Abstract
For >20 years, the enigmatic behavior of plant mitochondrial genomes has been well described but not well understood. Chimeric genes appear, and occasionally are differentially replicated or expressed, with significant effects on plant phenotype, most notably on male fertility, yet the mechanisms of DNA replication, chimera formation, and recombination have remained elusive. Using mutations in two important genes of mitochondrial DNA metabolism, we have observed reproducible asymmetric recombination events occurring at specific locations in the mitochondrial genome. Based on these experiments and existing models of double-strand break repair, we propose a model for plant mitochondrial DNA replication, chimeric gene formation, and the illegitimate recombination events that lead to stoichiometric changes. We also address the physiological and developmental effects of aberrant events in mitochondrial genome maintenance, showing that mitochondrial genome rearrangements, when controlled, influence plant reproduction, but when uncontrolled, lead to aberrant growth phenotypes and dramatic reduction of the cell cycle.
INTRODUCTION
The plant mitochondrial genome displays several features that are distinctive to the plant kingdom. These include the incorporation of foreign DNA sequences, a partially linearized genome, split gene sequences requiring RNA trans-splicing for proper expression, large, recombinationally active repeats, a stoichiometrically variable multipartite genome organization, and an unusually high incidence of illegitimate recombination leading to gene chimeras (Adams and Palmer, 2003; Knoop, 2004). The linear genome structure and T4 phage-like features (Backert and Borner, 2000) of the genome imply that components of the genome maintenance apparatus may have been acquired over plant evolution (Lang et al., 1999). Furthermore, the linear genome organization would be consistent with a recombination-mediated replication initiation system (Kreuzer, 2005).
More than 20 years of literature exists documenting the recombinogenic nature of the plant mitochondrial genome. This literature includes many examples of aberrant recombination induced by tissue culture conditions (Kanazawa et al., 1994) upon alloplasmic substitution, wide hybridization (Kaul, 1988; Dieterich et al., 2003), or spontaneously (Marienfeld and Newton, 1994; Newton et al., 1998). In many of these cases, the genomic rearrangement activity was detected as the induction or loss of cytoplasmic male sterility (CMS). CMS involves the inability of an otherwise phenotypically normal plant to shed viable pollen. In a population of hermaphrodites, male-sterile plants generally have an adaptive advantage due to increased seed production (Charlesworth, 2002), leading to a mating system known as gynodioecy (Darwin, 1877). The CMS phenotype varies widely, from homeotic floral morphological variants (Farbos et al., 2001; Linke et al., 2003) to premature tapetal breakdown (Levings, 1993), incomplete callose deposition (Abad et al., 1995), and gametophytic abortion (Lee et al., 1980). Likewise, the underlying mitochondrial rearrangements associated with CMS are each distinct (Schnable and Wise, 1998).
Several years ago it was demonstrated that commonly observed ghost bands detected by gel blot hybridization of plant mitochondrial DNA represented a population of substoichiometric genomic forms retained generation to generation within the mitochondrial population (Small et al., 1987). Under particular conditions, these substoichiometric DNA intermediates could be differentially amplified to high relative copy number, altering expression of the genes (including CMS mutations) encoded therein. The dramatic amplification or suppression of specific subgenomic molecules was referred to as substoichiometric shifting (SSS) and was subsequently shown to be under nuclear gene control (Mackenzie and Chase, 1990).
To further investigate the evolutionary implications and molecular mechanisms underlying the SSS process in plants, our laboratory identified two nuclear genes that regulate SSS in Arabidopsis thaliana. The first, designated MSH1 (formerly CHM), encodes a homolog of the Eschericha coli MutS mismatch repair component (Abdelnoor et al., 2003). The locus encodes a protein with six protein domains, one comprising a GIY-YIG homing endonuclease. This protein is highly conserved within the plant kingdom, and the endonuclease domain distinguishes it from homologs in the archaea, bacteria, and animals. A similar protein, with a different endonuclease domain, has also been found in the corals but not in any other animal species (Abdelnoor et al., 2006). Mutation of the MSH1 locus results in reproducible mitochondrial rearrangements in Arabidopsis (Martinez-Zapater et al., 1992; Sakamoto et al., 1996; Abdelnoor et al., 2003). The second gene is the focus of this study. Here, we have investigated the role of this E. coli RecA homolog in maintaining mitochondrial genome stability and its functional relationship to MSH1. Mutations at either locus result in SSS. We demonstrate the influence of nonreciprocal ectopic recombination activity on the SSS phenomenon, and we present the cellular and developmental implications of msh1 and recA3 mutant-associated mitochondrial genome instability in plants. We also present a model to explain the related phenomena of chimeric gene formation, illegitimate recombination to effect SSS, and mitochondrial DNA replication.
RESULTS
RECA3 Function in Arabidopsis Is Distinct from Other RecA Homologs
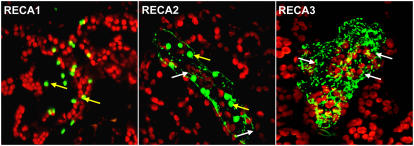
There exist at least three distinct RecA homologs in the Arabidopsis genome, located on chromosomes 1 (At1g79050), 2 (At2g19490), and 3 (At3g10140). A fourth homolog has also been identified (At3g32920) but appears to represent a pseudogene and is not described here. The three intact loci are designated RECA1, RECA2, and RECA3, respectively. Alignment of their predicted protein sequences to each other and to E. coli RecA revealed several distinct features within RECA3 as detailed in Figure 1. The three differences indicated are in very strongly conserved motifs (McGrew and Knight, 2003). Development of N-terminal gene fusions to green fluorescent protein (GFP) showed that RECA1 is a plastid-targeted protein, RECA2 is dual targeted to mitochondria and plastids, and RECA3 is targeted to mitochondria (Figure 2). RECA3 has previously been shown to be mitochondrially targeted by a different method and to partially complement the mutagen-sensitive phenotype of the E. coli recA− mutation (Khazi et al., 2003).
Figure 1.
Protein Sequence Alignment of the Three Organellar Arabidopsis RECA Proteins along with RecA from E. coli (GI:67471975).
Identical residues in all four are indicated by gray shading, and residues that are similar are indicated in light gray. Notable differences between RECA3 and the others are indicated by black shading and include the following changes in highly conserved motifs: (1) substitution of a Lys for a conserved Pro residue in the ATP binding/hydrolyzing P-loop, (2) a Lys-to-Pro substitution in the RecA signature motif involved in monomer–monomer interaction, and (3) a C-terminal deletion of the region of negatively charged residues that have been shown to be important in RecA strand exchange activity.
Figure 2.
Subcellular Localization of Arabidopsis RECA Proteins by Particle Bombardment of Targeting Presequence-GFP Fusions in Young Arabidopsis Leaf Cells.
DNA fragments representing the first 80 codons of each gene were amplified from genomic DNA and inserted into the binary vector pK7FWG2 (Karimi et al., 2002). These constructs were analyzed by particle bombardment of Arabidopsis leaves and confocal microscopy. Representative GFP-containing mitochondria are indicated by white arrows and the GFP-containing plastids by yellow arrows.
