Abstract
Cork (phellem) is a multilayered dead tissue protecting plant mature stems and roots and plant healing tissues from water loss and injuries. Cork cells are made impervious by the deposition of suberin onto cell walls. Although suberin deposition and cork formation are essential for survival of land plants, molecular studies have rarely been conducted on this tissue. Here, we address this question by combining suppression subtractive hybridization together with cDNA microarrays, using as a model the external bark of the cork tree (Quercus suber), from which bottle cork is obtained. A suppression subtractive hybridization library from cork tree bark was prepared containing 236 independent sequences; 69% showed significant homology to database sequences and they corresponded to 135 unique genes. Out of these genes, 43.5% were classified as the main pathways needed for cork biosynthesis. Furthermore, 19% could be related to regulatory functions. To identify genes more specifically required for suberin biosynthesis, cork expressed sequence tags were printed on a microarray and subsequently used to compare cork (phellem) to a non-suberin-producing tissue such as wood (xylem). Based on the results, a list of candidate genes relevant for cork was obtained. This list includes genes for the synthesis, transport, and polymerization of suberin monomers such as components of the fatty acid elongase complexes, ATP-binding cassette transporters, and acyltransferases, among others. Moreover, a number of regulatory genes induced in cork have been identified, including MYB, No-Apical-Meristem, and WRKY transcription factors with putative functions in meristem identity and cork differentiation.
Land plants have evolved lipophilic barriers that protect the internal living tissues from dehydration, injuries, and pathogens, and have evolved regulatory networks to adjust the barriers to the changing physiological and environmental conditions of the plant. Plant primary organs, such as young stems and leaves, are protected by the cuticle, a lipophilic extracellular polymer membrane composed of cutin and waxes. Secondary (mature) stems and roots, tubers, and healing tissues are protected by cork, a tissue with multiple layers of cells that are dead at maturity. Key compounds for cork impermeability are suberin, a complex polymer comprising both aliphatic and aromatic domains, and associated waxes. Cork is part of the plant constitutive defense system and contains secondary compounds such as triterpenoids and soluble phenylpropanoids that act on herbivores, microbes, and fungi.
Cork, or phellem, which is the technical term for cork, is formed by the phellogen (cork cambium). Cork formation involves proliferation and commitment of the phellogen derivatives, cell expansion and extensive deposition of suberin and waxes, and an irreversible program of senescence ending in cell death. The two best known and most studied examples of cork are the suberized skin of potato (Solanum tuberosum) tuber (Sabba and Lulai, 2002) and the bark of the cork tree (Quercus suber), from which bottle cork is obtained (Silva et al., 2005). In the cork tree, the phellogen forms a continuous layer of cells that envelops the tree trunk and produces, each year, a 2- to 3-mm thick layer of almost pure cork that adheres to that of the previous year (Caritat et al., 2000). The chemical composition of the cork tree bark has been widely analyzed by chemical fractionation (for review, see Silva et al., 2005). Although the amounts of the different components can show significant variations (Pereira, 1988; Lopes et al., 2001), on average it contains 15% extractives (7.5% waxes and 7.5% tannins), 41% aliphatic suberin (referred to as suberin in cork tree literature), 22% aromatic suberin (referred to as lignin in cork tree literature), 20% polysaccharides, and 2% ashes (Pereira, 1988). The monomeric composition of the aliphatic (suberin) fraction has been analyzed by Holloway (1983) and by Bento et al. (1998) and that of the aromatic (lignin) fraction by Marques et al. (1999) and Lopes et al. (2000), among others. Waxes have been analyzed by Castola et al. (2005) and mainly consist of terpenes and sterols. Tannins, mostly ellagitannins, and other soluble polyphenols have been analyzed by Cadahia et al. (1998) and Varea et al. (2001). Many of the enzymatic activities necessary for cork biosynthesis may be inferred from the chemical composition. Cork is a site for four main secondary metabolic pathways: acyl-lipids, phenylpropanoids, isoprenoids, and flavonoids. The acyl-lipids pathway is required for the biosynthesis of the linear long-chain compounds forming the aliphatic suberin domain, which share upstream reactions with cutin biosynthesis (Kolattukudy, 1981; Nawrath, 2002; Heredia, 2003; Kunst and Samuels, 2003). The phenylpropanoids pathway is needed for the biosynthesis of the cork aromatic components, which share the same basic reactions with wood lignin (Dixon et al., 2002; Boerjan et al., 2003). The isoprenoids pathway is needed for wax terpenes (Laule et al., 2003) and sterols (Benveniste, 2004), and the flavonoids pathway for tannins (Koes et al., 2005).
Suberin, the main cork component, is defined in literature as a complex biopolymer found in suberized cells that comprises an aliphatic cutin-like and an aromatic lignin-like domain (Bernards, 2002). The aliphatic domain is a glycerol-bridged polyester with associated esterified phenolics (Moire et al., 1999; Graca and Santos, 2006). The aromatic domain is a polyphenolic substance mostly composed of hydroxycinnamic acid derivatives and is presumably involved in linking the aliphatic domain to cell wall (Kolattukudy, 1980; Bernards and Lewis, 1998). The three-dimensional structure of suberin is not yet clear, although several tentative models have been presented (Kolattukudy, 1981, 2001; Lopes et al., 2000; Bernards, 2002). These models discuss possible linkages within the suberin and postulate linkages between suberin and the lignin/carbohydrate cell wall matrix. Only a few studies report enzymatic activities involved in suberization, and most of them deal with the aromatic metabolism. Cottle and Kolattukudy (1982) and Bernards and Razem (2001) demonstrated the induction in suberizing tissues of key enzymes capable of generating hydroxycinnamic acids and proposed a biosynthetic pathway for the aromatic domain based in lignin biosynthesis. Peroxidase activity associated with suberization was detected in potato tuber (Espelie et al., 1986; Bernards et al., 1999) and in tomato (Solanum lycopersicum) root tissue (Quiroga et al., 2000). The presence of a hydrogen peroxide (H2O2) generating system necessary for the peroxidase activity was indirectly demonstrated by Razem and Bernards (2003). Experimental evidence for enzymes involved in the metabolism of the aliphatic domain is limited to the in vitro demonstration of an ω-hydroxyacid oxidation (Agrawal and Kolattukudy, 1977), the demonstration of an elongase activity in maize (Zea mays) roots (Schreiber et al., 2005), and the isolation from potato tuber discs of a ferulate acyltransferase possibly involved in linking the aromatic monomer ferulate to the aliphatic domain (Lotfy et al., 1994, 1995). The biosynthetic pathway for the aliphatic monomers has been hypothesized (Kolattukudy, 2001; Bernards, 2002; Franke et al., 2005). It is accepted that the synthesis begins with the general fatty acid (FA) synthesis pathway giving rise to long-chain FAs (LCFAs), that the condensation of very LCFAs (VLCFAs) takes place in the endoplasmic reticulum, and that P450 monooxygenases catalyze most FA oxidation reactions. The three-dimensional polyester is thought to be achieved by esterification between FA, with glycerol acting as an important small cross-linker (Kolattukudy, 2001). However, transport of the aliphatic monomers and polymerization in the apoplast are still unknown.
Although cork and suberin are critical to the life of both herbaceous and woody plants, molecular genetic approaches are still lacking (Yephremov and Schreiber, 2005). Today, analysis of suberin biosynthesis and function should be conducted using suberin-defective mutants, which cannot be easily obtained with cork tree. A much better choice would be Arabidopsis (Arabidopsis thaliana); however, despite the fact that Arabidopsis synthesizes suberin and develops a phellem, only two mutants with altered suberin or defective phellem have been identified to date: elongation defective1, a pleiotropic mutant showing ectopic suberin deposition (Cheng et al., 2000), and, very recently, a knockout mutant for the glycerol-3-P acyltransferase 5 gene (GPAT5; Beisson et al., 2007).
Molecular genetic approaches to suberin are limited to the cloning and characterization of suberin-associated peroxidases in potato (Roberts and Kolattukudy, 1989), tomato (Quiroga et al., 2000), and muskmelon (Cucumis melo; Keren-Keiserman et al., 2004), but their role has not clearly been proven (Sherf et al., 1993; Lucena et al., 2003). Cuticle mutants have been identified for Arabidopsis, a fact that allowed researchers to identify a number of genes necessary for the synthesis and transport of cutin and wax (Jenks et al., 2002; Kunst and Samuels, 2003; Yephremov and Schreiber, 2005). Recently, a genome-wide study of the shoot epidermis of Arabidopsis (Suh et al., 2005) highlighted a series of new candidate genes relevant in cutin and wax synthesis. In lignin research, molecular genetic approaches have been widely used in the past. Global xylem transcript profiling has been reported for Arabidopsis (Ko et al., 2004; Ehlting et al., 2005) and for several tree species (Whetten et al., 2001; Kirst et al., 2004; Andersson-Gunneras et al., 2006). For a number of genes, involvement in lignin biosynthesis was demonstrated by forward and reverse genetic approaches (Anterola and Lewis, 2002; Sibout et al., 2005; Abdulrazzak et al., 2006). An excellent review (Carlsbecker and Helariutta, 2005) summarizes this knowledge of the molecular genetics of regulatory networks in xylem.
