Abstract
Arabinoxylans (AXs) are major components of graminaceous plant cell walls, including those in the grain and straw of economically important cereals. Despite some recent advances in identifying the genes encoding biosynthetic enzymes for a number of other plant cell wall polysaccharides, the genes encoding enzymes of the final stages of AX synthesis have not been identified. We have therefore adopted a novel bioinformatics approach based on estimation of differential expression of orthologous genes between taxonomic divisions of species. Over 3 million public domain cereal and dicot expressed sequence tags were mapped onto the complete sets of rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana) genes, respectively. It was assumed that genes in cereals involved in AX biosynthesis would be expressed at high levels and that their orthologs in dicotyledonous plants would be expressed at much lower levels. Considering all rice genes encoding putative glycosyl transferases (GTs) predicted to be integral membrane proteins, genes in the GT43, GT47, and GT61 families emerged as much the strongest candidates. When the search was widened to all other rice or Arabidopsis genes predicted to encode integral membrane proteins, cereal genes in Pfam family PF02458 emerged as candidates for the feruloylation of AX. Our analysis, known activities, and recent findings elsewhere are most consistent with genes in the GT43 families encoding β-1,4-xylan synthases, genes in the GT47 family encoding xylan α-1,2- or α-1,3-arabinosyl transferases, and genes in the GT61 family encoding feruloyl-AX β-1,2-xylosyl transferases.
All higher plants are believed to synthesize arabinoxylan (AX) or glucuronarabinoxylan (GAX) as a component of their cell walls. This hemicellulose may function in coating and cross-linking cellulose microfibrils (Carpita and Gibeaut, 1993). In primary cell walls, this cellulose and hemicellulose network is thought to be embedded in a protein and pectic matrix.
Xylan polysaccharides have a backbone of β-1,4-linked d-xylosyl residues, which in AX have α-l-arabinofuranosyl side chains attached through 1,3 and 1,2 linkages. GAX has single α-1,2-linked residues of GlcA or 4-O-methyl-GlcA attached to the xylosyl residue backbone in addition to arabinosyl substitutions. In grasses, Ara may be feruloylated and then further substituted with β-1,2-xylosyl residues (Wende and Fry, 1997a). The main hemicellulose in dicots is xyloglucan, a β-1,4-linked glucan with xylosyl side chain substitutions that can be further decorated with Gal and Fuc. However, some species show unusual characteristics; sugar beet (Beta vulgaris) cell walls appear to be almost devoid of hemicelluloses, including xylans and xyloglucan (Renard and Thibault, 1993).
The relative importance of the different polysaccharides of walls varies widely. In dicot primary walls, including Brassicas and Arabidopsis (Arabidopsis thaliana), the major hemicellulose is xyloglucan (Bacic et al., 1988; Carpita, 1996; Harris et al., 1997). However, in the type II walls of plants in the Poales order (grasses), such as rice (Oryza sativa), wheat (Triticum aestivum), and barley (Hordeum vulgare), and other commelinoid monocots, AX occurs as a major constituent of all primary and secondary cell walls (Wilkie, 1979). Hence, AX is a major component of the dietary fiber consumed by humans in cereal products and also has impact on their processing properties and quality for livestock feed. Furthermore, the exploitation of plant biomass for biofuels will depend largely on our ability to digest AX and other cell wall polymers. A key factor in digestibility is believed to be the feruloylation of AX and GAX in cereals, which allows possible cross-linking between AX chains and from AX to lignin (Grabber, 2005). Despite the huge economic and nutritional importance of AX, genes that encode the xylan synthase (which generates the xylan backbone), the arabinosyl transferases (which substitutes Ara onto the Xyl residues), and the AX feruloyl transferase (which substitutes ferulate onto these Ara residues) have not been identified.
Until recently, it was thought that the xylan synthase gene was likely to belong to the cellulose-synthase-like (Csl) family (Richmond and Somerville, 2000); however, this now seems less likely because members do not show xylan synthase activity when expressed in insect cells (Liepman et al., 2005). Recent progress in identifying the functions of key glycosyl transferase (GT) genes has been made by using their expression patterns as an initial screen. Brown et al. (2005) and Persson et al. (2005) used microarray data to identify genes that were coexpressed with secondary cell wall-specific cellulose synthase genes in Arabidopsis. These included several GT genes for which insertional mutants were shown to have altered cell wall composition. Aspeborg et al. (2005) used array data and Geisler-Lee et al. (2006) used numbers of ESTs from poplar (Populus spp.) to assess abundance of transcripts for enzymes in the CAZy database of carbohydrate-active enzymes (Coutinho et al., 2003) and relate these to cell wall metabolism in different tissues.
In this study, we also use EST counts, but specifically to prioritize candidates for the synthesis of AX and its side chains. Provided that ESTs are assigned to genes in a rigorous fashion, taking proper account of unreliable sequences and ambiguous top alignment hits, EST counts are a powerful means of assessing expression. Counts of ESTs from nonnormalized libraries have been shown to be correlated with other measures of expression, such as real-time reverse transcription-PCR (Boutanaev et al., 2002) and microarray and serial analysis of gene expression data (Haverty et al., 2004). There is some evidence that EST counts may tend to exaggerate the expression of abundant transcripts and underestimate the expression of rare transcripts (Haverty et al., 2004), but, for this study, which aims to rank candidates on relative expression, they are appropriate. They have the great advantage of being an open system representing the whole transcriptome, whereas public array data for the species considered here, with the exception of Arabidopsis, currently only represent a part. We employ a novel bioinformatics approach exploiting the wealth of public EST data available and linking these to the fully sequenced rice and Arabidopsis genomes. Due to the unstructured nature of EST libraries that have been submitted to public databases, precautions have to be taken to check for bias; however, because it would be expected that nearly every tissue would reflect the difference in cell wall composition between dicots and cereals, this is less of a problem in our analysis.