Analysis of T-DNA insertion mutations of the three RecA homologs obtained from the Salk Institute and Syngenta indicated that disruption of RECA1 or RECA2 resulted in lethality. However, disruption of the RECA3 locus resulted in viable and phenotypically normal plants displaying reproducible mitochondrial DNA rearrangements.
recA3 Mutations Lead to Mitochondrial Rearrangements Similar, but Not Identical, to Those Occurring in msh1 Mutants
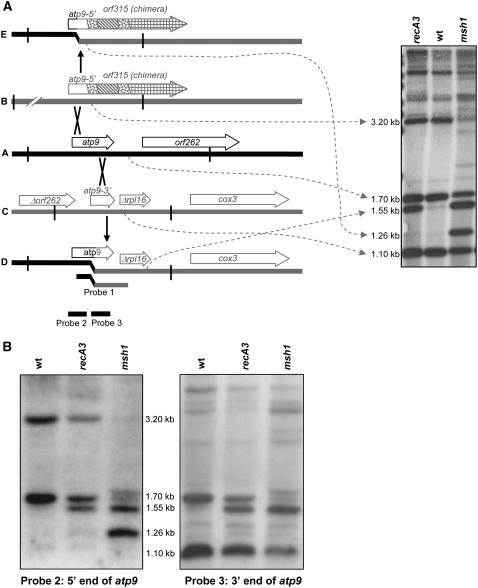
Heterozygous RECA3/recA3-1 mutant plants were self-crossed, and the homozygous progeny were identified by PCR using primers that specifically identified the T-DNA insertion. Mitochondrial DNA from at least eight independent recA3-1 homozygotes was digested with BamHI and hybridized with a probe encompassing the atp9-rpl16 junction sequence previously shown to be representative of substoichiometric shifting (Martinez-Zapater et al., 1992; Sakamoto et al., 1996; Abdelnoor et al., 2003). The blot displayed the novel bands shown in Figure 3A. Further hybridization with probes to other segments of these genes (Figure 3B) allowed the identification of these bands as indicated in Figure 3A. The molecules indicated as A, B, and C are all present in wild-type mitochondrial DNA, including the heterozygous RECA3/recA3-1 parents. Molecule A includes the full-length atp9 gene, while molecule B includes 177 bp of 5′ untranslated region and 158 bp of the coding sequence of atp9 (corresponding to amino acids 1 to 52) as part of a larger gene chimera called orf315 (Unseld et al., 1997). Molecule C includes only 63 bp of the coding sequence (corresponding to amino acids 66 to 85, plus the stop codon) and 186 bp of 3′ untranslated region (Forner et al., 2005). The two fragments of atp9 in molecules B and C do not overlap and therefore cannot undergo homologous recombination. However, each includes identical sequence to the full-length atp9 gene in molecule A and can recombine with it. In recA3-1 homozygotes, the only recombinant molecule observed is molecule D, a recombination product of molecules A and C. The reciprocal recombinant, with a predicted BamHI fragment size of 1.22 kb is not seen. Additional minor bands seen in Figure 3A are not yet fully characterized. The asymmetric accumulation of recombinant DNA molecules appears to be a hallmark of plant mitochondrial genome behavior (Fauron et al., 1995) and is now modeled in Arabidopsis by our group.
Figure 3.
Analysis of Asymmetric Recombination Associated with SSS.
(A) Predominant recombination events in recA3-1 and msh1-1 mutant plants. BamHI sites are indicated by vertical lines. The blot was hybridized with a probe spanning atp9 and Δrpl16 from molecule D (indicated as Probe 1). Dashed arrows indicate the BamHI fragments corresponding to the bands on the DNA gel blot. Molecules A and B are derived from the published sequence of the Arabidopsis ecotype C24 sequence (accession number NC_001284) (Unseld et al., 1997). The BamHI sites in molecule A are found at positions 278381 and 280053 and those in molecule B are at positions 20236 and 17037. Molecule C is derived from the sequence of BAC T17H1 (accession number AC007143) originating in ecotype Col-0 and also described by Forner et al. (2005). The BamHI sites are located at positions 57709 and 56613 in BAC T17H1. Molecule D is the result of recombination between molecules A and C anywhere in the 249 bp of identity in the 3′ region of atp9 and has been described by Sakamoto et al. (1996), although their explanation of the origin of this fragment was incorrect because they were not aware of molecule C. Molecule E is predicted by in silico recombination of molecules A and B within the 335-bp identity in the 5′ end of atp9.
(B) Additional hybridization analysis of the bands indicated in (A). BamHI digests were blotted and hybridized with probes representing the 5′ and 3′ ends of the atp9 gene, indicated as Probes 2 and 3 in (A). As expected, the 1.10-kb band does not hybridize to the 5′ end probe, and the 3.20- and 1.26-kb bands do not hybridize to the 3′ end probe.
This result is similar to that seen in msh1-1 homozygotes, which also display the nonreciprocal recombination product D and an additional band, corresponding to molecule E, the nonreciprocal recombination product of molecules A and B (Figure 3A). Although this pattern of bands was previously reported (Martinez-Zapater et al., 1992; Sakamoto et al., 1996; Abdelnoor et al., 2003), the identity of the fragments was not known. The reciprocal product of molecule E, with a predicted size of 3.61 kb, is not seen. Hybridization with multiple probes from the unique sequences in these regions confirmed the identity of the 1.26-kb band as configuration E and showed that it is not the reciprocal product of molecule D (Figure 3B). These results indicate that the recombination products seen in recA3-1/recA3-1 mutants are a subset of those seen in msh1-1/msh1-1 mutants, and in both mutants, only one of the two possible reciprocal recombinant products accumulates.
Genetic Analysis of RECA3 and MSH1 Action Suggests an Influence on de Novo Mitochondrial Recombination Activity
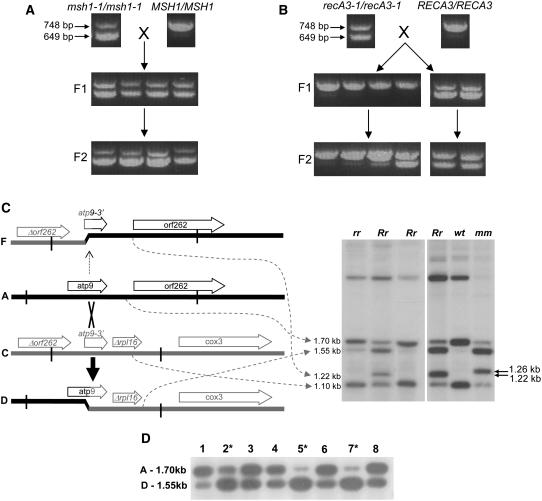
The mitochondrial genome rearrangements observed in msh1-1 mutants are completely stable and, once established, are permanent. Figure 4A indicates a cross of homozygous msh1-1 mutant plants pollinated with the wild type. A three-primer PCR assay was used to detect molecule A and the recombinant molecule D (see Figure 3A) in these crosses. The msh1-1/msh1-1 parent shows both molecules, and the MSH1/MSH1 pollen parent shows only molecule A. The F1 progeny inherit the mitochondrial genome from the maternal parent, as expected, but the nuclear genotype of MSH1/msh1-1 has not effected a reversal to the wild-type mitochondrial genome. Selfing these F1 plants results in completely stable maintenance of the maternally inherited cytoplasm, even in those F2 plants that are homozygous wild type. By contrast, pollination of homozygous recA3-1 mutants by the wild type results in F1 individuals, ∼80% of which display the wild-type pattern (Figure 4B). Thus, the RECA3 allele is capable of reversing the mutant mitochondrial genome pattern to the wild type in most cases. Selfing the reversed plants resulted in a 3:1 Mendelian ratio of wild-type to recA3/recA3 mitochondrial genomes, while selfing the F1 plants that retained molecule D showed that it continued to be retained in the F2.
Figure 4.
The Nature of recA3 Phenotype Reversibility.
(A) Mitochondrial SSS mediated by msh1-1. The pollen parent is indicated to the right. SSS was detected using a PCR-based assay as previously described (Sakamoto et al., 1996). A single forward primer is located in the 5′ end of the atp9 gene. Two different reverse primers were used: one in orf262 and the other in Δrpl16. The top 748-bp band corresponds to the amplification product from molecule A, and the bottom 649-bp band corresponds to the amplification product from molecule D.
(B) Mitochondrial SSS mediated by recA3-1. The PCR assay was as in (A).
