = 10 μm.
To determine if the SKD1-positive MVBs are involved in the endocytic pathway, we incubated transgenic GFP-SKD1 Arabidopsis seedlings with FM4-64, a fluorescent marker that binds the plasma membrane and is internalized by endocytosis. We incubated transgenic seedling roots in 2 μM FM4-64 for 5 min on ice to minimize endocytosis, rinsed the seedlings, and followed the internalization of the dye at room temperature for 30 min. After 10 min, most of the internalized FM4-64 dye colocalized with GFP-SKD1–positive compartments in root cells, suggesting that GFP-SKD1 is targeted to an endosomal multivesicular compartment (Figure 3G). We performed similar experiments with FM 5-95, which is a less lipophilic analog of FM4-64, and found similar internalization timing and colocalization with GFP-SKD1 (data not shown).
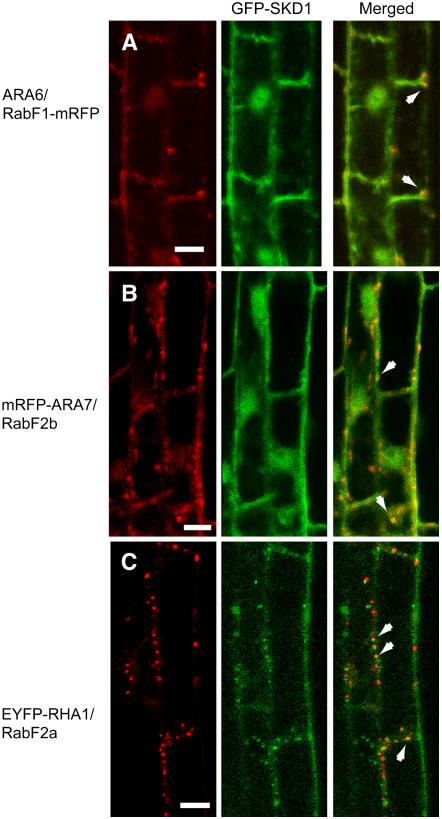
SKD1 Partially Colocalizes with the Endosomal Rab GTPases RHA1/RabF2a, ARA7/RabF2b, and ARA6/ RabF1 on MVBs
To further characterize the identity of the SKD1-positive endosomes, we analyzed the localization pattern of SKD1 in relationship to three endosomal Rab5-related GTPases, RHA1/RabF2a, ARA7/RabF2b, and ARA6/RabF1. We crossed the Pro35S:GFP-SKD1 plants with Arabidopsis lines expressing Pro35S:EYFP (enhanced yellow fluorescent protein)-RHA1/RabF2a, Pro35S:mRFP (monomeric red fluorescent protein)-ARA7/RabF2b, or Pro35S:ARA6/RabF1-mRFP. The colocalization analysis between GFP-SKD1 and the fluorescent fusions of RabGTPases was particularly difficult due to the strong cytosolic signal from SKD1-GFP. However, the careful analysis of the fluorescent signals in epidermal root cells showed partial colocalization of GFP-SKD1 with the three Rab GTPase fusion proteins on punctate structures, likely MVBs (Figures 4A to 4C). From the totality of recorded GFP-SKD1–positive compartments in epidermal cells from different lines, 79% of them were also labeled with EYFP-RHA1/RabF2a (n = 124), 74% were labeled with mRFP-ARA7/RabF2b (n = 142), and 69% were labeled with ARA6/RabF1-mRFP (n = 133).
Figure 4.
Coexpression of GFP-SKD1 and Fluorescent Fusion Proteins of Endosomal Rab GTPases in Arabidopsis.
(A) Partial colocalization of ARA6/RabF1-mRFP and GFP-SKD1 in Arabidopsis root epidermal cells.
(B) Partial colocalization of mRFP-ARA7/RabF2b and GFP-SKD1 in Arabidopsis root epidermal cells.
(C) Partial colocalization of EYFP-RHA1/RabF2a and GFP-SKD1 in Arabidopsis root epidermal cells.
Arrows indicate areas of colocalization. Bars = 10 μm.
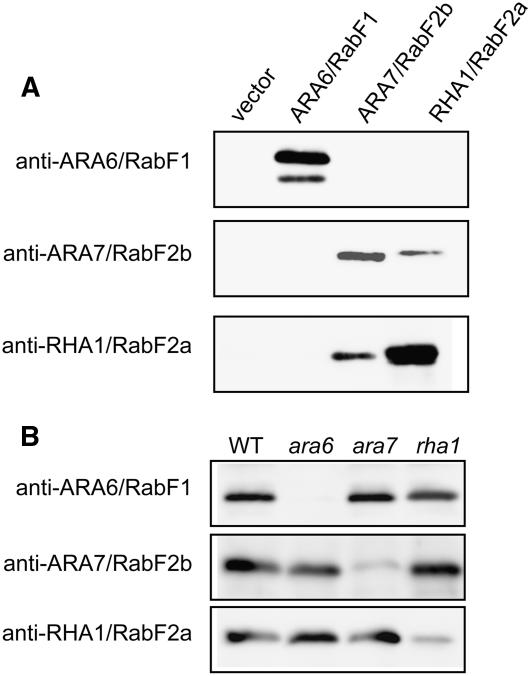
Since the expression of the GFP-SKD1 and the three endosomal Rab GTPase fluorescent fusion proteins was driven by the CaMV35S promoter, we decided to rule out protein mislocalization due to overexpression by immunolocalizing the native proteins in wild-type plants. We used previously characterized antibodies against ARA6/RabF1 (Ueda et al., 2001) and new antibodies against RHA1/RabF2a and ARA7/RabF2b. Since RHA1/RabF2a and ARA7/ RabF2b are 90% identical and 96% similar to each other, we tested the specificity of the three polyclonal antibodies on recombinant protein extracts from yeast cells expressing Arabidopsis RHA1/RabF2a, ARA7/RabF2b, and ARA6/RabF1 (Figure 5A). Whereas the anti-ARA6/RabF1 antibodies did not cross-react with the other two Rab5-related proteins, the other two antibodies recognized both RHA1/RabF2a and ARA7/RabF2b but with different affinities. The anti-ARA7/RabF2b showed higher affinity for ARA7/RabF2b, whereas the anti-RHA1/RabF2a showed higher affinity for RHA1/RabF2a. Neither the anti-ARA7/RabF2b nor the anti-RHA1/RabF2a antibodies recognized the ARA6/RabF1 protein. We further tested the specificity of the three antibodies on total protein extracts from wild-type Arabidopsis plants and knockout mutants with T-DNA insertions in RHA1/RabF2a, ARA7/RabF2b, and ARA6/RabF1 (Figure 5B). In all cases, the antibodies showed high specificity for the Rab5-related GTPases and did not recognize any other proteins in Arabidopsis.
Figure 5.
Characterization of Antisera against ARA6/RabF1, ARA7/RabF2b, and RHA1/RabF2a.
(A) Protein gel blots of recombinant Rab GTPases expressed in yeast with three polyclonal antibodies.
(B) Protein gel blots of total protein extracts from the wild type and ara6, ara7, and rha1 knockout Arabidopsis mutants.
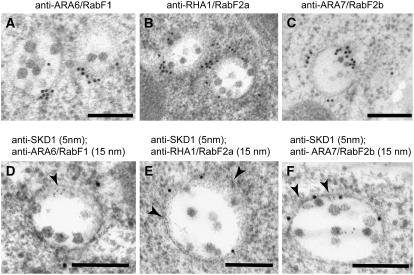
The three antibodies recognized epitopes on MVBs in plastic sections (Figures 6A to 6C). We then performed double immunogold labeling with antibodies against Arabidopsis SKD1 and against the three Rab5-type GTPases (Figures 6D to 6F). In all cases, the immunogold results support the partial colocalization of SKD1 and the three endosomal Rab GTPases on MVBs shown in the confocal analysis of the fluorescent proteins.
Figure 6.
Immunolocalization of Rab5-Like GTPases and SKD1 on High-Pressure Frozen/Freeze-Substituted Wild-Type Roots.
(A) to (C) Immunolocalization of ARA6/RabF1 (A), RHA1/RabF2a (B), and ARA7/RabF2b (C) with polyclonal antisera.
(D) to (F) Double immunolabeling of Rab5-related GTPases and SKD1. Arrowheads indicate SKD1 labeling (5-nm gold particles).
Bars = 200 nm.
The Expression of GFP-SKD1E232Q in Plant Cells Induces the Formation of Aberrant Endomembrane Compartments
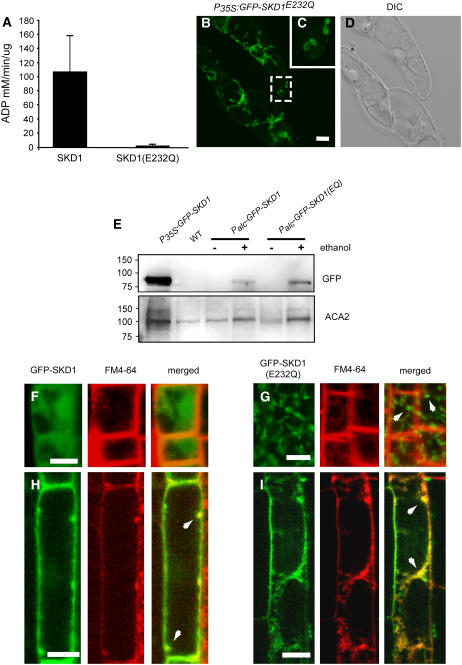
The expression of mutated versions of Vps4p/SKD1 that are not able to hydrolyze ATP in mammalian and yeast cells (SKD1E235Q and Vps4pE233Q, respectively) induces dominant-negative endosomal sorting defects. This mutation involves the change of a Glu residue to a Gln residue in a highly conserved region of the AAA ATPase domain, and it has been shown to block ATP hydrolysis in Vps4p and ice plant mc-SKD1 (Babst et al., 1997; Jou et al., 2006) and other AAA ATPases, such as N-ethyl-maleimide–sensitive factor (Whiteheart et al., 1994). To confirm that the equivalent mutation in At-SKD1 (i.e., E232Q) disrupts ATPase activity, we expressed recombinant SKD1 and SKD1E232Q in bacteria and tested the in vitro ATPase activity of both proteins by a colorimetric malachite green-based assay (see Methods). Whereas SKD1 was able to hydrolyze ATP in vitro, the ATPase activity of SKD1E232Q was seriously compromised, confirming that the mutation interferes with the ATPase activity of At-SKD1 (Figure 7A).
