Abstract
The widely distributed DEGP proteases play important roles in the degradation of damaged and misfolded proteins. Arabidopsis thaliana contains 16 DEGP-like proteases, four of which are located in the chloroplast. Here, we show that DEG5 and DEG8 form a hexamer in the thylakoid lumen and that recombinant DEG8 is proteolytically active toward both a model substrate (β-casein) and photodamaged D1 protein of photosystem II (PSII), producing 16-kD N-terminal and 18-kD C-terminal fragments. Inactivation of DEG5 and DEG8 resulted in increased sensitivity to photoinhibition. Turnover of newly synthesized D1 protein in the deg5 deg8 double mutant was impaired, and the degradation of D1 in the presence of the chloroplast protein synthesis inhibitor lincomycin under high-light treatment was slowed in the mutants. Thus, DEG5 and DEG8 are important for efficient turnover of the D1 protein and for protection against photoinhibition in vivo. The deg5 deg8 double mutant showed increased photosensitivity and reduced rates of D1 degradation compared with single mutants of deg5 and deg8. A 16-kD N-terminal degradation fragment of the D1 protein was detected in wild-type plants but not in the deg5 deg8 mutant following in vivo photoinhibition. Therefore, our results suggest that DEG5 and DEG8 have a synergistic function in the primary cleavage of the CD loop of the PSII reaction center protein D1.
INTRODUCTION
Photosystem II (PSII), which catalyzes light-dependent water oxidation and plastoquinone reduction in chloroplasts, is a large pigment-protein complex in the thylakoid membrane. PSII consists of >20 intrinsic and extrinsic proteins. The PSII reaction center complex is composed of the D1 and D2 proteins, the α- and β-subunits of cytochrome b559, and the PsbI protein. The D1 and D2 heterodimer binds all the essential redox components of PSII required to transfer electrons from the manganese cluster of the water-oxidizing complex to the plastoquinone pool (Nanba and Satoh, 1987). These include P680 (the primary electron donor), pheophytin (the primary electron acceptor), and both the first and second quinone acceptors (QA and QB). D1 acts as the electron donor to P680, YZ, and together with D2 is thought to bind the manganese cluster involved in water oxidation. Oxygen-evolving PSII complexes also contain the intrinsic chlorophyll a binding proteins (CP43 and CP47), the extrinsic oxygen-evolving complex (33-, 23-, and 17-kD proteins), and several low molecular mass proteins (Nelson and Yocum, 2006).
A distinct feature of PSII is that it is very sensitive to photoinhibition, the PSII reaction center D1 protein being the main target for light-induced damage among PSII proteins (for reviews, see Prasil et al., 1992; Aro et al., 1993). Damaged D1 proteins are degraded and subsequently replaced with newly synthesized copies (Kyle et al., 1984; Mattoo et al., 1984; Ohad et al., 1984; Adir et al., 1990). This efficient repair mechanism must be essential to maintain PSII in a functional state (Prasil et al., 1992; Aro et al., 1993). At least two different sites have been shown to be susceptible to light-induced impairment of electron transfer reactions (Prasil et al., 1992; Aro et al., 1993): one located on the donor side and the other on the acceptor side (Andersson and Styring, 1991; Barber and Andersson, 1992). Photoinhibition on the acceptor side has been proposed to be primarily induced by the overreduction of the primary acceptor, QA, to QAH2 and the subsequent release of QAH2 from PSII (Vass et al., 1992). Photoinhibition on the donor side has been shown to be due to photoinactivation of the electron transfer from YZ to P680 when the oxygen-evolving complex is inactivated (Eckert et al., 1991).
Photoinhibition on either the donor side or the acceptor side may trigger the damage and subsequent degradation of PSII proteins, particularly the D1 protein (Andersson and Aro, 2001; Huesgen et al., 2005). In acceptor-side photoinhibition, the primary cleavage of the D1 protein appears to occur in the stromal loop connecting the transmembrane helices D and E, based on the detection of 23-kD N-terminal and 10-kD C-terminal fragments in samples examined by various authors (Greenberg et al., 1987; Canovas and Barber, 1993; Shipton and Barber, 1994; Kanervo et al., 1998). In donor-side photoinhibition, cleavage in the luminal AB or CD loops has been reported. Cleavage at the luminal AB loops produces 10-kD N-terminal and 24-kD C-terminal fragments (Barbato et al., 1991; De Las Rivas et al., 1992), whereas cleavage at the CD loops results in the generation of 16-kD N-terminal and 18-kD C-terminal fragments (De Las Rivas et al., 1992; Shipton and Barber, 1994; Kettunen et al., 1996; Wiklund et al., 2001).
Although turnover of the D1 protein has been extensively studied, much effort has recently been made to identify the proteases responsible for the degradation of the photodamaged D1 protein. In vitro experiments have shown that DEGP2 (now renamed DEG2), which is a member of the DEGP/HTRA family of Ser proteases, is responsible for the primary cleavage between helices D and E of the D1 protein in a GTP-dependent manner (Spetea et al., 1999; Haußühl et al., 2001). The 23-kD fragment of the D1 protein has been found to be subsequently degraded by the ATP-dependent zinc metalloprotease FTSH (Lindahl et al., 2000). However, in vivo analysis of var2-2 Arabidopsis thaliana mutants, which lack FTSH2 (a membrane of the FTSH family) has suggested that FTSH2 may be responsible for the primary cleavage of the D1 protein under high-light treatment and that FTSH2 is required both for the efficient turnover of the D1 protein and protection against photoinhibition (Bailey et al., 2002). Furthermore, in Synechocystis sp PCC 6803, the inactivation of one FTSH2 homolog (srl0228) resulted in dramatic increases in the organism's sensitivity to high-light treatment. FTSH was shown to directly bind to PSII and to be involved in the early steps of D1 degradation (Silva et al., 2003). FTSH2 is also involved in the heat-induced primary cleavage of the D1 protein and the production of its corresponding fragments (Yoshioka et al., 2006).
DEG/HTR family proteases are ATP-independent Ser endopeptidases, which are present in both prokaryotes and eukaryotes (Gottesman, 1996; Pallen and Wren, 1997; Adam and Clarke, 2002; Clausen et al., 2002; Kieselbach and Funk, 2003; Huesgen et al., 2005). DEG/HTR proteases were initially identified in Escherichia coli, in which the family is represented by DEGP, DEGQ, and DEGS (Pallen and Wren, 1997; Clausen et al., 2002). DEGP has both molecular chaperone and proteolytic activity. The chaperone function dominates at low temperature, while it is proteolytically activated at elevated temperatures (Spiess et al., 1999). Misfolded or aberrant soluble and membrane proteins have been implicated as the physiological substrates of DEGP proteases in E. coli (Clausen et al., 2002). In the cyanobacterium Synechocystis 6803, there are three homologs of E. coli DEG proteases: DEGP/HTRA, DEGQ/HHOA, and DEGS/HHPB (Sokolenko et al., 2002; Kieselbach and Funk, 2003; Huesgen et al., 2005). Analysis of the double and triple deg mutants has demonstrated that the DEG proteases do not play an essential role in D1 turnover and repair in vivo, although they are required for photoprotection (Barker et al., 2006). In Arabidopsis, 16 genes coding for DEGP-like protease have been identified, four of which are located in the chloroplast according to proteomic data (Peltier et al., 2002; Schubert et al., 2002). DEG1, DEG5, and DEG8 were found in the thylakoid lumen, and DEG2 was peripherally attached to the stromal side of the thylakoid membrane. The presence of these DEG proteases in the thylakoid lumen suggests that they may be responsible for the degradation of both soluble luminal proteins and intrinsic membrane proteins with lumen-exposed loops connecting transmembrane helices. In vitro analysis of recombinant DEGP1, now renamed DEG1, has shown that DEG1 from Arabidopsis is able to degrade thylakoid lumen proteins, such as plastocyanin and the PsbO subunit of the PSII oxygen-evolving complex (Chassin et al., 2002). Size exclusion chromatography indicated that the purified DEG1 is present in both monomeric and hexameric forms, both of which are proteolytically active (Chassin et al., 2002). However, the functional roles of DEG5 and DEG8 proteases in chloroplasts have not been addressed either in vitro or in vivo to date.
Therefore, in the study reported here, we used both biochemical and genetic approaches to assess the physiological importance of DEG proteases in chloroplast function. First, we investigated the interactions between DEG5 and DEG8 and showed that DEG5 and DEG8 form a complex. Then, we overexpressed DEG5 and DEG8 in E. coli and characterized their proteolytic activity. We show that recombinant DEG8, but not DEG5, is proteolytically active toward both β-casein and PSII reaction center D1 protein. deg5, deg8, and deg5 deg8 mutants showed the increased sensitivity to photoinhibition. Moreover, degradation of the PSII reaction center D1 protein was slowed in the mutants under high irradiance, suggesting that DEG is involved in early stages of D1 degradation.
RESULTS
Protein Complex Formation of DEG5 and DEG8
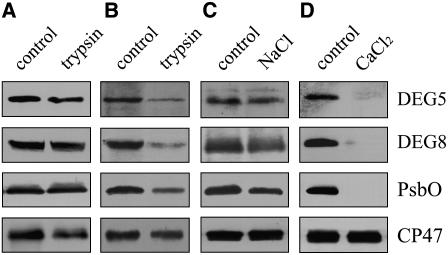
The subcellular location and membrane association of DEG5 and DEG8 were examined by immunoblot analysis with the polyclonal antisera raised against recombinant proteins of DEG5 and DEG8. Protease digestion assays of intact thylakoid membranes showed that DEG5 and DEG8 were protected from trypsin treatment, as is the oxygen-evolving luminal PsbO protein of PSII (Figure 1A). However, after sonication of the thylakoid membranes, DEG5, DEG8, and PsbO were all sensitive to trypsin digestion (Figure 1B). These results indicate that DEG5 and DEG8 are thylakoid lumen proteins, in accordance with the results of previous proteomic studies (Peltier et al., 2002; Schubert et al., 2002). After incubation of sonicated thylakoid membranes with 1 M NaCl, considerable amounts of DEG5 and DEG8 were detected in the membrane (Figure 1C), but both DEG5 and DEG8 were effectively removed by treatment with 1 M CaCl2 (Figure 1D). These results indicate that DEG5 and DEG8 are peripheral proteins associated with the membrane.
