Abstract
Gibberellin (GA) biosynthesis is regulated by feedback control providing a mechanism for GA homeostasis in plants. However, regulatory elements involved in the feedback control are not known. In this report, we show that a rice (Oryza sativa) YABBY1 (YAB1) gene had a similar expression pattern as key rice GA biosynthetic genes GA3ox2 and GA20ox2. Overexpression of YAB1 in transgenic rice resulted in a semidwarf phenotype that could be fully rescued by applied GA. Quantification of the endogenous GA content revealed increases of GA20 and decreases of GA1 levels in the overexpression plants, in which the transcripts of the biosynthetic gene GA3ox2 were decreased. Cosuppression of YAB1 in transgenic plants induced expression of GA3ox2. The repression of GA3ox2 could be obtained upon treatment by dexamethasone of transgenic plants expressing a YAB1-glucocorticoid receptor fusion. Importantly, we show that YAB1 bound to a GA-responsive element within the GA3ox2 promoter. In addition, the expression of YAB1 was deregulated in GA biosynthesis and signaling mutants and could be either transiently induced by GA or repressed by a GA inhibitor. Finally, either overexpression or cosuppression of YAB1 impaired GA-mediated repression of GA3ox2. These data together suggest that YAB1 is involved in the feedback regulation of GA biosynthesis in rice.
The plant hormone GA regulates development throughout the life cycle of the plant. The biosynthesis of GA is regulated by both developmental and environmental stimuli (for review, see Hedden and Phillips, 2000; Olszewski et al., 2002). The genes encoding GA-biosynthetic enzymes have been identified in numerous species and are most highly expressed in rapidly growing tissues or organs (Olszewski et al., 2002). The final steps in the biosynthesis of the bioactive GAs, from GA53 or GA12 to GA1 or GA4, respectively, are catalyzed in parallel pathways by GA 20-oxidase (GA20ox) and GA3ox, whereas the bioactive GA1 and GA4 and their immediate precursors (GA20 and GA9, respectively) are converted to inactive forms by GA2ox (Hedden and Phillips, 2000; Olszewski et al., 2002). These enzymes, which are particularly important for the control of bioactive GA levels, are encoded by small gene families (Hedden and Phillips, 2000; Sakamoto et al., 2004). Overexpression or suppression of GA20ox, GA3ox, or GA2ox genes in transgenic plants often results in altered concentrations of active GA, emphasizing the importance of these genes in regulating GA content (for review, see Hedden and Phillips, 2000). These genes are also important in GA homeostasis. Transcript levels of some GA3ox and GA20ox genes are reduced, while GA2ox transcripts are increased by elevated GA signaling or GA treatment (Hedden and Phillips, 2000; Olszewski et al., 2002), indicating that GA biosynthesis is controlled by feedback regulation through the activity of the GA response pathway. When this pathway is derepressed as a result of loss-of-function mutations in the DELLA proteins that act as negative regulators, bioactive GAs as well as GA20ox and/or GA3ox mRNA are present at lower levels than in the wild-type plants (Croker et al., 1990; Martin et al., 1996; Dill and Sun, 2001; Silverstone et al., 2001). Conversely, gain-of-function mutations in the repressors or loss-of-function mutations in the positive components of the GA response pathway often result in higher levels of bioactive GA and up-regulation of GA20ox and GA3ox gene expression (Fujioka et al., 1988; Talon et al., 1990; Xu et al., 1995; Cowling et al., 1998; Ueguchi-Tanaka et al., 2000; Amador et al., 2001).
No signaling components downstream of the DELLA proteins have been clearly identified as mediators of GA homoeostasis, although several transcriptional regulators have been shown to modify GA metabolic gene expression. For example, the KNOX I class of homeodomain proteins repress GA20ox gene expression in the shoot apical meristem (SAM) in a number of species (for review, see Hay et al., 2004), while the FUS3 transcription factor down-regulates GA3ox in Arabidopsis (Arabidopsis thaliana) embryos (Curaba et al., 2004; Gazzarrini et al., 2004; for review, see Fleet and Sun, 2005).
In rice (Oryza sativa), GA3ox and GA20ox are encoded by two and four genes, respectively (Sakamoto et al., 2004). Loss-of-function mutations such as d18 in GA3ox2 and sd1 in GA20ox2 result in dwarf or semidwarf plants (for review, see Sakamoto et al., 2004). These genes are expressed in rapidly growing regions, such as leaf primordia, young leaves, elongating internodes, developing anthers, and embryo (Kaneko et al., 2003), but not in the vegetative SAM. The expression of both genes is also detected in reproductive meristems and stamen primordia during floral organogenesis (Kaneko et al., 2003). Their expression pattern suggests that the enzymes encoded by rice GA3ox2 and GA20ox2 play a major role in GA biosynthesis during both vegetative and reproductive development. In most cases, their expression in rice tissues overlaps with that of GA signaling genes, indicating that GA is synthesized at the sites of perception (Kaneko et al., 2003). The deactivating enzyme GA2ox is also encoded by a small gene family, two members of which (GA2ox3 and GA2ox4) are broadly expressed in many organs or tissues (Sakamoto et al., 2004).
The plant genome encodes a large number of transcription factors, many of which are specific to plants. Among them, the YABBY family, which is characterized by the juxtaposition of two conserved domains (a C2C2 zinc finger domain toward the amino terminus and a putative helix-loop-helix “YABBY” domain conserved in HMG and other transcription factors toward the carboxyl terminus), is not found in yeast or animal cells (Bowman and Smyth, 1999; Golz and Hudson, 1999). The Arabidopsis genome contains at least six members of the YABBY gene family (FILAMENTOUS FLOWER or YABBY1 [YAB1]; YAB2; YAB3; INNER NO OUTER or YAB4; YAB5; and CRABS CLAW [CRC]), all of which show a polar expression pattern, being confined to the abaxial side of one or more aboveground lateral organs (Bowman, 2000). Loss of expression of Arabidopsis YABBY genes results in loss of polar differentiation of abaxial cell types in the lateral organs, whereas ectopic expression of family members in the adaxial regions of developing cotyledons and leaves is sufficient to induce abaxialization of the adaxial cells (Eshed et al., 1999; Sawa et al., 1999; Siegfried et al., 1999).
The rice genome also seems to contain six members of the YABBY family. Recently, it has been shown that a rice CRC-related YABBY gene, DL (the mutation of which induces a drooping leaf phenotype), is required for carpel specification and leaf midrib development (Yamaguchi et al., 2004). Interestingly, none of the YABBY genes studied in rice shows an adaxial/abaxial polar expression pattern in lateral organs (Jang et al., 2004; Yamaguchi et al., 2004; Dai et al., 2007). Alteration of their expression by mutation or by ectopic expression produces no change in the adaxial/abaxial polarity of the lateral organs. These results suggest that rice YABBY genes may have functions different from their Arabidopsis homologs.
In this work, we studied the expression pattern and regulatory function of rice YAB1 by transgenic approaches. Our data suggest that this gene is involved in the regulation of GA homeostasis in rice.