Figure 7.
Expression of GFP-SKD1E232Q.
(A) In vitro ATPase activity of recombinant SKD1 and SKD1E232Q measured by a malachite green-based colorimetric assay. The ATPase activities shown are mean values ± se of three repetitions.
(B) to (D) Expression of GFP-AtSKD1E232Q under the control of the CaMV35S promoter in tobacco BY2 cells.
(B) Projection of 70 confocal slices. Most of the fluorescent GFP signal comes from elongated membranous structures.
(C) Higher magnification view of the area indicated in (B).
(E) Protein gel blot analysis of expression levels of GFP-SKD1 proteins driven by the CaMV35S promoter and by the ethanol-inducible system. Antibodies against Arabidopsis Calcium ATPase2 (ACA2), an endoplasmic reticulum (ER)–localized protein, were used to test protein loading.
(F) and (G) Ethanol-induced expression of GFP-SKD1 (F) and GFP-SKD1E232Q (G) in Arabidopsis root meristematic cells incubated with FM4-64. Arrows indicate enlarged GFP-SKD1E232Q–positive compartments.
(H) and (I) Ethanol-induced expression of GFP-SKD1 (H) and GFP-SKD1E232Q (I) in Arabidopsis epidermal root cells incubated with FM4-64. Arrows indicate colocalization between GFP-SKD1E232Q and FM4-64 on enlarged organelles.
Bars = 10 μm in (B) and (F) to (I).
To test the effects of this mutation in the plant endomembrane system, we overexpressed GFP-At-SKD1E232Q in tobacco BY2 cells under the control of the CaMV35S promoter. We analyzed nine independently transformed BY2 lines and observed that the green fluorescent signal localizes to a convoluted membranous compartment but not to the cytoplasm (Figures 7B to 7D), consistent with the hypothesis that the ATP-bound form of SKD1 is recruited to endosomal membranes. These GFP-At-SKD1E232Q–positive compartments resemble enlarged endosomal/vacuolar hybrid compartments. However, all GFP-At-SKD1E232Q tobacco BY2 cell lines exhibited a very low growth rate and invariably died within a few weeks after transformation, suggesting that the overexpression of At-SKD1E232Q is lethal in plant cells.
We also attempted to isolate T-DNA lines for At-SKD1 to analyze loss-of-function mutant phenotypes. However, none of the analyzed lines showed altered gene expression (data not shown; see Methods for more information).
Ethanol-Inducible Expression of At-SKD1 and At-SKD1E232Q
To further analyze the effects of the E232Q mutation in At-SKD1, we attempted to transform Arabidopsis plants with the Pro35S:GFP-AtSKD1E232Q transgene but were unsuccessful, confirming our initial observation that the overexpression of SKD1E232Q causes a lethal phenotype in plants. We then expressed both GFP-SKD1 and GFP-SKD1E232Q under the control of an ethanol-inducible promoter (Sweetman et al., 2002). We exposed 1-week-old seedlings to ethanol vapors and observed the localization of the GFP signal in root cells 2 to 5 d after induction. Although expression levels were clearly heterogeneous in different areas of the root, we were able to observe both by immunoblot analysis and confocal imaging that the fusion proteins were expressed in the induced transgenic seedlings (Figures 7E to 7I). However, the levels of expression driven by the ethanol-inducible promoter were much lower than the expression levels in the Pro35S:GFP-SKD1 lines (Figure 7E). Confocal microscopy analysis of the ethanol-induced lines showed that whereas the GFP-SKD1 fusion protein is mostly cytoplasmic as in the Pro35S:GFP-SKD1 lines, the GFP-SKD1E232Q protein showed a wider distribution range, from mostly cytoplasmic to almost exclusively associated with enlarged membranous compartments both in meristematic and fully elongated root epidermal cells (Figures 7F to 7I). These enlarged GFP-SKD1E232Q–positive compartments were very similar to those observed in BY2 cells expressing Pro35S:GFP-SKD1E232Q. In epidermal cells, the enlarged GFP-SKD1E232Q–positive compartments stained with FM4-64 10 min after incubation (Figure 7I). In meristematic cells, the enlarged compartments also stained with FM4-64 but after much longer incubation times, likely due to a lower endocytosis rate in these cells. Since the expression levels were heterogeneous in different areas of the seedlings, we believe that the variation in the distribution pattern of GFP-SKD1E232Q is directly related to the expression level of the fusion protein and the amount of GFP-SKD1E232Q required for triggering the dominant-negative phenotype.
SKD1E232Q Induces MVB Enlargement and Interferes with the Pinching off of MVB Internal Vesicles
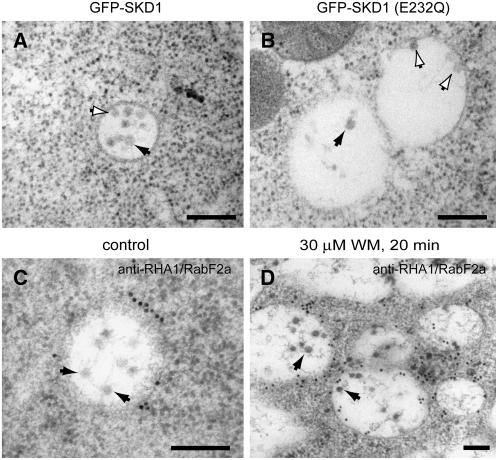
To understand the effects of the SKD1E232Q mutation at the ultrastructural level, we studied high-pressure frozen/freeze-substituted ethanol-induced seedlings. We found that whereas cells expressing GFP-SKD1 exhibit normal MVBs ∼280 nm in diameter and with numerous internal vesicles, cells expressing GFP-SKD1E232Q have a high proportion of enlarged MVBs with fewer internal vesicles (Table 1, Figures 8A and 8B). In both inducible lines, MVBs were labeled with anti-GFP antibodies (see Supplemental Figure 2 online), and only GFP-labeled MVBs were considered for the statistical analysis shown in Table 1. Inward budding profiles were frequently observed in the limiting membrane of both control and enlarged MVBs (Figure 8B). However, there was no significant difference in the diameter of the internal vesicles/budding profiles between MVBs in cells expressing either GFP-SKD1 or GFP-SKD1E232Q (P value = 0.132; see Table 1).
Table 1.
Quantitative Analysis of MVB Structural Features in Ethanol-Induced Seedlings Expressing GFP-SKD1 and GFP-SKD1E232Q
| MVB Diameter (in nm) | Free Vesicles/Cross Section | Inward Budding Profiles/MVB Cross Section | Inward Budding Profiles/Linear μm of Limiting Membrane | Diameter of Internal Vesicles (in nm) | |
|---|---|---|---|---|---|
| GFP-SKD1 | 285 (±45)an = 40 | 5.5 (±1.8) | 0.5 (±0.6) | 0.9 (±0.6) | 36.2 (±4.0) n = 74 |
| GFP-SKD1E232Q | 581 (±92)an = 32 | 1.3 (±1) | 1.2 (±1) | 0.6 (±0.5) | 37.5 (±3.3) n = 35 |
Difference in MVB diameter between the two lines is significant at P < 0.01.
Figure 8.
Electron Micrographs of MVBs in GFP-SKD1E232Q–Expressing Cells and in Wortmannin-Treated Cells.
(A) MVBs in control root cells expressing GFP-SKD1. Note the MVB luminal vesicles (black arrow) and an inward budding profile on the limiting membrane (white arrow).
(B) Enlarged MVBs in root cells expressing GFP-SKD1E232Q. Note the decrease in the number of luminal vesicles (black arrow). Numerous inward budding profiles are seen on the limiting membrane (white arrows).
(C) and (D) 30 μM wortmannin treatment on root cells and immunogold detection of RHA1/RabF2a.
(C) MVB in control cells. Arrows indicate luminal vesicles.
(D) Multivesicular compartments in wortmannin-treated cells with enlarged luminal vesicles (arrows).
Bars = 200 nm.
To compare the alteration in MVB architecture caused by SKD1E232Q to the effects of drugs known to affect endosome morphology and function, we also studied high-pressure frozen/freeze-substituted seedlings treated with 30 μM wortmannin, a fungal metabolite that specifically inhibits phosphatidylinositol 3-kinase (PI3) activity. Production of phosphatidylinositol 3-phosphate (PI3P) has been shown to be important for a variety of vesicle-mediated trafficking events, including endocytosis and sorting of receptors in multivesicular endosomes (Kjeken et al., 2001; Johnson et al., 2006). We found multivesicular compartments in wortmannin-treated root cells that ranged from ∼500 nm to 2 to 3 μm in diameter (Figure 8D). These last highly enlarged and pleiomorphic multivesicular organelles resemble MVB vacuolar hybrid compartments. To test if these compartments derived from MVBs, we performed immunogold experiments with endosomal markers. All multivesicular compartments in wortmannin-treated cells were labeled with the anti-RHA1/RabF2a antibodies, confirming their endosomal origin. In contrast with the MVBs seen in GFP-SKD1E232Q–expressing cells, the diameter of the internal vesicles in the wortmannin-induced compartments was significantly larger and more variable than in MVBs of control seedlings (54.95 ± 8.5 nm in wortmannin-treated MVBs versus 36.8 ± 3.5 in untreated control MVBs; n = 35; P < 0.05) (Figures 8C and 8D).