Figure 1.
Immunolocalization and Membrane Association of DEG5 and DEG8.
(A) and (B) Intact thylakoid membranes (A) and sonicated thylakoid membranes (B) from mature Arabidopsis leaves were incubated with trypsin for 15 min at 25°C.
(C) and (D) The sonicated thylakoid membranes were incubated with 1 M NaCl (C) or 1 M CaCl2 (D) for 30 min at 4°C. After these treatments, the thylakoid proteins were separated by SDS-PAGE and immunodetected with anti-DEG5, anti-DEG8, anti-PsbO, and anti-CP47 antibodies.
PsbO and CP47 were used as markers. Membrane preparations that had not been subjected to any of these treatments were used as controls. The experiments were repeated five times independently, and similar results were obtained. Results of a representative experiment are shown.
The functional forms of DEG/HTRA proteins have been shown to be oligomers (Gottesman, 1996; Pallen and Wren, 1997; Clausen et al., 2002). To check this possibility, thylakoid membranes were solubilized with n-dodecyl β-d-maltoside (DM), and the protein complexes were separated in a blue native (BN) gel. After the first-dimensional separation in the presence of Coomassie blue dye, the separated protein complexes were further subjected to denaturing SDS-PAGE and immunoblot analysis using specific antibodies (Figure 2). The immunoblots indicated the presence of dimeric PSII, monomeric PSII, and CP43-free PSII in the BN gel at positions corresponding to ∼460, 240, and 200 kD, respectively. A complex that migrated more slowly than the PSII dimer was also detected, probably corresponding to photosystem I–light-harvesting complex I (PSI-LHCI) because it reacted with PsaA/B antibody. Immunoblot analysis showed that both DEG5 and DEG8 migrate in a protein complex of ∼460 kD, which also corresponds to the PSII dimer (Figure 2). To further estimate the relative amounts of DEG5 and DEG8 in the immunoblots (Figure 2), we also calibrated the signals from them, based on the thylakoid membrane protein loadings. The relative protein levels in thylakoids were deduced from the strengths of the immunoblot signals from the thylakoid membrane protein with those of the expressed proteins in a dilution series (data not shown), which indicated that the ratio of the two proteins is ∼1:1.
Figure 2.
Immunodetection of Thylakoid Proteins Separated by Two-Dimensional Gel Electrophoresis.
Thylakoid membranes were separated in the first dimension in a BN gel followed by SDS-PAGE in the second dimension. The proteins resolved were immunodetected with anti-PsaA/B, anti-CP47, anti-CP43, anti-DEG5, and anti-DEG8 antibodies. Similar results were obtained in two additional independent experiments.
The above results suggest that either DEG5 and DEG8 form complexes or they are associated with PSII. To test these possibilities, pull-down experiments were performed with recombinant DEG5 and DEG8 proteins fused with N-terminal His tags. The purified His-DEG5 and His-DEG8 fusion proteins were incubated with DM-solubilized thylakoid membranes. After washing with the buffer described in Methods, the nickel-nitrilotriacetic acid agarose (Ni-NTA) resin-bound proteins were separated by SDS-PAGE and examined by immunoblot analysis (Figure 3). DEG5 was detected when His-DEG8 fusion proteins were used in the assay, but no protein was detected when only the solubilized thylakoid membrane and the resin were used (Figure 3A). Reciprocally, DEG8 was detected when His-DEG5 fusion protein was used (Figure 3B). However, CP47, a PSII core protein, was not detected no matter when either His-DEG5 or His-DEG8 fusion proteins were used (Figures 3A and 3B). Pull-down experiments were also performed using the samples after photoinhibitory treatments. CP47 was not detected using either His-DEG5 or His-DEG8 fusion proteins (Figures 3C and 3D). These results indicate that neither DEG5 nor DEG8 colocalize with PSII.
Figure 3.
Pull-Down Assay of the Interaction between DEG5 and DEG8.
Ten micrograms of DEG8 ([A] and [C]) and DEG5 ([B] and [D]) His-tag fusion proteins coupled to Ni-NTA resin were incubated with 100 μg of DM-solubilized thylakoid membranes without ([A] and [B]) and with ([C] and [D]) photoinhibitory treatments. Bound proteins were eluted, separated by SDS-PAGE, and immunodetected with the antisera directed against DEG5, DEG8, and CP47. Similar results were obtained in two additional independent experiments.
Proteolytic Activity of DEG5 and DEG8
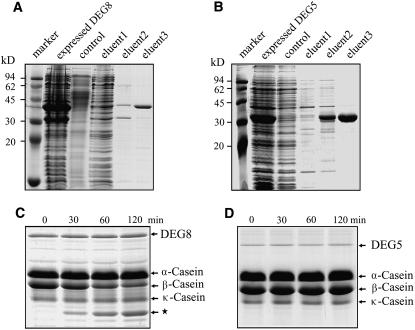
To examine whether the recombinant His-tagged DEG5 and DEG8 proteins were proteolytically active, we incubated them with a mixture of α-, β-, and κ-forms of casein. β-Casein is the preferred substrate for assaying bacterial DEGP activity under in vitro conditions (Lipinska et al., 1990). The results showed that β-casein was efficiently degraded by the fractions containing overexpressed DEG8, and the contents of α- and κ-forms remained unchanged under our experimental conditions (Figures 4A and 4C). More than 50% of the β-casein was degraded within the first 60 min. However, the fractions containing overexpressed DEG5 were not capable of degrading β-casein (Figures 4B and 4D). These results indicate that the recombinant DEG8 was proteolytically active.
Figure 4.
Purification and β-Casein Degradation Activity of DEG5 and DEG8.
The BL21 cells were harvested after the addition of isopropylthio-β-d-galactoside for 2 h, and the overexpressed DEG8 (A) and DEG5 (B) proteins were purified on a Ni-NTA agarose resin matrix. Samples were resolved by SDS-PAGE and stained with Coomassie blue. The purified proteins from eluent 3 were applied to a Sephadex G-75 column and eluted with 20 mM NaH2PO4, pH 7.8, for renaturation. The purified recombinant DEG8 (0.5 μg) (C) and DEG5 (0.5 μg) (D) proteins were incubated with a mixture of α-, β-, and κ-casein (15 μg) for 30, 60, or 120 min at 37°C. After terminating the reaction, the reaction mixtures were subjected to SDS-PAGE on a 12% (w/v) acrylamide gel. The locations of DEG5, DEG8, and α-, β-, and κ-forms of casein and degradation products (star) in the gel are indicated on the right. Similar results to those presented in (C) and (D) were obtained from three independent experiments. Results from a representative experiment are shown.
Physiological Target of DEG8
The results described above showed that the recombinant DEG8 was able to degrade β-casein. Next, we examined the physiological target of DEG8 protease. Since the DEGP protease has been suggested to degrade damaged or misfolded proteins (Gottesman, 1996; Pallen and Wren, 1997; Clausen et al., 2002), the thylakoid membranes used in this experiment were first subjected to high-light treatment (1600 μmol m−2 s−1) for 90 min at 0°C. Treatment with high light causes irreversible oxidative damage, which subsequently induces a conformational change in the D1 protein, making it vulnerable to proteolytic degradation (Prasil et al., 1992; Aro et al., 1993; Andersson and Aro, 2001). The thylakoid membranes isolated from high light–treated leaves were washed with CaCl2 to remove endogenous DEG proteases and incubated with or without recombinant DEG8 in the dark at 37°C. In the absence of recombinant DEG8, the D1 protein contents remained unchanged and no degradation products were detected (Figure 5A). However, when the recombinant DEG8 was added to isolated thylakoid membranes, the amount of full-length D1 protein decreased with time, and a polypeptide of ∼18 kD was immunodetected using a polyclonal antibody raised against the DE loop of the D1 protein (Figure 5B). Further immunoblot analysis with the antibody directed against the N-terminal 58 to 86 residues of the D1 protein also revealed a reduction in the abundance of the full-length D1 protein and the appearance of a 16-kD fragment (Figure 5C). When DEG8 was incubated with the thylakoid membranes that had not been treated with high light, the D1 protein content did not change (Figure 5D). Thus, DEG8 is proteolytically active toward damaged D1.
Figure 5.
Degradation of the Photodamaged D1 Protein by Recombinant DEG8.
(A) to (C) Thylakoid membranes isolated from plants subjected to high-light treatment were incubated without (A) or with purified recombinant DEG8 ([B] and [C]), and the degradation of D1 was analyzed by immunoblots using antibodies directed against the DE loop ([A] and [B]) and the N-terminal amino acid residues (58 to 86) of D1 (C).
(D) Thylakoid membranes from leaves that had not been subjected to high-light treatment were incubated with purified recombinant DEG8, and the thylakoid proteins were separated by SDS-PAGE and immunodetected with the antibody raised against the DE loop of the D1 protein.
(E) High light–treated thylakoid membranes (10 μg chlorophyll) were incubated with purified recombinant DEG8 (0.5 μg), and the thylakoid proteins were separated and immunodetected with anti-D1, anti-D2, anti-CP43, anti-CP47, anti-cytochrome f, anti-CF1β, anti-PsaA/B, anti-PsbO, and anti-LHCII antibodies.