We searched for genes that encode the enzymes responsible for AX synthesis making a minimal number of assumptions. These are that the enzymes must be integral membrane proteins, because AX is synthesized in the Golgi, and that the expression of the encoding genes should reflect the relative abundance of AX (Wilkie, 1979; McNeil et al., 1984; Bacic et al., 1988). The fact that polysaccharide synthases appear to occur at relatively low abundance (Dhugga, 2005) shows that no simple relationship exists between amounts of different enzyme types and their products, but it seems highly probable that expression of the same enzyme type should reflect the gross differences between species. Thus, we assumed that the expression of genes encoding AX synthetic enzymes will be high in absolute terms in monocots compared with other GT genes, and substantially higher than that for their putative orthologs in dicots. The complete complement of rice genes was linked to ESTs from rice, wheat, and barley and the complete complement of Arabidopsis genes to ESTs from Arabidopsis, soybean (Glycine max), Brassica spp., and potato (Solanum tuberosum) to identify genes that satisfy these criteria. The results clearly indicate that specific genes within the GT43, GT47, GT61, and PF02458 families are the most likely candidates to encode the enzymes that synthesize AX and its side chains.
RESULTS AND DISCUSSION
ESTs were taken from the dbEST database (Boguski et al., 1993) for a selection of cereal (rice, wheat, and barley) and dicot (Arabidopsis, soybean, Brassica spp., and potato) species, excluding those from normalized or subtractive libraries. To consolidate expression data from several species, cereal and dicot ESTs were mapped by sequence similarity onto the genes of rice and Arabidopsis, respectively. Rice and Arabidopsis genes were then organized into orthologous groups and the ratio of counts of the corresponding cereal ESTs to dicot ESTs in each group was calculated after normalization for the difference in the total number of cereal and dicot ESTs. Tables I and II summarize results from this procedure for all the genes predicted both to encode GT enzymes in the CAZy database (Coutinho et al., 2003) and to be integral membrane proteins in the Aramemnon database (Schwacke et al., 2003; the complete list from which Tables I and II are derived is available as Supplemental Table S1).
Table I.
Counts of cereal ESTs matching rice (Os) or dicot ESTs matching Arabidopsis (At) loci predicted to encode integral membrane GTs
Orthologous groups with more than 100 matching ESTs are shown and are ranked by the normalized ratio of cereal EST counts to dicot EST counts. The first rice locus and first Arabidopsis locus in each group are orthologs; the others are paralogs to these. Groups with normalized ratio >2.5 are shown.
| GT Family | Os Loci | Cereal EST Counts | Ortholog % Identity (Alignment Length) | At Loci | Dicot EST Counts | Normalized Ratioa | CAZy Family Annotation
|
|
|---|---|---|---|---|---|---|---|---|
| At Gene Name | Os Gene Name | |||||||
| 61 | Os02g04250 | 170 | 37% (488) | At3g18180 | 2 | Putative xylosyl transferases | ||
| + 4 paralogs | ||||||||
| Total | 489 | Total | 2 | 114.4 | ||||
| 61 | Os02g22480 | 88 | 44% (390) | At3g18170 | 14 | Putative xylosyl transferases | ||
| + 13 paralogs | ||||||||
| Total | 930 | Total | 14 | 31.1 | ||||
| 47 | Os01g70200 | 158 | 85% (406) | At1g27440 | 6 | IRX10, AtGUT1b | ||
| + 5 paralogs | At5g61840 | 33 | OsGUT1 | |||||
| Total | 748 | Total | 38 | 9.2 | ||||
| 31 | Os05g35274 | 260 | 64% (390) | At1g33430 | 16 | Putative-N-acetylhexosyaminyl transferases | ||
| Os01g65590 | 29 | |||||||
| Total | 289 | Total | 16 | 8.4 | ||||
| 2 | Os09g25490 | 475 | 80% (1,058) | At5g17420 | 31 | IRX3, AtCESA7b | OsCESA9 | |
| Total | 475 | Total | 31 | 7.2 | ||||
| 2 | Os10g32980 | 297 | 76% (1,051) | At5g44030 | 37 | IRX5, AtCESA4b | OsCESA7 | |
| Total | 297 | Total | 37 | 3.8 | ||||
| 2 | Os05g43530 | 21 | 71% (708) | At4g07960 | 18 | AtCslC12 | OsCslC7 | |
| + 3 paralogs | OsCslC1,9,10 | |||||||
| Total | 141 | Total | 18 | 3.7 | ||||
| 2 | Os01g54620 | 245 | 73% (1,009) | At4g18780 | 33 | IRX1, AtCESA8b | OsCESA4 | |
| Total | 245 | Total | 33 | 3.5 | ||||
| 2 | Os06g02180 | 154 | 80% (1,168) | At3g03050 | 29 | AtCslD3 | OsCslD2 | |
| + 11 paralogs | + 3 paralogs | AtCslD6,4,2 | OsCslF1-9, D1,3 | |||||
| Total | 407 | Total | 54 | 3.5 | ||||
| 43 | Os05g03174 | 62 | 42% (369) | At2g37090 | 16 | IRX9b | ||
| + 2 paralogs | Putative β-glucuronosyl transferases | |||||||
| Total | 111 | Total | 16 | 3.2 | ||||
| 77 | Os02g46120 | 6 | 54% (368) | At1g14590 | 16 | Putative α-xylosyl or α-1,3-galactosyl transferases | ||
| + 10 paralogs | + 4 paralogs | |||||||
| Total | 294 | Total | 43 | 3.2 | ||||
| 43 | Os10g13810 | 5 | 49% (351) | At1g27600 | 18 | Putative β-glucuronosyl transferases | ||
| + 3 paralogs | ||||||||
| Total | 120 | Total | 18 | 3.1 | ||||
| 61 | Os01g31370 | 26 | 55% (472) | At2g41640 | 22 | Putative β-xylosyl transferases | ||
| + 3 paralogs | + 5 paralogs | |||||||
| Total | 243 | Total | 38 | 3.0 | ||||
| 48 | Os06g08380 | 10 | 75% (1,924) | At2g13680 | 6 | 1,3-Glucan synthases (callose) | ||
| + 3 paralogs | At2g36850 | 21 | ||||||
| Total | 167 | Total | 27 | 2.9 | ||||
| 43 | Os06g47340 | 200 | 52% (489) | At5g67230 | 14 | Putative β-glucuronosyl transferases | ||
| Os04g55670 | 42 | At4g36890 | 31 | |||||
| Total | 242 | Total | 44 | 2.6 | ||||
| 64 | Os05g46260 | 90 | 54% (715) | At5g04500 | 18 | Putative α-N-acetylhexosaminyl transferases | ||
| Os03g48010 | 7 | |||||||
| Total | 97 | Total | 18 | 2.5 | ||||
Ratio is normalized by multiplying by the ratio of total counts for all loci of dicot ESTs to cereal ESTs, a factor of 0.468.
