Abstract
To elucidate the effect of high temperature on grain-filling metabolism, developing rice (Oryza sativa) ‘Nipponbare’ caryopses were exposed to high temperature (33°C/28°C) or control temperature (25°C/20°C) during the milky stage. Comprehensive gene screening by a 22-K DNA microarray and differential hybridization, followed by expression analysis by semiquantitative reverse transcription-PCR, revealed that several starch synthesis-related genes, such as granule-bound starch synthase I (GBSSI) and branching enzymes, especially BEIIb, and a cytosolic pyruvate orthophosphate dikinase gene were down-regulated by high temperature, whereas those for starch-consuming α-amylases and heat shock proteins were up-regulated. Biochemical analyses of starch showed that the high temperature-ripened grains contained decreased levels of amylose and long chain-enriched amylopectin, which might be attributed to the repressed expression of GBSSI and BEIIb, respectively. SDS-PAGE and immunoblot analysis of storage proteins revealed decreased accumulation of 13-kD prolamin, which is consistent with the diminished expression of prolamin genes under elevated temperature. Ripening under high temperature resulted in the occurrence of grains with various degrees of chalky appearance and decreased weight. Among them, severely chalky grains contained amylopectin enriched particularly with long chains compared to slightly chalky grains, suggesting that such alterations of amylopectin structure might be involved in grain chalkiness. However, among high temperature-tolerant and sensitive cultivars, alterations of neither amylopectin chain-length distribution nor amylose content were correlated to the degree of grain chalkiness, but rather seemed to be correlated to grain weight decrease, implying different underlying mechanisms for the varietal difference in grain chalkiness. The possible metabolic pathways affected by high temperature and their relevance to grain chalkiness are discussed.
High temperature during the grain-filling stage causes deleterious effects on the yield and quality of crop products (Peng et al., 2004). Temperature above certain growth-optimal temperatures impairs dry matter production, generally decreasing grain size in all major cereal crops, such as rice (Oryza sativa), wheat (Triticum aestivum), barley (Hordeum vulgare), and maize (Zea mays). Such small grains result in not only decreased yield but also low milling quality. For japonica cultivars of rice, temperatures higher than 26°C render chalky grain appearance as well as reduction of grain weight. Severely chalky brown rice grains are inferior for polishing quality and palatability. The chalky grains ripened under high temperature conditions resulted in lower yield after polishing and less sticky texture after cooking than translucent grains ripened under low temperature (Table I). Because the Japanese market prefers sticky rice, the rice grains with a chalky appearance are easily recognized and only achieve low sale prices, which is one of the recent problems for Japanese rice-producing farmers. To circumvent quantitative and qualitative losses of crop production by forthcoming global warming, studies concerning physiological effects of elevated temperature on grain filling are indispensable.
Table I.
Effect of grain-filling temperature on rice polishing yield and palatability
| Cultivar | Treatmenta | Average Temperature for 0–20 DAH | Grain Weight | Perfect Kernel Ratiob | Yield after Polishingc | Palatabilityd | Stickinessd |
|---|---|---|---|---|---|---|---|
| °C | mg | % | % | ||||
| ‘Koshihikari’ | NT | 26.5 | 20.7 | 69.7 | 90.6 | −0.42 | −0.21 |
| ET + house | 28.8 | 20.0 | 26.8 | 88.2 | −2.46 | −1.14 | |
| *** | *** | ||||||
| ‘Sasanishiki’ | NT | 27.7 | 20.5 | 56.6 | 86.3 | −0.91 | −0.42 |
| ET + house | 28.6 | 19.3 | 14.5 | 81.0 | −2.23 | −1.00 | |
| *** | *** |
NT, Normal transplanting (on April 26, 2005); ET, early transplanting (on May 19, 2005); house, covered by a plastic-film greenhouse after heading.
The ratio of translucent nonchalky grains measured by a grain-grading machine, ES-1000 (Shizuoka Seiki).
The ratio of the weight of white rice after constant polishing by Ricepal 31 (Yamamoto) to that of brown rice.
Palatability and stickiness of cooked rice determined by 57 tasting panelists comparing to two standard samples, ‘Koshihikari’ and ‘Kochihibiki’, for relative value 0 and −2, respectively. ***, Significant at 0.1% level as determined by t test.
It has been reported that high temperature at the milky stage of grain filling has the greatest influence on rice grain chalkiness (Tashiro and Wardlaw, 1991a), and the panicle is the most sensitive organ to high temperature (Sato and Inaba, 1973; Morita et al., 2004). There are known to be varietal differences in grain chalkiness among rice cultivars when ripened under a given temperature. The japonica varieties ‘Koshiibuki’ and ‘Tentakaku’ provide less chalky grains even when they are exposed to high temperature (here defined as high temperature tolerant), whereas ‘Hatsuboshi’ and ‘Sasanishiki’ produce severely chalky grains (high temperature sensitive). However, the varietal differences in grain chalkiness are poorly understood at the molecular level. Microscopic observation of the chalky part of high temperature-ripened grains revealed that loosely packed starch granules create air spaces between themselves to reflect light randomly (Tashiro and Wardlaw, 1991a; Zakaria et al., 2002). Thus, to determine the underlying biochemical mechanism for grain chalkiness, the effect of high temperature on starch synthesis in developing caryopses has been investigated so far.
Starch consists of amylose (linear α-1,4-polyglucan) and amylopectin (α-1,6-branched polyglucans) in rice grains. Although amylose synthesis is exclusively governed by granule-bound starch synthase (GBSS), amylopectin is synthesized via concerted reactions catalyzed by multiple isoforms of enzymes: soluble starch synthase (SS), starch branching enzyme (BE), and starch debranching enzyme (for review, see Nakamura, 2002). In particular, the relative balance of α-1,6-branch formation and the subsequent α-1,4-chain elongation, which are catalyzed by distinct BE and soluble SS isoforms, respectively, are important for determining the amylopectin fine structure. The effect of high temperature on these enzymes was extensively studied in wheat and maize, and activity and/or gene expression for soluble SS was impaired under elevated temperature (Hawker and Jenner, 1993; Singletary et al., 1994; Hurkman et al., 2003). In rice, high temperature resulted in a reduction of activity and gene expression for GBSSI and BEs (Umemoto and Terashima, 2002; Jiang et al., 2003), decrease of amylose content (Asaoka et al., 1989; Umemoto and Terashima, 2002), and increase of long chain of amylopectin (Asaoka et al., 1984; Umemoto et al., 1999).
Prior to the synthesis of amylose and amylopectin, Suc is loaded into endosperm cells by Suc transporters and converted to Glc-6-P via several reaction steps catalyzed by enzymes such as Suc synthase (SuSy) and then into ADP-Glc, the substrate for α-1,4-polyclucan synthesis, by ADP-Glc pyrophosphorylase (AGP). Import of ADP-Glc and Glc-6-P into amyloplasts is conducted by distinct transporters. For each enzyme and transporter involved, several isoforms are known to be differentially regulated. In maize, the activities of AGP and SuSy as well as soluble SS were reduced by high temperature (Singletary et al., 1994; Wilhelm et al., 1999).
To comprehend the effect of high temperature on the wide range of grain-filling metabolic steps simultaneously, proteomic approaches have been employed. The analyses using high temperature-exposed caryopses of rice or wheat revealed the induction of heat shock proteins (HSPs) as well as the repression of several starch metabolism-related enzyme proteins (Majoul et al., 2003; Lin et al., 2005). HSPs are molecular chaperones that stabilize proteins in their functional conformations and prevent the aggregation of heat-denatured proteins (for review, see Wang et al., 2004). However, these approaches provided information limited to the identification of enzymes or proteins expressed at relatively abundant levels. Because in developing caryopses some proteins such as debranching enzymes expressed at intermediate and low levels have been recently revealed to participate in grain-filling metabolism (Nakamura, 2002) and such low expression level proteins might be barely detectable by proteomic analyses, a more comprehensive expression analysis is required. Current progress on the rice genome project provides cDNA information covering approximately two-thirds of the genome (Kikuchi et al., 2003), and a microarray containing approximately one-half of the genes is available. Considering the minimal time and cost required, DNA microarrays are a powerful tool to comprehend the effect of high temperature on the expression of grain filling-related genes. In this study, we aimed to elucidate the effect of high temperature on metabolism during grain filling at the gene expression level and discussed its relevance to changes in accumulation of storage materials, such as starch and storage proteins, as well as grain chalkiness.
RESULTS
Rice Grains Ripened under High Temperature Showed Chalky Appearance and Low Amylose Content
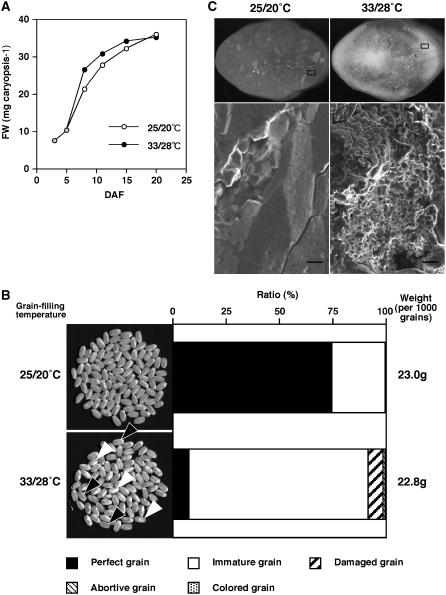
We intended to identify genes whose transcript levels are responsive to temperature during the grain-filling phase. Because gene expression intensity of field-grown plants is often influenced by numerous stresses from the environment, we raised plants in incubators under controlled illumination and temperature conditions to achieve highly reproducible grain filling. In the system, rice ‘Nipponbare’ plants exposed to high temperature (33°C/28°C) from 5 to 20 d after flowering (DAF) matured faster than those ripened under a control condition (25°C/20°C). After starting the temperature treatment at 5 DAF, the fresh weight of high temperature-treated caryopses increased faster than the control plants (Fig. 1A). However, the weight increase completely ceased by 15 DAF, while the weight of the control caryopses continued to increase until at least 20 DAF. At maturity, the grains exposed to high temperature were slightly lighter than the control grains. Weight of high temperature-treated grain was always lower than that of the control grain under the same conditions, although the reduction ratio fluctuated from 88.4% to 99.1% in each set of experiments (93.5% in average, n = 7). However, decrease of the weight was significant (P < 0.02, n = 7). The appearance of high temperature-ripened grains was severely chalky, such that 84% of grains were categorized as immature by an imaging analysis, while the control grains were mostly translucent (Fig. 1B). Furthermore, scanning electron microscopy of transverse sections revealed that endosperm of chalky grains ripened under high temperature contained loosely packed starch granules with large air spaces, while translucent grains ripened under normal temperature were filled with densely packed granules (Fig. 1C).
Figure 1.
Grain filling using plant growth incubators. A, Change in the fresh weight of rice caryopses developing under 33°C/28°C (black circles) or 25°C/20°C (white circles). Values are the mean of at least 50 grains. B, Appearance and weight of dehulled grains. The ratio of perfect (translucent), immature (mainly chalky), damaged, abortive, and colored grains was determined by a grain-grading machine, ES-1000 (Shizuoka Seiki), and is indicated from left to right of the bars in order. Because colored gains were few, the symbol is inside lines. Black and white arrowheads indicate almost translucent and severely chalky grains ripened under 33°C/28°C, respectively. C, Scanning electron micrographs of transverse sections of 25°C/20°C-ripened translucent (left) and 33°C/28°C-ripened chalky grains (right). Top and bottom, Light microscope and scanning electron microscope images, respectively. The areas indicated by boxes were analyzed by scanning electromicroscopy. Bars = 10 μm.
Amylose content is known to be influenced by ripening temperature. Determined by an iodine colorimetric method, amylose content in the ‘Nipponbare’ grains ripened in the 33°C/28°C incubator was 17.1%, which was lower than that of the control grains (18.8%; Table II). Total starch contents, the ratio of starch weight to kernel weight, in 25°C/20°C-ripened and 33°C/28°C-ripened grains were 66.8% ± 0.6% and 66.8% ± 1.4% (mean ± sd, n = 5), respectively, which are not significantly different. Thus, grains ripened under high temperature with incubators had light weight, chalky appearance, and low amylose content, which are common features in field-grown rice grains ripened under high temperature in natural conditions.
Table II.
Amylose content of rice grains of various cultivars ripened under high (33°C/28°C) and normal (25°C/20°C) temperature
| Cultivar | Amylose Content
|
Reduction Ratiob | ||
|---|---|---|---|---|
| 25°C/20°C | 33°C/28°C | |||
| % | % | |||
| ‘Nipponbare’ | 18.8 ± 0.39a | 17.1 ± 0.61 | *** | 91.0 |
| ‘Koshiibuki’ | 17.8 ± 0.49 | 15.1 ± 0.98 | *** | 84.6 |
| ‘Tentakaku’ | 19.7 ± 0.60 | 15.5 ± 0.80 | *** | 78.7 |
| ‘Sasanishiki’ | 18.4 ± 0.95 | 16.5 ± 1.13 | ** | 89.6 |
| ‘Hatsuboshi’ | 19.8 ± 1.11 | 15.0 ± 0.48 | *** | 75.9 |
Values are means ± sd of five replicates.
