Abstract
The C-terminal domain (CTD) of RNA polymerase II is phosphorylated during the transcription cycle by three cyclin-dependent kinases (CDKs): CDK7, CDK8, and CDK9. CDK9 and its interacting cyclin T partners belong to the positive transcription elongation factor b (P-TEFb) complexes, which phosphorylate the CTD to promote transcription elongation. We report that Arabidopsis thaliana CDK9-like proteins, CDKC;1 and CDKC;2, and their interacting cyclin T partners, CYCT1;4 and CYCT1;5, play important roles in infection with Cauliflower mosaic virus (CaMV). cdkc;2 and cyct1;5 knockout mutants are highly resistant and cdkc;2 cyct1;5 double mutants are extremely resistant to CaMV. The mutants respond normally to other types of plant viruses that do not replicate by reverse transcription. Expression of a reporter gene driven by the CaMV 35S promoter is markedly reduced in the cdkc;2 and cyct1;5 mutants, indicating that the kinase complexes are important for transcription from the viral promoter. Loss of function of CDKC;1/CDKC;2 or CYCT1;4/CYCT1;5 results in complete resistance to CaMV as well as altered leaf and flower growth, trichome development, and delayed flowering. These results establish Arabidopsis CDKC kinase complexes as important host targets of CaMV for transcriptional activation of viral genes and critical regulators of plant growth and development.
INTRODUCTION
The transcription of protein-coding genes in eukaryotes is performed by RNA polymerase II (RNAP II) and forms a so-called transcription cycle that includes preinitiation, initiation, promoter clearance, elongation, and termination (Sims et al., 2004). The transcription cycle starts with the assembly of the preinitiation complex at the promoter, which includes the general transcription factors TFIID, TFIIB, TFIIE, and TFIIH as well as RNAP II. Assembly of the preinitiation complex is followed by ATP-dependent melting of the double-stranded DNA (dsDNA) template and formation of an open complex between RNAP II and the DNA template. Once the open complex is established, transcription initiation occurs and RNAP II is allowed to clear the promoter and engaged to make the transition from initiation to elongation. Transcription elongation is a dynamic and highly regulated process that coordinates downstream events such as capping and splicing of primary transcripts. The final step in the cycle is transcription termination. At this stage, the mRNA is cleaved, polyadenylated, and transported to the cytoplasm for translation.
The transcription cycle involving RNAP II is accompanied by another cycling event: phosphorylation of the C-terminal domain (CTD) of the largest subunit of RNAP II (Sims et al., 2004). The RNAP II CTD contains multiple repeats of the heptapeptide sequence YSPTSPS (26 such repeats in yeast, 32 in Caenorhabditis elegans, 45 in Drosophila, and 52 in mammals). RNAP II in the preinitiation complex is unphosphorylated or hypophosphorylated, whereas transcription-competent RNAP II is heavily phosphorylated on its CTD, with Ser-2 and Ser-5 as the major modified residues. Phosphorylation of the CTD regulates the transition of RNAP II from initiation to elongation and the efficiency of elongation and pre-mRNA processing (capping, splicing, and polyadenylation).
Three cyclin-dependent kinases (CDKs), CDK7, CDK8, and CDK9, are involved directly in the phosphorylation of the CTD and in the regulation of different stages of the transcription cycle (Pinhero et al., 2004). CDK7 and its cyclin (Cyc) H partner are components of the general RNAP II transcription factor, TFIIH, involved in transcription initiation (Nigg, 1996). CDK8 and its CycC partner are also components of the RNAP II holoenzyme; they function to negatively regulate initiation by phosphorylation of the CTD of RNAP II and the CycH subunit of TFIIH (Akoulitchev et al., 2000). CDK9 and its CycT or CycK partner belong to the positive transcription elongation factor b (P-TEFb) (Marshall and Price, 1995).
In mammalian cells, several factors that function in elongation have been purified and identified, including P-TEFb, DSIF (for 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole–sensitive-inducing factor), and NELF (negative elongation factor). Once transcription is initiated, NELF interacts with DSIF to induce RNAP II pausing, resulting in arrested transcription (Sims et al., 2004). P-TEFb can overcome the effects of the DSIF/NELF complex and release RNAP II from its arrest. P-TEFb phosphorylates the CTD of RNAP II, thus catalyzing the transition from initiation to elongation of transcription (Sims et al., 2004). In humans, P-TEFb is recruited by the Tat protein of human immunodeficiency virus type I (HIV-1) to stimulate transcriptional initiation and elongation (Mancebo et al., 1997).
Arabidopsis thaliana contains a single gene, NRPB1 (At4g35800), encoding the largest subunit of RNAP II with a CTD containing 42 repeats of the YSPTSPS heptapeptide (Dietrich et al., 1990). CTD kinases and phosphatases that control the dynamic change of the CTD phosphoarray have also been identified in plants. In Arabidopsis, three genes encode D-type CDK7 proteins: CDKD;1, CDKD;2, and CDKD;3 (Shimotohno et al., 2003). CDKD;2 forms a stable complex with CycH;1 and exhibits high kinase activity toward the CTD of RNAP II. T-DNA insertion mutants for CDKD;1 and CDKD;3, however, exhibit no developmental defects throughout the life cycle under normal growth conditions, suggesting that the two CDK7 genes are not essential for plant development (Shimotohno et al., 2003). Arabidopsis contains a single gene encoding an E-type CDK8 protein (CDKE;1; At5g63610) with CTD kinase activity. CDKE;1, also known as HEN3, is required for the specification of stamen and carpel identities and for the proper termination of stem cells in the floral meristem (Wang and Chen, 2004). There are >20 genes encoding CTD phosphatase–like proteins (CPLs) in Arabidopsis, based on domain architecture identified in family member proteins of other eukaryotes (Koiwa et al., 2004). Genetic evidence indicates that CPL3 is a negative regulator of abiotic stress signaling (Koiwa et al., 2002). Thus, the dynamic regulation of CTD phosphorylation plays an important role in plant growth, development, and response to environmental conditions.
Proteins similar to C-type CDK9 have been reported in tomato (Solanum lycopersicum) (Joubes et al., 2001) and Medicago (Fulop et al., 2005). As with CDK9 genes from other organisms, expression of these plant CDK9 genes is not regulated by the cell cycle. Furthermore, the Medicago CDKC1/cyclin T1 kinase complex phosphorylates the CTD of RNAP II and restores the transcriptional activity of a HeLa nuclear extract depleted of endogenous CDK9 complexes (Fulop et al., 2005). Thus, plant CDK9/cyclin T complexes have a role in transcription similar to that of P-TEFb. Arabidopsis contains two genes encoding CDK9-like proteins, CDKC;1 (At5g10270) and CDKC;2 (At5g64960) (Menges et al., 2005), and five genes encoding cyclin T-like proteins, CYCT1;1 (At1g35440), CYCT1;2 (At4g19560), CYCT1;3 (At1g27630), CYCT1;4 (At4g19600), and CYCT1;5 (At5g45190) (Wang et al., 2004). Yeast two-hybrid assays and in vitro immunoprecipitation have shown that both CDKC;1 and CDKC;2 interact with CYCT1;3, suggesting that they form P-TEFb–like complexes (Barroco et al., 2003). However, the biological roles of the Arabidopsis CDKC complexes have not been genetically analyzed.
In this study, we generated loss-of-function mutants for the two CDKC and five CYCT1 genes from Arabidopsis for analysis of their roles in infection by Cauliflower mosaic virus (CaMV), a dsDNA virus that depends on host RNAP II for transcription. We also analyzed their phenotypes in leaf and flower growth, trichrome development, and flowering. Our results indicate that Arabidopsis P-TEFb–like kinase complexes are important host targets of CaMV for transcriptional activation of viral genes and critical regulators of plant growth and development.
RESULTS
Arabidopsis cdkc;2 and cyct1;5 Mutants Are Resistant to CaMV
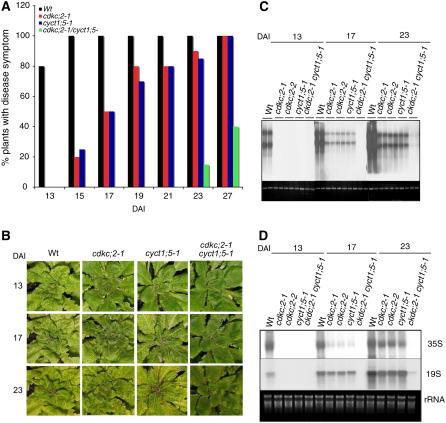
CaMV is a pararetrovirus whose DNA genome is replicated by the reverse transcription of an RNA intermediate. The CaMV genome consists of a circular dsDNA molecule of ∼8 kb that forms a minichromosome in the nucleus of the host cell. It is transcribed unidirectionally by the cellular RNAP II into two major capped and polyadenylated transcripts, the 35S and 19S RNAs, from their own promoters (Hull, 2002). Arabidopsis is a host to CaMV; therefore, it can be used as a model system for the identification of host factors important for infection. Because of the reliance of CaMV on host RNAP II to synthesize its viral RNA templates for reverse transcription and translation, we examined the role in CaMV infection of Arabidopsis CDKC;2, which encodes a homolog to the CDK9 subunit of P-TEFb (Barroco et al., 2003) (see Supplemental Figure 1 online). We identified two independent T-DNA insertion mutants for CDKC;2: cdkc;2-1, with a T-DNA insertion in the 10th intron; and cdkc;2-2, with a T-DNA insertion in the 8th exon (see Supplemental Figure 2 online). RNA gel blotting failed to detect CDKC;2 full-length transcripts of the expected size in either mutant. To determine the impact of the disruption of CDKC;2 on CaMV infection, we mechanically inoculated wild-type and mutant plants with partially purified CaMV virons and compared them for disease symptom development. In wild-type plants, disease symptoms (initially vein clearing and chlorosis, followed by leaf stunting and curling) was first observed at ∼10 to 12 d after inoculation (DAI); at 13 DAI, ∼80% of wild-type plants had developed the symptoms (Figure 1A). At these stages, however, no disease symptoms were observed in the cdkc;2 mutants (Figure 1A). At 15 DAI, 100% of wild-type plants but only 20% of the mutant plants developed disease symptoms (Figure 1A). During the remaining period up to 27 DAI, the percentages of the mutant plants that developed disease symptoms gradually increased and could reach up to 100% at 4 weeks after inoculation (Figure 1A). A similar delay in disease symptom development in the cdkc;2 mutants was observed when the plants were inoculated by particle bombardment of an infectious CaMV DNA clone with artificial long terminal repeats (pCa122) (Kobayashi et al., 2002) (Figure 1B).