Arabidopsis LIP5 Interacts with SKD1 and Increases Its ATPase Activity in Vitro
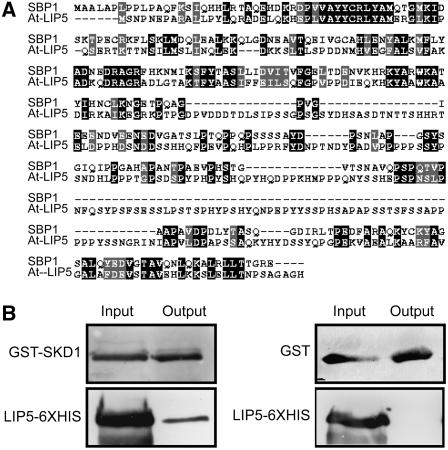
We performed a yeast two-hybrid screening using At-SKD1 as a bait. Amino acids 1 to 435 of the At-SKD1 protein were cloned in frame with the GAL4 DNA binding domain of the bait vector. The resulting bait vector was transformed into mating type A of strain PJ694 and tested for autoactivation of the β-galactosidase reporter gene. The yeast two-hybrid screen was conducted using a cDNA library from etiolated Arabidopsis (Col) seedlings. Approximately 18 million clones were screened via mating. Of these, 11 yeast lines tested positive (via selection on His dropout plus 1 mM 3-amino-1,2,4,-triazole and β-galactosidase assay) for interaction. Plasmids were rescued and analyzed via restriction digest. These isolated prey plasmids were retransformed into the alpha mating type of PJ694 and validated in a mating and selection assay with the Arabidopsis SKD1 bait, the empty bait vector, and unrelated baits. Out of seven confirmed positive clones, three were found to encode a predicted protein of 421 amino acids (At4g26750). This protein is 24% identical and 42% similar to murine SBP1/LIP5 (Figure 9A), which has been characterized as an SKD1 interactor (Fujita et al., 2004). As in mammals and yeast cells, there appears to be only one LIP5 gene in the Arabidopsis genome. We further tested this interaction by performing an in vitro pull-down assay between glutathione S-transferase (GST)-SKD1 and 6xHIS-LIP5 recombinant proteins expressed in bacteria. We found that GST-SKD1 but not GST alone binds to 6xHIS-LIP5 (Figure 9B).
Figure 9.
Arabidopsis LIP5 Interacts with SKD1.
(A) Protein alignment of murine SBP1 and Arabidopsis LIP5.
(B) In vitro pull-down assay to confirm interactions between LIP5 and GST-SKD1 but not between LIP5 and GST alone. Protein gel blots of recombinant proteins with the anti-GST and anti-6xHIS antibodies.
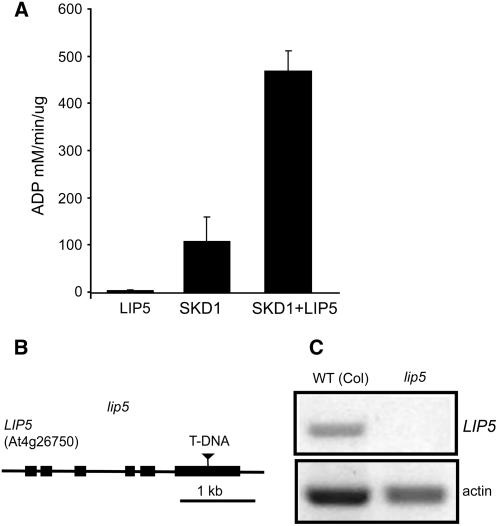
Vta1p, the yeast homolog of SBP1/LIP5, has been shown to be a positive regulator of Vps4p ATPase activity in vitro (Azmi et al., 2006; Lottridge et al., 2006). To determine how At-LIP5 influences SKD1 activity, we tested the in vitro ATPase activity of SKD1 in the presence or absence of LIP5. Whereas in the absence of LIP5, SKD1 hydrolyzes ∼106 mM of ATP per min/per μg, in the presence of recombinant LIP5, the hydrolysis rate shows a fourfold to fivefold increase (Figure 10A), indicating that LIP5 is a strong positive SKD1 regulator.
Figure 10.
LIP5 Function in Plants.
(A) In vitro ATPase activity of recombinant SKD1, LIP5, and SKD1+LIP5 measured in a malachite green-based colorimetric assay. The ATPase activities shown are mean values ± se of three repetitions.
(B) Position of the T-DNA insertion in the lip5 mutant.
(C) RT-PCR amplification of LIP5 and actin (control) from RNA extracts of wild-type and lip5 mutant plants.
Since blocking the ATPase activity of SKD1 seems to lead to lethal effects in plants, we asked whether mutant plants with no LIP5 protein show defects in development. We identified one T-DNA allele, lip5, in the SAIL T-DNA collection to study the loss-of-function phenotype. PCR amplification confirmed that lip5 contains a T-DNA insertion in the last exon of At4g26750 (Figure 10B). RT-PCR of RNA extracts of homozygous lip5 plants with primers flanking the insertion suggests that this is a null mutant (Figure 10C). Interestingly, lip5 mutant plants showed no apparent phenotypic defects in normal growth conditions, suggesting that basal levels of SKD1 activity are enough for plant cell function and development.
DISCUSSION
SKD1 Function May Play an Essential Role in Plant Endosomal Trafficking
The AAA ATPase protein superfamily is characterized by the presence of one or two copies of a highly conserved AAA ATPase module of ∼230 amino acid residues. AAA ATPases use the energy from ATP hydrolysis to induce conformational changes in other proteins or protein complexes causing unfolding or dissociation of substrate proteins (Hanson and Whiteheart, 2005). They are found in all organisms (Archaea, Eubacteria, and Eukaryota) and are essential for a variety of functions, such as vesicular transport, mitochondrial functions, peroxisome assembly, and proteolysis. The primary function of the AAA ATPase Vps4p/SKD1 appears to be the removal of the ESCRT machinery from the endosomal limiting membrane at one of the last steps in the MVB invagination pathway (Sachse et al., 2004; Babst, 2005). When Vps4p/SKD1 function is blocked, either in vps4Δ mutants or in cell lines expressing ATPase-deficient Vps4p/SKD1 proteins, both biosynthetic and endocytic pathways are highly compromised, and aberrant endosomes called class E compartments arise. The class E compartments that arise in mammalian cells when SKD1 function is blocked resemble enlarged MVBs with a reduced number of internal vesicles (Sachse et al., 2004), whereas in yeast, class E compartments are multicisternal structures (Babst et al., 1997; Nickerson et al., 2006). The mammalian class E compartment has been shown to be a hybrid organelle containing markers for Golgi/TGN, early and late endosomes, and lysosomes (Yoshimori et al., 2000; Fujita et al., 2003). However, neither in mammalian nor in yeast cells is Vps4p/SKD1 function required for cell viability. We have found that the overexpression of At-SKD1E232Q, an At-SKD1 form that is not able to hydrolyze ATP, causes dramatic alterations in the endomembrane system and cell death in plants. We also have evidence that the dominant-negative effect of SKD1E232Q is dosage dependent. Whereas high expression levels driven by the CaMV35S promoter result in a lethal phenotype, the lower expression levels achieved with the ethanol-inducible system caused enlargement of MVBs without compromising cell viability or plant development. The lethal phenotype caused by the overexpression of ATPase-deficient SKD1 in plants but not in yeast or mammalian cells suggests that SKD1 may play an essential role in plant cells.
In the halophyte M. crystallinum (ice plant), the expression of mc-SKD1 is salt induced and is especially high in cells engaged in secretory process, such as bladder cells in the leaves (Jou et al., 2004, 2006). However, most of the mc-SKD1 protein was found to localize to the ER and the TGN, not to endosomal compartments. This is hard to reconcile with numerous other reports that showed the localization of Vps4p/SKD1 primarily in the cytoplasm and on endosomes (Babst et al., 1997, 1998; Bishop and Woodman, 2000; Yoshimori et al., 2000; Scheuring et al., 2001; Fujita et al., 2003; Sachse et al., 2004; this study). The reason for this discrepancy may be due to either special SKD1 functions in halophytes or to the method used to detect mc-SKD1. Jou et al. used antibodies raised against the whole protein (Jou et al., 2006); therefore, they may have detected other AAA ATPases in their immunofluorescence studies.
SKD1 Function May Be Required for the Release of Internal Vesicles from the Limiting Membrane
The GFP-SKD1E232Q tobacco BY2 lines exhibit highly distorted membranous compartments, probably because of the high protein expression levels achieved with the CaMV35S promoter. Due to the compromised viability of the cells lines, we did not perform further experiments to study the changes in the endomembrane system. However, the controlled expression of the same fusion protein at lower levels in Arabidopsis plants with an ethanol-inducible system allowed us to study the effects of the SKD1E232Q mutation on MVBs. The reduction in the number of luminal vesicles in MVBs from ethanol-induced GFP-SKD1E232Q plants suggests that At-SKD1 may be involved in the formation of endosomal vesicles. The abundance of inward budding profiles in the limiting membrane of enlarged MVBs suggests that SKD1 may be involved in the fission/pinching off of the luminal vesicles but not in the initial vesiculation process of the limiting membrane. Moreover, the diameter of the MVB luminal vesicles in GFP-SKD1E232Q cells remains unaffected (Table 1), further suggesting that the initial steps in luminal vesicle formation do not depend on SKD1 activity. These results are in agreement with the structural studies performed in HeLa cells overexpressing the ATPase-deficient VPS4AE228Q protein (Sachse et al., 2004). In these cells, the number of endosomal inward budding profiles increased, and the authors hypothesized that the overexpression of VPS4AE228Q delays rather than completely inhibits the fission of inward membranes, increasing the chance of capturing vesiculation events in electron microscopy studies.
We also analyzed the MVB structure in wortmannin-treated root tips. Wortmannin is a potent inhibitor of PI3 kinase activity, blocking the phosphorylation of phosphatidylinositol to PI3P (Arcaro and Wymann, 1993; Wurmser and Emr, 1998). In mammalian and yeast cells, PI3P is highly enriched on luminal MVB vesicles (Gillooly et al., 2000). The synthesis of PI3P in endosomal membranes is mediated by the class III PI3 kinase VPS34, which is recruited to endosomal membranes by the myristoylated regulatory subunit Vps15p in yeast (Stack et al., 1995; Kihara et al., 2001) and p150 in mammals (Panaretou et al., 1997; Murray et al., 2002). Cultured mammalian cells treated with wortmannin develop enlarged endosomes/lysome hybrid compartments with a reduced number of luminal vesicles (Futter et al., 2001). Silencing of the human VPS34 gene leads to a similar phenotype (Johnson et al., 2006), suggesting that synthesis of PI3P is necessary for MVB luminal vesicle formation. In plants, most of the PI3P has been shown to localize to ARA7/RabF2b-positive endosomes (Voigt et al., 2005; Vermeer et al., 2006), and tobacco BY2 cells treated with 33 μM wortmannin for 1 h also showed distorted MVBs with less lumimal vesicles (Tse et al., 2004). We observed alterations in MVB architecture in Arabidopsis seedling roots treated with 30 μM wortmannin for 20 min. One of the most striking features of these wortmannin-treated MVBs is the presence of larger luminal vesicles than in control MVBs. This suggests that wortmannin is affecting early steps in the vesiculation process, upstream of SKD1 action, when the membrane budding areas are initially defined.