The ratio of amylose content of 33°C/28°C-treated rice to that of 25°C/20°C-treated rice. ** and ***, Significant at 0.5% and 0.1% level, respectively, as determined by Student's t test.
High Temperature Affected Expression of Genes Involved in Starch Metabolism, Storage Protein Synthesis, and Stress Responses
To investigate the metabolic alterations caused by high temperature during grain filling, genes whose transcript level was increased or decreased by high temperature were identified from developing caryopses harvested at 10 DAF by: (1) employing the Agilent rice 22-K oligo DNA microarray system; (2) screening of 6 × 104 cDNA clones by differential hybridization; or (3) cloning by PCR with subtracted cDNA libraries. Among 21,938 rice genes on the microarray, 45 genes were up-regulated more than 2-fold by high temperature during grain filling, while 39 were down-regulated to less than one-half the level of the control. In addition, five genes encoding storage proteins and one allergenic protein gene were isolated by differential screening, but none by the subtractive hybridization. Considering their annotation and homology information, all the genes isolated were categorized into carbohydrate-metabolizing enzymes/translocators, storage proteins/related enzymes, HSPs/factor, and other up-regulated or down-regulated genes, as summarized in Table III.
Table III.
Expression level of various genes in response to high temperature grain filling
The values of microarray and dot-blot hybridization are means of the data obtained from independent hybridizations (n = 2 and 4, respectively).
| Accession No. | Description | Gene Namea | Microarray
|
Dot-Blot Hybridization
|
RT-PCR
|
Identified as | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fold- Change | 25°C/20°C | 33°C/28°C | Fold- Change | 25°C/20°C | 33°C/28°C | Fold- Changeb | ||||
| Starch/carbohydrate-metabolizing enzymes | ||||||||||
| AK070431 | Rice waxy | GBSSI | 0.35 | 207,723 | 73,043 | 0.82 | 2,836 | 2,474 | 0.50 | |
| AK102058 | Rice GBSS II precursor | GBSSII | 0.61 | 938 | 576 | 0.65 | 1,247 | 863 | ||
| AK109458 | Rice sp. japonica soluble SS | SSI | 0.97 | 3,092 | 2,985 | 1.25 | ||||
| AK101978 | Rice putative soluble SS II-3 | SSIIa | 0.77 | 33,221 | 25,656 | 0.87 | ||||
| AK066446 | Rice soluble SS II-2 | SSIIb | 0.93 | 2,488 | 2,315 | |||||
| AK072339 | Rice soluble SS II-1 | SSIIc | 0.78 | 758 | 593 | |||||
| AK061604 | Unknown expressed protein | SSIIIa | 0.90 | 3,711 | 3,354 | 0.90 | ||||
| AK059368 | Wheat SS III | SSIIIb | 0.95 | 2,142 | 2,028 | |||||
| AK066808 | Wheat SS isoform IV; nuclear gene for plastid product | SSIVa | 1.11 | 986 | 1,097 | |||||
| AK067577 | Wheat SS isoform IV; nuclear gene for plastid product | SSIVb | 0.92 | 5,173 | 4,771 | |||||
| AK065121 | Rice (japonica cultivar group) BE-I | BEI | 1.06 | 12,259 | 12,946 | 0.82 | ||||
| AK108535 | Rice BE-3 | BEIIb | 0.41 | 127,584 | 52,108 | 0.81 | 2,298 | 1,970 | 0.59 | |
| AB093426 | Rice (japonica cultivar group) isoamylase | ISA1 | 0.94 | |||||||
| AK101554 | Maize isoamylase-type starch debranching enzyme ISO3 (iso3) | ISA3 | 0.93 | 307 | 284 | |||||
| AB012915 | Rice starch debranching enzyme | PUL | 0.83 | |||||||
| AK101744 | Rice α-amylase, clone pOS103 | Amy1A | 1.10 | 280 | 308 | 2.43 | ||||
| AK059671 | Rice Amyc2 α-amylase | Amy2A | 1.03 | 380 | 393 | |||||
| AK063988 | Rice α-amylase, clone pOS137 | Amy3A | 1.02 | 727 | 739 | |||||
| AK101358 | α-Amylase | Amy3C | 1.22 | 206 | 251 | |||||
| AK064124 | Rice α-amylase, clone pOS137 | Amy3D | 1.31 | 128 | 168 | 2.29 | ||||
| AK064300 | α-Amylase | Amy3E | 3.39 | 4,367 | 14,799 | 2.26 | ||||
| AK065152 | Arabidopsis putative α-amylase (At1g69830/T17F3_14) | Amy4A | 0.77 | 952 | 737 | |||||
| AK070300 | Arabidopsis At5g45300 putative β-amylase: RAFL16-68-D16 | (Bamy) | 0.97 | 9,573 | 9,321 | |||||
| AK067249 | Arabidopsis putative β-amylase (At3g23920) | (Bamy) | 0.93 | 1,430 | 1,329 | |||||
| AK068968 | Arabidopsis putative β-amylase (At3g23920) | (Bamy) | 1.18 | 1,365 | 1,617 | |||||
| AK100910 | Rice (japonica cultivar group) AGP large subunit | AGPL1 | 0.88 | 4,756 | 4,165 | 1.03 | ||||
| AK071497 | Rice AGP large subunit | AGPL2 | 0.69 | 71,750 | 49,582 | 0.79 | 1,833 | 1,522 | 0.74 | |
| AK069296 | Barley AGP large subunit (blpl) | AGPL3 | 0.48 | 842 | 407 | |||||
| AK121036 | Citrullus lanatus AGP large subunit 2 (wml2) | AGPL4 | 1.22 | 884 | 1,140 | |||||
| AK073146 | Rice AGP small subunit | AGPS1 | 0.57 | 9,680 | 5,565 | 0.91 | 1,124 | 1,079 | 0.67 | |
| AK071826 | Rice AGP 51-kD subunit | AGPS2a | 0.79 | 69,403 | 54,531 | 0.97 | 2,447 | 2,502 | ||
| AK103906 | Rice AGP 51-kD subunit | AGPS2b | 1.12 | 2,257 | 2,670 | 0.64 | ||||
| AK121403 | Rice (japonica cultivar group) OsCIN1 cell wall invertase | CIN1 | 1.05 | 1,217 | 1,347 | |||||
| AK072276 | Maize cultivar W22 inbred cell wall invertase 2 (Incw2) | CIN2 | 0.88 | 4,737 | 4,152 | |||||
| AK120779 | Rice (japonica cultivar group) cell wall invertase 3 | CIN3 | 0.67 | 1,207 | 854 | |||||
| AK069080 | Triticum monococcum extracellular invertase (exin1) | INV1 | 1.02 | 5,862 | 5,978 | |||||
| AK099312 | Rice (indica cultivar group) ‘IR64’ vacuolar invertase 1 | INV2 | 0.62 | 1,327 | 876 | |||||
| AK072245 | Lolium temulentum putative soluble acid invertase (inv1:4) | INV3 | 0.82 | 1,358 | 1,111 | |||||
| AK061240 | Rice (indica cultivar group) invertase (INV) | (INV) | 0.91 | 2,078 | 1,898 | |||||
| AK103334 | Rice (indica cultivar group) invertase (INV) | (INV) | 1.12 | 289 | 325 | |||||
| AK100373 | Lolium temulentum alkaline/neutral invertase | (INV) | 1.01 | 1,963 | 1,975 | |||||
| AK065130 | Allium cepa invertase | (INV) | 1.14 | 695 | 794 | |||||
| AK100334 | Bambusa oldhamii clone BSUS1b SuSy 2 | SuSy1 | 1.05 | 2,477 | 2,590 | |||||
| AK072074 | Rice SuSy | SuSy2 | 0.77 | 22,992 | 17,803 | 0.86 | 1,322 | 1,202 | 0.58 | |
| AK100306 | Wheat SuSy type 2 | SuSy3 | 0.57 | 32,652 | 18,686 | 0.93 | 1,451 | 1,422 | 0.81 | |
| AK099406 | Maize SuSy 3 | SuSy4 | 0.70 | 3,673 | 2,557 | 0.83 | 1,768 | 1,549 | ||
| AK065549 | Citrus unshiu CitSUSA SuSy | SuSy5 | 1.01 | 233 | 235 | |||||
| AK063304 | Arabidopsis putative SuSy (At4g02280) | SuSy6 | 0.95 | 359 | 341 | |||||
| AK071525 | Maize Suc-phosphatase (spp1) | (SPP) | 1.05 | 282 | 297 | |||||
| AK063330 | Aegilops speltoides Suc-phosphatase | (SPP) | 0.90 | 1,972 | 1,781 | |||||
| AK063433 | Barley Suc-phosphate synthase | (SPS) | 0.88 | 233 | 205 | |||||
| AK069527 | Maize Suc-phosphate synthase | (SPS) | 1.18 | 225 | 265 | |||||
| AK071732 | Saccharum officinarum SoSPS2 premature mRNA for Suc-phosphate synthase | (SPS) | 0.82 | 1,085 | 885 | |||||
| AK101676 | Oncidium ‘Goldiana’ Suc-phosphate synthase (SPS) | (SPS) | 0.99 | 1,147 | 1,139 | |||||
| AK065780 | Rice (japonica cultivar group) UGP, UDP-Glc pyrophosphorylase | UGP | 0.60 | 5,831 | 3,506 | 0.95 | ||||
| AK071248 | Rice sp. indica UDP-Glc pyrophosphorylase (UDPGase) | (UGP) | 0.77 | 467 | 358 | |||||
| AK112015 | Rice sp. indica UDP-Glc pyrophosphorylase (UDPGase) | (UGP) | 1.01 | 500 | 503 | |||||
| AK068061 | Arabidopsis clone C105270 putative Glc-6-P isomerase (At4g24620) | PGI-a | 0.70 | 6,872 | 4,792 | 0.64 | 810 | 545 | 0.84 | |
| AK068236 | Rice phospho-Glc isomerase (Pgi-b) | PGI-b | 0.74 | 2,226 | 1,656 | 1.06 | 947 | 1,061 | 0.91 | |
| AK100027 | Rice (japonica cultivar group) Suc transporter | SUT1 | 1.01 | 2,198 | 2,219 | 0.72 | ||||
| AK109461 | Barley Suc transporter 2 (sut2) | SUT2 | 1.05 | 1,318 | 1,381 | |||||
| AK071452 | Rice (japonica cultivar group) OsSUT3 Suc transporter | SUT3 | 1.30 | 288 | 376 | |||||
| AK065430 | Vitis vinifera putative Suc transporter (VvSUC12) | SUT4 | 0.91 | 512 | 467 | |||||
| AK073105 | Maize Suc transporter | SUT5 | 0.99 | 294 | 292 | |||||
| AK060577 | Rice GPT | GPT1 | 0.82 | 26,786 | 21,997 | 0.59 | 1,135 | 712 | 0.66 | |
| AK070124 | Pisum sativum GPT precursor, nuclear gene encoding plastid protein | GPT2 | 1.06 | 1,978 | 2,094 | |||||
| AK065618 | Rice GPT putative Glc-6-P/phosphate-translocator | (GPT) | 0.94 | 1,274 | 1,191 | |||||
| AK062253 | Rice GPT putative Glc-6-P/phosphate-translocator | (GPT) | 0.93 | 1,520 | 1,417 | |||||
| AK103471 | Solanum tuberosum brittle1 protein | BT1-1 | 1.20 | 820 | 986 | 1.47 | 782 | 1,219 | ||
| AK107368 | Maize brittle-1 protein (bt1) | BT1-2 | 0.89 | 188,078 | 166,841 | 0.92 | 2,043 | 1,993 | 0.70 | |
| AK063766 | Rice starch phosphorylase L isozyme | PHOL | 0.49 | 31,965 | 15,598 | 0.45 | 2,211 | 1,050 | ||
| AK103367 | Rice starch phosphorylase H isozyme | PHOH | 1.02 | 2,329 | 2,366 | |||||
| AK067082 | Arabidopsis 4-α-glucanotransferase (At2g40840) | DPE2 | 0.88 | 2,678 | 2,344 | |||||
| AK065739 | Rice sp. indica cyPPDKB | PPDKA | 1.53 | 2,107 | 3,221 | 1.26 | ||||
| AK068025 | Rice (japonica cultivar group) orthophosphate dikinase | PPDKB | 0.45 | 10,499 | 4,705 | 0.43 | ||||
| Storage proteins | ||||||||||
| AK108254 | Rice 10-kD prolamin | 10kD Pro | 1.01 | 37,957 | 38,427 | |||||
| AF194115 | Rice Prolamin 7 | Pro 7 | 0.72 | 8,422 | 6,379 | 0.57 | DS16 (down- regulated) | |||
| S39468 | Rice 13-kD prolamin | 13kD Pro | 0.85 | 7,761 | 6,967 | 0.42 | DS86 (down- regulated) | |||
| AK107785 | Rice prolamin, clone: λ RM9 | 16kD Pro | 1.39 | 143,851 | 200,016 | 1.24 | 4,740 | 6,212 | 1.03 | |
| AK107238 | Rice preproglutelin | Glu RG21 | 0.95 | 292,967 | 277,529 | |||||
