Abstract
The aim of this study was to investigate the relationship between the phosphorylation and activation states of phosphoenolpyruvate carboxykinase (PEPCK) and to investigate how the phosphorylation states of PEPCK and phosphoenolpyruvate carboxylase (PEPC) are coordinated in response to light intensity and CO2 concentration during photosynthesis in leaves of the C4 plant Guinea grass (Panicum maximum). There was a linear, reciprocal relationship between the phosphorylation state of PEPCK and its activation state, determined in a selective assay that distinguishes phosphorylated from nonphosphorylated forms of the enzyme. At high photon flux density and high CO2 (750 μL L−1), PEPC was maximally phosphorylated and PEPCK maximally dephosphorylated within 1 h of illumination. The phosphorylation state of both enzymes did not saturate until high light intensities (about 1,400 μmol quanta m−2 s−1) were reached. After illumination at lower light intensities and CO2 concentrations, the overall change in phosphorylation state was smaller and it took longer for the change in phosphorylation state to occur. Phosphorylation states of PEPC and PEPCK showed a strikingly similar, but inverse, pattern in relation to changes in light and CO2. The protein phosphatase inhibitor, okadaic acid, promoted the phosphorylation of both enzymes. The protein synthesis inhibitor, cycloheximide, blocked dark phosphorylation of PEPCK. The data show that PEPC and PEPCK phosphorylation states are closely coordinated in vivo, despite being located in the mesophyll and bundle sheath cells, respectively.
Plants with C4 photosynthesis elevate the CO2 concentration in the vicinity of Rubisco, thereby suppressing the oxygenase activity of Rubisco and photorespiration. The typical C4 process utilizes two leaf cell types. In the mesophyll cells, inorganic carbon is fixed by phosphoenolpyruvate carboxylase (PEPC) and converted to the C4 acids, malate and Asp. In the bundle sheath cells, these C4 acids are decarboxylated, releasing CO2 for fixation by Rubisco. A C3 compound is then returned to the mesophyll to complete the C4 cycle. Although PEPC acts as the carboxylase in the mesophyll of all C4 plants, the decarboxylases vary in different biochemical subgroups of C4 plants, which contain either NADP-malic enzyme, sometimes in tandem with phosphoenolpyruvate carboxykinase (PEPCK; Wingler et al., 1999), NAD-malic enzyme by itself, or NAD-malic enzyme in combination with PEPCK, as in Guinea grass (Panicum maximum; Hatch, 1987; Burnell and Hatch, 1988). These carboxylation and decarboxylation reactions must be regulated to accommodate the large and rapid changes in flux that occur during photosynthesis and must be coordinated if the C4 cycle is to operate efficiently (Leegood et al., 1996).
A number of factors contribute to the regulation of PEPC in plants. First, the enzyme is allosterically regulated by inhibitors such as malate and by activators such as sugar phosphates (Chollet et al., 1996). Second, the effects of metabolites are modulated by phosphorylation of the enzyme. Reversible, light-dependent phosphorylation of PEPC occurs in C4 and other plants, which leads to activation by decreased sensitivity to inhibition by malate and increased sensitivity to activation by Glc-6-P (Chollet et al., 1996). The activity of the kinase that mediates the phosphorylation of PEPC appears to be regulated solely by transcriptional control and protein turnover (Nimmo, 2003; Izui et al., 2004).
PEPCK is also subject to reversible phosphorylation in the leaves of some C4 plants, such as Guinea grass, although not in leaves of others, such as Urochloa panicoides (Walker et al., 1997). In Guinea grass, PEPCK is phosphorylated in darkened leaves and dephosphorylated in illuminated leaves (Walker and Leegood, 1996). Changes in PEPCK phosphorylation state lead to diurnal changes in its sensitivity to regulation by adenylates, which are likely to lead to its activation in illuminated leaves (Walker et al., 2002). Other than diurnal measurements of its activation state (Walker et al., 2002; Leegood and Walker, 2003), it is not known how the activation state of PEPCK is related to changes in the phosphorylation state or how changes in CO2 concentration and light intensity influence the activation state of PEPCK in the leaves of C4 plants.
A major deficiency in our understanding of the regulation of C4 photosynthesis is how carboxylation processes in the mesophyll and decarboxylation processes in the bundle sheath are coordinated. PEPC and PEPCK are both cytoplasmic enzymes that are active in the light. In some C4 plants, such as Guinea grass, light activation of PEPC occurs by phosphorylation and light activation of PEPCK occurs by dephosphorylation. The aim of this study was to investigate whether the phosphorylation states of PEPC and PEPCK are coordinated in response to light intensity and CO2 concentration during photosynthesis in leaves of Guinea grass.