(F) Purified recombinant DEG8 (1 μg protein) was added to heat-treated thylakoid membranes obtained following high-light illumination and incubated in the absence (control) or presence of the following protease inhibitors: 100 μg/mL PMSF, 1 μg/mL aprotinin, 1 μg/mL E64 [3-carboxy-trans-2,3-epoxypropyl-leucylamido(4-guanidino)butane], 1 μg/mL pepstatin, and 500 μg/mL EDTA. After these treatments, the thylakoid proteins were separated and immunodetected with specific antibodies raised against the DE loop of D1. These experiments were repeated three times independently, and similar results were obtained. Results from a representative experiment are shown.
To further elucidate the potential substrates of DEG8, we examined the steady state levels of other thylakoid membrane proteins in the presence of recombinant DEG8. Immunoblot analysis with anti-D2, -CP47, -CP43, -PsbO, -Cytf, -light-harvesting complex II (LHCII), -CF1β, and -PsaA/B antibodies showed that the levels of these proteins remained almost constant in the presence of recombinant DEG8 (Figure 5E).
To test the properties of DEG8, which as a DEGP protease belongs to the chymotrypsin-like family (Gottesman, 1996; Pallen and Wren, 1997; Clausen et al., 2002) and should therefore have chymotrypsin-like responses to protease inhibitors, we performed in vitro experiments with various inhibitors in the reaction mixture. To examine the effects of protease inhibitors on the activities of recombinant DEG8 toward photodamaged D1, the thylakoids isolated from high light–treated leaves were denatured at 90°C for 10 min to inactivate endogenous proteolytic activities. Phenylmethylsulfonyl fluoride (PMSF) and aprotinin, which are inhibitors of Ser proteases, strongly inhibited the degradation of the D1 protein (Figure 5F). Pepstatin, an inhibitor of aspartic protease, also had considerable inhibitory effect, while E-64 and EDTA, inhibitors of Cys and metallo proteases, had no effect on the proteolytic activity of DEG8 (Figure 5F). Thus, the results were consistent with expectations.
T-DNA Insertion Mutants of deg5, deg8, and deg5 deg8
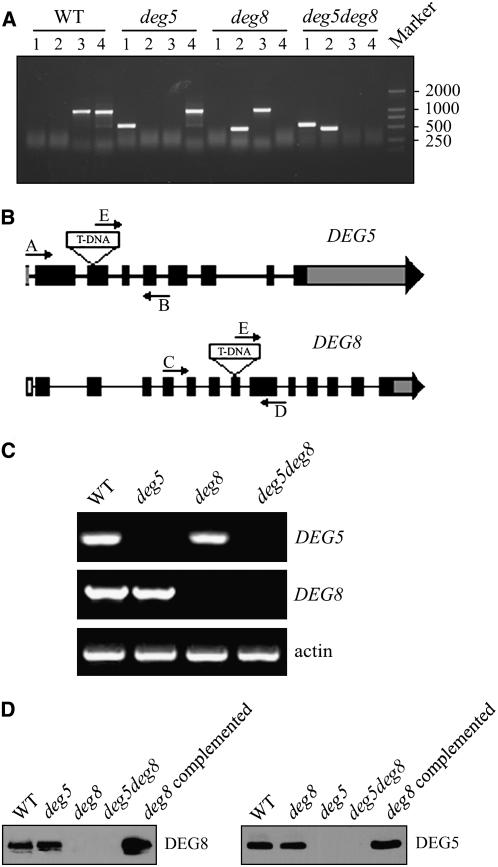
To study the in vivo function of DEG5 and DEG8, we obtained Arabidopsis lines from the SALK collections in which T-DNA was inserted into DEG5 and DEG8 (the mutants were named deg5 and deg8, respectively). The T-DNA insertion positions in these mutants were determined by PCR and subsequent sequencing of the amplified products (Figure 6A). The deg5 mutant contains a T-DNA insertion within the 429-bp sequence downstream of the ATG codon, and the deg8 mutant contains a T-DNA insertion within the 1560 bp downstream of the ATG codon (Figure 6B). RT-PCR analysis revealed that expression of the DEG5 and DEG8 genes was not detectable in the respective mutants compared with that in the wild-type plants (Figure 6C), and complementary immunoblot analyses detected virtually no DEG5 and DEG8 protein in them (Figure 6D). By contrast, the levels of DEG5 and DEG8 proteins in the complemented plants were comparable to those in the wild-type plants (Figure 6D).
Figure 6.
Identification of the deg5, deg8, and deg5 deg8 Mutants.
(A) PCR analysis of genomic DNA from the wild type and deg5, deg8, and deg5 deg8 mutants to confirm the homozygosity of the mutants. Lane 1, amplification with primers E and B; lane 2, amplification with primers E and D; lane 3, amplification with primers A and B; lane 4, amplification with primers C and D.
(B) Schematic diagram of the DEG5 and DEG8 genes (not to scale). Closed boxes represent exons. Gray boxes represent the untranslated regions of the genes. Arrows indicate the location of primers used for PCR analyses.
(C) RT-PCR analysis of DEG5 and DEG8 gene expression. RT-PCR was performed with the specific primers for DEG5, DEG8, or actin.
(D) Immunodetection of DEG5 and DEG8. Thylakoid membranes were isolated from the wild type, deg5, deg8, and deg5 deg8 mutants, the deg8 mutant complemented with DEG8 cDNA, and the deg5 mutant complemented with DEG5 cDNA and separated by SDS-PAGE. The proteins were immunodetected with anti-DEG5 and anti-DEG8 antibodies.
To further examine the genetic relationship between DEG5 and DEG8, we generated a deg5 deg8 homozygous double mutant. When grown under 120 μmol m−2 s−1 light, the growth rates of the deg5, deg8, and deg5 deg8 mutants were slightly reduced relative to the wild type (Figure 7). Chlorophyll fluorescence analysis of dark-adapted leaves of wild-type and mutant plants showed that the Fv/Fm ratio was very similar in the deg5 (0.82 ± 0.02), deg8 (0.81 ± 0.01), and deg5 deg8 (0.81 ± 0.01) mutants and the wild-type plants (0.82 ± 0.01). To analyze the phenotypes of the mutants during growth under high irradiance, we transferred wild-type and mutant plants that were grown initially at 120 μmol m−2 s−1 to a greenhouse with a maximum intensity of 1000 μmol m−2 s−1 at noon. Our results showed that when exposed to high light, the growth of the mutants was inhibited compared with the wild-type plants, most markedly in the deg5 deg8 mutant. Moreover, some of the mutant leaves exhibited symptoms of yellowing, while there was no obvious high-light effect in wild-type plants (Figure 7B).
Figure 7.
Phenotypes of deg5, deg8, and deg5 deg8 Mutants and Wild-Type Plants.
(A) Four-week-old plants grown under a photon flux density of 120 μmol m−2 s−1.
(B) Two-week-old plants grown in the growth chamber under a photon flux density of 120 μmol m−2 s−1 and then transferred to a greenhouse for another 2 weeks with maximum photon flux density at noon of ∼1000 μmol m−2 s−1. Similar results were obtained in two additional independent experiments.
Biochemical Characterization of Protein Composition of the Mutants
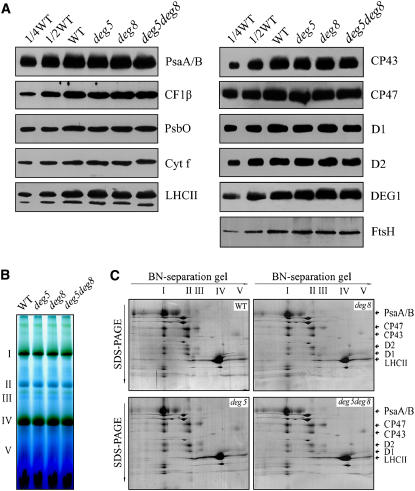
To examine the steady state levels of the thylakoid proteins, immunoblot analyses were performed with the antibodies raised against specific subunits of the photosynthetic protein complexes. The results showed that levels of the tested thylakoid proteins per unit of chlorophyll, including D1, D2, LHCII, PsbO, CP47, and CP43 (PSII proteins), PsaA/B of the PSI reaction center proteins, cytochrome f of the cytochrome b6f complex, and CF1β of ATP synthase, were not altered in the deg5, deg8, or deg5 deg8 mutants (Figure 8A). The levels of the DEG1 and FTSH proteins also remained unchanged in the mutants (Figure 8A). In addition, the thylakoid protein complexes were subjected to BN/SDS-PAGE analysis (Figures 8B and 8C). After the first-dimensional BN gel separation in the presence of Coomassie blue dye, five major chlorophyll-protein complexes were resolved, apparently representing monomeric PSI and dimeric PSII (band I), monomeric PSII (band II), CP43-free PSII (band III), and trimeric (band IV) and monomeric LHCII (band V) (Figure 8B). The abundance of PSI, PSII, and LHCII protein complexes were not changed in the mutant. Analyses of the two-dimensional SDS-PAGE gels after Coomassie blue staining confirmed the above results (Figure 8C).
Figure 8.
Analysis of Thylakoid Proteins from deg5, deg8, and deg5 deg8 Mutants and Wild-Type Plants.
(A) Immunoblot analysis of thylakoid proteins from deg5, deg8, and deg5 deg8 mutants and wild-type plants. Thylakoid proteins (1 μg chlorophyll) from the wild type and deg5, deg8, and deg5 deg8 mutants were separated by SDS-urea-PAGE and immunodetected with anti-D1, anti-D2, anti-CP43, anti-CP47, anti-PsbO, anti-LHCII, anti-PsaA/B, anti-cytochrome f, anti-DEG1, anti-CF1β, and anti-FTSH antibodies (which recognize the different isomers of FTSH equally well).
(B) Thylakoid membranes (10 μg chlorophyll) from wild-type and deg5, deg8, and deg5 deg8 mutant leaves were solubilized and separated by BN gel electrophoresis. The positions of protein complexes were identified with appropriate antibodies (see Guo et al., 2005).