| AK064478 | Rice glutelin, clone: λ RG55 | Glu RG55 | 1.60 | 131,410 | 209,827 | 1.38 | 5,903 | 8,618 | 0.95 | |
| AK107285 | Rice glutelin type I (clone pREE 103) | Glu type I | 1.15 | 241,921 | 277,110 | 1.17 | 10,526 | 13,054 | 0.78 | DS18 (up- regulated) |
| AK107314 | Rice glutelin type II (clone pREE 99) | Glu type II | 0.94 | 182,855 | 171,010 | 1.23 | 10,346 | 13,473 | 0.97 | DS72 (up- regulated) |
| AK107343 | Rice glutelin | Glu B-1 | 0.82 | 242,697 | 198,004 | 0.92 | 9,150 | 8,924 | 0.72 | DS91 (down- regulated) |
| AK107271 | Rice glutelin 1 (Gt22) | Glu A-3 | 0.86 | 250,840 | 215,299 | |||||
| AK105347 | Rice globulin-like protein, clone Ose710 | Glb-like | 2.60 | 8,580 | 22,322 | 2.63 | 1,231 | 3,418 | 0.94 | |
| AK107328 | Rice (japonica cultivar group) allergenic protein, clone RA14b | RAG2 | 1.66 | 200,925 | 333,657 | 1.55 | 4,564 | 7,464 | 1.20 | DS108 (up- regulated) |
| AK068268 | Rice PDI | PDI | 0.53 | 22,285 | 11,748 | 0.56 | 3,818 | 2,244 | 0.68 | |
| HSPs | ||||||||||
| AK105433 | Rice HSP (HSP101) | HSP101 | 1.91 | 1,091 | 2,082 | 1.39 | 576 | 846 | 1.32 | |
| AK063751 | Wheat HSP80-2 protein | HSP82 | 2.95 | 185 | 546 | 1.15 | 889 | 1,085 | 3.04 | |
| AK065431 | Spinacia oleracea heat shock C70 protein | HSP70 | 1.80 | 8,498 | 15,267 | 2.02 | 1,564 | 3,333 | 2.70 | |
| AK063618 | Rice HSP 26 | HSP26 | 6.72 | 2,007 | 13,497 | 1.40 | 657 | 974 | 2.63 | |
| AK063700 | Arabidopsis Columbia endomembrane-localized small HSP AtHSP22.0 | HSP22a | 1.62 | 3,597 | 5,825 | 0.59 | 683 | 424 | 6.73 | |
| AK107883 | Glycine max low-Mr HSP Hsp22.3 (Gmhsp22.3) | HSP22b | 2.69 | 156 | 419 | 1.57 | 1,004 | 1,665 | 1.99 | |
| AK071240 | Maize an 18-kD HSP | HSP16.9 | 2.34 | 3,268 | 7,661 | 1.76 | 861 | 1,605 | 1.42 | |
| AK106545 | Arabidopsis HSF 7 (hsf7) | HSF | 1.69 | 1,223 | 2,069 | 1.16 | 824 | 1,010 | 4.31 | |
| Up-regulated genes | ||||||||||
| AK063493 | Barley lipid transfer protein 7a2b | 5.26 | 913 | 4,804 | ||||||
| AK066733 | Avena fatua aldose reductase-related protein | 5.24 | 1,080 | 5,659 | ||||||
| AK073443 | Digitalis lanata lanatoside 15-O-acetylesterase | 4.42 | 1,736 | 7,680 | 1.45 | 955 | 1,462 | |||
| AK103890 | Maize ubiquitin/ribosomal protein S27a fusion protein | 4.04 | 285 | 1,150 | 1.10 | 1,542 | 1,789 | |||
| AK064058 | Unknown expressed protein | 3.43 | 433 | 1,486 | 1.87 | 623 | 1,230 | |||
| AK064356 | Rice class III chitinase homolog (OsChib3H-h), clone: S2321 | 3.36 | 972 | 3,264 | ||||||
| AK102970 | Rice pathogenesis-related thaumatin-like protein | 3.23 | 3,185 | 10,276 | ||||||
| AK107696 | Unknown expressed protein | 3.20 | 42,783 | 136,988 | ||||||
| AK105832 | Rice (japonica cultivar group) OsENOD93a early nodulin | 3.10 | 5,542 | 17,179 | 1.85 | 793 | 1,554 | |||
| AK069098 | Rice Ramy1 (ramy1) | 3.10 | 17,492 | 54,194 | 1.86 | 739 | 1,455 | |||
| AK110620 | Arabidopsis putative terminal Flower 1 protein (At1g18100) | 3.07 | 1,001 | 3,076 | 1.55 | 809 | 1,324 | |||
| AK073843 | Rice (japonica cultivar group) chitinase | 3.06 | 464 | 1,419 | 1.57 | 728 | 1,212 | |||
| AK105267 | Arabidopsis unknown protein (At5g01220) | 3.05 | 1,195 | 3,638 | 1.84 | 491 | 954 | |||
| AK062520 | Rice lectin-like protein | 2.94 | 786 | 2,308 | ||||||
| AK059202 | Unknown expressed protein | 2.89 | 8,256 | 23,886 | ||||||
| AK063504 | Wheat (cDNA I) EC protein | 2.87 | 15,008 | 43,048 | ||||||
| AK063126 | Rice glucanase (GLU) | 2.72 | 2,581 | 7,026 | 0.99 | 725 | 758 | |||
| AK107301 | Arabidopsis purple acid phosphatase (PAP20) | 2.66 | 4,062 | 10,796 | 1.50 | 861 | 1,364 | |||
| AK073083 | Rice embryo-specific (Ose705) mRNA | 2.60 | 92,487 | 240,067 | ||||||
| AK099481 | Rice U2 snRNP auxiliary factor, small subunit 35a | 2.52 | 1,060 | 2,674 | 1.42 | 723 | 1,087 | |||
| AK062619 | Unknown expressed protein | 2.52 | 998 | 2,515 | 1.97 | 554 | 1,157 | |||
| AK064494 | Unknown expressed protein | 2.48 | 10,200 | 25,337 | ||||||
| AK067257 | Rice Bowman-Birk proteinase inhibitor | 2.44 | 47,097 | 114,783 | ||||||
| AK059772 | Rice (japonica cultivar group) OsENOD93a early nodulin | 2.37 | 1,909 | 4,531 | ||||||
| AK061304 | Rice sp. japonica clone S2148_A putative glutathione S-transferase OsGSTU3 | 2.35 | 705 | 1,655 | 1.02 | 763 | 826 | |||
| AK070191 | Unknown expressed protein | 2.21 | 1,496 | 3,313 | ||||||
| AK063796 | Rice sp. japonica clone C63266_1A putative glutathione S-transferase OsGSTF5 | 2.21 | 756 | 1,672 | 1.26 | 670 | 892 | |||
| AK058507 | Nicotiana tabacum LIM-domain protein | 2.21 | 2,879 | 6,362 | ||||||
| AK108569 | Arabidopsis At5g24230 unknown protein, clone: RAFL21-45-L01 | 2.18 | 6,823 | 14,872 | ||||||
| AK108237 | Ananas comosus polyphenol oxidase | 2.13 | 1,577 | 3,361 | ||||||
| AK102261 | Arabidopsis clone 37751 mRNA | 2.12 | 1,542 | 3,277 | ||||||
| AK058291 | Barley xyloglucan endotransglycosylase-like protein (XEB) | 2.11 | 555 | 1,170 | ||||||
| AK067373 | Unknown expressed protein | 2.10 | 2,319 | 4,872 | ||||||
| AK099995 | Unknown expressed protein | 2.10 | 10,458 | 21,971 | ||||||
| AK061288 | Rice lipid transfer protein LPT IV | 2.08 | 34,166 | 70,911 | ||||||
| AK107215 | Rice (japonica cultivar group) allergenic protein, clone RA14c | 2.07 | 61,386 | 127,317 | ||||||
| AK100844 | Rice FK506 binding protein | 2.07 | 2,286 | 4,735 | ||||||
| AK063691 | Rice rab25 | 2.06 | 16,210 | 33,460 | 1.59 | 665 | 1,122 | |||
| AK102146 | Rice vacuolar H+-pyrophosphatase | 2.06 | 5,505 | 11,323 | ||||||
| AK062831 | Rice (japonica cultivar group) thionin Osthi1 | 2.04 | 666 | 1,360 | ||||||
| AK105316 | Rice early embryogenesis protein (OSE351) | 1.99 | 135,819 | 269,660 | ||||||
| AK108345 | Maize peroxidase (pox1) | 1.96 | 803 | 1,571 | 1.27 | 676 | 911 | |||
| AK063584 | Rice elicitor-responsive gene-3 (ERG3) | 1.95 | 1,660 | 3,240 | ||||||
| AK069446 | Rice (japonica cultivar group) catalase | 1.92 | 1,234 | 2,372 | 1.10 | 814 | 944 | |||
| AK063592 | Rice hydrophobic LEA-like protein | 1.87 | 5,710 | 10,657 | 1.08 | 773 | 886 | |||
| Down-regulated genes | ||||||||||
| AK059459 | Unknown expressed protein | 0.11 | 7,257 | 825 | ||||||
| AK107610 | Arabidopsis putative subtilisin Ser protease (At2g05920) | 0.22 | 3,145 | 697 | ||||||
| AK072149 | Anthoceros formosae chloroplast rps3, rpl22, ribosomal protein S3, ribosomal protein L22 | 0.24 | 1,238 | 294 | 0.15 | 1,002 | 163 | |||
| AK064209 | Lycopersicon esculentum RNA-directed RNA polymerase | 0.24 | 339 | 82 | ||||||
| AK066203 | Unknown expressed protein | 0.25 | 6,004 | 1,506 | 0.42 | 1,041 | 466 | |||
| AK107401 | Arabidopsis Ser carboxypeptidase (At5g36180) | 0.27 | 1,856 | 497 | 0.81 | 1,068 | 911 | |||
| AK061920 | Rice EN242 | 0.32 | 26,483 | 8,482 | 0.99 | 1,090 | 1,142 | |||
| AK073415 | Rice sp. japonica putative glutathione S-transferase OsGSTZ2 | 0.32 | 31,443 | 10,106 | ||||||
| AK062797 | A. formosae chloroplast Ycf4 protein | 0.36 | 2,275 | 828 | 0.21 | 808 | 179 | |||
| AK103804 | Arabidopsis At4g31120 putative kinase binding protein, clone: RAFL19-91-C03 | 0.38 | 1,461 | 557 | 0.43 | 1,239 | 561 | |||
| AK107707 | Unknown expressed protein | 0.39 | 348 | 136 | 0.62 | 1,359 | 892 | |||
| AK063467 | Arabidopsis Glu dehydrogenase 2 (GDH2) | 0.41 | 2,868 | 1,172 | 0.91 | 984 | 950 | |||
| AK107986 | Rice (japonica cultivar group) floral organ regulator 1 (FOR1) | 0.41 | 381 | 157 | 0.97 | 1,560 | 1,608 | |||
| AK101511 | Unknown expressed protein | 0.42 | 3,610 | 1,510 | 0.87 | 1,666 | 1,533 | |||
| AK103071 | Unknown expressed protein | 0.42 | 3,068 | 1,293 | 0.85 | 1,097 | 984 | |||
| AK107597 | Unknown expressed protein | 0.44 | 2,846 | 1,242 | 0.89 | 1,447 | 1,357 | |||
| AK064573 | Arabidopsis putative signal recognition particle protein (At5g61970) | 0.44 | 1,859 | 815 | ||||||
| AK101962 | Arabidopsis putative pectin methylesterase (At5g09760) | 0.45 | 4,964 | 2,217 | 0.81 | 1,400 | 1,196 | |||
| AK069206 | Rice OSK3 | 0.45 | 17,820 | 8,014 | ||||||
| AK059231 | Arabidopsis clone 6145 mRNA | 0.45 | 133,348 | 60,330 | ||||||
| AK058903 | Rice acyl carrier protein II | 0.46 | 10,006 | 4,634 | ||||||
| AK070669 | Arabidopsis putative subtilisin Ser protease ARA12 (At5g67360) | 0.46 | 1,531 | 712 | ||||||
| AK059234 | L. esculentum (‘Rutgers’) ribosomal protein S25 | 0.46 | 10,164 | 4,724 | ||||||
| AK058954 | Arabidopsis At4g21105 unknown protein, clone: RAFL19-89-M12 | 0.47 | 11,409 | 5,329 | 0.75 | 1,661 | 1,318 | |||
| AK108210 | Unknown expressed protein | 0.47 | 190,252 | 89,440 | ||||||
| AK062052 | Maize ribosomal protein L17 (rpl17) | 0.47 | 6,637 | 3,124 | ||||||
| AK060247 | A. formosae chloroplast rps7 ribosomal protein S7 | 0.47 | 20,809 | 9,826 | ||||||
| AK072845 | Rice putative histone deacetylase HD2 | 0.48 | 5,248 | 2,498 | 0.71 | 1,921 | 1,451 | |||
| AK100959 | Rice β-expansin (EXPB3) | 0.48 | 12,169 | 5,866 | ||||||
| AK062048 | Wheat Sec61p (sec61) | 0.48 | 6,665 | 3,216 | ||||||
| AK102932 | Arabidopsis putative C3HC4 zinc finger protein (At2g30580/T6B20.7) | 0.49 | 4,118 | 2,018 | 0.90 | 1,239 | 1,178 | |||
| AK059477 | Maize chloroplast large ribosomal protein 2 (rpl2) | 0.50 | 9,265 | 4,610 | ||||||
| AK071514 | Elaeis guineensis clone opsc112 protein disulfide isomerase | 0.50 | 49,301 | 24,556 | 0.86 | 1,291 | 1,177 | |||
| AK061951 | Lupinus luteus tRNA-Gln synthetase | 0.50 | 2,429 | 1,221 | ||||||
| AK072751 | Lolium perenne nucleoside diphosphate kinase (NDPK) | 0.52 | 30,084 | 15,533 | 0.74 | 2,059 | 1,602 | |||
Names in parentheses indicate tentative categories, not gene names.