Figure 1.
Enhanced Resistance of cdkc;2 and cyct1;5 Mutants to CaMV.
(A) Disease symptom development in mechanically inoculated plants. Forty wild-type, cdkc;2-1, and cyct1;5-1 single or double mutant plants were mechanically inoculated with partially purified CaMV virons (0.5 mg protein/mL), and plants with CaMV mosaic symptoms were scored at the indicated DAI.
(B) Disease symptom development in plants inoculated by bombardment of the infectious CaMV DNA clone pCa122. Photographs of representative plants were taken at the indicated DAI. Disease symptom development in the cdkc;2-2 mutant was the same as in the cdkc;2-1 mutant.
(C) Viral DNA accumulation. Wild-type and mutant plants were inoculated by bombardment of pCa122, and total DNA was isolated from leaf tissues harvested at the indicated DAI and probed with the P6 open reading frame of CaMV. Ethidium bromide staining of genomic DNA is shown as a loading control.
(D) Viral RNA accumulation. Total RNA was isolated from inoculated plants and first probed with the P3 open reading frame of CaMV for detection of the 35S RNA. The blot was stripped and reprobed with the P6 open reading frame of CaMV for detection of the 19S RNA. Ethidium bromide staining of rRNA is shown for the assessment of equal loading.
Accumulation of viral DNAs and RNAs was also examined in whole plants after inoculation by particle bombardment of pCa122. At 13 DAI, CaMV viral DNAs and RNAs were detected at high levels in wild-type plants but were undetected in the cdkc;2 mutants (Figures 1C and 1D). At 17 DAI, viral DNAs were detected in the mutants, but their levels were substantially lower than those in the wild-type plants (Figure 1C). Likewise, reduced levels of viral RNAs were detected at 17 DAI in the cdkc;2 mutants relative to those in the wild-type plants; the reduction was particularly pronounced for the full-length 35S RNA (Figure 1D). At 23 DAI, the viral RNAs in the mutants increased to relatively high levels but were still substantially lower than those in the wild-type plants (Figure 1D). Thus, similar to disease symptom development, the accumulation of CaMV DNAs and RNAs was substantially delayed and reduced as a result of the disruption of the host CDKC;2 gene.
The CDK9 subunit of P-TEFb forms functional kinase complexes with multiple cyclin T subunits (Peng et al., 1998). Arabidopsis contains five genes encoding cyclin T–like proteins (CYCT1;1 to CYCT1;5) (Wang et al., 2004). T-DNA insertion mutants were identified for CYCT1;1 to CYCT1;4 (see Supplemental Figure 2 online) from various collections, but they exhibited no altered phenotypes in response to CaMV infection. On the other hand, the cyct1;5-1 T-DNA insertion mutant was deficient in its capacity to support CaMV infections, a phenotype strikingly similar to that of the cdkc;2 mutants. As in the cdckc;2 mutants, disease symptom development was delayed by 3 to 4 d in the cyct1;5-1 mutant relative to that in wild-type plants after CaMV inoculation (Figures 1A and 1B). Accumulation of viral DNAs and RNAs was also similarly reduced in the cyct1;5-1 mutant relative to that in wild-type plants (Figures 1C and 1D). To confirm that the altered response of the cyct1;5-1 mutant to CaMV was due to the disruption of CYCT1;5, we performed genetic complementation of the mutant. A full-length CYCT1;5 cDNA clone was placed behind its native promoter and transformed into the cyct1;5-1 mutant. Transformants were identified through BASTA selection and analyzed for CaMV infection. Transformation of the mutant with CYCT1;5 resulted in complete recovery of the wild-type phenotype in CaMV response (see Supplemental Figure 3 online). This finding indicates that the enhanced resistance of the cyct1;5-1 mutant to CaMV is the result of the disruption of CYCT1;5.
To determine whether the mutants for the two genes have additive effects on CaMV resistance, we generated cdkc;2 cyct1;5 double mutants through genetic crosses. After CaMV infection, disease symptom development was further delayed in the double mutants compared with the cdkc;2 and cyct1;5 single mutants (Figures 1A and 1B). Thus, at ∼3 weeks after CaMV inoculation, almost 100% of the cdkc;2 and cyct1;5 single mutants developed disease symptoms, but only10 to 20% of the cdkc;2 cyct1;5 double mutant plants started to develop mild mosaic symptoms (Figures 1A and 1B). During the remaining period of the experiments, up to 4 weeks after inoculation, the percentage of the cdkc;2 cyct1;5 double mutant plants exhibiting symptoms increased, but 40 to 50% of the mutants failed to develop any symptoms at all (Figure 1A). Accumulation of viral DNA and RNA was also further delayed and reduced in the double mutants compared with the single mutants. At 17 DAI, viral DNAs and RNAs were detected in cdkc;2 and cyct1;5 single mutants, albeit at reduced levels relative to those in wild-type plants; no viral DNAs or RNAs were detected in the cdkc;2 cyct1;5 double mutants at this stage (Figures 1C and 1D). Even at the very late stages of infection (e.g., at 23 DAI), only trace amounts of viral DNAs and RNAs were detected in the cdkc;2 cyct1;5 double mutants (Figures 1C and 1D). These results indicate that the cdkc;2 cyct1;5 double mutants were extremely resistant to CaMV.
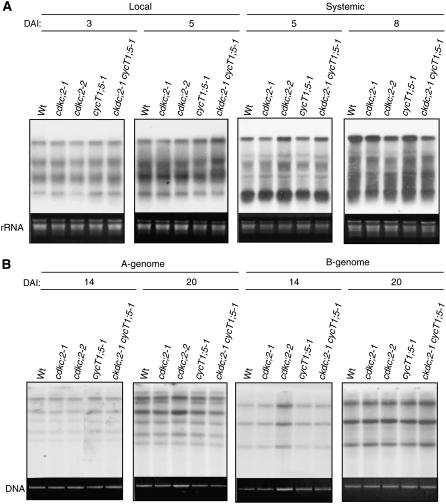
To determine whether the observed resistance of the cdkc;2 and cyct1;5 mutants to CaMV infection is specific to CaMV, we tested the mutants for their responses to a tobamovirus (tobacco mosaic virus isolate cg [TMV-cg], a RNA virus) (Ishikawa et al., 1991) and a geminivirus (Cabbage leaf curl virus [CaLCuV], a single-stranded DNA virus) (Turnage et al., 2002). After inoculation of three lower fully expanded leaves with purified TMV-cg virons, the cdkc;2 and cyct1;5 single and double mutants accumulated viral RNAs in both lower inoculated leaves and upper systemically infected leaves to levels similar to those in wild-type plants (Figure 2A). When infectious DNA clones of CaLCuV were inoculated through particle bombardment, these mutants also accumulated the same levels of viral DNAs as wild-type plants (Figure 2B). Thus, these mutants respond normally to TMV-cg and CaLCuV, two viruses that do not replicate through a reverse transcription mechanism.
Figure 2.
Response of Arabidopsis cdkc;2 and cyct1;5 Mutants to TMV-cg and CaLCuV.
(A) Accumulation of TMV-cg viral RNAs. Three leaves of 4-week-old Arabidopsis plants were inoculated with TMV-cg (5 μg/mL). The lower, inoculated leaves and upper, systemic infected leaves of five plants were collected at the indicated DAI for RNA isolation. RNA gel blot analysis was performed with 32P-labeled TMV-cg coat protein gene as a probe.
(B) Accumulation of CaLCuV DNA-A and DNA-B. Four-week-old Arabidopsis plants were bombarded with the infectious DNA clones of CaLCuV. Total DNA was isolated from infected leaves at the indicated DAI. DNA gel blot analysis was performed with 32P-labeled DNA fragments specific to the A or B components of CaLCuV.
