Abstract
An Arabidopsis thaliana leaf-variegated mutant yellow variegated2 (var2) results from loss of FtsH2, a major component of the chloroplast FtsH complex. FtsH is an ATP-dependent metalloprotease in thylakoid membranes and degrades several chloroplastic proteins. To understand the role of proteolysis by FtsH and mechanisms leading to leaf variegation, we characterized the second-site recessive mutation fu-gaeri1 (fug1) that suppressed leaf variegation of var2. Map-based cloning and subsequent characterization of the FUG1 locus demonstrated that it encodes a protein homologous to prokaryotic translation initiation factor 2 (cpIF2) located in chloroplasts. We show evidence that cpIF2 indeed functions in chloroplast protein synthesis in vivo. Suppression of leaf variegation by fug1 is observed not only in var2 but also in var1 (lacking FtsH5) and var1 var2. Thus, suppression of leaf variegation caused by loss of FtsHs is most likely attributed to reduced protein synthesis in chloroplasts. This hypothesis was further supported by the observation that another viable mutation in chloroplast translation elongation factor G also suppresses leaf variegation in var2. We propose that the balance between protein synthesis and degradation is one of the determining factors leading to the variegated phenotype in Arabidopsis leaves.
INTRODUCTION
Differentiation of plastids into chloroplasts in higher plants is a light-dependent and organ-specific process (Lopez-Juez and Pyke, 2005). Morphologically, it accompanies rapid formation of the thylakoid membrane where light-capturing antenna and protein complexes of photosynthetic electron transfer and ATP synthesis are assembled. Mutations that perturb chloroplast development and thylakoid formation have been isolated by forward and reverse genetic approaches (Leister, 2003). These mutations are often completely lethal and may result in embryonic or seedling lethality. Mutants also commonly have pale-colored or variegated leaves.
Leaf variegation is derived from a formation of sectors that contain either normal-appearing chloroplasts or abnormal plastids and thus represent a unique category of the chlorotic mutants (Kirk and Tilney-Bassett, 1978; Sakamoto, 2003; Aluru et al., 2006). Single-recessive mutations that cause variegated or striped patterns are known in many species (e.g., Wang et al., 2000; Curie et al., 2001; Yaronskaya et al., 2003). Since the variegation defects do not interfere with proper chloroplast development in green sectors, such variegated mutants are more useful for studying thylakoid biogenesis than pale and albino mutants. Characterization of the genes responsible for leaf variegation demonstrated that variegation occurs through various chloroplast functions (Sakamoto, 2003; Aluru et al., 2006). A fundamental question is how two types of cells, one with normal chloroplasts and the other with abnormal plastids, can form under the same genetic background. In other words, why aren't all of the cells evenly distributed with defective chloroplasts? White sectors in variegation may not simply represent dead tissues, since their phenotypes are apparently different from those of cell death mutants (e.g., Mach et al., 2001; Gray et al., 2002; Mateo et al., 2004). One interesting hypothesis is that forming variegated sectors itself may help plants adapt to environmental changes, since variegated leaves are frequently observed in nature (Tsukaya et al., 2004).
We have been studying leaf-variegated mutants yellow variegated1 (var1) and var2, which exhibit a typical leaf-variegated phenotype in Arabidopsis thaliana. The responsible genes VAR1 and VAR2 encode different FtsH metalloproteases (FtsH5 and FtsH2, respectively) in thylakoid membranes (Chen et al., 2000; Takechi et al., 2000; Sakamoto et al., 2002). FtsH is a membrane-anchored ATP-dependent protease that belongs to the AAA+ protein family. Members of the AAA+ family are universally found in living organisms (Neuwald et al., 1999; Ogura and Wilkinson, 2001; Ito and Akiyama, 2005). The ATP binding domain of AAA+ proteins forms a hexameric ring structure (Krzywda et al., 2002; Bieniossek et al., 2006). Arabidopsis has 12 FtsH homologs, and nine of them, including FtsH2 and FtsH5, are targeted to chloroplasts (Sakamoto et al., 2003). FtsH2 and FtsH5 are the most abundant among all chloropalstic FtsHs and are constitutively expressed at high levels. Several proteins were suggested to be the substrates of FtsHs (see Sakamoto, 2006).
In Arabidopsis, the variegated phenotype is specific to the loss of FtsH2/VAR2 and FtsH5/VAR1, whereas the knockouts of other FtsH genes do not show any visible phenotype (Sakamoto et al., 2003). FtsH2 and FtsH5 appear to form a major FtsH heterocomplex of ∼400 kD. There are two types of chloroplast FtsHs, A and B, represented by the pairs of FtsH1/FtsH5 and FtsH2/FtsH8, respectively (Sakamoto et al., 2003; Yu et al., 2004; Zaltsman et al., 2005b). Complementation analysis of var1 and var2 with different FtsH genes suggested that the two types are functionally distinguishable and are required for rescuing variegation (Yu et al., 2004, 2005). Genetic analysis of the double mutants ftsh1 fths5 and ftsh2 ftsh8 also supports the idea that the two types of FtsH are necessary for proper chloroplast development (Zaltsman et al., 2005b). Since the two types of FtsHs are conserved among higher plants (Garcia-Lorenzo et al., 2005; Yu et al., 2005) as well as unicellular algae (Minagawa and Takahashi, 2004; Misumi et al., 2005), this suggests that the heterogeneous composition of the FtsH complex is important. Currently, FtsH is proposed to play a dual role in chloroplasts. It is thought to function in the quality control of photosystems as a protease and also in the formation of thylakoid membranes through a yet ill-defined function (Chen et al., 2000; Takechi et al., 2000).
As a means to understand the mechanisms leading to leaf variegation, it would be logical to search for second-site mutations that suppress leaf variegation of var2. Indeed, the screening of var2 suppressors has been successfully performed by Park and Rodermel (2004), suggesting that several recessive mutations exist. In this study, we focused on one of the var2 suppressors that was isolated by our forward genetics screen. This mutation is single recessive to the variegated phenotype caused by var2. Interestingly, it also suppresses the variegation of var1 and var1 var2. The mutation was found to reside within a gene encoding a potential translation initiation factor 2 (IF2) in chloroplasts. We show evidence that this protein indeed acts as an IF2 in vivo. We also show that another viable mutation in a different gene encoding a translation elongation factor G suppresses the leaf variegation of var2. Thus, the variegated phenotype due to the lack of several FtsH proteases can be affected by reduced chloroplast translation. Based on these collective data, we propose that the balance between protein synthesis and degradation is an important factor in chloroplast maintenance and thylakoid development.
RESULTS
Isolation of Suppressor Mutants fu-gaeri from var2-6 Background
An ethyl methanesulfonate–mutagenized M2 population was generated from the var2-6 T-DNA insertional mutant. M2 seeds were sown on plates containing Murashige and Skoog (MS) media, and individuals that suppressed variegation were selected for further analysis. Among 10,000 M2 seeds that were tested, ∼20 putative plants that suppressed variegation (mutants were named fu-gaeri [fug], which stands for “recovery of variegation” in Japanese) were identified. In this study, we focused on one of the suppressor lines, which was designated sv2.52.
All of the F1 progeny from a cross between sv2.52 and var2 showed leaf variegation. In the F2 population, variegation and normal plants segregated at a 3:1 ratio (Table 1). These results demonstrate that the mutation responsible for suppressing variegation of var2 is single-recessive. As a result, this mutation was subsequently named fug1-1. We then crossed sv2.52 (abbreviated as var2-6 fug1-1 hereafter) with wild-type plants, and the segregation of variegation was scored (Table 2). In F2 progeny, we observed three major phenotypes: normal, variegated, and suppressor-like, which showed pale color in a few of the first true leaves. These three phenotypes segregated close to the ratio of 12:3:1, respectively, suggesting that fug1-1, by itself, does not cause a visible phenotype, at least under our screening conditions.
Table 1.
Segregation Analysis of a Cross between var2-6 fug1-1 × var2 Mutants
| Number of Progeny (%)
|
||||
|---|---|---|---|---|
| Cross | Generation | Normal | Variegated | χ2 (3:1) |
| var2-6 fug1-1 × var2-6 | F1 | 0 (0) | 3 (100) | |
| F2 | 84 (28) | 216 (72) | 0.230 | |
| var2-6 fug1-1 × var2-1 | F1 | 0 (0) | 3 (100) | |
| F2 | 17 (24.3) | 53 (75.7) | 0.890 | |
Table 2.
Segregation Analysis of a Cross between var2-6 fug1-1 × the Wild Type
| Number of Progeny (%)
|
||||||
|---|---|---|---|---|---|---|
| Cross | Generationa | Normal | Variegated | fug1-Likeb | Kms,c | χ2 (12:3:1) |
| var2-6 fug1-1 × Col | F1 (free) | 30 (100) | 0 (0) | 0 (0) | ||
| F2 (free) | 65 (76.5) | 15 (17.6) | 5 (5.9) | |||
| F2 (Km) | 31 (34.4) | 24 (26.7) | 4 (4.4) | 31 (34.4) | 0. 952 | |
| var2-6 fug1-1 × Ler | F1 (free) | 30 (100) | 0 (0) | 0 (0) | ||
| F2 (Km) | 5074 (48.4) | 1437 (12.2) | 353 (2.3) | 4285 (36.4) | ||
Scoring in each generation was performed with MS plates that were either supplemented with (Km) or without (free) kanamycin.
The fug1-like phenotype is represented by a pale-leaf phenotype (see text).
Kanamycin-sensitive plants bleached on kanamycin plates.