This gene was identified as being coexpressed with secondary cell wall-specific CESA genes in Arabidopsis (Brown et al., 2005).
Table II.
Results as for Table I, but for groups with normalized ratio <0.4
| GT Family | Os Loci | Cereal EST Counts | Ortholog % Identity (Alignment Length) | At Loci | Dicot EST Counts | Normalized Ratioa | CAZy Family Annotation
|
|
|---|---|---|---|---|---|---|---|---|
| At Gene Name | Os Gene Name | |||||||
| 34 | Os03g18820 | 47 | 74% (460) | At4g02500 | 42 | |||
| + 2 paralogs | At3g62720 | 53 | AtXT1 | |||||
| Total | 81 | Total | 95 | 0.4 | Xyloglucan α-xylosyl transferases | |||
| 1b | Os02g09510 | 42 | 42% (474) | At4g15480 | ||||
| + 3 paralogs | + 6 paralogs | |||||||
| Total | 62 | Total | 77 | 0.4 | ||||
| 2 | Os09g30130 | 56 | 49% (723) | At1g55850 | 69 | AtCslE1 | OsCslE6 | |
| + 2 paralogs | + 3 paralogs | AtCslG1-3 | OsCslE1,2 | |||||
| Total | 152 | Total | 194 | 0.4 | ||||
| 1b | Os02g09510 | 42 | 42% (474) | At4g15480 | 18 | |||
| + 6 paralogs | + 3 paralogs | |||||||
| Total | 167 | Total | 257 | 0.3 | ||||
| 8 | Os02g29530 | 59 | 69% (504) | At3g25140 | 100 | QUA1 | ||
| + 2 paralogs | At3g02350 | 66 | ||||||
| Total | 107 | Total | 166 | 0.3 | ||||
| 77 | Os03g38930 | 40 | 61% (638) | At2g35610 | 69 | Putative α-xylosyl or α-1,3-galactosyl transferases | ||
| Total | 40 | Total | 69 | 0.3 | ||||
| 31 | Os04g48950 | 29 | 57% (523) | At5g41460 | 8 | Putative N-acetylhexosaminyl transferases | ||
| + 3 paralogs | + 5 paralogs | |||||||
| Total | 45 | Total | 90 | 0.2 | ||||
| 2 | Os02g09930 | 52 | 72% (523) | At5g22740 | 163 | AtCslA02 mannan synthase | OsCslA1 | |
| + 2 paralogs | At4g13410 | 2 | AtCslA15 | OsCslA11, 12 | ||||
| Total | 82 | Total | 165 | 0.2 | ||||
| 47 | Os06g23420 | 4 | 48% (469) | At5g62220 | 79 | AtGT18 (MUR3 related) | ||
| Os04g48480 | 27 | |||||||
| Total | 31 | Total | 79 | 0.2 | ||||
| 1b | Os11g38650 | 89 | 48% (475) | At1g01420 | 21 | |||
| + 8 paralogs | + 8 paralogs | |||||||
| Total | 130 | Total | 343 | 0.2 | ||||
| 1b | Os06g09240 | 11 | 42% (457) | At5g17050 | 35 | |||
| Os07g05420 | 13 | + 3 paralogs | ||||||
| Total | 23 | Total | 81 | 0.1 | ||||
| 22 | Os01g11070 | 92 | 67% (572) | At1g16900 | 42 | Putative dolichyl-p-Man α-mannosyl transferases | ||
| + 4 paralogs | ||||||||
| Total | 92 | Total | 327 | 0.1 | ||||
| 2 | Os09g25900 | 9 | 70% (699) | At4g31590 | 61 | AtCslC05 | OsCslC2 | |
| Os08g15420 | 26 | + 3 paralogs | AtCslC06,8,4 | OsCslC3 | ||||
| Total | 35 | Total | 169 | 0.1 | ||||
Ratio is normalized by multiplying by the ratio of total counts for all loci in this set of dicot ESTs to cereal ESTs, a factor of 0.468.
The mature proteins encoded by these groups do not contain transmembrane domains, but are present because of the inclusive method used to predict integral membrane proteins.
The first criterion for selection of candidate genes involved in AX biosynthesis is that they should be highly expressed in cereals and, therefore, Tables I and II show only those orthologous groups with more than 100 associated ESTs. The second criterion is that they should be more highly expressed in grass species than dicot species due to the much greater prevalence of AX in the former; xylans make up about 5% of the primary cell wall in dicots compared to 20% in grasses (McNeil et al., 1984). The orthologous groups were therefore ranked in descending order of the normalized ratio of EST counts in cereals to EST counts in dicots. Table I shows the 16 orthologous groups where these normalized ratios were greater than 2.5. Interestingly, the table contains members of just seven GT families, corresponding in order of decreasing cereal expression bias to GT 61, 47, 31, 2, 43, 77, and 48. An orthologous group of genes that occurred high in the list is the GT2 family CslF genes, which encode mixed-linkage β-glucan synthases (Burton et al., 2006). Because mixed-linkage β-glucan is quite abundant in grass cell walls, but does not exist in dicots, this finding supports the suggestion that grass-specific synthases occur high in the ranked list. Because no exact corresponding genes to the CslF genes exist in Arabidopsis, the method groups them together in an orthologous group with the related CslD genes, which must encode enzymes with different activities.
Further evidence for the validity of the method came from inspection of the bottom of the ranking (i.e. those GT genes that are much more highly expressed in dicots than in cereals). These should encode the enzymes responsible for the cell wall components that are more abundant in dicots (e.g. xyloglucan and pectin; Bacic et al., 1988). Table II shows all the orthologous groups for which the normalized EST count ratio was below 0.4. As expected, this list includes genes encoding enzymes active in xyloglucan synthesis (AtXT1 genes in GT34 family; Bencur et al., 2005) and QUA1, which is implicated in the synthesis of the pectin homogalacturonan (Bouton et al., 2002). It is possible that different orthologous groups within the same GT family could substitute for one another in dicots and cereals; thus, one group would be highly expressed in cereals, the other in dicots. This can be ruled out for the GT61 and GT43 families for which all groups were more highly expressed in cereals and for the GT22 and GT34 families for which all groups are more highly expressed in dicots.