RESULTS
Sequence Analysis of Guinea Grass PEPCK
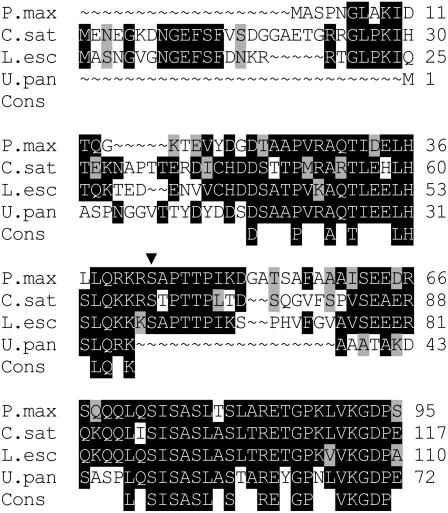
PEPCK cDNA from Guinea grass was sequenced (GenBank accession no. AAQ10076). The molecular mass predicted from the amino acid sequence is 70,682 D. Figure 1 shows a comparison of the derived N-terminal regulatory sequence with those of U. panicoides (a C4 grass in which PEPCK is not phosphorylated [Walker et al., 1997]) and of cucumber (Cucumis sativus) and tomato (Lycopersicon esculentum), in which PEPCK is known to be phosphorylated. Like the cucumber and tomato enzymes, PEPCK from Guinea grass contains a similar sequence (RKRS, residues 40–43), including a putative target Ser (indicated by the arrow) that is recognized by plant Ca2+-dependent protein kinase and mammalian cAMP-dependent protein kinases, but absent from PEPCK from U. panicoides (Leegood and Walker, 2003).
Figure 1.
N-terminal deduced amino acid sequence comparison and consensus sequence of PEPCK from Guinea grass, U. panicoides (GenBank accession no. S52988), tomato (AAG01894), and cucumber (S52637). Black shading indicates identical residues; gray shading indicates similarity between residues. The putative phosphorylated Ser is indicated by ▾.
Effect of Light and CO2 on Phosphorylation State of PEPC and PEPCK
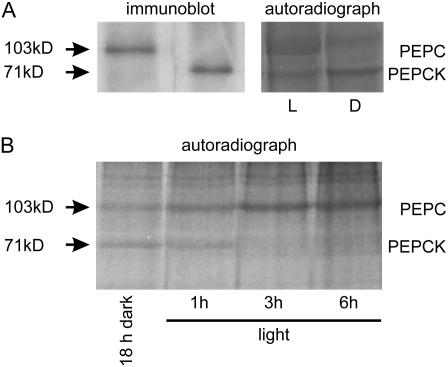
The phosphorylation states of PEPC and PEPCK were measured by illuminating leaves supplied with [33P] inorganic phosphate (Pi). PEPC (103 kD) and PEPCK (71 kD) proteins were identified on gels by immunoblotting, which were then compared with autoradiographs (Fig. 2A). There were reciprocal changes in the phosphorylation of PEPC, which increased during illumination, and the phosphorylation of PEPCK, which decreased during illumination (Fig. 2, A and B). The bands on the autoradiographs were then quantified by densitometry.
Figure 2.
Autoradiographs showing changes in the phosphorylation states of PEPC and PEPCK between light (1 h at a PFD of 400 μmol quanta m−2 s−1 in air) and dark (18 h; A) and during illumination after a period of 18-h darkness (B). Leaves were supplied with [33P] Pi and illuminated for up to 6 h (at a PFD of 400 μmol quanta m−2 s−1) in air. Extracts were subjected to SDS-PAGE. The phosphorylated bands for PEPC and PEPCK were identified by comparison with immunoblots using specific antisera (A).
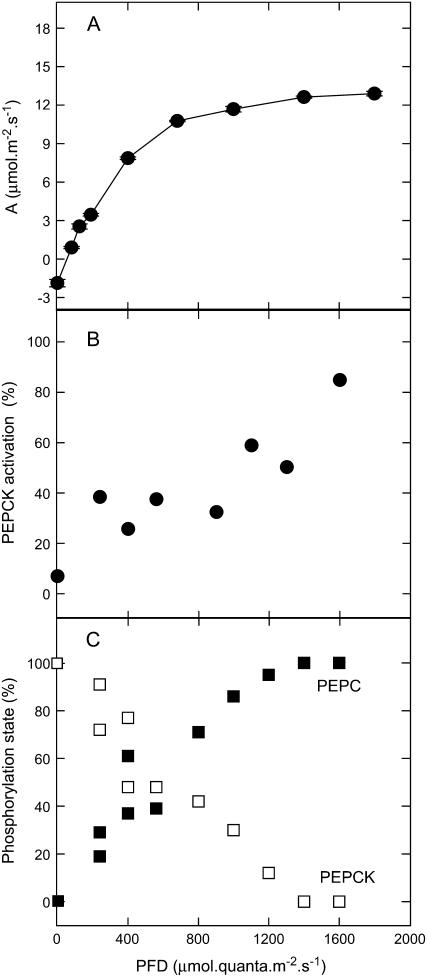
The relationship between the phosphorylation states of PEPC and PEPCK and the rate of photosynthesis in relation to light intensity is shown in Figure 3. Phosphorylation of PEPC and PEPCK showed a similar, but inverse, pattern (Fig. 3C). Like the rate of CO2 assimilation (Fig. 3A), the phosphorylation state of both enzymes did not saturate until high photon flux densities (PFDs; about 1,400 μmol quanta m−2 s−1) were reached. The relative activity (activation state) of PEPCK was determined in the carboxylation direction, but at a low concentration of PEP and by adding ATP, in addition to ADP, which distinguishes phosphorylated from nonphosphorylated forms of the enzyme (Walker et al., 2002). There was a linear, reciprocal relationship between the phosphorylation state and the activation state of PEPCK (Fig. 3B).
Figure 3.
