Abstract
Chain elongated, methionine (Met)-derived glucosinolates are a major class of secondary metabolites in Arabidopsis (Arabidopsis thaliana). The key enzymatic step in determining the length of the chain is the condensation of acetyl-coenzyme A with a series of ω-methylthio-2-oxoalkanoic acids, catalyzed by methylthioalkylmalate (MAM) synthases. The existence of two MAM synthases has been previously reported in the Arabidopsis ecotype Columbia: MAM1 and MAM3 (formerly known as MAM-L). Here, we describe the biochemical properties of the MAM3 enzyme, which is able to catalyze all six condensation reactions of Met chain elongation that occur in Arabidopsis. Underlining its broad substrate specificity, MAM3 also accepts a range of non-Met-derived 2-oxoacids, e.g. converting pyruvate to citramalate and 2-oxoisovalerate to isopropylmalate, a step in leucine biosynthesis. To investigate its role in vivo, we identified plant lines with mutations in MAM3 that resulted in a complete lack or greatly reduced levels of long-chain glucosinolates. This phenotype could be complemented by reintroduction of a MAM3 expression construct. Analysis of MAM3 mutants demonstrated that MAM3 catalyzes the formation of all glucosinolate chain lengths in vivo as well as in vitro, making this enzyme the major generator of glucosinolate chain length diversity in the plant. The localization of MAM3 in the chloroplast suggests that this organelle is the site of Met chain elongation.
Plants synthesize an almost uncountable number of secondary metabolites. More than 100,000 have been identified so far, which may represent only 10% of the actual total in nature (Schwab, 2003; Wink, 2003). Only a small fraction of this diversity is present in individual plant species, yet single species contain multiple types of secondary metabolites and many representatives of a single type. For example, Arabidopsis (Arabidopsis thaliana) is reported to produce over 170 secondary metabolites, including approximately 50 terpenes, 25 benzenoids, and 20 phenylpropanoids (D'Auria and Gershenzon, 2005). In the last few years, researchers have begun to identify the biochemical bases of this diversity. In some cases, biosynthetic enzymes catalyze multiple product formation (Köllner et al., 2004; Tholl et al., 2005) or have broad substrate specificities (Wan and Wilkins, 1994; Gang et al., 2002). These properties may allow the generation of a large variety of secondary metabolites with a relatively small amount of biochemical machinery.
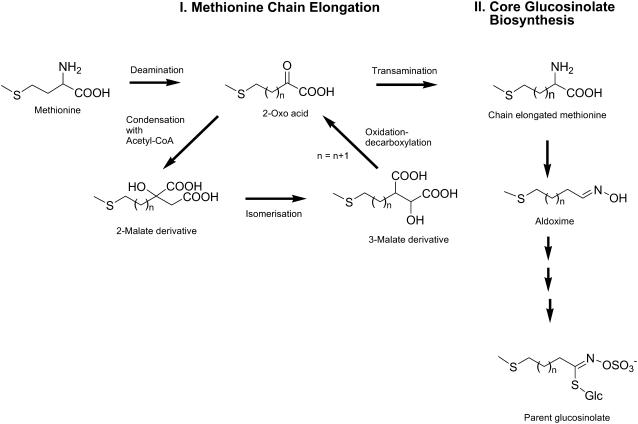
One of the largest groups of secondary metabolites in Arabidopsis, with 35 representatives, is the glucosinolates (Reichelt et al., 2002), which serve as antiherbivore defenses (Wittstock et al., 2003). These amino acid-derived compounds have variable side chains attached to a common core structure of a Glc residue linked via a sulfur atom to a (Z)-N-hydroximinosulfate ester (Fahey et al., 2001; Fig. 1). The structural diversity of glucosinolates arises from different precursor amino acids, variation in side chain length, and diverse patterns of secondary oxidation and esterification (Tokuhisa et al., 2004). The most abundant group of glucosinolates in Arabidopsis consists of those derived from Met. These are biosynthesized by the chain elongation of Met followed by construction of the core glucosinolate skeleton and side chain modifications (Wittstock and Halkier, 2002; Halkier and Gershenzon, 2006).
Figure 1.
Scheme of Met-derived glucosinolate biosynthesis in Arabidopsis. This process can be divided into the Met chain elongation cycle (I) and the biosynthesis of the core glucosinolate structure (II). The parent methylthioalkyl glucosinolates can undergo further side chain modifications.
Considerable attention has focused on chain elongation, because it is a major contributor to the diversity of glucosinolate content and, as the first phase of glucosinolate biosynthesis, is responsible for the diversion of amino acid flux from primary to secondary metabolism. Glucosinolates of six different chain lengths are produced in Arabidopsis. Studies with labeled Met and acetate have shown that elongation of the Met side chain involves a repetitive cycle of three reaction steps that result in the net addition of one methylene carbon for each turn of the cycle (Chisholm and Wetter, 1964; Lee and Serif, 1968, 1970; Graser et al., 2000). To enter the cycle, Met is first deaminated to the corresponding 2-oxo-acid, 4-methylthio-2-oxobutanoic acid. An aminotransferase involved in this step has been identified recently (Schuster et al., 2006). Elongation is initiated by condensation of the 2-oxo-acid with acetyl-CoA yielding a methylthioalkylmalate (MAM) intermediate. Subsequently, isomerization and oxidation-decarboxylation reactions yield a 2-oxo acid that is extended by one methylene group. This product can undergo additional cycles of elongation or it can be transaminated to form an elongated amino acid, which then enters the pathway for formation of the core glucosinolate structure. In the latter case, Met elongated by a single methylene group would result in the formation of 3-methylthiopropylglucosinolate, referred to as a C3 glucosinolate because of the three methylene groups in its side chain. C4 to C8 glucosinolates are the result of further chain elongations, with C8 representing the longest aliphatic glucosinolate type found in Arabidopsis.
The chain-length spectrum of aliphatic glucosinolates in Arabidopsis is critically influenced by whether the 2-oxo acid intermediates undergo further elongation or are diverted into glucosinolate core biosynthesis. Hence, we have focused on the elongation cycle enzymes at this important branching point, the MAM synthases, which catalyze the condensation of 2-oxo acid derivatives of Met with acetyl-CoA. The MAM synthases belong to a large family of enzymes that condense various 2-oxo acids with an acyl-CoA ester (Textor et al., 2004). One member of this group is isopropylmalate synthase (IPMS; EC 2.3.3.13), which condenses 2-oxo-isovalerate with acetyl-CoA, the chain elongation reaction in Leu biosynthesis.
Molecular studies on MAM synthases in Arabidopsis began with the identification of four candidate genes in the Columbia (Col-0) accession that showed similarity to the IPMS-encoded genes of other organisms (Campos de Quiros et al., 2000; Kroymann et al., 2001). Two of these candidate genes are immediately adjacent to each other at the GS-ELONG (glucosinolate elongation) locus on chromosome V (Magrath et al., 1994). Because this locus controls the dominant glucosinolate side chain length (C3 or C4), the genes were thought to encode MAM synthases. When heterologously expressed in Escherichia coli, one of these genes (At5g23010) was demonstrated to encode a protein that catalyzes the condensation reactions for two cycles of Met chain elongation (Kroymann et al., 2001; Textor et al., 2004) and so was named MAM1. Plant lines with mutations in MAM1 showed a decrease in C4 glucosinolates and a corresponding increase in C3 glucosinolates. However, the residual C3 and C4 glucosinolates in these lines, plus the continued accumulation of C5 to C8 glucosinolates, suggested the presence of additional MAM activities. Furthermore, MAM activity was detected in protein extracts of a mutant plant line (gsm 1-1 = TU1) lacking a functional MAM1 allele (Textor et al., 2004).
The residual MAM activity may be ascribed to the product of the second gene at the GS-ELONG locus (At5g23020), named MAM-L for MAM like, because an insertion mutation in the MAM-L locus showed alterations in the biosynthesis of long-chain aliphatic glucosinolates (Field et al., 2004). It is not likely that this MAM activity arises from either of the other two candidate genes in Arabidopsis with similarity to IPMS genes of other organisms. Both candidates, At1g74040 and At1g18500, have recently been shown to encode actual IPMSs by biochemical and molecular methods (de Kraker et al., 2006).
