Abstract
YABBY and WUSCHEL-LIKE HOMEOBOX (WOX) genes have been shown to play important roles in lateral organ formation and meristem function. Here, we report the characterization of functional relationship between rice (Oryza sativa) YAB3 and WOX3 in rice leaf development. Rice YAB3 is closely related to maize (Zea mays) ZmYAB14 and Arabidopsis (Arabidopsis thaliana) FILAMENTOUS FLOWER (FIL), whereas rice WOX3 is highly conserved with maize narrow sheath1 (NS1) and NS2 and Arabidopsis PRESSED FLOWER (PRS). In situ hybridization experiments revealed that the expression of both genes was excluded from the shoot apical meristem, but the transcripts were detected in leaf primordia, young leaves, and reproductive organs without any polar distribution. The function of the two genes was studied by both overexpression and RNA interference (RNAi) in transgenic rice. YAB3 RNAi induced twisted and knotted leaves lacking specialized structures such as ligule and auricles, while no phenotypic change was observed in YAB3 overexpression plants, suggesting that rice YAB3 may be required for leaf cell growth and differentiation. Overexpression of WOX3 repressed YAB3 and showed a YAB3 RNAi phenotype. The expression of class I KNOTTED-LIKE HOMEOBOX (KNOX) genes was ectopically induced in leaves of YAB3 RNAi or WOX3 overexpression plants. Data from inducible WOX3 expression and DNA-protein interaction assays suggested that WOX3 acted as a transcriptional repressor of YAB3. These data reveal a regulatory network involving YAB3, WOX3, and KNOX genes required for rice leaf development.
Most plant organs are formed during the postembryonic stages from the meristems. The shoot apical meristems (SAMs) are organized pools of undifferentiated or embryonic cells maintained by a dynamic balance between cell division and differentiation. The meristematic identity of cells in the SAM is correlated with the expression of specific regulatory genes. During the formation of leaf organ primordia, cells at the flanks of the SAM undergo a fundamental developmental transition from an indeterminate to a determinate cell fate as they are recruited into leaves. For the SAM to maintain its indeterminate function throughout the life of a plant, it is essential that cells recruited into lateral organs be constantly replenished. A growing list of transcription factor proteins are required to maintain, and possibly to establish, the SAM. KNOTTED1 (KN1) defines the first homeobox gene family to be isolated in plants and was identified from maize (Zea mays) gain-of-function mutants that produced knots, or outgrowths, of indeterminate tissue on the leaf (Vollbrecht et al., 1991). KN1-like homeobox (KNOX) genes that share the highest degree of sequence similarity with KN1 are expressed in overlapping domains within the SAMs of both monocot and dicot plants (for review, see Reiser et al., 2000; Hay et al., 2004). Loss-of-function mutation of KN1 in maize or of the closely related gene SHOOTMERISTEMLESS in Arabidopsis (Arabidopsis thaliana) results in failure to maintain a SAM (Long et al., 1996; Vollbrecht et al., 2000). Down-regulation of KNOX expression in leaf founder cells within the meristem marks a change in cell fate from meristem to leaf (Smith et al., 1992). Exclusion of KNOX expression from leaves is important for leaf development, as ectopic expression confers indeterminate features to Arabidopsis leaves, including ectopic meristems and a dramatic change in leaf shape (Chuck et al., 1996). At least two mechanisms exist to repress KNOX expression in Arabidopsis leaves. The myb transcription factor ASYMMETRIC LEAVES1 and ASYMMETRIC LEAVES2, a member of the LATERAL ORGAN BOUNDARIES family of transcriptional regulators, act together to repress expression of KNOX genes in the leaf (Byrne et al., 2000; Ori et al., 2000). Activity of the YABBY family of putative transcription factors also contributes to the exclusion of KNOX gene expression from Arabidopsis leaves (Kumaran et al., 2002).
The YABBY family transcription factors are characterized by a C2C2 zinc finger domain toward the amino terminus and a putative helix-loop-helix domain, also named YABBY, conserved in high mobility group transcription factors toward the carboxyl terminus (Bowman and Smyth, 1999; Golz and Hudson, 1999). There are at least six YABBY genes in Arabidopsis (FILAMENTOUS FLOWER [FIL] or YAB1, YAB2, YAB3; INNER NO OUTER [INO] or YAB4, YAB5; and CRABS CLAW [CRC]). All of these genes show a polar expression pattern and function to determine the abaxial cell fate of one or more above-ground lateral organs (Bowman, 2000). In addition to the repressive function of KNOX gene expression, Arabidopsis YABBY genes are mainly involved in the control of abaxial identity of lateral organs (Eshed et al., 1999; Siegfried et al., 1999). In contrast to Arabidopsis YABBY genes, YABBY gene family members that are closely related to FIL (ZmYAB9, ZmYAB14) are expressed on the adaxial side of incipient and developing leaf primordia (Juarez et al., 2004). These observations suggest that YABBY genes in monocots may have different functions in lateral organ formation compared to their homologs in Arabidopsis.
In rice (Oryza sativa), there are also at least six members in the YABBY gene family (Jang et al., 2004). Recently, the rice YAB1 has been shown to play a role in stamen and carpel development (Jang et al., 2004). The CRC-related rice YABBY gene DROOPING LEAF (DL) whose mutation induces a dl phenotype is required for carpel specification and leaf midrib development (Yamaguchi et al., 2004). In contrast to Arabidopsis and maize YABBY members, rice YAB1 and DL do not show any adaxial/abaxial polar expression pattern in lateral organs. Overexpression or mutation of these genes results in no adaxial/abaxial polarity change in lateral organs (Jang et al., 2004; Yamaguchi et al., 2004; Dai et al., 2007), suggesting at least these two rice YABBY members have different function from their Arabidopsis or maize homologs. However, the function of FIL or ZmYAB14/9 closely related rice YABBY genes was not known.
Arabidopsis WUSCHEL (WUS) gene functions in a more restricted set of cells in the SAMs to promote stem cell fate and is regulated by CLAVATA genes in a negative feedback loop (Mayer et al., 1998). The WUS and WUS-like homeobox genes (WOX) contain a WUS box toward the amino terminus in addition to the homeobox toward the carboxyl-terminus end. It has been suggested that Arabidopsis WOX genes play an important role in region-specific transcription programs early during embryogenesis and lateral organ development (Haecker et al., 2004). For instance, STIMPY/WOX9 is required for the growth of vegetative SAM in Arabidopsis (Wu et al., 2005), while PRESSED FLOWER (PRS/WOX3) is involved in the regulation of lateral axis-dependent sepal development as well as leaf stipules (Matsumoto and Okada, 2001; Nardmann et al., 2004). The maize duplicated narrow sheath1 (NS1) and NS2 genes, which are the immediate homologs of PRS, are suggested to function during recruitment of organ founder cells in a lateral domain of the SAM (Nardmann et al., 2004). Rice genome contains at least nine homologs of WOX genes, among which quiescent center-specific homeobox (QHB) has been found to function in root development (Kamiya et al., 2003).