Morphologies of var2 fug1 and fug1
Leaf morphologies of var2 fug1 and fug1 are shown in Figure 1 together with those of wild-type ecotype Columbia (Col) and var2. Under normal growth conditions, var2 grew slower than Col, whereas var2 fug1 grew comparably to var2-6 (Figures 1A and 1B). The only difference that we observed between var2 fug1 and Col was that at the onset of the formation of the first few true leaves, var2 fug1 showed a slightly paler color than those of Col. However, later on in plant development, this difference became indistinguishable. Analysis of leaf cross sections indicated that epidermal and mesophyll cells were organized similarly between Col, var2, and var2 fug1. However, cells in both var2 and var2 fug1 appeared smaller in size than those from Col, an effect that was likely due to their retarded growth (Figure 1C). Detailed examination by transmission electron microscopy revealed that chloroplast structures in var2 fug1 were comparable to those in Col, with developed granal stacks (Figure 1D). Leaf variegation, detected on true leaves but not on cotyledons of var2, looked completely suppressed in var2 fug1 (Figure 1A). Although detailed observations occasionally detected some dot-like white sectors toward the edge of leaves (Figure 1B), this pattern clearly differed from the variegation that was observed in var2. Otherwise, the overall leaf morphology and plant architecture looked similar between var2 and var2 fug1. In fug1-1 (without var2), we observed no visible phenotype compared with Col (Figure 1A).
Figure 1.
Leaf Phenotypes of the Variegated Mutant var2 and Its Suppressor Line fug1.
(A) Photographs of 5-week-old wild-type Col, var2-1, var2-1 fug1-1, fug1-3, and fug1-1 grown under normal conditions.
(B) Photographs of detached rosette leaves from Col (5 weeks old) and var2-6 fug1-1 (6 weeks old) and a second true leaf from Col, var2-6 fug1-1, and var2-6.
(C) Toluidine Blue–stained thin sections of a 3-week-old third true leaf from Col, var2-1, var2-1 fug1-1, and fug1-1.
(D) to (G) Electron micrographs of chloroplasts from 3-week-old leaves of var2-6 fug1-1 (D), Col (E), var2-6 fug1-1 (F), and var2-1 fug1-1 (G). Bars = 833 nm in (D), 1.4 μm in (E), 5.0 μm in (F), and 1.3 μm in (G).
Map-Based Cloning of fug1
F2 progeny showing kanamycin resistance, from a cross between var2-6 fug1-1 and Landsberg erecta (Ler), were subjected to PCR-assisted map-based cloning. Our initial mapping identified two chromosomal positions that were linked to var2-6 fug1-1. One of them, as expected, mapped to the bottom arm of chromosome 2, which corresponded to var2. The other linkage, likely corresponding to fug1, mapped to the top arm of chromosome 1. We designed new markers and narrowed the map position within ∼470 kb (5813 to 6283 Mb on chromosome 1; see Supplemental Figure 1A online), where 122 annotated genes were included.
To identify the fug1 locus within this region, we hypothesized that the protein encoded by fug1 may be targeted to chloroplasts. Among the annotated genes of this region were 23 genes that were predicted by the TargetP program to encode proteins with a chloroplast transit peptide at the N terminus. Nucleotide sequences of these genes were determined in var2-6 fug1-1, and we detected a single nucleotide substitution at the At1g17220 locus, which encodes the putative prokaryotic translation initiation factor 2 (cpIF2). A single C-to-T substitution at nucleotide +4437 was identified that resulted in an amino acid substitution from Leu to Phe (see Supplemental Figure 1B online). To examine whether the point mutation detected in At1g17220 is linked to the suppressor phenotype, we generated a derived cleaved amplified polymorphic sequence (dCAPS) marker to detect the polymorphism. PCR analysis of 100 plants with the fug1-like phenotype showed that all were homozygous for this mutation.
A T-DNA Insertion within At1g17220 Suppresses Leaf Variegation in var2
To confirm that At1g17220 corresponds to FUG1, we searched for T-DNA insertion mutants in the SIGnAL T-DNA express database (http://signal.salk.edu/cgi-bin/tdnaexpress). Two T-DNA lines for At1g17220 were found and characterized (Figure 2A). One line (termed fug1-2) contained a T-DNA insertion that disrupted the gene at +2517 in intron 5. The other line (termed fug1-3) contained an insertion upstream of the gene at −367, which presumably affects the promoter activity. Our initial attempts to isolate fug1-2 homozygous lines failed, and only heterozygous plants were isolated after analyzing seeds from selfed heterozygotes. Upon close examination of young siliques in heterozygotes with a dissection microscope, we identified defective embryos that looked white and that aborted during maturation (Figure 2B). The aborted/nonaborted embryos segregated in a 3:1 ratio (see Supplemental Table 1 online). Based upon these findings, we concluded that a T-DNA insertion in cpIF2 resulted in an embryo-lethal phenotype.
Figure 2.
Identification of the fug1 Alleles and RNAi Suppression Analysis.
(A) Schematic representation of the gene At1g17220, which corresponds to FUG1 (also shown as cpIF2). Locations of T-DNA insertions in two lines (fug1-2 and fug1-3) as determined by sequence analysis. Insertion sites are indicated by white arrowheads with the nucleotide positions (+2517 and −367, respectively; +1 corresponds to A of the initiation codon ATG in the genomic sequence). An amino acid substitution found in fug1-1 (+4437, L963F) is indicated by an asterisk. The region corresponding to a DNA fragment that was used for the RNAi suppression experiment is indicated by a red bar.
(B) Dissected young siliques from fug1-2 showing embryo lethality. Defective seeds detected in young siliques of fug1-2 are indicated by asterisks. Bars = 0.5 mm.
(C) Suppression of the var2 leaf variegation by fug1-3. Photographs of 5-week-old var2-1 (left) and var2-1 fug1-3 (right) are shown. The genotype of the var2-1 fug1-3 plants was determined by PCR with gene-specific primers (see Methods). Bars = 5 mm.
(D) Accumulation of FUG1 mRNA examined by RT-PCR. Primers for FtsH2(VAR2)+FtsH8, FUG1, and ACTIN2 were prepared, and RT-PCR was performed with total RNAs from 3-week-old plants as indicated above the gel. For each gene, two gels with different amplification cycles (shown on the right) are indicated. RT-PCR was repeated at least three times, and the representative result is shown.
(E) Accumulation of cpIF2 protein examined by immunoblot analysis. Total protein extracts from cotyledons of Col, var2-1, var2-1 fug1-1, fug1-1, and fug1-3 were separated by 7.5% SDS-PAGE and probed with anti-FUG1 (top), anti-FtsH2 (middle), and Lhcb1 (bottom). Samples were equally loaded based on total chlorophyll contents. We repeated immunoblot analysis at least three times, and the representative result is shown. The band of 105 kD corresponding to the mature size of cpIF2 is indicated by an open arrowhead. An E. coli extract from the strain carrying pTrc99AcpIF2Δ64 (see Figure 4) also detected the band of 105 kD (right). Additional bands detected by the E. coli extract (indicated by closed arrowheads) likely represent degradation products of cpIF2, since none of the signals were detected by cell extracts without expressing cpIF2 (data not shown).
(F) RNAi suppression of cpIF2 restores the formation of green sectors in true leaves. Schematic representation of the gene cassette used in RNAi (cpIF2-RNAi) is indicated on the top, consisting of the CaMV 35S promoter (Pro35S), part of β-glucuronidase gene (ΔGUS), and the nopaline synthase terminator (NOS). The DNA fragment specific for FUG1 inserted in the gene cassette (shown in [A]) is highlighted by two red arrows (Antisense and Sense). Photographs of a parental var2-1 (var2-1) and two cpIF2-RNAi transgenics (transformants) displaying enhanced green sectors are indicated on the bottom. Bars = 1 mm.
In fug1-3, however, we were able to successfully identify homozygous T-DNA insertion lines subsequent to PCR analysis of genomic DNA extracts (data not shown). To examine the expression of cpIF2, RT-PCR was performed using total RNA that was isolated from the homozygous fug1-3 and other controls. Accumulation of cpIF2 mRNA was substantially repressed in the homozygous fug1-3 line but not in var2-6 fug1-1 and fug1-1 (Figure 2D). We subsequently tested cpIF2 accumulation by immunoblot analysis with a rabbit polyclonal antibody that was raised against a truncated recombinant cpIF2 protein (see Methods). Our cpIF2 antibody recognized a band of 105 kD that corresponded to the mature size of cpIF2 (Figure 2E). This band also corresponded to the signals detected by an Escherichia coli cell extract in which a mature-sized cpIF2 was overexpressed. No bands were obtained from the immunoblot using E. coli negative control extracts (data not shown), indicating that our antibody did not cross-react with endogenous E. coli IF2. The amount of cpIF2 normalized by Lhcb1 decreased in fug1-3, and this reduction was in accordance to the results obtained from RT-PCR (20% to Col, averaged from three independent experiments). Thus, the T-DNA insertion in fug1-3 appears to have resulted in reduced expression of cpIF2, leading to a phenotype similar to that of fug1-1. In our next line of investigation, we crossed fug1-3 and var2-1 or var2-6, and the resulting F2 progeny were subjected to PCR analysis for the isolation of double mutants. In plants that were homozygous for both var2 and fug1-3, leaf variegation was suppressed (Figure 2C). Suppression of the var2 variegation with the two alleles fug1-1 and fug1-3 strongly suggested that FUG1 corresponds to cpIF2.