LIP5 Is Not Essential for Normal Plant Growth and Development
We have identified a putative LIP5/SBP1/Vta1p homolog in Arabidopsis that interacts with SKD1 both in yeast two-hybrid and in vitro pull-down assays. The fact that three independent clones identified by yeast two-hybrid screening encoded Arabidopsis LIP5 suggests that LIP5 is a strong SKD1 interactor. Vta1p and SBP1 interact with SKD1 through a highly conserved C-terminal domain to promote proper assembly of Vps4p/SKD1 oligomers and to increase Vps4p/SKD1 ATPase activity in vitro (Fujita et al., 2004; Scott et al., 2005b; Azmi et al., 2006; Lottridge et al., 2006). We have also shown that At-LIP5 is a strong positive regulator of At-SKD1 activity (Figure 10A), confirming the conservation of SKD1-LIP5 function in eukaryotes.
In yeast, the deletion on VTA1 causes different phenotypes in different genetic backgrounds, ranging from cells with no obvious endosomal defects to cells with class E compartments (Yeo et al., 2003; Shiflett et al., 2004; Azmi et al., 2006). In mammalian cells, depletion of LIP5 by gene silencing reduces degradation of the epidermal growth factor receptor that accumulates in enlarged endosomes (Ward et al., 2005). Although we have not studied the trafficking of specific endosomal cargo proteins in the Arabidopsis lip5 mutant, the normal development and growth of these mutant plants indicates that endosomal functions are not seriously compromised. These differences in the severity of the lip5/vta1Δ mutant phenotypes in mammalian cells, yeast, and plants may reflect differences in the intensity of endosomal trafficking. At least in yeast, Vps4p is still recruited to endosomes in the absence of Vta1p; therefore, basal Vps4p ATPase activity is expected to occur in vta1Δ cells (Azmi et al., 2006). Whereas this basal SKD1/Vps4p activity may be enough to assure MVB sorting in some cells/growth conditions, it may not be enough when the MVB trafficking pathway is upregulated. We are currently growing the Arabidopsis lip5 mutant plants in different environmental conditions that could demand higher levels of SKD1 activity and thus could reveal phenotypic alterations.
Are MVBs the Only Type of Endosomes in Plants?
Although the class E VPS machinery regulating endosomal sorting seems to be conserved across eukaryotes (Babst, 2005; Mullen et al., 2006; Spitzer et al., 2006; Winter and Hauser, 2006), the organization of the endosomal system seems to vary among organisms. In mammalian cells, the endosomal compartment consists of early/recycling, intermediate, and late endosomes. Early endosomes are tubulo-vesicular organelles enriched in Rab5, Rab4, and Rab21 GTPases (Simpson et al., 2004) and are the first compartment inside the cell where endocytosed molecules accumulate. Early endosomes form membranous projections rich in Rab4, which are believed to mature into recycling endosomes by separating from Rab5 endosomes and by recruiting Rab11 (Sonnichsen et al., 2000; De Renzis et al., 2002). Recycling receptors are sent back to the plasma membrane from both early and recycling endosomes. Early endosomes are believed to mature into intermediate endosomes at the time the inward budding process starts. In yeast, although the endosomal recycling function is present, no early endosomes have been unambiguously identified. The fact that the expression of Vps4p/SKD1 EQ proteins lead to two structurally different aberrant organelles in yeast and mammals further supports the notion that endosomes are not similar in all organisms.
In plants, although the endocytic/endosomal pathways are present (Geldner, 2003, 2004; Grebe et al., 2003; Baluska et al., 2004; Lee et al., 2004; Russinova et al., 2004; Tse et al., 2004; Aniento and Robinson, 2005; Haupt et al., 2005; Murphy et al., 2005; Takano et al., 2005; Voigt et al., 2005; Ortiz-Zapater et al., 2006; Robatzek et al., 2006), it is not completely clear how the plant endosomal system is organized. Whereas MVBs have been repeatedly observed in plant cells, the identification of typical tubulo-vesicular early endosomes has remained elusive. As mentioned above, mammalian early/recycling endosomes are enriched in specific Rab GTPases, such as Rab4, Rab5, Rab11, and Rab21. In plants, neither Rab4 nor Rab21 homologs have been identified (Ueda and Nakano, 2002; Vernoud et al., 2003), whereas the three Arabidopsis Rab5 homologs, ARA6/RabF1, RHA1/RabF2a, and ARA7/RabF2b, localize to multivesicular endosomes (this study), and three Rab11 plant homologs, Pra3, ARA4/RabA5c, and RabA4b, localize to the TGN (Ueda et al., 1996; Inaba et al., 2002; Preuss et al., 2004, 2006). Based on the transient expression of fluorescent fusion proteins of the three Rab5 homologs in Arabidopsis, two types of partially overlapping endosomal compartments have been defined: RHA1/RabF2a-ARA7/RabF2b-positive and ARA6/RaF1-positive endosomes (Ueda et al., 2004; Nielsen, 2005). RHA1/RabF2a-ARA7/RabF2b–positive endosomes seem to be involved in early steps of the endosomal pathway since they uptake FM4-64 before the ARA6/RaF1-positive endosomes do. GNOM, a GDP/GTP exchange factor for the ARF GTPases (Steinmann et al., 1999) that mediates the recycling of the auxin efflux carrier PIN1 from endosomes back to the plasma membrane (Geldner, 2003), appears to be important for functional integrity of RHA1/RabF2a-ARA7/RabF2b–positive endosomes but not for ARA6/RabF1-positive endosomes (Ueda et al., 2004). However, in this study, we have found that both types of endosomes are structurally alike (i.e., they are MVBs and are able to recruit SKD1). In addition, a plant homolog of the retromer subunit SNX1 (for sorting nexin 1), which is preferentially localized to early endosomes in mammalian cells and mediates recycling of receptors from endosomes back to the TGN (Seaman, 2005), colocalizes with both ARA7/RabF2b and ARA6/RabF1 but not with GNOM in plants (Jaillais et al., 2006). Three more plant retromer subunits, Arabidopsis VPS26, VPS29, and VPS39, have been unambiguously localized to MVBs (Oliviusson et al., 2006). In addition, our recent electron tomographic analysis of MVB formation in Arabidopsis embryos indicates that MVBs arise de novo from the fusion of Golgi/TGN vesicles and not from the maturation of early endosomes (Otegui et al., 2006).
Therefore, how many functionally different endosomes do plant cells have? Do plants have typical tubular early endosomes as mammalian cells do? Recent studies in plants indicate that the TGN itself or an immediate TGN derivative acts as an early endosome (Dettmer et al., 2006; Lam et al., 2007). Further evidence supporting the TGN as an early compartment in the plant endocytic pathway comes from electron microscopy studies tracking cationized ferritin in protoplasts. In these studies, ferritin deposits were first seen inside the cells (5 min after incubation) in Golgi stacks, TGN, and a compartment called “partially coated reticulum” (Joachim and Robinson, 1984; Tanchak et al., 1988; Galway et al., 1993). Ten minutes after incubation, ferritin was detected in MVBs. The partially coated reticulum was first described by Pesacreta and Lucas (1985) as a tubulo-reticular structure bearing clathrin coats. However, the partially coated reticulum is hard to differentiate from the TGN (Hillmer et al., 1988), and in fact, the partially coated reticulum and the TGN appear to incorporate cationized ferritin almost at the same time (Tanchak et al., 1988). The TGN has been shown to detach from the Golgi stacks and freely float in the cytoplasm in maize (Zea mays) root cells (Mollenhauer, 1971) and, more recently, in Pichia pastoris cells (Mogelsvang et al., 2003). Therefore, it is possible that the partially coated reticulum is in fact a TGN-derived compartment that acts as the first station in the endocytic pathway, receiving recently endocytosed material from the plasma membrane.
METHODS
Plant Material and Transformation
Seeds of Arabidopsis thaliana ecotype Columbia were sterilized in 50% bleach and 70% ethanol for 5 min and then washed with distillated water three times and germinated on Murashige and Skoog (MS)–agar plates supplemented with 20 μg/mL hygromycin. Plants were grown at 22 ± 3°C under 16-h-light/8-h-dark cycles. For induction, transgenic seeds were grown on MS-agar plates for 1 week. A piece of filter paper soaked with 50 μL of 70% ethanol was placed inside the plate. Seedlings were studied 3 to 5 d after induction.
The T-DNA insertional mutants lip5 (SAIL_854_F08), ara6 (SAIL_880-C07), ara7 (WiscDsLox355B06), and rha1 (SAIL_596-A03) were obtained from the ABRC. The following T-DNA lines with insertions in At-SKD1 were analyzed: GT_5_43384 (Exon Trapping Insert Consortium) and FLAG-450H05 and FLAG-461H04 (Institut National de la Recherche Agronomique).
Arabidopsis transformation was performed via floral dipping using Agrobacterium tumefaciens cultures (Clough and Bent, 1998).
Tobacco (Nicotiana tabacum) BY2 cells were cultured in a BY2 maintenance medium consisting of MS salts B1 (1 mg/L), inositol (0.1 g/L), 3% sucrose, KH2PO4 (180 mg/L), and 2,4-D (0.2 mg/L). All supplements were purchased from (Sigma-Aldrich). Cells were normally subcultured once a week by transferring 0.2 mL of old culture to 20 mL of fresh medium. Cultures were incubated in the dark at 26°C in a 120 rpm shaker. For transformation of BY2 cells, 5 mL of 3- to 5-d-old BY2 cell culture was mixed with 0.2 mL of overnight-transformed Agrobacterium culture and incubated at 26°C in a 120 rpm shaker for ∼30 h before plating on MS-agar plates containing carbenicillin (500 mg/L) and hygromycin (50 mg/L). After ∼20 d, stably transformed BY2 cell clones were picked and transferred to liquid MS medium supplemented with carbenicillin (500 mg/L) and hygromycin (50 mg/L).