The fold-change ratio for RT-PCR analysis is the ratio of cumulative expression levels for 8 to 30 DAF.
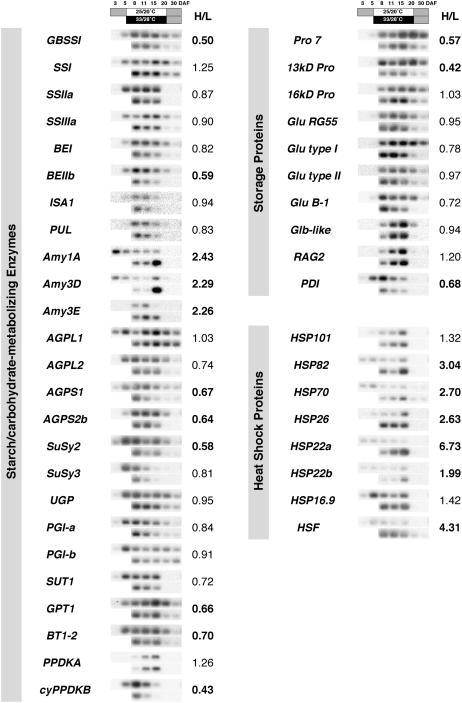
For carbohydrate metabolism, especially starch synthesis, the genes encoding major functional enzymes are predominantly known (Hirose and Terao, 2004; Ohdan et al., 2005), and available genes related to starch metabolism were included in the list of gene expression levels (Table III). For some genes whose full-length cDNA clones were available at the Rice Genome Resource Center, National Institute of Agrobiological Sciences (Tsukuba, Japan), but not included in the Agilent 22-K microarray, their expression levels were briefly checked by dot-blot hybridization. While most of carbohydrate enzymes showed moderate changes at high temperature (the ratio of expression level at high temperature to that of the control determined by the microarray analysis ranged around 0.80–1.00, 0.89 on average), transcription of GBSSI and BEIIb was strongly reduced to 35% and 41% of the control, respectively. In contrast, the expression of Amy3E was notably induced by high temperature.
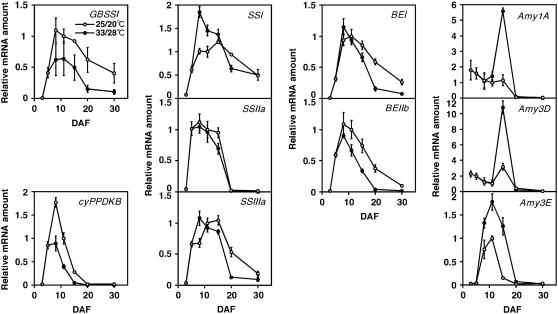
Because starch synthesis-related gene isoforms expressed in developing seeds have been identified by previous studies (Hirose and Terao, 2004; Ohdan et al., 2005), expression of the major seed-specific types of genes, including GBSSI, BEIIb, and Amy3E, was investigated for the time course of grain filling by semiquantitative reverse transcription (RT)-PCR analysis using gene-specific primers (Table IV). As shown in Figure 2, the expression of GBSSI and BEIIb, as well as AGPS1, AGPS2b, Susy2, GPT1, and BT1-2, was strongly diminished during grain filling at 33°C/28°C. Comparing the sum of densitometrically determined expression intensity values from 8 to 30 DAF to correct for the advancement of grain filling due to temperature variation, the total expression through the ripening period was reduced to 50% and 59% by high temperature for GBSSI and BEIIb, respectively. The quantification of the expression time course showed that GBSSI expression was reduced throughout the ripening phase, and that the transcription of BE genes, especially BEIIb, was more impaired than that of soluble SS genes (Fig. 3). In contrast, the expression of α-amylase genes was induced by high temperature and peaked at 15, 15, and 11 DAF for Amy1A, Amy3D, and Amy3E, respectively, and total expression from 8 to 30 DAF for high temperature was 2.43-, 2.29-, and 2.26-fold that for the control, respectively.
Table IV.
Gene-specific PCR primers and PCR cycles for semiquantitative RT-PCR amplification
The nucleotide sequences used for designing the PCR primers are shown by their accession number. The designed primer pairs yielded a strong single band for each gene.
| Enzyme/Property | Gene Name | Accession No. | Primer Pair | Amplicon Sizea | No. of Cycles | Synonymous to (Reference) |
|---|---|---|---|---|---|---|
| SS (granule-bound) | GBSSI | AK070431 | F: AACGTGGCTGCTCCTTGAA | 218 | 19 | Waxy (Okagaki, 1992) |
| R: TTGGCAATAAGCCACACACA | ||||||
| SS (soluble) | SSI | AK109458 | F: GGGCCTTCATGGATCAACC | 279 | 22 | |
| R: CCGCTTCAAGCATCCTCATC | ||||||
| SSIIa | AK101978 | F: GGCCAAGTACCAATGGTGAA | 272 | 23 | SSII-3 (Hirose and Terao, 2004; Jiang et al., 2004) | |
| R: GCATGATGCATCTGAAACAAAGC | ||||||
| SSIIIa | AK061604 | F: GCCTGCCCTGGACTACATTG | 334 | 23 | SSIII-2 (Hirose and Terao, 2004; Dian et al., 2005) | |
| R: GCAAACATATGTACACGGTTCTGG | ||||||
| BE | BEI | AK065121 | F: TGGCCATGGAAGAGTTGGC | 191 | 22 | |
| R: CAGAAGCAACTGCTCCACC | ||||||
| BEIIb | AK108535 | F: ATGCTAGAGTTTGACCGC | 261 | 20 | ae (Nishi et al., 2001); RBE3 (Mizuno et al., 1993) | |
| R: AGTGTGATGGATCCTGCC | ||||||
| Debranching enzyme | ISA1 | AB093426 | F: TGCTCAGCTACTCCTCCATCATC | 132 | 22 | Sugary-1 (Kubo et al., 1999) |
| R: AGGACCGCACAACTTCAACATA | ||||||
| PUL | AB012915 | F: ACCTTTCTTCCATGCTGG | 202 | 22 | ||
| R: CAAAGGTCTGAAAGATGGG | ||||||
| α-Amylase | Amy1A | AK101744 | F: GCGCCTGGTGTCAATCAGAA | 309 | 30 | |
| R: CGGATCGGATACAGCTCGTTG | ||||||
| Amy3D | AK119761 | F: TGCACGGCAAGGACTACAGC | 305 | 31 | ||
| R: CCAACGGTTACAAACTGCGTGA | ||||||
| Amy3E | AK064300 | F: GAGCACGCTGGACATCCTCA | 309 | 23 | ||
| R: GCTCGTACACATCTCGCAGCA | ||||||
| AGP (large subunit) | AGPL1 | AK100910 | F: ATGCAGTGCAGTGCGTCTTT | 183 | 23 | OsAPL3 (Akihiro et al., 2005) |
| R: ACTTCACTCGGGGCAGCTTA | ||||||
| AGPL2 | AK071497 | F: CGGGACCGTCATATAAAAGCA | 242 | 19 | OsAPL2 (Akihiro et al., 2005) | |
| R: TCCCATTCCAAAACAAACCA | ||||||
| AGP (small subunit) | AGPS1 | AK073146 | F: AGAATGCTCGTATTGGAGAAAATG | 258 | 22 | OsAPS1 (Akihiro et al., 2005) |
| R: GGCAGCATGGAATAAACCAC | ||||||
| AGPS2b | AK103906 | F: AACAATCGAAGCGCGAGAAA | 186 | 21 | Shrunken (Kawagoe et al., 2005); OsAPS2 (Akihiro et al., 2005) | |
| R: GCCTGTAGTTGGCACCCAGA | ||||||
| SuSy | SuSy2 | AK072074 | F: TTCAGCAGGAGAAGCCGTCAGC | 150 | 22 | RSus2 (Huang et al., 1996) |
| R: CCGGCGTTTATTTGAGGCAAGC | ||||||
| SuSy3 | AK100306 | F: CGGTGAAAAGAATGGGCAATG | 180 | 21 | RSus3 (Huang et al., 1996) | |
| R: CCATGAAAAGGCCAGAGCAT | ||||||
| UDP-Glc pyrophosphorylase | UGP | AK065780 | F: TCCTGGCCCGGTTTAAGTCA | 258 | 23 | |
| R: TGCCGAATGCACACGACAAT | ||||||
| Phospho-Glc isomerase | PGI-a | AK068061 | F: ATCCAGCACATGGCAGCAAA | 324 | 24 | |
| R: AAGGGCACGGGATGACAAGA | ||||||
| PGI-b | AK068236 | F: TGGGGAGTGGAACTGGGAAA | 237 | 26 | ||
| R: CAGAATATGCCGGCTCAACC | ||||||
| Suc transporter | SUT1 | AK100027 | F: AGTTCCGGTCGGTCAGCAT | 241 | 26 | |
| R: ACCGAGGTGGCAACAAAG | ||||||
| GPT | GPT1 | AK060577 | F: AGAAGGGATCCAGATGAAGAATG | 150 | 22 | |
| R: AACAAGAAACGAGCAACATAGACC | ||||||
| ADP-Glc translocator | BT1-2 | AK107368 | F: TGATTGTGCATGGGTGTGATG | 132 | 22 | |
| R: AACAGAGGAAATCGAATCCTACG | ||||||
| PPDK | PPDKA | AJ004965 | F: TTGCAAATGCAGAAACAACCAT | 410 | 26 | |
| R: TGCTGCTAGCCTTGCAATTG | ||||||
| cyPPDKB | D87952 | F: GCTCCGGCTCAATGTGCTCGT | 165 | 24 | ||
| R: CTCCGTCGACACCGTGAAC | ||||||
| chPPDKB | D87952 | F: CCAGGATGCCGTCGGTTTCGA | 368 | NAb | ||
| R: CTCCGTCGACACCGTGAAC | ||||||
| Prolamin | Pro 7 | AF194115 | F: GCGCAGCAGCTACAACTCCA | 206 | 15 | DS16 (this work) |
| R: TGAGCTTATTTTTAACTTCCGAACCA | ||||||
| 13kD Pro | S39468 | F: GGCCATAGCGCACCAGCTAC | 233 | 15 | DS86 (this work) | |
| R: TGTCACATACGATGATACCTGAGC | ||||||
| 16kD Pro | AK107785 | F: TTGCCAGGCTATTTGCACCA | 285 | 15 | ||
| R: CGAACAGCCAAAGACTATTCCAAA | ||||||
| Glutelin | Glu RG55 | AB016501 | F: CCAGCAACAATGCCAACCAG | 207 | 18 | |
| R: TCAGGCCTTGGAGCCTCAAC | ||||||
| Glu type I | AK107285 | F: TTCCGTGCTCTCCCAAATGA | 230 | 14 | DS18 (this work) | |
| R: TGGACAGTACATAGCAGCAAAACA | ||||||
| Glu type II | AK107314 | F: GGTGCATTCACTCCCCTCCA | 225 | 16 | DS72 (this work) | |
| R: CATTGGAACGGGAACACAAAAA | ||||||
| Glu B-1 | AK107343 | F: CGCCGTTCTGGAACGTCAAT | 316 | 14 | DS91 (this work) | |
| R: TTCTTGAGGCTTCGGGCTTG | ||||||
| Globulin | Glb-like | U45322 | F: CGAGAACGGCGAGAAGTGGT | 214 | 21 | |
| R: GCCCTTGCTGAAGCTCGACA | ||||||
| Allergen | RAG2 | AK107328 | F: GTCGACGACAGCTGGTGCAG | 313 | 16 | DS108 (this work) |
| R: TGCTTCCTGACAAATGAAAGCA | ||||||
| PDI | PDI | AK068268 | F: GGATGCAACTGCGAACGATG | 277 | 23 | |
| R: GTCAGGTCCCGTCTCCTCCA | ||||||
| HSP | HSP101 | AK105433 | F: GAGCTGGCCTACAGGGTGGA | 270 | 24 | |
| R: TTCAACGGAGACACCCCACA | ||||||
| HSP82 | AK063751 | F: AGCTCGGCCTCAACATCGAC | 309 | 30 | ||
| R: CGATGGCTTAGAAAGATGTACGC | ||||||
| HSP70 | AK065431 | F: GCCCCAAGATCGAGGAGGTC | 288 | 28 | ||
| R: CAAACAATGCGCACATGCAA | ||||||
| HSP26 | AK063618 | F: AGCGCAGCGTGAGCTCCTAC | 284 | 25 | ||
| R: TCATTCGCTCGTTCGCTGAG | ||||||
| HSP22a | AK063700 | F: CGGTCCTACGGCAGGTTCTG | 315 | 31 | ||
| R: CCGGAACATGTGCTCTGCAA | ||||||
| HSP22b | AK107883 | F: GAGGTTCTGGAGGCGGTTCA | 232 | 34 | ||
| R: GGTGTCGCCGATCACTATTCA | ||||||
| HSP16.9 | AK071240 | F: CGTGCAAGTACCTGCGGATG | 342 | 27 | ||
| R: TCGCATACGGCATACAGACCA | ||||||
| Heat shock transcription factor | HSF | AK106545 | F: TCCAGCTCCAGCCAAACGAT | 255 | 30 | |
| R: CTTGAACTTTCCGCCGCAAC |
Numbers indicate the size in bp of amplified fragments.