To reveal the genetic repertoire of cork cells and to identify genes likely to be related to suberin synthesis, we used a two-step strategy. First, by means of suppression subtractive hybridization (SSH), a library of ESTs preferentially induced in cork was obtained. Then, these ESTs were printed on a microarray and subsequently used for a global comparison between a suberin-producing (cork/phellem) and a non-suberin-producing (wood/xylem) tissue. Isolation of suberin genes in the cork tree is particularly attractive because of its exceptional capacity to produce suberin. Peeling of the external bark from the cork tree trunk allowed the harvesting of differentiating cork layers (Fig. 1A) and provided a highly enriched material for molecular investigations. In the following pages, we present an initial analysis of the genomics of cork cells in cork tree bark; as far as we know, this is the first global approach to cork and suberin molecular biology.
Figure 1.
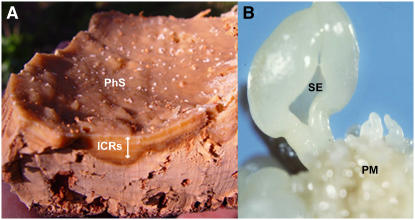
Cork (tester) and somatic embryo (driver) tissues used for SSH. A, Piece of cork showing the internal phellodermic surface (PhS) and, in the transversal section, the inner cork rings (ICRs) consisting of cells in different stages of suberization and dead cells that still contain a high amount of water. RNA used in our experiments was extracted from scrapings taken from the phellogenic surface comprising the inner cork rings. B, Proliferative mass (PM) obtained from cork tree somatic embryo (SE) cultures. [See online article for color version of this figure.]
RESULTS
Cork Subtractive Library
Suberin is a product of the secondary metabolism that is regulated in a tissue-specific manner. It was our intention to find candidate genes for cork and suberin biosynthesis; therefore, we chose as driver tissue for the SSH a fully undifferentiated tissue consisting of the proliferative mass obtained from cork tree somatic embryo cultures (Fig. 1B). The proliferative mass is a translucent, fully undifferentiated, nonvascularized tissue that develops in the hypocotyls of the recurrent somatic embryos. Cork tree somatic embryogenesis has been carefully characterized at anatomical and ultrastructural levels in previous works (Puigderrajols et al., 1996, 2001). Neither suberin deposition nor multilamellated cell walls could be detected in somatic embryos using optical and fluorescent microscopy techniques or by electron microscopy. The extraction of RNA from cork is difficult due to its high proportion of dead cells and phenols; however, using the protocol of Chang et al. (1993) that prevents oxidation of phenols, good quality RNA was obtained. A further mRNA purification step was added to reduce rRNA contamination and to increase SSH efficiency. Finally, the SSH products were cloned into pCR4-TOPO vector, resulting in a library of 975 cDNA clones. Subtraction efficiency was checked by cohybridizing tester (cork) and driver (embryo) RNAs on the cork tree microarray described in the “Materials and Methods” section. This control experiment confirmed a high yield of subtraction efficiency; 96% of reliable ESTs (coefficient of variation < 0.3) were up-regulated in cork (fold change [FC] > 1.5), and 75% of reliable ESTs showed signal intensity not significantly higher than background in embryo (data are shown in Supplemental Table S2).
Single-run sequencing of the library yielded 694 readable sequences longer than 100 bp. Of these, 579 grouped into 121 contiguous sequences (contigs) and 115 were single sequences (singletons). Thus, in total, 236 independent sequences were obtained. Sequence redundancy (100 × [1 − {contigs + singletons/readable sequences}]) was of 66%. BLASTX analysis (Altschul et al., 1990) showed that 69% of the independent sequences (71 singletons and 92 contigs) showed high sequence homology at the amino acid level to database sequences (e value < e−21) and that 31% (44 singletons and 29 contigs) could not be assigned to any gene ontology and were classified as no hits (e value > e−10). Sequences with the same GenBank entry were assumed to represent the same gene, and, thus, the library was found to contain 135 unique genes (121 with assigned function and 14 with unknown function). The genes with assigned function were manually grouped into functional categories using National Center for Biotechnology Information, The Arabidopsis Information Resource (TAIR), and published data. Categories were established taking into account the main metabolic and cellular processes leading to cork biosynthesis. Table I shows a selected list of genes with putative functions thought to be important for suberin biosynthesis and cork regulation grouped in functional categories. The complete list is given in Supplemental Table S1.
Table I.
Selected list of the more relevant candidate genes for suberin biosynthesis and cork regulation grouped in functional categories
The complete list of genes is reported on Supplemental Table S1. For each gene, the EST GenBank accession numbers and the putative molecular function are given. The putative functions were assigned based on the highest BLASTX score match (e value < 10−20) and not always are supported by biochemical data in plants. The cork to wood expression ratio is given as FC for genes with good evidence of being differential expressed (B > 3). N is the number of ESTs in the library.
| Functional Categories | EST GenBank Accession No. | Description | Best TAIR BLASTX | FC | N |
|---|---|---|---|---|---|
| Acyl lipids | |||||
| FA synthesis | EE743836 | Dihydrolipoamide S-acetyltransferase, putative | At3g25860 | 6 | 2 |
| FA synthesis | EE743871 | Biotin carboxyl carrier protein subunit | At5g15530 | 45 | 1 |
| FA elongation | EE743843 | PATE/FAT | At1g08510 | 21 | 5 |
| FA elongation | EE743810, EE743811 | LACS, putative | At1g49430 | 41 | 21 |
| FA elongation | EE743856 | ACL, putative | At5g49460 | 7 | 5 |
| FA elongation | EE743839 | KCS, putative | At5g43760 | 46 | 5 |
| FA elongation | EE743779, EE743679 | KCR, putative | At1g67730 | 21 | 2 |
| FA hydroxylation | EE743846, EE745211, EE743708 | FA ω-hydroxylase, CYP86A1 | At5g58860 | 38 | 6 |
| Lipid oxidation | EE743788, EE743799, EE743803 | LOX-1 | At3g22400 | 24 | 8 |
| Lipid oxidation | EE743678 | LOX-1 | At1g55020 | – | 1 |
| Lipid metabolism (putative) | EE743851 | GDSL-motif lipase/hydrolase, putative | At2g23540 | 49 | 14 |
| Lipid metabolism (putative) | EE743823, EE743824, EE743825 | GDSL-motif lipase/hydrolase-like protein | At5g37690 | 35 | 33 |
| Lipid metabolism (putative) | EE743815, EE743814 | GDSL-motif lipase/hydrolase-like protein | At5g22810 | 29 | 8 |
| Lipid metabolism (putative) | EE743858, EE743859 | GDSL-motif lipase/hydrolase family protein, similar to family II lipases | At1g74460 | 24 | 8 |
| Lipid metabolism (putative) | EE743686, EE743687 | GDSL-motif lipase/hydrolase family protein, putative APG protein | At4g26790 | 11 | 5 |
| Lipid metabolism (putative) | EE743698 | GDSL-motif lipase/hydrolase family protein, putative nodulin | At1g54790 | 3 | 1 |
| Lipid metabolism (putative) | EE743672 | Esterase/lipase/thioesterase family protein | At3g62860 | 33 | 1 |
| Lipid metabolism (putative) | EE743671 | Lipase class 3 family protein | At4g00500 | 5 | 1 |
| Glycerol ester synthesis | EE743864, EE743865 | GPAT | At3g11430 | 41 | 7 |
| Glycerol ester synthesis | EE743668 | GPAT | At1g01610 | 9 | 4 |
| Lipid transfer | EE743694 | ABC transporter protein (WBC subfamily), putative | At5g13580 | 32 | 1 |
| Phenylpropanoids | |||||
| Phenylpropanoid pathway | EE743744 | PAL, putative | At2g37040 | 19 | 3 |
| Phenylpropanoid pathway | EE743705, EE743706 | PAL | At3g53260/At2g37040 | 8 | 7 |
| Phenylpropanoid pathway | EE743662 | Cinnamate 4-hydroxylase, CYP73 | At2g30490 | 2 | 1 |
| Phenylpropanoid pathway | EE743863 | 4CL | At3g21240 | 3 | 1 |
| Phenylpropanoid pathway | EE743677 | 4CL | At1g51680 | – | 1 |
| Phenylpropanoid pathway | EE743816 | Cinnamoyl CoA reductase | At1g15950 | – | 3 |
| Phenylpropanoid derivatives | EE743676, EE743847 | F5H, CYP84A1, putative | At4g36220 | 11 | 2 |
| Acyltransferase | EE743861 | HCBT | At5g41040 | 32 | 27 |
| Acyltransferase | EE743848, EE745210, EE743849 | HCBT, putative | At5g41040 | 36 | 13 |
| Cross-linking/oxidase | EE743777, EE743766 | Diphenol oxidase laccase | At5g05390 | 8 | 16 |
| Cross-linking/oxidase | EE743857 | Laccase | At2g40370 | 9 | 6 |
| Cross-linking/oxidase | EE743784, EE743807 | Multicopper oxidase, putative | At5g05390 | 6 | 4 |
| Cross-linking/oxidase | EE743710 | Copper-containing amine oxidase | At4g12290 | 2 | 1 |
| Miscellaneous | |||||
| Monoxygenase | EE743874, EE745209, EE745208 | Cytochrome P450 protein, CYP72A14 | At3g14680 | 65 | 16 |
| Monoxygenase | EE743711, EE743700 | Cytochrome P450 protein, CYP72A15 | At3g14690 | 50 | 32 |
| Monoxygenase | EE743852, EE743841 | Cytochrome P450 protein, CYP72A7 | At3g14610 | 61 | 21 |