RESULTS
YAB1 Expression Pattern
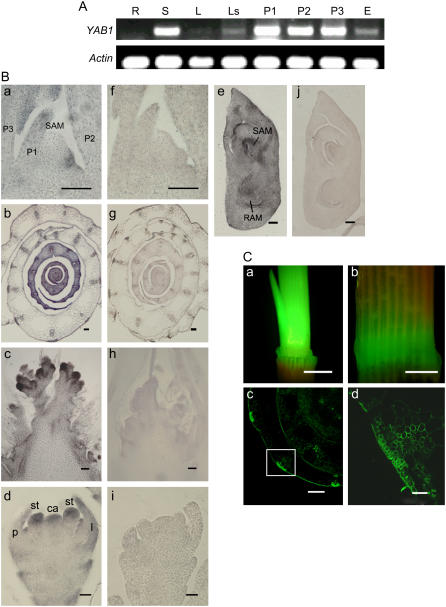
Reverse transcription (RT)-PCR analysis showed that YAB1 was expressed in both shoots and developing panicles (Fig. 1A). In addition, YAB1 expression was detected in leaf sheaths at a lower level (Fig. 1A). In situ hybridization experiments detected YAB1 mRNA in the shoot apex and in young leaves (Fig. 1B, a and b). In the shoot apex, YAB1 mRNA was detected only in leaf primordia but not in the SAM (Fig. 1B, a). The expression of YAB1 in young leaves did not show any adaxial/abaxial polar distribution but was enhanced in both the upper and lower epidermis and in cells surrounding the vascular bundles (Fig. 1B, b). YAB1 mRNA was also detected by in situ hybridization in rachis meristems and in lemma, palea, stamen, carpel primordia, and the embryonic axis (Fig. 1B, c–e). No adaxial/abaxial polar distribution of YAB1 transcripts in the stamen or carpel primordia was observed (Fig. 1B, d). In addition, transgenic rice plants containing the GFP gene, under the control of the YAB1 promoter region spanning from the initiation ATG codon to −1,662, showed GFP expression in elongating internodes and in leaf sheaths (Fig. 1C, a and b), in which GFP was detected mostly in sclerenchyma, vascular bundles and epidermal cells (Fig. 1C, c and d), confirming the in situ hybridization data (Fig. 1B, b). Therefore, YAB1 was expressed widely in many different tissue or cell types, mostly in rapidly dividing or elongation tissues or organs but not in the SAM.
Figure 1.
Expression pattern of YAB1. A, Detection of YAB1 transcript by semiquantitative RT-PCR in roots (R), shoots (S), mature leaves (L), leaf sheaths (Ls), and panicles at three developmental stages (P1–P3; P1, primary panicle branch primordia; P2, secondary branch primordia; P3, stamen and carpel primordia) and developing embryo (E). B, In situ hybridization detection of YAB1 transcripts in leaf primordia (a and f), young leaves (b and g), rachis meristem (c and h), floral meristem (d and i), and developing embryo (e and j) with antisense (a–e) or sense (f and g) cDNA of YAB1 as probes. Bars in a, f, c, and h = 5 μm; bars in e and j = 10 μm; bars in b, g, and d, I = 20 μm. C, Detection of GFP expression under the control of the YAB1 promoter (−1,662 to 1) in the elongating stem (a) and the leaf sheath (b). c, Cross section of sheaths showing GFP expression in sclerenchyma, vascular bundles, and epidermal cells. d, Enlargement of the boxed area in c. Bars in a and b = 1 cm, in c = 30 μm, in d = 100 μm. ca, Carpel primordium; l, lemma; p, palea; P1 to P3, leaf primordium; RAM, root apical meristem; st, stamen primordium.
Overexpression of YAB1 Resulted in Semidwarf Phenotype
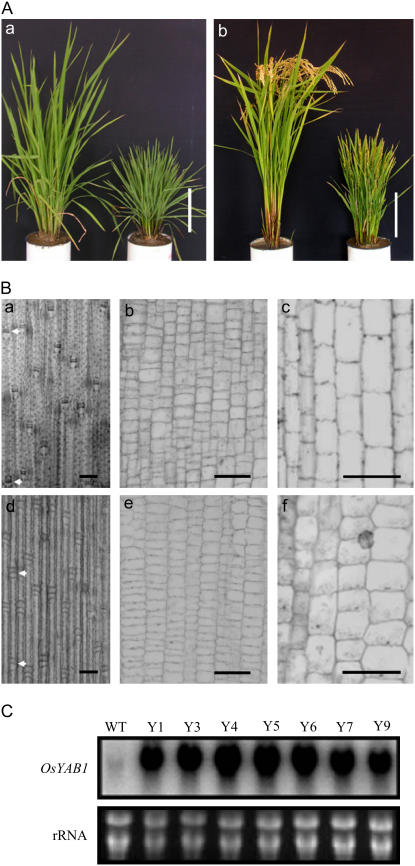
To study the function of YAB1, full-length cDNAs of the gene were isolated from a cDNA library of indica rice ‘Minghui 63.’ The indica cDNA presented a number of nucleotide substitutions resulting in seven amino acid changes of the protein compared to the published japonica sequence (Supplemental Fig. S1). This cDNA was cloned between the double cauliflower mosaic virus (CaMV) 35S promoter and the NOS (nopaline synthase) terminator in a rice transformation vector (see “Materials and Methods”). The construct was introduced into ‘Nipponbare’ by Agrobacterium-mediated transformation, and more than 30 independent transgenic T0 plants were produced. A semidwarf phenotype occurred in many of the transgenic lines, with plant height about one-third that of the wild type together with about 3 times more tillers (Fig. 2A; Tables I and II). Further investigation of the semidwarf transgenic plants indicated that elongation of all the internodes, except internode V, was affected (Table I). Stem epidermal, elongating, and intercalary meristem cells in the transgenic plants were shorter than in the wild type (Fig. 2B), indicating that cell elongation was reduced. No alteration of leaf shape and flowering time was observed. The overexpression plants were partially fertile. Transgenic plants showing phenotypic changes were subsequently analyzed in the T1 generation for cosegregation between the phenotype and the transgene. Eight semidwarf transgenic plants were analyzed by RNA-gel blots. As shown in Figure 2C, all of the semidwarf plants overexpressed the transgene. Southern-blot analysis of these lines indicated that they contained a single transgene insertion in the genome (Supplemental Fig. S2).
Figure 2.
Overexpression of YAB1 under the double CaMV 35S promoter induced semidwarf phenotypes in transgenic rice. A, Comparison between wild type and a YAB1-overexpressing line (Y4) at vegetative and reproductive stages. Bars = 15 cm. B, Overexpression of YAB1 affected cell elongation in stems. a and d, Epidermal cells. Arrows indicate the upper or lower limits of epidermal cells. Bars = 2 μm. b and e, Cells in the intercalary meristem. Bars = 10 μm. c and f, Cells in elongating zone. Bars = 10 μm. a to c, Wild type; d to f, YAB1-overexpressing plants. C, RNA gel-blot hybridization showing that all examined semidwarf lines (Y1, Y3–Y7, and Y9) showed overexpression of YAB1.
Table I.