Cloning of Arabidopsis SKD1 and Rab5-Related GTPases
Arabidopsis SKD1 cDNA was obtained by RT-PCR of RNA extracts from whole Arabidopsis seedlings. RNA extraction was performed with Trizol reagent (Invitrogen) and RT-PCR with the RT-PCR system kit from Promega. SKD1 was amplified using the forward primer 5′-CTCTAGAATGTACAGCAATTTCAAGG-3′ (SKD1-F) to generate an Xba1 site before the start codon and the reverse primer 5′-CGAGCTCTCAACCTTCTTCTCCAAAC-3′ (SKD1-R) to insert a SacI site after the stop codon. The coding region of GFP was amplified by PCR from the template pBin m-gfp5-ER (Haseloff et al., 1997) with the forward primer 5′-GAAAGTCGACATGAGTAAAGGAGAAG-3′ to introduce a SalI site before the start codon and the reverse primer 5′-CGAGCTCTCTAGATTTGTATAGTTCATCCAT-3′ to replace the HDEL ER retention signal and stop codon with an XbaI site. The SKD1 and GFP5 PCR-amplified fragments were cloned in frame in pBSII KS+ (Stratagene) and subsequently subcloned into pMZ215S (Matthew et al., 2000) using the SalI and SacI sites to obtain a expression cassette consisting of the CaMV35S promoter, the tobacco mosaic virus translational enhancer, the sequence encoding for the fusion GFP-SKD1, and the polyadenylation signal of nopaline synthase. The entire expression cassette was subcloned in the binary vector pCambia 1300 (Cambia) using the BamHI and EcoRI restriction sites. The E232Q mutation in Arabidopsis SKD1 was introduced using PCR techniques. First, two SKD1 fragments harboring the E232Q mutation were generated using the SKD1-F primer plus the reverse primer 5′-GTACCACACAAAGAATCTATCTGATCAACAAAAATAATCGAGGGAGCACTTTCACGGGCCATC-3′ (SKD1-R-EQ) and the SKD1-R primer plus the forward primer 5′-CCTCGATTATTTTTGTTGATCAGATAGATTCTTTGTGTGGTACACGTGGAGAAGGAAACGAGA-3′(SKD1-F-EQ). In a second PCR reaction, the two SKD1 fragments previously generated were used as template with the SKD1-F and SKD1-R primers to generate full-length SKD1E232Q.
To generate the ethanol-inducible system, we used the pbinSRNACatN binary vector (Sweetman et al., 2002) and subcloned SKD1 and SKD1E232Q after the AlcA promoter.
The plasmids for the transient expression of ARA6/RabF1 and ARA7/RabF2b tagged with mRFP (provided by Roger Y. Tsien, University of California, San Diego) were constructed as previously described (Ueda et al., 2004). Open reading frames (ORFs) of ARA6/RabF1 and ARA7/RabF2b were fused to the ORF of mRFP in the direction of 5′-ARA6/RabF1-mRFP-3′ and 5′-mRFP-ARA7/RabF2b-3′, respectively, and subcloned into the pHTS13, a derivative of pBSII KS+, which contains the CaMV35S promoter and the Nos terminator. The expression cassettes for 35S:ARA6/RabF1-mRFP and 35S:mRFP-ARA7/RabF2b were subcloned into pCambia1300 using the EcoRI and SalI restriction sites. The RHA1/RabF2a ORF was amplified using the forward primer 5′-CGGGATCCATGGCTACGTCTGGAAACAAGA-3′ and reverse primer 5′-GCTCTAGACTAAGCACAACA CGATGAACTC-3′ and inserted into pCambia 1300 with EYFP at the N terminus under the control of the CaMV35S promoter (Preuss et al., 2004).
The sequences of all constructs were confirmed by DNA sequencing. Binary vectors were introduced into Agrobacterium by electroporation.
Phylogenetic Analysis of Vps4p/SKD1 Amino Acid Sequences
Maximum parsimony analyses were performed in PAUP*4.0b10 (Swofford, 2001). Maximum parsimony heuristic searches used 100 random taxon addition replicates (holding one tree at each step) and tree-bisection-reconnection branch swapping. All characters were equally weighted, and gaps were treated as missing data. To estimate clade support, we conducted parsimony bootstrap analysis using 10,000 bootstrap replicates with simple taxon addition (holding one tree at each step) and tree-bisection-reconnection branch swapping.
Confocal Imaging of Fluorescent Proteins and FM4-64
For analyzing colocalization between GFP-SKD1 and FM4-64, 1-week-old Arabidopsis transgenic seedlings were placed in 2 μM FM4-64 (Invitrogen) in double distilled water for 5 min in ice. The seedlings were rinsed in fresh double distilled water and immediately imaged in a Zeiss 510 laser scanning confocal microscope equipped with a Meta detector (Zeiss). GFP was excited at 488 nm of an argon laser, and the emission was collected between 500 and 530 nm. FM4-64 was excited at 488 nm of an argon laser, and the emission was collected between 565 and 615 nm.
For imaging the lines expressing GFP-SKD1 and EYFP-RHA1/RabF2a, GFP was excited with a 458-nm wavelength of an argon line, and the emission collected using a 500- to 530-nm band-pass filter, whereas EYFP was excited at 514-nm wavelength, and the emission was collected using a 535- to 590-nm band-pass filter in a multitrack configuration.
For analyzing colocalization between GFP-SKD1 and the mRFP-Rab5 proteins, 1-week-old seedlings expressing both transgene constructs were used. GFP was excited at 488 nm, and the emission was collected between 500 and 530 nm. mRFP was excited at 543 nm, and the emission collected between 565 and 615 nm. Colocalization analysis was performed using ImageJ (http://rsb.info.nih.gov/ij/).
Images were edited using software provided by Zeiss for the 510 Meta and assembled for figures in Adobe Photoshop.
Antibodies
Peptide antibodies against three peptides of Arabidopsis SKD1 (IEYVKQAVHEDNAGNYNKAFP, ATRPKTKPKDGEGGGKDGE, and KTQDAMFFFKSPDGT) were raised in rabbits. The three peptides were injected in the same set of rabbits, so the resulting serum contained a mixture of the three peptide antibodies.
Anti-RabF1 antibody was raised in a rabbit using a GST-ARA6/RabF1 fusion protein as antigen as previously described (Ueda et al., 2001). For the production of anti-ARA7/RabF2b antibodies, the full-length ORF was amplified by PCR to generate BamHI sites at both ends and subcloned into pGEX-2T (GE Healthcare). This plasmid was introduced into Escherichia coli, and expression of recombinant protein, ARA7/RabF2b-GST, was induced by isopropylthio-β-galactoside. Recombinant ARA7/RabF2b-GST was purified from cell lysates by affinity chromatography on a glutathione-sepharose 4B column (GE Healthcare) and used to raise polyclonal antibodies in two rabbits. The antibodies obtained were purified by protein G affinity column chromatography (GE Healthcare). The cross-reactivity of these antibodies was tested for protein extracts from yeast cells expressing ARA6/RabF1, RHA1/RabF2a, and ARA7/RabF2b under the control of constitutive TDH3 promoter in pTU1, which is a derivative of pRS316 (Sikorsky and Hieter, 1989) containing the CMK1 terminator and the URA3 marker. Yeast lysates (10 μg protein per lane) were separated by SDS-PAGE and analyzed by immunoblotting. The anti-ARA6/RabF1 antibody was used at the dilution of ×200, and anti-RabF2b antibodies were used at ×1000 for immunoreaction.
For the production of the anti-RHA1/RabF2a antibodies, the RHA1/RabF2a ORF was PCR amplified and cloned into a pGEX vector for expression as GST fusion proteins. GST-RHA1/RabF2a was expressed in E. coli BL21 cells. The fusion protein was purified with glutathione-sepharose beads and used for experiments. During purification, RHA1/RabF2a was proteolytically cleaved from GST and eluted from the glutathione-sepharose beads. This cleaved RHA1/RabF2a protein was then used for antiserum production in rabbits. Prior to use, antibodies were affinity purified with recombinant RHA1/RabF2a fusion protein using standard antibody purification techniques (Preuss et al., 2004).
Commercial antibodies against GST (Sigma-Aldrich), 6xHis (Sigma-Aldrich), and GFP (Clontech) were used. The anti-ACA2 antibodies were provided by Jeffrey F. Harper (University of Nevada, Reno) (Hwang et al., 2000).
Electron Microscopy and Immunolabeling
Arabidopsis embryos and root tips and tobacco BY2 cells were high-pressure frozen/freeze-substituted for electron microscopy analysis as described previously (Otegui et al., 2002).
For immunolabeling, some high-pressure frozen samples were substituted in 0.2% uranyl acetate (Electron Microscopy Sciences) plus 0.2% glutaraldehyde (Electron Microscopy Sciences) in acetone at −80°C for 72 h and warmed to −50°C for 24 h. After several acetone rinses, these samples were infiltrated with Lowicryl HM20 (Electron Microscopy Sciences) during 72 h and polymerized at −50°C under UV light for 48 h. Sections were mounted on formvar-coated nickel grids and blocked for 20 min with a 5% (w/v) solution of nonfat milk in PBS containing 0.1% Tween 20. The sections were incubated in the primary antibodies (1:10 in PBS-Tween 20) for 1 h, rinsed in PBS containing 0.5% Tween 20, and then transferred to the secondary antibody (anti-rabbit IgG 1:10) conjugated to 15-nm gold particles for 1 h. Controls omitted either the primary antibodies or used the preimmune serum.
For double labeling experiments, plastic sections were first blocked with 5% (w/v) milk, incubated with the first primary antibody, followed by incubation in the goat anti-rabbit IgG conjugated to either 15- or 5-nm gold particles, as explained above. Following a fixation step (5% glutaraldehyde, 30 min) and a second blocking step with 5% milk, the grids were incubated with either no antisera or specific antisera for 1 h, followed by 1 h of incubation with anti-rabbit IgG linked directly to either 5- or 15-nm colloidal gold particles.