Reduced CaMV 35S Promoter Activity in the cdck;2 and cyct1;5 Mutants
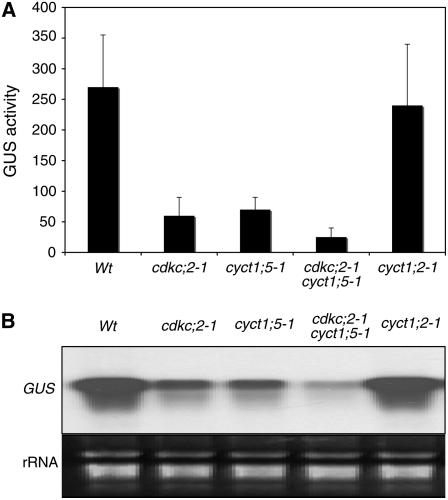
The CaMV 35S promoter, which directs the production of the 35S pregenomic RNA, is strongly active in plant cells in the absence of any viral protein (Odell et al., 1985). To test whether CDKC;2 and CYCT1;5 are important for the viral promoter activity, we transformed the cdkc;2 and cyct1;5 mutant plants with a construct containing a β-glucuronidase (GUS) reporter gene driven by the CaMV 35S promoter. As controls, the same reporter gene construct was also transformed into the wild type and the cyct1;2-1 insertion mutant, which responds normally to CaMV (see Supplemental Figure 2 online). To determine the transgene expression, we assayed GUS activity and GUS transcripts in 15 to 20 independent wild-type or mutant transformants. In the wild-type background, the transformants had an average of ∼265 units of GUS activity (Figure 3A) and accumulated high levels of GUS transcripts (Figure 3B). In the cdkc;2 and cyct1;5 single mutant backgrounds, the average GUS activities were ∼65 units, representing a fourfold reduction from those in the wild-type plants (Figure 3A). The reduced GUS activity in these single mutant transformants was correlated with reduced levels of GUS transcripts (Figure 3B). In the cdkc;2 cyct1;5 double mutant background, the average GUS activity was further reduced to ∼35 units, and the reduced GUS activity was correlated with the very low levels of GUS transcripts in the double mutant transformants (Figure 3). By contrast, both the GUS activities and GUS transcripts in the cyct1;2 mutant transformants were similar to those in the wild-type transformants (Figure 3). Thus, CDKC;2 and CYCT1;5 are required for the high CaMV 35S promoter activity.
Figure 3.
CDKC;2 and CYCT1;5 Dependence of the CaMV 35S Promoter Activity.
(A) GUS activities in the wild type, cdkc;2, and cyct1;5 mutants transformed with a GUS transgene driven by the CaMV 35S promoter. Average GUS activities were calculated from 15 to 20 independent T1 transformants. GUS activities are expressed in units (nanomoles of 4-methylumbelliferone per minute per milligram of total soluble protein).
(B) Accumulation of GUS transcripts. Total RNA was pooled from 15 to 20 T1 transformants and probed with the full-length GUS gene fragment.
Complete CaMV Resistance of the Non-Null cdkc;1 cdkc;2 and cyct1;4 cyct1;5 Double Mutants
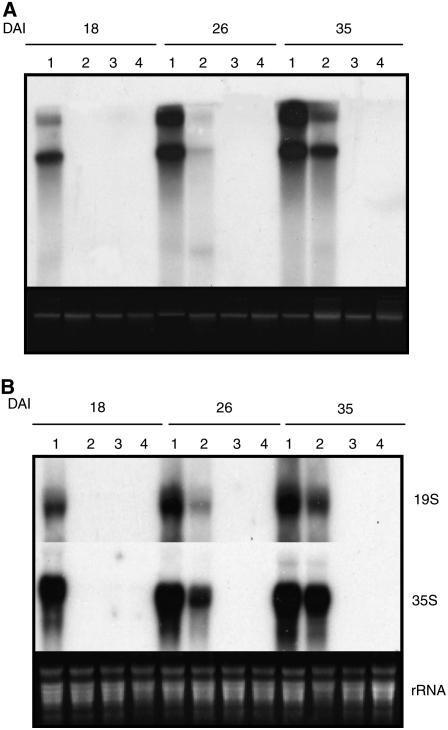
Although the cdkc;2 and cyct1;5 mutants are null, they behave like leaky mutants in that they are not completely resistant to CaMV and there is an additive effect between the two mutants in CaMV resistance (Figure 1). The leaky phenotypes of the null mutants are probably due to the presence of functionally related homologs. Arabidopsis CDKC;2 and CYCT1;5 are structurally closely related to CDKC1 and CYCT1;4, respectively (see Supplemental Figure 1 online). Because there is no knockout mutant for CDKC;1 and the double knockout mutant of cyct1;4−/− and cyct1;5−/− is embryo-lethal (see below), we transformed a CDKC;1 RNA interference (RNAi) construct into cdkc;2-1 and a CYCT1;4 RNAi construct into cyct1;5-1 to generate non-null double mutants. T1 transgenic plants (cdkc;2/CDKC;1 RNAi and cyct1;5/CYCT1;4 RNAi) with suppressed expression of the targeted genes were identified by RT-PCR (see Supplemental Figure 4 online) and tested for CaMV resistance. The non-null cdkc;2/CDKC;1 RNAi and cyct1;5/CYCT1;4 RNAi double mutants as well as wild-type plants and cdkc;2 cyct1;5 double knockout mutant plants were inoculated by bombardment with the infectious pCa122 CaMV DNA clone. At 18 DAI, viral DNAs and RNAs were observed in the wild type but not in any of the double mutants (Figure 4). At 26 and 35 DAI, however, significant levels of viral DNAs and RNAs were detected in the cdkc;2 cyct1;5 double mutant plants but not in the transgenic cdkc;2/CDKC;1 RNAi or cyct1;5/CYCT1;4 RNAi plants (Figure 4). Thus, loss of function of the non-null cdkc;2/CDKC;1 RNAi and cyct1;5/CYCT1;4 RNAi double mutants was completely resistant to CaMV.
Figure 4.
CaMV Resistance of the cdkc;2/CDKC;1 RNAi and cyct1;5/CYCT1;4 RNAi Mutants.
(A) Viral DNA accumulation. Wild-type (lane 1), cdkc;2 cyct1;5 (lane 2), cdkc;2/CDKC;1 RNAi (lane 3), and cyct1;5/CYCT1;4 RNAi (lane 4) plants were inoculated by bombardment of pCa122, and total DNA was isolated from leaf tissues harvested at the indicated DAI and probed with the P6 open reading frame of CaMV. Ethidium bromide staining of genomic DNA is shown as a loading control.
(B) Viral RNA accumulation. Total RNA was isolated from inoculated wild-type (lane 1), cdkc;2 cyct1;5 (lane 2), cdkc;2/CDKC;1 RNAi (lane 3), and cyct1;5/CYCT1;4 RNAi (lane 4) plants. The RNA gel blot was first probed with the P3 open reading frame of CaMV for detection of the 35S RNA. The blot was stripped and reprobed with the P6 open reading frame of CaMV for detection of the 19S RNA. Ethidium bromide staining of rRNA is shown for the assessment of equal loading.
CDKC;1 and CDKC;2 Interact with Multiple Cyclins and Are CTD Kinases
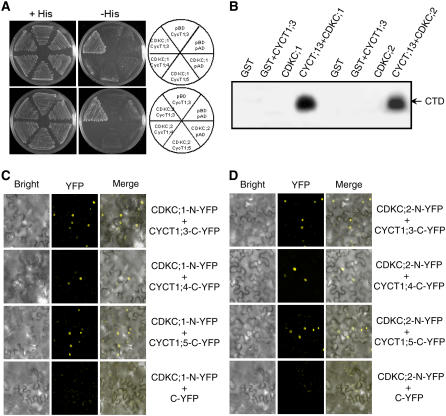
Based on established interactions of CDK9 kinases with CYCT subunits and the strikingly similar phenotypes of the cdkc and cyct1 mutants in CaMV resistance, it is likely that CDKC;1 and CDKC;2 form P-TEFb–like complexes with CYCT1;4 and CYCT1;5 in plant cells. We tested possible interactions between these proteins using yeast two-hybrid assays. From a previously reported yeast two-hybrid screen, CYCT1;3 is an interacting partner for both CDKC;1 and CDKC;2 (Barroco et al., 2003) and was included in the assays as a positive control. We confirmed that both CDKC;1 and CDKC;2 interacted with CYCT1;3 but failed to detect their interactions with CYCT1;4 or CYCT1;5 in the yeast two-hybrid assays (Figure 5A).
Figure 5.
CDKC;1 and CDKC;2 Interact with CYCT1 Proteins and Phosphorylate the CTD Peptide.
(A) CDKC;1 and CDKC;2 interact with CYCT1;3 but not CYCT1;4 or CYCT1;5 in yeast cells. The Gal4 DNA binding domain–CDKC;1 or –CDKC;2 fusion vector was cotransformed with various activation domain–CYCT1 fusion vectors into yeast cells and grown in nonselective (+His) or selective (−His) medium. The empty DNA binding (pBD) and activation (pAD) vectors were included as negative controls. Growth without added His (−His) demonstrates interaction between protein pairs.
(B) Recombinant glutathione S-transferase (GST), GST-tagged CDKC;1 and CDKC;2, and His-tagged CYCT1;3 proteins were expressed and purified from E. coli. Phosphorylation of the GST-tagged CTD of RNAP II by the indicated proteins was assayed in the presence of [γ-32P]ATP.
(C) and (D) BiFC analysis of CDKC;1 (C) and CDKC;2 (D) interactions in planta with CYCT1 proteins. Yellow fluorescence was observed in the nuclear compartment of N. benthamiana leaf epidermal cells, which results from complementation of the N-terminal part of the YFP fused with CDKC;2 (CDKC;2-N-YFP) with the C-terminal part of the YFP fused with CYCT1;3 (CYCT1;3-C-YFP) or CYCT;5 (CYCT1;5-YFP). No fluorescence was observed when CDKC;1-N-YFP or CDKC;2-N-YFP was coexpressed with unfused C-YFP. Bright-field images (left), YFP epifluorescence images (middle), and overlay images of plasmid autofluorescence and YFP epifluorescence (right) of the same cells are shown.