We investigated whether a large bias in the tissue types of libraries made for cereals and dicots could affect the results in Table I. Inspection of all the libraries used showed the most obvious differences to be (1) a larger proportion of seed libraries for cereals compared to dicots; and (2) a preponderance of tuber libraries for potatoes, presumably due to their economic importance. The procedure used to generate Table I was therefore repeated, but for two subsets, excluding (1) all seed libraries and (2) all root/tuber libraries. The results were very similar to those in Table I showing that they are not caused by a tissue bias (data summarized in Supplemental Table S3). Because dicots and cereals differ in primary cell wall AX content much more than in secondary cell walls, we might expect the differential expression of candidates for AX synthesis to be exaggerated in developing tissues. We investigated this by identifying a subset of ESTs annotated as from young or developing tissue; unfortunately, this subset is probably too small to test this and the results (Supplemental Table S3) were not conclusively different from Table I.
Not all plant GTs are in the CAZy database; bioinformatics approaches have identified new putative GTs, some of which have subsequently been experimentally confirmed (Egelund et al., 2004) and proteomic studies of Golgi-located proteins have also revealed more putative GTs (P. Dupree, unpublished data). We were also interested in genes responsible for the addition of ester-linked phenolic groups onto AX in grass species, particularly ferulate. For these reasons, we analyzed all rice genes that are not in CAZy, but are predicted either to encode integral membrane proteins or are in orthologous groups containing such genes. All genes that are highly expressed in cereals with more than 100 cereal ESTs and at least 20 in each cereal species were considered (Supplemental Table S2). Based on homology of domains present in these, no further genes likely to encode GT proteins emerged as candidates for AX synthesis. However, one of the most differentially expressed groups was a group of 12 rice loci from Pfam family PF02458, which is part of the CoA-acyl transferase superfamily. This family includes genes encoding hydroxycinnamoyl transferases (Yang et al., 1997), although none are currently known to act with sugar acceptors. Whereas the abundance of xylan differs quantitatively between grasses and dicots, the presence of feruloyl groups on AX appears to be an absolute difference because these are present in cell walls of all Gramineae, but have never been detected in dicots (Bacic et al., 1988). The high degree of differential expression and relatively low similarity between rice and Arabidopsis orthologs for the PF02458 group in Table III therefore appears consistent with these genes encoding enzymes that transfer feruloyl and perhaps other, rarer hydroxycinnamoyl residues onto AX. A complication is that this family also encodes acetyl transferases and AX is often heavily acetylated (Carpita, 1996); it is therefore possible the group may also be responsible for this activity. Nevertheless, these genes remain the strongest candidates for AX feruloyl transferase because no other acyl transferase group shows nearly as much differential expression between cereals and dicots (Supplemental Table S2).
Table III.
EST counts for orthologous group of genes in Pfam family PF02458, part of the CoA-acyl transferase superfamily
This group has the third highest normalized ratio in the set of genes predicted to encode membrane proteins (full set in Supplemental Table S2).
| Os Loci | Cereal EST Counts | Ortholog % Identity (Alignment Length) | At Loci | Dicot EST Counts | Normalized Ratioa | Known Activities in this Family (PFAM Annotation) |
|---|---|---|---|---|---|---|
| Os01g18744 | 48 | 40% (443) | At3g62160 | 6 | Anthranilate N-hydroxycinnamoyl/benzoyl transferase | |
| Os05g04584 | 61 | Deacetylvindoline 4-O-acetyl transferase | ||||
| Os05g08640 | 41 | Trichothecene 3-O-acetyl transferase | ||||
| Os06g39390 | 29 | |||||
| Os01g08380 | 96 | |||||
| Os06g39470 | 23 | |||||
| Os04g11810 | 7 | |||||
| Os05g19910 | 21 | |||||
| Os01g42870 | 93 | |||||
| Os01g42880 | 114 | |||||
| Os04g09590 | 4 | |||||
| Os01g09010 | 332 | |||||
| Total | 868 | Total | 6 | 59.0 |
Ratio is normalized by multiplying by the ratio of total counts for all loci of dicot ESTs to cereal ESTs in this set, a factor of 0.408.
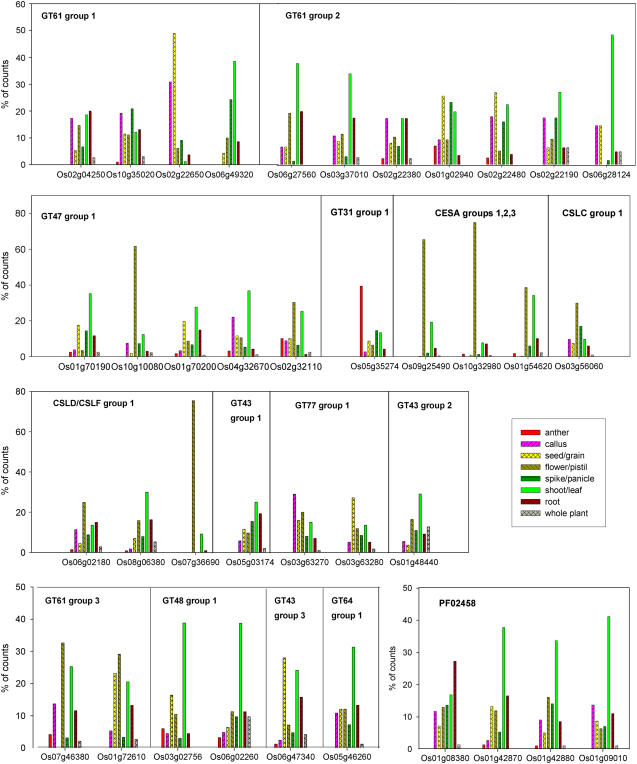
Further evidence for the functions of the candidates in Tables I and III can be derived from the distribution of cDNA libraries in which cereal ESTs occur. Because AX is prevalent in all primary and secondary cereal cell walls, the expectation is that functional groups representing AX biosynthetic genes should be highly expressed in all tissues, although the tissue specificity of individual genes within groups may vary. Figure 1 summarizes this information and shows that this criterion is met for most groups, but not for the three GT2 family cellulose synthases (CESA) that in dicots are specifically associated with secondary cell wall synthesis. The expression of these CESA genes was mostly in the flower/pistil/carpel category. Similarly, GT31 expression was mostly anther specific. Genes for which expression is mostly limited to only a few libraries can be regarded as less reliable; Table IV summarizes the information for highly expressed cereal genes in the orthologous groups of Table I and again shows that this applies most to the CESA genes for which >50% of the counts are contributed by only two to three libraries.
Figure 1.