To study transcriptional regulatory networks controlling rice shoot development, we underwent a research program to systematically study the expression and functional relationship of members of rice YABBY, WOX, and other putative transcription factor gene families in controlling shoot/leaf development. In this report, we present data on the expression and functional analysis of a rice YABBY gene (YAB3), which is closely related to ZmYAB14/9 and to FIL, and a NS1/2/PRS-related rice WOX gene (WOX3). We show that both genes are coexpressed in most lateral organ primordia and young leaves without adaxial/abaxial polarity. Down-regulation of YAB3 or overexpression of WOX3 induced knotted outgrowth of leaves that lack clear separation between the leaf sheath and the leaf blade, along with ectopic expression of KNOX genes in leaves of the transgenic plants. This phenotype suggests that rice YAB3 is required for cell differentiation during leaf development. Expression and DNA-binding studies suggest that WOX3 functions as a transcriptional repressor of YAB3. Together, our data reveal a transcriptional regulatory hierarchy required for rice leaf development.
RESULTS
YAB3 Was Expressed in the Leaf and Floral Organ Primordia
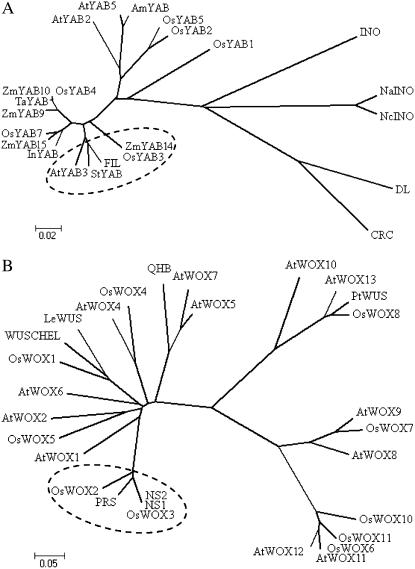
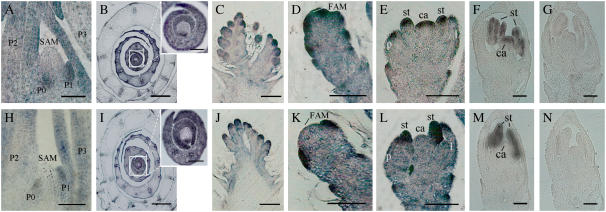
Phylogenic analysis of YABBY proteins showed that rice YAB3 was highly conserved with ZmYAB14 (Juarez et al., 2004). The monocot proteins were mostly closely related to FIL among the Arabidopsis YABBY members (Fig. 1A). In situ hybridization experiments with a gene-specific region of the rice YAB3 cDNA as the probe revealed that the YAB3 transcripts were detected in leaf primordia and in young leaves but excluded from the SAM. In the young leaves, YAB3 transcripts were homogenously distributed (Fig. 2). During panicle development, YAB3 transcripts were detected initially in the floret meristems and then in the primordia of the floral organs, including palea, lemma, stamens, and carpel, and later were restricted to the stamens and carpel. Unlike Arabidopsis YABBY genes and maize ZmYAB14/9, rice YAB3 did not show any polar expression patterns in the floral organs. This expression pattern suggested that rice YAB3 may have a different function.
Figure 1.
Phylogeny analysis of YABBY and WOX families. Neighbor-joining trees of YABBY (A) and WOX (B) proteins from rice (Os), Arabidopsis (At), Antirrhinum majus (Am), maize (Zm), Triticum aestivum (Ta), Solanum tuberosum (St), Ipomoea nil (In), Nymphaea alba (Na), Nymphaea colorata (Nc), and Solanum lycopersicum (Le). GenBank accession numbers are shown in Table I.
Figure 2.
In situ detection of YAB3 and WOX3 transcripts. A to G, Sections hybridized with YAB3 antisense (A–F) or sense (G) probes. H to N, Sections hybridized with WOX3 antisense (H–M) or sense (N) probes. A and H, Longitudinal sections of SAM. B and I, Transverse sections of SAM; insets, enlarged views of the central areas of the apexes. C and J, Developing panicle longitudinal sections. D to F and K to M, Longitudinal sections of florets at different development stages. The midrib regions of leaf primordia (A and H) are designated by plastochron (P) numbers, such that the leaf incipient primordium is labeled P0, the next older leaf is labeled P1, and so on. FAM, Floral apical meristem; st, stamen; ca, carpel; l, lemma; p, palea. Bar = 100 μm.
YAB3 Was Nucleus Localized
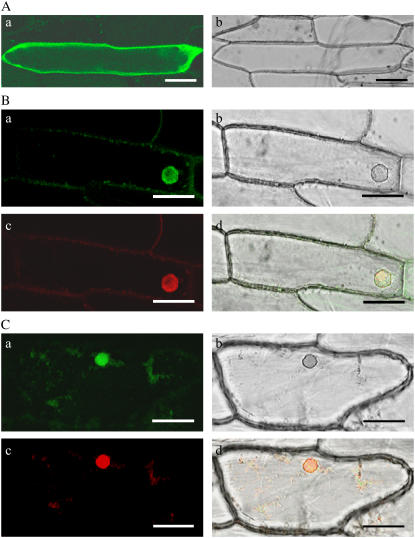
To study the subcellular localization of YAB3, the full-length YAB3 coding sequence was translationally fused to the GFP-coding sequence under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The GFP alone controlled by the 35S promoter served as a control. The constructs were introduced into the onion (Allium cepa) cells using the particle bombardment method. The GFP expression was examined with confocal microscopy 36 h after bombardment. GFP alone was located throughout the cells (Fig. 3A), while the YAB3-GFP fusion protein was localized in the nuclei of the onion cells (Fig. 3B).
Figure 3.
Nuclear localization of YAB3 and WOX3 GFP alone was localized in cytosol of onion skin cells (A). YAB3-GFP (B) and WOX3-GFP (C) are nucleus localized. a, GFP images. b, Transmission images. c, DAPI staining. d, Merged images. Bar = 50 μm.