Suppression of var2 Variegation by RNA Interference of cpIF2
To conclude that the fug1 locus was indeed responsible for the suppression of var2 leaf variegation, it was necessary to complement the mutant locus. However, we had difficulty cloning a full-length cDNA (data not shown; Campos et al., 2001). Instead of complementing fug1 by its own cDNA, we performed RNA interference (RNAi) suppression analysis, using a part of the cDNA as described below. An RNAi construct (cpIF2-RNAi) was generated in which a pair of the DNA fragments specific for cpIF2 (shown red in Figure 2A) was placed in an inverted orientation under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Figure 2F). We introduced cpIF2-RNAi into var2-1 mutants by Agrobacterium tumefaciens–mediated transformation and obtained four transformants. Although these transformants were selected by kanamycin resistance and grew very slowly, they developed true leaves that contained less white sectors (Figure 2F). This phenotype differed from var2-1, in which white sectors dominated the first true leaves. These results are comparable to our previous observations in fug1-1 and fug1-3 and further supported our conclusion that reduced expression of cpIF2 suppresses leaf variegation.
In addition to less-variegated leaves, two of the four transformants showed a severe phenotype with white cotyledons and died after developing a few true leaves (data not shown). The white cotyledon phenotype looked similar to that observed in snowy cotyledon1 (sco1). This particular mutation has been shown to result from a defect in translation elongation factor G in chloroplasts (cpEF-G; Albrecht et al., 2006; see below). We therefore considered that a strong repression of chloroplast translation can lead to improper chloroplast development in cotyledons.
Properties of cpIF2
FUG1 (cpIF2) encompasses 5335 bp (including 5′ and 3′ untranslated regions [UTRs] predicted from a full-length cDNA U12290) and consists of 12 exons. It encodes a protein of 1026 amino acids with an estimated molecular mass of 110 kD and shows significant sequence similarity (65.8%) to IF2 isolated from common bean (Phaseolus vulgaris) (Campos et al., 2001; see Supplemental Figure 2 online). In E. coli, protein synthesis from mRNA initiates by recruiting the initiation complex with 30S ribosomes and fMet-tRNAmet. Three proteins, IF1, IF2, and IF3, are involved in this process (reviewed in Ramakrishnan, 2002). The initiation complex then recruits 50S ribosomes to form the translation machinery so that elongation can proceed. IF2 plays a central role in binding fMet-tRNAmet with 30S ribosomes and subsequently forms a large complex with 50S ribosomes in a GTP-dependent manner. An E. coli IF2-1 (the largest isoform) is characterized by six domains, and cpIF2 retains a conserved GTP binding motif included in domain IV. An amino acid substitution (Leu to Phe) in fug1-1 occurred in a residue conserved in most of the known IF2s (Leu or Ile). Interestingly, the mutation occurred within the particular domain (VI-2) that was suggested to play a role in binding to fMet-tRNAmet (Mortensen et al., 1998; reviewed in Laursen et al., 2005). The mutation in fug1-1 seems to affect accumulation of cpIF2 indirectly, since a reduction of cpIF2 was observed by immunoblot analysis in fug1-1 and var2-1 fug1-1 (Figure 2E).
cpIF2 Is the Only IF2 in Chloroplasts
Based on genomic information, there only appears to be two prokaryotic IF2 homologs in Arabidopsis (see Supplemental Figures 2 and 3 online). One of these, cpIF2, which was identified in this study, is more similar to common bean IF2 than the other Arabidopsis homolog (encoded in At4g11160 and designated as mtIF2). Phylogenetic analysis of IF2s from other organisms (shown in Figure 3B) indicated that cpIF2 and mtIF2 belong to a different clade consisting of plant IF2 homologs. The mtIF2 clade appeared more related to mitochondrial IF2s. To examine the actual organellar localization of cpIF2 and mtIF2 in vivo, we conducted a transient assay using Arabidopsis protoplasts derived from mesophylls and protein fusions to green fluorescent protein (GFP). DNA fragments corresponding to putative N-terminal targeting signals from both IF2s were fused in frame to GFP and were expressed in protoplasts under the control of the CaMV 35S promoter (Figure 3A). When the N-terminal signal from cpIF2 was used, GFP signals were exclusively colocalized with chloroplasts. By contrast, the N-terminal signal from mtIF2 did not deliver GFP into chloroplasts. On the contrary, we instead found rod-shaped GFP signals colocalized with MitoTracker orange stain. These results indicated that, as expected from phylogenetic analysis, cpIF2 is chloroplastic and mtIF2 is mitochondrial. The embryo-lethal phenotype of the null mutation fug1-2 is thus explained by the fact that cpIF2 is the only IF2 in chloroplasts.
Figure 3.
GFP Transient Assay and Phylogenetic Analysis.
(A) Cellular localization of cpIF2 as revealed by a GFP transient assay. Protoplasts prepared from leaf tissues were transformed with N-terminal GFP fusion constructs that contained a putative targeting sequence from either cpIF2 (cpIF2TP-GFP) or from mtIF2 (mtIF2TP-GFP). A GFP construct that lacked the targeting signals was also transformed as a negative control (GFP). Images of bright-field (right), GFP fluorescence (middle), and chlorophyll autofluorescence (left) are shown. To ascertain mitochondrial localization, protoplasts expressing mtIF2-GFP were stained by a 50 nM Mito-Tracker Orange dye, and the signals were detected simultaneously with chlorophyll autofluorescence. Bars = 20 μm.
(B) A phylogenetic tree of prokaryotic IF2 homologs. Bootstrap values (percentage) are shown in red, and expected distances as counts of amino acid substitutions per site are shown in black at the nodes.
In Vivo Complementation of IF2 Deficiency in E. coli by cpIF2
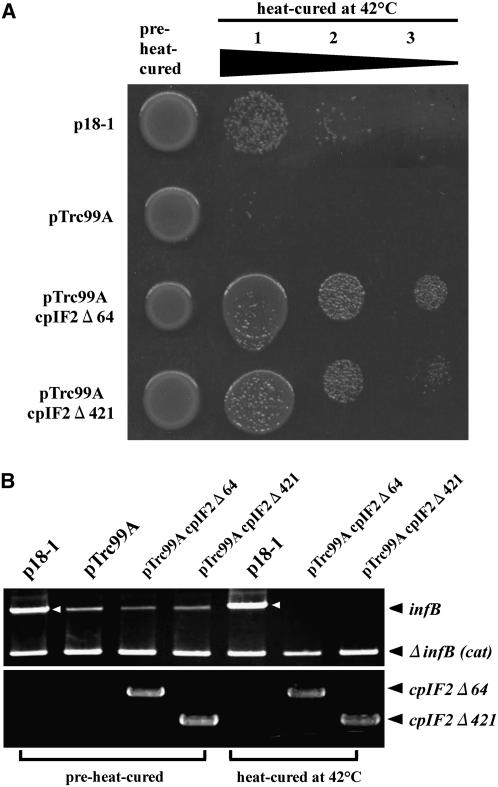
To test if cpIF2 is indeed involved in translation, we conducted an in vivo complementation experiment of an E. coli mutant, SL598R. Because IF2 is essential and no null mutants are available, an active form of infB (encoding IF2) in SL598R is provided by thermosensitive lysogen, and the chromosomal infB is deleted. Heat-curing at 42°C leads to the loss of IF2, and this strain would no longer survive unless an additional IF2 activity is supplied (Laalami et al., 1991). Partial FUG1 cDNAs, corresponding to a mature cpIF2 without its transit peptide (cpIF2Δ64) or a truncated IF2 lacking 421 N-terminal residues but retaining the GTPase domain (cpIF2Δ421), were cloned into an expression vector pTrc99A and were introduced into SL598R. Immunoblot analysis of cell extracts from SL598R (cpIF2Δ64) with the anti-cpIF2 antibodies revealed that cpIF2 is properly expressed at the protein level (Figure 2E). As shown in Figure 4A, the resulting strains were capable of surviving after heat-curing, whereas the strain with the empty plasmid pTrc99A did not. Using PCR analysis, we were able to conclude with certainty that the surviving cells from SL598R (cpIF2Δ64) and SL598R (cpIF2Δ421) completely lost endogenous infB after heat-curing but retained cpIF2 (Figure 4B). These results demonstrate that cpIF2 is capable of functionally complementing E. coli IF2 and that it acts in the process of translation.
Figure 4.
Complementation of the E. coli infB Null Mutation by Arabidopsis cpIF2.
(A) The E. coli SL598R mutant strain, containing a functional infB in a thermosensitive lambda lysogen, was transformed with the four plasmids (indicated on the left). An empty pTrc99A vector was used as a negative control. The positive control plasmid (p18-1) is a pBR322 derivative and carries a wild-type infB. Each cell line, before and after heat-curing, was spotted on ampicillin plates, and photographs were taken after overnight incubation at 30°C. Rows 1 to 3 represent a series of cell dilutions (rows 2 and 3 are 10-fold and 50-fold dilutions of row 1, respectively) that were plated after heat-curing at 42°C for 1 h. The two truncated cpIF2 proteins that were used in this experiment (cpIF2Δ64 and cpIF2Δ421) are described in Results. We considered that the low recovery of the strain carrying p18-1 compared with others is due to the difference of vectors used.
(B) PCR was conducted to verify the loss of infB due to the heat-curing treatment. Genomic DNA was isolated from cell lines before or after heat-curing, and samples were subjected to PCR analysis using primers specific for infB (top panel) and cpIF2 (bottom panel). The bands corresponding to an active form of infB (infB), an inactive form of infB [ΔinfB (cat)], cpIF2Δ64, and cpIF2Δ421 are indicated on the left. The bands in the p18-1 lanes shown by white arrowheads derive from infB cloned in p18-1.