(C) BN-PAGE–separated thylakoid proteins were further separated by SDS-urea-PAGE gel. Names of the proteins resolved by the second-dimension SDS-PAGE, previously identified, are indicated by arrows. These experiments were repeated three times independently, and similar results were obtained. Results from a representative experiment are shown.
Photoinhibition in deg5, deg8, and deg5 deg8 Mutants
The in vitro experiments described above showed that DEG5 and DEG8 form a complex and that the PSII reaction center D1 protein is a potential physiological target of DEG8 (Figures 2 and 5). To determine whether DEG5 and DEG8 are involved in photoinhibition and repair processes, PSII activity was measured, in terms of the maximal efficiency of PSII photochemistry (Fv/Fm), in the wild-type and deg5, deg8, and deg5 deg8 T-DNA insertion mutants under high-light illumination (1600 and 800 μmol m−2 s−1). In the absence of lincomycin, Fv/Fm declined in wild-type leaves to ∼55% of the dark-adapted values within 2 h of illumination at a light intensity of 1600 μmol m−2 s−1 and to 83% at a light intensity of 800 μmol m−2 s−1 (Figures 9A and 9B). The corresponding declines in the mutants were considerably stronger: Fv/Fm fell to ∼40 and 30% of dark-adapted values in the single and double mutants, respectively, after 2 h in 1600 μmol m−2 s−1 light (Figure 9A) and to ∼70 and 60% of dark-adapted values after 2 h treatment with 800 μmol m−2 s−1 light (Figure 9B). These results clearly demonstrate that photosensitivity was higher in the mutants. In the presence of lincomycin, the decline in wild-type leaves was more rapid and continued until Fv/Fm values approached ∼10% of the dark-adapted values (Figures 9A and 9B). Since lincomycin blocks the repair of PSII by inhibiting de novo protein synthesis in the chloroplast, the decline in Fv/Fm reflects the rate of photodamage to PSII. Interestingly, in the presence of lincomycin, the decline in Fv/Fm in the deg5, deg8, and deg5 deg8 mutants was similar to that observed in wild-type leaves during the same photoinhibitory light treatment (Figure 9C), suggesting that wild-type and mutant leaves have similar rates of PSII photoinhibition.
Figure 9.
Photoinactivation of PSII Activity under High-Light Illumination.
(A) and (B) The maximal photochemical efficiency of PSII (Fv/Fm) was measured for detached leaves from the wild-type (squares), deg5 (circles), deg8 (triangles), and deg5 deg8 (diamonds) plants during exposure to irradiance of 1600 μmol m−2 s−1 (A) or 800 μmol m−2 s−1 (B) in the absence of lincomycin.
(C) Maximal photochemical efficiency of PSII (Fv/Fm) was measured for detached leaves from wild-type (squares), deg5 (circles), deg8 (triangles), and deg5 deg8 (diamonds) plants during exposure to irradiance of 1600 μmol m−2 s−1 in the presence of lincomycin.
(D) Recovery of maximum photochemical efficiency of PSII after photoinhibition. Leaves of wild-type (squares), deg5 (circles), deg8 (triangles), and deg5 deg8 (diamonds) plants were illuminated to induce ∼50% photoinhibition of PSII, and the restoration of maximal photochemical efficiency was subsequently followed at an irradiance of 20 μmol m−2 s−1. Time 0 represents dark-adapted samples before light and lincomycin treatment. Values shown are averages ± se of at least six replicated experiments.
To further assess the capacity of the mutants to recover from photoinhibition, we monitored changes in Fv/Fm values in wild-type and mutant leaves after high-light treatment. The high-intensity irradiance treatment applied in this experiment resulted in ∼50% reductions in the Fv/Fm values for both wild-type and mutant leaves. As shown in Figure 9D, mutant leaves recovered from photoinhibition slightly more slowly than wild-type leaves. After a 6-h recovery period, the Fv/Fm values of wild-type and deg5 deg8 mutant leaves had reached 93 and 86% of their dark-adapted values, respectively (Figure 9D).
PSII Protein Degradation in Vivo
To establish whether the high susceptibility of the deg5, deg8, and deg5 deg8 mutants to photoinhibition is related to PSII protein turnover, we analyzed the PSII protein contents of the thylakoid membranes in wild-type and mutant plants under high-light illumination in the presence of lincomycin. In the presence of this inhibitor, the contents of the PSII reaction center D1 protein gradually declined in deg5, deg8, deg5 deg8, and wild-type plants under high-light illumination, and the rate of reduction was slower in the mutants (Figure 10A). Four hour of illumination at 1600 μmol m−2 s−1 resulted in losses of ∼90% of D1 protein in wild-type plants, 65% in both deg5 and deg8 mutants, and ∼35% in deg5 deg8 mutants during the same period of irradiance (Figure 10A). The contents of another PSII reaction center protein, D2, and the PsbO protein of the oxygen-evolving complex were also measured following the same treatment as described for D1 and were found to be relatively stable in both the wild-type and mutant plants (Figures 10B and 10C). It is interesting to note that the D2 protein is stable following the loss of D1 in wild-type plants (Figure 10B). It is unclear what the reason is underlying this response.
Figure 10.
Immunoblot Analysis of Thylakoid Proteins after Photoinhibition.
Thylakoid membranes were isolated from wild-type, deg5, deg8, and deg5 deg8 leaves that had been exposed to high-light illumination in the presence of lincomycin for 0.5, 1, 2, or 4 h. The thylakoid proteins were separated by SDS-PAGE and immunodetected with specific antibodies directed against D1 (A), D2 (B), and PsbO (C) proteins. X-ray films were scanned and analyzed using the AlphaImager 2200 documentation and analysis system. Percentage protein levels shown below the lanes were estimated by comparison with levels found in corresponding samples taken at time 0. Similar results were obtained in two additional independent experiments.
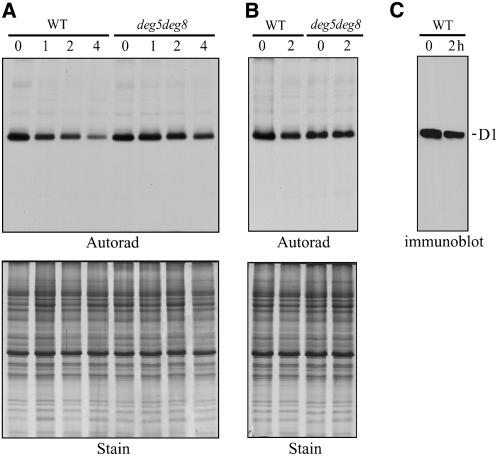
To assess D1 protein turnover more directly, the leaves of wild-type and deg5 deg8 double mutant plants were pulse-labeled in the presence of 120 μmol m−2 s−1, and the degradation of D1 was subsequently monitored in the absence of lincomycin under high light. As shown in Figure 11, the D1 protein was the most heavily labeled of the thylakoid proteins, and the rate of D1 synthesis was slightly lower in the mutant than in the, plants (Figures 11A and 11B). However, in wild-type leaves, the half-life of the degradation of newly synthesized D1 was ∼2 h, while in the deg5 deg8 mutant, little loss of signal from the D1 protein was detected, so the degradation of newly synthesized D1 was slower (Figure 11A). Thus, in the mutant, the turnover of D1 was impaired compared with the wild type. After pulse labeling for 30 min, D1 degradation was also examined under high light in the presence of lincomycin to avoid the possible effect of protein synthesis on protein degradation. Again, however, the degradation of newly synthesized D1 was slower in the deg5 deg8 mutant than in the wild type (Figure 11B).
Figure 11.
Synthesis and Degradation of Thylakoid Proteins.
(A) Pulse and chase analysis of thylakoid membrane proteins. Leaves of the wild type and deg5 deg8 mutants were labeled for 30 min at 120 μmol m−2 s−1, followed by a chase of 1, 2, or 4 h at 1600 μmol m−2 s−1. Thylakoid membranes with equal amounts of chlorophyll were isolated and separated by SDS-PAGE. The resolved proteins were visualized by Coomassie blue staining (Stain) and autoradiography (Autorad).
(B) Degradation of thylakoid proteins. Leaves of the wild type and deg5 deg8 mutants were labeled for 30 min at 120 μmol m−2 s−1, lincomycin was added, and the samples were chased for 2 h at 1600 μmol m−2 s−1. Thylakoid proteins were separated by SDS-PAGE, stained with Coomassie blue (Stain), and visualized by autoradiography (Autorad).
(C) Immunoblot analysis with D1 DE-loop antibodies. Thylakoid membranes were isolated from wild-type leaves that had been exposed to high-light treatment in the presence of lincomycin for 2 h. The thylakoid membrane proteins were separated by SDS-PAGE and immunodetected with anti-D1 DE-loop antibodies. The location of the D1 protein is indicated at the right. Similar results were obtained in two additional independent experiments.
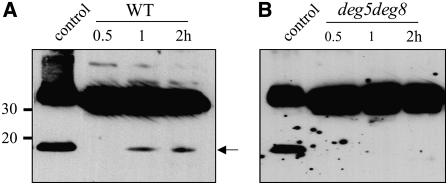
To detect photoinhibition-induced degradation fragments of the D1 protein in intact leaves, the Arabidopsis leaves were exposed to high-light treatment and then thylakoid membranes were isolated and subjected to immunoblot analysis with the antibodies directed against the N-terminal 58 to 86 residues of the D1 protein. The 16-kD D1 degradation fragments were detected with these antibodies in samples from wild-type plants following in vivo photoinhibition (Figure 12A) but not in samples from the deg5 deg8 mutant (Figure 12B).
Figure 12.
Immunodetection of the Degradation Products of the D1 Protein in the Thylakoid Membranes Isolated from High Light–Treated Arabidopsis Leaves.