| Monoxygenase | EE743868 | Cytochrome P450-like protein, CYP86B2 | At5g08250 | 23 | 2 |
| Monoxygenase | EE743831 | Cytochrome P450 family protein, CYP87A2 | At1g12740 | 30 | 10 |
| Monoxygenase | EE743830 | Cytochrome P450, CYP72A59 | At3g14680 | 49 | 6 |
| Acyltransferase | EE743818, EE743866 | BAHD acyltransferase, putative | At1g24430 | 48 | 28 |
| Transfer protein | EE743658 | ABC transporter like protein (ATH subfamily) | At2g40090 | 79 | 1 |
| Stress | |||||
| Reactive oxygen species scavenging | EE743659 | Ascorbate peroxidase, putative | At3g09640 | 67 | 1 |
| Regulatory Proteins | |||||
| Transcription factor | EE743870 | MYB-related transcription factor | At5g52660 | 60 | 1 |
| Transcription factor | EE743680 | R2R3-MYB transcription factor | At3g49690 | 3 | 1 |
| Transcription factor | EE743809 | WRKY transcription factor | At2g46130 | 28 | 4 |
| Transcription factor | EE743827 | NAM-like protein | At3g18400 | 16 | 5 |
| Transcription factor | EE743667 | NAM-like protein | At5g13180 | 3 | 1 |
| Transcription factor | EE743828 | SQUAMOSA promoter-binding protein like | At2g47070 | 6 | 3 |
| Transcription regulator | EE743879 | AS1-interacting KH protein | At3g29390 | 5 | 1 |
| Peptide receptor | EE743817 | PSKR, Leu-rich repeat receptor kinase | At2g02220 | 7 | 2 |
| Phosphatase activity | EE743872 | Protein phosphatase 2C, putative | At4g31750 | 4 | 1 |
| Signal transduction | EE743878 | Annexin, chain A | At1g35720 | 27 | 1 |
| Signal transduction | EE743733 | Calmodulin-binding family protein | At3g59690 | – | 2 |
| Ethylene forming | EE743812 | ACC oxidase | At1g05010 | – | 2 |
| Ethylene forming | EE743887 | ACC oxidase, putative | At2g19590 | – | 1 |
| Regulated proteolysis | EE743670 | Major surface glycoprotein like | At5g42620 | 6 | 1 |
| Regulated proteolysis | EE743819 | Cys proteinase | At4g39090 | 81 | 1 |
| Regulated proteolysis | EE743884 | 26S proteasome regulatory subunit, putative | At5g09900 | 91 | 1 |
| Regulated proteolysis | EE743682 | 26S proteasome regulatory particle non-ATPase subunit12 | At1g64520 | 4 | 1 |
| Regulated proteolysis | EE743666 | Ubiquitin-fusion degradation protein like | At5g15400 | 4 | 1 |
| Regulated proteolysis | EE743674 | RING-H2 zinc finger protein, ubiquitin ligase, putative | At1g72220 | – | 1 |
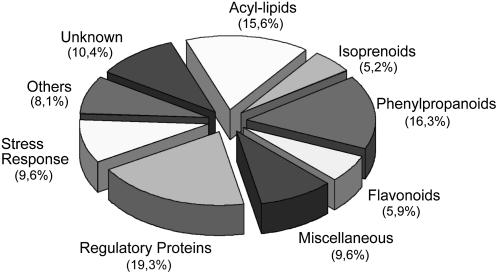
The relative contribution of the genes to the different categories is shown in Figure 2. Acyl lipids, isoprenoids, phenylpropanoids, and flavonoids, the four categories that represent the major pathways for the synthesis of cork chemical components, amounted to 43.5% of the genes. The regulatory proteins category, which includes transcriptional regulation, signal transduction, and regulated proteolysis-related genes, amounted to 19% of the genes. The category stress, which combines genes related to detoxifying enzymes and cell wall strengthening, amounted to 9.5%; and the category unknown, which groups those genes with no assigned biological function, amounted to 10%. The genes not fitting into any of the above classes were grouped according to their annotations into two different categories named miscellaneous and others. The miscellaneous category, which includes genes compatible with the main pathways leading to cork biosynthesis but whose substrates have not been characterized, amounted to 9%. The genes in others are not further discussed in this article. On the other hand, as can be observed in Table I and Supplemental Table S1, the number of ESTs (N, redundancy) showed remarkable differences among the genes. In SSH libraries, although SSH should, in principle, decrease the frequency of abundant transcripts while increasing the probability of rare transcripts, genes both differentially and strongly expressed become overrepresented (Ranjan et al., 2004). Therefore, with all precautions, we can hypothesize that genes showing a high redundancy are preferentially and strongly expressed in cork. This is the case of genes encoding long-chain acyl CoA synthase (LACS), hydroxycinnamoyl-CoA/benzoyl transferase (HCBT), and cytochromes of the CYP72A subfamily, among others.
Figure 2.
Relative contribution of the cork library genes to the different functional categories used in this work.
Differential Screening between Phellem and Xylem by Microarray Hybridization
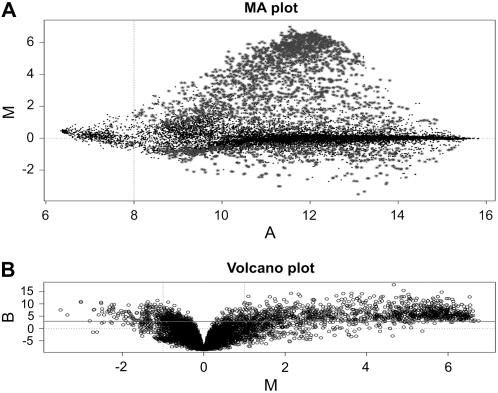
Cork (phellem) and wood (xylem) tissues have common features: both originate from secondary (cambial) meristems and both synthesize aromatic polymers. However, only cork tissue produces suberin and associated waxes. Therefore, to identify genes mostly related to suberin synthesis, the cork ESTs from our library were printed on a microarray (for details, see “Materials and Methods”) and hybridized to cork and wood tissue. For hybridization, cork and wood RNA was obtained from field-grown cork trees during the vegetative season when both cambial layers are in full activity. Three independent cork trees (biological replicates) were sampled and a dye swap for each biological sample (technical replicates) was performed. Microarray data were lowess normalized to account for intensity-dependent differences between channels. After normalization, dye swap replicates showed no strong deviations from linearity (Fig. 3A), proving low dye bias. The comparison between the biological and technical replicates showed a high degree of interarray reproducibility, with Pearson's correlation coefficients ranging from 0.95 to 0.98 (Supplemental Fig. S1). To select those genes with good evidence of being differentially expressed, we used a Volcano plot (B/M, odds versus ratio; Fig. 3B) and established a cutoff of B > 3. FC values for genes with B > 3 are given in Table I and Supplemental Table S1; all data can be found in Supplemental Table S2.
Figure 3.
Plots illustrating the quality criteria applied in microarray hybridization. Average data over six hybridizations of cork versus wood are shown. A, MA plot: expression ratio versus intensity. Bigger dots correspond to ESTs differentially expressed. No dye bias can be observed in this plot. B, Volcano (BM) plot: odds of differential expression versus ratio. Genes with log odds greater than 3 (over the solid horizontal line) are considered as differentially expressed. Note that most spots are cork up-regulated.
The great majority of the library genes were up-regulated in cork (B > 3, FC > 2). Regarding the main cork biosynthetic pathways, genes within the acyl lipids and isoprenoid categories, more relevant for suberin and wax biosynthesis, showed much higher FC values than genes within the phenylpropanoids and flavonoids categories, more relevant for aromatic compounds biosynthesis. The fact that most genes in the phenylpropanoids category were up-regulated in cork could indicate that specific paralogs are induced in this tissue. This hypothesis is supported, for instance, by two 4-coumarate: CoA ligase 1 (4CL)-coding genes, with only one paralog differentially expressed in cork. Quite the opposite, the two paralogs coding HCBT are both strongly cork up-regulated (FC = 37 and FC = 32, respectively). Because such a strong cork induction suggests a specific role in cork synthesis, HCBT could be a key enzyme for synthesis of phenylpropane derivatives characteristic of suberin, such as feruloyltyramine. Most genes of the acyl-lipids category were strongly up-regulated in cork. This applies to genes possibly involved in the synthesis of suberin monomers, such as the ω-hydroxylase CYP86A1 or the β-ketoacyl-CoA synthase (KCS), and enzymes that catalyze ester bonds, such as GPAT. The putative lipases/esterases, including GDSL-motif putative lipases, were highly cork up-regulated, and, although the lipase function of these proteins has not been proven in plants, a possible lipase role in cork cannot be discarded. Moreover, interestingly, the highest FC values within the functional categories were shown by genes of the miscellaneous category. This is a remarkable result taking into account that the miscellaneous category contains genes encoding enzymes, such as cytochrome P450s, transporters, and one putative acyltransferase, which may catalyze reactions important in the biosynthesis of suberin or other cork chemical components.
With regard to the stress, regulatory proteins, and others categories, changes of the FC within each category showed diverse behaviors. Two genes involved in regulated proteolysis (ubiquitin/26S proteasome regulatory subunit, FC = 91; Cys proteinase, FC = 81) were the two most phellem up-regulated genes in the library. It should also be noted that some transcription factors (MYB, FC = 60; WRKY, FC = 28; and No-Apical-Meristem [NAM], FC = 16) and some signal transduction genes (protein kinase, FC = 31; calcium-binding annexin, FC = 27) exhibited high FC values. On the other hand, all genes of the unknown category showed strong cork up-regulation, a fact pointing to possible phellem-specific functions for these genes.