Lengths of the internodes and relative contribution of each internode (in parentheses) to the total culm in YAB1-overexpressing lines in comparison with wild-type plants
The average values were calculated from measures of at least 20 plants ±sd.
| Lines | Length of the Internodes
|
Total Culm | |||||
|---|---|---|---|---|---|---|---|
| P | I | II | III | IV | V | ||
| cm | |||||||
| WT | 19.8 ± 1.0 (25.3) | 32.6 ± 2.9 (41.6) | 13.4 ± 1.0 (17.1) | 7.9 ± 1.3 (10.1) | 3.8 ± 1.7 (4.8) | 0.9 ± 0.3 (1.1) | 78.4 ± 3.4 |
| Y1 | 16.3 ± 2.7 (27.6) | 27.8 ± 2.3 (42.8) | 8.3 ± 1.2 (14.1) | 4.3 ± 0.8 (7.3) | 1.7 ± 0.6 (3.0) | 0.7 ± 0.2 (1.2) | 59.1 ± 2.8 |
| Y3 | 11.2 ± 0.6 (32.0) | 11.4 ± 2.1 (32.6) | 4.8 ± 1.2 (13.7) | 4.0 ± 1.3 (11.4) | 2.6 ± 0.7 (7.4) | 1.0 ± 0.4 (2.9) | 35.0 ± 4.0 |
| Y4 | 14.1 ± 2.0 (29.2) | 20.2 ± 2.0 (41.8) | 7.7 ± 2.0 (15.9) | 3.7 ± 1.4 (7.7) | 2.0 ± 1.0 (4.1) | 0.6 ± 0.1 (1.3) | 48.3 ± 4.0 |
Table II.
Comparison of average tiller number in transgenic and wild-type plants
The average values were calculated from measures of at least 20 plants ±sd.
| Wild Type | Y1 | Y3 | Y4 |
|---|---|---|---|
| 18.7 ± 4.0 | 61.1 ± 24 | 68.2 ± 19 | 67.0 ± 25 |
The Dwarf Phenotype of YAB1-Overexpressing Plants Could Be Suppressed by Exogenous GA Treatment
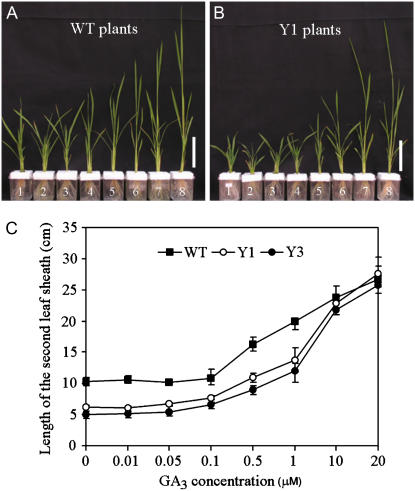
To determine whether or not the semidwarf phenotype of the YAB1 overexpression plants was caused by GA deficiency, the response of the plants to exogenous GA was measured. Four-week-old (at four-leaf stage) transgenic and wild-type plants were transferred to a hydroponic culture medium supplemented with 10 μm paclobutrazol (an inhibitor of GA biosynthesis) for 1 d to reduce the endogenous GA to background levels, before adding GA3 at different final concentrations ranging from 0.01 to 20 μm. Plant heights were measured 10 d after GA application. The data presented in Figure 3 showed that at 10 μm, GA could fully rescue the dwarf phenotype of the YAB1 overexpression plants.
Figure 3.
Rescue of semidwarf phenotype by GA3 at different concentrations. Wild-type (A) and YAB1 overexpression plants (B) at four-leaf stage were transferred to hydroponic culture media containing 10 μm paclobutrazol (an inhibitor of GA biosynthesis) for 1 d, then supplemented with GA3 at the concentration of 0 (1), 0.01 (2), 0.05 (3), 0.1 (4), 0.5 (5), 1 (6), 10 (7), or 20 μm (8). The lengths of the second leaf sheath were measured 10 d after the treatment (C). Squares, white circles, and black circles represent wild type, Y1, and Y3, respectively. Error bars represent sd from at least 20 plants. [See online article for color version of this figure.]
Endogenous GA Contents in the Overexpression Plants
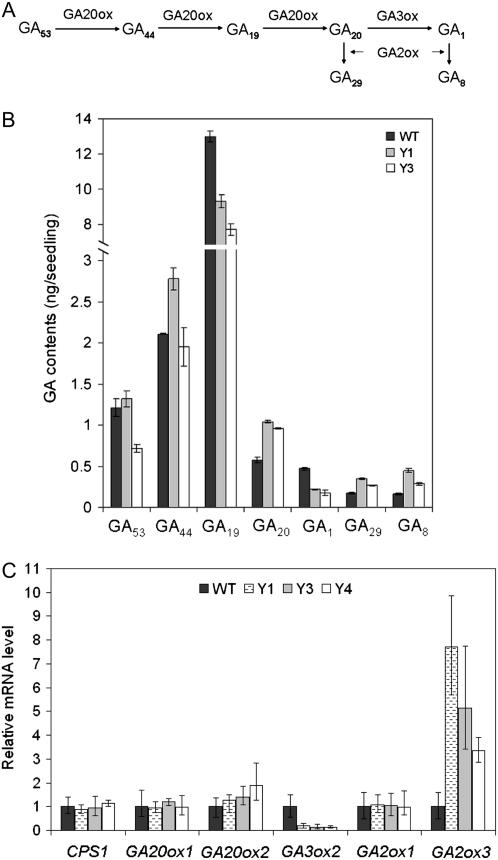
It has been shown that GA1 is the major bioactive GA in the vegetative organs in rice (Kobayashi et al., 1988). Using gas chromatography-mass spectrometry, we measured endogenous levels of different 13-hydroxylated GAs within the GA1 metabolic pathway (Fig. 4A) in both transgenic and wild-type seedlings. The data in Figure 4B shows that in the overexpression lines, the GA1 content decreased to about one-half that of the wild type, whereas the levels of GA20 (precursor of GA1), GA8 (deactivated form of GA1), and GA29 (deactivated form of GA20) increased about 2-fold. Changes in the levels of more upstream precursors (GA53 and GA19) also occurred, including a slight decrease in GA19 content in the transgenic lines.
Figure 4.
Overexpression of YAB1 altered GA accumulation and GA metabolic gene expression. A, Schematic representation of 13-hydroxylated GA metabolic pathway, involving GA20ox, GA3ox, and GA2ox. B, Quantification of 13-hydroxylated GAs in four-leaf-stage seedlings. Data are means ± sd (n = 3). C, Quantification of transcripts of six GA metabolic genes by real-time RT-PCR. The PCR signals were normalized with that from actin transcripts. Transcript levels from wild type were set at 1. Data are means ± sd (n = 3).
Overexpression of YAB1 Altered GA Metabolic Gene Expression
Changes in the GA content in the YAB1 overexpression lines may be due to altered expression of one or more of the genes encoding GA metabolic enzymes. We compared expression of six representative genes encoding four enzymes, ent-copalyl diphosphate synthase (CPS), GA3ox, GA20ox, and GA2ox, in young leaves of transgenic and wild-type plants by quantitative RT-PCR. As shown in Figure 4C, mRNA levels of GA3ox2 in the YAB1-overexpressing lines (Y1, Y3, and Y4) were reduced to 10% to 20% of the wild-type levels. In contrast, GA2ox3 was induced in the overexpression lines. It is suggested that GA3ox2 makes major contributions to the biosynthesis of GA1 during vegetative development (Kaneko et al., 2003). The expression results are consistent with the observed increases of GA20 (substrate for GA3ox), GA29, and GA8 (products of GA2ox) and decrease of GA1 (product of GA3ox and substrate for GA2ox) in the overexpression plants.