Protein Gel Blot Analysis
Total protein extracts were obtained by grinding tissues in protein extraction buffer (20 mM Tris-HCl, pH 7.5, 5 mM EDTA, and 2 μM β-mercaptoethanol) with protease inhibitors (Roche). The extracts were spun for 10 min at 4°C and the supernatant was loaded on SDS-PAGE gel with loading buffer. SDS-PAGE gel electrophoresis and blotting were done according to manufacturer's manual. The immune detection signals were visualized with the ECL Plus system (GE Healthcare).
In Vitro Interaction Assay
The LIP5 ORF was PCR amplified using the forward primer 5′-AAAGAATTCATGTCGAACCCAAACG-3′ and the reverse primer 5′-AAAGTCGACTCAGTGACCGGCACC-3′ from LIP5 cDNA obtained from the ABRC (clone U13054) and cloned into pET for expression of 6xHIS-LIP5 using the EcoRI and SalI sites added by the primers. The SKD1 and SKD1E232 coding regions were PCR amplified using the forward primer 5′-AAAGGATCCATGTACAGCAATTTCAAGGAAC-3′ and the reverse primer 5′-AAAGAATTCTCAACCTTCTTCTCCAAACTC-3′ from the plasmids previously generated for the expression of GFP-tagged proteins. The resulting products were cloned into the pET and pGEX plasmids at the BamHI and EcoRI sites to express 6xHis- and GST-tagged proteins, respectively. The resulting vectors were transformed into E. coli BL21 cells via electroporation and sequenced to confirm the expected sequence. GST fusion proteins were purified as explained above for the recombinant Rab GTPases with glutathione-sepharose beads. 6xHis-tagged proteins were purified using nickel-nitrilotriacetic acid agarose beads (Qiagen) according to the manufacturer's instructions.
For the interaction assay, 10 μL of each crude protein extract was added to 430 μL B/R buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 2 mM ATP, 2 mM CaCl2, 2mM MgCl2, 5 mM β-mercaptoethanol, 0.2% Nonidet P-40, and 5% glycerol) containing 50 μL of a 50% glutathione-sepharose bead slurry. The assay was incubated for 30 min at 4°C. The glutathione-sepharose beads were then rinsed three times with 0.5 mL B/R buffer and resuspended in SDS-PAGE loading buffer for analysis.
ATPase Assay
ATPase activity was measured by a modified malachite green-based colorimetric method (Carter and Karl, 1982; Kagami and Kamiya, 1995). Recombinant 6xHis-SKD1, 6xHis-SKD1E232Q, and 6xHIS-LIP5 were used for the assay. Protein concentration was determined with a Bradford assay kit (Bio-Rad) according to the manufacturer's instructions. ATPase reactions with 4 μg of each protein in a total volume of 20 μL were performed at 21°C in 20 mM HEPES buffer, pH 7.4, containing 150 mM KCl, 5 mM MgCl2, 5% glycerol, and 2 mM β-mercaptoethanol. Reactions were initiated by the addition of 1 mM ATP and stopped 20 min later by the addition of the malachite green reagent. A calibration curve was calculated using dilutions of K2HPO4 as a standard.
Wortmannin Treatment
One-week-old Arabidopsis seedlings grown on MS-agar plates were placed in 1.5 mL of 0.25× MS liquid media and allowed to equilibrate for 1 h. Seedlings were then transferred to 0.25× MS containing 30 μM wortmannin (Sigma-Aldrich), prepared from a 30 mM wortmannin stock in DMSO. Control seedlings were placed in a 0.25× MS solution containing 0.1% DMSO with no wortmannin. After 20 min, seedlings were transferred to fresh 0.25× MS and immediately processed by high-pressure freezing.
Yeast Two-Hybrid Screen
The yeast two-hybrid screen (Fields and Song, 1989) was performed at the Molecular Interaction Facility (University of Wisconsin) using yeast strains developed by Philip James (Durfee et al., 1993). Libraries were in pACT and pACT2 (Durfee et al., 1993) or pGAD-T7Rec (BD Biosciences) prey vectors. Amino acids 1 to 435 of the Arabidopsis SKD1 (At2g27600) protein were cloned in frame with the GAL4 DNA binding domain of bait vector pBUTE (a kanamycin-resistant version of GAL4 bait vector pGBDUC1). The resulting bait vector was sequenced to confirm an in-frame fusion, then transformed into mating type A of strain PJ694 and tested for autoactivation of the β-galactosidase reporter gene. The yeast two-hybrid screen was conducted using a cDNA library from etiolated Arabidopsis (Col) seedlings. Approximately 18 million clones were screened via mating. Of these, 11 yeast wells tested positive (via selection on histidine dropout plus 1 mM 3-amino-1,2,4-triazole and β-galactosidase assay) for interaction. Plasmids were rescued and analyzed via restriction digest. These isolated prey plasmids were retransformed into the alpha mating type of PJ694 and validated in a mating and selection assay with the Arabidopsis SKD1 bait, the empty bait vector, and unrelated baits. Seven clones were positive (grew in interaction selection media and β-galactosidase positive) in the validation test and subsequently were identified via sequencing.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are At2g27600 (SKD1), At4g26750 (LIP5), At3g54840 (ARA6/RabF1), At5g45130 (RHA1/RabF2a), and At4g19640 (ARA7/RabF2b).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Protein Gel Blot Analysis of Total Protein Extracts from Wild-Type and GFP-SKD1 Transgenic Lines of Tobacco BY2 Cells and Arabidopsis.
Supplemental Figure 2. Immunolocalization of GFP on MVBs of Control Root Cells Expressing GFP-SKD1 and GFP-SKD1E232Q.
Supplementary Material
Acknowledgments
We thank David Rancour (University of Wisconsin) for his help with the expression of recombinant proteins and the ATPase assay, Kathleen Dana (University of Colorado, Boulder) for her help with the transformation of BY2 cells, and Margaret Koopman (University of Wisconsin) for her assistance with the phylogenetic analysis. We also thank Andy Greenland (Syngenta, Norwich, UK) for the generous gift of the pbinSRNACatN plasmid, Roger Tsien (University of California, San Diego) for the mRFP marker, and Jeffrey F. Harper (University of Nevada, Reno) for ACA2 antibodies. This work was supported by a National Science Foundation grant (MCB-0619736) and a grant from the Antorchas Foundation (Argentina, 14022-14) to M.S.O. and a National Institutes of Health grant (GM065505) to G.O.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Marisa S. Otegui (otegui@wisc.edu).
Online version contains Web-only data.
References
- Aniento, F., and Robinson, D.G. (2005). Testing for endocytosis in plants. Protoplasma 226 3–11. [DOI] [PubMed] [Google Scholar]
- Arcaro, A., and Wymann, M.P. (1993). Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: The role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem. J. 296 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi, I., Davies, B., Dimaano, C., Payne, J., Eckert, D., Babst, M., and Katzmann, D.J. (2006). Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 172 705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M. (2005). A protein's final ESCRT. Traffic 6 2–9. [DOI] [PubMed] [Google Scholar]
- Babst, M., Sato, T.K., Banta, L.M., and Emr, S.D. (1997). Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16 1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., Wendland, B., Estepa, E.J., and Emr, S.D. (1998). The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17 2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska, F. (2002). F-actin-dependent endocytosis of cell wall pectins in meristematic root cells. Insights from brefeldin A-induced compartments. Plant Physiol. 130 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska, F., Samaj, J., Hlavacka, A., Kendrick-Jones, J., and Volkmann, D. (2004). Actin-dependent fluid-phase endocytosis in inner cortex cells of maize root apices. J. Exp. Bot. 55 463–473. [DOI] [PubMed] [Google Scholar]
- Bishop, N., and Woodman, P. (2000). ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell 11 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K., Lottridge, J., Helliwell, S.B., Goldthwaite, L.M., Luzio, J.P., and Stevens, T.H. (2004). Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic 5 194–210. [DOI] [PubMed] [Google Scholar]
- Carter, S.G., and Karl, D.W. (1982). Inorganic phosphate assay with malachite green: An improvement and evaluation. J. Biochem. Biophys. Methods 7 7–13. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- De Renzis, S., Sonnichsen, B., and Zerial, M. (2002). Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat. Cell Biol. 4 124–133. [DOI] [PubMed] [Google Scholar]
- Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y.-D., and Schumacher, K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe, P., Mathur, J., Hulskamp, M., and Gadella, T. (2005). Microtubule plus-ends reveal essential links between intracellular polarization and localized modulation of endocytosis during division-plane establishment in plant cells. BMC Biol. 3 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee, T., Becherer, K., Chen, P.L., Yeh, S.H., Yang, Y., Kilburn, A.E., Lee, W.H., and Elledge, S. (1993). The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7 555–569. [DOI] [PubMed] [Google Scholar]
- Fields, S., and Song, O. (1989). A novel genetic system to detect protein-protein interactions. Nature 20 245–246. [DOI] [PubMed] [Google Scholar]