NA, Not amplified.
Figure 2.
Expression of genes related to starch metabolism, storage protein synthesis, and heat stress response in developing caryopses, as revealed by semiquantitative RT-PCR analysis. The time-course profiles for ripening under 25°C/20°C and 33°C/28°C are shown in the top and bottom rows for each gene, respectively. The developmental stage of the caryopsis is indicated by DAF. The expression levels were quantified densitometrically, and the ratio of accumulation of the transcript levels for 8 to 30 DAF of 33°C/28°C to that of 25°C/20°C (H/L) is indicated for each gene.
Figure 3.
Comparison of expression of genes for SSs, BEs, and α-amylases, and cyPPDKB. The expression levels were determined by semiquantitative RT-PCR analysis and densitometry, and the value of 25°C/20°C, 11 DAF was set to “1” for each gene. Results from four independent PCRs are shown with error bars (sd).
Down-regulation of a pyruvate orthophosphate dikinase (PPDK) gene, PPDKB, by high temperature treatment was revealed by the microarray analysis (Table III). Rice has two PPDK-encoding genes, PPDKA and PPDKB, and PPDKB has dual promoter sites, producing two distinct transcripts, cytosolic-type cyPPDKB and chloroplastic-type chPPDKB (Imaizumi et al., 1997). Time-course expression analysis by RT-PCR showed that expression of cyPPDKB was repressed by high temperature to 43% of the normal temperature level, while PPDKA was slightly induced (Figs. 2 and 3). No transcript for chPPDKB was detected in the developing caryopses.
Expression of genes encoding seed-specific storage proteins was differentially affected by rising temperature during grain filling. Expression of the genes for 13-kD prolamin, namely Pro-7 and 13kD Pro, increased and peaked at 15 to 20 DAF. However, under 33°C/28°C ripening, the expression was strongly attenuated at 20 DAF, resulting in a reduction of the total 8- to 30-DAF expression level to 57% and 42%, respectively (Fig. 2). In contrast, glutelin genes, most of which were expressed intensively 11 to 15 DAF, were less affected by high temperature. Furthermore, transcription of a gene for protein disulfide isomerase, PDI, which is reported to be necessary for precise sorting of storage proteins to protein bodies (Takemoto et al., 2002), was impaired to 68% of the control by high temperature.
HSP or heat shock factor (HSF) genes were also induced by high temperature. While induction ratios ranged from 1.62 to 6.72 in the microarray analysis and from 1.32 to 6.73 in the RT-PCR analysis (Table III), low-Mr HSPs, such as HSP26 and HSP22a, and a putative transcription factor gene (HSF) homologous to RHSF2 were most prominently induced (Fig. 2).
For other responsive genes, many genes encoding pathogenesis-related defense proteins, such as lipid transfer proteins, chitinases, thaumatin, glucanase, proteinase inhibitor, and thionin, were notably up-regulated more than 2-fold by high temperature in the microarray analysis (Table III).
Because the sample lot used for microarray and hybridization screenings is different from that used for RT-PCR analysis, it is difficult to compare the gene expression levels obtained by microarray, dot-blot hybridization, and RT-PCR analyses to each other. However, a similar tendency (correlation coefficient [R] = 0.668 for 67 available gene expression data sets; Table III) was observed for change ratio of gene expression between data from microarray and dot-blot hybridization, for which the same RNA sample set was used. In contrast, data from RT-PCR analysis showed less correlation to microarray data (R = 0.424 for 38 available data sets) and dot-blot hybridization data (R = 0.072 for 29 available data sets), probably because of difference in analyzed time point (cumulation of 8–30 DAF versus 10 DAF) as well as difference in used sample lots.
Grains Ripened under High Temperature Contained Amylopectin with Longer Side Chains
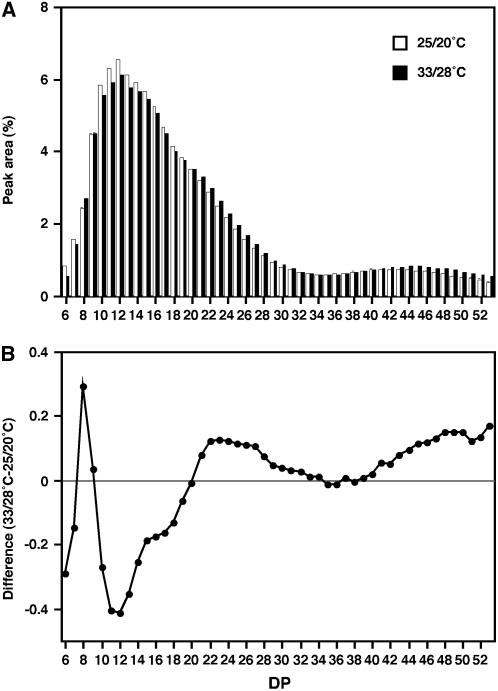
Because the reduction of expression of starch BE genes (BEI and BEIIb) under high temperature might be accompanied by structural alterations of amylopectin, the distribution of amylopectin side chains by length was elucidated for the ‘Nipponbare’ rice by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) analysis. As shown in Figure 4, rising temperature during the grain-filling period resulted in a decrease of short side chains of degree of polymerization (DP) 10 to 19 and increase of relatively longer side chains of DP 21 to 32 and DP more than 41.
Figure 4.
Comparison of the chain-length profile of amylopectin in ‘Nipponbare’ grain ripened under different temperatures. A, Chain-length profile. Debranched amylopectin extracted from 25°C/20°C-ripened (white bars) or 33°C/28°C-ripened (black bars) grains were analyzed by HPAEC-PAD, and the relative peak area of the chromatogram is shown for the individual DP. The data is the mean ± sd of five independent measurements. B, Difference in the chain-length distribution of amylopectin. The difference in the relative peak area in A between 33°C/28°C-ripened and 25°C/20°C-ripened grains is shown in the DP range of 6 to 53.
13-kD Prolamin Had Reduced Accumulation in Grains Ripened under High Temperature
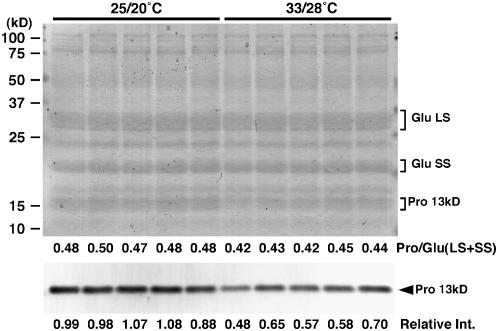
Among various seed storage proteins, the expression of 13-kD prolamin genes was most remarkably decreased under high temperature ripening (Fig. 2). SDS-PAGE analysis of total protein in mature grains also revealed that high temperature-ripened grains contained relatively less 13-kD prolamin, while they accumulated an apparently similar amount of both large and small subunits of glutelin compared with the control grains (Fig. 5). Furthermore, an immunoblot analysis using a 13-kD prolamin-specific antibody showed that approximately 50% to 70% amount of the protein accumulated in high temperature-exposed grains compared to the control.
Figure 5.
SDS-PAGE and immunoblot analyses of seed storage proteins. Total protein was extracted from mature grains ripened under 25°C/20°C or 33°C/28°C, and five independent extracts for each were separated on a SDS-polyacrylamide (18%) gel, followed by Coomassie Brilliant Blue staining or immunoblot detection with a polyclonal antibody raised against purified 13-kD prolamin. Large and small subunits of glutelin and 13-kD prolamin are indicated on the right side, and their relative amount was quantified densitometrically. For the SDS-PAGE, the ratio of 13-kD prolamin amount to the sum of glutelin large and small subunits is shown at the bottom, and, for the immunoblot, the relative amount of 13-kD prolamin is shown.
Among High Temperature-Ripened Grains, the Chalky Grains Contained Amylopectin Enriched with Longer Side Chains Compared to the Translucent Grains
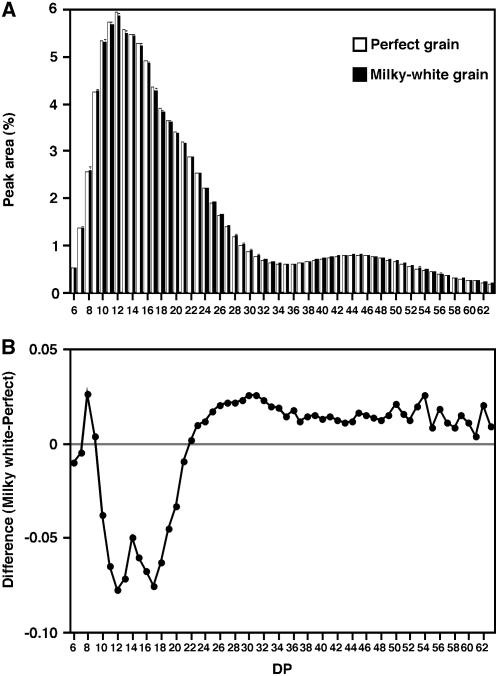
As shown in Figure 1B, grains with various degrees of chalkiness were obtained by ripening under high temperature. To elucidate the differences in the biochemical constituents between chalky and translucent grains both ripened under 33°C/28°C, the severely chalky, milky-white grains (indicated by white arrowheads in Fig. 1B) and less chalky, almost translucent grains (indicated by black arrowheads) were selected by hand and measured separately for amylopectin side chain length distribution and amylose content. Although the amylose contents of translucent perfect grains (16.4% ± 0.66%) and severely chalky grains (17.1% ± 0.80%) were not significantly different (P > 0.10), the amylopectin extracted from chalky grains consisted of a slightly smaller proportion of short side chains (DP 10–21) and larger proportion of long chains (DP > 23) than that from translucent grains (Fig. 6).
Figure 6.
Comparison of the chain-length profile of amylopectin in perfect (translucent) and milky-white (severely chalky) grains ripened under 33°C/28°C. A, Chain-length profile. Debranched amylopectin extracted from ‘Nipponbare’ perfect (white bars) or milky-white (black bars) grains was analyzed by HPAEC-PAD, and the relative peak area of the chromatogram is shown for the individual DP. The data is the mean ± sd of five independent measurements. B, Difference in the chain-length distribution of amylopectin. The difference in the relative peak area in A between milky-white and perfect grains is shown in the DP range of 6 to 63.
Difference in the Extent of Chalkiness among japonica Rice Cultivars Was Apparently Not Related to Amylose Content and Amylopectin Structure
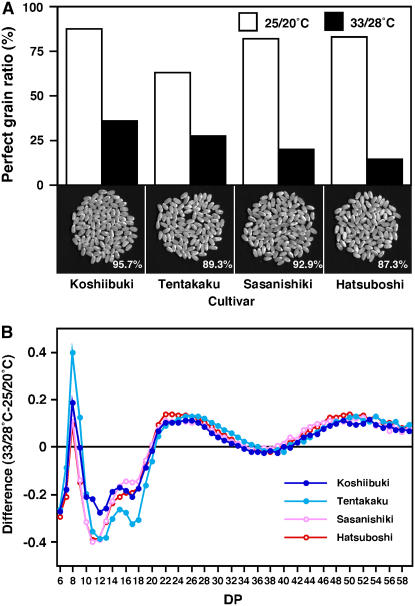
To determine whether the differences in the extent of chalky grain occurrence caused by high temperature among cultivars are attributed to alteration of starch composition or not, the correlation between chalky grain ratio, grain weight, amylose content, and amylopectin side chain length distribution was examined. ‘Koshiibuki’ and ‘Tentakaku’, which are high temperature-tolerant cultivars, contained more translucent perfect grains even when exposed to high temperature, while high temperature-sensitive ‘Hatsuboshi’ and ‘Sasanishiki’ contained more chalky grains (Fig. 7A). Decreases in grain weight were slight for ‘Koshiibuki’ (the ratio of 33°C/28°C-treated to 25°C/20°C-treated; 95.7%) and ‘Sasanishiki’ (92.9%), but larger for ‘Tentakaku’ (89.3%) and ‘Hatsuboshi’ (87.3%). Reductions of amylose content were 84.6%, 78.7%, 89.6%, and 75.9% for ‘Koshiibuki’, ‘Tentakaku’, ‘Sasanishiki’, and ‘Hatsuboshi’, respectively (Table II). Alteration in length distribution of amylopectin side chains by high temperature is shown in Figure 7B, and the cumulative values of the decrease in the proportion (%) of side chains of DP 10 to 19 and increase in the proportion (%) of side chains of DP 22 to 31, here tentatively designated the elongation index, were 2.82, 4.04, 3.21, and 3.59 for ‘Koshiibuki’, ‘Tentakaku’, ‘Sasanishiki’, and ‘Hatsuboshi’, respectively. For the expression of the genes described above in response to high temperature, these cultivars showed quantitatively similar induction/repression profiles to those observed for ‘Nipponbare’ (data not shown). As shown in Table V, the extent of reduction in amylose content and amylopectin side chain elongation index were apparently not related to the reduction of translucent perfect grains (R = 0.061 and 0.051, respectively) and seemed rather to be correlated to grain weight reduction (R = 0.795 and −0.836, respectively).