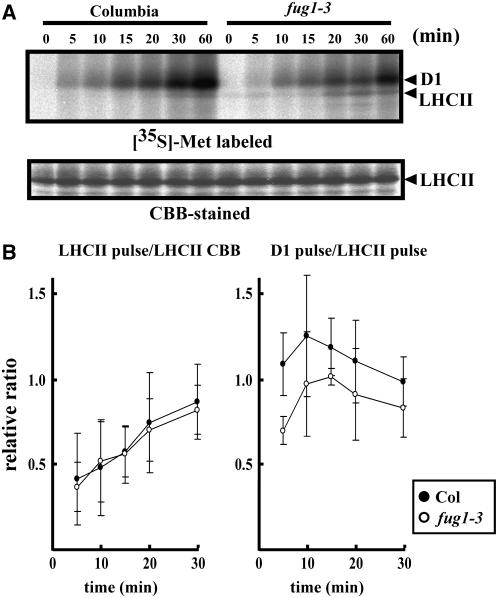
An in Vivo Labeling Experiment Suggests That Synthesis of D1 Protein Is Reduced in fug1
Next, we examined the influence of the fug1 mutation in chloroplast protein synthesis. To accomplish this, in vivo labeling of chloroplast proteins was performed by infiltrating [35S]Met into mature leaves of Col and fug1-3 mutants. After infiltration, leaves were exposed under moderate light conditions (150 μmol/m2/s) to initiate protein synthesis. Labeled proteins (Figure 5A) were separated and analyzed by SDS-PAGE. The results showed that the bands corresponding to the photosystem II (PSII) reaction center D1 protein and the light-harvesting complex II (LHCII) protein were very rapidly labeled, as previously reported (e.g., Aro et al., 1993). These data reflect the fact that D1 turns over very rapidly (Schuster et al., 1988; Aro et al., 1994). We therefore measured D1 synthesis as an index of chloroplast translation. D1 synthesis in fug1 is also interesting to test, since FtsH2 has been shown to participate in the repair cycle of PSII by degrading photodamaged D1 (Lindahl et al., 2000; Bailey et al., 2002).
Figure 5.
Protein Synthesis in Col and fug1-3 Studied by in Vivo Protein Labeling.
(A) In vivo labeling of thylakoid membrane proteins in Col and fug1-3. Fully expanded mature leaves (∼7 weeks old under 12-h daylength) from Col and fug1-3 were thoroughly infiltrated with solution containing [35S]Met and were transferred under a fluorescent bulb (150 μmol/m2/s) for a period of incubation (0, 5, 10, 15, 20, 30, and 60 min). Thylakoid membranes were separated by urea SDS-PAGE and stained by Coomassie blue (CBB). The bands corresponding to D1 and LHCII, confirmed by immunoblot analysis, are indicated by arrowheads in each panel. We repeated this experiment more than three times, and the representative result is shown.
(B) Quantification of the LHCII and D1 protein labeling. Ratios of the LHCII radiolabel over the amount of LHCII detected by Coomassie blue (left) or over the LHCII radiolabel (right) was plotted at each time point (bars indicate sd; n = 3). To normalize values from three independent experiments, the ratio of Col at 60 min was adjusted as 1, and the relative ratios are indicated.
First, we normalized labeled LHCIIs based on the steady state level of LHCII (Figure 5B, left panel). This revealed that no significant difference between Col and fug1-3 was detectable in the rate of LHCII synthesis that proceeded in cytosol. Next, we used labeled LHCII as an internal control and normalized D1 synthesis. The fug1-3 mutant was found to reproducibly display a lower D1 synthesis rate than Col, although the labeled intensity varied in each experiment and the difference was not statistically evident (Figure 5B, right panel). Nevertheless, the rate we observed was significantly decreased at a very early time point (5 min), suggesting that fug1 indeed affected the D1 protein synthesis rate. A similar result was also obtained when D1 signals were normalized by the steady state LHCII (data not shown). As a result, it is reasonable to consider that cpIF2 most likely acts on chloroplast translation. It should be noted that fug1-3 is a leaky mutation that may have little effect on chloroplast translation. Because of the rapid turnover of D1, the effect of fug1-3 may be masked at a labeling time of >10 min.
Chloroplast Protein Accumulation in var2 fug1 and fug1
Immunoblot analysis was performed to examine the extent of the fug1 mutation in steady state levels of photosystems. Proteins isolated from Percoll gradient-purified chloroplasts of Col and the mutants were subjected to immunoblotting using antibodies raised against FtsH2, D1, PsbO, PsaF, and Lhcb1 (Figure 6A; proteins were equally loaded based on total chlorophyll content). When normalized by Lhcb1, the steady state levels of D1, PsbO, and PsaF were not significantly altered between Col, var2-1, and the mutants containing fug1 (Figure 6B). These data are in good agreement with the fact that fug1 had very little effect on D1 synthesis in vivo with a longer labeling time. These results show that photosystems and other complexes, in spite of a possible reduced synthesis rate at the onset of development, accumulate normally in mature chloroplasts of var2 fug1 and fug1.
Figure 6.
Steady State Accumulation of Chloroplast Proteins.
(A) Chloroplasts were purified by a Percoll step gradient from each plant line as indicated below the immunoblot. Leaves at the similar age in Figure 5 were subjected to chloroplast purification experiments. Proteins from the chloroplasts were equalized by total chlorophyll content and separated on SDS-PAGE. Results of immunoblots using anti-FtsH2 (VAR2), anti-D1 (PsbA), anti-PsbO, anti-PsaF, and anti-Lhcb1 antibodies are indicated.
(B) Relative amounts of proteins estimated by immunoblot analysis. Signals of immunoblots were quantified by NIH Image, normalized by the signals of Lhcb1, and calibrated based upon the protein amount in Col (bars indicate sd; n = 3).
Sensitivity of fug1 and var2 fug1 to High Light
Loss of FtsH2 in var2 has been shown to affect the repair cycle of PSII; consequently, var2 is unable to acclimate to high light stress (Bailey et al., 2002; Sakamoto et al., 2002). This was proven by the fact that green sectors of var2 leaves, when irradiated by high light, exhibited an irreversible decrease in PSII activity (measured by Fv/Fm, the maximum quantum yield of PSII photochemistry). We tested this photosensitivity in fug1 and var2 fug1. Young leaves of fug1 (3 weeks old, grown in MS plates), irrespective of coexistence with var2, exhibited Fv/Fm measurements that were significantly lower than those of Col and var2 even prior to high-light irradiation (Figure 7A, left panel). This implies that, despite the lack of a visible phenotype, fug1 affects photosynthetic activity at an early stage of leaf development. By contrast, fully expanded rosette and cauline leaves (5 weeks old, grown in soil) showed similar Fv/Fm levels between Col and fug1. Interestingly, in mature leaves the high-light sensitivity of var2 appeared to be suppressed by fug1. This effect was not restricted to a particular area but was observed in most of the leaf (Figure 7B). We therefore considered that the suppression of leaf variegation is partly associated with increased resistance of PSII to photoinhibition.
Figure 7.
Effects of fug1 and var2 fug1 on the Sensitivity to High Light.
(A) The ratio of variable to maximum fluorescence (Fv/Fm) measured in detached leaves of Col, var2-1, and var2-1 fug1-1 using the FluorCam 700MF (bars indicate sd; n = 5 from different plants). Leaves were exposed to high light (800 μmol/m2/s for 240 min), and Fv/Fm was calculated. Results from leaves that were harvested from plants grown under normal light (70 μmol/m2/s) in MS plates or soil are shown in left and right panels, respectively.
(B) Digital images of Fv/Fm. The values were measured by the FluorCam 700MF at time 0 and 240 min in detached leaves and were visualized by pseudocolor index as indicated on the bottom. Typical images from 3-week-old leaves (left) and from 5-week-old cauline leaves (right) are shown. Bars = 5 mm.
fug1 Suppresses Leaf Variegation of var1 and var1 var2
To further assess the effect of fug1 on the loss of FtsH, we crossed var2-1 fug1-1 to var1-1. Consequently, we isolated plants having the genotypes of var1-1 fug1-1 and var1-1 var2-1 fug1-1. The var1 mutant lacks FtsH5, and the degree of leaf variegation is less in var1 than in var2 (Sakamoto et al., 2002). Similarly to var2, coexistence of fug1 with var1 suppressed leaf variegation (Figure 8A). It was previously shown that var1 and var2 act synergistically and the double mutant exhibits highly variegated leaves and sterility (Sakamoto et al., 2002; Zaltsman et al., 2005b). To our surprise, when the triple mutant var1 var2 fug1 was generated, it again suppressed leaf variegation and the first few true leaves looked green. Although the triple mutant showed a slight variegation, similar to the extent of var1, it was able to grow normally and produce seeds at later stages of development. These results suggest that fug1 can suppress variegation induced by FtsH deficiency, regardless of which particular FtsH gene is defective.
Figure 8.
Suppression of Leaf Variegation by var1 var2 fug1 and var2 sco1.
(A) Leaf variegation of var1 and var1 var2 was suppressed by fug1. Photographs of a parental var1-1 (top left, 4 weeks) and var1-1 var2-1 double mutant (bottom left, 5 weeks) are shown, together with var1-1 fug1-1 double mutant (middle, 5 weeks; right, 10 weeks) and var1-1 var2-1 fug1-1 triple mutant (middle, 5 weeks; right, 10 weeks). Bars = 1 mm in left panel and 5 mm in right panels.