Tissue distribution of cereal EST counts for rice loci with 50 or more cereal ESTs in orthologous groups from Table I. Only counts from libraries with tissue information are included and data are presented as a percentage of the total of these for each locus.
Table IV.
Library information and coexpression data for individual rice loci from orthologous groups shown in Table I with at least 50 matching cereal ESTs
| Locus | GT Family | No. Libraries with Counts | No. Libraries to Give 50% of Countsa | UDP-GDH Correlationb | UDP-GlcAdc Correlationc | PF02458 Group Correlationd |
|---|---|---|---|---|---|---|
| Os02g04250 | GT61 | 55 | 11 | −0.30 | −0.16 | 0.48** |
| Os10g35020 | GT61 | 46 | 10 | −0.20 | −0.02 | 0.39** |
| Os02g22650 | GT61 | 27 | 4 | −0.07 | −0.12 | 0.12 |
| Os06g49320 | GT61 | 34 | 8 | −0.12 | −0.05 | 0.34** |
| Os02g22480 | GT61 | 41 | 11 | 0.04 | 0.11 | 0.26* |
| Os06g28124 | GT61 | 25 | 4 | −0.02 | −0.13 | −0.03 |
| Os02g22380 | GT61 | 38 | 9 | 0.07 | 0.11 | 0.24 |
| Os06g27560 | GT61 | 35 | 6 | −0.22 | −0.06 | 0.63** |
| Os02g22190 | GT61 | 29 | 9 | −0.19 | −0.09 | 0.12 |
| Os01g02940 | GT61 | 33 | 6 | −0.15 | −0.12 | −0.04 |
| Os03g37010 | GT61 | 55 | 9 | 0.26* | 0.34* | 0.27* |
| Os01g70200 | GT47 | 55 | 14 | 0.63** | 0.49** | 0.14 |
| Os01g70190 | GT47 | 71 | 15 | 0.22 | 0.43** | 0.01 |
| Os02g32110 | GT47 | 37 | 10 | −0.15 | −0.10 | −0.10 |
| Os04g32670 | GT47 | 41 | 8 | −0.22 | −0.16 | 0.06 |
| Os10g10080 | GT47 OsGUT1 | 39 | 2 | −0.12 | −0.13 | −0.07 |
| Os05g35274 | GT31 | 76 | 11 | −0.04 | −0.13 | −0.10 |
| Os09g25490 | GT2 OsCESA9 | 32 | 3 | −0.15 | −0.12 | 0.07 |
| Os10g32980 | GT2 OsCESA7 | 29 | 2 | −0.13 | −0.11 | 0.03 |
| Os03g56060 | GT2 OsCslC9 | 34 | 7 | 0.08 | 0.00 | −0.02 |
| Os01g54620 | GT2 OsCESA4 | 39 | 3 | −0.09 | −0.01 | 0.11 |
| Os06g02180 | GT2 OsCslD2 | 49 | 6 | −0.15 | −0.07 | 0.30* |
| Os07g36690 | GT2 OsCslF2 | 10 | 2 | −0.10 | −0.08 | 0.07 |
| Os08g06380 | GT2 OsCslF6 | 53 | 13 | 0.33* | 0.49** | 0.31* |
| Os05g03174 | GT43 | 29 | 7 | 0.07 | −0.02 | 0.11 |
| Os03g63270 | GT77 | 38 | 8 | −0.07 | 0.09 | 0.24 |
| Os03g63280 | GT77 | 22 | 5 | 0.03 | −0.02 | −0.06 |
| Os01g48440 | GT43 | 27 | 7 | 0.29* | 0.43** | 0.14 |
| Os07g46380 | GT61 | 27 | 6 | −0.20 | −0.14 | 0.41** |
| Os01g72610 | GT61 | 32 | 8 | −0.20 | −0.24 | 0.41** |
| Os06g02260 | GT48 | 35 | 12 | 0.24* | 0.37** | 0.23 |
| Os03g02756 | GT48 | 34 | 10 | 0.08 | 0.13 | −0.07 |
| Os06g47340 | GT43 | 74 | 18 | 0.35** | 0.33* | 0.16 |
| Os05g46260 | GT64 | 34 | 8 | −0.16 | −0.04 | 0.16 |
The minimal number of libraries required to give 50% of the total counts for this locus. If a large proportion of the counts arise from a few libraries, this may indicate that this is not representative of generally high expression in cereals.
Correlation coefficients for normalized counts for each gene with normalized counts across 63 cereal libraries. Counts were normalized by dividing by total number of ESTs in each library to correct for variation due to library size. *, Significant at P < 0.05; **, significant at P < 0.01.
bCorrelations with sum of counts for three UDP-Glc dehydrogenase genes.
Correlations with sum of counts for three UDP-GlcAdc genes.
Correlations with sum of counts for genes in Pfam family PF02458 shown in Table III.
Genes encoding xylan synthase and xylan arabinosyl transferase might be expected to be coexpressed with UDP-GlcA decarboxylase (UDP-GlcAdc), which is responsible for generating the substrates for AX synthesis: both UDP-Xyl and, via an epimerase, UDP-Ara (Zhang et al., 2005). Whereas UDP-Xyl and UDP-Ara also provide the Xyl and Ara present in other polysaccharides, such as xyloglucan, arabinogalactan of arabinogalactan proteins, and arabinan side chains in pectin, the greater abundance of AX in grass cell walls may be expected to result in a correlation for ESTs in cereal libraries. For such correlations, it is possible to look at individual loci separately to gain information on which are the best candidates within groups. There is significant correlation between the expression of UDP-GlcAdc and one gene locus from the GT61 family, two GT47 loci, OsCslF6, two GT43 loci, and a GT48 locus (Table IV). All these loci also have significant or near-significant correlation with UDP-Glc dehydrogenase, which is responsible for the synthesis of UDP-GlcA. Coexpression between PF02458 genes and these genes was also examined. This showed highly significant correlations with six GT61 loci and less significant correlations with two more GT61 and two GT2 loci (Table IV). If the PF02458 genes do encode feruloyl transferases, this argues for close association in function between these GT61 genes and feruloylation.