Figure 7.
Comparison of high temperature-tolerant (‘Koshiibuki’ and ‘Tentakaku’) and -sensitive (‘Sasanishiki’ and ‘Hatsuboshi’) cultivars. A, Appearance grade of grains matured under 25°C/20°C (white bar) and 33°C/28°C (black bar). The ratio of perfect (translucent) grain was determined by a grain-grading machine, ES-1000 (Shizuoka Seiki). Appearance of grains ripened under 33°C/28°C is shown in the photograph with the ratio of reduction of grain weight to that of the 25°C/20°C-treated control. B, Difference in the chain-length distribution of amylopectin. For the respective cultivars, the difference in the relative peak area on the HPAEC-PAD chromatogram between 33°C/28°C-ripened and 25°C/20°C-ripened grains is shown in the DP range of 6 to 59.
Table V.
Correlation between grain quality, weight, amylose content, and amylopectin structure
| Cultivar
|
Correlation Coefficient R to Perfect Grain Reduction Ratio | Correlation Coefficient R to Grain Weight Reduction Ratio | ||||
|---|---|---|---|---|---|---|
| ‘Koshiibuki’ | ‘Tentakaku’ | ‘Sasanishiki’ | ‘Hatsuboshi’ | |||
| Perfect grain reduction ratio (%)a | 41.2 | 43.9 | 24.4 | 17.7 | – | – |
| Grain weight reduction ratio (%)a | 95.7 | 89.3 | 92.9 | 87.3 | 0.431 | – |
| Amylose content reduction ratio (%)a | 84.6 | 78.7 | 89.6 | 75.9 | 0.061 | 0.795 |
| Amylopectin side chain elongation indexb | 2.82 | 4.04 | 3.21 | 3.59 | 0.051 | −0.836 |
The ratio of the value of 33°C/28°C-treated rice to that of 25°C/20°C-treated rice.
The total value cumulative decrease in the proportion of side chains of DP 10 to 19 and increase in the proportion of side chains of DP 22 to 31 in Figure 7B.
DISCUSSION
For better understanding of the effect of high temperature on the various metabolic processes of filling grains, it is important to establish the proper conditions of growth and high temperature treatment, which reproduce accurately what is observed in high temperature-ripened rice grains under field conditions. It is widely accepted in the field-grown rice that chalky grain appearance (Tashiro and Wardlaw, 1991a), decreased grain weight (Tashiro and Wardlaw, 1991b), and low amylose content (Asaoka et al., 1989; Umemoto and Terashima, 2002) are the typical symptoms caused by the grain filling under high temperature. In this study, we could reproduce all of these three symptoms in the high temperature plot (Fig. 1; Table II) as well as comparable grain weight and maturity to those of field-grown rice in the control plot, which is probably attained by the enhanced illumination and extensive management of plant growth including tiller number.
The comprehensive screening using microarray and conventional differential hybridization methods revealed that many genes were responsive at the transcriptional level to elevated temperature during the milky stage of grain filling, which is the most heat-sensitive phase leading to the production of chalky grains. The chalky appearance has been reported to be associated with the development of numerous air spaces between loosely packed starch granules and a change in light refraction (Tashiro and Wardlaw, 1991a; Zakaria et al., 2002). Actually, chalky grains ripened under high temperature in this study were observed to have similar ultra-microstructure (Fig. 1C). The expression level of genes encoding starch- or carbohydrate-metabolizing enzymes and translocators was diminished to 89% of the control on average by exposure to high temperature (Fig. 2). Among them, the transcription of GBSSI and genes for BE, which are indispensable for the production of starch components, amylose and amylopectin, respectively, was remarkably repressed throughout the grain-filling phase, whereas genes for α-amylase, a starch-consuming enzyme, were transiently induced by high temperature (Fig. 3). Reduction of amylose content by high temperature (Table II) can be explained by the repression in expression of GBSSI, whose encoding protein and activity also has been reported to be down-regulated (Hirano and Sano, 2000; Umemoto and Terashima, 2002). Decreased expression of BE genes, especially BEIIb, with relatively less changed or increased expression of soluble SS genes, might provide amylopectin with fewer but more elongated branches. Amylopectin in high temperature-ripened grains was found to be rich in middle and long side chains compared to the control (Fig. 4), which is consistent with a previous observation using rice plants grown under natural light (Umemoto et al., 1999). Down-regulation of the transcription of GBSSI and BE genes as well as the activity of their product enzymes was also reported for indica rice (Jiang et al., 2003), suggesting conservation of these responses to high temperature within rice subspecies.
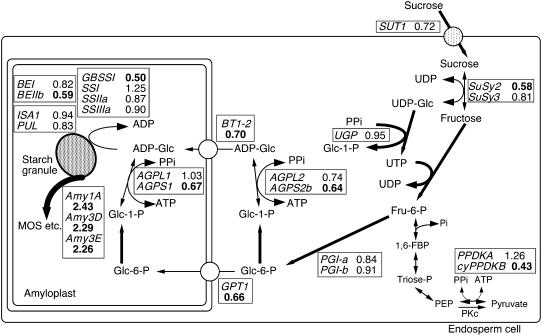
Starch metabolism-related genes reportedly expressed during the milky stage of grain filling, as well as other genes for carbohydrate-metabolizing enzymes or translocators whose strong expression in 10-DAF caryopses was revealed by microarray analysis, were subjected to time-course expression analysis by semiquantitative RT-PCR. The cumulative gene expression levels from 8 to 30 DAF were determined as total potential contribution to de novo starch synthesis. By placing the calculated ratios of the level of high temperature-treated caryopses to that of the control beside each reaction, a metabolic map was tentatively depicted for starch-related metabolism to indicate reaction steps responsive to elevated temperature at the transcriptional level (Fig. 8). This map allowed a global comprehension regarding the effects of high temperature on starch-related metabolism and revealed that several genes other than those described above, such as small subunit of AGP (AGPS1 and AGPS2b), ADP-Glc translocator (BT1-2), GPT1, and SuSy2, were down-regulated. Signal peptide-dependent targeting of α-amylases Amy1A, Amy3D, and Amy3E to plastids has been reported in rice seedling leaves and suspension-cultured cells (Chen et al., 2004; Asatsuma et al., 2005). However, whether similar localization occurs in developing caryopses or not remains to be determined.
Figure 8.
Ratio of cumulative expression level during 8 to 30 DAF of starch metabolism-related genes. The cumulative expression levels during 8 to 30 DAF were determined by semiquantitative RT-PCR and densitometry for grain filling under 25°C/20°C and 33°C/28°C, as described in the legend of Figure 2, and the ratio of cumulative transcript level for 33°C/28°C to 25°C/20°C is shown for the respective genes encoding enzymes/translocators on the starch-metabolizing pathway. The genes induced >1.5-fold and the corresponding reaction step are indicated in bold and with a thick arrow, while the genes repressed to <0.70-fold and the corresponding steps are indicated in bold and with thin arrows. The products of AGPL1 and AGPS1 genes have been estimated to be localized in amyloplasts, while those of AGPL2 and AGPS2b in the cytosol (Akihiro et al., 2005; Ohdan et al., 2005). The pathways described in the text are solely depicted. MOS, Maltooligosaccharide; PKc, cytosolic pyruvate kinase.
PPDKB has been recently identified as the causative gene for floury endosperm-4 (flo-4) mutation, whose grains showed a chalky appearance phenotype (Kang et al., 2005). High temperature impaired expression of cytosolic-type transcript of the gene cyPPDKB (Figs. 2 and 3), while no transcript for chPPDKB was detected in the caryopses, suggesting that decreased expression of cyPPDKB is one of the possible factors triggering for grain chalkiness, although the underlying mechanism of how down-regulation of the enzyme renders floury chalky endosperm is unclear.
Production of storage proteins, the other major constituent of mature rice grains, was also affected by high temperature during ripening. Among them, 13-kD prolamin, one of the major storage proteins, specifically decreased in response to high temperature for both transcripts and protein, while glutelins were less affected (Figs. 2 and 5). A previous proteomic approach had revealed up-regulation of Prolamin 7, a 13-kD prolamin, in developing caryopses harvested 9 DAF (Lin et al., 2005). This discrepancy might be attributed to differences in the analyzed samples. Because grain maturation progresses faster under high temperature (Fig. 1A), comparison of snap-shot images of expression level at a given early stage of grain filling, such as 9 DAF, corresponding to the onset of storage material synthesis, might not be consistent with that of the cumulative expression level throughout the whole ripening period as described in this research. Actually, under the high temperature conditions in our study, Prolamin 7 transiently showed higher expression level at 8 DAF than the control (Fig. 2).
Genes for various species of HSPs, typical molecular chaperones, were also induced by ripening under high temperature (Fig. 2). Among them, HSP101 is the orthologous gene of Arabidopsis (Arabidopsis thaliana) HSP101, whose heterogeneous overexpression in rice plants has been reported to confer thermotolerance to the plant (Katiyar-Agarwal et al., 2003). However, whether rice HSP101 could contribute to physiological protection of the developing caryopses against elevated temperature or not remains to be determined. In Arabidopsis, heat shock-inducible transcription factors HSF1 and HSF3 govern the expression of arrays of HSPs and regulate thermotolerance (Prändl et al., 1998; Lohmann et al., 2004). Whether HSF, a putative HSF strongly induced by high temperature ripening, has a similar function or not remains unclear.
To elucidate the causative biochemical factors for the chalky appearance of high temperature-ripened grains with regard to starch components, severely chalky grains and almost translucent grains, both of which were harvested from high temperature-exposed ‘Nipponbare’ plants, were compared for amylose content and amylopectin chain-length distribution. The chalky grains were shown to have amylopectin consisting of more elongated side chains than the translucent grains (Fig. 6), although the amylose content was not significantly different. The alteration of amylopectin side chain distribution is reminiscent of the amylose-extender (ae) mutant, which is deficient in BEIIb gene. Grains of the ae mutant have amylopectin lacking in short side chains of DP 6 to 16 and enriched in long chains of DP more than 19 and with a severely chalky opaque appearance (Nishi et al., 2001). The chalkiness of the mutant was recovered by introducing the wild-type BEIIb transgene in an expression level-dependent manner (Tanaka et al., 2004). Considering that the expression of the BEIIb gene was repressed under high temperature ripening, one possible explanation for grain chalkiness is the production of ae-like amylopectin by down-regulation of BEIIb expression. Chalky appearance of the ae mutant, whose severity changed in a manner dependent on Ae allele dosage or expression level, became prominent when the BEIIb protein amounts were lower than approximately 50% of the wild-type level (Nishi et al., 2001; Tanaka et al., 2004). In this study, ripening under high temperature reduced expression of BEIIb to 59% of the control level (Fig. 2). The extent of alteration of amylopectin side chain length distribution by high temperature was almost similar to that reported for the ae grains with moderate BEIIb protein levels. Thus, the critical expression level of the changes for the chalky appearance might be estimated around one-half of the level under the normal temperature, and reduced BEIIb expression as well as other perturbed metabolic factors would result in grain chalkiness under high temperature. However, the underlying mechanism of how the ae-like amylopectin renders chalky grain appearance is not clear at this time, and the precise relevance between the down-regulation of BEIIb and chalky appearance under high temperature remains to be determined. In Chlamydomonas, synthesis of amylopectin long chains by GBSSI has been reported (Delrue et al., 1992; Maddelein et al., 1994). Altered expression of GBSSI as well as BEIIb might be involved in the amylopectin structural modification in high temperature-exposed caryopses.
Furthermore, to clarify the underlying mechanisms for varietal differences in the degree of grain chalkiness, correlations between the extent of grain chalkiness, reduction of amylose content, and degree of amylopectin side chain elongation were investigated using cultivars ranging from high temperature tolerant (‘Koshiibuki’ and ‘Tentakaku’) to high temperature sensitive (‘Sasanishiki’ and ‘Hatsuboshi’). However, the reduction of amylose content and amylopectin side chain elongation by high temperature was not correlated to the grain chalkiness, but rather correlated to grain weight reduction (Table V), suggesting that the difference in the extent of grain chalkiness between high temperature-tolerant and -sensitive cultivars might be attributed to neither the reduction of amylose content nor the alteration in amylopectin structure. However, these biochemical alterations of starch components, amylose and amylopectin, were shown to be conserved among japonica rice cultivars.