- Fujita, H., Umezuki, Y., Imamura, K., Ishikawa, D., Uchimura, S., Nara, A., Yoshimori, T., Hayashizaki, Y., Kawai, J., Ishidoh, K., Tanaka, Y., and Himeno, M. (2004). Mammalian class E Vps proteins, SBP1 and mVps2/CHMP2A, interact with and regulate the function of an AAA-ATPase SKD1/Vps4B. J. Cell Sci. 117 2997–3009. [DOI] [PubMed] [Google Scholar]
- Fujita, H., Yamanaka, M., Imamura, K., Tanaka, Y., Nara, A., Yoshimori, T., Yokota, S., and Himeno, M. (2003). A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: Class E vps phenotype in mammalian cells. J. Cell Sci. 116 401–414. [DOI] [PubMed] [Google Scholar]
- Futter, C.E., Collinson, L.M., Backer, J.M., and Hopkins, C.R. (2001). Human VPS34 is required for internal vesicle formation within multivesicular endosomes. J. Cell Biol. 155 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway, M., Rennie, P., and Fowke, L. (1993). Ultrastructure of the endocytotic pathway in glutaraldehyde-fixed and high-pressure frozen/freeze-substituted protoplasts of white spruce (Picea glauca). J. Cell Sci. 106 847–858. [DOI] [PubMed] [Google Scholar]
- Geldner, N. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal re-cycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230. [DOI] [PubMed] [Google Scholar]
- Geldner, N. (2004). The plant endosomal system—Its structure and role in signal transduction and plant development. Planta 219 547–560. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Friml, J., Stierhof, Y.-D., Jurgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413 425–428. [DOI] [PubMed] [Google Scholar]
- Gillooly, D., Morrow, I., Lindsay, M., Gould, R., Bryant, N., Gaullier, J., Parton, R., and Stenmark, H. (2000). Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19 4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe, M., Xu, J., Mobius, W., Ueda, T., Nakano, A., Geuze, H.J., Rook, M.B., and Scheres, B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13 1378–1387. [DOI] [PubMed] [Google Scholar]
- Gruenberg, J., and Stenmark, H. (2004). The biogenesis of multivesicular bodies. Nat. Rev. Mol. Cell Biol. 5 317–323. [DOI] [PubMed] [Google Scholar]
- Hanson, P.I., and Whiteheart, S.W. (2005). AAA+ proteins: Have engine, will work. Nat. Rev. Mol. Cell Biol. 6 519–529. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt, S., Cowan, G.H., Ziegler, A., Roberts, A.G., Oparka, K.J., and Torrance, L. (2005). Two plant-viral movement proteins traffic in the endocytic recycling pathway. Plant Cell 17 164–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, E.M., and Lamb, C.J. (1991). Arabinogalactan-rich glycoproteins are localized on the cell surface and in intravacuolar multivesicular bodies. Plant Physiol. 98 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke, L., and Dunn, R. (2003). Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19 141–172. [DOI] [PubMed] [Google Scholar]
- Hillmer, S., Freundt, H., and Robinson, D.G. (1988). The partially coated reticulum and its relationship to the Golgi apparatus in higher plant cells. Eur. J. Cell Biol. 47 206–212. [Google Scholar]
- Hurley, J.H., and Emr, S.D. (2006). The ESCRT complexes: Structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35 277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I., Sze, H., and Harper, J.F. (2000). A calcium-dependent protein kinase can inhibit a calmodulin-stimulated Ca2+ pump (ACA2) located in the endoplasmic reticulum of Arabidopsis. Proc. Natl. Acad. Sci. USA 97 6224–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba, T., Nagano, Y., Nagasaki, T., and Sasaki, Y. (2002). Distinct localization of two closely related Ypt3/Rab11 proteins on the trafficking pathway in higher plants. J. Biol. Chem. 277 9183–9188. [DOI] [PubMed] [Google Scholar]
- Jaillais, Y., Fobis-Loisy, I., Miege, C., Rollin, C., and Gaude, T. (2006). AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443 106–109. [DOI] [PubMed] [Google Scholar]
- Joachim, S., and Robinson, D.G. (1984). Endocytosis of cationic ferritin by bean leaf protoplasts. Eur. J. Cell Biol. 34 212–216. [PubMed] [Google Scholar]
- Johnson, E.E., Overmeyer, J.H., Gunning, W.T., and Maltese, W.A. (2006). Gene silencing reveals a specific function of hVps34 phosphatidylinositol 3-kinase in late versus early endosomes. J. Cell Sci. 119 1219–1232. [DOI] [PubMed] [Google Scholar]
- Jou, Y., Chiang, C.-P., Jauh, G.-Y., and Yen, H.E. (2006). Functional characterization of ice plant SKD1, an AAA-Type ATPase associated with the endoplasmic reticulum-Golgi network, and its role in adaptation to salt stress. Plant Physiol. 141 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jou, Y., Chou, P., He, M., Hung, Y., and Yen, H. (2004). Tissue-specific expression and functional complementation of a yeast potassium-uptake mutant by a salt-induced ice plant gene mcSKD1. Plant Mol. Biol. 54 881–893. [DOI] [PubMed] [Google Scholar]
- Jürgens, G. (2004). Membrane trafficking in plants. Annu. Rev. Cell Dev. Biol. 20 481–504. [DOI] [PubMed] [Google Scholar]
- Kagami, O., and Kamiya, R. (1995). Nonradiactive method for ATPase assays. Methods Cell Biol. 47 147–150. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Odorizzi, G., and Emr, S.D. (2002). Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3 893–905. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Stefan, C.J., Babst, M., and Emr, S.D. (2003). Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, A., Noda, T., Ishihara, N., and Ohsumi, Y. (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjeken, R., Mousavi, S., Brech, A., Griffiths, G., and Berg, T. (2001). Wortmannin-sensitive trafficking steps in the endocytic pathway in rat liver endothelial cells. Biochem. J. 357 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzer, A.M., Brandizzi, F., Neumann, U., Paris, N., Moore, I., and Hawes, C. (2004). AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J. Cell Sci. 117 6377–6389. [DOI] [PubMed] [Google Scholar]
- Lam, S.K., Siu, C.L., Hillmer, S., Jang, S., An, G., Robinson, D.G., and Jiang, L. (2007). Rice SCAMP1 defines clathrin-coated, trans-Golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19 296–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, G.-J., Sohn, E.J., Lee, M.H., and Hwang, I. (2004). The Arabidopsis Rab5 homologs Rha1 and Ara7 localize to the prevacuolar compartment. Plant Cell Physiol. 45 1211–1220. [DOI] [PubMed] [Google Scholar]
- Lin, Y., Kimpler, L.A., Naismith, T.V., Lauer, J.M., and Hanson, P.I. (2005). Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J. Biol. Chem. 280 12799–12809. [DOI] [PubMed] [Google Scholar]
- Lottridge, J.M., Flannery, A.R., Vincelli, J.L., and Stevens, T.H. (2006). Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc. Natl. Acad. Sci. USA 103 6202–6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew, T.Z., Steven, R.T., and Kathleen, J.D. (2000). Accumulation of a thermostable endo-1,4-β-D-glucanase in the apoplast of Arabidopsis thaliana leaves. Mol. Breed. V6 37–46. [Google Scholar]
- Miaczynska, M., Pelkmans, L., and Zerial, M. (2004). Not just a sink: Endosomes in control of signal transduction. Curr. Opin. Cell Biol. 16 400–406. [DOI] [PubMed] [Google Scholar]
- Mogelsvang, S., Gomez-Ospina, N., Soderholm, J., Glick, B.S., and Staehelin, L.A. (2003). Tomographic evidence for continuous turnover of Golgi cisternae in Pichia pastoris. Mol. Biol. Cell 14 2277–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer, H.H. (1971). Fragmentation of mature dictyosome cisternae. J. Cell Biol. 49 212–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday, G.K., Peer, W.A., and Murphy, A.S. (2003). Vesicular cycling mechanisms that control auxin transport polarity. Trends Plant Sci. 8 301–304. [DOI] [PubMed] [Google Scholar]
- Mullen, R.T., McCartney, A.W., Flynn, C.R., and Smith, G.S.T. (2006). Peroxisome biogenesis and the formation of multivesicular peroxisomes during tombusvirus infection: A role for ESCRT? Can. J. Bot. 84 551–564. [Google Scholar]
- Murk, J.L.A.N., Humbel, B.M., Ziese, U., Griffith, J.M., Posthuma, G., Slot, J.W., Koster, A.J., Verkleij, A.J., Geuze, H.J., and Kleijmeer, M.J. (2003). Endosomal compartmentalization in three dimensions: Implications for membrane fusion. Proc. Natl. Acad. Sci. USA 100 13332–13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A.S., Bandyopadhyay, A., Holstein, S.E., and Peer, W.A. (2005). Endocytotic cycling of PM proteins. Annu. Rev. Plant Biol. 56 221–251. [DOI] [PubMed] [Google Scholar]
- Murray, J.T., Panaretou, C., Stenmark, H., Miaczynska, M., and Backer, J.M. (2002). Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic 3 416–427. [DOI] [PubMed] [Google Scholar]
- Nara, A., Mizushima, N., Yamamoto, A., Kabeya, Y., Ohsumi, Y., and Yoshimori, T. (2002). SKD1 AAA ATPase-dependent endosomal transport is involved in autolysosome formation. Cell Struct. Funct. 27 29–37. [DOI] [PubMed] [Google Scholar]
- Nickerson, D.P., West, M., and Odorizzi, G. (2006). Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 175 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, E. (2005). Rab GTPases in plant endocytosis. In Plant Endocytosis, J. Samaj, F. Baluska, and D. Menzel, eds (Berlin: Springer-Verlag), pp. 177–196.