Down-Regulation of YAB3 Induced Defects in Leaf and Flower Development
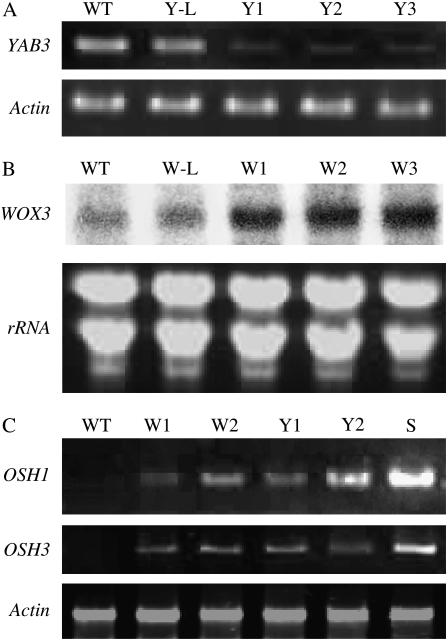
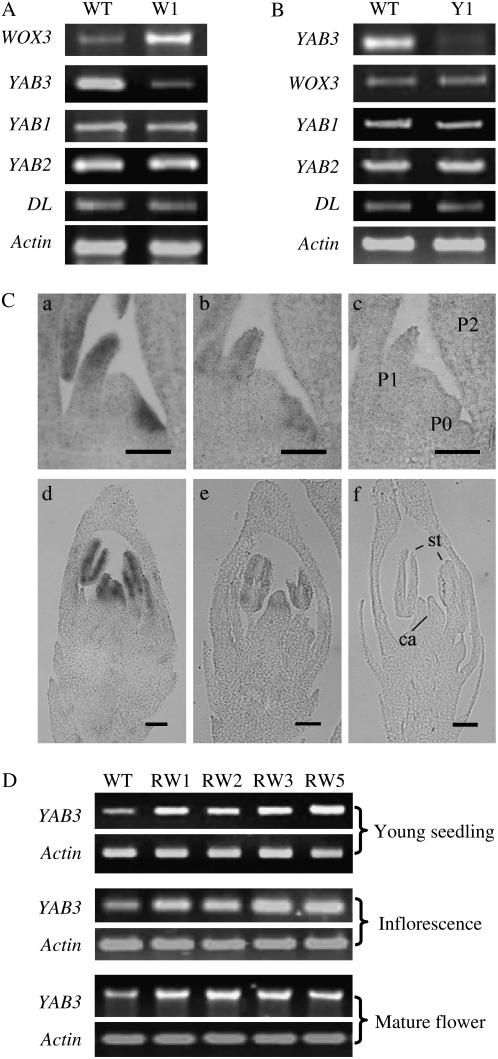
To study the developmental function of YAB3, we used a gene-specific region from the 3′ end of the cDNA (see “Materials and Methods”) to construct a double-strand RNA producing or RNA interference (RNAi) vector. The construct was introduced into ‘Zhonghua 11’ by Agrobacterium-mediated transformation. More than 30 independent T0 transgenic plants were produced for each construct. Most of the transgenic plants produced aberrant twisted leaves displaying knotted outgrowth. The specialized rice leaf structures such as ligule, lamina joint, and auricles that are located at the junction between the leaf blade and the leaf sheath in the wild type were absent in the phenotypic transgenic plants (Fig. 4). Examination of cross sections revealed no alteration of the leaf adaxial/abaxial polarity (Supplemental Fig. S1). All the phenotypic plants showed a decrease of YAB3 transcripts (Fig. 5A).
Figure 4.
Shoot phenotypes induced by YAB3 RNAi or by WOX3 overexpression. A, Wild-type leaf at abaxial (a) and adaxial (b) views showing the structures of ligule (l), auricle (au), and lamina joint (lj). bl, Blade; s, sheath. B, Phenotypes in YAB3 RNAi (a–d) and WOX3 (e–h) overexpressing plants. a and e, Profile of the transgenic plants. b and f, Enlarged view of the parts with twisted leaves in transgenic plants. Insets, junctions between leaf blades and sheaths with ligules and auricles missing, indicated by triangles. c and g, Knots in the lower parts of some transgenic leaves; arrows indicate the position of the knots. d and h, Knots in the upper parts of transgenic leaves, arrows indicate the position of the knots. The knotted regions in c, d, g, and h are enlarged (insets).
Figure 5.
Expression analysis of YAB3, WOX3, and KNOX genes in YAB3 RNAi and WOX3 overexpression plants. A, YAB3 transcript levels revealed by RT-PCR in wild-type (WT), a nonphenotypic (Y-L), and three phenotypic (Y1 to Y3) YAB3 RNAi transgenic plants. B, WOX3 transcript levels revealed by northern blots in wild-type (WT), a nonphenotypic (W-L), and three phenotypic (W1–W3) WOX3 overexpression transgenic plants. C, Transcript levels of rice KNOX genes OSH1 and OSH3 revealed by RT-PCR in leaves from wild-type (WT), phenotypic YAB3 RNAi (Y1 and Y2), and WOX3 overexpression plants (W1 and W2). Shoot apex mRNA was used as a positive control (S).
The twisted and knotted YAB3 RNAi leaf phenotype was reminiscent of the knotted leaves in kn1 mutants in maize, which is a dominant mutation causing ectopic expression of KN1 in leaves. In addition, overexpression of rice class I KNOX genes also induces knots on leaves and diffusion of the blade-sheath boundary (Sentoku et al., 2000). Reverse transcription (RT)-PCR analysis showed that two tested rice KNOX genes (OSH1 and OSH3) were indeed ectopically expressed in leaves of YAB3 RNAi plants (Fig. 5C), suggesting that YAB3 had a function to repress KNOX genes in rice leaves.
The panicle development seemed normal. However, most florets were degenerated, but a few florets remained normal and eventually gave rise to seeds. Further investigation of the severely degenerated florets revealed that there was no recognizable anther structure and that ovules seemed not to exist (Supplemental Fig. S2). Florets with a less severe phenotype had smaller anthers than the wild type, which produced very few mature pollens (Supplemental Fig. S2).
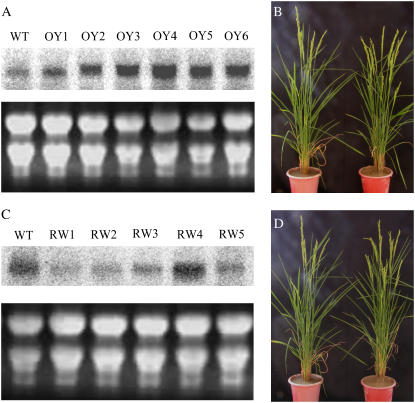
To study whether the overexpression of YAB3 had any effect on rice growth, we made a transgenic plant with a construct in which the maize ubiquitin promoter was used to direct the expression of the YAB3 cDNA. Among more than 40 transgenic lines obtained, no visible phenotype was observed. Northern-blot analysis showed most tested transgenic plants overexpressed YAB3 (Fig. 6A).