Thylakoid membranes were isolated from wild-type (A) and deg5 deg8 (B) leaves that had been exposed to high-light illumination for 1 and 2 h. The thylakoid membrane proteins were separated by SDS-PAGE and immunodetected with antibodies raised against the N-terminal residues (58 to 86) of the D1 protein. Control indicates samples containing recombinant DEG8 and high light–treated thylakoid membranes. The degradation fragment of the D1 protein is indicated with an arrow. Similar results were obtained in two additional independent experiments.
DISCUSSION
DEG5 and DEG8 Form a Complex
The BN gel and pull-down analyses showed that Arabidopsis DEG5 and DEG8 form a protein complex in the lumen (Figures 2 and 3), similar to DEG proteases in other organisms. In E. coli, the functional unit of DEG protease is trimeric, and larger DEG complexes, ranging from tetramers to dodecamers, are also formed (Clausen et al., 2002). BN gel analysis showed that Arabidopsis DEG5, DEG8, and PSII complex comigrated in the gel (Figure 2). In addition, the pull-down experiments using His-DEG5 and His-DEG8 fusion proteins further revealed that DEG5 and DEG8 interacted when the thylakoid membranes were solubilized with DM (Figure 3). Immunoblot analysis also confirmed that PSII was not associated with DEG5 and DEG8 (Figure 3). These results suggest that DEG5 and DEG8 were present as heterocomplexes. The apparent molecular mass of these complexes was estimated to be ∼460 kD, while the molecular mass of DEG5 is ∼32 kD and that of DEG8 is ∼42 kD, according to SDS-PAGE analysis. Estimates of the relative levels of DEG5 and DEG8 in the second-dimensional SDS-PAGE after separation in BN gels suggest that the DEG5-to-DEG8 ratio is ∼1:1. Thus, these results indicate that these DEG proteins are present in the thylakoid lumen as hexamers.
A common characteristic of DEG protease is the presence of PDZ domains, which are known to regulate protein–protein interactions, such as the assembly of multimeric protein complexes and protease-substrate recognition (Clausen et al., 2002; Schlieker et al., 2004). Deletion of one or two PDZ domains will generate smaller oligomers, suggesting that the PDZ domains are essential for the maintenance of the oligomeric protein complex. PDZ has also been found to play essential roles in the regulation of protease activity (Clausen et al., 2002). DEG8 contains one PDZ domain, while DEG5 does not contain any obvious PDZ domain (Kieselbach and Funk, 2003; Huesgen et al., 2005). Thus, formation of protein complexes between DEG5 and DEG8 may have implications for their substrate recognition and regulation of their activity.
DEG5 and DEG8 Are Required for Efficient PSII Repair under High-Light Illumination
We have used deg5, deg8, and deg5 deg8 double mutants of Arabidopsis to study the physiological role of the DEG proteases in the repair cycle of PSII. PSII activity in these deg mutants showed increased sensitivity to high irradiance; the increases were stronger in the deg5 deg8 double mutant than in the deg5 and deg8 mutants (Figure 9). Moreover, the rate of PSII photoinhibition was the same in the mutants and wild-type plants in the presence of the chloroplast protein synthesis inhibitor lincomycin (Figure 9). We suggest therefore that impairment of PSII repair cycle is responsible for the enhanced sensitivity of PSII to photoinhibition in the mutants. Thus, the high-light susceptibility of the mutants can be mainly attributed to impairment of the reactivation of damaged PSII during illumination.
The repair of PSII is initiated by the degradation of the photodamaged D1 protein, which is subsequently replaced with new D1 copies (Aro et al., 1993). In the presence of a protein synthesis inhibitor, degradation of the D1 protein is slowed down compared with the absence of inhibitor. It has been suggested that the synthesis and incorporation of newly synthesized D1 into the PSII complex and the degradation of damaged D1 are mutually dependent during the turnover process (Komenda and Barber, 1995). Thus, impairment of the repair cycle could result from impairment of either D1 synthesis or D1 degradation. Our results clearly demonstrated that the degradation of the D1 protein was slower in the mutants following lincomycin and high-light treatment than in the wild-type plants (Figures 10 and 11). The pulse-labeling experiments also revealed that rates of D1 protein synthesis were slightly reduced in the mutants (Figure 11). Thus, our results suggest that DEG5 and DEG8 may be involved in the initial cleavage of photodamaged D1 protein in vivo (Figures 10 to 12), which is consistent with the in vitro biochemical characterization (Figure 5). Furthermore, the levels of another PSII reaction center protein, D2, and the oxygen-evolving PsbO protein remained relatively stable in the mutant and wild-type plants following treatments with lincomycin and high light (Figure 10). These results indicate that DEG5 and DEG8 play specific roles in D1 protein degradation.
Our results showed that DEG proteases are involved in the repair of PSII and protection against photoinhibition (Figures 7 and 9 to 11). During growth under high irradiance, these mutants also showed high-light sensitivity in terms of growth and bleaching, and the extent was most pronounced in the deg5 deg8 mutant (Figure 7B). However, no apparent difference was observed in growth between wild-type and mutant plants under light conditions of 120 μmol m−2 s−1 (Figure 7A). In addition, the mutants' pigment composition was similar to that of the wild type (data not shown), and their Fv/Fm ratios in the mutants were high. Also, the rates of photodamage in the wild-type and mutant plants in the presence of lincomycin were similar (Figure 9). Examination of the capacity of the mutant to recover from photoinhibition revealed that Fv/Fm ratios in the mutants recovered following the high-light treatment, albeit slightly more slowly than in the wild type (Figure 9D). Thus, since the degradation of D1 by DEG5 and DEG8 in the mutants is impaired, no apparent differences observed in Fv/Fm ratios and plant fitness among the mutants and wild-type plants could be due to increased turnover of damaged D1 by proteases other than the DEG proteases.
D1 Is the Physiological Target of DEG5 and DEG8
Bacterial DEG proteases have been implicated as key players in the control of protein quality in response to a variety of stresses. Besides their function as molecular chaperones, DEG proteases are also responsible for the degradation of abnormal periplasmic proteins (Clausen et al., 2002). A number of proteins, such as colicin A lysis protein, K88 and K99 pilin subunits, MalS, and PapA pilin, have been described as physiological substrates for E. coli DEGP (Kim and Kim, 2005). However, the physiological substrates of the cyanobacterial and plant DEG proteases remain largely unknown. Inactivation of the three DEG proteases of Synechocystis 6803 resulted in increased sensitivity to photoinhibition. However, in the double mutant, the D1 protein degradation rate was similar between the wild type and mutant. Therefore, the DEG proteases in Synechocystis may not play an essential role in D1 turnover and PSII repair in vivo (Barker et al., 2006)
The involvement of a Ser-type protease in the degradation of photodamaged D1 has been biochemically characterized (Virgin et al., 1991). Our biochemical characterization of DEG8's properties also provides direct evident that it has Ser protease activity. The aspartic peptidase inhibitor we tested also had inhibitory effects on DEG8 activity (Figure 5). These findings suggest that the characteristic catalytic triad (Ser, His, and Asp) of DEG/HTR Ser proteases may be present in DEG8. The detection of 16-kD N-terminal and 18-kD C-terminal D1 fragments in in vitro experiments (Figure 5) indicates that DEG8 catalyzes the primary cleavage of the damaged D1 protein at the luminal loop connecting the transmembrane helices C and D. Moreover, D1 is the specific physiological target of DEG8 because the photosynthetic proteins tested were not degraded by DEG8 under these conditions (Figure 5). The function of DEG8 in the degradation of photodamaged D1 points to the possibility that DEG proteases colocalize with PSII. However, neither DEG5 nor DEG8 was found to be associated with PSII (Figure 7). Thus, it can be speculated that either DEG only transiently interacts with photodamaged D1 or the interaction between DEG and PSII cannot be detected in pull-down experiments.
In Arabidopsis, FTSH proteases form protein complexes (Sakamoto et al., 2003; Yu et al., 2004, 2005; Zaltsman et al., 2005a), and two types of closely related pairs of these proteases are present: type A (FTSH1 and FTSH5) and type B (FTSH2 and FTSH8). The FTSH proteins are interchangeable within a pair but not between pairs. Inactivation of one pair of duplicated genes will result in the loss of the products of the other duplicated genes (Sakamoto et al., 2003; Yu et al., 2005; Zaltsman et al., 2005a). Thus, the genes within a pair are functionally redundant.
However, the protein level of DEG8 remained unchanged in the mutants lacking DEG5 and vice versa (Figure 6). This finding, and several other observations outlined below, suggests that DEG5 and DEG8 have a synergistic function in the degradation of D1 and PSII turnover in vivo. The extent of sensitivity to photoinhibition in deg5 and deg8 mutants was similar, and the deg5 deg8 double mutant showed higher sensitivity to high light than the deg5 and deg8 single mutants (Figure 9). The degradation of D1 protein in the mutants was slowed under high-light illumination. The double mutant displayed a 35% decrease in D1 protein content, and the single deg5 and deg8 mutants displayed 65% reductions after high-light treatment for 4 h (Figure 10). It is possible that proteolytic activity was partly retained in the mutants lacking either DEG5 or DEG8, although the proteolytic activity of recombinant DEG5 was not detected in vitro. Immunoblot analysis detected 16-kD N-terminal degradation fragments of the D1 protein after in vivo photoinhibition in samples from wild-type plants, originating from cleavage between transmembrane helices C and D. However, no such degradation fragments were detected in the deg5 deg8 mutant (Figure 12). The D1 proteolysis products detected in vivo resemble the degradation fragments either from in vitro donor-side photoinhibition (De Las Rivas et al., 1992) or from the incubation of the high light–treated thylakoid membranes with recombinant DEG8 (Figure 5). These results suggest that donor-side photoinhibition may occur in vivo. In fact, similar results have been observed in pumpkin (Cucurbita pepo) and pea (Pisum sativum) (Shipton and Barber, 1994; Kettunen et al., 1996). Thus, DEG5 and DEG8 form a protein complex in the lumen that is involved in the primary cleavage of the luminal loop connecting the CD loop in vivo. As noted above, these findings collectively suggest that DEG5 and DEG8 play synergistic rather than redundant roles in the degradation of D1 and PSII turnover in vivo.