In this article, we describe an extensive biochemical characterization of the MAM-L enzyme, showing that it does indeed account for the remaining MAM activity in Arabidopsis. In addition to in vitro studies of the heterologously expressed enzymes with an extensive series of substrates, we determined the subcellular location of MAM-L and the relative steady-state levels of the transcript in various tissues. Furthermore, we determined glucosinolate content in mutant plant lines where the levels of MAM-L and MAM1 enzyme activity have been decreased through genetic alterations at the MAM loci. We propose that MAM-L be renamed MAM3 based on the biochemical properties of the encoded enzyme demonstrated here, the existing rules for Arabidopsis gene nomenclature, and the prior naming of MAM1 and MAM2 (Kroymann et al., 2003). To minimize confusion, Table I summarizes the disparate nomenclature used in previous publications for the five members of the MAM/IPMS gene family in Arabidopsis.
Table I.
Nomenclature for Arabidopsis (Col-0) genes of the IPMS/MAM synthase gene family used in recent publications
| Arabidopsis Genome Initiative Locus Code | N-Terminal Sequence | Bacterial Artificial Chromosomes | Kroymann et al. (2001) | Junk and Mourad (2002) | Field et al. (2004) | This Article |
|---|---|---|---|---|---|---|
| At1g18500 | MASSLLR | F15H18 | – | EST116C2T7 | MAML-4 | IPMS1 |
| At1g74040 | MESSILK | F2P9.9 | – | IMS1 | MAML-3 | IPMS2 |
| At5g23010 | MASSLLT | CT20O7 | MAM1 | IMS3 | MAM1/L | MAM1 |
| –a | MASSLLTb | – | – | – | – | MAM2 |
| At5g23020 | MASLLLT | MYJ24 | MAM-L | IMS2 | MAM1/L | MAM3 |
Col-0 ecotype lacks MAM2. The gene would be located between At5g23000 and At5g23010 (MAM1) on the Col-0 genome (Kroymann et al., 2003).
Landsberg erecta.
RESULTS
MAM3 Has MAM Synthase Activity
The open reading frame (ORF) of the MAM3 gene without the 5′ sequence encoding a putative plastid-targeting peptide was cloned from the Col-0 accession into a plasmid construct containing the T7 viral promoter and a C-terminal polyhistidine peptide and expressed in the E. coli BL21(DE3) strain. The recombinant protein was detected in the bacterial extract as a prominent band of the predicted size of 53 kD by SDS-PAGE analysis. This extract exhibited MAM synthase activity with 4-methylthio-2-oxobutanoic acid, the 2-oxo acid derived from Met, and 14C-labeled acetyl-CoA. Purification of the recombinant polyhistidine-containing protein by application to a nickel-nitrilotriacetic acid agarose (Ni-NTA) affinity chromatography resin and elution with l-His resulted in an active enzyme fraction that was more than 90% pure as judged by SDS-PAGE with Coomassie staining.
Basic characterization of MAM3 was performed by incubation with 4-methylthio-2-oxobutanoic acid and 14C-labeled acetyl-CoA as substrates. The assay products were separated by radio-HPLC and the identity of the product, 2-(2′-methylthioethyl)-malate, was confirmed by liquid chromatography-mass spectroscopy (MS) in comparison with an authentic 2-(2′-methylthioethyl)-malate standard (Textor et al., 2004). The recombinant enzyme had a pH optimum at pH 8.0 with half maxima at pH 6.3 and 9.2 (range tested, pH 5.5–10 at intervals of 0.5). The temperature optimum was 32°C, with 70% activity at 25°C and 39°C (range tested, 22°C–40°C at intervals of 2°C). As in the case of MAM1 (Textor et al., 2004), MAM3 activity was dependent on the presence of a divalent cation with Mn2+ giving highest activity. If the MAM3 activity in the presence of Mn2+ is set to 100, then the addition of Fe2+ (59), Co2+ (41), Mg2+ (39), and Ca2+ (39) resulted in moderate activity, while that of Zn2+ (18), Mo2+ (17), Ni2+ (14), and Cu2+ (1) resulted in low activity. Consistent with this divalent cation dependency, product formation was inhibited 50% by 10 μm EDTA and completely by 40 μm EDTA. Unlike MAM1, the addition of ATP did not promote MAM3 activity; instead, activity was reduced 50% by an ATP concentration of 18 mm. Similarly, there was no requirement for dithiothreitol (DTT), which was necessary at 1 mm for the activity of MAM1.
MAM3 Catalyzes All the Condensation Reactions of Glucosinolate Chain Elongation in Arabidopsis in Vitro But with Different Efficiencies
To investigate the ability of MAM3 to catalyze the various condensation reactions of the chain elongation cycle of Arabidopsis glucosinolate biosynthesis, the recombinant enzyme was incubated with a series of 2-oxo acids of different chain lengths and substrate analogs where the sulfur atom is substituted with a methylene group. Arabidopsis requires six different substrates for condensation reactions to produce the six different chain length glucosinolates (C3–C8) that it accumulates. The products of MAM3 activity were identified by liquid chromatography-MS and confirmed in comparison to authentic standards (see Supplemental Fig. S1). The reactions catalyzed include all six condensations predicted to occur in Arabidopsis using either the natural substrates or the nonsulfur analogs. The enzyme was unable to catalyze the reaction with the longest compound, 2-oxo-dodecanoic acid, the analog of the substrate leading to C9 glucosinolates, which are not observed in Arabidopsis (Table II).
Table II.
Suitability of 2-oxo acids as substrates for MAM3-mediated condensation with acetyl-CoA
| 2-Oxo Acid Substrate | Structure | Resulting Glucosinolate | Suitabilitya |
|---|---|---|---|
| 4-Methylthio-2-oxobutanoate | 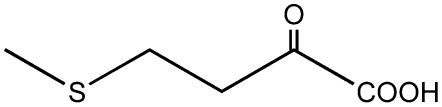 |
C3 | + |
| 2-Oxo-hexanoate | 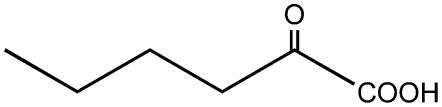 |
C3 | + |
| 5-Methylthio-2-oxopentanoate | 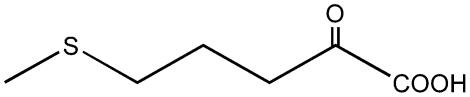 |
C4 | + |
| 2-Oxo-heptanoate |  |
C4 | + |
| 6-Methylthio-2-oxohexanoate |  |
C5 | + |
| 2-Oxo-octanoate | 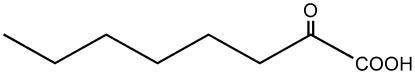 |
C5 | + |
| 2-Oxo-nonanoate | 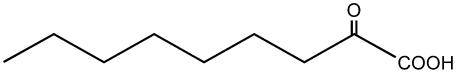 |
C6 | + |
| 8-Methylthio-2-oxooctanoate | 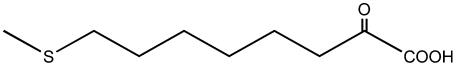 |
C7 | + |
| 2-Oxo-decanoate |  |
C7 | + |
| 9-Methylthio-2-oxononanoate | 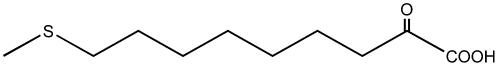 |
C8 | + |
| 2-Oxo-undecanoate |  |
C8 | + |
| 2-Oxo-dodecanoate | 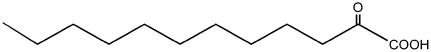 |
C9 | − |
+, Condensation product formed; −, condensation product not formed.
To compare the substrates, kinetic studies were done with the native substrates available (Table III), all of which exhibited standard Michaelis-Menten kinetics. The Km values obtained ranged from 932 μm for 4-methylthio-2-oxobutanoic acid to 81 μm for 9-methylthio-2-oxononanoic acid, suggesting increased substrate affinity with increasing chain length. The kcat (turnover number) values were highest with the medium chain length substrate (6-methylthio-2-oxohexanoic acid) and lower for the shorter substrates (4-methylthio-2-oxobutanoic acid and 5-methylthio-2-oxopentanoic acid) and the longer substrates (8-methylthio-2-oxooctanoic acid and 9-methylthio-2-oxononanoic acid). The resulting specificity constants (kcat/Km) for the substrate series reflect this trend, also indicating that the intermediate chain length substrates are catalytically most efficient and the longest substrates are the least efficient. The Km for acetyl-CoA was 2.3 mm and the kcat value was 3.0 s−1.