Relationship between the CO2 assimilation rate (A), the activation state of PEPCK (B), and the phosphorylation states of PEPCK (□) and of PEPC (▪; C) during illumination of Guinea grass leaves. For measurements of phosphorylation state, leaves were preincubated with [33P] Pi, as described in “Materials and Methods.” Samples were taken, leaves darkened for 18 h, then illuminated at various light intensities for 2 h. The activation state of PEPCK was measured using a selective assay, as described in “Materials and Methods.”
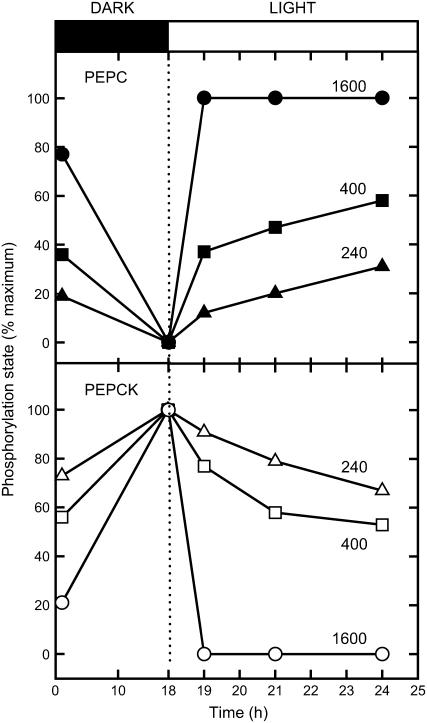
The effects of illumination at three different light intensities on the phosphorylation states of PEPC and PEPCK are shown in Figure 4. During the preceding dark pretreatment, PEPC was dephosphorylated and PEPCK was phosphorylated. The phosphorylation state at the end of the dark period was taken as the reference point for determination of the relative phosphorylation state. At high PFD, PEPC was maximally phosphorylated and PEPCK maximally dephosphorylated within 1 h of illumination. At lower PFDs, the overall change in phosphorylation state was smaller and it took longer for the change in phosphorylation state to occur.
Figure 4.
Responses of the phosphorylation states of PEPC and PEPCK to changes in PFD during illumination of Guinea grass leaves. Leaves were preincubated with [33P] Pi as described in “Materials and Methods.” Samples were taken, leaves darkened for 18 h, then illuminated at the light intensities indicated (μmol quanta m−2 s−1) for up to a further 6 h.
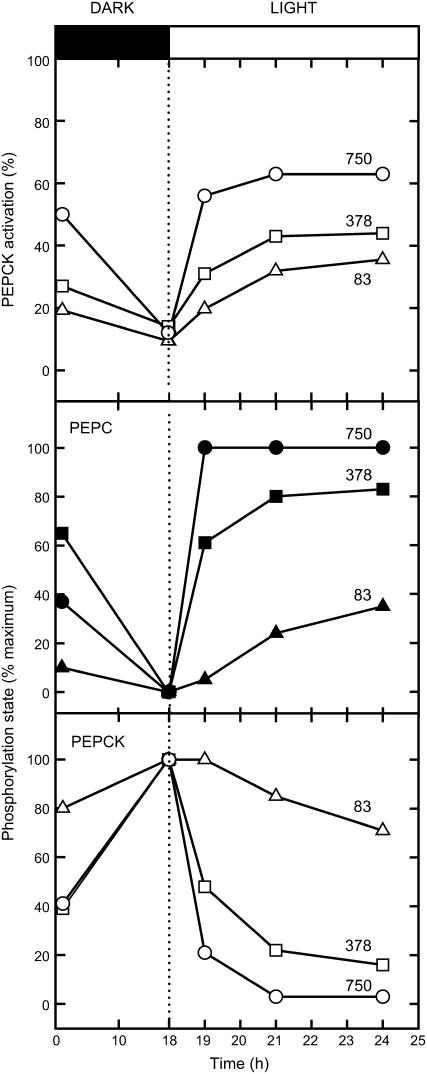
The effects of illumination at three CO2 concentrations on the phosphorylation states of PEPC and PEPCK are shown in Figure 5. As for Figure 4, PEPC was dephosphorylated and PEPCK was phosphorylated during the preceding dark pretreatment. At a high CO2 concentration (750 μL L−1, approximately twice ambient CO2 concentration), PEPC was maximally phosphorylated within 1 h of illumination and PEPCK maximally dephosphorylated within 3 h. At ambient (378 ppm) and below ambient (83 ppm) CO2, the overall change in phosphorylation state of both enzymes was smaller and it took longer for the change in phosphorylation state to occur. After illumination at different CO2 concentrations for differing periods, the PEPCK activation state showed a similar inverse relationship to the phosphorylation state as seen in Figure 3.
Figure 5.
Responses of the phosphorylation states of PEPC and PEPCK and the activation state of PEPCK to changes in CO2 concentration during illumination of Guinea grass leaves. Leaves were preincubated with [33P] Pi as described in “Materials and Methods.” Samples were taken, leaves darkened for 18 h, then illuminated at the CO2 concentrations indicated (in μL L−1) for up to a further 6 h. The PFD was 400 μmol quanta m−2 s−1. The activation state of PEPCK was measured using a selective assay, as described in “Materials and Methods.”