(C) Recombinant molecules in recA3-1 mutants before and after pollination by the wild type. BamHI digests are hybridized with Probe 1 from Figure 3A; rr indicates homozygous recA3-1 mutants, while the two lanes marked Rr represent separate progeny from pollination of recA3-1/recA3-1 plants with RECA3/RECA3 pollen. Molecules A and C are identical to those shown in Figure 3A. Crossing over between molecules A and C in recA3-1 mutant plants results in accumulation of molecule D (as also seen in Figure 3A). The reciprocal recombinant is molecule F. The panel on the right is a separate experiment confirming that the 1.22-kb band seen in recA3-1 mutants (molecule F) is distinct from the 1.26-kb band (molecule E) previously seen in msh1-1 mutants.
(D) DNA gel blot analysis of different recA3-1/recA3-1 plants. BamHI digestion, blotting, and hybridizations were as in (C). Plants with higher stoichiometry of molecule D are indicated by asterisks.
Gel blot analysis revealed the underlying basis for the two different forms among the RECA3/recA3-1 F1 plants. These plants demonstrate one of two patterns of mitochondrial DNA. While most of the F1 progeny lose molecule D, reversing the mitochondrial atp9 recombination event that had been induced in the mutants, some retain it (Figure 4C). Those plants that retain molecule D also acquire a 1.22-kb molecule designated F (Figure 4C). Although similar in size to the 1.26-kb band seen in msh1-1 mutants, it is not the same fragment. The 1.26-kb band does not hybridize to the 3′ end of atp9, while the 1.22-kb band represented as molecule F does not hybridize to the 5′ end of atp9. Further analysis of molecule F by PCR and hybridization revealed that it is the reciprocal recombinant of molecule D, as indicated in the figure.
Eight recA3-1/recA3-1 plants are analyzed in Figure 4D. All of these plants have both molecules A and D, assayed by DNA gel blot analysis. However, there are different stoichiometries of these two molecules: plants 2, 5, and 7 show recombinant molecule D in higher stoichiometry than parental molecule A, while plants 1, 3, 4, 6, and 8 contain molecule A in higher copy than molecule D. When these plants were pollinated with the wild type, the results were striking. All the progeny of the plants predominant for molecule D retained molecule D (40 F1 progeny tested from nine different siliques). However, 95% of the progeny from the plants predominant for molecule A lost molecule D (211 out of 223 total progeny from 31 different siliques). The exceptions appeared sporadically, with no more than two seeds in any silique. We suggest that when molecule D is less abundant in a plant than molecule A, reintroduction of the wild-type RECA3 allele causes the loss of molecule D, but when molecule D is the most abundant configuration, it is stable and reciprocal recombination occurs to produce F. The basis for this threshold effect is unknown but could be due to the partitioning of organelles during cell division, known as cytoplasmic sorting, resulting in homoplasmy (Birky, 1983).
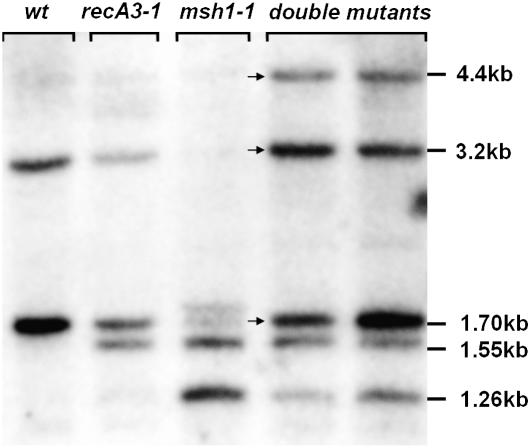
A similar phenomenon occurs in msh1 mutants. This was first observed by Sakamoto et al. (1996), but the data could not be interpreted completely because they based their analysis on DNA sequence from ecotype C24 (Unseld et al., 1997), while they were using plants of ecotype Landsberg erecta (Ler). Although C24 includes the configurations indicated in Figure 3A as A and B, Ler has only configuration D. When the msh1 mutation is introduced into Ler, the Δrpl16 pseudogene in molecule D recombines with a complete copy of rpl16 elsewhere in the genome to produce two novel fragments reported by Sakamoto et al. (1996), which are reciprocals of each other. Of particular significance, the accumulation of both reciprocal recombinants occurred only after msh1 mutant plants were pollinated by the wild type. Unlike the maternal distorted leaf phenotype reported by Sakamoto et al. (1996) from these crosses, there is no obvious phenotypic change emerging from crosses of the recA3-1 mutant. This distinction is likely because msh1 mutants undergo additional rearrangements, including loss of some DNA configurations, that are irreversible (e.g., 3.20- and 4.40-kb fragments in Figure 7).
Figure 7.
DNA Gel Blot Analysis of the Double Mutant.
BamHI digests of DNA from Col-0 and from homozygous recA3, msh1, and double mutant plants were blotted and hybridized with Probe 2 from Figure 3A. Two different double mutant plants are shown. Arrows indicate stoichiometric differences in the double mutant compared with either of the single mutants.
RECA3 and MSH1 Have Enhanced Expression in Reproductive Tissues
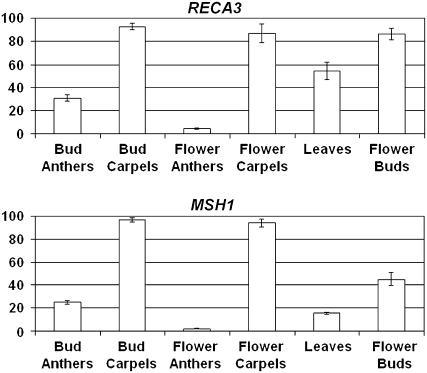
Previous experiments to track the developmental timing of substoichiometric shifting (Johns et al., 1992; S.A. Mackenzie, unpublished data) have suggested that nuclear influence on mitochondrial genome configuration may be most pronounced during or immediately preceding flowering. We used quantitative real-time RT-PCR to detect mRNA levels of both RECA3 and MSH1 in roots, mature leaves, young leaves, and unopened flower buds. RNA samples were also prepared from dissected stamens and carpels taken from both unopened flower buds and fully opened flowers. Although both RecA3 and Msh1 transcript levels were nearly undetectable in young roots and mature leaves (data not shown), Figure 5 shows that the genes are expressed in young leaves and flower buds, with levels highest in carpel tissues. In all tissues but ovules, the mRNA levels of MSH1 were lower than RECA3 (Figure 5; data not shown). The data suggest that functional specialization of RECA3 and MSH1 in higher plants may have involved not only adaptations in gene structure but also in gene expression pattern.
Figure 5.
Expression of RECA3 and MSH1.
Quantitative real-time RT-PCR analysis of RECA3 and MSH1 gene expression. Levels of RNA in the tissues indicated were assayed as described by Livak and Schmittgen (2001), using ubiquitin as an internal standard. Tissues were taken from 6-week-old plants. The data represent means of four replicates ± SE.
RECA3 and MSH1 Function in Distinct Pathways
Both recA3 and msh1 mutants undergo mitochondrial genome changes in highly similar but nonidentical patterns, leading us to question whether these two proteins carry out similar functions within the plant mitochondrion. The homologies to bacterial RecA and MutS might suggest that RECA3 is involved in recombination and that MSH1 is involved in mismatch repair. However, we find no evidence for defects in mismatch repair in msh1 mutants, and there is ample precedent for thinking that a MutS homolog does not function in mismatch repair. In yeast, the MutS homolog MSH2 promotes symmetric recombination events in meiosis (Hoffmann et al., 2005), and in mammalian cells, the MSH3/MSH5 heterodimer has been found to bind to Holliday junctions during double-strand break repair–mediated recombination (Snowden et al., 2004). Recent results in yeast suggest that MSH1 is not involved in mismatch repair but is involved in stability of the genome and maintenance of homoplasmy (Sia and Kirkpatrick, 2005). The differences observed between msh1 and recA3 mutants in mitochondrial rearrangement patterns and reversibility of phenotype suggest that the genes act in two overlapping but distinct pathways. To test this hypothesis, we generated msh1-1 recA3-1 double mutants.