Analysis of YAB1 Cosuppression Plants
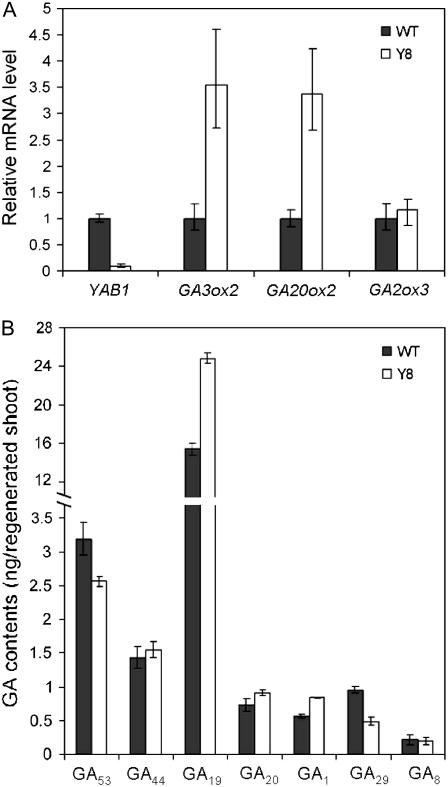
Examination of transgene copy numbers by Southern blots revealed that two lines had at least two insertions of the YAB1 transgene (Supplemental Fig. S2). These lines were found to contain lower YAB1 mRNA levels compared to the wild type (Supplemental Fig. S3A), suggesting that expression of both endogenous and transgenic YAB1 was being suppressed, as might be anticipated, because multiple copy transgenes have a high probability of inducing cosuppression and gene silencing (Schubert et al., 2004). The expression of two other YABBY genes (YAB2 and DL) was not changed (Supplemental Fig. S3A). The cosuppression was therefore specific to YAB1. Examination of GA metabolic gene expression in cosuppression line Y8 by quantitative RT-PCR showed a clear increase in GA3ox2 transcript levels compared to the wild type (Fig. 5A). However, the expression of GA2ox3 was not altered (Fig. 5A). The results confirmed by RNA gel-blot hybridization (Supplemental Fig. S3B) suggested that YAB1 was required for the expression of GA3ox2 but not for GA2ox3. The induction of GA2ox3 in YAB1 overexpression plants (Fig. 4C) might be achieved by an indirect mechanism (discussed later). In addition, GA20ox2 mRNA levels were also increased (Fig. 5A; Supplemental Fig. S3B). Analysis of 13-hydoxylated GAs in plants showed slight increases in GA19, GA20, and GA1 contents and reductions in GA53 and GA29 contents in the cosuppression plants (Fig. 5B). The changes in GA contents are consistent with the transcript levels of GA20ox2, GA3ox2, and GA2ox3 present in line Y8.
Figure 5.
Cosuppression of YAB1 altered GA biosynthetic gene expression and GA levels. A, Relative transcript levels by real-time RT-PCR of GA3ox2, GA20ox2, and GA2ox3 transcript levels between wild-type and Y8 plants after normalization with ACTIN transcripts. B, Quantification of 13-hydraxylated GA forms. Data are means ± sd (n = 3).
The cosuppression plants did not show the slender phenotype, probably because of only slight increase of GA1 or effects on other developmental aspects (see below) induced by the cosuppression. Instead, leaf blades of the cosuppression plants drooped outwards, forming an acute angle with the leaf sheaths (Fig. 6, A and B). In addition, leaves were rolled toward the abaxial side, forming a cylinder-like structure (Fig. 6D). Transverse sections showed increases in the number of the bulliform cells situated between the vasculatures on the adaxial side of the leaf blade (Fig. 6F). Bulliform cells deposited in the adaxial side of the leaf blade are responsible for the leaf rolling that may occur during drought stress in rice. These cells in Y8 plants had a rectangular or triangular shape rather than the spherical form observed in the wild-type plants. Other cells in the leaves appeared normal, with no alteration in adaxial-abaxial identity observed in the arrangement of the leaf inner structure. These observations suggested that the rolled leaf phenotype may be due to the increases in bulliform cell number and that YAB1 may have a function in bulliform cell division and differentiation. The cosuppression plants were sterile due to defects in anther and carpel formation (Fig. 6H).
Figure 6.
Cosuppression of YAB1 induces rolled leaves and abnormalities in flower development. A, Wild-type rice at heading stage. B, Y8 plant at the heading stage with rolled leaves dropped, forming an acute angle with the sheaths. C, Lower part of a rolled leaf. D, Upper part of a rolled leaf. E and F, Comparison of a leaf section in Y8 (F) with wild type (E) showing altered bulliform cells in Y8. G and H, Comparison of Y8 (H) stamens and carpels to wild type (G). Bar = 20 cm in A and B, 4 cm in C and D, 100 μm in E and F, and 5 mm in G and H. bc, Bulliform cells; ad, adaxial side; ab, abaxial side.
YAB1 Is Directly Involved in the Regulation of GA3ox2
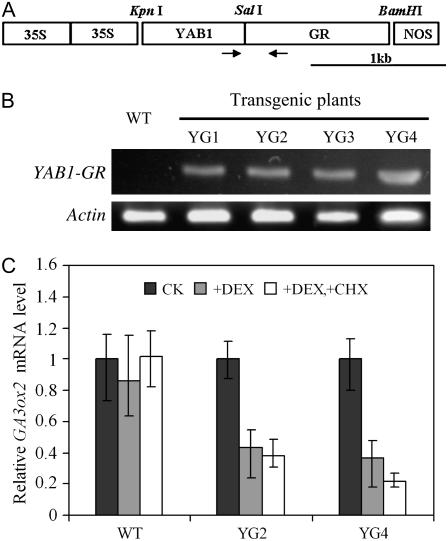
To determine whether YAB1 directly regulated the expression of the GA metabolic genes, we produced transgenic rice producing YAB1 fused with the glucocorticoid receptor (GR) that is used to sequestrate the fusion protein in the cytoplasm (Aoyama and Chua, 1997; Fig. 7A). Four analyzed lines that showed transgene expression (Fig. 7B) were assayed with dexamethasone (DEX), which induces conformational changes in GR allowing the fusion protein to be targeted into the nucleus. DEX treatment induced repression of GA3ox2 expression in the transgenic plants (Fig. 7C). Furthermore, we assayed the DEX induction in the absence or presence of cycloheximide (CHX), which blocks de novo protein synthesis. The presence of DEX and DEX plus CHX had similar effects on the repression of GA3ox2 transcript levels (Fig. 7C). In the wild-type plants, the expression of GA3ox2 did not change significantly after treatment.
Figure 7.