(B) Another mutation, sco1, which is defective in translation elongation in chloroplasts, suppressed the leaf variegation of var2. Photographs of 4-week-old sco1 (left) and 5-week-old var2-1 sco1 double mutant (right) are shown. Bars = 1 mm in left panel and 5 mm in right panel.
Suppression of var2 by a Novel Mutation sco1 Deficient in Chloroplast Translation
The results obtained so far clearly indicated that the impact of fug1 on leaf variegation is attributable to the reduction of chloroplast translational activity but not of IF2 activity per se. This raised a possibility that not only fug1, but also other mutations related to chloroplast translation such as sco1, may act as a suppressor of var2. It was shown that sco1 is a viable mutation with white cotyledons and results from a point mutation in cpEF-G (Albrecht et al., 2006). The mutation largely affects seedling and cotyledon development, presumably because chloroplast protein synthesis is particularly important at this stage. We crossed sco1 with var2 to generate the double mutant. In their F2 population, plants showing the sco1 phenotype (white cotyledon) and homozygous for sco1 and var2 were obtained. These plants developed normal true leaves and were fertile (Figure 8B), suggesting that sco1 is epistatic to var2. More importantly, we clearly demonstrated that sco1 indeed suppressed the leaf variegation of var2. Here again, we emphasize that the white cotyledons observed in sco1 resembled those in the transgenic plants obtained by cpIF2-RNAi. All together, we consider that protein synthesis in chloroplasts plays an important role in seedling and cotyledon development, strengthening our assumption that a slight reduction of chloroplast translation suppresses leaf variegation caused by the loss of FtsHs.
DISCUSSION
Identification of cpIF2 as a Suppressing Factor of Leaf Variegation
Characterization of one of the var2 suppressors, fug1-1, reported in this study led us to identify cpIF2, an indispensable factor for chloroplast translation. A wide array of evidence was compiled that supported the supposition that FUG1 encodes cpIF2. Namely, collective data from map-based cloning, genetic analysis of fug1-1, isolation of a T-DNA knockdown strain (fug1-3), and RNAi analysis of cpIF2 all supported this claim. Since the influence of the amino acid substitution that was detected in cpIF2 was unclear, we cannot completely rule out the possibility that fug1-1 actually has a different mutation tightly linked to cpIF2 that suppressed leaf variegation. Nevertheless, all the other experiments collectively provide compelling evidence that a reduction of cpIF2 can indeed suppress leaf variegation in var2.
Translational machinery in chloroplasts originates from its bacterial origin, but its components are encoded in nuclear and chloroplast genomes (Manuell et al., 2004; Laursen et al., 2005). Trans-acting factors involved in translation of chloroplast mRNAs have been characterized in Chlamydomonas reinhardtii (reviewed in Manuell et al., 2004). Despite these findings, little is known about general chloroplast translation factors, such as IFs (Laursen et al., 2005). IF2 and IF3 have been characterized in chloroplasts of Euglena (e.g., Ma and Spremulli, 1990; Lin et al., 1994). In this study, GFP transient assays confirmed that Arabidopsis contains two prokaryotic IF2s (cpIF2 and mtIF2) that reside in chloroplast and mitochondria. The essential function of cpIF2 as a sole chloroplastic IF2 was also demonstrated by the null mutation fug1-2.
As evidenced by white cotyledons detected in our cpIF2-RNAi transformants and sco1 mutants, high levels of chloroplast translation may be required for cotyledon and seedling development. In accordance with these observations, microarray database analysis revealed that most green tissues accumulate FUG1 mRNAs, with the highest level in cotyledons. Our immunoblot analysis also revealed that cpIF2 accumulates at high levels in cotyledons and young leaves (Figure 2E; our unpublished data). In addition, we showed that photosynthetic activity measured by Fv/Fm can be affected in fug1, particularly at relatively young leaves grown under MS plates. Like other translation factors, it is possible that cpIF2 might be necessary at an early stage of chloroplast development.
The Balance between Protein Synthesis and Degradation Determines Leaf Variegation
FtsH2 (VAR2) and FtsH5 (VAR1) are the major components of heteromeric FtsH complexes. Despite their similarity to one another, recent observations suggest that FtsH2 and FtsH5 perform distinct functions (Yu et al., 2005; Zaltsman et al., 2005b). FtsH2/FtsH5 represent most FtsHs present in thylakoid membranes, whereas a closely related pair of isoforms, FtsH8/FtsH1, seem to play compensatory roles. It was shown that fug1 suppresses not only the variegation of var2 but also of var1 and var1 var2. The triple mutant var1 var2 fug1 did not fully recover the variegation but exhibited significant green leaves. Surprisingly, in the triple mutant, a severe growth defect of var1 var2 double mutants was suppressed to recover fertility, implying a substantial impact of fug1 on the effect of FtsH deficiency. Here, we demonstrate that fug1 is epistatic to var1 and var2 in recovering leaf variegation. The possibility that cpIF2 directly interacts FtsH2 and FtsH5 can be excluded, since var1 and var2 alleles used in this study are nonsense mutants (Chen et al., 2000; Takechi et al., 2000; Sakamoto et al., 2002): normal FtsH2/FtsH5 proteins are not produced under these backgrounds (data not shown). As a result, it is concluded that leaf variegation is suppressed by an indirect effect, most likely by an altered translational activity.
We were interested to determine if the suppression of leaf variegation in var results from reduced translational activities in chloroplasts. Another viable mutation, sco1, in which cpEF-G is mutated by an amino acid substitution, allowed us to test this hypothesis. Our results clearly showed that sco1 acts as a suppressor, as it is epistatic to var2 (it recovered variegation but showed white cotyledons). Similarly to cpIF2, cpEF-G encoded in SCO1 appears to be a sole EF-G in chloroplasts. Although the actual role of cpEF-G in chloroplast translation elongation is still unknown, the amino acid substitution in sco1 was predicted to compromise the binding of cpEF-G to ribosomal complexes. Profound effects of sco1 in seedling development suggest that, together with cpIF2 and other translation factors, cpEF-G is essential for chloroplast biogenesis at an early state. EF-G contains multi-GTPase domains that are required for interacting with 70S ribosomes. Thus, our identification of two suppressors, acting on the different steps of chloroplast translation, directed us to conclude that suppression of leaf variegation is mediated by a general reduction in translation.
FtsHs participate in the ATP-dependent degradation of various plastid proteins, including Rieske FeS (Ostersetzer and Adam, 1997), LHCII proteins (Żelisko et al., 2005), and D1 (Lindahl et al., 2000; Bailey et al., 2002; Komenda et al., 2006). Taken together with our presented data, we propose that the balance between protein synthesis and degradation is one of the determinant factors that leads to leaf variegation, at least in case of var mutants. It is not currently clear whether variegation is associated with balancing a particular chloroplastic protein or not. In this study, we showed that D1 protein synthesis is affected in fug1. In addition, FtsH2 has been shown to degrade D1 protein in a light-dependent manner (Bailey et al., 2002). At least D1 protein levels can be altered by fug1, although it has little effect on the steady state level and acts only transiently. D1 is constantly damaged by exposure to light. In the younger leaves, the effect of fug1 on D1 synthesis is presumed to be larger than that indicated by in vivo labeling at a later stage. By limiting the amount of photodamaged D1 accumulating at the early stage of leaf development due to the loss of FtsH2, fug1 may prevent leaf variegation in var mutants.
It should be noted that fug1 showed a reduced PSII activity when grown in MS plates. By contrast, it showed a PSII level comparable to Col when mature leaves grown in soil were used. Moreover, the sensitivity of var2 to high light appeared to become normal when var2 and fug1 are combined. Although we cannot fully explain how this is accomplished, our observations imply that the balance between protein synthesis and degradation also influences the repair cycle of PSII. In chloroplasts, translation may be a prerequisite for initial biogenesis of chloroplasts, as evidenced by the reduced Fv/Fm in relatively young leaves of fug1. By contrast, at the stage where chloroplasts are fully expanded and matured, translation plays a rather critical role in the maintenance of chloroplasts, such as in the PSII repair. It is not clear whether the high-light sensitivity that is observed in green sectors of variegated mutants is also the cause of chloroplast deficiency in the white sectors. Further studies are necessary to clarify this point.
Suppressors of Leaf Variegation and Evolutional Implication
We initially assumed that characterization of suppressors in var2 would enable us to identify which novel factors or pathways, if any, regulate the formation of leaf variegation. Although this was not the case, our data suggest that variegation is associated with various chloroplast redundant activities. The observed suppression of leaf variegation by fug1 and clpc2 is similar in this sense. Park and Rodermel (2004) have reported that mutation or inactivation of ClpC2, a subunit comprising an ATPase complex of the Clp protease, leads to a suppression of variegation in var2. Loss of ClpC2 itself does not cause any visible phenotype, whereas loss of another isomer ClpC1 results in the decrease of Fv/Fm and a chlorotic phenotype (Sjögren et al., 2004; Clarke et al., 2005; Kovacheva et al., 2005). ClpC is related to FtsH in that both are AAA proteins (containing an ATP binding motif) and comprise a hexameric ring structure. ClpC acts as a chaperone subunit of the Clp protease in chloroplasts (reviewed in Adam et al., 2006; Sakamoto, 2006). The loss of ClpC2 is not lethal because of the presence of another homolog, ClpC1. However, the absence of ClpC2 likely alters the overall Clp activity that is required for protein quality control in chloroplasts. These results imply that some redundant function in chloroplast biogenesis/maintenance may act to suppress leaf variegation of var. Conversely, it may mean that the formation of leaf variegation requires maximal levels of chloroplast activities.