Validation of Differential Expression of Genes by Reverse Transcription-PCR
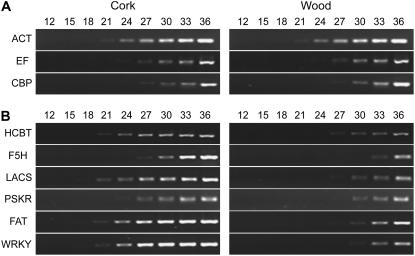
We used the reverse transcription (RT)-PCR with incremental cycle numbers to validate the cork-to-wood gene expression ratios measured by the microarray. The transcript abundance was analyzed for six relevant candidate genes having moderate to high FC values. The genes selected for validation were: HCBT, ferulate 5-hydroxylase (F5H), LACS, palmitoyl-acyl carrier protein thioesterase (PATE/FAT), WRKY transcription factor (WRKY), and phytosulfokine receptor (PSKR). As control, the transcript levels of three constitutive genes (actin, elongation factor, and cap-binding protein) were measured to verify that equal amounts of cDNA were used for both tissues (Fig. 4A). Gene-specific oligonucleotides (Supplemental Table S3) were used in PCR reactions containing equal amounts of both cork and wood cDNAs as templates. Products of incremental cycle numbers were subsequently analyzed. The difference in cycle numbers required for equal amplification of the corresponding PCR product in cork and wood, respectively, was used to estimate levels of differences in expression within the two tissues. The cDNA was obtained from the three biological replicates used in the microarray hybridization. The possible contamination by genomic DNA was excluded using actin primers specifically designed to differentiate genomic DNA from cDNA.
Figure 4.
RT-PCR analysis of transcripts differentially expressed between cork and wood after incremental PCR cycles. Equal amounts of cork and wood cDNA were used as template for each PCR reaction. PCR products were analyzed at each cycle number indicated. Note that amplification products of housekeeping genes (A) show no differences between both tissues, whereas target genes (B) exhibit moderate to more pronounced differences.
Results of RT-PCR with incremental cycle numbers (Fig. 4B) confirmed the differential expression of all six selected genes. Amplification products of their transcripts exhibited differences of three to nine cycles, which correspond to the cork-to-wood gene expression ratios measured by the microarray.
DISCUSSION
We report a collection of cork genes potentially important for cork biosynthesis and differentiation based on sequence homology and microarray comparison. This list includes a set of genes possibly involved in the biosynthesis, transport, and polymerization of suberin that, in general, agrees with the biosynthetic pathway suggested by Kolattukudy (2001), Bernards (2002), and Franke et al. (2005). The list also contains a number of putative cork regulatory genes that might be of particular interest, considering the lack of knowledge in this field. Finally, a number of genes with unknown function strongly induced in cork appeared in this study. Although this work does not prove the involvement of the candidate genes in cork differentiation, on the basis of this study, direct experimental approaches can be designed.
In the two following sections, we discuss the putative roles of a set genes potentially relevant for suberin biosynthesis and the regulation of cork differentiation, considering available data on homologous (best BLASTX hit) and related genes.
Candidate Genes for Suberin
Synthesis of Aliphatic Monomers
Dihydrolipoamide S-acetyltransferase and biotin carboxyl carrier protein are enzymes involved in de novo FA biosynthesis, a step necessary for the synthesis of LCFA and VLCFA. VLCFAs are precursors of waxes and some suberin monomers. Their up-regulation in cork may be due to a higher demand for acyl chains in this tissue. PATE/FAT and LACS are enzymes involved in the export of FA and LCFA from the chloroplast, a required step for the synthesis of VLCFA. Genes encoding FAT and LACS enzymes are required for normal wax and cuticle development (Bonaventure et al., 2003; Schnurr et al., 2004). Cytochromes P450 catalyze several key reactions in the synthesis of the aliphatic monomers. CYP86A1 is a hydroxylase that has capacity for ω-hydroxylation of C12-C18 FA (Benveniste et al., 1998). Members of the CYP86A family, LACERATA and ATT1, are involved in cuticle and cutin synthesis (Wellesen et al., 2001; Xiao et al., 2004). Lipoxygenases (LOXs) catalyze the addition of molecular oxygen to polyunsaturated FA and have been related to epoxydation of cutin monomers by the LOX/peroxygenase pathway (Blee and Schuber, 1993; Lequeu et al., 2003). FA elongase complexes in the endoplasmic reticulum catalyze chain elongation required for the biosynthesis of VLCFA. Two genes encoding condensing enzymes of these complexes are present in our library, a KCS and a β-ketoacyl CoA reductase (KCR). These two genes show, respectively, high similarity to At5g43760 (91% similarity) and At1g67730 (79% similarity), which have been identified as possible candidates for cuticular wax biosynthesis (Costaglioli et al., 2005). Mutations in KCS (KCS1 [Todd et al., 1999]; FIDDLEHEAD [Yephremov et al., 1999; Pruitt et al., 2000]; CUT1 [Millar et al., 1999]; CER6 [Fiebig et al., 2000]) and KCR (Dietrich et al., 2005) have a pronounced effect on cuticular wax deposition. ATP-citrate-lyase (ACL) produces the acetyl-CoA needed for chain elongation. Down-regulation of cytosolic ACL reduces cuticular wax deposition (Fatland et al., 2005).
In a transcriptome approach, Suh et al. (2005) identified different members of the FAT, LACS, KCS, KCR, and CYP86A gene families as candidates for waxes and cutin biosynthesis in Arabidopsis.
Transport of Aliphatic Materials
ATP-binding cassette (ABC) transporters are membrane proteins known for their function of translocating a broad range of substances across biological membranes, including lipids, sterols, and drugs. The library contains a cork up-regulated ABC transporter showing 78% similarity to AtWBC6. This transporter was put within the category acyl-lipids metabolism, because close members of this family are related to lipid transfer (Otsu et al., 2004) and the export of cuticular wax (CER5/AtWBC12; Pighin et al., 2004). Transporters of this subfamily are up-regulated in the stem epidermis of Arabidopsis (Suh et al., 2005). The library also contains a highly cork up-regulated ABC transporter, classed in “Miscellaneous,” which belongs to the ATH subfamily. AtABC1, the only member of this subfamily investigated in plants, encodes a chloroplast protein putatively involved in light signaling (Moller et al., 2001). However, in mammals, members of this subfamily translocate a wide range of substrates, including terpenes (Sun et al., 1999; Sanchez-Fernandez et al., 2001).
Assembly of the Aliphatic Polyester
The polymerization of the suberin glycerol polyester is poorly understood but is thought to take place in the apoplast by esterification of the carboxyl groups of α,ω-diacids and ω-hydroxyacids with the alcohol groups of either glycerol or ω- and midchain hydroxy FA (Kolattukudy, 2001). Our library contains two putative GPATs that catalyze the formation of ester bonds. Although the known plant GPATs were previously thought to be involved in membrane and storage lipid synthesis, the analysis of the transcriptome of the stem epidermis suggested a role of these enzymes in the synthesis of aliphatic polyesters (Suh et al., 2005), which has now been confirmed by analysis of the gpat5 mutant (Beisson et al., 2007).
Synthesis of Aromatic Monomers
Genes encoding phenylpropanoid-related enzymes that act upstream in the synthesis of aromatic compounds are represented in the library. Most of them catalyze biosynthetic reactions, leading to the phenylpropanoid precursors of the aromatic part of suberin (see Bernards, 2002). Some of these enzymes, such as Phe ammonia-lyase (PAL) and cinnamate 4-hydroxylase, are key enzymes for the regulation of the phenylpropanoids pathway. Other enzymes, such as F5H, are thought to be important for the synthesis of the hydroxycinnamic acids typical of suberin (Bernards and Lewis, 1998). Two genes, both strongly up-regulated in phellem, code for HCBTs, a family of enzymes capable of catalyzing the synthesis of N-hydroxycinnamoyl amides such as feruloyltyramine (Yang et al., 1997). Feruloyltyramine is found in potato wound suberin (Negrel et al., 1996) and some evidence supports the hypothesis that it is also a component of the aromatic suberin in cork tree (Marques et al., 1999). HCBTs belong to the BAHD acyltransferase superfamily (D'Auria, 2006), discussed below. On the other hand, the biosynthesis of hydroxycinnamic acid amides and their subsequent polymerization in the plant cell wall is generally accepted as a plant defense response (Facchini et al., 2002).
Assembly of the Aromatic Polymer
Despite the fact that peroxidase-mediated coupling was proposed for the assembly of the aromatic monomers in suberizing cells and that several suberin-associated class III peroxidases have been described (Kolattukudy, 2001; Bernards, 2002), no class III peroxidase was found in our cork library. However, cell wall peroxidase activity could be developed by other proteins present in the library, like the apoplastic annexin (classed in the regulatory proteins category), which has peroxidase activity (Gorecka et al., 2005). The extracellular H2O2 needed for the peroxidase catalytic activity could be provided by the copper-containing amine oxidase (Rea et al., 2004). Conversely, this study underscores three genes coding for laccases, which are extracellular oxidases with capacity for coupling phenylpropanoids. A role for laccases in lignin synthesis has been proposed (Kiefer-Meyer et al., 1996; LaFayette et al., 1999; Ranocha et al., 2002; Ehlting et al., 2005) and a similar function in the synthesis of the aromatic part of suberin can be hypothesized (Liang et al., 2006).