Influence of Dithiothreitol and Inhibitors on Phosphorylation and Activation States of PEPC and PEPCK
The effects of dithiothreitol (DTT) and inhibitors on the phosphorylation state and activation states of PEPC and PEPCK were investigated by feeding them directly to the cut ends of leaves via the transpiration stream (Table I). This means that they were supplied directly to the bundle-sheath cells from the xylem. Access to the mesophyll cells would then occur after diffusion through the bundle-sheath cells. The phosphorylation state is expressed as a percentage of the maximum in the water control in the dark for PEPCK or in the light for PEPC. The activation state of PEPC was measured by comparing its activity in the presence and absence of 1 mm malate, which inhibits the dephosphorylated, dark form of the enzyme to a greater degree (Echevarria et al., 1994).
Table I.
Effects of DTT and inhibitors on the phosphorylation and activation states of PEPC and PEPCK in intact leaves of Guinea grass
For measurements of the phosphorylation state, leaves were preincubated with [33P] Pi, as described in “Materials and Methods.” Samples were taken, leaves darkened for 18 h, and then supplied with each chemical. Activation states were determined using spectrophotometric assays, as described in “Materials and Methods.” Data are means ± se of three samples. The phosphorylation state for the control (supplied only with water) is shown as 100% in the light for PEPC and 100% in the dark for PEPCK. Activation state is shown as a percentage of the maximal enzyme activity measured. *, P is significantly different (at 5% level) when compared to the control.
| Treatment | Phosphorylation State
|
Activation State
|
||||||
|---|---|---|---|---|---|---|---|---|
| PEPCK
|
PEPC
|
PEPCK
|
PEPC
|
|||||
| Light 2 h | Dark 2 h | Light 2 h | Dark 2 h | Light 2 h | Dark 2 h | Light 2 h | Dark 2 h | |
| % water control | % maximum | |||||||
| Control | 31 ± 11 | 100 ± 0 | 100 ± 0 | 0 | 62 ± 9 | 27 ± 7 | 71 ± 2 | 6 ± 4 |
| 10 mm DTT | 26 ± 7 | 71 ± 26 | 29 ± 7* | 0 | 77 ± 10 | 40 ± 4 | 38 ± 4* | 4 ± 1 |
| 250 μm cycloheximide | 116 ± 55 | 15 ± 5* | 17 ± 1* | 7 ± 1 | 44 ± 6 | 66 ± 11* | 12 ± 5* | 14 ± 2 |
| 50 nm okadaic acid | 165 ± 16* | 171 ± 21* | 99 ± 2 | 60 ± 16* | 16 ± 2* | 16 ± 0 | 88 ± 3* | 48 ± 4* |
Okadaic acid, which inhibits the activity of protein phosphatase1 (PP1) and PP2A (Cohen et al., 1990), significantly increased phosphorylation of PEPCK above the water control in both light and dark and thereby decreased the activation state of PEPCK. Okadaic acid significantly increased phosphorylation of PEPC in the dark, thereby increasing its activation state. There was also a marginal increase in the PEPC activation state in the light, but this was not accompanied by an increase in the phosphorylation state. Feeding an inhibitor of protein synthesis, cycloheximide, significantly decreased phosphorylation and the activation state of PEPC in the light and decreased phosphorylation and increased the activation state of PEPCK in the dark. Feeding DTT significantly decreased the phosphorylation and activation states of PEPC in the light (Table I).
DISCUSSION
Phosphorylation of PEPCK is a regulatory mechanism that contributes to its inactivation in the dark in certain C4 and Crassulacean acid metabolism plants (Leegood and Walker, 2003). The data in Figures 3 and 5 establish that the phosphorylation and activation states of PEPCK are strongly inversely related and that the activation state of PEPCK can be used as a measure of the phosphorylation state in vivo. It is also clear from a comparison of the data in Table I that there is a positive correlation between the activation and phosphorylation states of PEPC.
These results show that regulation of PEPC and PEPCK by phosphorylation in vivo is a factor determining their activity that can operate over a wide range of CO2 concentrations and light intensities up to full sunlight. There is strong correlation between phosphorylation/activation and the rate of photosynthetic CO2 fixation. Phosphorylation of these two enzymes is not, therefore, merely acting as an on-off switch between light and dark. The response of phosphorylation to changes in light intensity or CO2 concentration takes from minutes to hours and is therefore likely to complement regulation of the activity of these enzymes by metabolites. Moreover, the rapidity of the response depends upon the magnitude of the change in light intensity or CO2 concentration. Previous measurements of the change in malate sensitivity of PEPC as a function of light intensity in another C4 plant, sorghum, are similar to those in Figure 4, with both the amplitude and initial velocity of activation varying with incident light (Bakrim et al., 1992). Somewhat surprisingly, light and CO2 response curves have not been measured for many enzymes of C4 photosynthesis or simultaneously for multiple enzymes, although previous measurements of the kinetics of activation of PEPC in the C4 plants maize (Zea mays), sorghum, and Salsola soda suggest similar kinetics of light activation and dark inactivation, with each process taking 1 to 2 h (Karabourniotis et al., 1983; Nimmo et al., 1987; Jiao and Chollet, 1988; Bakrim et al., 1992). This slow activation of PEPC and PEPCK by light compares with the very short times (5–10 min) required to fully activate NADP-malate dehydrogenase via thioredoxin (Johnson and Hatch, 1970) or pyruvate, Pi dikinase, by phosphorylation (Burnell and Hatch, 1985) in maize leaves or comparably short times to activate thioredoxin-linked enzymes and Rubisco in the leaves of C3 plants (e.g. Sassenrath-Cole and Pearcy, 1994).