To establish double mutant plants on an unaltered mitochondrial genome, we used recA3-1/recA3-1 mutants as females in crosses to msh1-1/msh1-1 mutants. The resulting double heterozygotes (RECA3 msh1-1/recA3-1 MSH1) had reversed mitochondrial genome configurations resembling the wild type. These plants were selfed, and the homozygous double mutants were identified by genotyping. These plants all had a distinct slow growth, small size, and delayed flowering phenotype. However, to ensure that the mitochondrial genome in these plants had not been altered by passage through a recA3-1 homozygote, these double mutant plants were used to pollinate Columbia-0 (Col-0). This produced plants that appeared completely normal, had inherited wild-type mitochondrial DNA from Col-0, and were heterozygous for both recA2-1 and msh1-1. The F1 progeny were allowed to self, and double mutants were again identified by genotyping. The double mutants obtained in this cross inherited the Col-0 cytoplasm, so the effect of simultaneous inactivation of both RECA3 and MSH1 on the mitochondrial genome could be assessed. All double mutant plants (>50 plants evaluated) were highly pronounced in phenotype, as shown in Figure 6, with slower growth rate, small size and delayed flowering (Figure 6A), reduced mitotic index (Figure 6B), reduced fertility, and low seed set (Figure 6C). The second generation plants, derived by selfing, were even more pronounced in phenotype. The mitochondrial genome of the double mutants appeared to consist of the Col-0 wild-type pattern superimposed on the msh1-associated pattern, together with the recA3-associated pattern (Figure 7). These observations support the hypothesis that these two genes act in distinct but overlapping pathways, and their simultaneous loss results in a higher degree of genome instability.
Figure 6.
Dual Distribution of MshI and RecA3 Conditions a Distinct Phenotype.
(A) Phenotype of msh1-1/msh1-1, recA3-1/recA3-1, and double mutant plants. Eight-week-old plants are shown. Col-0, recA3-1/recA3-1, msh1-1/msh1-1, and recA3-1 msh1-1/recA3-1 msh1-1 double mutant plants were planted together and grown under identical conditions.
(B) Mitotic index determination. Two median longitudinal sections per root and two roots per group were examined. Mitotic profiles were recorded for root apical meristem within 80 μm. The mean mitotic indices are shown and were 7.75% for Col-0 (i), 4.75% for msh1 (ii), 5.5% for recA3 (iii), and 0.75% for the double mutant (iv). Bar = 10 μm.
(C) Abnormal flower morphology of recA3 msh1 double mutant plants. The stigma appears to be developed and receptive before the pollen matures. Fewer pollen grains are seen in the double mutants than in Col-0.
DISCUSSION
Plant Mitochondrial SSS Is Controlled by at Least Two Nuclear Genes That Have Undergone Specialization
We have presented evidence to suggest that two nuclear genes, RECA3 and MSH1, are uniquely adapted in plants to control mitochondrial genome maintenance. While both genes have maintained sequence features strongly reminiscent of their bacterial counterparts, several modifications have occurred in both genes that appear to be conserved in plants. We have described some of the distinguishing features of the RECA3 protein sequence in this study and recently reported novel features of MSH1, including well-conserved protein domains and an unusual fusion with a GIY-YIG endonuclease (Abdelnoor et al., 2006). Other essential recombination controls within the mitochondrial and plastid genomes appear to be performed by at least two additional recA homologs in Arabidopsis.
The SSS Phenomenon and Its Control Appear to Be Timed with the Transition to Reproduction in Plants
Specialization of RECA3 and MSH1 functions may also derive from their developmental regulation. Both loci appeared to be expressed at relatively low levels in most plant tissues but were enhanced in expression during flower development. This timing may correspond to that at which SSS occurs (Johns et al., 1992) and appears consistent with the point at which RECA3 effects SSS reversal or reciprocal recombination in this study.
The RNA interference–mediated suppression of MSH1 expression in tobacco (Nicotiana tabacum) and tomato (Solanum lycopersicum) results in a condition of CMS (Sandhu et al., 2007). This observation suggests a direct relationship between MSH1 expression, mitochondrial genome stability, and the microsporogenesis process. The association of unusual mitochondrial chimeric sequences with the breakdown of pollen development has long been documented (Schnable and Wise, 1998; Hanson and Bentolila, 2004) but, to date, with little understanding of the direct relationship of mitochondrial genome structure to pollen abortion. More detailed investigation of the role of enhanced MSH1 and RECA3 expression during plant reproduction may provide important insight to this long-held question.
While loss of MSH1 or RECA3 functions individually can result in a fairly mild and sustainable plant phenotype, loss of the two functions simultaneously has profound effects on the plant, particularly with regard to growth rate and reproduction. More detailed analysis of the msh1-1 recA3-1 double mutants is currently underway to understand cell cycle and redox status of these plants, but it is clear that while the influence of SSS may be predominantly at the reproductive stage, the mitochondrial genome instability effected in the double mutant influences all stages of plant growth and development.
The SSS Process Appears to Be Associated with Nuclear Regulation of de Novo, Mitochondrial, Nonhomologous Recombination: A Proposed Model
The identification of RECA3 and MSH1 has made possible a much more detailed investigation of the SSS process by permitting its direct induction. The reproducible events that occur in recA3 and msh1 mutants allow us to infer their normal function, and detailed analysis of mitochondrial genomic regions participating in SSS has permitted us to model the process.
Our model links mitochondrial DNA replication, the generation of chimeric genes, and the nonreciprocal recombination between short repeated sequences resulting in altered stoichiometry and expression of the chimeras. We propose that double-strand breaks are central to all these processes. When double-strand breaks occur, they can be repaired in a number of ways (Aguilera, 2001; Bleuyard et al., 2006; Preston et al., 2006). One possibility is nonhomologous end joining (NHEJ), resulting in chimeric genes. For example, orf315 includes fragments from at least three genes, including atp9, orf153b, and two noncontiguous fragments of orf262 (Figure 3A, molecule B). Whether this chimera was formed by a series of events over time or whether it was assembled from scrambled fragments of DNA all at once is unknown. Another example is the 1790-bp insertion upstream of the cox3 gene in Col-0 (Forner et al., 2005). This insertion includes at least six noncontiguous fragments of the genome joined together (Figure 3A, molecule C). We do not observe de novo appearance of chimeric genes in recA3 or msh1 mutants, suggesting that these proteins are not involved in NHEJ. However, the chimeras that are already present in the genome are substrates for the recombination events that we observe in the mutants.
In wild-type Arabidopsis, repeats of up to 560 bp are not recombinationally active, while repeats of 6.5 and 4.2 kb are (Unseld et al., 1997). There are no repeats between 560 and 4.2 kb in size. This pattern is consistent with other species (Kubo et al., 2000; Notsu et al., 2002; Ogihara et al., 2005; Sugiyama et al., 2005). The DNA sequence identity of these repeats is evidence of frequent gene conversion events at both short and long repeats. However, in wild-type plants, gene conversion is only accompanied by crossovers at the long repeats and not at the short repeats. The aberrant events we and Sakamoto et al. (1996) have observed in msh1 and recA3 mutants are crossovers at two of the short repeats, those including fragments of the atp9 and rpl16 genes. We conclude that both short and long repeats are subject to frequent strand invasion, heteroduplex, and DNA synthesis events, resulting in gene conversion, and that the normal functions of RECA3 and MSH1 are to prevent crossing over at short repeats by directing these intermediates exclusively into gene conversion events. There must be a size threshold that allows recombination at the 4.2- and 6.5-kb repeats but not at the shorter repeats, as has been found in yeast (Inbar et al., 2000). This process is conceptually similar to the suppression of homeologous recombination at repetitive sequences in nuclear genomes (Nicholson et al., 2000; Goldfarb and Alani, 2005; Mieczkowski et al., 2006; Yang et al., 2006). MutS homologs have also been implicated in this process in nuclei (Li et al., 2006; Weinstock et al., 2006).