Table III.
Kinetic parameters for various 2-oxo acids with recombinant MAM3a
| Substrate | Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|
| μm | nmol min−1mg−1 | s−1 | m−1s−1 | |
| 4-Methylthio-2-oxobutanoic acid (→C3 glucosinolates) | 932 ± 56 | 1,448 ± 299 | 1.3 ± 0.3 | 1,380 |
| 5-Methylthio-2-oxopentanoic acid (→C4 glucosinolates) | 476 ± 199 | 1,495 ± 679 | 1.3 ± 0.6 | 2,730 |
| 6-Methylthio-2-oxohexanoic acid (→C5 glucosinolates) | 463 ± 210 | 2,869 ± 768 | 2.5 ± 0.7 | 5,400 |
| 8-Methylthio-2-oxooctanoic acid (→C7 glucosinolates) | 253 ± 123 | 364 ± 143 | 0.3 ± 0.1 | 1,280 |
| 9-Methylthio-2-oxononanoic acid (→C8 glucosinolates) | 81 ± 21 | 31 ± 8 | 0.03 ± 0.01 | 370 |
| 2-Oxoisovalerate | 1,000 ± 200 | 199 ± 38 | 0.18 ± 0.03 | 200 |
| Pyruvate | 8,600 ± 4,300 | 191 ± 48 | 0.17 ± 0.04 | 23 |
| Acetyl-CoA | 2,300 ± 1,200 | 3,344 ± 1428 | 3.0 ± 1.3 | 1,300 |
Data presented are means ± sds of at least five replicates per substrate.
MAM3 Also Catalyzes Reactions with Other 2-Oxo Acids
Because the MAM3 enzyme has about 50% amino acid identity with Arabidopsis proteins predicted to encode IPMSs (that catalyze a step in Leu biosynthesis), it may also have this catalytic capability. The recombinant protein was tested with 2-oxoisovalerate (3-methyl-2-oxobutanoate), the substrate for the Leu biosynthetic reaction, and other 2-oxo acids. MAM3 was able to convert 2-oxoisovalerate (to isopropylmalate) and pyruvate (to citramalate), but based on their kinetic parameters, these were less preferred substrates than the Met-derived 2-oxo acids involved in glucosinolate formation (Table III). The enzyme also converted 4-methyl-2-oxopentanoate and 5-methyl-2-oxohexanoate to their malate derivatives with specific activities of 422 ± 56 and 1,070 ± 294 nmol min−1 mg−1, respectively. These reactions represent the condensation reactions of a chain elongation cycle predicted for Leu-derived glucosinolates, which have been identified in Arabidopsis ecotypes (Kliebenstein et al., 2001; Reichelt et al., 2002). Additionally, the enzyme converted 3-methyl-2-oxopentanoate with a specific activity of 86 ± 9 nmol min−1 mg−1 to a product, representing the chain elongation reaction for Ile-derived glucosinolates, not yet described in Arabidopsis. Finally, a reaction product was formed with phenylpyruvate (specific activity of 22 ± 4 nmol min−1 mg−1), which represents the condensation reaction leading to phenylethylglucosinolate, a compound known from Arabidopsis (Reichelt et al., 2002; Brown et al., 2003). A prior quantitative trait loci mapping study that implicated the GS-Elong locus (that includes MAM1, MAM2, and MAM3) in controlling Phe elongation for glucosinolate biosynthesis (Kliebenstein et al., 2001) supports the role of MAM3 in catalyzing reaction with phenylpyruvate in vivo.
To confirm the ability of MAM3 to catalyze IPMS activity in vivo, the auxotrophic E. coli strain CV512(DE3), which is lacking IPMS activity, was transformed with vector constructs containing either the E. coli leuA gene (coding for the endogenous IPMS) or MAM3 from Arabidopsis. If MAM3 had IPMS activity in vivo, it should complement the mutation by restoring the ability of CV512(DE3) to grow on a minimal medium without supplemental Leu. Under such conditions, growth was not observed with the negative control CV512(DE3), whereas the positive control, strain CV512(DE3) with the leuA construct, showed growth within 2 d when incubated at 37°C and within 3 d when incubated at 28°C. No complementation was observed with the strain containing the MAM3 construct when the cells were grown at 37°C, but colony growth was observed at 28°C after 3 d of culture. Hence, the Arabidopsis MAM3 was able to complement the mutant in the gene encoding IPMS and restored autotrophic growth.
MAM3 Is Targeted to the Chloroplasts
To learn more about subcellular localization of MAM3, an antibody was raised against the purified enzyme. The antibody was carefully tested for cross-reactivity with the other members of the MAM/IPMS family of Arabidopsis (Col-0) and showed weak cross-reactivity with only MAM1 (Supplemental Fig. S2). Immunolocalization experiments with leaf tissue of Arabidopsis (Col-0) performed with the anti-MAM3 serum confirmed that MAM3 is targeted to the chloroplasts (Fig. 2) as predicted on the basis of its N-terminal sequence (Kroymann et al., 2001). Because the immediately preceding enzyme of the Met chain elongation cycle, the aminotransferase, was recently shown to be localized in the cytosol (Schuster et al., 2006), this implies that the 2-oxo acid substrate of MAM3 is imported from the cytosol into chloroplasts.
Figure 2.
Immunocytochemical localization of MAM3 protein in Arabidopsis leaves. Cross section of leaves were probed with anti-MAM3 antibody (A and C) or with preimmune serum (E) followed by a fluorescence-labeled secondary antibody. A strong green fluorescence label within chloroplasts in A and C is indicative of the MAM3 protein. Chloroplasts are defined by positive 4,6-diamidino-2-phenylindole staining (B) of the same section as shown in A and by starch granules visualized in D by the differential interference contrast image of C. Negative control performed by treatment with preimmune serum did not exhibit any label (E). Bars = 20 μm in A, B, E, and 5 μm in C and D.
MAM3 Mutants Lack Long-Chain Glucosinolates and Their Production Can Be Restored by Ectopic Expression of MAM3
To understand the role of MAM3 in determining the glucosinolate profile in planta, we characterized two mutant lines of the Arabidopsis Col-0 accession, one with a nucleotide alteration in the MAM3 gene and the other with a T-DNA insertion, resulting in changes in MAM3 gene expression.
The mutant line TU3 (gsm2-1) had been isolated and described briefly by Haughn et al. (1991) as lacking C6, C7, and C8 aliphatic glucosinolates in the leaves. We confirmed these results in seeds but found a minor amount of C6 in leaves (Fig. 3). The absence of the longer aliphatic glucosinolates segregated as a recessive trait. Based on our biochemical characterization of MAM3, we considered it likely that the gsm2-1 mutation was in this gene. DNA sequence comparisons of wild-type and mutant MAM3 clones revealed a base substitution in gsm2-1 at position 788 relative to the predicted start of the ORF of the MAM3. The G to A transition, consistent with the alkylating activity of ethylmethanesulfonate used to generate the mutant population, results in a missense mutation converting a Gly to a Glu codon.
Figure 3.
Glucosinolate profile of leaves (A) and seeds (B) of MAM3 mutants compared to Col-0 wild type. Glucosinolates were purified as desulfoglucosinolates, fractionated by reverse-phase HPLC, and individually identified and quantified. Individual Met-derived glucosinolates are grouped according to their chain length (no. of methylene carbons in the R group, C2–C8), while indole glucosinolates are depicted separately. Data show the means and ses of three replicate samples.
To demonstrate that this mutation in MAM3 was responsible for the altered glucosinolate profile, two different methods were used. First, the mutant enzyme G263E was created by site-directed mutagenesis. When this enzyme was tested for MAM activity using the same conditions as for the wild-type enzyme, no activity could be found. Second, the gsm2-1 mutant line was used for the transgenic expression of a gene construct consisting of the 35S promoter and the ORF of MAM3. Leaves from eight randomly selected individuals of a T2 segregating population were analyzed for levels of the endogenous MAM3 and MAM1 transcripts, the introduced MAM3 transcript, and glucosinolate content. Three of these individuals (1–3; Table IV) had detectable levels of the introduced MAM3 transcript and between 12 and 22 μmol/g dry weight combined of C6, C7, and C8 glucosinolates. These levels of long-chain, Met-derived glucosinolates were about 5-fold higher than those detected in wild-type Arabidopsis and demonstrate complementation of the gsm2-1 mutant phenotype. In one individual, in which no transcript from the introduced MAM3 gene was detectable by reverse transcriptase (RT)-PCR, there were also no detectable long-chained aliphatic glucosinolates (data not shown). The remaining four individuals exhibited 5-fold lower levels of the introduced transcript than those observed in the three complemented individuals and showed aberrant sizes of endogenous MAM1 and MAM3 transcripts. These lines lacked long-chain, Met-derived glucosinolates (data not shown) and had reduced (4–19 μmol/g dry weight) levels of all aliphatic glucosinolates.