In summary, the evidence above suggests that groups of genes in the families GT61, GT47, GT2 CslC, GT43, GT77, GT48, or GT64 are candidates for AX biosynthesis. Of these, the groups of GT61, GT47, and GT43 families of inverting GTs are the best candidates because they all have both high cereal EST totals for each family of over 400 and also widespread expression in different tissues. The single group of GT2 CslC genes with cereal counts of 141 is not as highly expressed. Similarly, the GT48 group is not as highly expressed (167 cereal ESTs) and the known activity for this family is callose synthesis. The GT77 and GT64 families differ from the others in Table I because they encode enzymes while retaining GT activity, whereas xylan synthase and xylan arabinosyl transferase require inverting activities, so that it seems unlikely that genes in either of these families have a direct role in AX biosynthesis.
Recent experimental results also provide independent support for the hypothesis that GT43 genes encode xylan synthase enzymes. The Arabidopsis knockout mutant of At2g37090, irx9, has an irregular xylem phenotype and markedly decreased Xyl content in its secondary cell walls (Brown et al., 2005) and this change has been suggested to be due to altered xylan (Brown et al., 2005; Bauer et al., 2006). The rice ortholog of IRX9, Os05g03174, is in the GT43 group with the greatest cereal-to-dicot ratio (Table I). The activity of members of the GT43 family has been established in mammals as β-1,3-glucuronosyl transferases, which transfer GlcA to a terminal Gal residue on glycoprotein from UDP-GlcA (Kitagawa et al., 1998). A recent structural and mutational analysis identified key residues that are important for binding the UDP-GlcA molecule in human enzymes (Fondeur-Gelinotte et al., 2006). One of these residues (R156) is conserved in all animal GT43 enzymes, but not in plants (Fondeur-Gelinotte et al., 2006). Interestingly, it appears possible from the structure that this Arg residue stabilizes the carboxylate group in GlcA; its absence in plant enzymes therefore suggests that UDP-Xyl, which is very similar in structure to UDP-GlcA, but lacks this carboxyl group, may be the donor rather than UDP-GlcA.
The known activity for GT61 gene products, including one cloned from Arabidopsis (Strasser et al., 2000), is glycoprotein β-1,2-d-xylosyl transferase, and a closely related gene (PttGT61A) was expressed in poplar tissues during secondary cell wall formation (Aspeborg et al., 2005). However, this particular GT61 orthologous group is not differentially expressed between dicots and cereals and those GT61 genes that do show much greater expression in cereals (Table I) are on different branches within the family. The results in Table IV suggest a close relationship in the expression of these genes with feruloylation. Feruloylated Ara residues on AX are frequently further substituted with β-1,2-d-xylosyl residues in all grass species tested (Wende and Fry, 1997a). These xylosyl residues can be terminal or the start of longer oligosaccharide side chains (Wende and Fry, 1997a, 1997b). It seems probable that the GT61 genes encode the xylosyl transferases responsible for adding these residues to the feruloyl Ara. Consistent with this is the fact that the GT61 groups are the most highly differentially expressed of any GT family (Table I) because this activity would be expected to be absent in dicots.
A GT47 gene family member in Arabidopsis, At2g35100, encodes a putative arabinan α-1,5-arabinosyl transferase (Harholt et al., 2006). The orthologous group for this gene is not highly expressed in cereals, but a GT47 group is highly expressed in cereals and may encode an AX arabinosyl transferase (Table I). A mutant in a Nicotiana plumbaginifolia ortholog to this GT47 group, NpGUT1, was associated with loss of GlcA (although Ara was also decreased) in pectin (Iwai et al., 2002). However, the Arabidopsis ortholog IRX10 (At1g27440) is coexpressed with secondary cell wall-specific CESA and the mutant irx10 has an irregular xylem phenotype (Brown et al., 2005), suggesting a different role than pectin synthesis. Also, poplar orthologs of NpGUT1 and IRX10 (PttGT47A, PttGT47D) were highly expressed during secondary wall formation (Aspeborg et al., 2005). Neither irx10 nor a knockout of a second GT47 gene with a similar phenotype (irx7) has decreased Ara content in their stem cell walls (Brown et al., 2005) and the major hemicellulose in poplar secondary cell walls is glucuronoxylan with little Ara substitution (Aspeborg et al., 2005). We nevertheless judge that the most likely function of these genes in cereals is to transfer Ara residues to the xylan backbone in the synthesis of AX. The higher differential expression of the GT47 group compared to GT43 groups (Table I) is consistent with this hypothesis because arabinosyl substitution of xylan is more common in grasses compared to dicots (Bacic et al., 1988).
In conclusion, the analysis here provides strong support for particular genes within the GT43, GT47, GT61, and PF02458 families being responsible for the synthesis of AX and its side chains. The most likely activities based on the arguments above and some particularly promising candidate genes for investigation are summarized in Table V. The novel approach using EST counts or other transcript abundance data applied here to reach these hypotheses could be readily extended to look for candidate genes responsible for other interspecific differences in plant biochemistry.
Table V.
Putative function of cereal genes in families identified in Tables I and III
Example locus is chosen on the basis of high expression and coexpression with other genes to be one of the most promising candidates.
| Family | Example Locus | Putative Function |
|---|---|---|
| GT61 | Os06g27560 | Feruloyl-AX β-1,2-xylosyl transferase |
| GT47 | Os01g70190 | Xylan α-1,2 or α-1,3-arabinosyl transferase |
| GT43 | Os06g47340 | β-1,4-Xylan synthase |
| PF02458 | Os01g09010 | AX feruloyl transferase |
MATERIALS AND METHODS
Coding nucleotide and protein sequences for all Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) genes were obtained from The Arabidopsis Information Resource (version 6; Rhee et al., 2003) and The Institute for Genomic Research (release 4; Ouyang et al., 2007), respectively. All public ESTs for Arabidopsis, Brassica spp., soybean (Glycine max), potato (Solanum tuberosum), rice, wheat (Triticum aestivum), and barley (Hordeum vulgare) were obtained from the dbEST division of GenBank (Boguski et al., 1993). From these, those which made any reference to normalized or subtractive libraries in the complete GenBank entry were excluded from the analysis because numbers of ESTs in such libraries no longer reflect abundance. About 11% of the ESTs were excluded for this reason, leaving a total of 3.4 million ESTs used in the study.