We further suggest that strand-invasion events at the long repeats are initiation sites for recombination-dependent replication (RDR). RDR requires coordination of the DNA ends to ensure that reciprocal and symmetric events occur, thus replicating the complete genome (Stohr and Kreuzer, 2002; Shcherbakov et al., 2006). This process is not disrupted by mutations in MSH1 or RECA3, although it may be disrupted by mutations in the other RecA homologs, which would explain their lethality. We propose that RECA3 and MSH1 are components of a surveillance mechanism that directs conversion events between short repeats, while allowing RDR to be initiated at long repeats. Several steps in the pathway leading to full recombinant formation could be affected, including branch migration, second-strand capture, replication fork establishment, or Holliday junction resolution. If an aberrant event occurs and the asymmetric recombinant molecule replicates efficiently, the stoichiometry of various genes in the mitochondrial genome will change, often with profound effects on the plant. This is the SSS process.
By contrast, when recA3 or msh1 mutations render the recombination surveillance system nonfunctional, homeologous and asymmetric events occur, shifting the stoichiometry of various segments of the genome. We propose that the events reproducibly observed in the mutants resemble low frequency, sporadic events that occur in wild populations to effect substoichiometric shifting and CMS. Consistent with this hypothesis, we can detect both recombination products in mutant plants and even in wild-type plants by PCR (data not shown), but these molecules are rare, as indicated by their apparent absence on blots.
This model applies whether the genomic molecules are circular or linear. Strand invasion events that lead to RDR could result in theta replication, rolling circle replication, or more complex T4-like events, depending on the molecules involved and their topologies. Still not understood are the detailed mechanisms that result in differential replication or instability of some of the recombination products, evident as asymmetric recombination within the recA3 and msh1 mutants during de novo generation of these novel recombinant molecules.
A remaining question is why there are so few distinct recombination events in the mutants. We suggest that double-strand breakage hot spots likely exist in a few locations, which lead to gene conversion in the wild type and recombination in the mutants. Other aberrant events may occur sporadically at additional locations in the mutants, but if they are infrequent they would be lost by cytoplasmic sorting. Thus, only the most frequent aberrant events will be seen, unless large populations or many generations are examined. Alternatively, localized cruciforms in the DNA, localized melting due to high transcriptional activity, or stalling replication forks could account for the sites of DNA exchange, although any of these alternative events would likewise need to occur at high frequency at specific locations.
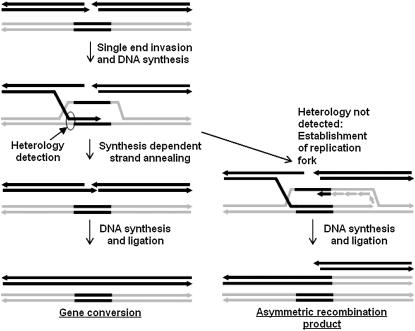
A model for the double-strand break–mediated events is diagrammed in Figure 8. Following strand invasion at a short repeat to establish a D loop, DNA synthesis will convert any mutations, thus maintaining the sequence identity of the short repeats. Branch migration will allow heterology to be detected, resulting in unwinding of the invading strand and resulting in synthesis-dependent strand annealing to complete gene conversion of the short repeat. On the other hand, if heterology is not detected, establishment of a replication fork will result in production of one of two possible reciprocal recombination products. Normally rare, this becomes frequent in the mutants. Why the same nonreciprocal recombinant is always produced is unknown. It may be due to asymmetry in the processing of the breakage site leading to the initial strand invasion event, asymmetry in resolution of the intermediates, or differential replication of the products. Some of these have been described in other systems (Villemure et al., 1997, 2003; Hunter and Kleckner, 2001).
Figure 8.
Model for Outcomes of Double-Strand Breaks at Short Repeats.
Two different molecules are indicated in black and red, with a segment of the black molecule repeated within the red. Invasion of the 3′ end to form a D loop, and extension by DNA polymerase leads to two possibilities. Rejection of the invading strand from the D loop and annealing to the other broken end will result in gene conversion. A replication fork could also form at the D loop, resulting in asymmetric recombination.
There are several possible fates for the broken end that has not invaded. It could be degraded, undergo NHEJ, or invade another DNA molecule (Stohr and Kreuzer, 2002; Shcherbakov et al., 2006). If it invades the asymmetric recombination product and initiates replication, regeneration of the parental molecule would occur. If it invades the recombinant molecule, the reciprocal recombinant would be produced. In the mutants, this evidently occurs at a low frequency, to account for the occasional appearance of the reciprocal recombination product in RECA3/recA3-1 backcross heterozygotes. One possible explanation is that the two broken ends are normally coordinated, leading to gene conversion at short repeats and establishment of two replication forks at long repeats to initiate RDR. If RECA3 and MSH1 are involved in selecting which pathway the intermediate follows, then disruption at either locus could lead to establishment of only one replication fork due to poor detection of heterologies, poor coordination of the ends, or both, resulting in the asymmetric recombination products seen in the mutants.
Our model predicts a hotspot for breakage events at atp9. It is interesting that the two defective copies of atp9 in the mitochondrial genome represent the 5′ and 3′ ends of the gene, with no overlap between the sequences (Figure 3A, molecules B and C). These are likely the result of breakage events near the middle of the gene, followed by NHEJ to produce the defective copies now fixed in the population. Recombination between other short repeats in the genome (including some very near the atp9 gene) has not been observed in msh1 or recA3 mutants, again suggesting a hot spot for initiation of strand invasion events in the atp9 gene.
The Acquisition of Aberrant Repeats in the Genome and the SSS Process They Mediate Likely Comprise Major Components of Plant Mitochondrial Genome Evolution
The evolutionary origin of recombinogenic, defective copies of mitochondrial genes such as atp9 or rpl16 is not known, but the differences in the complement of atp9 repeats present in the three Arabidopsis ecotypes Col-0, C24, and Ler (Forner et al., 2005), suggest relatively recent origins followed by homoplasmy. Variation in the presence of repeats that facilitate this type of illegitimate recombination in Arabidopsis suggests not only that the recombination activity associated with SSS can be acquired or lost but that we now have the opportunity to learn what physiological or developmental features are gained or lost by the plant in the process.
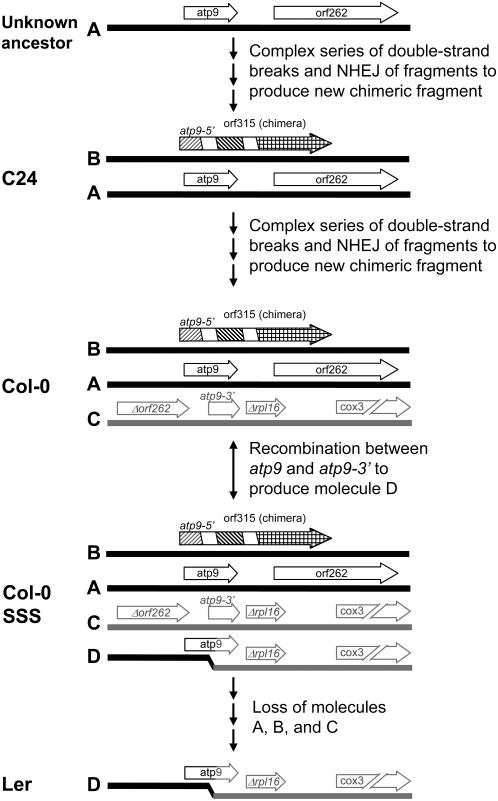
Using the atp9 and rpl16 repeats as indicators of mitochondrial genome evolution in Arabidopsis, one can hypothesize that double-strand breakage events leading to novel repeated sequences within the genome represents a key step in major mitochondrial genome transitions. Ecotype C24 represents a mitochondrial form prior to the introduction of such a repeat configuration (Figure 9), and Col-0 represents a mitochondrial type following synthesis and fixation of the novel atp9 chimera near cox3 (Forner et al., 2005). Low frequency illegitimate recombination at these repeats provides the dynamic of SSS that is evident in the Col-0 ecotype. However, it appears that the next evolutionary step in this transition is an irreversible loss of many of these molecules, which has apparently occurred within the Ler mitochondrial genome.