Table IV.
Glucosinolate content (μmol g−1 dry weight) in leaves of gsm2-1 and lines transformed with 35S∷MAM3a
| Line | C3 | C4 | C5 | C6 | C7 | C8 | ΣCi |
|---|---|---|---|---|---|---|---|
| Col-0 wild type | 2.7 | 16.3 | 0.5 | 0.3 | 0.4 | 2.2 | 22.4 |
| gsm2-1 | 2.5 | 18.2 | 0.6 | 0.2 | n.d. | n.d. | 21.5 |
| T2 Line 1 | 2.3 | 6.5 | n.d. | 1.0 | 3.2 | 8.6 | 21.6 |
| T2 Line 2 | 3.9 | 8.0 | n.d. | 1.6 | 6.2 | 15.9 | 35.5 |
| T2 Line 3 | 4.1 | 6.4 | n.d. | 1.1 | 3.9 | 10.1 | 25.7 |
Presented are analyses of Col-0 wild type, the gsm2-1 parent line, and three lines randomly selected from a T2 segregating population that had detectable levels of the introduced MAM3 transcript. n.d., Not detected.
The Salk_007222 line (gsm2-2) is reported to contain a T-DNA insertion in the sixth intron of the MAM3 gene. The report was confirmed by PCR using gsm2-2 genomic DNA as a template with oligonucleotide primer pairs derived from the T-DNA insert and MAM3 gene sequences (data not shown). The mutation generated a recessive phenotype consisting of a 90% reduction in C8 glucosinolates in gsm2-2 leaves relative to wild type (Fig. 3). The seed of gsm2-2 exhibited reductions in the levels of both C7 and C8 glucosinolates, although in both gsm2-1 and gsm2-2, the relative proportion of C7 and C8 to total glucosinolates was somewhat higher in the seeds than in the leaves.
MAM3 mutants lacked only long-chain glucosinolates even though in vitro, the enzyme is capable of carrying out the condensation reactions necessary for forming the full range of chain lengths of Met-derived glucosinolates occurring in Arabidopsis. This apparent discrepancy is likely due to the presence of MAM1 (Kroymann et al., 2001; Textor et al., 2004). To learn more about MAM3 activity in vivo, we measured the glucosinolate profiles of a T-DNA insertion line (gsm1-3) in which the MAM1 gene is disrupted. All glucosinolate chain lengths were evident with a large reduction in C4 glucosinolates accompanied by a commensurate increase in C3 glucosinolates as well as a slight increase in C8 glucosinolates (Fig. 4). A very similar phenotype was described before for a MAM1 missense mutant (Haughn et al., 1991; Kroymann et al., 2001). To determine whether MAM3 mutations affect nonaliphatic glucosinolates as well, we examined the correlations among the propyl, butyl, and indolic classes. Across all MAM3 mutants and the wild type, there was a positive correlation between propyl and indolic glucosinolates (r = 0.84, P < 0.05) and a negative correlation between butyl and indolic glucosinolates (r = −0.88, P < 0.05), indicating there might be metabolic cross talk between the accumulation of aliphatic and indole glucosinolates.
Figure 4.
Glucosinolate profile of leaves (A) and seeds (B) of the MAM1 insertion mutant gsm1-3 compared to Col-0 wild type. Aliphatic glucosinolates sum up to 22.4/87.2 μmol g−1 dry weight (leaf/seed) in wild type and 27.3/96.9 μmol g−1 dry weight in the mutant. Glucosinolates were isolated, identified, and quantified as explained in the Figure 3 legend.
The Organ Expression Profile of MAM3 Transcripts Is Different from That of MAM1
To identify the patterns of MAM gene expression in wild-type Arabidopsis Col-0 and the MAM3 mutant lines, transcript levels for both MAM1 and MAM3 were determined relative to the transcript levels for the actin gene ACT8 using RT-PCR on RNA extracted from root, leaf, flower, and silique tissues.
In wild-type plants, the highest levels of expression of MAM3 transcript were observed in the roots, followed by the mature leaves, expanding leaves, and siliques (Fig. 5). Under the cycle conditions used, transcript was not detected in stems (data not shown) and flowers. This profile differs from the pattern of MAM1 transcript accumulation, where the highest levels were found in the expanding leaves, followed by mature leaves, flowers, roots, and siliques. The transcript profile for the MAM3 mutant, gsm2-1, was similar to the wild-type profile as would be expected for a single base substitution mutation in the middle of an exon. For the gsm2-2 line, with an insertion in MAM3, transcript levels for MAM3 were lower than wild type in all tissues. The reduction, but not elimination, of wild-type transcript levels is consistent with an insertion in the intron of MAM3, because it is likely that a low level of processing of the heteronuclear RNA occurs to remove the intron containing the T-DNA insert, resulting in a detectable level of wild-type mRNA.
Figure 5.
Steady-state mRNA transcript levels of MAM1 and MAM3 in tissues of Col-0 wild-type and MAM3 mutant lines determined by RT-PCR. DNA fragments for individual transcripts of ACT8 (actin), MAM1, and MAM3 were generated by PCR amplification of products from RT activity primed by oligo(dT) hybridization to the RNA. The PCR products are shown after separation on 1% agarose gels stained with ethidium bromide. Tissue source: 1, roots; 2, mature leaves; 3, expanding leaves; 4, flowers; and 5, siliques.
DISCUSSION
The aliphatic glucosinolates in Arabidopsis are a large group of Met-derived secondary metabolites (Halkier and Gershenzon, 2006). Their structural diversity arises in large part from the variable length of their side chains. Chain elongation of Met proceeds by a repetitive three-step cycle in which the first step, the condensation of a 2-oxo acid with acetyl-CoA (Fig. 1), is catalyzed by enzymes known as MAM synthases (Kroymann et al., 2001; Falk et al., 2004; Textor et al., 2004; Benderoth et al., 2006). Based on mutant analyses, it was recently suggested that MAM3 (MAM-L) catalyzes the condensation reactions leading to long-chain glucosinolates (Field et al., 2004). However, in this study, we demonstrate that MAM3 actually carries out all six condensation reactions leading to chain elongation in planta and, thus, shapes the complete aliphatic glucosinolate profile of Arabidopsis.
MAM3 Has a Very Broad Substrate Specificity for 2-Oxo Acids
The enzymatic properties of MAM3 heterologously expressed in E. coli were generally similar to those previously described for the Arabidopsis MAM1 (Textor et al., 2004) and the MAM enzymes from other species like Eruca sativa (Falk et al., 2004), Arabidopsis lyrata, and Boechera stricta (Benderoth et al., 2006) in the pH and temperature optima and the requirement for a divalent cation. However, in contrast to MAM1, MAM3 was not activated by ATP, had no DTT requirement, and had a Km for acetyl-CoA (2.3 mm) that was nearly 10-fold higher than that of MAM1 (245 μm) and the E. sativa enzyme (340 μm). In comparing the amino acid sequences of MAM3 and MAM1 (the gene encoding the E. sativa enzyme has not yet been isolated), no ATP-binding site motifs are apparent that might explain the different effects of this nucleotide. However, there are differences between MAM3 and MAM1 in the number (11 versus 9) of Cys residues that might result in differences in the number of disulfide bridges present in the enzyme and, thus, in its DTT requirement. Currently, nothing is known about the secondary and tertiary structure of the MAM enzymes. The crystal structure of a distantly related protein, the IPMS of Mycobacterium tuberculosis, does not have any disulfide bridges, and the enzyme shows no requirement for DTT for activity (Koon et al., 2004a, 2004b).