In conclusion, we elucidated the effect of elevated temperature on grain-filling metabolism at the gene expression level and revealed down-regulation of several genes for starch or storage protein synthesis (such as GBSSI, BEIIb, and prolamin) and up-regulation of genes for starch-consuming α-amylases and HSPs. Especially, down-regulation of 13-kD prolamin and cyPPDKB (flo-4) and up-regulation of α-amylase genes were, to our knowledge, newly identified in this study, which is probably attained by excellent reproducibility of the incubator system as well as high sensitivity of microarray analysis. However, the activity of several starch-metabolizing enzymes, including BEIIb, has been recently reported to be interdependently controlled by posttranslational modifications such as protein phosphorylation and redox modulation (Tetlow et al., 2004). Actually, expression of several redox reaction-related genes, such as PDIs (AK068268 and AK071514) and glutathione S-transferases (AK061304, AK063796, and AK073415), was changed by high temperature in the microarray analysis (Table III). Analyses of such posttranslational regulation are required to understand the effect on starch metabolism. To clarify the precise mechanism for chalky grain occurrence, further research such as gene functional analyses using transgenic plants is necessary along with the determination of key enzyme activities and quantification of their metabolite levels. According to recent studies, ectopic overexpression of α-amylase genes resulted in grains with decreased weight and chalky appearance even ripened under ambient temperature (Asatsuma et al., 2006), and antisense suppression of Suc transporter gene SUT1 decreased the grain weight in the transgenic rice plant (Scofield et al., 2002). Expression of α-amylase genes and SUT1 was up-regulated and down-regulated by high temperature ripening, respectively (Fig. 2), suggesting that these alterations in gene expression might contribute to grain weight reduction and/or grain chalkiness. The grain-filling system using incubators offers a suitable experimental model for the evaluation of transgenic plants with highly reproducible grain filling achieved because restricted time and space are available for growing transgenic plants in many laboratories. The analyses using transgenic plants are currently in progress.
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa sp. japonica) ‘Nipponbare’, ‘Koshiibuki’, ‘Tentakaku’, ‘Sasanishiki’, and ‘Hatsuboshi’ were grown at 27°C/22°C (12-h-day/12-h-night cycles) until flowering in a plant incubator (model FLI-301NH; Eyela) equipped with a sodium lamp, which allows continuous illumination at an intensity of 560 μmol photons m−2 s−1. The relative humidity, which is an important factor for vigorous growth and grain filling, was approximately 50% to 70%, corresponding to 5 to 16 hPa of vapor pressure deficit. Six plants were grown in a plastic container (15 × 10 × 6 cm) filled with 600 mL of rice nursery culture soil (containing 0.15 g each of nitrogen, phosphate, and potassium), and each plant was restricted to the main culm by the removal of tillers as they developed. The number of days from sowing to heading was 62 d for ‘Nipponbare’ and 75 d for other cultivars. Approximately 15 to 20 d before heading, 3 g of a fertilizer (containing 0.18 g nitrogen, 0.24 g phosphate, 0.18 g potassium, and 0.06 g magnesium) was supplied per container. Each spikelet was marked on the flowering day for subsequent sampling. Five DAF, the plants were transferred to either a 33°C/28°C or 25°C/20°C chamber for high temperature or control treatment, respectively, and then the temperature was maintained at 25°C/20°C from 20 DAF to maturity. During the course of grain filling, developing caryopses were detached from the ear, weighed, immediately frozen in liquid nitrogen, and stored at −80°C until use. On 40 and 45 DAF, caryopses attached on the upper half of ears were dehulled, weighed, and photographed, and appearance quality was determined by a rice grain image analyzer (model ES-1000; Shizuoka Seiki), which can distinguish between perfect, immature (mainly chalky), damaged (cracked), abortive, and colored grains. The heat stress treatment experiment of ‘Nipponbare’ was repeated twice: (1) For each of the high temperature-treated and control plots, approximately 1,000 developing caryopses from 40 plants were sampled at 10 DAF and used in bulk for microarray analysis and hybridization screenings; and (2) 50 to 200 caryopses from five plants were sampled at various developmental stages and used for gene expression analysis by RT-PCR. For scanning electron microscopy, grains were cut transversely with a razor blade. The fractured surfaces were viewed with a Keyence VE-7800 scanning electron microscope at an accelerating voltage of 12 kV. For amylose content measurement and HPAEC-PAD analysis of amylopectin, 10 dehulled brown rice grains were polished to remove the embryo and pericarp by a test mill Pearlest (Kett) and crushed with a hammer; the resulting powder was used.
Oligo DNA Microarray Analysis and Dot-Blot Hybridization Analysis
A rice 22-K custom oligo DNA microarray kit (Agilent Technologies) that contains 21,938 oligonucleotides synthesized based on the sequence data of the rice full-length cDNA project (Kikuchi et al., 2003) was used. Total RNA was extracted from developing caryopses that were harvested from both 25°C/20°C- and 33°C/28°C-treated plants 10 d after heading using the method of Chang et al. (1993), but with the polyvinylpyrrolidone and spermidine omitted from the extraction buffer. The yield and RNA purity were determined spectrophotometrically. Integrity was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA (200 ng) was labeled with Cy3 or Cy5 using an Agilent Low RNA Input Fluorescent Linear Amplification kit (Agilent Technologies). Fluorescently labeled targets were hybridized to Agilent rice 22-K custom oligo DNA microarrays. Hybridization and wash processes were performed according to the manufacturer's instructions, and hybridized microarrays were scanned using an Agilent Microarray Scanner (Agilent Technologies). Feature Extraction software (Agilent Technologies) was employed for the image analysis and data extraction processes.
For several genes responsive to high temperature grain filling, the change of gene expression level was further confirmed by dot-blot hybridization. Ten nanograms of plasmid clone containing the full-length cDNA, which was obtained from the Rice Genome Resource Center, National Institute of Agrobiological Sciences (Tsukuba, Ibaraki, Japan), was denatured by boiling and blotted onto a nylon membrane (positively charged; Roche Diagnostics) using a dot blotter (model AE-6190; ATTO). Polyadenylated [poly(A+)] RNA was isolated from 25°C/20°C- or 33°C/28°C-treated caryopses using Oligotex-dT30 Super (Roche Diagnostics) and used for the synthesis of digoxigenin-labeled single-strand cDNA probes. RT was conducted at 42°C for 1 h in a 20-μL labeling reaction mixture containing 1 μg of poly(A+) RNA, 50 mm Tris-HCl, pH 8.3, 75 mm KCl, 3 mm MgCl2, 10 mm dithiothreitol, 0.5 mm each of dNTP except for 0.325 mm dTTP, 0.025 μg/μL oligo(dT)12-18 primer, 0.175 mm digoxigenin-dUTP, and 200 units of SuperScript II reverse transcriptase (Invitrogen). Hybridization was performed in high SDS buffer, and the blot was finally washed in 0.1× SSC and 0.1% SDS at 65°C. Detection was conducted using CDP-Star (Roche Diagnostics) according to the manufacturer's instructions. The signal intensities were determined with a Fluor-S Max imaging system and Quantity-One analysis software (Bio-Rad) and corrected by subtracting the local background.
Screening of High Temperature Grain Filling-Responsive Genes by Differential Hybridization and Subtractive Hybridization
For differential hybridization, poly(A+) RNA was purified from caryopses that were harvested from both 25°C/20°C- and 33°C/28°C-treated plants 10 d after heading. Complementary DNA was synthesized from each RNA and inserted into the Uni-ZAP XR vector (Stratagene) according to the manufacturer's instructions. Approximately 3 × 104 unamplified recombinant phages from each library were differentially screened with radioactively labeled single-strand cDNA probes synthesized from poly(A+) RNA of caryopses harvested from either 33°C/28°C-treated plants (plus probe) or 25°C/20°C-treated plants (minus probe). A 25-μL labeling reaction mixture contained 1 μg of poly(A+) RNA, 50 mm Tris-HCl, pH 8.3, 75 mm KCl, 3 mm MgCl2, 4 mm dithiothreitol, 0.8 mm each of dNTP except for 4.8 μm dCTP, 0.02 μg/μL oligo(dT)12-18 primer, 200 μCi of [α-32P]dCTP, and 200 units of SuperScript III reverse transcriptase (Invitrogen). After incubation at 42°C for 1 h, unincorporated nucleotides were separated by a Sephadex G-50 spin column (Roche Diagnostics), and labeled probe DNA was used for hybridization. A total of 24 plates (12 for the 25°C/20°C-treatment library and 12 for the 33°C/28°C-treatment library), each containing approximately 2.5 × 103 clones, were used to prepare pairs of replica nylon membranes (Hybond-N; GE Healthcare Bio-Science), one of which was hybridized with the plus probe and the other with the minus probe. Hybridization was performed at 65°C for 16 h in 6× SSC containing 0.1% SDS, 0.1% Ficoll, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin, and 100 μg/mL salmon sperm DNA. Filters were washed twice in 2× SSC and 0.1% SDS at 25°C and then five times in 0.1× SSC and 0.1% SDS at 65°C. The filters were exposed to XAR film (Kodak) with an intensifying screen at −80°C for 20 h. Among 6 × 104 clones screened, 20 hybridized differentially to the two probes. These cDNA clones were excised with helper phage and recircularized to generate subclones in the pBluescript SK− phagemid vector according to the manufacturer's instructions (Stratagene). Sequencing analysis revealed that they encoded six distinct genes. The clones containing the longest cDNA fragments, designated as DS16, DS18, DS72, DS86, DS91, and DS108, were selected for further analysis.
PCR-select subtractive hybridization was conducted using the same set of cDNAs prepared from 25°C/20°C-treated and 33°C/28°C-treated caryopses according to the manufacturer's instructions (BD Biosciences).
Semiquantitative RT-PCR Analysis
Total RNA was prepared from developing caryopses as described above. The isolated RNA (5 μg) was reverse-transcribed (SuperScript II; Invitrogen) using an oligo(dT)12-18 primer. An aliquot of the first-strand cDNA mixture corresponding to 6.25 ng of total RNA was used as a template. The semiquantitative PCR (10 μL total volume) was done using 0.25 units of Taq polymerase (ExTaq; TaKaRa). Thermocycling time and temperature were as follows: 98°C for 2 min, followed by an appropriate number of cycles of 97°C for 30 s, 55°C for 30 s, 72°C for 1 min, and a final extension period of 72°C for 2 min. The numbers of PCR cycles in the linear range of DNA amplification for each gene and designed gene-specific primers are listed in Table IV. Amplified bands were cloned and sequenced to confirm that they were fragments of the intended gene. The amplified DNA fragments were separated on a 2.0% (w/v) agarose gel, transferred to a nylon membrane (Hybond-N+; GE Healthcare Bio-Science), hybridized with specific cDNA probes amplified from the corresponding cDNA clone, and visualized by the AlkPhos Direct Labeling and Detection system (GE Healthcare Bio-Science) following the manufacturer's instructions. The signal intensities were determined with a Fluor-S Max imaging system and Quantity-One analysis software (Bio-Rad). For the GBSSI, SSI, SSIIa, SSIIIa, BEI, BEIIb, Amy1A, Amy3D, Amy3E, and cyPPDKB reactions, the experiment was repeated four times using the same cDNA sources, and the average mean and sd values of relative transcript abundance were calculated.
Determination of Amylose Content by Iodine Colorimetric Analysis
Apparent amylose content was measured by an iodine colorimetric method (Juliano, 1971). Twenty milligrams of polished rice powder was gelatinized by treatment with 0.1 mL of 95% ethanol and 0.9 mL of 1 m NaOH and stood for 10 min in boiling water. After the addition of 5 mL of distilled water, the solution was homogenized and filled up to 10 mL with distilled water. An aliquot (1 mL) of the solution was taken and added by 0.2 mL of 1 m acetic acid, 0.2 mL of 0.2% (w/v) I2, 2% (w/v) KI, and 8.6 mL of distilled water. After incubation at 27°C for 20 min, A620 was measured using a model DU7400 spectrophotometer (Beckman Coulter). Apparent amylose content was estimated by the method of Juliano (1971) from the base calibration line, which was obtained from the absorbance values by changing the ratio of potato amylose (Sigma, Type III) and waxy rice powder in the iodine solution.
Determination of the Distribution of α-1,4-Glucan Chain Length of α-Polysaccharides by HPAEC-PAD
Five milligrams of polished rice powder was suspended in 5 mL of methanol and boiled for 10 min. The homogenate was centrifuged at 8,000g for 3 min. The precipitated polyglucan was washed twice with 5 mL of distilled water, resuspended in a further 5 mL of distilled water, and then boiled for 60 min. An aliquot (1 mL) of the gelatinized polyglucan sample was added to 50 μL of 600 mm sodium acetate buffer, pH 4.6, and 10 μL of 2% (w/v) NaN3, and hydrolyzed by adding 5 μL of Pseudomonas amyloderamosa isoamylase (295 units, Hayashibara Biochemical Laboratories). After incubation at 40°C for 24 h, the hydroxyl groups of the debranched glucans were reduced with 3 mg of sodium borohydride under alkaline conditions for 20 h by the method of Nagamine and Komae (1996). The precipitate was dried in vacuo at room temperature. The reduced isoamylolysate sample was dissolved in 100 μL of 1 m NaOH for 60 min and diluted with 900 μL of distilled water. A 25-μL aliquot of the preparation was injected into a BioLC (model DX-500, Dionex) equipped with a pulsed amperometric detector and a CarboPac PA-1 column (4 mm × 25 cm). Size fractionation of α-1,4-glucans was performed with a linear gradient of sodium acetate (0–450 mm) in 0.1 m NaOH at a flow rate of 1 mL min−1.