YAB1 is directly involved in the transcriptional regulation of GA3ox2. A, Schematic representation of construct in which YAB1 is fused to the GR. Arrows indicate the positions of primers used in B. B, RT-PCR detection of YAB1-GR transcripts in four independent rice transgenic lines. C, Effects of DEX on the transcription of GA3ox2 in the presence of CHX (10 μm). CHX was added 1 h prior the addition of DEX. Samples were harvested after 2-h treatment. Data normalized with ACTIN transcripts are means from three biological repeats ±sd. The values from nontreated samples were assessed as 1.
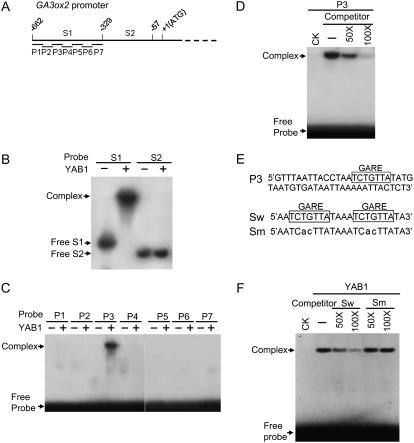
To know whether YAB1 could bind to the GA3ox2 promoter, yeast one-hybrid assays revealed that that YAB1 interacted with a 0.8-kb promoter region of GA3ox2 (not shown). This promoter fragment was subsequently cut into two segments (Fig. 8A) that were used for gel shift assays with Escherichia coli-produced YAB1 protein. The region from −662 to −329 was bound by YAB1 protein (Fig. 8B). Double-stranded oligonucleotides corresponding to seven overlapped regions of the DNA fragment were tested for binding to YAB1. One of the oligonucleotides (P3) retained the specific YAB1-binding activity (Fig. 8, C and D). Sequence analysis (with software PLANTCARE) revealed that this 50-bp region contained a GA-responsive element (GARE; 5′-TAACAGA; Fig. 8E). To test whether this element was responsible for the binding of YAB1, binding assays with oligonucleotides containing a pair of the wild-type or substituted GARE sequences showed that YAB1 bound specifically to the wild-type GARE (Fig. 8F).
Figure 8.
YAB1 binds to a GARE in GA3ox2 promoter. Gel shift binding assays with E. coli produced rice indica YAB1 protein to GA3ox2 promoter regions. A, Schematic representation of the GA3ox2 promoter regions with indications relative to the ATG codon. B, Binding assays with GA3ox2 promoter fragments S1 and S2 in the presence (+) or absence (−) of YAB1 protein. C, Binding assays with seven overlapping double-stranded oligonucleotides corresponding to S1 in the presence (+) or absence (−) of YAB1 protein. D, YAB1-binding assays with 32P-labeled P3 in the presence or absence (−) of cold P3 at 50 to 100 m excesses as indicated. E, Top, P3 contains a GARE. Bottom, Sequences of oligonucleotides corresponding to a doublet of the wild-type (Sw) or mutated (Sm) GARE used in F. F, YAB1-binding assays to 32P-labeled Sw in the presence or absence (−) of Sw or Sm at 50 to 100 m excesses as indicated. Positions of binding complexes and free probes are indicated by arrows.
These data together suggested a direct involvement of YAB1 in regulating the expression of GA3ox2.
GA Treatment Transiently Induced YAB1 Expression
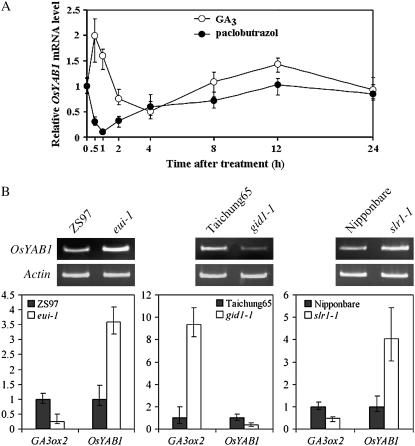
The expression pattern of YAB1 in shoot and floral meristems or in elongating internodes was similar to that of GA biosynthetic genes (Fig. 1; Kaneko et al., 2003). Therefore, we investigated whether GA regulated the expression of YAB1. Rice seedlings at the four-leaf stage were sprayed with 50 μm GA3, and the aerial parts were harvested at different time points for quantitative RT-PCR analysis. As shown in Figure 9A, the YAB1 mRNA level was increased 2-fold only 30 min after GA treatment and subsequently decreased to the control level at 2 h after the treatment. These data suggested that GA transiently induced the expression of YAB1. Interestingly, the expression of YAB1 increased again from 8 to 12 h after the treatment, suggesting that additional regulators were involved in YAB1 expression (M. Dai and D.-X. Zhou, unpublished data). These results were confirmed by RNA gel-blot hybridization (Supplemental Fig. S4).
Figure 9.
GA transiently induced expression of YAB1 in seedlings. A, Real-time RT-PCR quantification of YAB1 transcripts isolated from seedlings treated with 10 μm paclobutrazol or with 50 μm GA3. Data normalized with actin transcripts are means from three biological repeats ±sd. B, YAB1 mRNA levels in eui-1, gid1-1, and slr1-1 compared to the respective wild-type plants (‘ZS97’, ‘Taichung65’, and ‘Nipponbare’) measured by both semiquantitative and real-time RT-PCR in which YAB1 and GA3ox2 mRNA levels are normalized with ACTIN mRNA levels (assessed as 1). Data are means from three biological repeats ±sd.
We also investigated the effect of a low endogenous GA content on expression of YAB1 by treating wild-type plants with the GA biosynthesis inhibitor paclobutrazol. Plants were harvested for RNA extraction at the same time points as for the GA treatment, and YAB1 transcript was analyzed by real-time quantitative RT-PCR. As shown in Figure 9A, paclobutrazol treatment produced the opposite YAB1 expression profile as obtained after GA3 treatment.
YAB1 Was Deregulated in eui, gid1, and slr1 Mutants
To confirm the observations in Figure 9A, we tested the YAB1 mRNA levels in rice GA biosynthesis (eui) and signaling (gid1 and slr1) mutants. The eui mutants accumulate biologically active gibberellins and show repression of GA3ox2 (Zhu et al., 2006). In eui-1, the expression of YAB1 was induced in agreement with the repression of GA3ox2 in the same mutant (Fig. 9B; Zhu et al., 2006). The gid1-1 mutants affecting GA reception have a GA-insensitive dwarf phenotype and show accumulation of bioactive GA but increased expression of GA biosynthetic genes sd1 (GA20ox2; Ueguchi-Tanaka et al., 2005) and GA3ox2 (Fig. 9B), indicating disruption of the feedback regulation. In this mutant, the expression of YAB1 is repressed (Fig. 9B). slr1-1 is a loss-of-function mutation of the rice DELLA gene that encodes a GA-signaling repressor (Ikeda et al., 2001). In slr1-1, the YAB1 expression was induced (Fig. 9B). The expression of GA3ox2 in eui, gid1, and slr1 showed an opposite expression pattern compared to YAB1 (Fig. 9B).These data indicate GA signaling is required for GA-mediated repression of YAB1 and YAB1 is a downstream negative feedback regulator of GA3ox2.