DTT and inhibitors were fed to leaves to ascertain how these affected the phosphorylation or activation states of PEPC and PEPCK. The effect of DTT was examined because sulfhydryl groups may play a role in the regulation of PEPCK in vitro (Walker et al., 1997) and of PEPC in isolated C4 mesophyll cells (Pierre et al., 2004). Feeding DTT to leaves had no significant effect on PEPCK phosphorylation, but it did significantly decrease both the phosphorylation state and the activation state of PEPC. Although it has been suggested that PEPC kinase from maize and Flaveria trinervia may be redox activated by thioredoxin (Saze et al., 2001; Tsuchida et al., 2001), this would be expected to lead to activation of PEPC rather than the observed inactivation.
Cycloheximide and okadaic acid both had a profound effect on the phosphorylation of PEPC and PEPCK. Okadaic acid is an inhibitor of PP2A, which dephosphorylates PEPC and PEPCK (Walker and Leegood, 1995; Nimmo, 2003). Treatment with this inhibitor therefore promoted the phosphorylation of both enzymes. Cycloheximide is an inhibitor of protein synthesis, which prevents the synthesis of PEPC kinase, which phosphorylates PEPC (Jiao et al., 1991; Bakrim et al., 1992). Accordingly, it blocked the activation of PEPC. A similar strong inhibition of the dark phosphorylation of PEPCK by cycloheximide suggests that the PEPCK kinase might also be regulated by protein turnover.
Overall, the data show that there was a tight reciprocal relationship between the phosphorylation states of PEPC and PEPCK in relation to both light intensity and CO2 concentration. Light saturation curves for the phosphorylation state of both enzymes were similar, as was the extent of phosphorylation/dephosphorylation at different CO2 concentrations. These data suggest that the factors that regulate the phosphorylation of both enzymes are similar. PEPC kinase is the smallest known protein kinase with no regulatory domains, but is under exquisite transcriptional control and is also controlled by protein turnover (Nimmo, 2003). There is also little evidence to suggest that PEPC phosphatase activity is regulated by metabolites (Nimmo, 2003). However, there is evidence from in vitro studies that metabolites, such as malate and Glc-6-P, can affect the susceptibility of PEPC to phosphorylation (Bakrim et al., 1992; Wang and Chollet, 1993; Echevarria et al., 1994). Coregulation of phosphorylation of PEPC and dephosphorylation of PEPCK would therefore have to occur either at this level or at the level of transcription of the respective kinases and/or phosphatases. Previous studies have implied that the process of PEPC phosphorylation is coordinated with Calvin cycle activity. It has been suggested that such coordination could occur either through changes in metabolites that accompany changes in the rate of C4 photosynthesis or through changes in energy charge (Bakrim et al., 1992; Jiao and Chollet, 1992; Giglioli-Guivarc'h et al., 1996). Giglioli-Guivarc'h et al. (1996) suggested that glycerate-3-P from the bundle sheath might act as a message that triggers cytosolic acidification in the mesophyll cells, which leads to a signaling cascade that activates PEPC kinase (see also Bakrim et al., 2001; Osuna et al., 2004). However, evidence contrary to the involvement of the Calvin cycle comes from studies of a maize mutant that lacks a functional Calvin cycle, but nevertheless retains effective light regulation of PEPC and its kinase (Smith et al., 1998).
We attempted to investigate the regulation, by metabolites, of the phosphorylation status of PEPCK in bundle-sheath strands isolated by the method of Hatch and Kagawa (1976). However, in these, PEPCK was readily proteolytically cleaved, which removes the regulatory N-terminal phosphorylation site (Walker et al., 1997) and therefore precludes measurements of its phosphorylation state. Rapid proteolysis of PEPCK in crude extracts and the inability of protease inhibitors to prevent it (Walker et al., 1997) also render measurement of PEPCK kinase activity extremely difficult. We also fed metabolites to leaves. The major influences on the phosphorylation/activation states of both enzymes were the C3 metabolites, dihydroxyacetone-P, glycerate-3-P, and PEP, with dihydroxyacetone-P significantly decreasing the activation/phosphorylation of PEPC in the light, while these metabolites all significantly decreased the activation state of PEPCK in the light (data not shown). However, it is not known how much metabolite interconversion occurred in these experiments or what the cytosolic concentrations of potentially regulatory metabolites were in the mesophyll and bundle sheath.
Clearly, further studies are needed of the effects that metabolites have on the expression and activity of the kinases and phosphatases that can regulate PEPC. The kinase and phosphatase for PEPCK also need to be identified and characterized and regulation of their activity and expression by metabolites studied. This would then give clues as to how these enzymes are coregulated, not only in Guinea grass, but also in some Crassulacean acid metabolism plants and in many other tissues of C3 plants in which both enzymes occur in the cytosol of the same cells and in which they are both regulated by phosphorylation (Walker and Leegood, 1996; Leegood and Walker, 2003). In addition, the regulatory properties of intact, unproteolyzed PEPCK (Burnell, 1986; Chen et al., 2002) need to be compared in C4 plants in which it is phosphorylated, such as Guinea grass, and in other C4 plants, such as U. panicoides, that lack PEPCK phosphorylation and in which coordination of PEPC and PEPCK must still occur.