Figure 9.
Hypothetical Pathway Leading to Different Genomic Molecules in the Different Ecotypes.
At the top is indicated a hypothetical ancestral mitochondrial genome containing only molecule A (labels are the same as in Figures 3 and 4C) containing the atp9 gene and orf262. A series of complex breaking and rejoining events results in molecule B, containing the chimeric gene known as orf315. This is the configuration found in ecotype C24. A further series of complex rearrangements results in molecule C, the configuration found in Col-0. Recombination between molecules B and C occurs to produce molecule D, a configuration indicated as Col-0 SSS. This configuration may occur spontaneously or reproducibly in the recA3 and msh1 mutants. This configuration is also reversible. Molecule D now includes a fully functional copy of the atp9 gene, so loss of molecule A will not be lethal. Loss of molecules A, B, and C results in the configuration seen in ecotype Ler.
It should be feasible to test for evidence of such mitochondrial genome transitions within other plant species. While in the case of Arabidopsis, these genomic shifts do not appear to confer any obvious phenotypic changes to the plant, one presumes that in natural populations, where CMS may be induced or suppressed in the process, such genomic shifts could provide important adaptive advantages (Mackenzie, 2005). It is likely no coincidence that RNA interference suppression of MSH1 in two other plant species gives rise to a CMS phenotype together with mitochondrial rearrangement (Sandhu et al., 2007).
The plant growth phenotype of recA3-1 msh1-1 double mutants suggests that these two genes control different aspects of homology surveillance. The ectopic recombination events seen in the single mutants are distinct, and the double mutants seem to suffer both sets of ectopic recombination. This suggests that while mitochondrial genes may exist in a few different linkage relationships on different molecules in the single mutants, they apparently exist in a large number of linkage relationships, topologies, and stoichiometries in the double mutants. This genomic complexity could interfere with replication efficiency, perhaps accounting for the observed reduction in mitotic index and plant growth rate (Mandal et al., 2005; McBride et al., 2006). The biochemical details of RECA3 and MSH1 functions that lead to prevention of homeologous recombination are important future questions.
METHODS
Arabidopsis thaliana Growth
Seeds were plated on 0.5× Murashige and Skoog media and cold treated at 4°C for 5 d to synchronize germination. The seed plates were then transferred to a growth chamber with 16 h of daylight at 22°C. Three-week-old seedlings were transplanted to soil (Metro Mix 360) and kept in 8 h of daylight at 24°C until they started bolting, after which the plants were moved to 16 h of daylight at 24°C. Alternatively, cold-treated seeds were sown directly on soil (Metro Mix 360) and kept in growth chambers at 8 h of daylight at 24°C for 5 to 6 weeks and then transferred to a growth chamber with 16 h of daylight at 24°C. Antibiotic selection was performed by adding kanamycin at 50 μg/mL or glufosinate at 5 μg/mL to the germination plates.
Mutants
Two types of msh1 alleles have been identified. Point mutations have been previously referred to as chm1-1, chm1-2, and chm1-3, while T-DNA insertion mutations have also been isolated (Redei, 1973; Martinez-Zapater et al., 1992). We previously showed that the MSH1 gene is encoded at the CHM locus (Abdelnoor et al., 2003). The MSH1 gene is named by the standard convention of naming mutS homologs, so we propose renaming the alleles as follows. The point mutations previously called chm1-1, chm1-2, and chm1-3 are now known as msh1-1, msh1-2, and msh1-3, respectively. The T-DNA insertion obtained from the Salk Institute (Salk_041951) is now known as msh1-T1.
T-DNA insertions for all three recA homologs were also obtained from the Salk Institute: recA1-1 (Salk_013488), recA1-2 (Salk_072979), and recA1-3 (Salk_020949). For RECA2, the alleles obtained were recA2-1 (Salk_076237), recA2-2 (Salk_068655), and recA2-3 (Salk_118134). Two alleles of RECA3 were obtained: recA3-1 (Sail_252_C06) and recA3-2 (Salk_146388).
Molecular Biology Procedures
Total genomic DNA was extracted from mature plants (leaves, stems, and flowers) using the DNeasy Plant Mini Kit (Qiagen). One leaf DNA for PCR genotyping was prepared according to published procedures (Martinez-Zapater and Salinas, 1997). Total RNA was isolated using TRIzol reagent according to the manufacturer's protocol (Invitrogen) and purified using the RNeasy Mini Kit (Qiagen).
Total genomic DNA samples were digested with the restriction enzyme BamHI according to the manufacturer's instructions. Agarose gel electrophoresis, DNA transfer to Hybond N+ nylon membrane (0.45 μm; Amersham), extraction of DNA fragments, labeling of DNA, and filter hybridizations were conducted according to procedures described previously (Janska and Mackenzie, 1993). Autoradiography was performed by exposure to x-ray film (Kodak) with intensifying screens (Pickett) at −70°C.
Genotypes of msh1-1 plants were verified by the SNAP method as described (Drenkard et al., 2000), using the primers msh1-1WF (5′-TTAAAAGATGGAAACCTCAACTGGGAGATGTTAC-3′), msh1-1MF (5′-TTAAAAGATGGAAACCTCAACTGGGAGATGTTAT-3′) and msh1-1Rev (5′-TGTGAGTAAGCTTGAAATTCAAAAAGGATTGTGTAC-3′). Genotypes of recA3-1 plants were verified using PCR to detect the T-DNA insertion with primers 252LP (5′-GATTCCATTAGCCATGAACCGA-3′), 252RP (5′-TCTTTCCAGATGCTTCTTTTCCG-3′), and LB3 (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′). The 5′-atp9 probe was generated using primers atp9F1 (5′-CCCAAAATGGCATAACCAAATGATTG-3′) and atp9R1 (5′-AAAATCAATAGGTGCCGGAGCTGCT-3′), and the 3′-atp9 probe was generated using primers atp9F2 (5′-ATGCTTATAGCATTTATACTTTCGTGGTCGC-3′) and atp9R2 (5′-GAAGGTTTCCATTCAGTCTCATAAAGCAAG-3′).
RT-PCR
Total RNA was extracted as described above. Equal amounts of RNA from different tissues were first converted to cDNA and then used for quantitative RT-PCR using the SYBR GreenER Two-Step quantitative RT-PCR kit for the iCycler (Invitrogen). Quantitative RT-PCR data collection and analysis were done using the iCycler iQ software (version 3.1) from Bio-Rad. Each sample was run in triplicate and the results averaged; the entire experiment was repeated.
Primers RealRecA3F2 (5′-ATCTAACATGCATTTCCCGCACGC-3′) and RealRecA3R2 (5′-TGGACGCAGACATTGAGACCACTT-3′); RealMsh1F1 (5′-TCATGCGTGTATGTG ATGCGGAGA-3′) and RealMsh1R1 (5′-ACTTGACCCTTGCAGTCCTTCCTT-3′); and RealUbqF1 (5′-CACCATTGACAACGTCAAGGCCAA-3′) and RealUbqR1 (5′-CACGCAGACGCAAGACCAAATGAA-3′) were used for real-time analysis of RECA3, MSH1, and ubiquitin, respectively.