Deregulation of YAB1 Impaired GA-Mediated Repression of GA3ox2
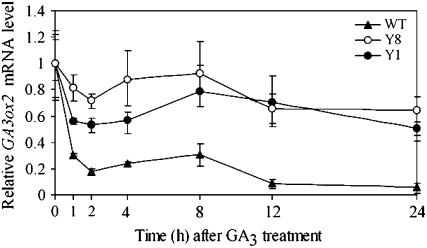
Our results suggested that YAB1 may function as a negative regulator of GA3ox2. To know whether YAB1 played a role in mediating GA-dependent down-regulation of GA3ox2 expression, both YAB1 overexpression and cosuppression plants along with wild type at the four-leaf stage were sprayed with 50 μm GA3 and harvested at different time points for RNA extraction and quantitative RT-PCR analyses. In wild-type plants, the applied GA rapidly (1–2 h) suppressed the expression of GA3ox2 to a much lower level (about a 5-fold decrease). In YAB1 cosuppression plants (Y8), the applied GA had little effect on the expression of GA3ox2, suggesting that YAB1 is required for GA-mediated GA3ox2 repression. However, overexpression of YAB1 also attenuated the GA-induced repression of GA3ox2 (Fig. 10). The later observation could be explained by an already repressed expression level of GA3ox2 in YAB1 overexpression plants. Applied GA had less effect on its repression than in wild-type plants. These data suggested that YAB1 is required for the GA-mediated GA3ox2 repression.
Figure 10.
Deregulation of YAB1 impaired GA-mediated repression of GA3ox2. Relative mRNA levels of GA3ox2 in wild-type (WT, triangles), YAB1 overexpression (Y1, black circles), and YAB1 cosuppression (Y8, white circles) plants treated by GA3 and harvested at different time points. The mRNA levels were measured by quantitative RT-PCR. The values at time point zero were assessed arbitrarily as 1. Data normalized with ACTIN transcripts are means from three biological repeats ±sd.
DISCUSSION
In this work, we present several lines of evidence that YAB1 is involved in the feedback regulation of GA biosynthesis. In contrast to the SAM-expressed KNOX genes that exclude the expression of GA20ox from the SAM (for review, see Hay et al., 2004), the expression domains of YAB1 overlap with those of GA3ox2, GA20ox2, and GA-signaling genes, which are expressed in diverse organs or tissues of rice, including embryo, shoots, and floral meristems (Fig. 1; Kaneko et al., 2003). This suggests that YAB1 have a function in modulating GA metabolic gene expression or GA signaling throughout the plant. The semidwarf phenotype, which can be suppressed by applied GA, and the changes in GA metabolic gene expression and GA levels induced by both overexpression and cosuppression of YAB1 support a regulatory function for YAB1 in GA biosynthesis (Figs. 2–4). Importantly, the expression of YAB1 is GA responsive, being rapidly and transiently induced by GA or repressed by inhibition of GA biosynthesis (Fig. 9), suggesting that YAB1and GA form part of a regulatory loop in GA-responsive tissues. Finally, YAB1 binds to the promoter of GA3ox2 and is required for GA-mediated repression of the gene (Figs. 7, 8, and 10). These data together would support a feedback regulatory scenario; the rapid activation of YAB1 by GA subsequently induces the repression of GA3ox2 and consequently decreases bioactive GA levels, leading to the repression of YAB1 itself. In addition, the effects of overexpression and cosuppression of YAB1 on GA contents is relatively moderated (about 2-fold changes; Figs. 4 and 5) in favor of a feedback regulatory function of the gene in GA biosynthesis.
Function of YAB1 in the Regulation of GA3ox2, GA2ox3, and GA20ox2
We have shown that overexpression of YAB1 induces repression of GA3ox2 and, consequently, accumulation of its substrate GA20 and a reduction in the amount of its bioactive product GA1 (Fig. 4). Accordingly, cosuppression of YAB1 results in increased GA3ox2 expression (Fig. 5). The experiments with the inducible YAB1 construct (Fig. 7) showed that the repression of GA3ox2 was obtained upon induction of the YAB1 activity. In addition, YAB1 interacts directly with a GARE sequence within the GA3ox2 promoter (Fig. 8). These data, together with the requirement of a normal expression level of YAB1 for GA-mediated GA3ox2 repression (Fig. 10), indicate that YAB1 is directly involved in the regulation of GA3ox2.
In contrast, YAB1 overexpression induces GA2ox3, which is consistent with the accumulation of the 2-oxidized products GA8 and GA29 (Fig. 4). However, the cosuppression of YAB1 did not decrease the expression of GA2ox3 (Fig. 5; Supplemental Fig. S2), which helps to explain why there was only a slight increase of GA1 accumulation and there was no typical slender phenotype. This discrepancy suggests that YAB1 is not involved in GA2ox3 regulation at natural expression levels in wild-type plants or that there is a baseline level of GA2ox3 expression regulated independently of YAB1. It is also possible that a reduction of GA2ox3 expression in the absence of YAB1 might be compensated for by feedback induction due to the increase of the active GA1 in the cosuppression plants. However, failure to detect any YAB1-binding activity of the GA2ox3 promoter where no GARE was found (not shown) suggests that YAB1 is not directly involved in the regulation of the gene. Conversely, overexpression of YAB1 does not significantly alter GA20ox2 expression (Fig. 4), but cosuppression of YAB1 induces the expression of GA20ox2 (Fig. 5). We favor the hypothesis that the expression of GA20ox2 is indirectly induced in the cosuppression plants through an independent feedback mechanism required for coactivation of both of the key biosynthetic genes, GA3ox2 and GA20ox2, because there was no direct binding of YAB1 to GA20ox2 promoter that contains no GARE sequence (not shown). Therefore, our data show that YAB1 is a direct regulator of GA3ox2, but its deregulation has indirect impact on the other key GA metabolic gene expression.
YAB1 Is Involved in the Negative Feedback Regulation of GA Homeostasis
GA biosynthesis is controlled by feedback regulation through the activity of the GA response pathway. DELLA proteins that act as negative regulators of the pathways are required for GA feedback regulation, as mutations in DELLA genes affect GA metabolic gene expression and bioactive GA homeostasis. GA induces DELLA protein degradation. It is suggested that DELLA proteins may directly or indirectly repress transcriptional activators of GA signaling pathways. However, no transcriptional regulator downstream of the DELLA proteins has been so far clearly identified as a mediator of GA homoeostasis, although it is shown that the tobacco (Nicotiana tabacum) protein REPRESSION OF SHOOT GROWTH (RSG), a bZIP transcription factor, binds to and activates the transcription of the early GA biosynthetic gene ent-kaurene oxidase. Overexpression of a dominant negative form of RSG results in a GA-deficient dwarf phenotype and prevents activation of an NtGA20ox gene in transgenic tobacco, suggesting that RSG plays a role in feedback regulation of NtGA20ox (Fukazawa et al., 2000). However, it is not known if RSG is directly involved in the regulation of NtGA20ox. Our present data show that YAB1 is likely a mediator of GA homeostasis downstream of the DELLA protein, as its expression is dependant on GA signaling and is derepressed in loss-of-function mutants of the rice DELLA gene (slr; Fig. 9) and deregulation of the expression impairs GA3ox2 expression (Fig. 10).