The most striking difference between MAM3 and other MAM enzymes studied to date is the much broader substrate specificity of MAM3. This enzyme accepts all six of the ω-methylthio-2-oxo acids employed in aliphatic glucosinolate chain elongation reactions in Arabidopsis. MAM1 uses only the three shortest of this series, 4-methylthio-2-oxobutanoate, 5-methylthio-2-oxopentanoate, and, at low levels, 6-methylthio-2-oxohexanoate (Textor et al., 2004; Benderoth et al., 2006). MAM2 from the Arabidopsis accession Landsberg erecta, the E. sativa MAM, and the MAMs of other species investigated so far are even more specialized and use only the very shortest ω-methylthio-2-oxo acid, 4-methylthio-2-oxobutanoate, as a substrate (Falk et al., 2004; Benderoth et al., 2006). MAM3 also accepts a variety of other 2-oxo acid substrates showing substantial activity with pyruvate and 2-oxoisovalerate, with kcat values in the same range as for the substrates involved in Met chain elongation. Though these characteristics were determined for a recombinant protein lacking the plastid-targeting sequence and expressed heterologously in E. coli, they are likely to be representative of the characteristics of the native protein. Previous results show that the properties of recombinant MAM1 expressed in an analogous manner corresponded very closely to those of the native MAM protein purified from Arabidopsis (Textor et al., 2004).
In vitro MAM3 converts pyruvate to citramalate, a metabolite previously identified in Arabidopsis (Fiehn et al., 2000), and so this reaction may be catalyzed by MAM3 in planta as well. MAM3 also converts 2-oxoisovalerate to isopropylmalate, an important branch-point reaction of Leu biosynthesis. The IPMS activity of MAM3 is not surprising given that this protein shares over 50% sequence identity at the amino acid level with the products of the two genes shown to serve as IPMSs in this species (Kroymann et al., 2001; Field et al., 2004; de Kraker et al., 2006). However, it is not clear if MAM3 participates in Leu biosynthesis in planta. Previous work showed that MAM3 expression could complement an E. coli mutant (leuA) lacking IPMS (Junk and Mourad, 2002). However, Field et al. (2004) failed to observe this complementation. In light of these contradictory results, we repeated the experiment and did observe functional complementation in the presence of the MAM3 when expression was done at 28°C but not at 37°C. These results, together with the chloroplast localization of MAM3, provide support for the ability of this enzyme to catalyze condensation with 2-oxoisovalerate in vivo. IPMS activity in spinach (Spinacia oleracea) is reported to be localized in chloroplasts (Hagelstein and Schultz, 1993). However, because the specific activity of the IPMSs for 2-oxoisovalerate is at least 40 times higher than those of MAM3, it appears very unlikely that this enzyme is active as IPMS under normal growth conditions (de Kraker et al., 2006). Final proof of whether or not MAM3 can function in Leu formation in Arabidopsis requires further investigations with lines carrying mutations in both IPMS genes.
MAM3 Catalyzes the Formation of All Glucosinolate Chain Lengths in Vivo as Well as in Vitro
Our investigations of Arabidopsis lines with altered MAM3 or MAM1 expression levels demonstrated that the broad specificity of the MAM3 enzyme in vitro is also realized in vivo. Given that the only two other MAM-like proteins in Arabidopsis (ecotype Col-0), the IPMSs, are highly unlikely to carry out a MAM reaction in vivo (de Kraker et al., 2006), the best measure of the catalytic abilities of MAM3 in vivo is the glucosinolate profile of the MAM1 insertion mutant, gsm1-3 (Fig. 4). Although a low level of MAM1 transcript is detectable in this mutant (see Supplemental Fig. S3), the near identity of the glucosinolate phenotype with the previously described MAM1 missense mutant (Kroymann et al., 2001) suggests that it is a virtual knockout. According to the glucosinolate profile of gsm1-3, MAM3 activity results in a leaf glucosinolate profile of all chain lengths C3 to C8 dominated by C3 and C8.
The scope of MAM1 catalytic activity in vivo, on the other hand, should be observed most clearly in the MAM3 knockout mutants. We found that the T-DNA insertion line (gsm2-2) had reduced C7 and C8 glucosinolates, a finding also reported previously for another insertion line of this gene, which additionally lacked C6 (Field et al., 2004). Consistent with these results, a line (gsm2-1) with a missense mutation in MAM3 lacked any long-chain glucosinolates and this phenotype was complemented by ectopic expression of MAM3. The short chain length glucosinolates that are still present in these MAM3 mutants (with C4 being dominant just as in wild type) must therefore be produced by MAM1.
The wild-type glucosinolate profile is clearly the result of the interplay of the two MAM enzymes, with MAM3 creating the basic profile consisting of all chain lengths from C3 to C8 but dominated by C3 and C8 glucosinolates. In wild-type plants, this profile becomes modified by the actions of MAM1, which inverts the C3 to C4 ratio and thus makes C4 glucosinolates the most dominant type in Arabidopsis Col-0. It is noteworthy that a quantitative trait loci mapping approach to discovering the locus responsible for changing the C3 to C4 ratio first identified the location of MAM1 (Magrath et al., 1994; Campos de Quiros et al., 2000; Kroymann et al., 2001). Final proof of the in vivo roles of MAM1 and MAM3 would be facilitated by analysis of a line mutated in both of these genes. Unfortunately, we failed to obtain such a double mutant either from crosses between lines carrying the single mutations or from attempts to cosuppress both genes.
MAM3 Is a Major Generator of Glucosinolate Structural Diversity
The structural diversity of aliphatic glucosinolates in Arabidopsis is due in large part to the variety of chain lengths present as well as the different types of secondary modifications that occur to the side chain (Halkier and Gershenzon, 2006). The variety of chain lengths formed is a direct consequence of the nature of the elongation process in which additional methylene groups are added one at a time to the parent Met skeleton in a cyclic process so that intermediates of all chain lengths in the series C3 to C8 are made. The broad substrate specificity of MAM3, which catalyzes the chain elongation reaction of the elongation cycle, is what facilitates this stepwise elongation process and so makes the variety possible.
Broad substrate specificity is a hallmark of many enzymes of secondary metabolism and contributes to the enormous diversity of this class (e.g. Gang et al., 2002). For example, many enzymes of the core glucosinolate biosynthetic pathway have broad tolerance for various aliphatic, aromatic, and indole side chains (Piotrowski et al., 2004; Halkier and Gershenzon, 2006), although the CYP79 (Chen et al., 2003; Mikkelsen et al., 2003) and CYP83 (Naur et al., 2003) series are more class specific. This allows many different types of glucosinolates to be made with the same set of enzymes. Similar low substrate specificities are known for enzymes of alkaloid and phenylpropanoid formation (Frick and Kutchan, 1999; Gang et al., 2002). Our knowledge of the enzymology of secondary metabolism is beginning to shed light on the biochemical bases of the enormous structural diversity observed. The next challenge will be to explain the functional significance of such diversity in the life of the plant.
MATERIALS AND METHODS
Chemicals and Plant Lines
Unless specified, all chemical reagents including enzyme substrates and authentic standards for reaction products were obtained from Aldrich, Fluka, Merck, or Sigma. For the enzyme assays, [1-14C]acetyl-CoA was purchased from Amersham Biosciences or Hartmann. The following chemicals were custom synthesized as indicated: 5-methylthio-2-oxopentanoic acid, 6-methylthio-2-oxohexanoic acid, and 9-methylthio-2-oxononanoic acid (Applichem); 8-methylthio-2-oxooctanoic acid (Hochschule Zittau). Synthesis of 2-oxoheptanoic acid was described previously (Falk et al., 2004). 2-Oxononanoic acid, 2-oxodecanoic acid, 2-oxoundecanoic acid, 2-oxododecanoic acid, and 5-methyl-2-oxohexanoic acid were synthesized by a Claisen condensation between diethyl oxalate and the respective aliphatic (Cn − 1) ethyl ester, yielding a diethyl 2-alkyl-3-oxosuccinate product that was subsequently hydrolyzed and decarboxylated under reflux with hydrochloric acid (Schreiber, 1956; Nakamura et al., 1988). Experimental conditions were essentially the same as described for the synthesis of 2-oxo-heptanoic acid (Falk et al., 2004), except that the 2-oxo acid product was not isolated from the residual oil by vacuum distillation but by crystallization with potassium carbonate (Roxburgh, 1997). Hence, the reaction mixture resulting from refluxing with HCl was three times extracted with diethyl ether (3 × 50 mL) and the combined layers were dried with sodium sulfate. After removal of the solvent under reduced pressure, the residual oil was redissolved in twice its volume of diethyl ether and potassium carbonate was added slowly up to the point where no more gas bubbles were released. The supernatant was decanted and the crystals formed rinsed with ether over a Büchner funnel. The crystals were suspended in 50 mL of diethyl ether and 1 m of HCl was added slowly to accommodate gas release up to pH 3. After shaking, the ether layer was separated, dried with sodium sulfate, and evaporated to yield the pure 2-oxo acid. The products were verified by MS and NMR. Overall yields were 9% to 12%.