- Oliviusson, P., Heinzerling, O., Hillmer, S., Hinz, G., Tse, Y.C., Jiang, L., and Robinson, D.G. (2006). Plant retromer, localized to the prevacuolar compartment and microvesicles in Arabidopsis, may interact with vacuolar sorting receptors. Plant Cell 18 1239–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Zapater, E., Soriano-Ortega, E., Marcote, M.J., Ortiz-Masia, D., and Aniento, F. (2006). Trafficking of the human transferrin receptor in plant cells: Effects of tyrphostin A23 and brefeldin A. Plant J. 48 757–770. [DOI] [PubMed] [Google Scholar]
- Otegui, M.S., Capp, R., and Staehelin, L.A. (2002). Developing seeds of Arabidopsis store different minerals in two types of vacuoles and in the endoplasmic reticulum. Plant Cell 14 1311–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M.S., Herder, R., Schulze, J.M., Jung, R., and Staehelin, L.A. (2006). The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18 2567–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M.S., Noh, Y.S., Martinez, D., Vila Petroff, M., Staehelin, L.A., Amasino, R.M., and Guiamet, J.J. (2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41 831–844. [DOI] [PubMed] [Google Scholar]
- Paciorek, T., Zazimalova, E., Ruthardt, N., Petrasek, J., Stierhof, Y.-D., Kleine-Vehn, J., Morris, D.A., Emans, N., Jurgens, G., Geldner, N., and Friml, J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435 1251–1256. [DOI] [PubMed] [Google Scholar]
- Panaretou, C., Domin, J., Cockcroft, S., and Waterfield, M.D. (1997). Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150·PtdIns 3-kinase complex. J. Biol. Chem. 272 2477–2485. [DOI] [PubMed] [Google Scholar]
- Paris, N., Stanley, C.M., Jones, R.L., and Rogers, J.C. (1996). Plant cells contain two functionally distinct vacuolar compartments. Cell 85 563–572. [DOI] [PubMed] [Google Scholar]
- Pesacreta, T., and Lucas, W. (1985). Presence of a partially coated reticulum in angiosperms. Protoplasma 125 173–184. [Google Scholar]
- Preuss, M.L., Serna, J., Falbel, T.G., Bednarek, S.Y., and Nielsen, E. (2004). The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16 1589–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss, M.L., Schmitz, A.J., Thole, J.M., Bonner, H.K.S., Otegui, M.S., and Nielsen, E. (2006). A role for the RabA4b effector protein PI-4Kβ1 in polarized expansion of root hair cells in Arabidopsis thaliana. J. Cell Biol. 172 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg, C., Bache, K.C., Gillooly, D.J., Madshus, I.H., Stang, E., and Stenmark, H. (2002). Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4 394–398. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., and Pelham, H.R.B. (2001). Sorting of proteins into multivesicular bodies: Ubiquitin-dependent and ubiquitin-independent targeting. EMBO J. 20 5176–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek, S., Chinchilla, D., and Boller, T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D.G., and Hinz, G. (1999). Golgi-mediated transport of seed storage proteins. Seed Sci. Res. 9 267–283. [Google Scholar]
- Robinson, D.G., Oliviusson, P., and Hinz, G. (2005). Protein sorting to the storage vacuoles of plants: A critical appraisal. Traffic 6 615–625. [DOI] [PubMed] [Google Scholar]
- Russell, M.R.G., Nickerson, D.P., and Odorizzi, G. (2006). Molecular mechanisms of late endosome morphology, identity and sorting. Curr. Opin. Cell Biol. 18 422–428. [DOI] [PubMed] [Google Scholar]
- Russinova, E., Borst, J.-W., Kwaaitaal, M., Cano-Delgado, A., Yin, Y., Chory, J., and de Vries, S.C. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16 3216–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse, M., Strous, G.J., and Klumperman, J. (2004). ATPase-deficient hVPS4 impairs formation of internal endosomal vesicles and stabilizes bilayered clathrin coats on endosomal vacuoles. J. Cell Sci. 117 1699–1708. [DOI] [PubMed] [Google Scholar]
- Samaj, J., Read, N.D., Volkmann, D., Menzel, D., and Baluska, F. (2005). The endocytic network in plants. Trends Cell Biol. 15 425–433. [DOI] [PubMed] [Google Scholar]
- Scheuring, S., Röhricht, R.A., Schöning-Burkhardt, B., Beyer, A., Müller, S., Abts, H.F., and Köhrer, K. (2001). Mammalian cells express two VPS4 proteins both of which are involved in intracellular protein trafficking. J. Mol. Biol. 312 469–480. [DOI] [PubMed] [Google Scholar]
- Scott, A., Chung, H., Gonciarz-Swiatek, M., Hill, G., Whitby, F., Gaspar, J., Holton, J., Viswanathan, R., Ghaffarian, S., Hill, C., and Sundquist, W. (2005. b). Structural and mechanistic studies of VPS4 proteins. EMBO J. 24 3658–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A., Gaspar, J., Stuchell-Brereton, M.D., Alam, S.L., Skalicky, J.J., and Sundquist, W.I. (2005. a). Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. USA 102 13813–13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, M.N.J. (2005). Recycle your receptors with retromer. Trends Cell Biol. 15 68–75. [DOI] [PubMed] [Google Scholar]
- Shen, B., Li, C., Min, Z., Meeley, R.B., Tarczynski, M.C., and Olsen, O.-A. (2003). Sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein. Proc. Natl. Acad. Sci. USA 100 6552–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiflett, S.L., Ward, D.M., Huynh, D., Vaughn, M.B., Simmons, J.C., and Kaplan, J. (2004). Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J. Biol. Chem. 279 10982–10990. [DOI] [PubMed] [Google Scholar]
- Shimada, T., Koumoto, Y., Li, L., Yamazaki, M., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2006). AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47 1187–1194. [DOI] [PubMed] [Google Scholar]
- Shope, J.C., DeWald, D.B., and Mott, K.A. (2003). Changes in surface area of intact guard cells are correlated with membrane internalization. Plant Physiol. 133 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth, L.E., Muday, G.K., King, E.J., Benton, G., Kim, S., Metcalf, K.E., Meyers, L., Seamen, E., and Van Norman, J.M. (2006). SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18 1396–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorsky, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silady, R.A., Kato, T., Lukowitz, W., Sieber, P., Tasaka, M., and Somerville, C.R. (2004). The gravitropism defective 2 mutants of Arabidopsis are deficient in a protein implicated in endocytosis in Caenorhabditis elegans. Plant Physiol. 136 3095–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, J.C., Griffiths, G., Wessling-Resnick, M., Fransen, J.A.M., Bennett, H., and Jones, A.T. (2004). A role for the small GTPase Rab21 in the early endocytic pathway. J. Cell Sci. 117 6297–6311. [DOI] [PubMed] [Google Scholar]
- Sonnichsen, B., De Renzis, S., Nielsen, E., Rietdorf, J., and Zerial, M. (2000). Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 149 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer, C., Schellmann, S., Sabovljevic, A., Shahriari, M., Keshavaiah, C., Bechtold, N., Herzog, M., Muller, S., Hanisch, F.-G., and Hulskamp, M. (2006). The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133 4679–4689. [DOI] [PubMed] [Google Scholar]
- Stack, J.H., DeWald, D.B., Takegawa, K., and Emr, S.D. (1995). Vesicle-mediated protein transport: Regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Gaweiler, L., Palme, K., and Jurgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Surpin, M., and Raikhel, N. (2004). Traffic jams affect plant development and signal transduction. Nat. Rev. Mol. Cell Biol. 5 100–109. [DOI] [PubMed] [Google Scholar]
- Sweetman, J.P., Chu, C., Qu, N., Greenland, A.J., Sonnewald, U., and Jepson, I. (2002). Ethanol vapor is an efficient inducer of the alc gene expression system in model and crop plant species. Plant Physiol. 129 943–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. (2001). PAUP*b10: Phylogenetic Analysis Using Parsimony. (Sunderland, MA: Sinauer Associates).
- Takano, J., Miwa, K., Yuan, L., von Wiren, N., and Fujiwara, T. (2005). Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 102 12276–12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu, H., Jee, J.G., Ohno, A., Goda, N., Fujiwara, K., Tochio, H., Shirakawa, M., and Hiroaki, H. (2005). Structural characterization of the MIT domain from human Vps4b. Biochem. Biophys. Res. Commun. 334 460–465. [DOI] [PubMed] [Google Scholar]
- Tanchak, M.A., Rennie, P., and Fowke, L. (1988). Ultrastructure of the partially coated reticulum and dictyosomes during endocytosis by soybean protoplasts. Planta 175 433–441. [DOI] [PubMed] [Google Scholar]
- Tse, Y.C., Mo, B., Hillmer, S., Zhao, M., Lo, S.W., Robinson, D.G., and Jiang, L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16 672–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, T., Anai, T., Tsukaya, H., Hirata, A., and Uchimiya, H. (1996). Characterization and subcellular localization of a small GTP-binding protein (Ara-4) from Arabidopsis: Conditional expression under control of the promoter of the gene for heat-shock protein HSP81-1. Mol. Gen. Genet. 250 533–539. [DOI] [PubMed] [Google Scholar]
- Ueda, T., and Nakano, A. (2002). Vesicular traffic: An integral part of plant life. Curr. Opin. Plant Biol. 5 513–517. [DOI] [PubMed] [Google Scholar]
- Ueda, T., Uemura, T., Sato, M.H., and Nakano, A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40 783–789. [DOI] [PubMed] [Google Scholar]
- Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a plant unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20 4730–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajjhala, P.R., Wong, J.S., To, H.-Y., and Munn, A.L. (2006). The β domain is required for Vps4p oligomerization into a functionally active ATPase. FEBS J. 273 2357–2373. [DOI] [PubMed] [Google Scholar]
- Vermeer, J.E.M., van Leeuwen, W., Tobena-Santamaria, R., Laxalt, A.M., Jones, D.R., Divecha, N., Gadella, T.W.J., and Munnik, T. (2006). Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47 687–700. [DOI] [PubMed] [Google Scholar]
- Vernoud, V., Horton, A.C., Yang, Z., and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt, B., et al. (2005). Actin-based motility of endosomes is linked to the polar tip growth of root hairs. Eur. J. Cell Biol. 84 609–621. [DOI] [PubMed] [Google Scholar]
- Ward, D.M., Vaughn, M.B., Shiflett, S.L., White, P.L., Pollock, A.L., Hill, J., Schnegelberger, R., Sundquist, W.I., and Kaplan, J. (2005). The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 280 10548–10555. [DOI] [PubMed] [Google Scholar]
- Whiteheart, S.W., Rossnagel, K., Buhrow, S.A., Brunner, M., Jaenicke, R., and Rothman, J.E. (1994). N-ethylmaleimide-sensitive fusion protein: A trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J. Cell Biol. 126 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, V., and Hauser, M.-T. (2006). Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 11 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser, A.E., and Emr, S.D. (1998). Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 17 4930–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, K.-S., Jin, U.-H., Kim, J., Song, K., Kim, S.J., Hwang, I., Lim, Y.P., and Pai, H.-S. (2003). Molecular characterization of NbCHMP1 encoding a homolog of human CHMP1 in Nicotiana benthamiana. Mol. Cells 17 255–261. [PubMed] [Google Scholar]
- Yeo, S.C.L., et al. (2003). Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae. J. Cell Sci. 116 3957–3970. [DOI] [PubMed] [Google Scholar]
- Yoshimori, T., Yamagata, F., Yamamoto, A., Mizushima, N., Kabeya, Y., Nara, A., Miwako, I., Ohashi, M., Ohsumi, M., and Ohsumi, Y. (2000). The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell 11 747–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.