Binding of YABBY Proteins to GARE
The GARE (TAACAGA) motif has been identified as a cis-regulatory element involved in the GA induction of rice α-amylase genes (Gubler and Jacobsen, 1992). A MYB-type transcription factor named GAMYB initially identified from barley (Hordeum vulgare) and later from rice binds to GARE and transactivates the transcription from the promoters (Gubler et al., 1995, 1999; Sutoh and Yamauchi, 2003). GA-induced activation of α-amylase genes as well as a number of other hydrolase genes requires an additional cis-element (5′-CAACTC, called CARE) in addition to GARE (Gubler and Jacobsen, 1992; Cercós et al., 1999; Sutoh and Yamaochi, 2003). No CARE sequence was found in the GA3ox2 promoter that is repressed by GA, suggesting that the GARE motif has either an activating or a repressing function, depending on promoter context or on the cognate binding factors. To our knowledge, no DNA-binding property has been described for YABBY proteins that belong to the zinc finger superfamily of transcription factors. In this study, we show that rice YAB1 binds to the GARE motif within the GA3ox2 promoter and represses transcription of the gene. It has been previously shown that in barley, a zinc finger protein, HRT (Hordeum repressor of transcription), represses GA-inducible gene expression and also binds to the GARE motif (Raventós et al., 1998).
At this stage, it is not known whether the other members of YABBY proteins bind to GARE. Arabidopsis YABBY genes are expressed in the abaxial domain of the leaf and are required for specifying adaxial/abaxial polarity of the leaf, in addition to contributing to repression of KNOX1 expression in the leaf primordium. Previous results and data from this study have shown that the rice YABBY genes are not expressed in a polar manner and that deregulation of expression by mutation or by transgenics did not affect the leaf adaxial/abaxial polarity (Jang et al., 2004; Yamaguchi et al., 2004; Dai et al., 2007), indicating that rice YABBY genes have distinct developmental function. The CRC-related rice DL gene is required for midrib formation in the leaf in addition to its requirement for flower development (Yamaguchi et al., 2004). The overexpression of YAB1 did not induce deformation of leaves, whereas cosuppression of YAB1 induced rolled leaf phenotype, but the midrib seemed to be normal (Fig. 6). The leaf phenotype observed in Y8 plants is unlikely to be related to DL and other YABBY genes, whose expression was not affected in Y8 plants. Therefore, we suggest that rice YAB1 has distinct functions from the other YABBY members.
It has to be pointed out that the YAB1 cDNA previous studied by Jang et al. (2004) was isolated from a japonica variety and the one of our study was isolated from an indica variety. The transgenic studies of the japonica YAB1 do not reveal similar phenotypes shown in this study. There are seven changes of amino acid sequences between the two sequences, of which four are located in the zinc finger domain (Supplemental Fig. S1). It is not known at this stage whether the phenotypic differences were due to the sequence changes.
MATERIALS AND METHODS
Materials
Rice (Oryza sativa spp. japonica) ‘Nipponbare’ was used in this study. The full-length cDNAs of YAB1 (accession no. AF098752) were isolated from a normalized cDNA library of the elite indica rice ‘Minghui 63’ (Zhang et al., 2005).
Transgene Constructs and Rice Transformation
The binary vector for transformation was constructed based on pCAMBIA1301 (CAMBIA) and pRTL2 (Mason et al., 1992). A HindIII fragment with a double CaMV 35S enhancer/promoter and the NOS A terminator from pRTL2 was inserted into pCAMBIA1301. The new vector was named as pDS1301. For overexpression of YAB1, the full-length cDNA of the gene was inserted into pDS1301digested with BamHI and KpnI. To create the YAB1-GR fusion construct, the full-length coding sequence of YAB1 was amplified by PCR. The stop codon of YAB1 was removed and replaced with a SalI site. A KpnI site was added to the end of the forward primer. The amplified fragment was inserted into the plasmid pSport1 (CLONTECH), which served as a medium vector by SalI and KpnI. The primers used in PCR were YAB1 (GR)-F (5′-GGTACCCAATCGATCTCTTCGGTGAG) and YAB1 (GR)-R (5′-GTCGACAAGTTGCTGCCGCCGCCGCC). In the same way, the DNA fragment encoding the steroid-binding domain of the mouse GR was amplified using the plasmid pBI-ΔGR as template. The PCR primers were GR-F (5′-GTCGACAGATCCTGAAGCTCGAAAAAC) with a SalI adaptor and GR-R (5′-GGATCCACCGGCAACAGGATTCAATG) with a BamHI adaptor. The amplified GR fragment was inserted downstream to and in frame with YAB1 in pSport1. The fused YAB1-GR sequence was then cut from pSport1 and inserted into p1301DS digested with KpnI and BamHI. The chimeric protein sequence was (YAB1, amino acids 1–169)-Leu-Ser-Thr-Asp-Pro-(GR, amino acids 507–794). Agrobacterium tumefaciens (strain EHA105)-mediated transformation of rice plants was conducted according to Hiei et al. (1994).
RNA Isolation, RNA Gel Blot, and RT-PCR
For expression analysis of YAB1, total RNA was extracted from different vegetative organs and panicles at different developmental stages using an RNA extraction kit (TRIzol Reagent, Invitrogen). For assaying expression of transgenes, total RNA was extracted from the flag leaves of the wild-type and transgenic plants. To quantify transcripts of GAox genes, total RNA was extracted from 1-month-old rice seedlings (four-leaf stage). For quantitative real-time PCR analysis, 4 μg total RNA was treated first with 2 U DNase I (Invitrogen) and then reverse transcribed in a total volume of 20 μL with 0.5 μg oligo(dT)15, 0.75 mm dNTPs, 10 mm dithiothreitol (DTT), and 200 U SuperScript II RNase H– reverse transcriptase (Invitrogen). RT-PCR was performed using gene-specific primers (see below) in a total volume of 25 μL with 1.5 μL of the RT reactions, 0.25 μm gene-specific primers, and 12.5 μL SYBR Green Master mix (Applied Biosystems) on a 7500 real-time PCR machine (Applied Biosystems) according to the manufacturer's instructions. The rice actin1 gene was used as the internal control. All primers were annealed at 58°C and run 45 cycles. The relative expression level of each gene in transgenic plants was compared to that in wild-type ones, after normalization using the actin1 cDNA level and averaging over three replicates. The primers used in RT-PCR for YAB1 were: YAB1-F (5′-CTTGCTCCTTTTCACCAAGC-3′) and YAB1-R (5′-ATGAGCCCAGTTCTTTGCAG-3′). The primers used in GA synthetic gene expression analysis were: CPS1-F (5′-GCGTGCATTTTCGAACCAA-3′), CPS1-R (5′-TTGGCCAGCACTGACACTCT-3′); GA20ox1-F (5′-GCCACTACAGGGCCGACAT-3′), GA20ox1-R (5′-TGGTTGCAGGTGACGATGAT-3′); GA20ox2-F (5-′CCAATTTTGGACCCTACCGC-3′), GA20ox2-R (5′-GAGAGAAGCCCAACCCAACC-3′); GA3ox2-F (5′-TCCTCCTTCTTCTCCAAGCTCAT-3′), GA3ox2-R (5′-GAAACTCCTCCATCACGTCACA-3′); GA2ox1-F (5′-TGACGATGATGACAGCGACAA-3′), GA2ox1-R (5′-CCATAGGCATCGTCTGCAATT-3′); and GA2ox3-F (5′-TGGTGGCCAACAGCCTAAAG-3′), GA2ox3-R (5′-TGGTGCAATCCTCTGTGCTAAC-3′). The actin1 primers were: Actin1-F (5′-TGCTATGTACGTCGCCATCCAG-3′) and Actin1-R (5′-AATGAGTAACCACGCTCCGTCA-3′).