To test whether Arabidopsis CDKC;1 and CDKC;2 are CTD kinases, we expressed their corresponding genes as well as CYCT1;3 in Escherichia coli and purified their recombinant proteins. CYCT1;3 was chosen here because it interacts with CDKC;1 and CDKC;2 both in yeast and in vitro (Figure 5A) (Barroco et al., 2003). When assayed in the absence of CYCT1;3, the recombinant CDKC proteins were not able to phosphorylate the RNAP II heptapeptide (Figure 5B). When the recombinant CDKC proteins were assayed with CYCT1;3, phosphorylation of the heptapeptide was observed (Figure 5B). These results support the notion that Arabidopsis CDKC;1 and CDKC;2 are cyclin-dependent CTD kinases.
CDKC;1 and CDKC;2 might form P-TEFb–like kinase complexes with CYCT1;4 and CYCT1;5 in plant cells but not in yeast cells because formation of the complexes requires activation or accessory proteins not present in yeast cells. To test this, we examined their interactions in planta using the bimolecular fluorescence complementation (BiFC) assay (Walter et al., 2004) in Agrobacterium tumefaciens–infiltrated tobacco (Nicotiana benthamiana) leaves. CDKC;1 and CDKC;2 were fused to the N-terminal yellow fluorescent protein (YFP) fragment and CYCT1;4 and CYCT1;5 were fused to the C-terminal YFP fragment. CYCT1;3 interacts with CDKC;1 and CDKC;2 in yeast cells and was also fused to the C-terminal YFP fragment as a positive control. As expected, when the fused CDKC;1-N-YFP or CDKC;2-N-YFP was coexpressed with CYCT1;3-C-YFP in tobacco leaves, a strong BiFC signal was detected in the nuclear compartment of the transformed cells (Figures 5C and 5D). When the same CDKC;1-N-YFP or CDKC;2-N-YFP was coexpressed with CYCT1;4-C-YFP or CYCT1;5-C-YFP, a strong BiFC signal was also observed in the nuclear compartment (Figures 5C and 5D). Control experiments in which CDKC;1-N-YFP or CDKC;2-N-YFP was coexpressed with unfused C-YFP protein did not show any fluorescence (Figures 5C and 5D). These experiments provide strong evidence that CDKC;1 and CDKC;2 form complexes with CYCT1;4 and CYCT1;5 in plant cells.
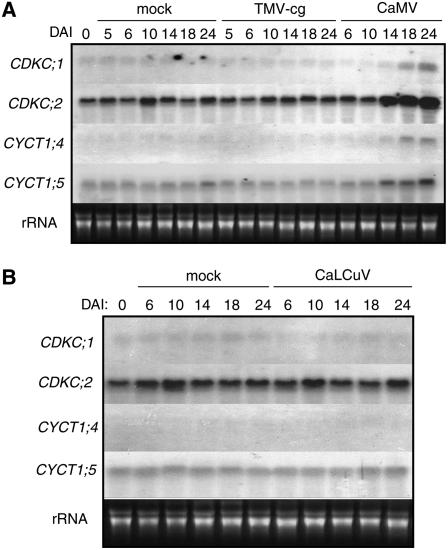
Induced Expression of CDKC and CYCT1 Genes by CaMV Infection
Because of their roles in CaMV infection, we analyzed the expression of these CDKC and CYCT1 genes in CaMV-infected plants. Fully expanded lower leaves of 4-week-old Arabidopsis plants were mechanically inoculated with CaMV. As described above, it takes ∼10 to 13 d after inoculation to first detect significant levels of CaMV viral DNA and observe symptom development in upper systemically infected leaves (Figure 1). As shown in Figure 6, the levels of transcripts for CDKC;1, CDKC;2, CYCT1;4, and CYCT1;5 were quite constant during the first 10 DAI but increased substantially at 14, 18, and 24 DAI. Neither mock inoculation nor TMV-cg or CaLCuV inoculation induced expression of the genes (Figure 6). Thus, the four genes were induced specifically by CaMV infection.
Figure 6.
Induced Expression of CDKC and CYCT1 Genes by CaMV.
Top, 4-week-old Arabidopsis plants were mechanically inoculated with TMV-cg or partially purified CaMV virions, with mock-inoculated (buffer only) plants used as controls. Bottom, plants were bombarded with the infectious DNA clones of CaLCuV or with gold particles (mock) as a control. Total RNA was isolated at the indicated DAI and probed with 32P-labeled CDKC and CYCT1 gene-specific DNA fragments.
Embryonically Lethal Null cyct1;4 cyct1;5 Double Mutants
To test the functional redundancy of CYCT1;4 and CYCT1;5, we initially attempted to generate cyct1;4-1 cyct1;5-1 double mutants through genetic crossing. About 120 F2 seedlings from cyct1;4+/− cyct1;5+/− F1 plants were genotyped by PCR but no cyct1;4−/− cyct1;5−/− homozygous double mutants were identified, although cyct1;4+/− cyct1;5−/− and cyct1;4−/− cyct1;5+/− plants were readily obtained. The absence of cyct1;4−/− cyct1;5−/− double homozygous mutants from the crosses could be the result of a gametophytic defect and/or a postfertilization, embryonic defect. To test possible gametophytic lethality, we conducted reciprocal crosses between wild-type and cyct1;4−/− cyct1;5+/− plants and genotyped the resultant F1 plants by PCR. As shown in Table 1, the F1 progeny from the reciprocal crosses comprised ∼50% cyct1;4+/− cyct1;5+/+ plants and 50% cyct1;4+/− cyct1;5+/− plants. We also conducted a genetic cross using a cyct1;4+/− cyct1;5−/− plant as the pollen donor and a cyct1;4−/− cyct1;5+/− plant as the pollen recipient. A total of 96 F1 progeny were analyzed and found to have a similar number of cyct1;4+/− cyct1;5+/−, cyct1;4−/− cyct1;5+/−, and cyct1;4+/−/cyct1;5−/− plants (Table 1). These results indicate that double mutations of cyct1;4 and cyct1;5 can be cotransmitted through both types of gametophytes; therefore, failure to obtain cyct1;4 cyct1;5 double homozygous mutants was due to zygote synthetic lethality. To confirm that the double homozygous mutations cause embryo lethality, we examined the seeds within the siliques of cyct1;4−/− cyct1;5+/− and cyct1;4+/− cyct1;5−/− plants. As shown in Figure 7A, ∼25% of seeds from the siliques of both genotypes were aborted. The small, brown, aborted seeds and transparent embryos suggest that developmental arrest occurs during embryogenesis.
Table 1.
Genetic Analysis of the Lethal Phenotype of the cyct1;4 cyct1;5 Double Mutant
| Cross
|
Number of Plants
|
||||||
|---|---|---|---|---|---|---|---|
| Female | Male | Total | cyct1;4+/− cyct1;5+/+ | cyct1;4+/− cyct1;5+/− | cyct1;4−/− cyct1;5+/− | cyct1;4+/− cyct1;5−/− | cyct1;4−/− cyct1;5−/− |
| Wild type | cyct1;4−/− cyct1;5+/− | 40 | 19 | 21 | 0 | 0 | 0 |
| cyct1;4−/− cyct1;5+/− | Wild type | 41 | 21 | 20 | 0 | 0 | 0 |
| cyct1;4−/− cyct1;5+/− | cyct1;4+/− cyct1;5−/− | 96 | 0 | 34 | 30 | 32 | 0 |
Figure 7.
Growth Phenotypes of Double Mutants.
(A) Morphology of seeds from wild-type, cyct1;4−/− cyct1;5−/+, and cyct1;4−/+ cyct1;5−/− plants. The aborted seeds from siliques of mutant plants are either pale or brown and shrunken.
(B) Morphology of 6-week-old wild-type and transgenic cdkc;2-1 mutant plants expressing a RNAi construct for CDKC;1. The extent of reduction in the size of the RNAi mutants was correlated with the extent of reduction in the level of CDKC;1 transcripts, as determined by RT-PCR (data not shown).
(C) Morphology of wild-type and transgenic cyct1;5 mutant plants expressing a RNAi construct for CYCT1;4. The altered leaf growth in the RNAi mutants was observed only in mutant plants with reduced levels of CYCT1;4 transcripts, as determined by RT-PCR (data not shown).
(D) Scanning electron microscopy images of rosette leaf surfaces of wild-type and cyct1;5/CYCT1;4 RNAi mutant plants. Bars = 50 μm.
Altered Growth and Development from Loss of Function of Both CDKC;1 and CDKC;2 or CYCT1;4 and CYCT1;5
Both cdkc;2/CDKC;1 RNAi and cyct1;5/CYCT1;4 RNAi mutants showed clear differences in leaf development from the wild type (Figures 7B and 7C). The rosette leaves of the mutants were smaller and more curled and twisted than those of wild-type plants. The extent of the abnormal leaf growth of the mutant plants correlated with the degree of decrease of the transcripts for the targeted CDKC;1 or CYCT1;4 in the respective mutants (data not shown).