Authentic standards for the reaction products, hexylmalate and nonanylmalate, were synthesized from methylheptanoate and methyldecanoate, respectively, using essentially the three-step procedure previously described (Chapple et al., 1988) for the synthesis of 2-methylthioethylmalate. However, hydrolysis of the nitrile groups to yield the carboxylic groups present in the alkylmalate products was done under more severe conditions by boiling in 66% sulfuric acid under reflux for 2 h (Vogel and Furniss, 1996). The 80-mL reaction mixture was then poured into 100 mL of crushed ice and extracted with ethyl acetate (3 × 70 mL). The ethyl acetate layers were combined and the solvent removed under reduced pressure. The brown residue was dissolved in 70 mL diethyl ether and extracted with 5% aqueous sodium carbonate (3 × 50 mL). The aqueous layers were combined, acidified, and extracted with diethyl ether (3 × 50 mL). The combined ether layers were dried and after removal of the solvent, the residue was crystallized in chloroform-hexane (2:1), yielding the pure alkylmalate product in 6% to 7% overall yield. The products were verified by MS and NMR.
The Arabidopsis (Arabidopsis thaliana) lines used in this study were obtained from the Arabidopsis Biological Resource Center and include (stock no.): Col-0 (CS3879), gsm1-3 (S057539), gsm2-1 (=TU3, CS2228), and gsm2-2 (S007222).
RNA Extraction and Preparation for RT-PCR and MAM3 Cloning
Total RNA was isolated from all tissues but siliques with Trizol (Invitrogen) according to the manufacturer's instructions. Total RNA was extracted from siliques using a hot borate procedure modified from Wan and Wilkins (Wan and Wilkins, 1994).
RT reactions were done with Moloney murine leukemia virus-RT (Promega) using the reagents and instructions provided. In brief, 2 μg of total RNA was incubated with 0.5 μg of specific or dT12 to 18 oligonucleotide primers (Invitrogen) at 65°C for 5 min and cooled to 4°C. Enzyme reaction buffer and 200 units of Moloney murine leukemia virus-RT was added to the RNA-primer mix and allowed to incubate at 42°C for 1 h. The reaction mix was heated to 70°C for 10 min and stored at −80°C until use.
To isolate the MAM3 ORF, a cDNA preparation was generated by a RT reaction with RNA extracted from roots of the Col-0 wild-type accession and primed with 2MAMLb (see Supplemental Table S1). In addition, a total DNA extract of a 1- to 2-kb size-selected cDNA phage library prepared from Col-0 leaf RNA (by J. Shockey, Washington State University) was screened by PCR using Pfu DNA polymerase (Stratagene). Products were obtained using the oligonucleotide primers, 1MAMLa and 2MAMLb (see Supplemental Table S1), which include the start and stop codons, respectively, of the gene predicted at Arabidopsis Genome Initiative locus At5g23020. Both preparations yielded identical MAM3 ORFs.
To prepare MAM3 for heterologous expression in bacteria, a putative chloroplast transit peptide and cleavage site were identified in the deduced amino acid sequence of MAM3 by a neural network search algorithm (Emanuelsson et al., 2000; http://www.cbs.dtu.dk/services/ChloroP/). The oligonucleotide primers 1MAM3h and 2MAM3k (see Supplemental Table S1) were designed to generate a truncated clone of the ORF of MAM3 lacking the predicted transit peptide sequence of 51 amino acids. This product was cloned into the pBAD-vector (Invitrogen). However, only a limited amount of protein was obtained after induced expression. Thus, MAM3 was amplified by PCR from the pBAD construct with the primers MAMLex1ff and MAMLex1rv (see Supplemental Table S1). The resulting 1,359-bp fragment was cloned into the pCRT7/CT-Topo vector (Invitrogen) using the pCRT7 Topo TA cloning kit (Invitrogen). The resulting MAM3 gene construct lacked the first 153 nucleotides of the ORF corresponding to the predicted transit peptide at the N terminus and included nucleotide sequence coding for a hexahistidine peptide at the C terminus.
Overexpression, Protein Purification, Antibody Production, and Site-Directed Mutagenesis
Escherichia coli strain BL21(DE3) (Studier et al., 1990) containing the MAM3 construct was grown at 37°C on Luria-Bertani medium (Invitrogen) in the presence of ampicillin (100 μg/mL) to an optical density at 600 nm of 0.6, induced with 1 mm isopropyl-β-d-thiogalactoside for 2 h and harvested. Cells were resuspended in buffer (50 mm Tris, pH 8.0, 1 mm MgCl2) and disrupted by sonication (Bandelin Sonoplus HD2070). The extract was clarified by centrifugation (6,500g, 10 min) and the supernatant mixed with Ni-NTA agarose (Qiagen). Enzyme purification was carried out in a batch procedure. After two washing steps (sonication buffer, sonication buffer with 50 mm l-His), MAM3 was eluted from the resin with 50 mm Tris, pH 8.0, buffer containing 1 mm MgCl2 and 200 mm l-His. Neither the presence of l-His (3–30 mm in the final assay depending on dilution) nor possible Ni2+ leakage from the Ni-NTA agarose column had any influence on MAM3 activity, as determined by comparison of a portion of direct NTA eluate to a fully desalted eluate redissolved in 50 mm Tris, pH 8.0. Hence, the NTA eluate was used directly for characterization. When investigating ion dependency (including the experiments with EDTA) and determining kinetic parameters, MAM3 was purified without Mg2+ in the buffers and still retained partial activity, which could be fully restored with cation addition. Although our previous work with MAM1 and a MAM3-containing fraction showed these to be monomers (Textor et al., 2004), the recombinant enzyme was present in a partially dimeric form based on analysis by gel filtration, probably due to the poly-His tag. However, there was no difference in substrate specificity between the two forms.
For antibody production, MAM3 purified as described above was subjected to gel filtration on Superdex 200 (Amersham). MAM3 eluted as a dimer and was used to raise antibodies in rabbits. The anti-MAM3 antibody was purified from the crude antiserum by affinity chromatography with MAM3 coupled to a matrix (Davids Biotechnology).
Site-directed mutagenesis of MAM3 was performed with the QuikChange Site-Directed Mutagenesis kit (Stratagene) using the primer pair Mut5ff/rv (see Supplemental Table S1).
Condensation Reaction Enzyme Assay
The enzyme assay for the condensation reaction between acetyl-CoA and 2-oxo acids was carried out in general as previously described (Kroymann et al., 2001). The standard assay contained 100 mm Tris, pH 8.0, 4 mm MnCl2, 1 mm acetyl-CoA, 9 to 17 μm [1-14C]acetyl-CoA (50 μCi/mL, 47 mCi/mmol), 3 mm of the 2-oxoacid, and 10 to 20 μg of Ni-NTA agarose-purified enzyme in a total volume of 250 μL. Despite the variation in chain length, all substrates dissolved completely in the assay buffer. For enzyme kinetics, the assay was incubated at 32°C, the optimal temperature of the enzyme, and stopped by the addition of 750 μL ethanol.
Assay products except for those from long-chain 2-oxo acid substrates were analyzed by ion exclusion HPLC (Nucleogel ion-300 OA, Macherey and Nagel) and detected by a flow-through radioactivity monitor (Radiomatic 500TR, Packard) as described (Kroymann et al., 2001). Assays with long 2-oxo acid substrates (8-methylthio-2-oxooctanoic acid, 9-methylthio-2-oxononanoic acid, 2-oxononanoic acid, and its longer derivatives) were additionally analyzed on a reverse-phase column (SupelcosilLC-18-DB, Supelco, 25 cm × 2.1 mm, 5 μm) eluted with a mixture of 0.1% formic acid (solvent A) and acetonitrile (solvent B) at 0.25 mL/min with the following program: 0 min, 20% B; 25 min, 70% B; 26 min, 100% B; total run time 37 min.