The loss of FtsH2 has been considered to determine the physiological state of chloroplasts in a cell-autonomous fashion (Chen et al., 2000; Takechi et al., 2000). Above the threshold, cells are destined to have normal-appearing chloroplasts, whereas below the threshold, the cells are capable of forming white sectors. The threshold appears to depend on developmental and environmental states. The number of white sectors decreases in the latter leaves, and expression of FtsH proteins other than FtsH2 is upregulated in the green sectors (Yu et al., 2004; Zaltsman et al., 2005a). Examination of the variegated pattern suggests that the fate of cells is determined at an early stage of plastid differentiation. It is probable that the decreased protein synthesis in fug1, which principally acts to inhibit full chloroplast function, lowered the threshold levels and allowed most cells to develop normal chloroplasts.
Since we have isolated many putative suppressor mutants, the suppression of leaf variegation in var appears to occur frequently (Park and Rodermel, 2004). The genetic data presented in this study may enable us to understand how various types of leaf variegation evolve in nature. Recent works in recessive mutations, such as leaf variegation and stripes, reveal that leaf variegation can occur through various reasons. One of the principals, as revealed by var1 var2, is that variegation is related to redundant biological functions. In this study, we present data that demonstrate the possibility that many viable mutations accumulated in the genome, either caused by gene redundancy or by silent mutations, have a potential to affect the development of white sectors. Thus, the accumulation of such mutations may contribute to diversification of variegated patterns that are commonly observed among house plants. Because we frequently observe leaf variegation, not only from genetic mutations but also from physiological changes such as pathogen infection, it is possible that variegation might be advantageous for plant survival under rugged environmental conditions. Further studies are required for elucidating the physiological determinants and biological significance of variegation.
METHODS
Plant Materials, Growth Conditions, and Genotyping by PCR
Arabidopsis thaliana ecotype Col or Ler was used as a source of wild type plant material. Alleles of var1, var2, and sco1 that were used in this study were described previously (Sakamoto et al., 2002; Albrecht et al., 2006). Suppressors were identified by screening M2 seeds from var2-6 that were mutagenized by 0.1% (v/v) ethyl methanesulfonate. Unless specifically noted, plants were germinated and grown on 0.7% (w/v) agar plates containing MS medium with Gamborg's vitamins (Sigma-Aldrich). These plates were supplemented with 1.5% (w/v) sucrose, and plants were maintained under 12-h light (∼60 μmol/m2/s) at 22°C. For further analysis, plants were transferred to soil after 4 weeks. Genotypes of var2-6 were determined by PCR using primers KT203, KT301, and TL-3 as previously described (Takechi et al., 2000). All of the primers used in this study are listed in Supplemental Table 2 online. T-DNA insertion lines corresponding to fug1-2 and fug1-3 (stock numbers SALK030159 and SAIL209_E08, respectively) were identified by searching SIGnAL T-DNA Express (http://signal.salk.edu/cgi-bin/tdnaexpress). Seed stocks were kindly provided by the ABRC. Verifications for T-DNA insertion were performed by PCR using primers TL-3, H, and O for fug1-2 and primers LB2, GFP-R, and pre-17220 for fug1-3. Validation for the position of T-DNA insertion sites within FUG1 in the individual plant lines was determined by sequence analysis. The genotype of var2-1 was determined by dCAPS-assisted PCR using primers dCAPS(2-1) and KT304 and digestion with Tsp45I. The genotype of fug1-1 was determined by dCAPS-assisted PCR using primers dCAPS(Xba)-R2 and N and digestion with XbaI. The genotype of var1-1 was determined by sequencing as previously described (Sakamoto et al., 2002). Isolation of the double mutants (var1-1 fug1-1, var2-1 fug1-1, var2-1 fug1-3, and var2-6 fug1-3) and the triple mutant var1-1 var2-1 fug1-3 was performed by crosses with the appropriate parent stocks. Genotypes for the F2 generation were determined by PCR/sequencing as described above.
Map-Based Cloning
F2 seedlings from a cross between var2-6 fug1-1 and Ler were used for mapping the fug1 locus. Approximately 2-week-old plants showing the suppressor-like phenotype (see text) were selected on MS plates containing 50 mg/L kanamycin. Genomic DNA was isolated from seedlings of the selected plants, and the ones showing homozygosity for var2-6 were used for mapping. Linkage analysis was performed by cleaved amplified polymorphic sequence (CAPS) and simple sequence length polymorphism (SSLP) markers, whose information is available at The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org). For fine mapping of fug1, we generated dCAPS markers based on the polymorphisms between Col and Ler that are available at the Monsanto Arabidopsis Polymorphism Collection. Generated markers are indicated in Supplemental Figure 1A online, and the primers for each marker are shown in Supplemental Table 2 online. All of the information on genomic sequences and annotated genes within the mapped chromosomal region was obtained from TAIR.
RNAi Suppression Analysis
The RNAi gene cassette (consisting of CaMV 35S promoter:XbaI-SalI-ΔGUS-SpeI-XhoI:NOS terminator) was generated according to Chuang and Meyerowitz (2000). This cassette was introduced into the binary vector pBI121 to generate pBI121-ΔGUS. The DNA fragment of cpIF2 for RNAi suppression analysis was PCR amplified using a combination of primer pairs: At1g17220 RNAi-SpeI and At1g17220 RNAi-XhoI for sense fragment or At1g17220 RNAi-SalI and At1g17220 RNAi-XbaI for antisense fragment). The PCR fragment was digested with appropriate restriction enzymes and inserted into pBI121-ΔGUS. Agrobacterium tumefaciens–mediated transformation of var2-1 plants was performed as previously described (Takechi et al., 2000), with the exception that transformants were selected on MS plates containing 25 mg/L kanamycin.
GFP Transient Assay and Microscopy Observation
A putative transit peptide of cpIF2 and mtIF2 was predicted to reside within the first 63 and 88 N-terminal residues, respectively. The cpIF2 cDNA sequences corresponding to the entire 5′ UTR (300 bp) and 76 N-terminal residues were PCR amplified and fused in frame into the expression vector p35S-sGFP as previously described (Sakamoto et al., 2003). The mtIF2 cDNA sequences corresponding to the entire 5′ UTR (50 bp) and 99 N-terminal residues were generated by RT-PCR and fused in frame into the p35S-sGFP. Primers used for PCR amplification (17220-GFP-F and 17220-GFP-R for cpIF2 and 11160-GFP-F and 11160-GFP-R for mtIF2) are listed in Supplemental Table 2 online. GFP transient assays were performed using Arabidopsis protoplasts prepared from leaves (n > 10). For observing mitochondrial signals, protoplasts were infused with a 50 nM aqueous solution of Mito-Tracker Orange dye (Molecular Probes) for 10 min. Leaf sections were prepared from a central area of the third true leaf of 24-d-old Col and mutant plants that were grown on MS plates. Thin slices of the leaf tissues were fixed in FAA (45% [v/v] ethanol, 5% [v/v] acetic acid, and 1.8% [v/v] formaldehyde), dehydrated by a dilution series of ethanol, and embedded in Technovit 7100 resin (Kulzer). Tranverse sections (3 μm) containing mesophyll cells were stained with 1% (w/v) Toluidine Blue (in 1% [w/v] boric acid). Sections were visualized with an Olympus BX51 light microscope that was equipped with a DP70 digital camera (Olympus). Transmission electron microscopy was used to examine the ultrastructure of chloroplasts according to previously described methods (Takechi et al., 2000).
Nucleic Acid Extractions and RT-PCR Analysis
For the mapping of fug1, seedlings of individual F2 plants showing the suppressor phenotypes were ground in a micocentrifuge tube containing DNA buffer (200 mM Tris-HCl, 250 mM NaCl, 25 mM EDTA, and 5% [w/v] SDS, pH 7.5). Samples were subsequently centrifuged at 15,000g for 10 min, and nucleic acids in the supernatant were precipitated by 0.6 volume of isopropanol. Segregation analysis was performed using the isolated DNAs with a pair of primers for CAPS and SSLP as described above. PCR and sequencing analyses were also performed as described previously (Sakamoto et al., 2002). Primers that were used for sequencing the fug1 locus (F, H, I, N, O, Q, and R) are listed in Supplemental Table 2 online. Total RNA was extracted from 3-week-old leaves using a Qiagen RNeasy plant mini kit and reverse transcribed using the Takara RNA PCR kit (AMV) version 3.0 according to the manufacturer's instructions. We added 0.5 μg of total RNA to 10 μL of reverse transcription reaction, and 1 μL of the reaction was used for 25 μL of PCR reaction with 25 cycles consisting of denaturing at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. Primers amplifying cDNAs of FtsH2 and FtsH8 (primers KT212 and KT207), FUG1 (primers F and I), and ACTIN2 (primers actin2-F and actin2-R) are listed in Supplemental Table 2 online. The products were viewed by ethidium bromide staining after agarose gel electrophoresis. Linearity of the PCR reaction was monitored by comparing relative amounts of PCR products after 20, 22, and 25 cycles.