Linkages between Aliphatic and Aromatic Units
Esterified ferulates are very important suberin monomers (Adamovics et al., 1977; Bernards and Lewis, 1992; Lotfy et al., 1994). Some members of the BAHD acyltransferase superfamily catalyze the esterification of hydroxycinnamates with FAs (Lotfy et al., 1995). BAHD acyltransferases are a large superfamily of enzymes showing in vitro high catalytic versatility and wide substrate specificity (D'Auria, 2006). The phellem up-regulated transferase classed in miscellaneous shows high homology at the amino acid level with several members of the BAHD superfamily (e value < e−30). Interestingly, CER2/Glossy2, a gene whose knockout leads to a wax mutant, encodes a BAHD member (Kunst and Samuels, 2003; D'Auria, 2006). Conversely, as previously discussed, the HCBT genes that also belong to this superfamily are more probably related to synthesis of aromatic amides.
In summary, our cork library contains a set of structural enzymes that are probably good candidates for the synthesis of the aliphatic and aromatic monomers of the suberin and also provides putative candidates for the assembly of the polymer. One interesting observation is the relative importance of the cytochrome P450 superfamily in the cork library. The abundance of P450 monooxygenases and of oxidases reflects the high complexity of synthesizing the cork polymeric matrix and indicates that cork cell metabolism must generate reactive oxygen species in high amounts. This corresponds to previous observations showing that cork cells suffer from fairly high oxidative stress (Pla et al., 1998, 2000). Here, we found that ascorbate peroxidase, a crucial enzyme for H2O2 detoxifying in plant cells (Davletova et al., 2005), was strongly up-regulated in cork.
Candidate Genes for Regulation of Cork Formation
Only very limited knowledge is available about the hormonal control of cork formation. The ethylene-forming enzyme aminocyclopropane-carboxylate (ACC) oxidase was highly expressed in cork and wood, without showing significant differences between both tissues. Although the possible role of ethylene in these tissues is unclear (Andersson-Gunneras et al., 2003; Lulai and Suttle, 2004), it could be a common regulator in cork and wood. Phytosulfokines (PSKs) are peptide hormones that induce cell dedifferentiation and reentrance into the cell cycle (Matsubayashi et al., 2002). The presence of a cork up-regulated PSK receptor kinase suggests a possible role of PSKs in phellem regulation. On the other hand, enzymes acting on lipid catabolism such as LOX1 and putative lipases (GDSL-motif proteins) could be involved in the synthesis of wound hormones, such as jasmonic acid (Wasternack et al., 2006). Interestingly, the library contains a highly phellem up-regulated annexin. This gene shows 89% similarity to At1g35720, an Arabidopsis annexin that senses the Ca2+ signal elicited by ABA and transmits it downstream in the signaling pathway (Lee et al., 2004).
Phellogen derivatives undergo very rapidly phases of cell division, cell expansion, bulk suberin deposition, and cell death marked by the complete autolysis of the cells. Regulated proteolysis is required during programmed cell death and for the switch from one developmental phase to another, a process that requires removing preexisting regulatory networks (Sullivan et al., 2003; Vierstra, 2003). Genes encoding regulated proteolysis, such as a Cys protease and a Ub/26S proteasome system, were highly induced in cork. Cys proteases have been involved in programmed cell death (Rojo et al., 2004). Moreover, Cys proteases and Ub/26S proteasome genes are among the most expressed genes during fiber cell death (Moreau et al., 2005).
We have found five transcription factors related to meristem identity that could play a key role in the maintenance of the phellogenic identity of cells or promote their differentiation into phellem cells. One of them is a R2R3 MYB transcription factor involved in axillary meristem identity in Arabidopsis (Muller et al., 2006). Another one is an asymmetric leaves 1-interacting protein, which could be important for establishing cell fate in leaf development (Phelps-Durr et al., 2005). Two ESTs encode proteins of the NAM family (Souer et al., 1996). Finally, a SQUAMOSA/APETALA1 transcription factor, which is a floral meristem identity gene (Mandel and Yanofsky, 1995), was also highly up-regulated. On the other hand, the library also contains a WRKY transcription factor that is highly induced in cork. WRKY transcription factors are a large family of plant-specific regulators that mainly control senescence, stress, and defense responses (Eulgem et al., 2000). WRKY factors modulate gene expression by binding to W boxes of some stress-induced genes, including P450s (Mahalingam et al., 2003; Narusaka et al., 2004).
In conclusion, a number of interesting regulatory candidate genes for cork regulation have been identified, although much more work is needed to elucidate their function.
MATERIALS AND METHODS
Plant Material and Tissue Harvesting
Cork (phellem) and wood (xylem) tissues were harvested from 15- to 20-year-old field-grown cork trees (Quercus suber) at Peratallada (Girona, Spain) during the growing season. External bark (cork bark) was removed and, using sterile scalpels, the exposed phellem tissue was harvested. Thus, fractions rich in differentiating phellem were obtained (Fig. 1A). Wood was obtained after removing the internal bark (secondary phloem) and fractions enriched in differentiating xylem were harvested as described above. Harvested samples of cork and wood were immediately frozen in liquid nitrogen and stored at −80°C. To prevent genetic and environmental variability, both cork and wood samples cohybridized in the array were obtained from the same tree specimens.
As a source of somatic embryos, a cork tree recurrent embryogenic line maintained in a medium free of plant growth regulators was used (Puigderrajols et al., 1996). Macronutrients were those from Schenk and Hildebrant medium (Schenk and Hildebrant, 1972), and micronutrients and vitamins were from Murashige and Skoog medium (Murashige and Skoog, 1962), including 3% (w/v) Suc. The culture was solidified with 0.6% (w/v) agar, and the pH was adjusted to 5.7. The cultures were incubated in a growth chamber at 25°C and a 16-/8-h photoperiod at 50 μmol m−2 s−1 was provided by cool-white plus Grolux fluorescent lamps. Subcultured cotyledonary stage embryos showing signs of secondary embryogenesis were picked, their hypocotyledonary region dissected, immediately frozen in liquid nitrogen, and stored at −80°C.
RNA Extraction and Double-Stranded cDNA Synthesis
Total RNA was extracted from cork and wood tissue as described by Chang et al. (1993) and from somatic embryo as described by Martin et al. (1993). Remaining traces of genomic DNA were removed by TURBO DNAse treatment (Ambion) and a further purification step was performed with the CleanUp protocol of RNeasy Plant Mini kit (Qiagen). RNA quality and purity were checked by formamide-formaldehyde denaturing agarose gel electrophoresis spectrophotometry using Nanodrop and Bioanalyzer 2100 (Agilent). To obtain poly(A+) RNA, 150 μg of total RNA was purified using Oligotex mRNA kit (Qiagen). The quality and yield of the mRNA was checked by denaturing agarose gel electrophoresis and RiboGreen RNA quantitation reagent (Molecular Probes). Double-stranded cDNA was synthesized from 300 ng of mRNA using primers from the PCR-Select cDNA Subtraction kit (CLONTECH), Superscript II, Escherichia coli DNA ligase, E. coli DNA polymerase, RNase H, and T4 DNA polymerase from Invitrogen, all according to manufacturer's recommendations.
cDNA Subtractive Library Construction (SSH)
The cork tree phellem subtractive library was made applying the SSH technique (Diatchenko et al., 1996, 1999) and using the PCR-Select cDNA Subtraction kit (CLONTECH). To isolate ESTs preferentially induced in phellem, cDNA from cork tissue as tester and cDNA from somatic embryo as driver were used. Two rounds of subtractive hybridization and PCR amplification according to CLONTECH instructions were performed. To improve the yield of long ESTs, products from five secondary PCRs were pooled and size selected on an agarose gel electrophoresis in three fractions: 150 to 800 bp, 800 to 1,200 bp, and 1,200 to 2,000 bp. Each cDNA fraction was concentrated and purified using Minelute PCR Purification kit (Qiagen), cloned into the pCR4-TOPO T/A cloning vector, and transformed in TOP10 competent cells (Invitrogen). Kanamycin-resistant colonies were picked and grown overnight in liquid Luria-Bertani medium containing kanamycin at 37°C and 320 rpm. Finally, glycerol was added to grown cultures to 15% final concentration and clones were stored at −80°C in 96-deep-well plates.
Sequencing and EST Analysis
Plasmids were isolated from overnight-grown bacterial cultures using a standard alkaline lysis protocol with SDS in 96-well format. Inserted fragments were amplified by PCR using M13 oligonucleotides. Reactions were carried out in a final volume of 100 μL containing 100 μm dNTPs mix, 4 μm M13 forward primer (GTAAAACGACGGCCAG), 4 μm M13 reverse primer (CAGGAAACAGCTATGAC), 0.02 units μL−1 DyNAzyme (Finnzymes), 2.5 mm MgCl2, and 50 to 100 μg plasmid template and using PTC220 Multicycler (Dyad). PCR was done for 1 cycle at 95°C for 2 min, 35 cycles at 95°C for 45 s, 50°C for 2 min, 72°C for 1 min, and an additional cycle at 72°C for 6 min. Liquid handling steps in plasmid preparation as well as for PCR set up were carried out using a liquid handler robot RSP 200 (Tecan). All PCR products were separated on agarose gels. Gel images were analyzed to verify the amplicon length and quality using GelMaster software (Bajla et al., 2005). Single-run sequencing was carried out by MWG-Biotech AG using ABI capillary sequencers. Raw sequences and base confidence scores were obtained from chromatogram files using the base-calling program Phred (Ewing and Green, 1998; Ewing et al., 1998). Poly-A tails were removed with Trimest, a tool from the EMBOSS package. Vector masking and trimming as well as adaptor removal were performed with cross match (http://www.phrap.org). Sequences with less than 100 bases after trimming were discarded. Remaining sequences were assembled using StackPack (SANBI, http://www.sanbi.ac.za/Dbases.html) with the standard configuration (for initial clustering, a similarity cutoff value of 96 matching bases over a window size of 100 bp compared in words of length 6). Assembled sequences were annotated by using the StackPack Annotation Module (Universite Bordeaux 2, http://cbi.labri.fr). With StackPack Annotation Module, the consensus sequences and singletons were blasted locally against the following databases: SWISSPROT (ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/swissprot.gz), NR (nonredundant protein sequence database; ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz), and NT (nucleotide sequence database; ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nt.gz). EST sequences with an e value > 10−10 were considered as no hit and excluded from further analysis. Functional assignment was conducted manually by analyzing the BLASTX- and BLASTN-annotated ESTs, where SWISSPROT hits were favored over NR hits and NR hits were favored over NT hits.