We examined cyct1;5/CYCT1;4 RNAi rosette leaves using scanning electron microscopy. A majority of adaxial trichomes of cyct1;5/CYCT1;4 RNAi leaves had two branches instead of the three branches found in typical trichomes of wild-type leaves. In addition, the adaxial surface of cyct1;5/CYCT1;4 RNAi leaves was rough and showed unusual cell and stomata shapes (Figure 7D)
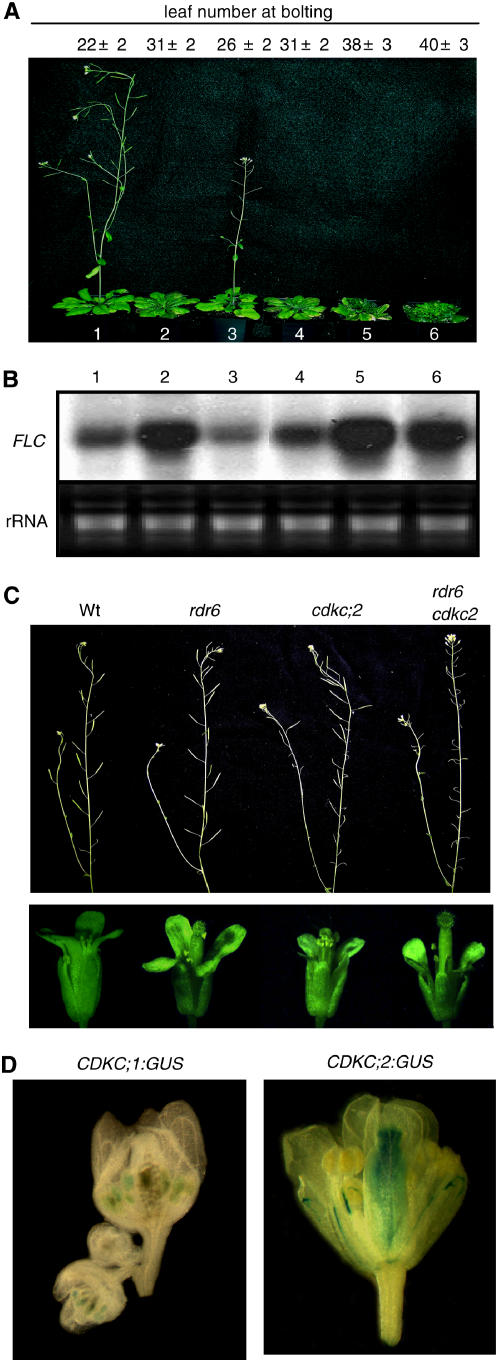
Loss of function of the CDKC and CYCT1 genes also resulted in delayed flowering. Compared with wild-type plants, flowering of the cyct1;5 mutant was slightly but significantly delayed when grown under a medium (12-h) photoperiod (Figure 8A). The cdkc;2 single mutants and the cdkc;2 cyct1;5 double mutants, on the other hand, showed substantially delayed flowering and had increased numbers of rosette leaves at the time of bolting (Figure 8A). The most extensive delay in flowering, however, was observed in the non-null cdkc;2/CDKC;1 RNAi and cyct1;5/CYCT1;4 RNAi double mutants, which had almost twice as many rosette leaves as wild-type plants at the time of bolting under the growth conditions (Figure 8A).
Figure 8.
Phenotypes of Flowering and Flower Development of Double Mutants.
(A) Plants of the wild type (plant 1), cdkc;2 (plant 2), cyct1;5 (plant 3), cdkc;2 cyct1;5 (plant 4), cdkc;2/CDKC;1 RNAi (plant 5), and cyct1;5/CYCT1;4 RNAi (plant 6) mutants grown under a 12-h-light/12-h-dark photoperiod. The photograph was taken 8 weeks after germination.
(B) FLC transcripts in 5-week-old plants of the wild type (lane 1), cdkc;2 (lane 2), cyct1;5 (lane 3), cdkc;2 cyct1;5 (lane 4), cdkc;2/CDKC;1 RNAi (lane 5), and cyct1;5/CYCT1;4 RNAi (lane 6) mutants grown under a 12-h-light/12-h-dark photoperiod.
(C) Siliques (top panel) and flower buds (bottom panel) of wild-type, rdr6-11, cdkc;2-1, and rdr6 cdkc;2 mutant plants.
(D) GUS activities in flower buds of stably transgenic Arabidopsis plants expressing CDKC;1:GUS (left) or CDKC;2:GUS (right). These images are representative results of multiple flowers buds examined from five independent transgenic lines for each construct.
The MADS box protein encoded by FLOWERING LOCUS C (FLC) functions as a repressor and is one of the central regulators of flowering in Arabidopsis. To examine the possible involvement of FLC in the delayed flowering of the mutants, we performed RNA gel blotting to compare mutant plants with wild-type plants for the levels of FLC transcripts. The FLC transcript was significantly elevated in the cdkc;2, cdkc;2/CDKC;1 RNAi and cyct1;5/cyCYCT1;4 RNAi mutants, which all displayed delayed flowering phenotypes (Figures 8A and 8B). The FLC transcript level was normal or slightly reduced in the cyct1;5 single mutant, which exhibited only a slight delay in flowering. In the cdkc;2 cyct1;5 double mutants, the FLC transcript level was only slightly higher than that in wild-type plants but substantially lower than that in the cdkc;2 mutants, even though the double mutants flowered at least at late as the cdkc;2 mutants (Figures 8A and 8B).
Functional Interaction of CDKC;2 with RNA-DEPENDENT POLYMERASE6 in the Regulation of Carpel Growth
P-TEFb has been shown to coordinate transcription with capping and 3′ end formation of primary transcripts (Ni et al., 2004; Adamson et al., 2005). Defects in capping and 3′ end formation were recently linked with RNA-DEPENDENT POLYMERASE6 (RDR6)–mediated RNA silencing in Arabidopsis (Gazzani et al., 2004; Herr et al., 2006; Luo and Chen, 2007). These observations prompted us to examine possible functional interactions between CDKC and RDR6. We crossed cdkc;2 with rdr6-11 (Peragine et al., 2004) and found that double mutants exhibited similar phenotypes of slightly distorted rosette leaves and delayed flowering found in the parental rdr6-11 and cdkc;2 mutants (data not shown). However, unlike their parental single mutants, which set numbers of seeds similar to those of wild type when grown in normal conditions, the fertility of the rdr6-11 cdk;2 double mutants was reduced by >80% (Figure 8C). The reduced fertility appeared to be a consequence of elongated carpels, such that pollen released from the anthers is unable to reach the stigma surface (Figure 8C). When manually fertilized, however, the double mutants could produce seeds, supporting our hypothesis that the mechanical defects in the coordinated growth of filaments and styles were mainly responsible for the drastically reduced fertility.
To explore the rather specific effect on carpel elongation associated with the disruption of CDKC;2 in the rdr6 mutant background, we examined possible tissue-specific expression of CDKC;2 by analyzing the expression of a GUS reporter gene driven by the CDKC;2 promoter in transgenic plants. For comparison, transgenic plants harboring the GUS reporter gene driven by the CDKC;1 promoter were also generated and analyzed. In vegetative tissues, GUS expression was detected in all tissues, but the staining was particularly intense in the vascular veins of leaves for both CDKC;1 and CDKC;2 (data not shown). In flowers, we detected the highest CDKC;2 expression in the carpels, particularly in the stigma and style (Figure 8D). By contrast, in flowers of transgenic plants expressing a GUS reporter gene driven by the CDKC;1 promoter, the highest GUS activities were detected in stamens, particularly in anthers (Figure 8D).
DISCUSSION
Role of Arabidopsis CDKC Complexes in CaMV Infection
CaMV was the first plant virus to be found to contain DNA instead of RNA as genetic material (Shepherd et al., 1968, 1970) and has been studied extensively, particularly with regard to the roles of viral regulatory sequences and proteins involved in the transcription, replication, movement, and pathogenesis of the virus (Kiss-Laszlo and Hohn, 1996; Kobayashi et al., 2002; Stavolone et al., 2005). In addition, the CaMV 35S promoter has been explored extensively for the expression of heterologous genes in transgenic plants. Despite this progress, little information is available about plant host factors targeted by the virus for the activation and regulation of transcription. Here, we report that Arabidopsis knockout mutants for CDKC;2 and CYCT1;5 are highly resistant to CaMV and the cdkc;2 cyct1;5 double mutant is extremely resistant to CaMV (Figure 1). By contrast, these mutants respond normally to TMV-cg and CaLCuV (Figure 2). Expression of Arabidopsis CDKC;2 and CYCT1;5 is induced by infection with CaMV but not with TMV-cg or CaLCuV (Figure 6), and CDKC;2 can phosphorylate the CTD of RNAP II in the presence of an interacting cyclin subunit (Figure 5B). These results strongly suggest that the CDKC;2/CYCT1;5 complex functions as a P-TEFb and plays an important role in infection with CaMV.
There are two CDK9-like (CDKC;1 and CDKC;2) and five CYCT1-like (CYCT1;1 to CYCT1;5) genes in Arabidopsis (Barroco et al., 2003; Wang et al., 2004). When T-DNA insertion or Ds-tagged lines for these genes were analyzed for CaMV infection, only those for CDKC;2 and CYCT1;5 displayed enhanced resistance (Figure 1). These results suggest that CaMV utilizes specific members of these small protein families for the activation of transcription. However, although the cdkc;2 and cyct1;5 mutants are null based on the absence of their respective transcripts, they exhibited a leaky phenotype in that they were not completely resistant to CaMV (Figure 1). The leaky phenotypes are due to partial functional redundancy of the related CDKC and CYCT1 proteins. When the expression of CDKC;1 and CYCT1;4 was inhibited by RNAi in the respective cdkc;2 and cyct1;5 mutant backgrounds, we observed complete resistance to CaMV in the resultant non-null double mutants (Figure 4). Thus, CaMV may utilize preferentially, but not exclusively, CDKC;2 and CYCT1;5 for transcription activation. A high but not absolute specificity of targeting of similar host factors by CaMV might have advantages for the virus. Although the cdkc;2 and cyct1;5 mutants have largely normal growth and development, the cyct1;4 cyct1;5 double mutant had a lethal phenotype. CaMV may have evolved to exploit the functional redundancy of these closely related proteins by preferentially targeting some of these transcription factors to avoid competition with the transcription of host genes, thereby reducing or avoiding a selection pressure from the host. On the other hand, different members of a gene family often exhibit differential cell- or tissue-specific expression patterns. If the targeting specificity were too stringent, CaMV transcription would be blocked in certain cells or tissues where genes encoding targeted factors are not expressed or are expressed at very low levels. An extremely high specificity would also limit the host range of the virus if the host factors required for viral transcription differed significantly in their structures, even among closely related plant species.