The general enzymatic properties of MAM3 and kinetic data for acetyl-CoA were determined over a range of 0.05 to 4 mm with 2-oxo-4-methylthiobutanoic acid as the cosubstrate at a 3-mm concentration. The assays for kinetic analysis of the 2-oxo acid substrates all contained 1 mm acetyl-CoA and a variable concentration of the 2-oxo acid ranging from 0.2% to 300% of the determined Km value. Divalent cations were tested with either Cl− or SO42− as counter ions by direct addition to the enzyme assay in 4-mm concentration. All assays were conducted in the linear range with respect to time and protein concentration and were repeated at least five times per substrate. Protein was quantified according to the method of Bradford using bovine serum albumin (BSA) as a standard. Michaelis-Menten kinetic parameters were determined using the EKI3 software program (Tuebingen University), which uses a nonlinear regression method described by Wilkinson (Wilkinson, 1961).
Bacterial Complementation
E. coli CV512 (F+ leuA371; Somers et al., 1973), which is deficient in IPMS, was used for complementation studies. This strain is able to grow on M9-minimal medium with Glc as the carbon source when supplemented with casaminoacids (Difco) or transformed with a construct containing a functional IPMS. To ensure expression of T7 polymerase-driven constructs, the isopropyl-β-d-thiogalactoside-inducible T7-polymerase gene was introduced into strain CV512 by the λDE3 lysogenization kit (Novagen) giving CV512(DE3). A pET28a (Novagen) construct containing the E. coli DH5α (Hanahan, 1983) IPMS gene leuA was used for complementation studies as a positive control. The leuA gene was amplified by PCR from genomic DNA of DH5a with the primer pair IPMEff/IPMErv (see Supplemental Table S1) and cloned into pET28a using the BamHI/XhoI restriction sites provided by the primers. Complementation efficiency was tested at two different incubation temperatures, 28°C and 37°C.
Immunocytochemistry
Freshly harvested rosette leaves of Arabidopsis ecotype Col-0 were fixed in phosphate-buffered saline (135 mm NaCl, 3 mm KCl, 1.5 mm KH2PO4, 8 mm Na2HPO4) containing 4% (w/v) paraformaldehyde and 0.1% (v/v) Triton X-100, embedded in polyethylene glycol and cut as described by Hause et al. (1996). Sections of 2-μm thickness were labeled with the rabbit anti-MAM3 antibody diluted 1:500 in phosphate-buffered saline containing 1% (w/v) acetylated BSA and 4.5% (w/v) BSA. Subsequently, an anti-rabbit-IgG antibody conjugated with AlexaFluor 488 (Molecular Probes) was used according to the supplier's instructions. Standard washes followed each antibody incubation. Sections were counterstained with 0.1 μg mL−1 4,6-diamidino-2-phenylindole (Sigma) and analyzed using an Axioscop2 (Carl Zeiss) equipped with the proper filter combinations. Micrographs were taken with a CCD camera (Sony) and processed through the Photoshop 8.0.1 program (Adobe).
Extraction of Genomic DNA
Genomic DNA was extracted from expanding leaves using the abbreviated protocol of Rogers and Bendich (1985). About 5 mg of young leaf tissue was collected in a 1.5-mL microfuge tube and homogenized with 10 μL of 2× cetyltrimethylammonium bromide solution (2% cetyltrimethylammonium bromide [w/v], 100 mm Tris, pH 8.0, 20 mm EDTA, 1.4 m NaCl, 1.0% polyvinylpyrrolidone [40,000 molecular weight]) using a micropestle. The sample was incubated at 65°C for 1 to 2 min, cooled briefly on ice, and extracted with 10 μL of chloroform-isoamyl alcohol (24:1). Then, about 5 to 10 μL of water was added, and the samples were centrifuged at 11,000g. The upper phase was recovered and 0.1 to 0.5 μL was used per 20 to 100 μL PCR.
RT-PCR
cDNAs for ACT8, MAM1, and MAM3 were generated by RT reactions of 2 μg of RNA with dT12 to 18 oligonucleotide primers. For each PCR reaction, 20-μL solutions were prepared consisting of 1× PCR buffer, 0.2 mm dNTP, 0.5 μm each primer (see Supplemental Table S1 for specific primer pairs), 16 ng of template RNA, and 0.7 units of Taq Polymerase (Promega). The reactions were subjected to an initial thermal denaturation of 94°C for 2 min, followed by 28 cycles at 94°C for 30 s, 53°C for 30 s, and 72°C for 30 s, and ending with an incubation at 72°C for 2 min. Amplicon formation was determined to be linear for these reaction conditions. Reaction products were fractionated along with DNA standards (Low Mass Ladder, Invitrogen) by electrophoresis on 1% agarose gels and visualized by UV fluorescence after incubation of gels in 0.5 μg/mL ethidium bromide for 15 min. Band fluorescence was normalized with a gel documentation system (GeneGenius, Synoptics) against ACT8 product (GeneTools Analysis Software Version 3.02, Synoptics). PCRs for each RT reaction and oligonucleotide primer pair were done at least four times and each RT reaction was done in duplicate.
Cloning of MAM3 for Plant Expression and Transformation
The ORF of the MAM3 gene was cloned into pCR-Blunt II-TOPO (Invitrogen). A clone was identified that had wild-type sequence and was oriented such that the XhoI restriction site was at the 5′ end and the KpnI restriction site was at the 3′ end of the ORF. The ORF was cloned into the primary pART7 vector (Gleave, 1992) at the XhoI and KpnI sites between the 35S promoter of Cauliflower mosaic virus and the octopine synthase transcriptional terminator. The expression construct was excised and transferred to the binary vector pART27 (Gleave, 1992). The Agrobacterium tumefaciens strain GV3101 was transformed with the plasmid using spectinomycin as the selectable marker (Holsters et al., 1978). Arabidopsis plants were inoculated with the transformed A. tumefaciens using a modification of the vacuum infiltration method of Bechtoldt et al. (1993).
Glucosinolate Extraction, Identification, and Quantification
Glucosinolate extraction and purification were done as previously described (Brown et al., 2003). The desulfoglucosinolate fractions were separated by HPLC on an Agilent HP1100 system using a C-18, fully end capped, reverse-phase column (LiChrospher RP-18, 250- × 4.6-mm i.d., 5-μm particle size, Chrompack). Procedures for the identification and quantification of individual desulfoglucosinolates were described previously (Brown et al., 2003).
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers At1g49240 (AC8), At5g23010 (MAM1), At5g23020 (MAM3, former MAM-L), At1g18500 (IPMS1), and At1g74040 (IPMS2).
Supplemental Data
The following materials are available in the on-line version of this article.
Supplemental Figure S1. Chromatograms and MS data of MAM3 reaction products.
Supplemental Figure S2. Western blot showing the specificity of anti-MAM3.
Supplemental Figure S3. Semiquantitative RT-PCR of steady-state transcript levels for genes AC8, MAM1, and MAM3 in the leaves of wild-type Col-0 (wt) and the mutant line gsm1-3.
Supplemental Table S1. Oligonucleotide primers used in this study.
Supplementary Material
Acknowledgments
We thank Nadine Gerth and Katrin Luck for technical assistance, Stefan Bartram for advice on substrate synthesis, and Ales Svatos for MS of the reaction products.