Construction of cDNA Microarray
The cDNA inserts of 975 clones from the cork oak (Quercus spp.) phellem SSH library genes were amplified as described above and printed on a microarray. Along with cork ESTs, 2,298 poplar (Populus spp.) ESTs (Dejardin et al., 2004), 2,096 oak ESTs, and 1,167 potato (Solanum tuberosum) ESTs from the PICME repository (http://www.picme.at) were added and used to normalize intensity data (Supplemental Table S2). For orientation, 48 Cy3 and 48 Cy5 labeled oligos were spotted as guide dots. Spotting buffer and human clones were used as negative controls. Controls and amplified ESTs were spotted twice side by side on Corning GAPS II glass slides using a contact spotter (Omnigrid/Gene-Machines). The spotting was performed with 48 SMP3 pins obtaining 17 × 18 spots per block. During spotting, the relative humidity was set to 45%. After printing, slides were scanned, cross-linked at 65 mJ, and stored at room temperature in the dark.
Differential Screening by Microarray Hybridization
For hybridization, 20 μg of total RNA of each tissue were reverse transcribed using Superscript II (Invitrogen) and dNTP mix containing aminoallyl-dUTP (CLONTECH). Purification of cDNA, coupling of fluorescent dye, and probes purification were performed using Atlas Glass Fluorescent Labeling kit (CLONTECH) following manufacturer's recommendations. We used Cy3 and Cy5 Mono-Reactive Dye packs (Amersham). After labeling, the probes were quantified using a Nanodrop spectrophotometer. Microarray hybridization was performed using a humidity hybridization chamber (In-Slide-Out, Boeckel Scientific). Briefly, slides were prehybridized with 5× SSC, 0.1% (w/v) SDS, 0.1 mg/mL bovine serum albumin for 1 h at 42°C, followed by a wash in 0.1× SSC for 5 min at 25°C, two washes in water for 30 s, and dried by centrifugation at 1,600 rpm for 2 min. Then, slides were hybridized overnight (18 h) at 37°C in DIG Easy Hyb (Roche) containing labeled probes, yeast (Saccharomyces cerevisiae) tRNA (0.1 mg/mL), and salmon sperm DNA (0.1 mg/mL). After hybridization, slides were washed three times in 0.1% (w/v) SDS, 1× SSC for 10 min at 50°C, and washed four to six times in 1× SSC. Then, slides were dried by centrifuging at 500 rpm for 5 min and immediately scanned using the DNA Microarray Scanner (Agilent) at 10-μm resolution and 100% laser intensity and photomultiplier tube settings.
Microarray images were quantified using GenePix 6.0 (Axon) software. Only spots with signal intensities twice above the local background, not saturated and not flagged by GenePix, were considered reliable and used for subsequent analysis. Extracted intensities were subtracted from the local background and the log2 ratios were normalized in an intensity-dependent fashion by print-tip lowess. For phellem to xylem hybridization, normalized log2 ratios were scaled between arrays to make all data comparable. Statistically significant differences in gene expression were determined by computing a Bayesian statistic using all log2 ratios from replicate hybridizations. ESTs were considered as differentially expressed when their Bayesian statistic B was higher than 3. All quantitative and statistical analyses were performed using MMARGE tool, a web implementation of the Limma package in the R environment. Relative fold-change data were recalculated using xylem as the reference tissue. For phellem to embryo hybridization, ESTs showing log2 ratio coefficient of variation lower than 0.3 (calculated among duplicated spots) were considered as reliable.
RT-PCR with Incremental Cycle Numbers
To verify the difference of expression levels between cork and wood, equal amounts of cDNA were used for PCR with gene-specific oligonucleotides in 100-μL reactions. For each tissue sample, single-stranded cDNA was synthesized from 1 μg total RNA using the Superscript II (Invitrogen) in a 20-μL reaction. Then, cDNA was 2.5-fold diluted and 1 μL of diluted cDNA was used as template. Primers were designed with Primer3 software (Rozen and Skaletsky, 2000); sequences are shown on Supplemental Table S3. PCR conditions were 2.5 mm MgCl2, 0.2 mm each dNTP, 0.5 μm each forward and reverse oligonucleotide, and 0.05 units μL−1 Eurotaq polymerase (Euroclone). Thermal cycling parameters were as follows: 1 cycle at 95°C for 3 min and 36 cycles at 95°C for 30 s, 57°C for 30 s, and 72°C for 1 min, using a T-Gradient thermocycler (Biometra). Aliquots of 10 μL were taken every three cycles from cycle 12 to 36 and analyzed by agarose gel electrophoresis stained with ethidium bromide as described (Lê et al., 2005). Control amplifications were performed with specific primers to actin, elongation factor, and cap-binding protein. Target amplifications were performed with specific primers to HCBT, WRKY, LACS, F5H, PSKR, and FAT. To discard possible genomic DNA contaminations we designed the actin primers complementary to two exons separated by an intron of 94 bp using Populus genomic DNA sequence.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EE743887, EE743657 to EE743884, and EE745207 to EE745213.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison between biological and technical replicates.
Supplemental Table S1. List of unigenes from the cork subtractive library classified into functional categories and their expression pattern in the cork to wood microarray comparison.
Supplemental Table S2. Experimental data for all ESTs represented on the microarray together with the detailed results of the hybridization experiments.
Supplemental Table S3. Primers used for semiquantitative PCR.
Supplementary Material
Acknowledgments
The authors thank Drs. L. Sumoy and J. Lozano for the efficiency of its microarray service as well as their kind suggestions along this work (Microarray Laboratory, Centre de Regulació Genòmica, Barcelona). The authors also thank Prof. L. Schreiber and Dr. R. Franke (University of Bonn) for discussions and very helpful comments on the manuscript, and Prof. S. Prat (Centro Nacional de Biotecnología, Madrid), Dr. E. Domínguez (Institut de Biologia Molecular de Barcelona), and Dr. G. Mir (Universitat de Girona) for their support and fruitful experimental advice. The authors are also very grateful to Dr. M. Toribio (IMIDRA, Madrid) for providing somatic embryos and Mr. J. Casellas (Germans Casellas S.A.) and E. Juliol (Universitat de Girona) for providing cork material. S.F. performed the sequencing, annotation, and printing of ESTs. This work is part of the PhD thesis of M.S. and O.S.