Database Search and Phylogenetic Analysis
All of the information on the annotated genes was obtained from TAIR. TargetP (http://www.cbs.dtu.dk/services/TargetP/) was used to predict chloroplast targeting. For generating a phylogenetic tree, IF2 homologs were detected by BLASTp using the entire amino acid sequence of cpIF2 as a query in the National Center for Biotechnology Information database website (http://www.ncbi.nlm.nih.gov/BLAST). According to annotations, we excluded homologs that are related to but apparently different from IF2, such as elongation factors. Homologs that do not contain the GTP binding domain are also excluded. Multiple alignment of the homologs was performed by GENETYX software version 7 (GENETYX) with the default parameters (gap penalty: gap insert −12 [≤ 0], gap extend −4 [≤ 0]; amino acid similarity matrix: PAM250), and the alignment was manually adjusted within the GTP binding domain (GXXXXGKT and GIT) by partial multiple alignment function. For constructing phylogenetic trees, the neighbor-joining method in the GENETYX software was used, and the bootstrap analysis with 1000 replicates was performed to test the confidence of topology. The tree was shown as rooted in GENETYX, and the distance between the operational taxonomic units at the root region is partitioned into half. Multiple alignment of the following IF2 homologs shown in Figure 3B is indicated in Supplemental Figure 3 online: Arabidopsis 1 and 2, Oryza sativa 1 and 2, Ostreococcus tauri, Saccharomyces cerevisiae, Candida albicans SC5314, Schizosaccharomyces pombe, Mus musculus, Rattus norvegicus, Bos taurus, Homo sapiens, Pan troglodytes, Yersinia pestis CO92, Escherichia coli K12, Bacillus subtilis subsp strain 168, Phaseolus vulgaris, Euglena gracilis, Prochlorococcus marinus strain MIT 9312, Guillardia theta, and Porphyra yezoensis.
Complementation of an Escherichia coli infB Null Mutation
Complementation of the E. coli strain SL598R was performed as described by Laalami et al. (1991). The principle is based upon a thermosensitive lambda lysogen containing the infB gene in the SL598R strain. SL598R contains an insertion of a cat gene cassette that interrupts the chromosomal infB gene. This disruption effectively eliminates IF2 function, and cells are unable to grow without lambda lysogen under permissive temperatures. However, under nonpermissive temperatures (42°C), cells are unable to survive unless a homologous or heterologous IF2 can complement IF2 function. The cDNA fragments corresponding to a mature-sized cpIF2 (lacking its transit peptide of 64 N-terminal residues) or to a truncated version of IF2 (lacking 421 N-terminal residues) were PCR amplified by the following primers: At1g17220 TP-XbaI and At1g17220 SalI-R2 or At1g17220 Cam-XbaI and At1g17220 SalI-R2. The resulting fragments were cloned into the expression vector pTrc99A and were termed as pTrc99AcpIF2Δ64 and pTrc99AcpIF2Δ421, respectively. These plasmids, in addition to control plasmids (pTrc99A and p18-1 containing an intact infB gene), were independently transformed into SL598R. After heat-stressing cells at 42°C, cell survivors were subjected to PCR analysis to confirm the absence of endogenous infB (using primers E. coli infB-F and E. coli infB-R).
In Vivo Labeling of Chloroplast Proteins
Chloroplast proteins were labeled in vivo using intact Arabidopsis leaves (fully expanded leaves, 7 to 8 weeks old under 12-h-light/12-h-dark condition) with a slightly modified method as previously described by Aro et al. (1993). We prepared 5 mg of a square-cut leaflet from Col and fug1-3, and two pieces were used to isolate total thylakoid proteins at each time point. The leaflets were placed in a glass vial containing 0.1 mCi/mL [35S]Met in 0.2% Tween 20, and the vial was sealed with a rubber cap. A syringe needle was pierced through the rubber cap, and radiolabels were infiltrated into the leaflets by repeated pressuring and depressuring with the syringe for ∼30 s. Leaflets were immediately placed on wet filter papers, and leaflets were irradiated with light by being placed under a fluorescence bulb (150 mmol/m2/s). To isolate thylakoid proteins, two pieces of leaflets were homogenized in ice-cold extraction buffer (50 mM HEPES, pH 7.6, 0.3 M mannitol, 10 mM NaCl, and 5 mM MgCl2). Homegenates were centrifuged at 4500g for 5 min, and pellets were subsequently washed with wash buffer (same as extraction buffer but with 0.1 M mannitol), and proteins were separated by SDS-PAGE. Thylakoid pellets were solubilized in SDS sample buffer (0.67 M Tris-HCl, pH 6.8, 2.3% [w/v] SDS, 5% [v/v] 2-mercaptoethanol, 10% [v/v] glycerol, and 0.08% [w/v] bromophenol blue) and denatured at 100°C for 2 min. The samples were separated by 14% (w/v) SDS-PAGE containing 7 M urea. The gels were stained by Coomassie Brilliant Blue R 250, destained, dried, and placed on an imaging plate (Fuji Photo Film). Radiolabels were detected by the BAS 1000 image analyzer (Fuji Photo Film).
Protein Extraction and Immunoblot Analysis
To isolate chloroplasts, 2 g of fully expanded leaves (7 to 8 weeks old under 12-h-light/12-h-dark condition) were ground in a blender with homogenization buffer (330 mM mannitol, 50 mM HEPES, and 2 mM Na2EDTA). Homogenates were then filtered through gauze, and the recovered extracts were centrifuged at 600g for 2 min. The recovered pellet was resuspended in 1 mL of homogenization buffer and was overlaid on three Percoll gradients (10, 40, and 80%) and centrifuged at 2500g for 15 min. The sediment at the interface between the 40 and 80% gradient, which represents intact chloroplasts, was recovered and used for further analysis. Chloroplast proteins (equivalent to 3 μg chlorophyll) were separated by SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane (ATTO). Immunoblot analysis was performed as described previously (Sakamoto et al., 2003). To generate a polyclonal antibody against cpIF2, a DNA fragment corresponding to the 336 C-terminal amino acids of cpIF2 was amplified by PCR and directly cloned into the expression vector pCR T7/NT-TOPO (Invitrogen). The truncated cpIF2 was expressed in the E. coli strain BL21-CodonPlus (DE)-RIL. The truncated protein was resolved with SDS-PAGE, and the recombinant protein band was excised and used for the generation of polyclonal antibodies as previously described (Sakamoto et al., 2003).
Chlorophyll Fluorescence Analysis
The FluorCam700MF (Photon Systems Instruments) was used according to the instructions provided by the manufacturer for measuring chlorophyll fluorescence emission. The maximum yield of chlorophyll fluorescence in the dark-adapted state (Fm) was induced by an 800-ms pulse of saturating white light (1200 μmol/m2/s). Variable fluorescence Fv, which is defined as the difference Fm – Fo (the minimum fluorescence in dark-adapted state), combined with maximum fluorescence Fm are used to calculate the maximum quantum yield of PSII photochemistry (Fv/Fm). Prior to the measurement, plants were maintained in the dark for 10 min, and the initial Fv/Fm values were recorded based on measuring Fm and Fo. Plants were then exposed to high light (800 μmol/m2/s) for 4 h using a halogen light source.
Accession Numbers
Accession numbers of IF2 homologs are as follows: Arabidopsis 1 and 2 (At4g11160 and At1g17220), O. sativa 1 and 2 (Os09g0515500 and Os05g0575300), O. tauri (CR954209.4), S. cerevisiae (X58379.1), C. albicans SC5314 (XM_711829.1), S. pombe (AL034353.2), M. musculus (BX284634.8), R. norvegicus (BC079421.1), B. taurus (NM_174393.2), H. sapiens (NM_001005369.1), P. troglodytes (XM_515483.1), Y. pestis CO92 (NC_003143.1), E. coli K12 (NC_000913.2), B. subtilis subsp Subtilis strain 168 (NC_000964.2), P. vulgaris (AF324244.1), E. gracilis (AF109129), Nostoc sp PCC7120 (BC_003272.1), Synechocystis sp PCC6803 (BA000022.2), P. marinus strain MIT 9312 (NC_007577.1), G. theta (NC_000926.1), and P. yezoensis (NC_007932.1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Segregation of Normal and Defective Seeds in the Young Siliques of Plants Heterozygous for fug1-2.
Supplemental Table 2. List of the Primers Used in This Study.
Supplemental Figure 1. Schematic Representation of Map-Based Cloning of the FUG1 Locus.
Supplemental Figure 2. Multiple Sequence Alignment of IF2 Homologs.
Supplemental Figure 3. Multiple Alignment Used for the Phylogenetic Tree in Figure 3B.
Supplementary Material
Acknowledgments
We thank Rie Hijiya and Chieko Hattori for their technical assistance. We also thank the ABRC for providing us with the T-DNA lines. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (16085207) and by the Oohara Foundation (to W.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Wataru Sakamoto (saka@rib.okayama-u.ac.jp).
Online version contains Web-only data.