Previous in vitro studies have shown that DEG2 at the stroma-exposed thylakoid membrane is involved in the primary cleavage of photodamaged PSII reaction center D1 protein at the stromal loop connecting transmembrane helices D and E (Haußühl et al., 2001). Growing evidence suggests that FTSH, a membrane-integral ATP-dependent protease, is also involved in the primary cleavage of photodamaged D1 protein, probably at its DE loop (Bailey et al., 2002; Silva et al., 2003; Yoshioka et al., 2006). Arabidopsis mutants with reduced levels of DEG1 also contain reduced amounts of FTSH, and FTSH2 mutants contain very low levels of DEG1 (Zaltsman et al., 2005b; Kapri-Pardes et al., 2007). DEG1 has been suggested to function together with FTSH and DEG2 in cleaving lumen-exposed regions of the D1 protein (Kapri-Pardes et al., 2007). Thus, it seems reasonable to speculate that DEG could cooperate with FTSH in efficiently cleaving the multiple transmembrane D1 proteins from both sides of the thylakoid membrane. Similar cooperative degradation of proteins in the inner mitochondrial membrane by proteases acting at both sides of the membrane has been reported previously (Leonhard et al., 2000).
METHODS
Plant Materials
Wild-type and mutant Arabidopsis thaliana (ecotype Columbia) plants were grown in soil under short-day conditions (10-h-light/14-h-dark cycles) with a photon flux density of 120 μmol m−2 s−1 at a constant temperature of 22°C. The T-DNA insertion lines SALK-099162 (DEG5; At4g18370) and SALK-004770 (DEG8; At5g39830) were obtained from the ABRC (Ohio State University). The T-DNA insertion lines were confirmed, by PCR and sequencing, using the following gene-specific and T-DNA–specific primers: LP (5′-TGGGAGTCCACAAAATATTGG-3′) and RP (5′-TTCCCTTCCTTGCTAAATCTTG-3′) for DEG5; LP (5′-AACTGTTTTCCCAGCTGGAC-3′), RP (5′-CTATTTCCCGGGTTAATGG-3′), and T-DNA LB (5′-TGGTTCACGTAGTGGGCCATCG-3′) for DEG8. The DNA fragments around the sites of the T-DNA insertions were amplified and cloned into the plasmid vector pGEM-T (Promega), and the precise location of the T-DNA insertion was determined by sequencing. Homozygous plants (identified by PCR analyses) for the T-DNA insertions were obtained, and then single mutants were backcrossed to the wild-type plants three times before generating double mutants to segregate out additional mutations. A deg5 deg8 double mutant was obtained by crossing single mutants of deg5 and deg8 and screening the F2 population by PCR using the primers described above.
Nucleic Acid Preparation and Analysis
Total RNA was isolated from 100-mg samples of fresh plant leaves using Trizol reagent, and DNA was isolated as described by Liu et al. (1995). The expression of DEG5 and DEG8 was determined by RT-PCR analysis. Equal RNA loading in each sample was confirmed by RT-PCR analysis of actin cDNA using the following primers: sense primer (5′-AACTGGGATGATATGGAGAA-3′) and antisense primer (5′-CCTCCAATCCAGACACTGTA-3′).
Complementation of deg5 and deg8
To complement the deg5 and deg8 mutations, cDNAs containing the coding regions of DEG5 and DEG8 were amplified by PCR using the following primers, which include SacI and BamHI restriction sites at their 5′ ends to facilitate cloning: sense 5′-GGATCCGCCATGACCATGGCTCTTGCTTC-3′ and antisense 5′-GAGCTCTTCCTCTTGCAGGACATCTTGCC-3′ for DEG5; sense 5′-GGATCCCGACTGGTCTCAGAGTTTTCTCG-3′ and antisense 5′-GAGCTCTCTCAAAGGGGGAAAAAACAGAG-3′ for DEG8. The PCR products were cloned into SacI and BamHI sites of pSN1301 under the control of the cauliflower mosaic virus 35S promoter. The constructs of pSN1301-DEG5 and pSN1301-DEG8 were transformed into Agrobacterium tumafaciens strain C58 via electroporation and introduced into homozygous deg5 and deg8 plants, respectively (Clough and Bent, 1998). The transformant plants were selected on medium containing half-strength Murashige and Skoog salt mix, 50 μg/mL hygromycin, and 0.8% agar. The resistant plants were transferred to soil to grow to maturity, and their transgenic status was further confirmed by PCR and immunoblot analyses.
Chlorophyll Fluorescence Measurements
Fluorescence measurements were performed using a portable fluorometer (PAM-2000; Walz). Before measurement, leaves were dark-adapted for 15 min. The maximum photochemical efficiency of PSII was then determined from the ratio of variable (Fv) to maximum (Fm) fluorescence (Fv/Fm = (Fm − Fo)/Fm) (Kitajima and Butler, 1975). All the above measurements were performed in a dark room in stable ambient conditions.
Photoinhibition and Recovery Treatments
Detached leaves, floating adaxial side up on water, were illuminated with a photon flux density of 1600 or 800 μmol m−2 s−1. To examine the role of chloroplast-encoded protein synthesis in the susceptibility of leaves to photoinhibition, detached leaves were incubated with their petioles submersed in 1 mM lincomycin solution at an irradiance of 20 μmol m−2 s−1 for 3 h prior to photoinhibitory light treatment. The temperature was maintained at 22°C during photoinhibition and recovery treatments.
To investigate the effects of high irradiance on plant growth, we transferred 2-week-old whole Arabidopsis plants grown in a growth chamber under a photon flux density of 120 μmol m−2 s−1 to a greenhouse under sunlight illumination for another 2 weeks. In the greenhouse, the maximum photon flux density at noon was ∼1000 μmol m−2 s−1, and average day/night temperatures were 25/22°C.
Thylakoid Membrane Preparation
Thylakoid membranes were prepared according to standard methods (Zhang et al., 1999). Briefly, the leaves were homogenized in an ice-cold isolation buffer containing 400 mM sucrose, 50 mM HEPES-KOH, pH 7.8, 10 mM NaCl, and 2 mM MgCl2 and filtered through two layers of cheesecloth. The filtrate was centrifuged at 5000g for 10 min. The thylakoid pellets were washed with isolation buffer, recentrifuged, and finally suspended in isolation buffer. The chlorophyll contents were determined spectrophotometrically as described by Porra et al. (1989). The resulting thylakoid membrane preparations were either used immediately or frozen in liquid N2 and stored at −70°C before use.
BN-PAGE, SDS-PAGE, and Protein Identification
BN-PAGE was performed essentially as described previously (Schägger et al., 1994). The thylakoid membranes were washed with 0.33 M sorbitol and 50 mM BisTris-HCl, pH 7.0, and solubilized with 1% (w/v) DM in 20% glycerol and 25 mM BisTris-HCl, pH 7.0, at 0.5 mg chlorophyll/mL for 10 min at 4°C. After centrifugation at 12,000g for 10 min, the supernatant was combined with one-tenth volume of 5% Serva blue G in 100 mM BisTris-HCl, pH 7.0, 0.5 M 6-amino-n-caproic acid, and 30% (w/v) glycerol and applied to 0.75-mm-thick 6 to 12% acrylamide gradient gels. For two-dimensional analysis, excised BN-PAGE lanes were soaked in SDS sample buffer and 5% β-mercaptoethanol for 60 min and layered onto 1-mm-thick 15% SDS polyacrylamide gels containing 6 M urea (Laemmli, 1970). After electrophoresis, the proteins were transferred to nitrocellulose membranes, probed with specific antibodies, and visualized by the enhanced chemiluminescence method. X-ray films were scanned and analyzed using an AlphaImager 2200 documentation and analysis system (Alpha Innotech). To estimate the molecular weights of the localized proteins, a mixture of standard proteins (Amersham Pharmacia Biotech) containing protein molecular mass standards of 669 kD (thyroglobulin), 440 kD (ferritin), 232 kD (catalase), 140 kD (lactate dehydrogenase), and 66 kD (BSA) was separated in a parallel gel.
Antiserum Production
The nucleotide sequences encoding the soluble part of DEG1, DEG5, and DEG8 were amplified by PCR using the follow primers: sense 5′-GGGCTAGCCCAGTTGATACAGTTGGTGG-3′ and antisense 5′-GAAGCTTCCGGCTTTGGTTCGAGAGTTAC-3′ for DEG1; sense 5′-GGGCTAGCGTCAATCTCTTCCAGAAAAC-3′ and antisense 5′-GGGAGCTCAATCGCAAAGCAACTTTGAC-3′ for DEG5; sense 5′-CCCGCTAGCGATTCAAAAGGGAACTTG-3′ and antisense 5′-CCCGCAAGCTTCACCTACACTGTACTCATCC-3′ for DEG8. The resulting DNA fragments of DEG1 and DEG8 were cleaved with NheI and HindIII and those of DEG5 with NheI and SacI. After cleavage, the fragments were fused in frame with the N-terminal His affinity tag of pET28a, and the resulting plasmids were transformed into Escherichia coli strain BL21 (DE3). The BL21 cells were harvested after the application of 0.4 mM isopropylthio-β-d-galactoside for 6 h and resuspended in 500 mM NaCl and 20 mM NaH2PO4, pH 7.8. After incubation for 30 min at 4°C in the presence of lysozyme at a final concentration of 1 mg/mL, the bacterial lysates were sonicated 30 times for 10 s each. The overexpressed His-DEG1, His-DEG5, and His-DEG8 fusion proteins in inclusion bodies were centrifuged at 3000g for 30 min, and the pellets were solubilized in 500 mM NaCl, 6 M urea, and 20 mM NaH2PO4, pH 7.8. The fusion proteins were purified on a Ni-NTA agarose resin matrix, and polyclonal antibodies were raised in rabbit with the purified antigens.