This work was supported by the Spanish Ministerio de Ciencia y Tecnología (grant no. AGL2003–00416), by Ministerio de Educación y Ciencia (FPI grant to O.S.), by the European Social Funds and the Departament d'Universitats, Investigació i Societat de la Informació of Catalonia (FI and BE grants to M.S.), and by the European Forest Genomic Network (STSM to M.S. and O.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (http://www.plantphysiol.org) is: Mercè Figueras (merce.figueras@udg.es).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abdulrazzak N, Pollet B, Ehlting J, Larsen K, Asnaghi C, Ronseau S, Proux C, Erhardt M, Seltzer V, Renou J, et al (2006) A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol 140 30–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamovics JA, Johnson G, Stermitz FR (1977) Ferulates from cork layers of Solanum tuberosum and Pseudotsuga menziesii. Phytochemistry 16 1089–1090 [Google Scholar]
- Agrawal VP, Kolattukudy PE (1977) Biochemistry of suberization: ω-hydroxyacid oxidation in enzyme preparations from suberizing potato tuber disks. Plant Physiol 59 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Andersson-Gunneras S, Hellgren J, Bjorklund S, Regan S, Moritz T, Sundberg B (2003) Asymmetric expression of a poplar ACC oxidase controls ethylene production during gravitational induction of tension wood. Plant J 34 339–349 [DOI] [PubMed] [Google Scholar]
- Andersson-Gunneras S, Mellerowicz E, Love J, Segerman B, Ohmiya Y, Coutinho P, Nilsson P, Henrissat B, Moritz T, Sundberg B (2006) Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J 45 144–165 [DOI] [PubMed] [Google Scholar]
- Anterola A, Lewis N (2002) Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61 221–294 [DOI] [PubMed] [Google Scholar]
- Bajla I, Hollander I, Fluch S, Burg K, Kollar M (2005) An alternative method for electrophoretic gel image analysis in the GelMaster software. Comput Methods Programs Biomed 77 209–231 [DOI] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB (2007) The acyltransferase GPAT5 is required for synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19 351–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento M, Pereira H, Cunha M, Moutinho A, van den Berg K, Boon J (1998) Thermally assisted transmethylation gas chromatography mass spectrometry of suberin components in cork from Quercus suber L. Phytochem Anal 9 75–87 [Google Scholar]
- Benveniste I, Tijet N, Adas F, Philipps G, Salaun J, Durst F (1998) CYP86A1 from Arabidopsis thaliana encodes a cytochrome P450-dependent fatty acid omega-hydroxylase. Biochem Biophys Res Commun 243 688–693 [DOI] [PubMed] [Google Scholar]
- Benveniste P (2004) Biosynthesis and accumulation of sterols. Annu Rev Plant Biol 55 429–457 [DOI] [PubMed] [Google Scholar]
- Bernards M (2002) Demystifying suberin. Can J Bot 80 227–240 [Google Scholar]
- Bernards MA, Fleming WD, Llewellyn DB, Priefer R, Yang X, Sabatino A, Plourde GL (1999) Biochemical characterization of the suberization-associated anionic peroxidase of potato. Plant Physiol 121 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards MA, Lewis NG (1992) Alkyl ferulates in wound healing potato tubers. Phytochemistry 31 3409–3412 [DOI] [PubMed] [Google Scholar]
- Bernards MA, Lewis NG (1998) The macromolecular aromatic domain in suberized tissue: a changing paradigm. Phytochemistry 47 915–933 [DOI] [PubMed] [Google Scholar]
- Bernards MA, Razem FA (2001) The poly(phenolic) domain of potato suberin: a non-lignin cell wall bio-polymer. Phytochemistry 57 1115–1122 [DOI] [PubMed] [Google Scholar]
- Blee E, Schuber F (1993) Biosynthesis of cutin monomers: involvement of a lipoxygenase peroxygenase pathway. Plant J 4 113–123 [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54 519–546 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Salas J, Pollard M, Ohlrogge J (2003) Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 15 1020–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadahia E, Conde E, de Simon B, Garcia-Vallejo M (1998) Changes in tannic composition of reproduction cork Quercus suber throughout industrial processing. J Agric Food Chem 46 2332–2336 [Google Scholar]
- Caritat A, Gutierrez E, Molinas M (2000) Influence of weather on cork-ring width. Tree Physiol 20 893–900 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Helariutta Y (2005) Phloem and xylem specification: pieces of the puzzle emerge. Curr Opin Plant Biol 8 512–517 [DOI] [PubMed] [Google Scholar]
- Castola V, Marongiu B, Bighelli A, Floris C, Lai A, Casanova J (2005) Extractives of cork (Quercus suber L.): chemical composition of dichloromethane and supercritical CO2 extracts. Ind Crop Prod 21 65–69 [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11 113–116 [Google Scholar]
- Cheng J, Lertpiriyapong K, Wang S, Sung Z (2000) The role of the Arabidopsis ELD1 gene in cell development and photomorphogenesis in darkness. Plant Physiol 123 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costaglioli P, Joube J, Garcia C, Stef M, Arveiler B, Lessire R, Garbay B (2005) Profiling candidate genes involved in wax biosynthesis in Arabidopsis thaliana by microarray analysis. Biochim Biophys Acta 1734 247–258 [DOI] [PubMed] [Google Scholar]
- Cottle W, Kolattukudy P (1982) Biosynthesis, deposition, and partial characterization of potato suberin phenolics. Plant Physiol 69 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria JC (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9 331–340 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Zhong S, Oliver D, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin A, Leple JC, Lesage-Descauses MC, Costa G, Pilate G (2004) Expressed sequence tags from poplar wood tissues: a comparative analysis from multiple libraries. Plant Biol (Stuttg) 6 55–64 [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YFC, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, et al (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93 6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lukyanov S, Lau Y, Siebert P (1999) Suppression subtractive hybridization: a versatile method for identifying differentially expressed genes. Methods Enzymol 303 349–380 [DOI] [PubMed] [Google Scholar]
- Dietrich C, Perera M, Yandeau-Nelson M, Meeley R, Nikolau B, Schnable P (2005) Characterization of two GL8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J 42 844–861 [DOI] [PubMed] [Google Scholar]
- Dixon R, Achnine L, Kota P, Liu C, Reddy M, Wang L (2002) The phenylpropanoid pathway and plant defence: a genomics perspective. Mol Plant Pathol 3 371–390 [DOI] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, et al (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42 618–640 [DOI] [PubMed] [Google Scholar]
- Espelie KE, Franceschi VR, Kolattukudy PE (1986) Immunocytochemical localization and time course of appearance of an anionic peroxidase associated with suberization in wound-healing potato tuber tissue. Plant Physiol 81 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 199–206 [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8 186–194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8 175–185 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Hagel J, Zulak KG (2002) Hydroxycinnamic acid amide metabolism: physiology and biochemistry. Can J Bot 80 577–589 [Google Scholar]
- Fatland B, Nikolau B, Wurtele E (2005) Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-citrate lyase in Arabidopsis. Plant Cell 17 182–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig A, Mayfield J, Miley N, Chau S, Fischer R, Preuss D (2000) Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L (2005) Apoplastic polyesters in Arabidopsis surface tissues: a typical suberin and a particular cutin. Phytochemistry 66 2643–2658 [DOI] [PubMed] [Google Scholar]
- Gorecka KM, Konopka-Postupolska D, Hennig J, Buchet R, Pikula S (2005) Peroxidase activity of annexin 1 from Arabidopsis thaliana. Biochem Biophys Res Commun 336 868–875 [DOI] [PubMed] [Google Scholar]
- Graca J, Santos S (2006) Linear aliphatic dimeric esters from cork suberin. Biomacromolecules 7 2003–2010 [DOI] [PubMed] [Google Scholar]
- Heredia A (2003) Biophysical and biochemical characteristics of cutin, a plant barrier biopolymer. Biochim Biophys Acta 1620 1–7 [DOI] [PubMed] [Google Scholar]
- Holloway P (1983) Some variations in the composition of suberin from the cork layers of higher-plants. Phytochemistry 22 495–502 [Google Scholar]
- Jenks MA, Eigenbrode SD, Lemieux B (2002) Cuticular waxes of Arabidopsis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD [DOI] [PMC free article] [PubMed]
- Keren-Keiserman A, Tanami Z, Shoseyov O, Ginzberg I (2004) Peroxidase activity associated with suberization processes of the muskmelon (Cucumis melo) rind. Physiol Plant 121 141–148 [DOI] [PubMed] [Google Scholar]
- Kiefer-Meyer MC, Gomord V, O'Connell A, Halpin C, Faye L (1996) Cloning and sequence analysis of laccase-encoding cDNA clones from tobacco. Gene 178 205–207 [DOI] [PubMed] [Google Scholar]
- Kirst M, Myburg A, De Leon J, Kirst M, Scott J, Sederoff R (2004) Coordinated genetic regulation of growth and lignin revealed by quantitative trait locus analysis of cDNA microarray data in an interspecific backcross of eucalyptus. Plant Physiol 135 2368–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Han K, Park S, Yang J (2004) Plant body weight-induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole-transcriptome profiling. Plant Physiol 135 1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10 236–242 [DOI] [PubMed] [Google Scholar]
- Kolattukudy P (1980) Bio-polyester membranes of plants: cutin and suberin. Science 208 990–1000 [DOI] [PubMed] [Google Scholar]
- Kolattukudy P (1981) Structure, biosynthesis, and biodegradation of cutin and suberin. Annu Rev Plant Physiol 32 539–567 [Google Scholar]
- Kolattukudy PE (2001) Polyesters in higher plants. Adv Biochem Eng Biotechnol 71 1–49 [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels A (2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42 51–80 [DOI] [PubMed] [Google Scholar]
- LaFayette PR, Eriksson KE, Dean JF (1999) Characterization and heterologous expression of laccase cDNAs from xylem tissues of yellow-poplar (Liriodendron tulipifera). Plant Mol Biol 40 23–35 [DOI] [PubMed] [Google Scholar]
- Laule O, Furholz A, Chang H, Zhu T, Wang X, Heifetz P, Gruissem W, Lange B (2003) Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 100 6866–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê Q, Gutierrez-Marcos JF, Costa LM, Meyer S, Dickinson HG, Lorz H, Kranz E, Scholten S (2005) Construction and screening of subtracted cDNA libraries from limited populations of plant cells: a comparative analysis of gene expression between maize egg cells and central cells. Plant J 44 167–178 [DOI] [PubMed] [Google Scholar]