MATERIALS AND METHODS
Plant Material
Seeds of Guinea grass (Panicum maximum; Herbiseed) were grown in a plant growth chamber at a light intensity of 1,500 μmol quanta m−2 s−1 with a 12-h photoperiod (30°C day, 20°C night, 60% humidity). Nutrient solution was applied once weekly in the form of Miracle-Gro (ICI). The third or fourth leaf was harvested at midday from plants at 5 weeks of age. For photosynthesis measurements, attached leaves were used.
cDNA Sequence Determination
Total RNA was purified from 40-d-old Guinea grass using an RNeasy plant mini kit (Qiagen). Two sets of degenerate primers were designed complementary to regions highly conserved between maize (Zea mays), Urochloa panicoides, and Arabidopsis (Arabidopsis thaliana) PEPCK genes: forward 1, GGCGTCATGCACTACCTCAT and reverse 1, CACGGCACGCCGTTGAT; forward 2, GCAGTACGCTGGGGAGAT and reverse 2, CCTTGTACGCCGGCCTTGT. Reverse transcription-PCR was carried out using a SuperScriptII RNase H− reverse transcriptase kit (Gibco-BRL). PCR products of expected sizes were gel extracted using the QIAquick gel extraction kit (Qiagen) and sequenced.
To obtain the full-length cDNA sequence, 5′ and 3′ RACE-PCR was carried out using a FirstChoice RLM-RACE kit (Ambion) and primers GGACCAGCCGGTGTTGACAA and CGTAGCAACCTCCCTCAA as 5′ RACE outer and inner primers, and forward 2 and forward 1 (above) as outer and inner primers for 3′ RACE. PCR fragments were sequenced and the full-length cDNA amplified. The complete sequence was submitted to GenBank (accession no. AAQ10076) and compared to databases using BLASTx and BLASTn algorithms and homologous sequences aligned using BioEdit (Hall, 1999).
In Vivo Phosphorylation Assay
For the experiments in which light intensity and CO2 concentration were changed, terminal 4-cm portions of Guinea grass were excised, placed in a 1-mL cuvette containing 160 μL of water and 40 μCi (4 μL) of [33P] Pi (specific activity 148 TBq mmol−1; ICN Biomedicals) and illuminated (1,600 μmol quanta m−2 s−1) at 25°C to 30°C for 2 h, at which point about 90% of the solution had been taken up. A further 160 μL of water and 4 μL of [33P] Pi were then added and the above procedure repeated. Leaf portions were then supplied with 400 μL of water and incubated in the dark overnight (18 h). For varying light intensity, one leaf portion was placed at each light intensity for up to 6 h. For varying CO2 concentrations, leaf portions were incubated in a glass chamber at a light intensity of 400 μmol quanta m−2 s−1 for up to 6 h. After incubation, a 2-cm portion of the basal leaf was cut and frozen in liquid N2. Each complete experiment in Figures 3 to 5 was repeated at least three times on different days, yielding similar results.
For feeding experiments, a similar feeding procedure for [33P] Pi was followed. One-centimeter pieces of leaves were excised and placed in a multiwell plate (Corning), each well containing 160 μL of water and 4 μL (40 μCi) of [33P] Pi and illuminated for 2 h, after which a further 160 μL of water were added and the above procedure repeated. Leaf portions were then supplied with 200 μL water and incubated overnight (18 h) in the dark. Leaf portions were transferred to a new multiwell plate containing 160 μL of a solution containing DTT or inhibitors (at pH 7.0; in triplicate) and illuminated at 1,600 μmol quanta m−2 s−1 for 2 h, or illuminated for 2 h and then darkened for 2 h. The leaf pieces were then transferred immediately into liquid N2.
Tissue was homogenized with 5 volumes of ice-cold 200 mm Bicine-KOH (pH 9.8), 50 mm DTT, then clarified by centrifugation at 20,000g for 3 min. Supernatants were added to an equal volume of SDS-PAGE solubilization buffer (62.5 mm Tris-HCl [pH 6.8], 10% [v/v] glycerol, 5% [w/v] SDS, 5% [v/v] 2-mercaptoethanol, 0.002% [w/v] bromophenol blue), placed at 100°C for 3 min, centrifuged at 20,000g for 3 min, and supernatants analyzed by SDS-PAGE.
SDS-PAGE, Autoradiography, and Immunoblotting
SDS-PAGE was performed using a 4.7% T/2.7% C stacking gel and a 10.5% T/2.7% C resolving gel. After electrophoresis, polypeptides were fixed in gels by immersion in 50% (v/v) methanol and 12% acetic acid. Polypeptides were visualized by colloidal Coomassie Blue G-250 (Sigma). For immunoblotting, transfer of polypeptides from an SDS-PAGE gel to Immobilon P membrane (Sigma) was done in a Pharmacia Multiphor apparatus. Immunoreactive polypeptides were visualized using an antiserum raised to purified Guinea grass PEPCK or Amaranthus edulis PEPC in conjunction with an ECL kit (Amersham). Autoradiography of dried gels was performed at −80°C using Kodak Biomax MR film (VWR), using intensifying screens for 96 to 168 h. Dried films were quantified by a densitometer (Vilber Lourmat). Comparison of autoradiographs and immunoblots enabled the identification of bands at 103 kD for PEPC and at 71 kD for PEPCK in Guinea grass (Walker et al., 1997). The degree of phosphorylation of PEPC or PEPCK in treatments was expressed as a percentage of the phosphorylation of the overnight dark sample for all experiments other than the substrate feeding experiments in which water was used as the control.