P-TEFb as an Evolutionarily Conserved Target of Retroviruses and Pararetroviruses
In humans, P-TEFb is targeted by HIV-1 and is required for its transcription and replication (Mancebo et al., 1997). A recent study reported that human T-lymphotropic virus type I, another complex retrovirus, recruits P-TEFb to stimulate viral gene transcription (Zhou et al., 2006). No such close link of P-TEFb has been reported with other animal DNA viruses (such as adenoviruses and herpes viruses) that also depend on host RNAP II for transcription (Flint and Shenk, 1997). Likewise, the Arabidopsis cdkc;2 and cyct1;5 mutants are resistant to CaMV (Figure 1) but respond normally to CaLCuV (Figure 2), DNA geminiviruses whose genes are also transcribed by host RNAP II (Hanley-Bowdoin et al., 2000). Thus, P-TEFb appears to be an evolutionally conserved target of complex retroviruses and pararetroviruses for transcription activation. This remarkable conservation most likely reflects a fundamental mechanism unique to the transcription of these viruses.
Although human P-TEFb is not known to play a critical role in the transcription of any human DNA virus, its overexpression in human cells can greatly activate the in vivo activity of the cytomegalovirus promoter (Peng et al., 1998). Thus, P-TEFb has an ability to enhance the activity of promoters from other DNA viruses; therefore, it is intriguing that no animal DNA virus is known to recruit this host factor for transcription activation. Unlike retroviruses and pararetroviruses, the products of transcription of most DNA viruses are used primarily as templates for translation; therefore, these viruses can utilize not only transcriptional mechanisms but also a number of posttranscriptional mechanisms that mediate RNA splicing or RNA stability to maximize the production of viral gene products. In retroviruses and pararetroviruses, the full-length genomic or pregenomic RNA is multifunctional; it is utilized as a template for translation and as a template for genome replication through reverse transcription (Kiss-Laszlo and Hohn, 1996). This RNA must be stringently regulated to achieve the appropriate balance for translation or replication. While posttranscriptional mechanisms such as splicing are necessary for gene expression of retroviruses and pararetroviruses, they would be counterproductive for replication of the virus by depleting the full-length viral RNA templates required for reverse transcription. As a result, retroviruses and pararetroviruses may have to rely more on transcriptional mechanisms, such as those mediated by P-TEFb, to enhance the production of genomic or pregenomic RNA.
The long terminal repeat of HIV-1 has minimal promoter activity in the absence of the viral Tat protein (Brady and Kashanchi, 2005). The CaMV 35S promoter, on the other hand, is strongly active in plant cells in the absence of any viral protein (Odell et al., 1985). By analyzing the expression of a reporter gene driven by the CaMV 35S promoter, we have shown that CDKC;2 and CYCT1;5 are required for the high CaMV 35S promoter activity (Figure 3). Thus, the CDKC;2/CYCT1;5 kinase complex might be recruited directly to the viral promoter for transcription activation. Since neither CDKC;2 nor CYCT1;5 is known to bind DNA, their recruitment by the 35S promoter is probably mediated by cellular DNA binding transcription factors that recognize regulatory cis-acting DNA sequences in the CaMV 35S promoter. It should also be pointed out that although the CaMV 35S promoter alone can direct the synthesis of full-length transcripts from heterologous transgenes in transgenic plants, additional viral factors may be required for efficient synthesis of the much longer 35S RNA during CaMV infection. These unidentified viral factors could function as additional recruiters of the cellular CDKC/CYCT1 complex to stimulate transcription, particularly transcription elongation.
Role of Arabidopsis CDKC Complexes in Plant Growth and Development
Although highly resistant to CaMV, the cdkc;2, cyct1;5, and cdkc;2 cyct1;5 mutants exhibit largely normal growth and development. The relatively mild phenotypes of the mutants in plant growth and development were apparently due to the presence of functionally redundant homologs. CYCT1;5 is structurally most closely related to CYCT1;4, and the cyct1;4 cyct1;5 double mutant was embryo-lethal (Table 1, Figure 7A), indicating that they are essential for plant growth. Based on the severe phenotypes of the non-null cdkc;2/CDKC;1 RNAi mutants, the two CDKC kinases most likely have overlapping, essential roles as well. Both the cdkc;2/CDKC;1 RNAi and cyct1;5/CYCT1;4 RNAi double mutants had altered leaf and trichrome growth compared with wild-type plants (Figure 7). The mutants also flowered late (Figure 8A) and produced drastically reduced numbers of seeds (data not shown). These results indicate that the CDKC complexes have important roles in both plant growth and development.
The most notable phenotype of the cdkc;2 mutants was significantly delayed flowering (Figure 8A). Flowering was further delayed in the cdkc;2/CDKC;1 RNAi and cyct1;5/CYCT1;4 RNAi double mutants (Figure 8A). Delayed flowering of the mutants was associated with enhanced levels of FLC transcripts (Figure 8B). In Arabidopsis, early flowering and repression of FLC are observed in the mutants for VIP4, ELF7, and ELF8 that encode homologs for subunits of PAF, a yeast transcription elongation complex containing Paf1, Cdc73, Ctr9, Rtf1, and Leo1 (Zhang and van Nocker, 2002; He et al., 2004; Oh et al., 2004). In yeast, PAF is linked to histone 3 Lys-4 methylation by the Set1 methyltransferase in actively transcribed genes (Krogan et al., 2003). ELF7 and ELF8 (homologs of Paf1 and Ctr9, respectively) are required for the enhancement of histone 3 Lys-4 methylation in FLC chromatin (He et al., 2004). More recent studies have revealed that the association of PAF with RNAP II is mediated by DISF, a transcription elongation factor known to interact both physically and functionally with P-TEFb (Qiu et al., 2006). Therefore, the observed regulation of Arabidopsis flowering time and FLC expression by genes encoding subunits of PAF and P-TEFb may be mechanistically linked. Intriguingly, while mutations of genes encoding subunits of PAF cause repression of FLC and early flowering (Zhang and van Nocker, 2002; He et al., 2004; Oh et al., 2004), mutations of genes encoding subunits of P-TEFb lead to elevated levels of FLC transcript and delayed flowering (Figure 8). These genetic data suggest that the actions of these transcription factors in the regulation of transcription elongation may be more complex than currently understood. Such complex actions have been well illustrated by the contradictory observations with DISF, which acts as a negative elongation factor in vitro but actually facilitates elongation in vivo (Sims et al., 2004). Apparently, DISF is required for transcriptional pausing, but upon phosphorylation of its SPT5 subunit by P-TEFb it can be converted into a positive elongation factor (Sims et al., 2004). DISF interacts with both PAF and P-TEFb, and its dual roles in transcription elongation may explain the opposite roles of Arabidopsis PAF-like and P-TEFb–like complexes in the regulation of flowering.
CDKC;2 and its interacting CYCT1;5 partner have an additive role in CaMV infection, based on the more resistant phenotype of the cdkc;2 cyct1;5 double mutant than the cdkc;2 and cyct1;5 single mutants (Figure 1). No such additive role was obvious in the regulation of flowering, since the flowering of the cdkc;2 cyct1;5 double mutants was similarly delayed as that of the cdkc;2 single mutants (Figure 8A). In fact, CDKC;2 appeared to be antagonized by its interacting CYCT1;5 partner in the regulation of FLC expression, based on the observation that FLC transcript levels were elevated in the cdkc;2 mutants but not in the cdkc;2 cyct1;5 double mutants (Figure 8B). It is possible that the role of a CDKC kinase is regulated by its specific CYCT partner and that disruption of a CYCT1 gene may cause a CDKC to associate with a different CYCT1 to form a complex with an altered function.
The altered phenotypes in the cdkc and cyct1 mutants likely resulted from the inhibited expression of targeted genes important for plant growth and development. In addition, factors affecting transcription elongation can also affect the processing of primary transcripts, such as capping, splicing, and 3′ end formation of mRNA (Sims et al., 2004). Recent genetic studies in Arabidopsis indicate that transcripts defective in capping or 3′ end formation are targeted by RDR6-mediated RNA silencing pathways (Gazzani et al., 2004; Herr et al., 2006). Through genetic analysis, we observed that CDKC;2 and RDR6 act cooperatively in the regulation of Arabidopsis carpel growth (Figure 8C). CDKC;2 is highly expressed in carpels, and its disruption may lead to the production of truncated or defective transcripts that could be targeted by RDR6-mediated RNA silencing. In the cdkc;2 rdr6 double mutants, the truncated or defective transcripts may accumulate, leading to abnormal carpel growth through unknown mechanisms.
METHODS
Plant Growth Conditions
Arabidopsis thaliana and Nicotiana benthamiana plants were grown in a growth chamber at 22°C with 120 μE·m−2·s−1 light intensity and a photoperiod of 12 h of light and 12 h of dark.