This work was supported by the German National Science Foundation (grant no. GE 1126/1–3), by the Max Planck Society, and by Virginia Polytechnic Institute.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jonathan Gershenzon (gershenzon@ice.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bechtold N, Ellis J, Pelletier G (1993) In-planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316 1194–1199 [Google Scholar]
- Benderoth M, Textor S, Windsor AJ, Mitchell-Olds T, Gershenzon J, Kroymann J (2006) Positive selection driving diversification in plant secondary metabolism. Proc Natl Acad Sci USA 103 9118–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62 471–481 [DOI] [PubMed] [Google Scholar]
- Campos de Quiros H, Magrath R, McCallum D, Kroymann J, Schnabelrauch D, Mitchell-Olds T, Richard M (2000) α-Keto acid elongation and glucosinolate biosynthesis in Arabidopsis thaliana. Theor Appl Genet 101 429–437 [Google Scholar]
- Chapple CCS, Decicco C, Ellis BE (1988) Biosynthesis of 2-(2′-methylthio)ethylmalate in Brassica carinata. Phytochemistry 27 3461–3463 [Google Scholar]
- Chen S, Glawischnig E, Jorgensen K, Naur P, Jorgensen B, Olsen CE, Hansen CH, Rasmussen H, Pickett JA, Halkier BA (2003) CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J 33 923–937 [DOI] [PubMed] [Google Scholar]
- Chisholm MD, Wetter LR (1964) Biosynthesis of mustard oil glucosides. IV. The administration of methionine-C14 and related compounds to horseradish. Can J Biochem 42 1033–1040 [DOI] [PubMed] [Google Scholar]
- D'Auria JC, Gershenzon J (2005) The secondary metabolism of Arabidopsis thaliana: growing like a weed. Curr Opin Plant Biol 8 308–316 [DOI] [PubMed] [Google Scholar]
- de Kraker JW, Luck K, Textor S, Tokuhisa JG, Gershenzon J (2006) Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol 143 970–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, Von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P (2001) The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56 5–51 [DOI] [PubMed] [Google Scholar]
- Falk KL, Vogel C, Textor S, Bartram S, Hick A, Pickett JA, Gershenzon J (2004) Glucosinolate biosynthesis: demonstration and characterization of the condensing enzyme of the chain elongation cycle in Eruca sativa. Phytochemistry 65 1073–1084 [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dormann P, Altmann T, Trethewey RN, Willmitzer L (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18 1157–1161 [DOI] [PubMed] [Google Scholar]
- Field B, Cardon G, Traka M, Botterman J, Vancanneyt G, Mithen R (2004) Glucosinolate and amino acid biosynthesis in Arabidopsis. Plant Physiol 135 828–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick S, Kutchan TM (1999) Molecular cloning and functional expression of O-methyltransferases common to isoquinoline alkaloid and phenylpropanoid biosynthesis. Plant J 17 329–339 [DOI] [PubMed] [Google Scholar]
- Gang DR, Beuerle T, Ullmann P, Werck-Reichhart D, Pichersky E (2002) Differential production of meta hydroxylated phenylpropanoids in sweet basil peltate glandular trichomes and leaves is controlled by the activities of specific acyltransferases and hydroxylases. Plant Physiol 130 1536–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20 1203–1207 [DOI] [PubMed] [Google Scholar]
- Graser G, Schneider B, Oldham NJ, Gershenzon J (2000) The methionine chain elongation pathway in the biosynthesis of glucosinolates in Eruca sativa (Brassicaceae). Arch Biochem Biophys 378 411–419 [DOI] [PubMed] [Google Scholar]
- Hagelstein P, Schultz G (1993) Leucine synthesis in spinach chloroplasts: partial characterization of 2-isopropylmalate synthase. Biol Chem Hoppe Seyler 374 1105–1108 [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57 303–333 [DOI] [PubMed] [Google Scholar]
- Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166 557–580 [DOI] [PubMed] [Google Scholar]
- Haughn GW, Davin L, Giblin M, Underhill EW (1991) Biochemical genetics of plant secondary metabolites in Arabidopsis thaliana: the glucosinolates. Plant Physiol 97 217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Demus U, Teichmann C, Parthier B, Wasternack C (1996) Developmental and tissue-specific expression of JIP-23, a jasmonate-inducible protein of barley. Plant Cell Physiol 37 641–649 [DOI] [PubMed] [Google Scholar]
- Holsters M, de Waele D, Depicker A, Messens E, Van Montagu M, Schell J (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163 181–187 [DOI] [PubMed] [Google Scholar]
- Junk DJ, Mourad GS (2002) Isolation and expression analysis of the isopropylmalate synthase gene family of Arabidopsis thaliana. J Exp Bot 53 2452–2454 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Gershenzon J, Mitchell-Olds T (2001) Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T (2001) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, Schnee C, Gershenzon J, Degenhardt J (2004) The variability of sesquiterpenes emitted from two Zea mays cultivars is controlled by allelic variation of two terpene synthase genes encoding stereoselective multiple product enzymes. Plant Cell 16 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koon N, Squire CJ, Baker EN (2004. a) Crystal structure of LeuA from Mycobacterium tuberculosis, a key enzyme in leucine biosynthesis. Proc Natl Acad Sci USA 101 8295–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koon N, Squire CJ, Baker EN (2004. b) Crystallization and preliminary X-ray analysis of alpha-isopropylmalate synthase from Mycobacterium tuberculosis. Acta Crystallogr D Biol Crystallogr 60 1167–1169 [DOI] [PubMed] [Google Scholar]
- Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T (2003) Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci USA 100 14587–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T (2001) A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol 127 1077–1088 [PMC free article] [PubMed] [Google Scholar]
- Lee CJ, Serif GS (1968) 2-Amino-6-(methylthio)caproic acid, a methionine homolog and precursor of progoitrin. Biochim Biophys Acta 165 569–571 [PubMed] [Google Scholar]
- Lee CJ, Serif GS (1970) Precursor role of [14C, 15N]-2-amino-6-(methylthio)caproic acid in progoitrin biosynthesis. Biochemistry 9 2068–2071 [DOI] [PubMed] [Google Scholar]
- Magrath R, Bano F, Morgner M, Parkin I, Sharpe A, Lister C, Dean C, Turner J, Lydiate D, Mithen R (1994) Genetics of aliphatic glucosinolates. I. Side chain elongation in Brassica napus and Arabidopsis thaliana. Heredity 72 290–299 [Google Scholar]
- Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA (2003) Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol 131 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Inoue K, Ushio K, Oka S, Ohno S (1988) Stereochemical control on yeast reduction of α-keto esters: reduction by immobilized bakers' yeast in hexane. J Org Chem 53 2589–2593 [Google Scholar]
- Naur P, Petersen BL, Mikkelsen MD, Bak S, Rasmussen H, Olsen CE, Halkier BA (2003) CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol 133 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M, Schemenewitz A, Lopukhina A, Muller A, Janowitz T, Weiler EW, Oecking C (2004) Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J Biol Chem 279 50717–50725 [DOI] [PubMed] [Google Scholar]
- Reichelt M, Brown PD, Schneider B, Oldham NJ, Stauber EJ, Tokuhisa J, Kliebenstein DJ, Mitchell-Olds T, Gershenzon J (2002) Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 59 663–671 [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5 69–76 [DOI] [PubMed] [Google Scholar]
- Roxburgh CJ (1997) Protocol for purifying α-ketocarboxylic acids. Aldrichim Acta 30 74 [Google Scholar]
- Schreiber J (1956) D'une methode de preparation générale des acides α cétoniques aliphatiques. B Soc Chim Fr 10 1361–1363 [Google Scholar]
- Schuster J, Knill T, Reichelt M, Gershenzon J, Binder S (2006) Branched-chain aminotransferase4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 18 2664–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab W (2003) Metabolome diversity: too few genes, too many metabolites? Phytochemistry 62 837–849 [DOI] [PubMed] [Google Scholar]
- Somers JM, Amzallag A, Middleton RB (1973) Genetic fine structure of the leucine operon of Escherichia coli K-12. J Bacteriol 118 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorf JW (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185 60–89 [DOI] [PubMed] [Google Scholar]
- Textor S, Bartram S, Kroymann J, Falk KL, Hick A, Pickett JA, Gershenzon J (2004) Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain elongation cycle. Planta 218 1026–1035 [DOI] [PubMed] [Google Scholar]
- Tholl D, Chen F, Petri J, Gershenzon J, Pichersky E (2005) Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J 42 757–771 [DOI] [PubMed] [Google Scholar]
- Tokuhisa J, de Kraker J-W, Textor S, Gershenzon J (2004) The biochemical and molecular origins of aliphatic glucosinolate diversity in Arabidopsis thaliana. In JT Romeo, ed, Secondary Metabolism in Model Systems, Recent Advances in Phytochemistry, Vol 38. Elsevier Science, Amsterdam, pp 19–38
- Vogel AI, Furniss BS (1996) Vogel's Textbook of Practical Organic Chemistry. Longman Press, Harlow, UK
- Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223 7–12 [DOI] [PubMed] [Google Scholar]
- Wilkinson GN (1961) Statistical estimations in enzyme kinetics. Biochem J 80 324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink M (2003) Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 64 3–19 [DOI] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7 263–270 [DOI] [PubMed] [Google Scholar]
- Wittstock U, Kliebenstein DJ, Lambrix V, Reichelt M, Gershenzon J (2003) Glucosinolate hydrolysis and its impact on generalist and specialist insect herbivores. In JT Romeo, ed, Integrative Phytochemistry: from Ethnobotany to Molecular Ecology, Recent Advances in Phytochemistry, Vol 37. Elsevier Science, Amsterdam, pp 101–125
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.