Figure 6.
Analysis of YAB3 overexpression and WOX3 RNAi plants. A, Expression analysis of YAB3 in wild-type (WT) and five independent YAB3 overexpression transgenic lines (OY1–OY6). B, Comparison of wild type (left) to the YAB1 overexpression transgenic line OY6 (right). C, Expression analysis of WOX3 in wild-type (WT) and five independent WOX3 RNAi transgenic plants (RW1–RW5). D, No phenotypic change observed in RW1 (right) compared to wild type (left). [See online article for color version of this figure.]
WOX3 Expression Pattern Overlapped with That of YAB3
Sequence analysis showed that rice WOX3 was highly conserved with maize NS1/NS2 genes that have been suggested to be orthologs of Arabidopsis PRS (Nardmann et al., 2004; Fig. 1). Both NS1/NS2 and PRS are expressed in meristematic foci and in the margins of lateral organ primordia. However, in situ hybridization experiments revealed that WOX3 had a different expression pattern than NS1/NS2 and PRS, which was actually similar to that of YAB3 in both vegetative shoots and in developing panicles, except in the carpel primordium where WOX3 transcripts were not detected (Fig. 2). WOX3 protein was also targeted into the nucleus of transiently transfected onion cells (Fig. 3C).
Overexpression of WOX3 Repressed YAB3
Similarly, we intended to study the developmental function of WOX3 by the transgenic approaches. Both overexpression and RNAi transgenic plants were obtained. Most of the WOX3 overexpression plants showed a similar leaf phenotype as the YAB3 RNAi plants (Fig. 4B). The WOX3 overexpression plants also showed ectopic expression of KNOX genes OSH1 and OHS3 (Fig. 5). However, no phenotypic alteration was observed in the WOX RNAi populations (Fig. 6). The phenotypic resemblance induced by the two transgenes suggested that there might be a regulatory relationship between WOX3 and YAB3. RT-PCR revealed that the expression of YAB3 was repressed in leaves of the WOX3 overexpression plants (Fig. 7A). The repression seemed to be specific to YAB3, because the expression of YAB1, YAB2, or DL was not altered (Fig. 7A). Down-regulation of YAB3 by RNAi had no effect on the expression of WOX3 or other YABBY genes (Fig. 7B). In situ hybridization experiments showed that overexpression of WOX3 reduced the YAB3 transcript levels in the shoot apexes and florets (Fig. 7C). Conversely, down-regulation of WOX3 by RNAi induced the expression of YAB3 in seedlings, inflorescences, and florets (Fig. 7D). However, there was no induction of YAB3 in flag (mature) leaves where YAB3 was not expressed (data not shown), suggesting that the silencing of YAB3 in mature leaves involves additional mechanisms.
Figure 7.
Expression analysis of WOX3 and YAB3 in the transgenic and wild-type plants. A, Semiquantitative RT-PCR analysis of four rice YABBY gene expression in WOX3 overexpression line W1 compared to wild type. Rice actin1 transcripts were measured as controls. B, Semiquantitative RT-PCR analysis of WOX3 and the YABBY genes in the YAB3 RNAi line Y1 compared to wild type. Rice actin1 transcripts were used as control. C, In situ hybridization to detect YAB3 transcripts in wild type (a and d) and WOX3 overexpression (b and e) shoot apexes (top) and florets (bottom). A shoot apex and a floret hybridized with a sense probe of YAB3 were used as controls (c and f). D, Expression analysis of YAB3 in wild-type and WOX3 RNAi 1-week-old seedlings, inflorescences, and florets. Rice actin1 transcripts were measured and used as controls.
WOX3 Was Involved in Direct Repression of YAB3
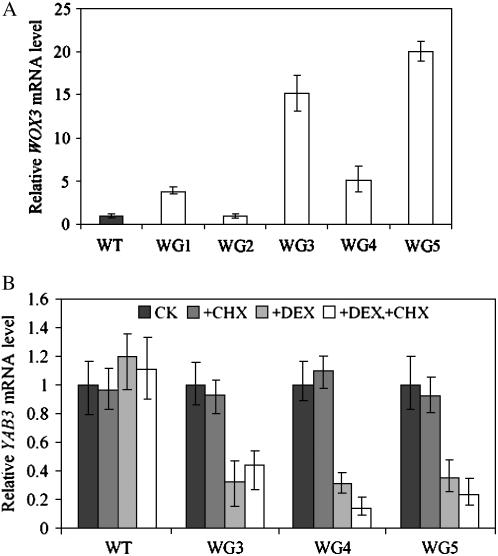
To confirm whether WOX3 could directly repress the expression of YAB3, we used the glucocorticoid receptor (GR)-inducible system to artificially regulate the activity of WOX3 in vivo. The translational stop codon of WOX3 was removed and replaced with a 288-amino acid segment of the mouse GR that contains the ligand-binding domain. The chimeric protein under the control of the double enhancers of the CaMV 35S promoter was introduced into rice plants by Agrobacterium-mediated transformation.
RT-PCR analysis showed that several analyzed transgenic plants overproduced the WOX3-GR mRNA (Fig. 8A). Three of the transgenic lines (WG3–WG5) were selected for further analysis. Siblings from each transgenic line were divided into three groups: one was treated with dexamethasone (DEX), the second was treated first with cycloheximide (CHX) for 1 h then with DEX, and the third had no treatment, together with the wild-type plants treated similarly. Leaves were harvested for RNA extraction and analyzed by quantitative real-time PCR to examine the transcript levels of YAB3. The results showed that the DEX treatment induced the repression of YAB3 by 2- to 5-fold in the presence or absence of CHX, while the expression of YAB3 in wild-type plants was not significantly affected by the treatments (Fig. 8B). These data indicated that the WOX3 was directly involved in the repression of YAB3 transcription.
Figure 8.
Activation of WOX3 directly repressed the expression of YAB3. A, Identification of transgenic lines expressing the WOX3-GR fusion by real-time RT-PCR. B, Relative expression levels of YAB3 in wild-type and three WOX3-GR transgenic plants treated with or without DEX and/or CHX. The YAB3 transcript levels were normalized with the actin1 mRNA levels, and that of the wild-type control was assessed as 1. Bars are sd ± three biological repeats.