GFP Constructs
The binary vector pK7FWG2 (Karimi et al., 2002) was used for GFP fusion constructs. DNA fragments from the three recA homologs were amplified from genomic DNA, cloned into pENTR-D, and transferred into pK7FWG2 with LR clonase following the manufacturer's instructions (Invitrogen). Forward primers corresponding to the annotated AUG and reverse primers corresponding to codon 80 were as follows: RECA1 forward primer 5′-CACCATGGATTCACAGCTAGTCTTGTCT-3′ and reverse primer 5′-GCGGTCGAGGAAGCGGGAATCAGCGTC-3′; RECA2 forward primer 5′-CACCATGGCGAGGATTCTCCGAAACGTT-3′ and reverse primer 5′-ATACATTATGGACCCTTTGCCAAATGA-3′; and RECA3 forward primer 5′-CACCATGGGTCGACTCTCATGGGCC-3′ and reverse primer 5′-GTCTTTATCAAAATCACCTGAAAGCTG-3′.
Transient Transformation and GFP Expression Assay
In separate experiments, 7 μg of DNA from individual constructs was delivered into 4-week-old leaves of Arabidopsis thaliana (ecotype Col-0) using tungsten particles and the Biolistic PDS-1000/He system (Bio-Rad). Particles were bombarded into Arabidopsis leaves using 900 p.s.i. rupture discs under a vacuum of 26 inches of Hg. After bombardment, Arabidopsis leaves were allowed to recover for 18 to 22 h on Murashige and Skoog medium plates at 22°C in 16 h of light before assaying GFP expression. Localization of GFP expression was conducted by laser scanning confocal microscopy using 488- and 633-nm excitation and two-channel measurement of emission: 522 nm (green/GFP) and 680 nm (red/chlorophyll).
Histology
For mitotic index determination, roots were collected from germinating seedlings and fixed in 2.5% glutaraldehyde in 0.1 M potassium phosphate buffer, pH 7.0, for 2 h. The roots were washed three times with the same buffer followed by postfixation in 1% OsO4 at 4°C overnight. After extensive washing in buffer, roots were dehydrated through a graded ethanol series and embedded individually in Spurr's epoxy resin (Electron Microscopy Science). Serial longitudinal sections were cut with an ultramicrotome at 1-μm thickness. Sections were stained with 0.5% (w/v) toluidine blue and counterstained with 0.05% (w/v) basic fuchsin. Two median longitudinal sections per root and two roots per group were examined. Mitotic profiles were recorded for root apical meristem within 80 μm, excluding root cap. Cell counting and photography were done with an Olympus AX70 equipped with an Optronics digital camera and MagnaFire 2.1 software (Optronics). Statistical analysis was performed using SPSS 12.
Bioinformatics
The multiple sequence alignment of the RecA proteins was done using Invitrogen's Vector NTI software. The ClustalW algorithm was used for generating the alignment.
Accession Numbers
Arabidopsis Genome Initiative (AGI) locus identifiers for the genes mentioned in this article are MSH1, At3g24320, The Arabidopsis Information Resource (TAIR) accession 2087193; and RECA3, AGI locus At3g10140, TAIR accession 2100038.
Acknowledgments
We thank the ABRC, the Salk Institute, Syngenta, and the Nottingham Arabidopsis Stock Centre for providing mutant seeds. We also thank Guichuan Hou and the Center for Biotechnology Microscopy Core Facility for histology and confocal microscopy services. Tom Clemente generously provided raillery. This work was supported in part by National Science Foundation Grant IOB-0417172 to S.A.M. and A.C.C. and National Science Foundation Grant MCB-0323377 to S.A.M.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sally A. Mackenzie (smackenzie2@unl.edu).
Open Access articles can be viewed online without a subscription.
References
- Abad, A.R., Mehrtens, B.J., and Mackenzie, S.A. (1995). Specific expression in reproductive tissues and fate of a mitochondrial sterility-associated protein in cytoplasmic male-sterile bean. Plant Cell 7 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelnoor, R., Christensen, A., Mohammed, S., Munoz-Castillo, B., Moriyama, H., and Mackenzie, S. (2006). Mitochondrial genome dynamics in plants and animals: Convergent gene fusions of a MutS homologue. J. Mol. Evol. 63 165–173. [DOI] [PubMed] [Google Scholar]
- Abdelnoor, R.V., Yule, R., Elo, A., Christensen, A.C., Meyer-Gauen, G., and Mackenzie, S.A. (2003). Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA 100 5968–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K., and Palmer, J. (2003). Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 29 380–395. [DOI] [PubMed] [Google Scholar]
- Aguilera, A. (2001). Double-strand break repair: Are Rad51/RecA–DNA joints barriers to DNA replication? Trends Genet. 17 318–321. [DOI] [PubMed] [Google Scholar]
- Backert, S., and Borner, T. (2000). Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 37 304–314. [DOI] [PubMed] [Google Scholar]
- Birky, C.W., Jr. (1983). The partitioning of cytoplasmic organelles at cell division. Int. Rev. Cytol. Suppl. 15 49–89. [DOI] [PubMed] [Google Scholar]
- Bleuyard, J.-Y., Gallego, M., and White, C. (2006). Recent advances in understanding of the DNA double-strand break repair machinery of plants. DNA Repair (Amst.) 5 1–12. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D. (2002). What maintains male-sterility factors in plant populations? Heredity 89 408–409. [DOI] [PubMed] [Google Scholar]
- Darwin, C.R. (1877). The Different Forms of Flowers on Plants of the Same Species. (London: John Murray).
- Dieterich, J.-H., Braun, H.-P., and Schmitz, U. (2003). Alloplasmic male sterility in Brassica napus (CMS ‘Tournefortii-Stiewe’) is associated with a special gene arrangement around a novel atp9 gene. Mol. Genet. Genomics 269 723–731. [DOI] [PubMed] [Google Scholar]
- Drenkard, E., Richter, B.G., Rozen, S., Stutius, L.M., Angell, N.A., Mindrinos, M., Cho, R.J., Oefner, P.J., Davis, R.W., and Ausubel, F.M. (2000). A simple procedure for the analysis of single nucleotide polymorphisms facilitates map-based cloning in Arabidopsis. Plant Physiol. 124 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farbos, I., Mouras, A., Bereterbide, A., and Glimelius, K. (2001). Defective cell proliferation in the floral meristem of alloplasmic plants of Nicotiana tabacum leads to abnormal floral organ development and male sterility. Plant J. 26 131–142. [DOI] [PubMed] [Google Scholar]
- Fauron, C., Casper, M., Gao, Y., and Moore, B. (1995). The maize mitochondrial genome: Dynamic, yet functional. Trends Genet. 11 228–235. [DOI] [PubMed] [Google Scholar]
- Forner, J., Weber, B., Wietholter, C., Meyer, R.C., and Binder, S. (2005). Distant sequences determine 5′ end formation of cox3 transcripts in Arabidopsis thaliana ecotype C24. Nucleic Acids Res. 33 4673–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb, T., and Alani, E. (2005). Distinct roles for the Saccharomyces cerevisiae mismatch repair proteins in heteroduplex rejection, mismatch repair and nonhomologous tail removal. Genetics 169 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, M., and Bentolila, S. (2004). Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16 (suppl.): S154–S169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, E., Eriksson, E., Herbert, B., and Borts, R. (2005). MLH1 and MSH2 promote the symmetry of double-strand break repair events at the HIS4 hotspot of Saccharomyces cerevisiae. Genetics 169 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, N., and Kleckner, N. (2001). The single-end invasion: An asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106 59–70. [DOI] [PubMed] [Google Scholar]
- Inbar, O., Liefshitz, B., Bitan, G., and Kupiec, M. (2000). The relationship between homology length and crossing over during the repair of a broken chromosome. J. Biol. Chem. 275 30833–30838. [DOI] [PubMed] [Google Scholar]
- Janska, H., and Mackenzie, S.A. (1993). Unusual mitochondrial genome organization in cytoplasmic male sterile common bean and the nature of cytoplasmic reversion to fertility. Genetics 135 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns, C., Lu, M., Lyznik, A., and Mackenzie, S. (1992). A mitochondrial DNA sequence is associated with abnormal pollen development in cytoplasmic male sterile bean plants. Plant Cell 4 435–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa, A., Tsutsumi, N., and Hirai, A. (1994). Reversible changes in the composition of the population of mtDNAs during dedifferentiation and regeneration in tobacco. Genetics 138 865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kaul, M.L.H. (1988). Male Sterility in Higher Plants. (Berlin: Springer).