Pull-Down Assays
Fragments encoding full-length DEG5 and DEG8 were generated by PCR using the respective cDNAs as templates and the following primers: sense 5′-GGGCTAGCGCATTGGAGCAATTCAAAGA-3′ and antisense 5′-GGGAGCTCAATCTGTCTCTATAAGCTG-3′ for DEG5; sense 5′-GCTAGCCTTGGTGATCCATCCGTTGC-3′ and antisense 5′-AAGCTTTGAACTTTTCTCTTCCAATG-3′ for DEG8. The PCR products of DEG5 and DEG8 were cut with NdeI and SacI, and NdeI and HindIII, respectively. The resulting fragments were inserted into pET28a plasmids and transformed into BL21. The BL21 cells were harvested after the addition of 0.4 mM isopropylthio-β-d-galactoside for 2 h, and the overexpressed proteins were purified on a Ni-NTA agarose resin matrix according to Chassin et al. (2002). The purified proteins were applied to a Sephadex G-75 column and eluted with 20 mM NaH2PO4, pH 7.8, for renaturation. The identities of the expressed proteins were confirmed by immunoblot analyses with specific antibodies.
Ten micrograms of each recombinant fusion protein was coupled to 100 μL of a 50% suspension (v/v) of Ni-NTA beads in equilibration buffer for 60 min at 4°C. After the thylakoid membranes (100 μg chlorophyll) had been solubilized with 1% (w/v) DM in 20% glycerol (w/v), 25 mM BisTris-HCl, pH 7.0, and 1 mM PMSF for 15 min at 4°C, the solubilized membrane was centrifuged at 10,000g for 10 min and the supernatant was incubated with His-DEG5 and His-DEG8, which had been coupled to the Ni-NAT resin. After incubation overnight with constant rotation at 4°C, the beads were washed five times with ice-cold 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 1 mM EDTA buffer, and the bound proteins were eluted with SDS-PAGE sample buffer. The eluted proteins were resolved by SDS-PAGE and subjected to immunoblot analyses.
Immunolocalization Studies
The locations of the DEG5 and DEG8 proteins were studied by protease protection experiments (Meurer et al., 1998). The Arabidopsis thylakoid membrane samples (0.1 mg chlorophyll/mL) were each sonicated three times for 15 s on ice. Trypsin was then added to a final concentration of 50 μg/mL, and the mixtures were incubated at 25°C for 15 min. After this incubation, the thylakoid membranes were solubilized with SDS sample buffer, and the proteins were separated by SDS-PAGE and immunodetected with specific antibodies.
The association of DEG5 and DEG8 with the thylakoid membrane was investigated following the procedure of Itzhaki et al. (1998). Thylakoid membranes were suspended to a final concentration of 0.1 mg chlorophyll/mL in 10 mM HEPES-KOH, pH 8.0, 10 mM MgCl2, 330 mM sorbitol, and 1 mM PMSF supplemented with either 1 M NaCl or 1 M CaCl2. Membrane preparations without supplements were used as controls. These suspensions were sonicated three times for 15 s on ice. After treatments, the membranes were pelleted at 100,000g for 2 h at 4°C, quickly washed with isolation buffer, and used for SDS-PAGE and immunoblot analyses.
Proteolytic Degradation Assays
The proteolytic activity of DEG5 and DEG8 was assayed in standard reaction mixtures including 0.5 μg of purified DEG5 and DEG8 and 15 μg mixtures of casein (α-, β-, and κ-casein) (Sigma-Aldrich) in a total volume of 30 μL. The mixtures were incubated for 0, 30, 60, and 120 min at 37°C and subjected to SDS-PAGE. After electrophoresis, the gels were stained with Coomassie Brilliant Blue G 250.
The proteolytic activity of DEG5 and DEG8 toward thylakoid proteins was examined using the same reaction mixture, except that casein was replaced by thylakoids, which had been isolated from wild-type Arabidopsis plants kept either in darkness or in high light at 1600 μmol m−2 s−1 for 90 min at 4°C, sonicated on ice, and washed with 1.0 M CaCl2 to reduce endogenous DEG8 activity before the addition of recombinant DEG8 (0.5 μg). After incubation of recombinant protein with the thylakoid membrane at 37°C in the dark as described by Haußühl et al. (2001), the degradation of D1 protein and other photosynthetic proteins was examined by immunoblot analysis.
To classify the DEG8 protease, thylakoid membranes isolated from plants that had been subjected to high-light treatment were incubated at 90°C for 10 min to inactivate the endogenous proteases. Purified recombinant DEG8 protein (1.0 μg) was added to the thylakoid membrane preparations (10 μg chlorophyll), which were then incubated in the presence or absence (controls) of the following protease inhibitors: 100 μg/mL PMSF, 1 μg/mL aprotinin, 1 μg/mL pepstatin, 1 μg/mL E64 [3-carboxy-trans-2,3-epoxypropyl-leucylamido(4-guanidino)butane], and 500 μg/mL EDTA.
In Vivo Labeling of Chloroplast Proteins
In vivo protein labeling was performed essentially as described by Meurer et al. (1998). Briefly, 4-week-old leaves were preincubated in 20 μg/mL cycloheximide for 30 min and radiolabeled with 1 μCi/μL 35S-Met (specific activity >1000 Ci/mmol; Amersham Pharmacia Biotech) at a light intensity of 120 μmol m−2 s−1 in the presence of 20 μg/mL cycloheximide for 30 min at 22°C. To study the turnover of D1 protein, the leaves were washed twice with 10 mM cold Met and further incubated in the presence of cold Met under high irradiance (1600 μmol m−2 s−1) for 1, 2, or 4 h after labeling. To examine the degradation of D1 protein, lincomycin was added after pulse labeling and the leaves were subjected to high-light illumination (1600 μmol m−2 s−1) for 2 h. After washing twice with homogenization buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 2 mM EDTA), thylakoid membranes were isolated and the proteins were separated by SDS-PAGE. For autoradiography, gels were stained, dried, and exposed to x-ray film.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are At4g18370 (DEG5) and At5g39830 (DEG8).
Acknowledgments
We thank Eva-Mari Aro and Zach Adam for providing the antibodies. We also thank the ABRC for Arabidopsis seeds. This research is supported by the Major State Basic Research Development Program (2006CB910300), the National Natural Science Foundation of China (30500037), and the Frontier Project of the Knowledge Innovation Engineering of the Chinese Academy of Sciences (KJCX2-SW-w29).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Lixin Zhang (zhanglixin@ibcas.ac.cn).
References
- Adam, Z., and Clarke, A.K. (2002). Cutting edge of chloroplast proteolysis. Trends Plant Sci. 7 451–456. [DOI] [PubMed] [Google Scholar]
- Adir, N., Shochat, S., Inoue, Y., and Ohad, I. (1990). Mechanism of the light-dependent turnover of the D1 protein. J. Biol. Chem. 265 12563–12568. [PubMed] [Google Scholar]
- Andersson, B., and Aro, E.-M. (2001). Photodamage and D1 protein turnover in photosystem II. In Regulation of Photosynthesis, B. Andersson and E.-M. Aro, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 377–393.
- Andersson, B., and Styring, S. (1991). Photosystem 2: Organization, function and acclimation. In Current Topics in Bioenergetics, Vol. 16, C.P. Lee, ed (New York: Academic Press), pp. 2–81.