DISCUSSION
Function of YABBY Genes in Rice Leaf Development
In Arabidopsis, the function of YABBY genes has been well studied and shown to promote abaxial cell fate in the lateral organs (Bowman, 2000; Sieber et al., 2004; Lee et al., 2005; Meister et al., 2005). Arabidopsis YABBY gene transcripts are detectable in the abaxial domains of lateral organs when primordia emerge and begin to differentiate from the meristem (Bowman, 2000). However, maize YABBY members ZmYAB14 and ZmYAB9 are shown to be expressed in the adaxial domain of leaves (Juarez et al., 2004). It is suggested that the maize YABBY genes play a role in lateral leaf outgrowth rather than specifying adaxial cell fate (Juarez et al., 2004). However, no loss-of-function phenotype of these genes is available to support the hypothesis. In this work, we report that the expression of rice YAB3 that is closely related to ZmYAB14 (Fig. 1A) did not show such an adaxial/abaxial polar expression pattern in leaves or reproductive organs (Fig. 2). Other studied rice YABBY genes including YAB1 and DL, which are closely related to Arabidopsis YAB2 and CRC, respectively, also have no polar expression in lateral organs (Jang et al., 2004; Yamaguchi et al., 2004; Dai et al., 2007). Likely, the function of rice YABBY genes in regulating leaf development may differ from its Arabidopsis and maize homologs. Analysis of RNAi transgenic plants revealed that rice YAB3 had a function in leaf development. The YAB3 RNAi plants do not show any abaxial-adaxial polarity change (Supplemental Fig. S1) but produce twisted and knotted leaves lacking differentiated tissues such as ligule, auricles, and distinction of the leaf sheath from the leaf blade, while the width of the leaf sheath and blade was not reduced (Fig. 4). The phenotypic alterations suggest that YAB3 is required for the promotion of differentiation during leaf organogenesis. This correlates with ectopic induction of class I KNOX genes OSH1 and OSH3 in YAB3 RNAi leaves (Fig. 5), suggesting that the phenotype may be induced by the KNOX ectopic expression, as it has been shown that overexpression of rice KNOX genes induces similar phenotypes (Sentoku et al., 2000). These results also suggest that YAB3 contributes to the exclusion of KNOX expression from the leaf tissues, as do Arabidopsis FIL and YAB3. This aspect of YABBY function seems to be conserved between rice YAB3 and the Arabidopsis homologs. Loss-of-function mutation of another rice YABBY gene DL does not produce the YAB3 RNAi phenotype (Yamaguchi et al., 2004), suggesting that the two rice YABBY genes have a distinct function in the regulation of KNOX gene expression and leaf development. In addition, overexpression of YAB3 does not induce any morphological alteration (Fig. 6), which differs also from other rice YABBY genes. For instance, overexpression of rice YAB1 induces defects in floral organ development (Jang et al., 2004), while DL ectopic expression plants produce curled leaf blades, forming a cylinder-like structure (Yamaguchi et al., 2004). These observations reinforce the suggestion that rice YABBY genes have distinct functions and that there is not much functional redundancy between them.
Rice WOX3 Developmental Function
Rice WOX3 is situated in the same branch as maize NS1/NS2 genes in the phylogeny tree, which are the immediate homologs of Arabidopsis PRS. PRS is expressed at the lateral regions of lateral organs at very early stages (Matsumoto and Okada, 2001). The expression of NS1/NS2 is also detected in meristematic foci and in the margins of lateral organ primordia (Nardmann et al., 2004). Loss-of-function mutations in the maize and Arabidopsis genes induce defects or deletion in leaf and sepal, respectively. Overexpression of PRS produced multicellular bulges with trichomes on the stem and on the peduncle (Matsumoto and Okada, 2001). It is suggested that there is a conserved function between NS and PRS during the recruitment of lateral founder cells from shoot meristems. During the formation of a maize leaf, founder cell recruitment begins on one flank of the SAM, which will form the central domain of the leaf, and proceeds toward the opposite flank, during which the function of NS/PRS initiates from lateral foci and recruits founder cells in a lateral meristematic domain (Nardmann et al., 2004). However, the rice WOX3 transcripts were detected in relatively larger areas in leaf primordia, young leaves, and floral meristems (Fig. 2). This suggests that the function of rice WOX3 might not be conserved with NS1/NS2 or PRS in leaf development. The transgenic phenotypes support this hypothesis, as overexpression of WOX3 induced deformed leaves in rice, while down-regulation of rice WOX3 produced no visible phenotype, despite an induction of YAB3 (Figs. 4 and 6). In addition, leaf founder cell recruitment is correlated with the repression of KNOX genes in simple leaf plants. Higher NS1/NS2/PRS activity would repress KNOX expression. However, rice WOX3 overexpression induced ectopic expression of KNOX genes in transgenic leaves. This reinforces the hypothesis that rice WOX3 is not involved in lateral founder cell recruitment during leaf formation.
No phenotype induced by down-regulation of WOX3 suggests that other WOX genes (i.e. WOX2 that is situated in the same branch as WOX3 in the tree; Fig. 1A) may act redundantly with WOX3 to function independently of YAB3/KNOX regulation. Among the rice WOX genes, only QHB has been functionally characterized. QHB is expressed in the central cells of the quiescent center and is required for the root apical meristem function (Kamiya et al., 2003). However, ectopic expression of QHB also affects normal shoot and leaf development in rice (Kamiya et al., 2003). The malformed leaves induced by QHB overexpression are much like the leaves of WOX3 overexpression or YAB3 RNAi plants, suggesting that basic regulatory functions, such as DNA binding and transcriptional repression, of these WOX proteins might have been conserved during evolution, while their natural developmental functions diverge at least partially as a result of their differential expression patterns.
Regulatory Relationship between WOX3 and YAB3 in Rice Leaf Development
Although WOX and YABBY genes have been studied in different species, the functional relationship between the two families has not been established. Our data provide evidence that rice WOX3 functioned as a repressor of YAB3. Overexpression of WOX3 represses YAB3, leading to similar phenotypic changes as induced by YAB3 RNAi (Figs. 4 and 7). Conversely, repression of WOX3 by RNAi induces the expression of YAB3 (Fig. 7). Experiments with inducible WOX3 expression suggest that WOX3 directly regulated YAB3 (Fig. 8). It has been determined that the target sequence of the WUS, the founding member of WOX, is TTAATGG in the intron of AGAMOUS gene (Lohmann et al., 2001). This sequence is also recognized by rice QHB (Kamiya et al., 2003), suggesting that the TTAATGG sequence may be the consensus WOX-binding site. Sequence analysis of YAB3 locus identified a homologous sequence within the forth intron of the gene. Experiments with yeast (Saccharomyces cerevisiae) one-hybrid assays showed that WOX3 could interact with a segment of 230 bp of the YAB3 gene compressing the putative WOX-binding site (Supplemental Fig. S3). In addition, the WOX3 protein produced in and purified from Escherichia coli cells could bind to the consensus-binding site of the YAB3 gene in gel-shift assays (Supplemental Fig. S4). The WOX3-binding motif within YAB3 is identical to the WUS site in AGAMOUS, while the flanking sequences are divergent. The WUS site within the AGAMOUS is immediately flanked by the LEAFY-binding site and both of them are required to activate the gene ( Lohmann et al., 2001). We speculate that additional cis-elements in the vicinity of the WOX3-binding site may exist in YAB3 to define specificity of the regulation. The WOX3-binding motif is not found in the other rice YABBY genes, in agreement with the observation of no alteration of expression of the other YABBY genes, suggesting that WOX3 specifically regulates YAB3 among the rice YABBY genes. However, whether the binding site within the YAB3 gene was responsible for the repression by WOX3 awaits further analysis.