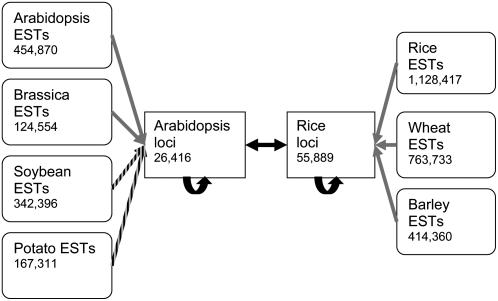
The relationship between rice and Arabidopsis genes and mapping of ESTs to genes was achieved with the BLAST program suite (Altschul et al., 1997) as shown in Figure 2. The rice-Arabidopsis relationship was defined by protein similarity; all gene models (putative splice variants) were compared, but only the highest scoring alignment was used for each locus. Orthologous pairs of rice and Arabidopsis genes were defined as those where the other locus was the top BLASTp hit from the other genome in both directions and were above an alignment score of 200 bits. All nonorthologous loci were defined as paralogs of the closest matching orthologous locus in the same genome above a threshold score of 200 bits. This resulted in 10,855 orthologous groups containing a total of 30,990 rice and 23,581 Arabidopsis loci, leaving 24,899 rice loci designated as having no ortholog. ESTs were assigned to the most similar locus of the appropriate species by sequence similarity (Fig. 2). In some cases, there were multiple EST-loci relationships because of identical alignment scores or ambiguities as to the best match due to multiple aligned regions. In these cases, equal fractions of the EST count were assigned to each locus. EST counts for splice variants were summed to give a single value per locus. Results were obtained and processed with custom Perl scripts employing Bioperl (Stajich et al., 2002) modules and stored in a MySQL database.
Figure 2.
Scheme for mapping of ESTs to genes by sequence similarity. Arabidopsis-rice loci relationships were defined by protein similarity searches (black arrows; BLASTp with bits score >200). The closest matching rice or Arabidopsis gene for every EST was identified by nucleotide similarity search (grey arrow; BLASTn with E < 10−5) or translated nucleotide similarity search (striped arrow; BLASTx with E < 10−5). Number of sequences are indicated.
Library information for ESTs was derived to look at tissue distribution and coexpression of cereal genes. Because formal library information is rarely present in dbEST, library was instead defined by an identifier composed of the species, cultivar, tissue, and stage fields from each EST entry. Libraries where either tissue or stage information was missing were excluded from these analyses. Tissue distribution was examined by assigning to categories tissue fields containing the regular expressions anther, callus, seed|grain|kernel|caryopsis|embryo|endosperm, flower|pistil|carpel, ear|spike|panicle|inflorescence, shoot|stem|leaf|leaves, root|tuber, or whole plant|seedling. In addition to these mutually exclusive classifications, a developing subset was used in an attempt to identify those libraries where primary cell wall synthesis was likely to predominate over secondary cell wall synthesis; this was defined as ESTs where the tissue or stage fields contained developing|growing|immature|young|seedling. The number of ESTs from nonnormalized libraries present in the database for each species and tissue classification is shown in Table VI.
Table VI.
Numbers of ESTs present in the database categorized by species and tissue type
Tissue-type classifications were assigned according to the presence of particular terms within the tissue and/or stage fields of the EST entry.
| Rice | Wheat | Barley | Cereal Counts | Arabidopsis | Brassica | Soybean | Potato | Dicot Counts | |
|---|---|---|---|---|---|---|---|---|---|
| Endosperm | 10,027 | 19,030 | 6,836 | 2% | 9 | 0 | 0 | 0 | 0% |
| Anther | 14,218 | 45,401 | 0 | 3% | 0 | 7,682 | 0 | 0 | 1% |
| Callus | 173,689 | 11,292 | 17,641 | 9% | 2,317 | 0 | 0 | 6,299 | 1% |
| Seed/grain | 39,781 | 189,788 | 62,637 | 13% | 16 | 6,576 | 39,644 | 0 | 4% |
| Flower/pistil/carpel | 203,531 | 49,333 | 0 | 11% | 68,105 | 8,747 | 19,163 | 0 | 9% |
| Spike/panicle | 83,731 | 90,290 | 36,011 | 9% | 6,761 | 0 | 0 | 0 | 1% |
| Shoot/leaf | 314,806 | 185,577 | 188,351 | 30% | 8,194 | 6,290 | 64,122 | 58,384 | 12% |
| Root/tuber | 73,773 | 67,100 | 24,575 | 7% | 185,720 | 11 | 52,382 | 68,868 | 28% |
| Whole plant | 34,957 | 21,028 | 993 | 2% | 12,988 | 16,925 | 104,182 | 0 | 12% |
| Developing | 143,601 | 143,056 | 110,580 | 17% | 49,455 | 33,652 | 222,997 | 15,885 | 29% |
| Total | 1,128,417 | 776,574 | 401,813 | 100% | 454,870 | 129,336 | 342,406 | 17,4344 | 100% |
Coexpression between rice loci was calculated based on the correlation of the number of associated cereal ESTs across libraries. Only libraries that contained more than 10,000 ESTs in total were used to avoid the problem of excessive influence of small libraries after normalization. This gave rise to 63 libraries for the cereal EST counts. Correlations were calculated from counts per library divided by total counts in the library to correct for variation in library size (Haverty et al., 2004).
Prediction of transmembrane domains from sequence is still uncertain and the various software tools generate probability of their occurrence. The Aramemnon (Schwacke et al., 2003) database gives a consensus prediction from many tools according to a set of criteria. However, many proteins classified as nonmembrane in Aramemnon are predicted to be membrane by one or more of these tools. To define a set of rice loci encoding putative integral membrane proteins in an inclusive manner, all Arabidopsis and all rice genes defined as integral membrane proteins in the Aramemnon database, release 4, were taken and all orthologous groups (as defined above) in which any of these genes occurred identified. This led to a set of 3,071 orthologous groups containing a total 6,046 rice loci and a further 4,550 rice loci with no Arabidopsis orthologs. This was used as the global set for analysis. A subset was identified in which any of the rice or Arabidopsis genes in a group were present in the CAZy database (Coutinho et al., 2003) and identified as encoding putative GTs. This CAZy subset constituted 126 orthologous groups with a total of 323 rice loci and a further 56 rice loci with no orthologs and was the dataset used to generate the results in Tables I and II and Supplemental Table S1. The remainder of the global set was used to generate the results in Table III and Supplemental Table S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Complete dataset of EST counts for the CAZy set of loci from which the summaries in Tables I and II are derived.
Supplemental Table S2. Complete dataset of EST counts for the complete set of loci defined as encoding putative membrane proteins derived from Aramemnon, excluding those in the CAZy set.
Supplemental Table S3. Sensitivity of results in Table I to choosing subsets of ESTs based on tissue and stage annotation.