Activation States of PEPC and PEPCK
For measurement of PEPC and PEPCK activity, leaf samples were extracted in 5 volumes of ice-cold 200 mm Bicine-KOH (pH 9.8), 50 mm DTT. The carboxylation activity of PEPCK was measured in a continuous assay at 25°C, including 100 mm HEPES (pH 7.0), 100 mm KCl, 90 mm KHCO3, 0.5 mm PEP, 1.0 mm ADP, 5 μm MnCl2, 4 mm MgCl2, 0.14 mm NADH, 6 units mL−1 of malate dehydrogenase for optimum activity, and with 0.8 mm ADP and 0.2 mm ATP instead of 1.0 mm ADP to estimate its activation state (Walker et al., 2002). The activity of PEPC was measured in a continuous assay at 25°C containing 100 mm HEPES (pH 7.3), 0.8 mm PEP, 5 mm MgCl2, 4.8 mm KHCO3, 0.35 mm NADH, 5 units mL−1 malate dehydrogenase. For estimation of the PEPC activation state, 1 mm malate was included in the assay (Echevarria et al., 1994). Activation state was expressed as a percentage of the maximal enzyme activity measured. Activities in the assays ranged between 7.8 and 13.3 units g−1 fresh weight for PEPCK and 15.1 and 19.5 units g−1 fresh weight for PEPC.
Photosynthesis Measurements
Steady-state rates of photosynthesis were measured using a portable infrared gas analyzer (LCA4; Analytical Development Co.). Light was supplied through fiber optics by a Schott KL 1500 lamp (H. Walz). The maximal PFD achievable at the surface of the leaf was 1,800 μmol quanta m−2 s−1. The leaf temperature was measured with a copper-constantan thermocouple secured on the underside of the leaf (supplied with the infrared gas analyzer). For all measurements, the leaf was illuminated in the chamber until the maximal rate of photosynthetic assimilation was attained.
PEPCK sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AAQ10076, S52988, AAG01894, and S52637.
This work was supported by a research grant from the Biotechnology and Biological Sciences Research Council, United Kingdom (grant no. BBSRC 50/P14423).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard C. Leegood (r.leegood@shef.ac.uk).
Open Access articles can be viewed online without a subscription.
References
- Bakrim N, Brulfert J, Vidal J, Chollet R (2001) Phosphoenolpyruvate carboxylase kinase is controlled by a similar signaling cascade in CAM and C4 plants. Biochem Biophys Res Commun 286 1158–1162 [DOI] [PubMed] [Google Scholar]
- Bakrim N, Echevarria C, Crétin C, Arrio-Dupont M, Pierre JN, Vidal J, Chollet R, Gadal P (1992) Regulatory phosphorylation of Sorghum leaf phosphoenolpyruvate carboxylase: identification of the protein-serine kinase and some elements of the signal transduction cascade. Eur J Biochem 204 821–830 [DOI] [PubMed] [Google Scholar]
- Burnell JN (1986) Purification and properties of phosphoenolpyruvate carboxykinase from C4 plants. Aust J Plant Physiol 13 577–587 [Google Scholar]
- Burnell JN, Hatch MD (1985) Light-dark modulation of leaf pyruvate, Pi dikinase. Trends Biochem Sci 10 288–291 [Google Scholar]
- Burnell JN, Hatch MD (1988) Photosynthesis in phosphoenolpyruvate carboxykinase-type C4 plants: pathways of C4 acid decarboxylation in bundle sheath cells of Urochloa panicoides. Arch Biochem Biophys 260 187–199 [DOI] [PubMed] [Google Scholar]
- Chen Z-H, Walker RP, Acheson RM, Leegood RC (2002) Phosphoenolpyruvate carboxykinase assayed at physiological concentrations of metal ions has a high affinity for CO2. Plant Physiol 128 160–164 [PMC free article] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O'Leary MH (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 47 273–298 [DOI] [PubMed] [Google Scholar]
- Cohen P, Holmes CF, Tsukitani Y (1990) Okadaic acid: a new probe for the study of cellular regulation. Trends Biochem Sci 15 98–102 [DOI] [PubMed] [Google Scholar]
- Echevarria C, Pacquit V, Bakrim N, Osuna L, Delgado B, Arrio-Dupont M, Vidal J (1994) The effect of pH on the covalent and metabolic control of C4 phosphoenolpyruvate carboxylase from Sorghum leaf. Arch Biochem Biophys 315 425–430 [DOI] [PubMed] [Google Scholar]
- Giglioli-Guivarc'h N, Pierre J-N, Brown S, Chollet R, Vidal J, Gadal P (1996) The light-dependent transduction pathway controlling the regulatory phosphorylation of C4 phosphoenolpyruvate carboxylase in protoplasts from Digitaria sanguinalis. Plant Cell 8 573–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA (1999) Bio-Edit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41 95–98 [Google Scholar]
- Hatch MD (1987) C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochim Biophys Acta 895 81–106 [Google Scholar]
- Hatch MD, Kagawa T (1976) Photosynthetic activities of isolated bundle sheath cells in relation to differing mechanisms of C4 pathway photosynthesis. Arch Biochem Biophys 175 39–53 [DOI] [PubMed] [Google Scholar]
- Izui K, Matsumura H, Furumoto T, Kai Y (2004) Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu Rev Plant Biol 55 69–84 [DOI] [PubMed] [Google Scholar]