In vegetative shoots, the expression of the two genes overlaps. We speculate that a threshold of minimal expression level of YAB3 is required to promote leaf cell differentiation and that WOX3 has a function to control or limit the expression levels of YAB3 in leaf tissues contributing to maintain the undifferentiated state of dividing cells. This regulatory relationship might be required to maintain the balance between cell division and cell differentiation during the leaf growth. No phenotypic alteration observed in YAB3 overexpression or in WOX3 RNAi plants suggest that additional factors are required for YAB3 function in promoting leaf cell differentiation.
In summary, our data reveal distinct functions of rice YAB3 and WOX3 from their closely related homologs in Arabidopsis and in maize, which form a regulatory module of rice leaf development, in which WOX3 negatively regulates YAB3 that in turn represses KNOX genes in leaf tissues (Table I).
Table I.
Accession numbers of YABBY and WOX proteins used for the construction of the phylogenic trees
For Arabidopsis YABBY and WOX proteins, see Bowman (2000) and Haecker et al. (2004), respectively.
| Accession Nos. of YABBY Proteins
|
Accession Nos. of WOX Proteins
|
||
|---|---|---|---|
| Name of Protein | Accession No. | Name of Protein | Accession No. |
| OsYAB1 | BAF12697 | OsWOX1 | CAE04846 |
| OsYAB2 | ABF97910 | OsWOX2 | AAV44211 |
| OsYAB3 | BAF15337 | OsWOX3 | BAE48302 |
| OsYAB4 | BAF26935 | OsWOX4 | CAE04492 |
| OsYAB5 | BAF30318 | OsWOX5 | NP_915421 |
| DL | ABF94636 | OsWOX6 | ABF95709 |
| OsYAB7 | BAF09473 | OsWOX7 | NP_916815 |
| TaYAB | AAQ93323 | OsWOX8 | NP_915983 |
| ZmYAB9 | AAP79886 | QHB | BAB84412 |
| ZmYAB10 | AAP79887 | OsWOX10 | BAD05582 |
| ZmYAB14 | AAP79884 | OsWOX11 | BAF22586 |
| ZmYAB15 | AAP79885 | NS1 | Q70UV1 |
| AmYAB | AAS10178 | NS2 | Q6S3I3 |
| StYAB | AAR87498 | PtWUS | AAR83341 |
| InYAB | CAG17551 | LeWUS | Q84VT7 |
| NaINO | BAC82106 | ||
| NcINO | BAC82107 | ||
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa) spp. Japonica variety ‘Zhonghua 11’ was used in this study.
Gene Cloning
The cDNA fragments of WOX3 and YAB3 were amplified by RT-PCR. The PCR primers were designed based on two cDNA clones, AB218893 and AK070205, respectively. The sequences of the primers for WOX3 were FLWOX3-F (5′-GGTACCCTGAGGAGGATGCCTCAGAC-3′) and FLWOX3-R (5′-GGATCCATATTGGCAGTGGCACACAC-3′), and for YAB3 were FLYAB3-F (5′-GGTACCAGGATACGCGCCATGATGTC-3′) and FLYAB3-R (5′-GGATCCTGAGGCGTTAGAATGGAGTG-3′). The amplified WOX3 and YAB3 fragments were inserted into the T vector (Invitrogen) for sequencing.
Rice Transformation
The binary vector used in overexpression transformation was constructed based on pCAMIA1301 (CAMBIA) and pRTL2 (Mason et al., 1992; Kulakova et al., 1995). A HindIII fragment with a double CaMV 35S enhancer/promoter and the nopaline synthase A terminator from pRTL2 was inserted into pCAMIA1301. The new vector was named as p1301DS. For overexpression of WOX3 and YAB3, the full-length coding sequences of the genes were inserted into p1301DS digested with BamHI and KpnI. The vector used in RNAi transformation is the vector pDS1301 (Chu et al., 2006). The gene-specific fragments were amplified from the cDNAs of the two genes by using the following primer pairs: for WOX3, WOX3RNAi-F (5′-GGGACTAGTCCATGGCTGGTTCCAGAACCACAAGG-3′) and WOX3RNAi-R (5′-GGGGAGCTCCCTAGGCTGCAGCAATCTTCCTTGAG-3′); for YAB3, YAB3RNAi-F (5′-GGGACTAGTCCATGGTGCTCTTCAAGGACGGTCTC-3′) and YAB3RNAi-F (5′-GGGGAGCTCCCTAGGTATAAGAGGCAGCACGCACA-3′). The PCR fragments were sequenced and inserted into pDS1301 to produce double-strand RNA. To create WOX3-GR fusion protein, the full-length coding region of WOX3 was amplified by PCR. The stop codon of WOX3 was removed and replaced with a SalI site. Another adaptor (KpnI) was added to the end of the forward primer. The PCR product was inserted into the plasmid pSport1 (CLONTECH) in the SalI and KpnI sites. The primers used in PCR were WOX3 (GR)-F (5′-GGTACCGAGGATGCCTCAGACCCCTTCG-3′) and WOX3 (GR)-R (5′-GTCGACCAATTGGTGGAGGTGGAGCAAG-3′). In the same way, the DNA fragment that contains the steroid-binding domain of the mouse GR was amplified using the plasmid pBI-ΔGR (Lloyd et al., 1994) as template. The PCR primers were GR-F (5′-GTCGACAGATCCTGAAGCTCGAAAAAC-3′) with a SalI adaptor and GR-R (5′-GGATCCACCGGCAACAGGATTCAATG-3′) with a BamHI adaptor. The amplified GR fragment was inserted downstream to and in frame with WOX3 in pSport1. The fused WOX3-GR sequence then was cut down from pSport1 and inserted into p1301DS with KpnI and BamHI.