- Aro, E.M., Virgin, I., and Andersson, B. (1993). Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143 113–134. [DOI] [PubMed] [Google Scholar]
- Bailey, S., Thompson, E., Nixon, P.J., Horton, P., Mullineaux, C.W., Robinson, C., and Mann, N.H. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277 2006–2011. [DOI] [PubMed] [Google Scholar]
- Barbato, R., Friso, G., Giardi, M.T., Rigoni, F., and Giacometti, G.M. (1991). Breakdown of the photosystem II reaction center D1 protein under photoinhibitory conditions: Identification and localization of the C-terminal degradation products. Biochemistry 30 10220–10226. [DOI] [PubMed] [Google Scholar]
- Barber, J., and Andersson, B. (1992). Too much of a good thing: Light can be bad for photosynthesis. Trends Biochem. Sci. 17 61–66. [DOI] [PubMed] [Google Scholar]
- Barker, M., de Vries, R., Nield, J., Komenda, J., and Nixon, P.J. (2006). The DEG proteases protect Synechocystis sp .PCC 6803 during heat and light stresses but are not essential for removal of damaged D1 protein during the photosystem two repair cycle. J. Biol. Chem. 281 30347–30355. [DOI] [PubMed] [Google Scholar]
- Canovas, P.M., and Barber, J. (1993). Detection of a 10 kDa break-down product containing the C-terminus of the D1-protein in photoinhibited wheat leaves suggests an acceptor side mechanism. FEBS Lett. 324 341–344. [DOI] [PubMed] [Google Scholar]
- Chassin, Y., Kapri-Pardes, E., Sinvany, G., Arad, T., and Adam, Z. (2002). Expression and characterization of the thylakoid lumen protease DegP1 from Arabidopsis. Plant Physiol. 130 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, T., Southan, C., and Ehrmann, M. (2002). The HtrA family of proteases: Implications for protein composition and cell fate. Mol. Cell 10 443–455. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- De Las Rivas, J., Andersson, B., and Barber, J. (1992). Two sites of primary degradation of the D1 protein induced by acceptor or donor side photoinhibition in photosystem II core complexes. FEBS Lett. 301 246–252. [DOI] [PubMed] [Google Scholar]
- Eckert, H.J., Geiken, B., Bernarding, J., Napiwotzki, A., Eichler, H.J., and Renger, G. (1991). Two sites of photoinhibition of the electron transfer in oxygen evolving and Tris-treated PS II membrane fragments from spinach. Photosynth. Res. 27 97–108. [DOI] [PubMed] [Google Scholar]
- Gottesman, S. (1996). Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30 465–506. [DOI] [PubMed] [Google Scholar]
- Greenberg, B.M., Gaba, V., Mattoo, A.K., and Edelman, M. (1987). Identification of a primary in vivo degradation product of the rapidly-turning-over 32 kd protein of photosystem II. EMBO J. 6 2865–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J.K., Zhang, Z.Z., Bi, Y.R., Yang, W., Xu, Y.N., and Zhang, L.X. (2005). Decreased stability of photosystem I in dgd1 mutant of Arabidopsis thaliana. FEBS Lett. 579 3619–3624. [DOI] [PubMed] [Google Scholar]
- Haußühl, K., Andersson, B., and Adamska, I. (2001). A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 20 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen, P.F., Schuhmann, H., and Adamska, I. (2005). The family of Deg proteases in cyanobacteria and chloroplasts of higher plants. Physiol. Plant 123 413–420. [Google Scholar]
- Itzhaki, H., Naveh, L., Lindahl, M., Cook, M., and Adam, Z. (1998). Identification and characterization of DegP, a serine protease associated with the luminal side of the thylakoid membrane. J. Biol. Chem. 273 7094–7098. [DOI] [PubMed] [Google Scholar]
- Kanervo, E., Murata, N., and Aro, E.M. (1998). Massive breakdown of the photosystem II polypeptides in a mutant of the cyanobacterium Synechocystis sp. PCC 6803. Photosynth. Res. 57 81–91. [Google Scholar]
- Kapri-Pardes, E., Naveh, L. and Adam, Z. (2007). The thylakoid lumen protease Deg1 is involved in the repair of photosystem II from photoinhibition in Arabidopsis. Plant Cell 19 1039–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen, R., Tyystjarvi, E., and Aro, E.-M. (1996). Degradation pattern of photosystem II reaction center protein D1 in intact leaves. Plant Physiol. 111 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieselbach, T., and Funk, C. (2003). The family of Deg/HtrA proteases: From Escherichia coli to Arabidopsis. Physiol. Plant 119 337–346. [Google Scholar]
- Kim, D.Y., and Kim, K.K. (2005). Structure and function of HtrA family proteins, the key players in protein quality control. J. Biochem. Mol. Biol. 38 266–274. [DOI] [PubMed] [Google Scholar]
- Kitajima, M., and Butler, W.L. (1975). Excitation spectra for photosystem I and photosystem II in chloroplasts and the spectral characteristics of the distributions of quanta between the two photosystems. Biochim. Biophys. Acta 408 297–305. [DOI] [PubMed] [Google Scholar]
- Komenda, J., and Barber, J. (1995). Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and dependent on protein synthesis. Biochemistry 34 9625–9631. [DOI] [PubMed] [Google Scholar]
- Kyle, D.J., Ohad, I., and Arntzen, C.J. (1984). Membrane protein damage and repair: Selective loss of a quinone-protein function in chloroplast membranes. Proc. Natl. Acad. Sci. USA 81 4070–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Leonhard, K., Guiard, B., Pellecchia, G., Tzagoloff, A., Neupert, W., and Langer, T. (2000). Membrane protein degradation by AAA proteases in mitochondria: Extraction of substrates from either membrane surface. Mol. Cell 5 629–638. [DOI] [PubMed] [Google Scholar]
- Lindahl, M., Spetea, C., Hundal, T., Oppenheim, A.B., Adam, Z., and Andersson, B. (2000). The thylakoid FtsH protease plays a role in the light induced turnover of the photosystem II D1 protein. Plant Cell 12 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinska, B., Zylicz, M., and Georgopoulos, C. (1990). The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J. Bacteriol. 172 1791–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junction by thermal asymmetric interlaced PCR. Plant J. 8 457–463. [DOI] [PubMed] [Google Scholar]
- Mattoo, A.K., Hoffman-Falk, H., Marder, J.B., and Edelman, M. (1984). Regulation of protein metabolism: Coupling of photosynthetic electron transport to in vivo degradation of the rapidly metabolized 32-kilodalton protein of the chloroplast membranes. Proc. Natl. Acad. Sci. USA 81 1380–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer, J., Plücken, H., Kowallik, K.V., and Westhoff, P. (1998). A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 17 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanba, O., and Satoh, K. (1987). Isolation of a photosystem II reaction center consisting of D-1 and D-2 polypeptides and cytochrome b-559. Proc. Natl. Acad. Sci. USA 84 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, N., and Yocum, C.F. (2006). Structure and function of photosystem I and II. Annu. Rev. Plant Biol. 57 521–565. [DOI] [PubMed] [Google Scholar]
- Ohad, I., Kyle, D.J., and Arntzen, C.J. (1984). Membrane protein damage and repair: Removal and replacement of inactivated 32-kilodalton polypeptides in chloroplast membranes. J. Cell Biol. 99 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen, M., and Wren, B. (1997). The HtrA family of serine proteases. Mol. Microbiol. 26 209–221. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Emanuelsson, O., Kalume, D.E., Ytterberg, J., Friso, G., Rudella, A., Liberles, D.A., Soderberg, L., Roepstorff, P., von Heijne, G., and van Wijk, K.J. (2002). Central functions of the luminal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14 211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedemann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectrometry. Biochim. Biophys. Acta 975 384–394. [Google Scholar]
- Prasil, O., Adir, N., and Ohad, I. (1992). Dynamics of photosystem II: Mechanisms of photoinhibition and recovery process. In The Photosystems: Structure, Function and Molecular Biology, Vol. 11, J. Barber, ed (Amsterdam: Elsevier Science Publishers), pp. 295–348.
- Sakamoto, W., Zaltsman, A., Adam, Z., and Takahashi, Y. (2003). Coordinated regulation and complex formation of yellow variegated 1 and yellow variegated 2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger, H., Cramer, W.A., and von Jagow, G. (1994). Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217 220–230. [DOI] [PubMed] [Google Scholar]
- Schlieker, C., Mogk, A., and Bukau, B. (2004). A PDZ switch for a cellular stress response. Cell 117 417–419. [DOI] [PubMed] [Google Scholar]
- Schubert, M., Petersson, U.A., Haas, B.J., Funk, C., Schroder, W.P., and Kieselbach, T. (2002). Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 277 8354–8365. [DOI] [PubMed] [Google Scholar]
- Shipton, C.A., and Barber, J. (1994). In vivo and in vitro photoinhibition reactions generate similar degradation fragments of D1 and D2 photosystem-II reaction-centre proteins. Eur. J. Biochem. 220 801–808. [DOI] [PubMed] [Google Scholar]
- Silva, P., Thompson, E., Bailey, S., Kruse, O., Mullineaux, C.W., Robinson, C., Mann, N.H., and Nixon, P.J. (2003). FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp.PCC6803. Plant Cell 15 2152–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolenko, A., Pojidaeva, E., Zinchenko, V., Panichkin, V., Glaser, V.M., Herrmann, R.G., and Shestakov, S.V. (2002). The gene complement for proteolysis in the cyanobacterim Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr. Genet. 41 291–310. [DOI] [PubMed] [Google Scholar]
- Spetea, C., Hundal, T., Lohmann, F., and Andersson, B. (1999). GTP bound to chloroplast thylakoid membranes is required for light-induced, multienzyme degradation of the photosystem II D1 protein. Proc. Natl. Acad. Sci. USA 96 6547–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess, C., Beil, A., and Ehrmann, M. (1999). A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97 339–347. [DOI] [PubMed] [Google Scholar]
- Vass, I., Styring, S., Hundal, T., Koivuniemi, A., Aro, E.M., and Andersson, B. (1992). Reversible and irreversible intermediates during photoinhibition of photosystem II: Stable reduced QA species promote chlorophyll triplet formation. Proc. Natl. Acad. Sci. USA 89 1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin, I., Salter, A.H., Ghanotakis, D.F., and Andersson, B. (1991). Light-induced D1 protein degradation is catalyzed by a serine-type protease. FEBS Lett. 287 125–128. [DOI] [PubMed] [Google Scholar]
- Wiklund, R., Salih, G., Mäenpää, P., and Jansson, C. (2001). Engineering of the protein environment around the redox-active TyrZ in photosystem II. Eur. J. Biochem. 268 5356–5364. [DOI] [PubMed] [Google Scholar]
- Yoshioka, M., Uchida, S., Mori, H., Komayama, K., Ohira, S., Morita, N., Nakanishi, T., and Yamamoto, Y. (2006). Quality control of photosystem II. cleavage of reaction center D1 protein in spinach thylakoids by FtsH protease under moderate heat stress. J. Biol. Chem. 281 21660–21669. [DOI] [PubMed] [Google Scholar]
- Yu, F., Park, S., and Rodermel, S.R. (2004). The Arabidopsis FtsH metalloprotease gene family: Interchangeability of subunits in chloroplast oligomeric complexes. Plant J. 37 864–876. [DOI] [PubMed] [Google Scholar]
- Yu, F., Park, S., and Rodermel, S.R. (2005). Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol. 138 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman, A., Feder, A., and Adam, Z. (2005. b). Developmental and light effects on the accumulation of FtsH protease in Arabidopsis chloroplasts: Implications for thylakoid formation and photosystem II maintenance. Plant J. 42 609–617. [DOI] [PubMed] [Google Scholar]
- Zaltsman, A., Ori, N., and Adam, Z. (2005. a). Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell 17 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L.X., Paakkarinen, V., van Wijk, K.J., and Aro, E.M. (1999). Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem. 274 16062–16067. [DOI] [PubMed] [Google Scholar]