- Khazi, F.R., Edmondson, A.C., and Nielsen, B.L. (2003). An Arabidopsis homologue of bacterial RecA that complements an E. coli recA deletion is targeted to plant mitochondria. Mol. Gen. Genet. 269 454–463. [DOI] [PubMed] [Google Scholar]
- Knoop, V. (2004). The mitochondrial DNA of land plants: Peculiarities in phylogenetic perspective. Curr. Genet. 46 123–139. [DOI] [PubMed] [Google Scholar]
- Kreuzer, K. (2005). Interplay between DNA replication and recombination in prokaryotes. Annu. Rev. Microbiol. 59 43–67. [DOI] [PubMed] [Google Scholar]
- Kubo, T., Nishizawa, S., Sugawara, A., Itchoda, N., Estiati, A., and Mikami, T. (2000). The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for rRNA(Cys)GCA. Nucleic Acids Res. 28 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, B., Gray, M., and Burger, G. (1999). Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 33 351–397. [DOI] [PubMed] [Google Scholar]
- Lee, S.J., Earle, E.D., and Gracen, V.E. (1980). The cytology of pollen abortion in S cytoplasmic male-sterile corn anthers. Am. J. Bot. 67 237–245. [Google Scholar]
- Levings, C.S., III (1993). Thoughts on cytoplasmic male sterility in cms-T maize. Plant Cell 5 1285–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., McVety, S., Younan, R., Liang, P., Du Sart, D., Gordon, P.H., Hutter, P., Hogervorst, F.B.L., Chong, G., and Foulkes, W.D. (2006). Distinct patterns of germ-line deletions in MLH1 and MSH2: The implication of Alu repetitive element in the genetic etiology of Lynch syndrome (HNPCC). Hum. Mutat. 27 388. [DOI] [PubMed] [Google Scholar]
- Linke, B., Nothnagel, T., and Borner, T. (2003). Flower development in carrot CMS plants: Mitochondria affect the expression of MADS box genes homologous to GLOBOSA and DEFICIENS. Plant J. 34 27–37. [DOI] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta]CT method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Mackenzie, S., and Chase, C. (1990). Fertility restoration is associated with loss of a portion of the mitochondrial genome in cytoplasmic male-sterile common bean. Plant Cell 2 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, S.A. (2005). The mitochondrial genome of higher plants: A target for natural adaptation. In Diversity and Evolution of Plants, R.J. Henry, ed (Oxon, UK: CABI Publishers), pp. 69–80.
- Mandal, S., Guptan, P., Owusu-Ansah, E., and Banerjee, U. (2005). Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell 9 843–854. [DOI] [PubMed] [Google Scholar]
- Marienfeld, J.R., and Newton, K.J. (1994). The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 mitochondrial gene and is associated with reduced complex I function. Genetics 138 855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Zapater, J.M., Gil, P., Capel, J., and Somerville, C.R. (1992). Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell 4 889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Zapater, J.M., and Salinas, J. (1997). Arabidopsis Protocols. (Totowa, NJ: Humana Press).
- McBride, H.M., Neuspiel, M., and Wasiak, S. (2006). Mitochondria: More than just a powerhouse. Curr. Biol. 16 R551–R560. [DOI] [PubMed] [Google Scholar]
- McGrew, D., and Knight, K. (2003). Molecular design and functional organization of the RecA protein. Crit. Rev. Biochem. Mol. Biol. 38 385–432. [DOI] [PubMed] [Google Scholar]
- Mieczkowski, P.A., Lemoine, F.J., and Petes, T.D. (2006). Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst.) 5 1010–1020. [DOI] [PubMed] [Google Scholar]
- Newton, K.J., Mariano, J.M., Gibson, C.M., Kuzmin, E.V., and Gabay-Laughnan, S. (1998). Involvement of S2 episomal sequences in the generation of NCS4 deletion mutation in maize mitochondria. Dev. Genet. 19 277–286. [DOI] [PubMed] [Google Scholar]
- Nicholson, A., Hendrix, M., Jinks-Robertson, S., and Crouse, G.F. (2000). Regulation of mitotic homeologous recombination in yeast: Functions of mismatch repair and nucleotide excision repair genes. Genetics 154 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notsu, Y., Masood, S., Nishikawa, T., Kubo, N., Akiduki, G., Nakazono, M., Hirai, A., and Kadowaki, K. (2002). The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: Frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics 268 434–445. [DOI] [PubMed] [Google Scholar]
- Ogihara, Y., et al. (2005). Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 33 6235–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, C., Flores, C., and Engels, W. (2006). Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics 172 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redei, G.P. (1973). Extra-chromosomal mutability determined by a nuclear gene locus in Arabidopsis. Mutat. Res. 18 149–162. [Google Scholar]
- Sakamoto, W., Kondo, H., Murata, M., and Motoyoshi, F. (1996). Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell 8 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu, A., Abdelnoor, R., and Mackenzie, S. (2007). Transgenic induction of mitochondrial rearrangements for cytoplasmic male sterility in crop plants. Proc. Natl. Acad. Sci. USA 104 1766–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable, P., and Wise, R. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3 175–180. [Google Scholar]
- Shcherbakov, V.P., Plugina, L., Shcherbakova, T., Sizova, S., and Kudryashova, E. (2006). Double-strand break repair in bacteriophage T4: Coordination of DNA ends and effects of mutations in recombinational genes. DNA Repair (Amst.) 5 773–787. [DOI] [PubMed] [Google Scholar]
- Sia, E., and Kirkpatrick, D. (2005). The yeast MSH1 gene is not involved in DNA repair or recombination during meiosis. DNA Repair (Amst.) 4 253–261. [DOI] [PubMed] [Google Scholar]
- Small, I., Isaac, P., and Leaver, C. (1987). Stoichiometric differences in DNA molecules containing the atpA gene suggest mechanisms for the generation of mitochondrial genome diversity in maize. EMBO J. 6 865–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden, T., Acharya, S., Butz, C., Bernardini, M., and Fishel, R. (2004). hMSH3-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15 437–451. [DOI] [PubMed] [Google Scholar]
- Stohr, B., and Kreuzer, K. (2002). Coordination of ends during double-strand-break repair in bacteriophage T4. Genetics 162 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, Y., Watase, Y., Nagase, M., Makita, N., Yagura, S., Hirai, A., and Sugiura, M. (2005). The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: Comparative analysis of mitochondrial genomes in higher plants. Mol. Genet. Genomics 272 603–615. [DOI] [PubMed] [Google Scholar]
- Unseld, M., Marienfeld, J.R., Brandt, P., and Brennicke, A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15 57–61. [DOI] [PubMed] [Google Scholar]
- Villemure, J.-F., Abaji, C., Cousineau, I., and Belmaaza, A. (2003). MSH-2-deficient human cells exhibit a defect in the accurate termination of homology-directed repair of DNA double-strand breaks. Cancer Res. 63 3334–3339. [PubMed] [Google Scholar]
- Villemure, J.-F., Belmaaza, A., and Chartrand, P. (1997). The processing of DNA ends at double-strand breaks during homologous recombination: Different roles for the two ends. Mol. Gen. Genet. 256 533–538. [DOI] [PubMed] [Google Scholar]
- Weinstock, D.M., Richardson, C.A., Elliott, B., and Jasin, M. (2006). Modeling oncogenic translocations: Distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst.) 5 1065–1074. [DOI] [PubMed] [Google Scholar]
- Yang, D., Goldsmith, E.B., Lin, Y., Waldman, B.C., Kaza, V., and Waldman, A.S. (2006). Genetic exchange between homeologous sequences in mammalian chromosomes is averted by local homology requirements for initiation and resolution of recombination. Genetics 174 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]