RNA Isolation, RNA Gel Blot, and RT-PCR
For expression analysis of YAB genes, total RNA was extracted from seedlings of the wild-type and transgenic plants at four-leaf stage using an RNA extracting kit (TRIzol reagent, Invitrogen). For expression analysis of KNOX1 genes, total RNA was extracted from leaves of wild-type and transgenic plants. For semiquantitative RT-PCR analysis, 4 μg total RNA was treated first with 2 units DNase I (Invitrogen) and then reverse transcribed in a total volume of 20 μL with 0.5 μg oligo(dT)15, 0.75 mm dNTPs, 10 mm dithiothreitol, and 200 units SuperScripTM II RNase H− reverse transcriptase (Invitrogen). The primers in RT-PCR were: YAB1-F (5′-CTTGCTCCTTTTCACCAAGC-3′), YAB1-R (5′-ATGAGCCCAGTTCTTTGCAG-3′); YAB2-F (5′-CCCATGAGAGCAGCAAGAAG-3′), YAB2-R (5′-CGTCGACACTAGCTGCATGT-3′); DL-F (5′-CAAATGAGGGTAGCCCAAGA-3′), DL-R (5′-CTGAGCTGACGACGGTGATA-3′); OSH1-F (5′-CTACCTCGACTGCCAGAAGG-3′), OSH1-R (5′-CCATGTGCATCAATCTCAGG-3′); OSH3-F (5′- GCTGGAGCAGAAGCAGATCAA-3′), OSH3-R (5′-CCAAGCTGACTTTCCCTTTGG-3′); for WOX3 and YAB3, primers were the same ones used in gene cloning. Rice actin1 gene was used as the internal control. The actin1 primers were: Actin1-F (5′-TGCTATGTACGTCGCCATCCAG-3′) and Actin1-R (5′-AATGAGTAACCACGCTCCGTCA-3′). For northern-blotting analysis, 20 μg of total RNA was separated on 1.2% (w/v) denaturing agarose gel before being transferred to nylon membranes. WOX3 fragment amplified with gene-specific primers 5′-CGGCGTCAGCGTCGGCAACT-3′ and 5′-ACGTGTGTGTGCCACTGCCA-3′ and YAB3 gene-specific fragment amplified by primers 5′-CAAATCACCCAGCTGAAAGC-3′ and 5′-AATGACGGCACCTCATCAAG-3′ were labeled with 32P-dCTP using Random Primer kit (Invitrogen) and hybridized to the RNA blots.
In Situ Hybridization
Plant materials were fixed in formaldehyde acetic acid (50% ethanol, 5% acetic acid, and 3.7% formaldehyde) overnight at 4°C, dehydrated through a concentration grade of ethanol, cleared through a xylene series, then infiltrated through a series of paraffin, and finally embedded in 100% paraffin melted at 52°C to 54°C. Then 8- to 10-μm-thick microtome sections were mounted on RNase free glass slides.
The hybridization and immunological detection were performed according to De Block and Debrouwer (1993). The WOX3 probe was amplified with the primers used in RNAi, and YAB3 probe was the fragment used in northern blotting. The PCR fragments were inserted into pGEM T-vector (Promega) for sequencing and RNA transcription. The digoxigenin-labeled sense and antisense RNA probes were produced by T7 and Sp6 transcriptase, respectively. The reagents used in the experiments were purchased from Roche.
GFP Imaging
The vector used for nuclear localization analysis was constructed based on the vector of pCAMBIA1391Xb (CAMBIA). The GUS fragment of pCAMBIA1391Xb was replaced by a CaMV 35S promoter-GFP cassette. The coding regions of WOX3 and YAB3 were amplified using the primer pairs: WOX3NLS-F (5′-GGTACCCTGAGGAGGATGCCTCAGAC-3′), WOX3NLS-R (5′-GGATCCATTGGTGGAGGTGGAGCAAG-3′); YAB3NLS-F (5′-GGTACCGGATCACCTAGCTAGGATAC-3′), YAB3NLS-R (5′-GGTACCGAATGGAGTGACACCCATGC-3′). The amplified fragment was inserted upstream to and in frame with GFP. The fusion plasmid (5 μg) was coprecipitated with 3 mg of gold particles. The particles were resuspended in ethanol in a total volume of 60 μL and divided into five aliquots for bombarding onion (Allium cepa) epidermal cells using the PDS-1000 system (Bio-Rad) at 1,100 psi helium pressure. The expression of the fusion protein of WOX3, YAB3, and GFP in the onion epidermal cells was observed by a confocal microscope (Leica) 36 h after bombardment.
Yeast One-Hybrid Analysis
Yeast (Saccharomyces cerevisiae) one-hybrid analysis was performed by using a CLONTECH system. The partial forth intron of YAB3 amplified with primers was inserted into pHIS2. The full-length coding sequence of WOX3 was amplified from the full-length cDNA clone using the primer pairs: WOX3CDS-F (5′-GAATTCCTGAGGAGGATGCCTCAGAC-3′) and WOX3CDS-R (5′-GGATCCATATTGGCAGTGGCACACAC-3′). The PCR product was inserted into the vector of pGADT7-Rec2 to fuse with the GAL4 activation domain. The two constructs were sequenced and used to transform the yeast strain Y187 for one-hybrid analysis.
Chemical Treatments and Quantitative RT-PCR
DEX (Sigma) was dissolved in ethanol to 30 mm and stored at −20°C before use. Before the experiment, DEX was diluted with growth medium to a final concentration of 10 μm. The regenerants from the same callus line were divided into three parts: one was treated with DEX, one was treated first with CHX for 1 h then treated with DEX, and the third had no treatment. For treatment of the whole plants, roots of the regenerants were submerged in growth medium with or without 10 μm DEX and CHX. After 12 h of treatment, the aerial part of the plantlets was harvested for RNA extraction, RT-PCR, and quantitative real-time PCR analysis.
Quantitative PCR was performed in a total volume of 25 μL with 1.5 μL of the RT reactions, 0.25 μm gene-specific primers, and 12.5 μL SYBR Green Master mix (Applied Biosystems) on a 7500 real-time PCR machine (Applied Biosystems) according to the manufacturer's instructions. The rice actin1 gene was used as the internal control. All primers were annealed at 58°C and run 45 cycles. The relative expression level of each gene in transgenic plants was compared to that in wild-type ones, normalized using the actin1 cDNA level, and averaged over three replicates. The primers used in real-time PCR were YAB3rq-F (5′-ATACAACG CATCAAGGCAAGC-3′), YAB3rq-R (5′-CATCAGGCCAAAATGGATGTG-3′) and WOX3rq-F (5′-GCCTCAAGGAAGATTGCTGCA-3′), and WOX3rq-R (5′-ACGCATATGGCC GCAGAAA-3′).