References
- Adam, Z., Rudella, A., and van Wijk, K.J. (2006). Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9 234–240. [DOI] [PubMed] [Google Scholar]
- Albrecht, V., Ingenfeld, A., and Apel, K. (2006). Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: The impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 60 507–518. [DOI] [PubMed] [Google Scholar]
- Aluru, M.R., Yu, F., Fu, A., and Rodermel, S. (2006). Arabidopsis variegation mutants: New insights into chloroplast biogenesis. J. Exp. Bot. 57 1871–1881. [DOI] [PubMed] [Google Scholar]
- Aro, E.M., McCaffery, S., and Anderson, J.M. (1993). Photoinhibition and D1 protein degradation in peas acclimated to different growth irradiances. Plant Physiol. 103 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro, E.M., McCaffery, S., and Anderson, J.M. (1994). Recovery from photoinhibition in peas (Pisum sativum L.) acclimated to varying growth irradiances (role of D1 protein turnover). Plant Physiol. 104 1033–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, S., Thompson, E., Nixon, P.J., Horton, P., Mullineaux, C.W., Robinson, C., and Mann, N.H. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277 2006–2011. [DOI] [PubMed] [Google Scholar]
- Bieniossek, C., Schalch, T., Bumann, M., Meister, M., Meier, R., and Baumann, U. (2006). The molecular architecture of the metalloprotease FtsH. Proc. Natl. Acad. Sci. USA 103 3066–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, F., Garcia-Gomez, B.I., Solorzano, R.M., Salazar, E., Estevez, J., Leon, P., Alvarez-Buylla, E.R., and Covarrubias, A.A. (2001). A cDNA for nuclear-encoded chloroplast translational initiation factor 2 from a higher plant is able to complement an infB Escherichia coli null mutant. J. Biol. Chem. 276 28388–28394. [DOI] [PubMed] [Google Scholar]
- Chen, M., Choi, Y., Voytas, D.F., and Rodermel, S. (2000). Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 22 303–313. [DOI] [PubMed] [Google Scholar]
- Chuang, C.F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A.K., MacDonald, T.M., and Sjögren, L.L.E. (2005). The ATP-dependent Clp protease in chloroplasts of higher plants. Physiol. Plant 123 406–412. [Google Scholar]
- Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S.L., Briat, J.F., and Walker, E.L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409 346–349. [DOI] [PubMed] [Google Scholar]
- Garcia-Lorenzo, M., Żelisko, A., Jackowski, G., and Funk, C. (2005). Degradation of the main Photosystem II light-harvesting complex. Photochem. Photobiol. Sci. 4 1065–1071. [DOI] [PubMed] [Google Scholar]
- Gray, J., Janick-Buckner, D., Buckner, B., Close, P.S., and Johal, G.S. (2002). Light-dependent death of maize lls1 cells is mediated by mature chloroplasts. Plant Physiol. 130 1894–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K., and Akiyama, Y. (2005). Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 59 211–231. [DOI] [PubMed] [Google Scholar]
- Kirk, J.T.O., and Tilney-Bassett, R.A.E. (1978). The Plastids: Their Chemistry, Structure, Growth and Inheritance. (Amsterdam: Elsevier/North-Holland Biomedical Press).
- Komenda, J., Barker, M., Kuviková, S., de Vries, R., Mullineaux, C.W., Tichy, M., and Nixon, P.J. (2006). The FtsH protease, slr0228, is important for quality control of photosystem two in the thylakoid membrane of Synechocystis PCC 6803. J. Biol. Chem. 281 1145–1151. [DOI] [PubMed] [Google Scholar]
- Kovacheva, S., Bedard, J., Patel, R., Dudley, P., Twell, D., Rios, G., Koncz, C., and Jarvis, P. (2005). In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 41 412–428. [DOI] [PubMed] [Google Scholar]
- Krzywda, S., Brzozowski, A.M., Verma, C., Karata, K., Ogura, T., and Wilkinson, A.J. (2002). The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5 Å resolution. Structure 10 1073–1083. [DOI] [PubMed] [Google Scholar]
- Laalami, S., Putzer, H., Plumbridge, J.A., and Grunberg-Manago, M. (1991). A severely truncated form of translational initiation factor 2 supports growth of Escherichia coli. J. Mol. Biol. 220 335–349. [DOI] [PubMed] [Google Scholar]
- Laursen, B.S., Sorensen, H.P., Mortensen, K.K., and Sperling-Petersen, H.U. (2005). Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister, D. (2003). Chloroplast research in the genomic age. Trends Genet. 19 47–56. [DOI] [PubMed] [Google Scholar]
- Lin, Q., Ma, L., Burkhart, W., and Spremulli, L.L. (1994). Isolation and characterization of cDNA clones for chloroplast translation initiation factor-3 from Euglena gracilis. J. Biol. Chem. 269 9436–9444. [PubMed] [Google Scholar]
- Lindahl, M., Spetea, C., Hundal, T., Oppenheim, A.B., Adam, Z., and Andersson, B. (2000). The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Juez, E., and Pyke, K.A. (2005). Plastids unleashed: Their development and their integration in plant development. Int. J. Dev. Biol. 49 557–577. [DOI] [PubMed] [Google Scholar]
- Ma, L., and Spremulli, L.L. (1990). Identification and characterization of large, complex forms of chloroplast translational initiation factor 2 from Euglena gracilis. J. Biol. Chem. 265 13560–13565. [PubMed] [Google Scholar]
- Mach, J.M., Castillo, A.R., Hoogstraten, R., and Greenberg, J.T. (2001). The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. USA 98 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuell, A., Beligni, M.V., Yamaguchi, K., and Mayfield, S.P. (2004). Regulation of chloroplast translation: Interactions of RNA elements, RNA-binding proteins and the plastid ribosome. Biochem. Soc. Trans. 32 601–605. [DOI] [PubMed] [Google Scholar]
- Mateo, A., Muhlenbock, P., Rusterucci, C., Chang, C.C., Miszalski, Z., Karpinska, B., Parker, J.E., Mullineaux, P.M., and Karpinski, S. (2004). LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136 2818–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa, J., and Takahashi, Y. (2004). Structure, function and assembly of Photosystem II and its light-harvesting proteins. Photosynth. Res. 82 241–263. [DOI] [PubMed] [Google Scholar]
- Misumi, O., Matsuzaki, M., Nozaki, H., Miyagishima, S.Y., Mori, T., Nishida, K., Yagisawa, F., Yoshida, Y., Kuroiwa, H., and Kuroiwa, T. (2005). Cyanidioschyzon merolae genome. A tool for facilitating comparable studies on organelle biogenesis in photosynthetic eukaryotes. Plant Physiol. 137 567–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen, K.K., Kildsgaard, J., Moreno, J.M., Steffensen, S.A., Egebjerg, J., and Sperling-Petersen, H.U. (1998). A six-domain structural model for Escherichia coli translation initiation factor IF2. Characterisation of twelve surface epitopes. Biochem. Mol. Biol. Int. 46 1027–1041. [DOI] [PubMed] [Google Scholar]
- Neuwald, A.F., Aravind, L., Spouge, J.L., and Koonin, E.V. (1999). AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9 27–43. [PubMed] [Google Scholar]
- Ogura, T., and Wilkinson, A.J. (2001). AAA+ superfamily ATPases: Common structure–diverse function. Genes Cells 6 575–597. [DOI] [PubMed] [Google Scholar]
- Ostersetzer, O., and Adam, Z. (1997). Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: The possible role of the FtsH protease. Plant Cell 9 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S., and Rodermel, S.R. (2004). Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc. Natl. Acad. Sci. USA 101 12765–12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan, V. (2002). Ribosome structure and the mechanism of translation. Cell 108 557–572. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W. (2003). Leaf-variegated mutations and their responsible genes in Arabidopsis thaliana. Genes Genet. Syst. 78 1–9. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W. (2006). Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57 599–621. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Tamura, T., Hanba-Tomita, Y., and Murata, M. (2002). The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7 769–780. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Zaltsman, A., Adam, Z., and Takahashi, Y. (2003). Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, G., Timberg, R., and Ohad, I. (1988). Turnover of thylakoid photosystem II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur. J. Biochem. 177 403–410. [DOI] [PubMed] [Google Scholar]
- Sjögren, L.L., MacDonald, T.M., Sutinen, S., and Clarke, A.K. (2004). Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 136 4114–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takechi, K., Sodmergen, Murata, M., Motoyoshi, F., and Sakamoto, W. (2000). The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol. 41 1334–1346. [DOI] [PubMed] [Google Scholar]
- Tsukaya, H., Okada, H., and Mohamed, M. (2004). A novel feature of structural variegation in leaves of the tropical plant Schismatoglottis calyptrata. J. Plant Res. 117 477–480. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Duby, G., Purnelle, B., and Boutry, M. (2000). Tobacco VDL gene encodes a plastid DEAD box RNA helicase and is involved in chloroplast differentiation and plant morphogenesis. Plant Cell 12 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaronskaya, E., Ziemann, V., Walter, G., Averina, N., Börner, T., and Grimm, B. (2003). Metabolic control of the tetrapyrrole biosynthetic pathway for porphyrin distribution in the barley mutant albostrians. Plant J. 35 512–522. [DOI] [PubMed] [Google Scholar]
- Yu, F., Park, S., and Rodermel, S.R. (2004). The Arabidopsis FtsH metalloprotease gene family: Interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 37 864–876. [DOI] [PubMed] [Google Scholar]
- Yu, F., Park, S., and Rodermel, S.R. (2005). Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol. 138 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman, A., Feder, A., and Adam, Z. (2005. a). Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts–Implications for thylakoid formation and photosystem II maintenance. Plant J. 42 609–617. [DOI] [PubMed] [Google Scholar]
- Zaltsman, A., Ori, N., and Adam, Z. (2005. b). Two types of FtsH protease subunits are required for chloroplast biogenesis and Photosystem II repair in Arabidopsis. Plant Cell 17 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Żelisko, A., Garcia-Lorenzo, M., Jackowski, G., Jansson, S., and Funk, C. (2005). AtFtsH6 is involved in the degradation of the light-harvesting complex II during high-light acclimation and senescence. Proc. Natl. Acad. Sci. USA 102 13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.