- Lee S, Lee EJ, Yang EJ, Lee JE, Park AR, Song WH, Park OK (2004) Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 16 1378–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lequeu J, Fauconnier ML, Chammai A, Bronner R, Blee E (2003) Formation of plant cuticle: evidence for the occurrence of the peroxygenase pathway. Plant J 36 155–164 [DOI] [PubMed] [Google Scholar]
- Liang M, Haroldsen V, Cai X, Wu Y (2006) Expression of a putative laccase gene, ZmLAC1, in maize primary roots under stress. Plant Cell Environ 29 746–753 [DOI] [PubMed] [Google Scholar]
- Lotfy S, Negrel J, Javelle F (1994) Formation of omega-feruloyloxypalmitic acid by an enzyme from wound-healing potato-tuber disks. Phytochemistry 35 1419–1424 [Google Scholar]
- Lopes M, Barros A, Neto C, Rutledge D, Delgadillo I, Gil A (2001) Variability of cork from Portuguese Quercus suber studied by solid-state C-13-NMR and FTIR spectroscopies. Biopolymers 62 268–277 [DOI] [PubMed] [Google Scholar]
- Lopes M, Neto C, Barros A, Rutledge D, Delgadillo I, Gil A (2000) Quantitation of aliphatic suberin in Quercus suber L. cork by FTIR spectroscopy and solid-state C-13-NMR spectroscopy. Biopolymers 57 344–351 [DOI] [PubMed] [Google Scholar]
- Lotfy S, Javelle F, Negrel J (1995) Distribution of hydroxycinnamoyl-CoA-omega-hydroxypalmitic acid O-hydroxycinnamoyltransferase in higher-plants. Phytochemistry 40 389–391 [Google Scholar]
- Lucena M, Romero-Aranda R, Mercado J, Cuartero J, Valpuesta V, Quesada M (2003) Structural and physiological changes in the roots of tomato plants over-expressing a basic peroxidase. Physiol Plant 118 422–429 [Google Scholar]
- Lulai E, Suttle J (2004) The involvement of ethylene in wound-induced suberization of potato tuber (Solanum tuberosum L.): a critical assessment. Postharvest Biol Technol 34 105–112 [Google Scholar]
- Mahalingam R, Gomez-Buitrago A, Eckardt N, Shah N, Guevara-Garcia A, Day P, Raina R, Fedoroff NV (2003) Characterizing the stress/defense transcriptome of Arabidopsis. Genome Biol 4 R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF (1995) A gene triggering flower formation in Arabidopsis. Nature 377 522–524 [DOI] [PubMed] [Google Scholar]
- Marques AV, Pereira H, Meier D, Faix O (1999) Structural characterization of cork lignin by thiacidolysis and permanganate oxidation. Holzforschung 53 167–174 [Google Scholar]
- Martin W, Nock S, Meyer-Gauen G, Häger KP, Jensen U, Cerff R (1993) A method for isolation of cDNA-quality mRNA from immature seeds of a gymnosperm rich in polyphenolics. Plant Mol Biol 22 555–556 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Morita A, Sakagami Y (2002) An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296 1470–1472 [DOI] [PubMed] [Google Scholar]
- Millar A, Clemens S, Zachgo S, Giblin E, Taylor D, Kunst L (1999) CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moire L, Schmutz A, Buchala A, Yan B, Stark RE, Ryser U (1999) Glycerol is a suberin monomer: new experimental evidence for an old hypothesis. Plant Physiol 119 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller S, Kunkel T, Chua N (2001) A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev 15 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau C, Aksenov N, Lorenzo MG, Segerman B, Funk C, Nilsson P, Jansson S, Tuominen H (2005) A genomic approach to investigate developmental cell death in woody tissues of Populus trees. Genome Biol 6 R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D, Schmitz G, Theres K (2006) Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Narusaka Y, Narusaka M, Seki M, Umezawa T, Ishida J, Nakajima M, Enju A, Shinozaki K (2004) Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Mol Biol 55 327–342 [DOI] [PubMed] [Google Scholar]
- Nawrath C (2002) The biopolymers cutin and suberin. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–14 [DOI] [PMC free article] [PubMed]
- Negrel J, Pollet B, Lapierre C (1996) Ether-linked ferulic acid amides in natural and wound periderms of potato tuber. Phytochemistry 43 1195–1199 [Google Scholar]
- Otsu C, daSilva I, de Molfetta J, da Silva L, de Almeida-Engler J, Engler G, Torraca P, Goldman G, Goldman M (2004) NtWBC1, an ABC transporter gene specifically expressed in tobacco reproductive organs. J Exp Bot 55 1643–1654 [DOI] [PubMed] [Google Scholar]
- Pereira H (1988) Chemical composition and variability of cork from Quercus suber L. Wood Sci Technol 22 211–218 [Google Scholar]
- Phelps-Durr TL, Thomas J, Vahab P, Timmermans MC (2005) Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17 2886–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pighin J, Zheng H, Balakshin L, Goodman I, Western T, Jetter R, Kunst L, Samuels A (2004) Plant cuticular lipid export requires an ABC transporter. Science 306 702–704 [DOI] [PubMed] [Google Scholar]
- Pla M, Huguet G, Verdaguer D, Puigderrajols P, Llompart B, Nadal A, Molinas M (1998) Stress proteins co-expressed in suberized and lignified cells and in apical meristems. Plant Sci 139 49–57 [Google Scholar]
- Pla M, Jofre A, Martell M, Molinas M, Gomez J (2000) Large accumulation of mRNA and DNA point modifications in a plant senescent tissue. FEBS Lett 472 14–16 [DOI] [PubMed] [Google Scholar]
- Pruitt RE, Vielle-Calzada JP, Ploense SE, Grossniklaus U, Lolle SJ (2000) FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA 97 1311–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigderrajols P, Fernandez-Guijarro B, Toribio M, Molinas M (1996) Origin and early development of secondary embryos in Quercus suber L. Int J Plant Sci 157 674–684 [Google Scholar]
- Puigderrajols P, Mir G, Molinas M (2001) Ultrastructure of early secondary embryogenesis by multicellular and unicellular pathways in cork oak (Quercus suber L.). Ann Bot 87 179–189 [DOI] [PubMed] [Google Scholar]
- Quiroga M, Guerrero C, Botella M, Barcelo A, Amaya I, Medina M, Alonso F, de Forchetti S, Tigier H, Valpuesta V (2000) A tomato peroxidase involved in the synthesis of lignin and suberin. Plant Physiol 122 1119–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan P, Kao YY, Jiang H, Joshi CP, Harding SA, Tsai CJ (2004) Suppression subtractive hybridization-mediated transcriptome analysis from multiple tissues of aspen (Populus tremuloides) altered in phenylpropanoid metabolism. Planta 219 694–704 [DOI] [PubMed] [Google Scholar]
- Ranocha P, Chabannes M, Chamayou S, Danoun S, Jauneau A, Boudet AM, Goffner D (2002) Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol 129 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razem F, Bernards M (2003) Reactive oxygen species production in association with suberization: evidence for an NADPH-dependent oxidase. J Exp Bot 54 935–941 [DOI] [PubMed] [Google Scholar]
- Rea G, de Pinto MC, Tavazza R, Biondi S, Gobbi V, Ferrante P, De Gara L, Federico R, Angelini R, Tavladoraki P (2004) Ectopic expression of maize polyamine oxidase and pea copper amine oxidase in the cell wall of tobacco plants. Plant Physiol 134 1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E, Kolattukudy P (1989) Molecular cloning, nucleotide sequence, and abscisic-acid induction of a suberization-associated highly anionic peroxidase. Mol Gen Genet 217 223–232 [DOI] [PubMed] [Google Scholar]
- Rojo E, Martin R, Carter C, Zouhar J, Pan S, Plotnikova J, Jin H, Paneque M, Sanchez-Serrano JJ, Baker B, et al (2004) VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol 14 1897–1906 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132 365–386 [DOI] [PubMed] [Google Scholar]
- Sabba R, Lulai E (2002) Histological analysis of the maturation of native and wound periderm in potato (Solanum tuberosum L.) tuber. Ann Bot 90 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Fernandez R, Davies TG, Coleman JO, Rea PA (2001) The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem 276 30231–30244 [DOI] [PubMed] [Google Scholar]
- Schenk RU, Hildebrant AC (1972) Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant-cell cultures. Can J Bot 50 199–204 [Google Scholar]
- Schnurr J, Shockey J, Browse J (2004) The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber L, Franke R, Lessire R (2005) Biochemical characterization of elongase activity in corn (Zea mays L.) roots. Phytochemistry 66 131–138 [DOI] [PubMed] [Google Scholar]
- Sherf B, Bajar A, Kolattukudy P (1993) Abolition of an inducible highly anionic peroxidase-activity in transgenic tomato. Plant Physiol 101 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Seguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva S, Sabino M, Fernandes E, Correlo V, Boesel L, Reis R (2005) Cork: properties, capabilities and applications. Int Matr Rev 50 345–365 [Google Scholar]
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85 159–170 [DOI] [PubMed] [Google Scholar]
- Suh M, Samuels A, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F (2005) Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol 139 1649–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Shirasu K, Deng XW (2003) The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat Rev Genet 4 948–958 [DOI] [PubMed] [Google Scholar]
- Sun H, Molday RS, Nathans J (1999) Retinal stimulates ATP hydrolysis by purified and reconstituted ABCR, the photoreceptor-specific ATP-binding cassette transporter responsible for Stargardt disease. J Biol Chem 274 8269–8281 [DOI] [PubMed] [Google Scholar]
- Todd J, Post-Beittenmiller D, Jaworski J (1999) KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17 119–130 [DOI] [PubMed] [Google Scholar]
- Varea S, Garcia-Vallejo M, Cadahia E, de Simon B (2001) Polyphenols susceptible to migrate from cork stoppers to wine. Eur Food Res Technol 213 56–61 [Google Scholar]
- Vierstra RD (2003) The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci 8 135–142 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Stenzel I, Hause B, Hause G, Kutter C, Maucher H, Neumerkel J, Feussner I, Miersch O (2006) The wound response in tomato role of jasmonic acid. J Plant Physiol 163 297–306 [DOI] [PubMed] [Google Scholar]
- Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A (2001) Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different robes of fatty acid omega-hydroxylation in development. Proc Natl Acad Sci USA 98 9694–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetten R, Sun Y, Zhang Y, Sederoff R (2001) Functional genomics and cell wall biosynthesis in loblolly pine. Plant Mol Biol 47 275–291 [PubMed] [Google Scholar]
- Xiao F, Goodwin S, Xiao Y, Sun Z, Baker D, Tang X, Jenks M, Zhou J (2004) Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J 23 2903–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Reinhard K, Schiltz E, Matern U (1997) Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA:anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol Biol 35 777–789 [DOI] [PubMed] [Google Scholar]
- Yephremov A, Schreiber L (2005) The dark side of the cell wall: molecular genetics of plant cuticle. Plant Biosyst 139 74–79 [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H (1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11 2187–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.