Determination of Starch Content
Ten milligrams of polished rice powder was washed twice with 80% (v/v) ethanol at 80°C. After centrifugation at 10,000g for 5 min, the pellet was resuspended in distilled water, boiled for 16 h, digested with amyloglucosidase (Roche Diagnostics) for 20 min at 55°C, and the resultant Glc determined by an enzymatic method (Nakamura and Miyachi, 1982).
Protein Analysis by SDS-PAGE and Immunoblotting
Ten grains of dehulled brown rice were crushed with a hammer and homogenized in 4 mL of 62.5 mm Tris-HCl, pH6.8, 4% (w/v) SDS, 5% 2-mercaptoethanol, and 8 m urea. After centrifugation at 8,000g for 10 min, the supernatant was boiled for 10 min and an 8-μL aliquot of protein was separated on an 18% T, 0.45% C SDS-polyacrylamide gel, where T and C denote the percentage of both acrylamide and bisacrylamide, and the percentage of bisacrylamide relative to the total concentration, respectively. Proteins were stained with Coomassie Brilliant Blue R-250 and quantified densitometrically using Quantity-One analysis software (Bio-Rad).
For immunoblotting, a 0.01-μL aliquot of protein was separated on the same gel and transferred onto an Immun-Blot polyvinylidene difluoride membrane (Bio-Rad) by semidry electroblotting. The blot was incubated with rabbit antiserum that contained polyclonal antibodies raised against purified rice 13-kD prolamin (Furukawa et al., 2003; provided by Dr. T. Masumura, Kyoto Prefectural University). Immunoreactive proteins were detected by incubation with alkaline phosphatase-conjugated antibodies against rabbit IgG and visualized with 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium. The protein bands were quantified using Quantity-One analysis software (Bio-Rad).
Acknowledgments
We thank Y. Nagamura and R. Motoyama for technical help on microarray analysis, S. Seo and Y. Ohashi for invaluable advice on the procedures for differential screening, T. Umemoto and N. Aoki for technical suggestions concerning starch analysis, and T. Ishimaru and M. Kondo for critical advice throughout the study. T. Masumura is acknowledged for providing the prolamin-specific antibody. The rice seeds of ‘Koshiibuki’ and ‘Tentakaku’ were kindly provided by Niigata Agricultural Research Institute and Toyama Agricultural Research Center, respectively.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hiromoto Yamakawa (hy741220@affrc.go.jp).
Open Access articles can be viewed online without a subscription.
References
- Akihiro T, Mizuno K, Fujimura T (2005) Gene expression of ADP-glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA. Plant Cell Physiol 46 937–946 [DOI] [PubMed] [Google Scholar]
- Asaoka M, Okuno K, Hara K, Oba M, Fuwa H (1989) Effects of environmental temperature at the early developmental stage of seeds on the characteristics of endosperm starches of rice (Oryza sativa L.). Denpun Kagaku 36 1–8 [Google Scholar]
- Asaoka M, Okuno K, Sugimoto Y, Kawakami J, Fuwa H (1984) Effect of environmental temperature during development of rice plants on some properties of endosperm starch. Starch 36 189–193 [Google Scholar]
- Asatsuma S, Sawada C, Itoh K, Okito M, Kitajima A, Mitsui T (2005) Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol 46 858–869 [DOI] [PubMed] [Google Scholar]
- Asatsuma S, Sawada C, Kitajima A, Asakura T, Mitsui T (2006) α-Amylase affects starch accumulation in rice grains. J Appl Glycosci (1999) 53 187–192 [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11 113–116 [Google Scholar]
- Chen M-H, Huang L-F, Li H-m, Chen Y-R, Yu S-M (2004) Signal peptide-dependent targeting of a rice α-amylase and cargo proteins to plastids and extracellular compartments of plant cells. Plant Physiol 135 1367–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski J-M, Van den Koornhuyse N, Maddelein M-L, Fournet B, Ball S (1992) Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase activity accumulate a structurally modified amylopectin. J Bacteriol 174 3612–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dian W, Jiang H, Wu P (2005) Evolution and expression analysis of starch synthase III and IV in rice. J Exp Bot 56 623–632 [DOI] [PubMed] [Google Scholar]
- Furukawa S, Mizuma T, Kiyokawa Y, Masumura T, Tanaka K, Wakai Y (2003) Distribution of storage proteins in low-glutelin rice seed determined using a fluorescent antibody. J Biosci Bioeng 96 467–473 [DOI] [PubMed] [Google Scholar]
- Hawker JS, Jenner CF (1993) High temperature affects the activity of enzymes in the committed pathway of starch synthesis in developing wheat endosperm. Aust J Plant Physiol 20 197–209 [Google Scholar]
- Hirano H-Y, Sano Y (2000) Comparison of waxy gene regulation in the endosperm and pollen in Oryza sativa L. Genes Genet Syst 75 245–249 [DOI] [PubMed] [Google Scholar]
- Hirose T, Terao T (2004) A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 220 9–16 [DOI] [PubMed] [Google Scholar]
- Huang JW, Chen JT, Yu WP, Shyur LF, Wang AY, Sung HY, Lee PD, Su JC (1996) Complete structures of three rice sucrose synthase isogenes and differential regulation of their expressions. Biosci Biotechnol Biochem 60 233–239 [DOI] [PubMed] [Google Scholar]
- Hurkman WJ, McCue KF, Altenbach SB, Korn A, Tanaka CK, Kothari KM, Johnson EL, Bechtel DB, Wilson JD, Anderson OD, et al (2003) Effect of temperature on expression of genes encoding enzymes for starch biosynthesis in developing wheat endosperm. Plant Sci 164 873–881 [Google Scholar]
- Imaizumi N, Ku MSB, Ishihara K, Samejima M, Kaneko S, Matsuoka M (1997) Characterization of the gene for pyruvate, orthophosphate dikinase from rice, a C3 plant, and a comparison of structure and expression between C3 and C4 genes for this protein. Plant Mol Biol 34 701–716 [DOI] [PubMed] [Google Scholar]
- Jiang H, Dian W, Liu F, Wu P (2004) Molecular cloning and expression analysis of three genes encoding starch synthase II in rice. Planta 218 1062–1070 [DOI] [PubMed] [Google Scholar]
- Jiang H, Dian W, Wu P (2003) Effect of high temperature on fine structure of amylopectin in rice endosperm by reducing the activity of the starch branching enzyme. Phytochemistry 63 53–59 [DOI] [PubMed] [Google Scholar]
- Juliano BO (1971) A simplified assay for milled-rice amylose. Cereal Sci Today 16 334–340 [Google Scholar]
- Kang H-G, Park S, Matsuoka M, An G (2005) White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C4-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J 42 901–911 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Agarwal M, Grover A (2003) Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol Biol 51 677–686 [DOI] [PubMed] [Google Scholar]
- Kawagoe Y, Kubo A, Satoh H, Takaiwa F, Nakamura Y (2005) Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J 42 164–174 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, et al (2003) Collection, mapping and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379 [DOI] [PubMed] [Google Scholar]
- Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y (1999) The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol 121 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SK, Chang MC, Tsai YG, Lur HS (2005) Proteomic analysis of the expression of proteins related to rice quality during caryopsis development and the effect of high temperature on expression. Proteomics 5 2140–2156 [DOI] [PubMed] [Google Scholar]
- Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F (2004) Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Genet Genomics 271 11–21 [DOI] [PubMed] [Google Scholar]
- Maddelein M-L, Libessart N, Bellanger F, Delrue B, D'Hulst C, Van den Koornhuyse N, Fontaine T, Wieruszeski J-M, Decq A, Ball S (1994) Toward an understanding of the biogenesis of the starch granule: determination of granule-bound and soluble starch synthase functions in amylopectin synthesis. J Biol Chem 269 25150–25157 [PubMed] [Google Scholar]
- Majoul T, Bancel E, Triboï E, Hamida JB, Branlard G (2003) Proteomic analysis of the effect of heat stress on hexaploid wheat grain: characterization of heat-responsive proteins from total endosperm. Proteomics 3 175–183 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Kawasaki T, Shimada H, Satoh H, Kobayashi E, Okumura S, Arai Y, Baba T (1993) Alteration of the structural properties of starch components by the lack of an isoform of starch branching enzyme in rice seeds. J Biol Chem 268 19084–19091 [PubMed] [Google Scholar]
- Morita S, Shiratsuchi H, Takanashi J, Fujita K (2004) Effect of high temperature on grain ripening in rice plants: analysis of the effects of high night and high day temperatures applied to the panicle and other parts of the plant. Jpn J Crop Sci 73 77–83 [Google Scholar]
- Nagamine T, Komae K (1996) Improvement of a method for chain-length distribution analysis of wheat amylopectin. J Chromatogr 732 255–259 [Google Scholar]
- Nakamura Y (2002) Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol 43 718–725 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Miyachi S (1982) Effect of temperature on starch degradation in Chlorella vulgaris 11th cells. Plant Cell Physiol 31 303–309 [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H (2001) Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol 127 459–472 [PMC free article] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB Jr, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56 3229–3244 [DOI] [PubMed] [Google Scholar]
- Okagaki RJ (1992) Nucleotide sequence of a long cDNA from the rice waxy gene. Plant Mol Biol 19 513–516 [DOI] [PubMed] [Google Scholar]
- Peng S, Huang J, Sheehy JE, Laza RC, Visperas RM, Zhong X, Centeno GS, Khush GS, Cassman KG (2004) Rice yields decline with higher night temperature from global warming. Proc Natl Acad Sci USA 101 9971–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prändl R, Hinderhofer K, Eggers-Schumacher G, Schöffl F (1998) HSF3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermotolerance when overexpressed in transgenic plants. Mol Gen Genet 258 269–278 [DOI] [PubMed] [Google Scholar]
- Sato K, Inaba K (1973) High temperature injury of ripening in rice plant. II. Ripening of rice grains when the panicle and straw were separately treated under different temperature. Proc Crop Sci Soc Japan 42 214–219 [Google Scholar]
- Scofield GN, Hirose T, Gaudron JA, Upadhyaya NM, Ohsugi R, Furbank RT (2002) Antisense suppression of the rice sucrose transporter gene, OsSUT1, leads to impaired grain filling and germination but does not affect photosynthesis. Funct Plant Biol 29 815–826 [DOI] [PubMed] [Google Scholar]
- Singletary GW, Banisadr R, Keeling PL (1994) Heat stress during grain filling in maize: effects on carbohydrate storage and metabolism. Aust J Plant Physiol 21 829–841 [Google Scholar]
- Takemoto Y, Coughlan SJ, Okita TW, Satoh H, Ogawa M, Kumamaru T (2002) The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol 128 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Fujita N, Nishi A, Satoh H, Hosaka Y, Ugaki M, Kawasaki S, Nakamura Y (2004) The structure of starch can be manipulated by changing the expression levels of starch branching enzyme IIb in rice endosperm. Plant Biotechnol J 2 507–516 [DOI] [PubMed] [Google Scholar]
- Tashiro T, Wardlaw IF (1991. a) The effect of high temperature on kernel dimensions and the type and occurrence of kernel damage in rice. Aust J Agric Res 42 485–496 [Google Scholar]
- Tashiro T, Wardlaw IF (1991. b) The effect of high temperature on the accumulation of dry matter, carbon and nitrogen in the kernel of rice. Aust J Plant Physiol 18 259–265 [Google Scholar]
- Tetlow IJ, Morell MK, Emes MJ (2004) Recent developments in understanding the regulation of starch metabolism in higher plants. J Exp Bot 55 2131–2145 [DOI] [PubMed] [Google Scholar]
- Umemoto T, Nakamura Y, Satoh H, Terashima K (1999) Differences in amylopectin structure between two rice varieties in relation to the effects of temperature during grain-filling. Starch 51 58–62 [Google Scholar]
- Umemoto T, Terashima K (2002) Activity of granule-bound starch synthase is an important determinant of amylose content in rice endosperm. Funct Plant Biol 29 1121–1124 [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9 244–252 [DOI] [PubMed] [Google Scholar]
- Wilhelm EP, Mullen RE, Keeling PL, Singletary GW (1999) Heat stress during grain filling in maize: effects on kernel growth and metabolism. Crop Sci 39 1733–1741 [Google Scholar]
- Zakaria S, Matsuda T, Tajima S, Nitta Y (2002) Effect of high temperature at ripening stage on the reserve accumulation in seed in some rice cultivars. Plant Prod Sci 5 160–168 [Google Scholar]