Identification and Genotyping of Knockout Mutants
The cdkc;2-1 (SALK_149280), cdkc;2-2 (SALK_029546), cyct1;2-1 (SALK_041740), cyct1;3-1 (SALK_064430), cyct1;4-1 (SALK_139324), and cyct1;5-1 (SALK_021004) T-DNA insertion mutants were identified from the Salk Arabidopsis T-DNA insertion population (Alonso et al., 2003). cyct1;3-2 (Flag_396B05) is a T-DNA insertion line obtained from the Institute of Agronomic Research at Versailles, France (Samson et al., 2002). The cyct1;1-1 (Pst20464) mutant is a Ds transposon-tagged line obtained from the RIKEN Genomic Science Center in Yokohama, Japan (Kuromori et al., 2004). Homozygous lines for the mutants were identified by PCR using primers corresponding to sequences flanking the insertion sites (see Supplemental Table 1 online).
Virus Infection
Mechanical inoculation of Arabidopsis plants with partially purified CaMV virons was performed as described (Yu et al., 2003b). To inoculate plants by particle bombardment, gold particles (1 μm, 320 μg) were coated with 2 μg of pCa122 plasmid DNA (Kobayashi et al., 2002) and bombarded into Arabidopsis plants as described (Xu et al., 2006). Inoculation of Arabidopsis plants with TMV-cg or CaLCuV was performed as described (Turnage et al., 2002; Yu et al., 2003a).
DNA and RNA Gel Blot Analyses
Total DNAs from virus-infected leaves were isolated as described (Yu et al., 1999). Total RNA was isolated from frozen plant materials as described (Zheng et al., 2006). DNA and RNA separation, blotting, and hybridization were performed using standard procedures (Sambrook et al., 1989).
Recombinant Protein Production and Kinase Activity Assays
The CDKC;1 and CDKC;2 coding sequences were PCR-amplified and cloned into pGEX-4T-1 (Amersham Biosciences) to generate in-frame fusions with the GST tag sequence. The CYCT1;3 coding sequence was PCR-amplified and cloned into pET-32a (Novagen) to generate His tag sequences. To generate GST fusion proteins containing two repeats of the YSPTSPS heptapeptide, two primers (5′-CGGGATCCTACTCCCCGACCTCCCCGTCCTACTCCCCGACCTCCCCGTCTTATGCCCCAGAATTTCCG-3′ and 5′-CGGAAATTCTGGGGCATAAGACG-3′) were annealed and extended by the Klenow enzyme to form a dsDNA fragment. The fragment was digested with BamHI/EcoRI and inserted into pGEX-4T-1. The resulting plasmids were introduced into Escherichia coli BL21 cells. Induction of recombinant protein production and purification were performed following the protocols recommended by the manufacturers.
CTD phosphorylation reactions were assembled on ice in a kinase buffer (25 mM Tris-Cl, pH 7.8, 15 mM MgCl2, and 1 mM DTT) containing 2 μg of recombinant GST-tagged CDKC;1 or CDKC;2 protein with or without 2 μg of His-tagged CYCT1;3 recombinant protein. The reactions were initiated by adding 1 μg of GST-CTD substrate protein and 2.5 μCi of [γ-32P]ATP and terminated by the addition of SDS loading buffer after 30 min of incubation at room temperature. The reaction mixtures were separated by 12.5% SDS-PAGE, and CTD phosphorylation was detected by autoradiography.
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed with the GAL4 system. The CDKC;1 and CDKC;2 coding sequences were amplified and cloned into pBD-GAL4 to generate DNA binding domain fusion partner proteins, and CYCT1;3, CYCT1;4, and CYCT1;5 coding sequences were inserted into pAD-GAL4 to form activation domain fusion proteins. All PCR-mediated cloned constructs were verified by sequencing. For interaction analysis, two combinatory constructs were transformed simultaneously into the YRG-2 yeast strain (Stratagene) and tested for His3+, Trp+, and Leu+ auxotrophy and LacZ reporter activity (β-galactosidase assay) according to the manufacturer's protocols.
BiFC Assays
DNA sequences for the N-terminal 173–amino acid EYFP (N-YFP) and the C-terminal 64–amino acid (C-YPF) fragments were PCR-amplified and cloned into the plant expression vectors pOCA30 (Chen and Chen, 2002) and pFGC5941 to generate pOCA-N-YFP and pFGC-C-YFP, respectively. The Arabidopsis CDKC;1 and CDKC;2 coding sequences were inserted into pOCA-N-YFP to generate the N-terminal in-frame fusions with N-YFP, whereas CYCT1;3, CYCT1;4, and CYCT1;5 were introduced into pFGC-C-YFP to form C-terminal in-frame fusions with C-YFP. The resulting clones were verified through sequencing. The plasmids were introduced into Agrobacterium tumefaciens (strain GV3101), and infiltration of N. benthamiana was performed as described previously (Walter et al., 2004). Confocal microscopy was performed using a Bio-Rad MRC-1024 laser scanning confocal imaging system.
Reporter Gene Assays of the CaMV 35S Promoter Activity
A GUS gene construct driven by the CaMV 35S promoter was generated by inserting the GUS gene into the plant transformation vector pKMB (Mylne and Botella, 1998). This construct was introduced into Agrobacterium and then transformed into Arabidopsis plants using the floral dip procedure (Clough and Bent, 1998). Primary transformants were identified by BASTA resistance in soil. GUS activity was measured using 4-methylumbelliferyl-β-d-glucuronide as substrate (Jefferson et al., 1987).
Generation of Transgenic RNAi Lines
To generate cdkc;2/CDKC;1 RNAi mutants, an ∼300-bp fragment corresponding to the 3′ untranslated sequence of CDKC;1 was amplified by PCR using primers 5′-atcggatccatttaaatCTGCCCACATATGAATCATCAC-3′ (italics are BamHI and SwaI sites, and uppercase letters correspond to the sequence of the genes to be amplified; sense primer) and 5′-atcggcgcgccactagtATCACATTAAATGTAAG-3′ (italics are AscI and SpeI sites; antisense primer), and the PCR product was then cloned into appropriate restriction sites of the pFGC5941 RNAi vector (http://www.chromdb.org/fgc5941.html). The resultant construct was transformed into the cdkc;2-1 mutant background by the Agrobacterium-mediated floral dip method. The cyct1;5/CYCT1;4 RNAi mutants were generated with the same strategy using an ∼350-bp CYCT1;4 fragment amplified by PCR with the primers 5′-atcggcgcgcctctagaCGAGATTCTTGATGGAAACAACTTG-3′ (italics are AscI and XbaI sites; sense primer) and 5′-atcatttaaatggatccGAGACCCTAAATCCTATGGGATGG-3′ (italics are SwaI and BamHI sites; antisense primer). The construct was transformed directly into the cyct1;5-1 mutant background. Seeds from Agrobacterium-treated plants were grown directly in soil, and transformants were selected for BASTA resistance. Suppressed expression of the targeted genes was analyzed by RT-PCR or RNA gel blotting.
RT-PCR Analysis
Total RNA was extracted and subjected to reverse transcription with SuperScript III reverse transcriptase (Invitrogen). Two CDKC;1-specific primers (5′-CCGGGCTTCACTATTGTCAT-3′ and 5′-GAACCAAGGCATCTTGGAAA-3′) were used to amplify a 374-bp product from CDKC;1; two CYCT1;4-specific primers (5′-AAACCGTGTTCCTGCATCTC-3′ and 5′-GGAAAATGTGCACCTGCTTT-3′) were used to amplify a 327-bp product from CYCT1;4; and two UBQ10-specific primers (5′-TCAATTCTCTCTACCGTGATCAAGATGCA-3′ and 5′-GGTGTCAGAACTCTCCACCTCAAGAGTA-3′) were used to amplify a 320-bp product from UBQ10. PCR conditions were as follows: 94°C for 3 min; followed by 30 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min.
Light and Electron Microscopy
A Nikon SMZ-U stereomicroscope was used to observe siliques and floral structures from wild-type and mutant plants. For scanning electron microscopy, rosette leaves of wild type and mutant plants were cryoprepared with liquid nitrogen. Samples were then examined by NOVA nano-scanning electron microscopy (FEI) operating at 3 kV and ∼14-mm working distance.
Promoter–GUS Analysis
The ∼1.3-kb promoter regions of the CDKC;1 and CDKC;2 genes were PCR-amplified from the genomic DNA and cloned into a modified plant expression vector that contains a promoterless GUS gene. The promoter–GUS constructs were transformed into Arabidopsis plants using the Agrobacterium-mediated floral dip method. Histochemical staining of GUS activity was performed as described previously (Jefferson et al., 1987).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: CDKC;1 (At5g10270), CDKC;2 (At5g64960), CYCT1;1 (At1g35440), CYCT1;2 (At4g19560), CYCT1;3 (At1g27630), CYCT1;4 (At4g19600), CYCT1;5 (At5g45190), FLC (At5g10140), NRPB1 (At4g35800), and UBQ10 (At4g05320).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. PCR Primers Used in Knockout Mutant Screening.
Supplemental Figure 1. Sequence Comparison of Arabidopsis CDKC and CYCT1 Proteins.
Supplemental Figure 2. Diagram of Arabidopsis Genes and T-DNA Insertion or Transposon-Tagging Mutants.
Supplemental Figure 3. Genetic Complementation of the cyct1;5 Mutant.
Supplemental Figure 4. RT-PCR Analysis of CDKC;1 and CYCT1;4 Transcripts from RNAi Plants.