Supplementary Material
Acknowledgments
We thank Dr. Sue Welham, Rothamsted Research, for statistical advice and Dr. Rainer Schwacke, University of Cologne, for help with use of the Aramemnon database.
This work was supported by a grant-in-aid from the Biotechnology and Biological Sciences Research Council of the United Kingdom to Rothamsted Research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rowan A.C. Mitchell (rowan.mitchell@bbsrc.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspeborg H, Schrader J, Coutinho PM, Stam M, Kallas A, Djerbi S, Nilsson P, Denman S, Amini B, Sterky F, et al (2005) Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol 137 983–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacic A, Harris PJ, Stone BA (1988) Structure and function of plant cell walls. In J Preiss, ed, The Biochemistry of Plants, Vol 14. Academic Press, San Diego, pp 297–372
- Bauer S, Vasu P, Persson S, Mort AJ, Somerville CR (2006) Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc Natl Acad Sci USA 103 11417–11422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencur P, Steinkellner H, Svoboda B, Mucha J, Strasser R, Kolarich D, Hann S, Kollensperger G, Glossl J, Altmann F, et al (2005) Arabidopsis thaliana beta 1,2-xylosyltransferase: an unusual glycosyltransferase with the potential to act at multiple stages of the plant N-glycosylation pathway. Biochem J 388 515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, Lowe TMJ, Tolstoshev CM (1993) dbEST—Database for expressed sequence tags. Nat Genet 4 332–333 [DOI] [PubMed] [Google Scholar]
- Boutanaev AM, Kalmykova AI, Shevelyou YY, Nurminsky DI (2002) Large clusters of co-expressed genes in the Drosophila genome. Nature 420 666–669 [DOI] [PubMed] [Google Scholar]
- Bouton S, Leboeuf E, Mouille G, Leydecker MT, Talbotec J, Granier F, Lahaye M, Hofte H, Truong HN (2002) Quasimodo1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14 2577–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Stone BA, Newbigin EJ, Bacic A, Fincher GB (2006) Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-d-glucans. Science 311 1940–1942 [DOI] [PubMed] [Google Scholar]
- Carpita NC (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47 445–476 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary-cell walls in flowering plants—consistency of molecular-structure with the physical-properties of the walls during growth. Plant J 3 1–30 [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328 307–317 [DOI] [PubMed] [Google Scholar]
- Dhugga KS (2005) Plant Golgi cell wall synthesis: from genes to enzyme activities. Proc Natl Acad Sci USA 102 1815–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J, Skjot M, Geshi N, Ulvskov P, Petersen BL (2004) A complementary bioinformatics approach to identify potential plant cell wall glycosyltransferase-encoding genes. Plant Physiol 136 2609–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondeur-Gelinotte M, Lattard V, Oriol R, Mollicone R, Jacquinet JC, Mulliert G, Gulberti S, Netter P, Magdalou J, Ouzzine M, et al (2006) Phylogenetic and mutational analyses reveal key residues for UDP-glucuronic acid binding and activity of beta 1,3-glucuronosyltransferase I (GlcAT-I). Protein Sci 15 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunneras S, et al (2006) Poplar carbohydrate-active enzymes: gene identification and expression analyses. Plant Physiol 140 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabber JH (2005) How do lignin composition, structure, and cross-linking affect degradability? A review of cell wall model studies. Crop Sci 45 820–831 [Google Scholar]
- Harholt J, Jensen JK, Sorensen SO, Orfila C, Pauly M, Scheller HV (2006) ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol 140 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PJ, Kelderman MR, Kendon MF, McKenzie RJ (1997) Monosaccharide compositions of unlignified cell walls of monocotyledons in relation to the occurrence of wall-bound ferulic acid. Biochem Syst Ecol 25 167–179 [Google Scholar]
- Haverty PM, Hsiao LL, Gullans SR, Hansen U, Weng ZP (2004) Limited agreement among three global gene expression methods highlights the requirement for non-global validation. Bioinformatics 20 3431–3441 [DOI] [PubMed] [Google Scholar]
- Iwai H, Masaoka N, Ishii T, Satoh S (2002) A pectin glucuronyltransferase gene is essential for intercellular attachment in the plant meristem. Proc Natl Acad Sci USA 99 16319–16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa H, Tone Y, Tamura J, Neumann KW, Ogawa T, Oka S, Kawasaki T, Sugahara K (1998) Molecular cloning and expression of glucuronyltransferase I involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. J Biol Chem 273 6615–6618 [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K (2005) Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci USA 102 2221–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil M, Darvill AG, Fry SC, Albersheim P (1984) Structure and function of the primary-cell walls of plants. Annu Rev Biochem 53 625–663 [DOI] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al (2007) The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res 35 D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei HR, Milne J, Page GP, Somerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard C, Thibault JF (1993) Structure and properties of apple and sugar-beet pectins extracted by chelating-agents. Carbohydr Res 244 99–114 [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen GH, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR (2000) The cellulose synthase superfamily. Plant Physiol 124 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flugge UI, Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JGR, Korf I, Lapp H, et al (2002) The bioperl toolkit: Perl modules for the life sciences. Genome Res 12 1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Mucha J, Mach L, Altmann F, Wilson IBH, Glossl J, Steinkellner H (2000) Molecular cloning and functional expression of beta 1,2-xylosyltransferase cDNA from Arabidopsis thaliana. FEBS Lett 472 105–108 [DOI] [PubMed] [Google Scholar]
- Wende G, Fry SC (1997. a) 2-O-β-d-xylopyranosyl-(5-O-feruloyl)-l-arabinose, a widespread component of grass cell walls. Phytochemistry 44 1019–1030 [Google Scholar]
- Wende G, Fry SC (1997. b) O-feruloylated, O-acetylated oligosaccharides as side-chains of grass xylans. Phytochemistry 44 1011–1018 [DOI] [PubMed] [Google Scholar]
- Wilkie KCB (1979) The hemicelluloses of grasses and cereals. Adv Carbohydr Chem Biochem 36 215–264 [Google Scholar]
- Yang Q, Reinhard K, Schiltz E, Matern U (1997) Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA: anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol Biol 35 777–789 [DOI] [PubMed] [Google Scholar]
- Zhang QS, Shirley N, Lahnstein J, Fincher GB (2005) Characterization and expression patterns of UDP-D-glucuronate decarboxylase genes in barley. Plant Physiol 138 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.