- Jiao J, Chollet R (1992) Light activation of maize phosphoenolpyruvate carboxylase protein-serine kinase activity is inhibited by mesophyll and bundle sheath-directed photosynthesis inhibitors. Plant Physiol 98 152–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J-A, Chollet R (1988) Light/dark regulation of maize leaf phosphoenolpyruvate carboxylase by in vivo phosphorylation. Arch Biochem Biophys 261 409–417 [DOI] [PubMed] [Google Scholar]
- Jiao J-A, Echevarria C, Vidal J, Chollet R (1991) Protein turnover as a component in the light/dark regulation of phosphoenolpyruvate carboxylase protein-serine kinase activity in C4 plants. Proc Natl Acad Sci USA 88 2712–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HS, Hatch MD (1970) Properties and regulation of leaf nicotinamide-adenine dinucleotide phosphate-malate dehydrogenase and “malic” enzyme in plants with the C4-dicarboxylic acid pathway of photosynthesis. Biochem J 119 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabourniotis G, Manetas Y, Gavalas NA (1983) Photoregulation of phosphoenolpyruvate carboxylase in Salsola soda L. and other C4 plants. Plant Physiol 73 735–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC, von Caemmerer S, Osmond CB (1996) Metabolite transport and photosynthetic regulation in C4 and CAM plants. In DT Dennis, DH Turpin, DD Layzell, DK Lefebvre, eds, Plant Metabolism. Longmans, London, pp 341–369
- Leegood RC, Walker RP (2003) Regulation and roles of phosphoenolpyruvate carboxykinase in plants. Arch Biochem Biophys 414 204–210 [DOI] [PubMed] [Google Scholar]
- Nimmo GA, McNaughton GAL, Fewson CA, Wilkins MB, Nimmo HG (1987) Changes in the kinetic properties and phosphorylation state of phosphoenolpyruvate carboxylase in Zea mays leaves in response to light and dark. FEBS Lett 213 18–22 [Google Scholar]
- Nimmo HG (2003) Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Arch Biochem Biophys 414 189–196 [DOI] [PubMed] [Google Scholar]
- Osuna L, Coursol S, Pierre J-N, Vidal J (2004) A Ca2+-dependent protein kinase with characteristics of protein kinase C in leaves and mesophyll cell protoplasts from Digitaria sanguinalis: possible involvement in the C4-phosphoenolpyruvate carboxylase phosphorylation cascade. Biochem Biophys Res Commun 314 428–433 [DOI] [PubMed] [Google Scholar]
- Pierre J-N, Prieto J-L, Gadal P, Vidal J (2004) In situ phosphoenolpyruvate carboxylase activity and kinetic properties in isolated Digitaria sanguinalis mesophyll cells. Photosynth Res 79 349–355 [DOI] [PubMed] [Google Scholar]
- Sassenrath-Cole G, Pearcy RW (1994) Regulation of photosynthetic induction state by the magnitude and duration of low light exposure. Plant Physiol 105 1115–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Ueno Y, Hisabori T, Hayashi H, Izui K (2001) Thioredoxin-mediated reductive activation of a protein kinase for the regulatory phosphorylation of C4-form phosphoenolpyruvate carboxylase from maize. Plant Cell Physiol 42 1295–1302 [DOI] [PubMed] [Google Scholar]
- Smith LH, Langdale JA, Chollet R (1998) A functional Calvin cycle is not indispensable for the light activation of C4 phosphoenolpyruvate carboxylase kinase and its target enzyme in the maize mutant bundle sheath defective2-mutable1. Plant Physiol 118 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida Y, Furumoto T, Izumida A, Hata S, Izui K (2001) Phosphoenolpyruvate carboxylase kinase involved in C4 photosynthesis in Flaveria trinervia: cDNA cloning and characterization. FEBS Lett 507 318–322 [DOI] [PubMed] [Google Scholar]
- Walker RP, Acheson RM, Técsi LI, Leegood RC (1997) Phosphoenolpyruvate carboxykinase in C4 plants: its role and regulation. Aust J Plant Physiol 24 459–468 [Google Scholar]
- Walker RP, Chen Z-H, Acheson RM, Leegood RC (2002) Effects of phosphorylation on phosphoenolpyruvate carboxykinase from the C4 plant, Guinea grass. Plant Physiol 128 165–172 [PMC free article] [PubMed] [Google Scholar]
- Walker RP, Leegood RC (1995) Purification and phosphorylation in vitro and in vivo, of phosphoenolpyruvate carboxykinase from cucumber cotyledons. FEBS Lett 362 70–74 [DOI] [PubMed] [Google Scholar]
- Walker RP, Leegood RC (1996) Phosphorylation of phosphoenolpyruvate carboxykinase in plants: studies in plants with C4 photosynthesis and Crassulacean acid metabolism and in germinating seeds. Biochem J 317 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-H, Chollet R (1993) Partial purification and characterization of phosphoenolpyruvate carboxylase protein-serine kinase from illuminated maize leaves. Arch Biochem Biophys 304 496–502 [DOI] [PubMed] [Google Scholar]
- Wingler A, Walker RP, Chen Z-H, Leegood RC (1999) Phosphoenolpyruvate carboxykinase is involved in the decarboxylation of aspartate in the bundle sheath of maize. Plant Physiol 120 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]