Supplementary Material
Acknowledgments
We thank Thomas Hohn (University of Basel, Basel, Switzerland) and Seiji Tsuge (Kyoto Prefectural University, Kyoto, Japan) for kindly providing the plasmid pCa122 and Dominique Niki Robertson (North Carolina State University, Raleigh) for CaLCuV plasmids. We also thank Chang-Deng Hu (Purdue University, West Lafayette, IN) for providing EYFP plasmids, Scott Poethig (University of Pennsylvania, Philadelphia) for rdr6-11 seeds, and the Arabidopsis Biological Resource Center at Ohio State University, the Institute of Agronomic Research (Versailles, France), and the RIKEN Genomic Science Center (Yokohama, Japan) for T-DNA insertion or Ds-tagged mutants. We are grateful to Debby Sherman for help with scanning electron microscopy. This is Journal Paper 2007-18103 of the Purdue University Agricultural Research Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Zhixiang Chen (zhixiang@purdue.edu).
Online version contains Web-only data.
References
- Adamson, T.E., Shutt, D.C., and Price, D.H. (2005). Functional coupling of cleavage and polyadenylation with transcription of mRNA. J. Biol. Chem. 280 32262–32271. [DOI] [PubMed] [Google Scholar]
- Akoulitchev, S., Chuikov, S., and Reinberg, D. (2000). TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407 102–106. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Barroco, R.M., De Veylder, L., Magyar, Z., Engler, G., Inze, D., and Mironov, V. (2003). Novel complexes of cyclin-dependent kinases and a cyclin-like protein from Arabidopsis thaliana with a function unrelated to cell division. Cell. Mol. Life Sci. 60 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, J., and Kashanchi, F. (2005). Tat gets the “green” light on transcription initiation. Retrovirology 2 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., and Chen, Z. (2002). Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Dietrich, M.A., Prenger, J.P., and Guilfoyle, T.J. (1990). Analysis of the genes encoding the largest subunit of RNA polymerase II in Arabidopsis and soybean. Plant Mol. Biol. 15 207–223. [DOI] [PubMed] [Google Scholar]
- Flint, J., and Shenk, T. (1997). Viral transactivating proteins. Annu. Rev. Genet. 31 177–212. [DOI] [PubMed] [Google Scholar]
- Fulop, K., Pettko-Szandtner, A., Magyar, Z., Miskolczi, P., Kondorosi, E., Dudits, D., and Bako, L. (2005). The Medicago CDKC;1-CYCLINT;1 kinase complex phosphorylates the carboxy-terminal domain of RNA polymerase II and promotes transcription. Plant J. 42 810–820. [DOI] [PubMed] [Google Scholar]
- Gazzani, S., Lawrenson, T., Woodward, C., Headon, D., and Sablowski, R. (2004). A link between mRNA turnover and RNA interference in Arabidopsis. Science 306 1046–1048. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin, L., Settlage, S.B., Orozco, B.M., Nagar, S., and Robertson, D. (2000). Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 35 105–140. [PubMed] [Google Scholar]
- He, Y., Doyle, M.R., and Amasino, R.M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr, A.J., Molnar, A., Jones, A., and Baulcombe, D.C. (2006). Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. USA 103 14994–15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, R. (2002). Matthews' Plant Virology. (San Diego, CA: Academic Press).
- Ishikawa, M., Obata, F., Kumagai, T., and Ohno, T. (1991). Isolation of mutants of Arabidopsis thaliana in which accumulation of tobacco mosaic virus coat protein is reduced to low levels. Mol. Gen. Genet. 230 33–38. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubes, J., Lemaire-Chamley, M., Delmas, F., Walter, J., Hernould, M., Mouras, A., Raymond, P., and Chevalier, C. (2001). A new C-type cyclin-dependent kinase from tomato expressed in dividing tissues does not interact with mitotic and G1 cyclins. Plant Physiol. 126 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., and Hohn, T. (1996). Pararetro- and retrovirus RNA: Splicing and the control of nuclear export. Trends Microbiol. 4 480–485. [DOI] [PubMed] [Google Scholar]
- Kobayashi, K., Tsuge, S., Stavolone, L., and Hohn, T. (2002). The cauliflower mosaic virus virion-associated protein is dispensable for viral replication in single cells. J. Virol. 76 9457–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa, H., et al. (2002). C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc. Natl. Acad. Sci. USA 99 10893–10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa, H., Hausmann, S., Bang, W.Y., Ueda, A., Kondo, N., Hiraguri, A., Fukuhara, T., Bahk, J.D., Yun, D.J., Bressan, R.A., Hasegawa, P.M., and Shuman, S. (2004). Arabidopsis C-terminal domain phosphatase-like 1 and 2 are essential Ser-5-specific C-terminal domain phosphatases. Proc. Natl. Acad. Sci. USA 101 14539–14544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., Dover, J., Wood, A., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Ryan, O.W., Golshani, A., Johnston, M., Greenblatt, J.F., and Shilatifard, A. (2003). The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol. Cell 11 721–729. [DOI] [PubMed] [Google Scholar]
- Kuromori, T., Hirayama, T., Kiyosue, Y., Takabe, H., Mizukado, S., Sakurai, T., Akiyama, K., Kamiya, A., Ito, T., and Shinozaki, K. (2004). A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 37 897–905. [DOI] [PubMed] [Google Scholar]
- Luo, Z., and Chen, Z. (2007). Improperly terminated, unpolyadenylated mRNA of sense transgenes is targeted by RDR6-mediated RNA silencing in Arabidopsis. Plant Cell 19 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo, H.S., Lee, G., Flygare, J., Tomassini, J., Luu, P., Zhu, Y., Peng, J., Blau, C., Hazuda, D., Price, D., and Flores, O. (1997). P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, N.F., and Price, D.H. (1995). Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270 12335–12338. [DOI] [PubMed] [Google Scholar]
- Menges, M., de Jager, S.M., Gruissem, W., and Murray, J.A. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41 546–566. [DOI] [PubMed] [Google Scholar]
- Mylne, J., and Botella, J.R. (1998). Binary vectors for sense and antisense expression of Arabidopsis ESTs. Plant Mol. Biol. Rep. 16 257–262. [Google Scholar]
- Ni, Z., Schwartz, B.E., Werner, J., Suarez, J.R., and Lis, J.T. (2004). Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell 13 55–65. [DOI] [PubMed] [Google Scholar]
- Nigg, E.A. (1996). Cyclin-dependent kinase 7: At the cross-roads of transcription, DNA repair and cell cycle control? Curr. Opin. Cell Biol. 8 312–317. [DOI] [PubMed] [Google Scholar]
- Odell, J.T., Nagy, F., and Chua, N.H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313 810–812. [DOI] [PubMed] [Google Scholar]
- Oh, S., Zhang, H., Ludwig, P., and van Nocker, S. (2004). A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Zhu, Y., Milton, J.T., and Price, D.H. (1998). Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12 755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhero, R., Liaw, P., Bertens, K., and Yankulov, K. (2004). Three cyclin-dependent kinases preferentially phosphorylate different parts of the C-terminal domain of the large subunit of RNA polymerase II. Eur. J. Biochem. 271 1004–1014. [DOI] [PubMed] [Google Scholar]
- Qiu, H., Hu, C., Wong, C.M., and Hinnebusch, A.G. (2006). The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol. Cell. Biol. 26 3135–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, R.J., Bruening, G.E., and Wakeman, R.J. (1970). Double-stranded DNA from cauliflower mosaic virus. Virology 41 339–347. [DOI] [PubMed] [Google Scholar]
- Shepherd, R.J., Wakeman, R.J., and Romanko, R.R. (1968). DNA in cauliflower mosaic virus. Virology 36 150–152. [DOI] [PubMed] [Google Scholar]
- Shimotohno, A., Matsubayashi, S., Yamaguchi, M., Uchimiya, H., and Umeda, M. (2003). Differential phosphorylation activities of CDK-activating kinases in Arabidopsis thaliana. FEBS Lett. 534 69–74. [DOI] [PubMed] [Google Scholar]
- Sims, R.J., III, Belotserkovskaya, R., and Reinberg, D. (2004). Elongation by RNA polymerase II: The short and long of it. Genes Dev. 18 2437–2468. [DOI] [PubMed] [Google Scholar]
- Stavolone, L., Villani, M.E., Leclerc, D., and Hohn, T. (2005). A coiled-coil interaction mediates cauliflower mosaic virus cell-to-cell movement. Proc. Natl. Acad. Sci. USA 102 6219–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnage, M.A., Muangsan, N., Peele, C.G., and Robertson, D. (2002). Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 30 107–114. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Wang, G., Kong, H., Sun, Y., Zhang, X., Zhang, W., Altman, N., DePamphilis, C.W., and Ma, H. (2004). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135 1084–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., and Chen, X. (2004). HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18 1310–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D., Fan, B., MacFarlane, S.A., and Chen, Z. (2003. a). Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant Microbe Interact. 16 206–216. [DOI] [PubMed] [Google Scholar]
- Yu, D., Xie, Z., Chen, C., Fan, B., and Chen, Z. (1999). Expression of tobacco class II catalase gene activates the endogenous homologous gene and is associated with disease resistance in transgenic potato plants. Plant Mol. Biol. 39 477–488. [DOI] [PubMed] [Google Scholar]
- Yu, W., Murfett, J., and Schoelz, J.E. (2003. b). Differential induction of symptoms in Arabidopsis by P6 of cauliflower mosaic virus. Mol. Plant Microbe Interact. 16 35–42. [DOI] [PubMed] [Google Scholar]
- Zhang, H., and van Nocker, S. (2002). The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J. 31 663–673. [DOI] [PubMed] [Google Scholar]
- Zheng, Z., Qamar, S.A., Chen, Z., and Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48 592–605. [DOI] [PubMed] [Google Scholar]
- Zhou, M., Lu, H., Park, H., Wilson-Chiru, J., Linton, R., and Brady, J.N. (2006). Tax interacts with P-TEFb in a novel manner to stimulate human T-lymphotropic virus type 1 transcription. J. Virol. 80 4781–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.