Histological Observation
Leaves and flowers were harvested from the wild-type and transgenic plants. The procedures of dehydration, clearing, infiltration, and embedding were carried out as mentioned above. The sections were deparaffinized in xylene, rehydrated through a graded ethanol series, and dried before staining with fast green (for leaves) and hematoxylin (for flowers).
Electrophoretic Mobility Shift Assay
To produce the WOX3 protein, the full-length WOX3 cDNA was inserted into the pET-32a expression vector (Novagen) and expressed in Rosetta-gami (DE3) Escherichia coli cells (Novagen). The tagged protein was purified with B-PER 6× His Spin Purification kit (Pierce) and dosed by using the Bradford reagents. The YAB3 intron DNA S1 (including the putative WUS-binding site, TTAATGG) was produced by annealing of two oligonucleotides: 5′-GAGCAACATTAATGGTCAGGTT-3′ and 5′-GTGCAACCTGACCATTAATGTT-3′. The S2 DNA, which is similar to S1 but lacks a WUS site, was generated by annealing of the oligonucleotides: 5′-GAGCAACATCAGGTT-3′ and 5′-GTGCAACCTGATGTT-3′. The double-stranded oligonucleotides were labeled with 32P-dCTP using the Klenow fragment. DNA-binding reactions were performed at room temperature for 20 min in 10 mm Tris, pH 7.5, 50 mm NaCl, 1 mm dithiothreitol, 1 mm EDTA, 1 mm MgCl2, 5% (v/v) glycerol, and 50 mg L−1 poly(dI-dC) poly(dI-dC) (Amersham Pharmacia Biotech) in the presence of about 50 ng of the WOX3 protein and 1 ng of the probes and separated on 6% polyacrylamide gels in the Tris-glycin (0.3% Tris, 1.88% glycin) buffer.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers BAF15337 (YAB3) and BAE48302 (WOX3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transverse-section of leaves from wild-type (A and B), YAB3 RNAi (C and D), and WOX3 overexpression (E and F) plants.
Supplemental Figure S2. Floral phenotypes in YAB3 RNAi- and WOX3-overexpression plants.
Supplemental Figure S3. One-hybrid analysis of binding of WOX3 to YAB3.
Supplemental Figure S4. Gel-shift analysis of WOX3 binding to YAB3 gene.
Supplementary Material
Acknowledgments
We thank Li Xianhua for technical assistance and Xu Chaiguo for rice field management.
This work was supported by the National Special Key Program of Rice Functional Genomics and by the National Natural Science Foundation of China.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Dao-Xiu Zhou (dao-xiu.zhou@u-psud.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Bowman JL (2000) The YABBY gene family and abaxial cell fate. Curr Opin Plant Biol 3 17–22 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126 2387–2396 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971 [DOI] [PubMed] [Google Scholar]
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, et al (2006) Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev 20 1250–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S (1996) KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou D-X (2007) The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol 144 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block M, Debrouwer D (1993) RNA-RNA in situ hybridization using digoxigenin-labeled probes: the use of high-molecular-weight polyvinyl alcohol in the alkaline phosphatase indoxyl-nitroblue tetrazolium reaction. Anal Biochem 215 86–89 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209 [DOI] [PubMed] [Google Scholar]
- Golz JF, Hudson A (1999) Plant development: YABBYs claw to the fore. Curr Biol 9 R861–863 [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131 657–668 [DOI] [PubMed] [Google Scholar]
- Hay A, Craft J, Tsiantis M (2004) Plant hormones and homeoboxes: bridging the gap? Bioessays 26 395–404 [DOI] [PubMed] [Google Scholar]
- Jang S, Hur J, Kim SJ, Han MJ, Kim SR, An G (2004) Ectopic expression of OsYAB1 causes extra stamens and carpels in rice. Plant Mol Biol 56 133–143 [DOI] [PubMed] [Google Scholar]
- Juarez MT, Twiggl RW, Timmermans MCP (2004) Specification of adaxial cell fate during maize leaf development. Development 131 4533–4544 [DOI] [PubMed] [Google Scholar]
- Kamiya N, Nagasaki H, Morikami A, Sato Y, Matsuoka M (2003) Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J 35 429–441 [DOI] [PubMed] [Google Scholar]
- Kulakova AN, Stafford TM, Larkin MJ, Kulakov LA (1995) Plasmid pRTL1 controlling 1-chloroalkane degradation by Rhodococcus rhodochrous NCIMB13064. Plasmid 33 208–217 [DOI] [PubMed] [Google Scholar]
- Kumaran MK, Bowman JL, Sundaresan V (2002) YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14 2761–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Baum SF, Alvarez J, Patel A, Chitwood DH, Bowman JL (2005) Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell 17 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW (1994) Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science 266 436–439 [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 793–803 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69 [DOI] [PubMed] [Google Scholar]
- Mason HS, Lam DM, Arntzen CJ (1992) Expression of hepatitis B surface antigen in transgenic plants. Proc Natl Acad Sci USA 89 11745–11749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N, Okada K (2001) A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev 15 3355–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815 [DOI] [PubMed] [Google Scholar]
- Meister RJ, Oldenhof H, Bowman JL, Gasser CS (2005) Multiple protein regions contribute to differential activities of YABBY proteins in reproductive development. Plant Physiol 137 651–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J, Ji J, Werr W, Scanlon MJ (2004) The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131 2827–2839 [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S (2000) Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532 [DOI] [PubMed] [Google Scholar]
- Reiser L, Sanchez-Baracaldo P, Hake S (2000) Knots in the family tree: evolutionary relationships and functions of knox homeobox genes. Plant Mol Biol 42 151–166 [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Liu YG, Shibata D, Kanaya E, Morita EH, Okada K (1999) FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev 13 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentoku N, Sato Y, Matsuoka M (2000) Overexpression of rice OSH genes induces ectopic shoots on leaf sheaths of transgenic rice plants. Dev Biol 220 358–364 [DOI] [PubMed] [Google Scholar]
- Sieber P, Petrascheck M, Barberis A, Schneitz K (2004) Organ polarity in Arabidopsis: NOZZLE physically interacts with members of the YABBY family. Plant Physiol 135 2172–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128 [DOI] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S (1992) A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116 21–30 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Reiser L, Hake S (2000) Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127 3161–3172 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S (1991) The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350 241–243 [DOI] [PubMed] [Google Scholar]
- Wu X, Dabi T, Weigel D (2005) Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr Biol 15 436–440 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY (2004) The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.