For semiquantitative RT-PCR analysis, 2 μg of total RNA was reverse-transcribed in a total volume of 20 μL with 0.5 μg oligo(dT)15, 0.75 mm dNTPs, 10 mm DTT, and 100 U SuperScript II RNase H− reverse transcriptase (Invitrogen). PCR was performed in a total volume of 20 μL with 1 μL of the RT reactions, 0.2 μm gene-specific primers, and 1 U rTaq (TaKaRa). A total of 28 to 30 cycles was performed. The primers used in RT-PCR for YAB1 were: YAB1-F (5′-CTTGCTCCTTTTCACCAAGC-3′) and YAB1-R (5′-ATGAGCCCAGTTCTTTGCAG-3′).
For northern-blotting analysis, 20 μg of total RNA was separated on 1.2% (w/v) denaturing agarose gel before being transferred to nylon membranes. Gene-specific probes were labeled with 32P-dCTP using a Random Primer kit (Invitrogen) and hybridized to the RNA blots. The probes were amplified by PCR using the following primers: GA3ox2-F (5′-CGACCTCTTCCACATCCTCACC-3′) and GA3ox2-R (5′-CGTCGGTGGAGACCATCTTGAG-3′); GA2ox3-F (5′-TTCTTCGTCAACGTCGGCGACTCGTTGC-3′) and GA2ox3-R (5′-TCTCAAACTGGGCCAGCCTGTTGTCTCC-3′); and GA20ox2-F (5′-GAGACCCTCTCCTTCGGCTTC-3′) and GA20ox2-R (5′-CATGGCGGGTAGTAGTTGCAC-3′).
GA Quantification
The growing shoots with shoot apex of 4-week-old seedlings of YAB1 overexpression transgenic and wild-type rice lines were harvested for quantitative GA analysis. After harvesting, the samples were immediately plunged into liquid N2, freeze-dried, and ground into fine powder using a mill ball. The extraction and purification were carried out as previously described (Coles et al., 1999) with some modifications. Briefly, samples were extracted by stirring overnight at 4°C in 80% (v/v) methanol-water (100 mL) containing [17-2H2]GAs (5–45 ng, depending on anticipated endogenous GAs contents) as internal standards, and 833 Bq each of the following tritiated GA standards: [1,2-3H2]GA1, [1,2-3H2]GA4, 16,17-dihydro[15,16,17-3H4]GA19, and [1,2,3-3H3]GA20. After filtration, the residue was reextracted with methanol (100 mL) for 2 h and refiltrated. The combined methanol extracts were evaporated almost to dryness under reduced pressure at 4°C. The residue was resuspended in water, adjusted to pH 8.0 (1 m KOH), and purified by QAE Sephadex A-25 (Pharmacia) anion-exchange column and C18 Solid Phase Extraction cartridge (500 mg; Thermo). The dried GAs were methylated twice with excess diazomethane, then dissolved in ethyl acetate. After partitioning against water, the organic phase was put through NH2 Solid Phase Extraction cartridge (Altech Associates) and evaporated to dryness in vacuo. GAs were resolved by reverse-phase HPLC and fractions combined based on the location of tritiated GA1, GA4, GA19, and GA20. Samples were analyzed as methyl ester trimethylsilyl ethers using a Finnigan GCQ gas chromatography-mass spectrometry system in selected ion monitoring mode with instrument parameters described previously (Coles et al., 1999). Characteristic ions (mass-to-charge ratio value) monitored by mass spectrometer under selected ion monitoring mode were as follows: GA8/[17-2H2]GA8, 594/596, 448/450; GA1/[17-2H2]GA1, 506/508, 448/450; GA29/[17-2H2]GA29, 506/508, 375/377; GA20/[17-2H2]GA20, 418/420, 375/377; GA44/[17-2H2]GA44, 432/434, 373/375; GA19/[17-2H2]GA19, 434/436, 374/376; and GA53/[17-2H2]GA53, 448/450, 389/391. The concentrations of GAs in the original extracts were determined from previously established calibration curves of the peak area ratios for unlabeled and deuterated GAs plotted against varying molar ratios of the two compounds. The same stock solutions of labeled GAs were used for production of the calibration curves.
In Situ Hybridization
Plant materials were fixed in FAA (50% ethanol, 5% acetic acid, 3.7% formaldehyde) overnight at 4°C, dehydrated through a concentration grade of ethanol, cleared through a xylene series, then infiltrated through a series of paraffin, and finally embedded in 100% paraffin melted at 52°C to 54°C. Then 10- to 15-μm-thick microtome sections were mounted on RNase free glass slides.
The YAB1 probe was amplified using the gene-specific primers as mentioned above. The PCR fragments were inserted into pBlueScript SK–. The digoxigenin-labeled sense and anti-sense RNA probes were produced by T7 and T3 transcriptase, respectively. The reagents used in the experiments were purchased from Roche.
GFP Imaging
For the construction of the fusion between the YAB1 promoter and GFP, the GUS sequence in pCAMBIA1381Xb (CAMBIA) was replaced by the GFP gene. The YAB1 promoter was amplified from ‘Nipponbare’ genomic DNA and inserted into the constructed pCAMBIA1381Xb-GFP at the EcoRI and BamHI sites. The primers used in PCR amplification were YAB1prom-F (5′-GGGAATTCTGATCCTGTAGCCCCATCTC-3′) and YAB1prom-R (5′-GGGAGCTCCATGCTCCGATGTAAACTGG-3′) with an EcoRI adaptor at one end and a BamHI adaptor at the other. Different tissues were harvested at different developmental stages of the transgenic plants, then fixed and sectioned for observation.
Southern-Blotting Analysis
Genomic DNA was extracted from young seedlings. A total of 4 μg of DNA was digested with BamHI overnight at 37°C, separated on 1% (w/v) agarose gel, then transferred to a nylon membrane and hybridized according to standard protocols.
Chemical Treatments
DEX (Sigma) was dissolved in ethanol to 30 mm and stored at −20°C before use. Before experiment, DEX was diluted with growth medium to a final concentration of 10 μm. Transgenic siblings from the same lines were divided into three parts: one was treated with DEX, one was treated first with CHX for 1 h then with DEX, and the third received no treatment but an equal volume of ethanol was added to the medium. After 12 h of treatment, the aerial parts of the plants were harvested for RNA extraction, RT-PCR, and quantitative PCR analysis.
Yeast One-Hybrid Analysis
Yeast one-hybrid analysis was performed by using a CLONTECH system. The promoter fragment of GA3ox2 was amplified by PCR, which spanned from the initiation ATG codon to −862 and inserted into pHIS2. The primers used were: GA3ox2P-F (5′-GAATTCAGTGGCTGCATGTGAGAATG-3′), GA3ox2P-R (5′-GAGCTCCTGGCAGAAAGCCAGTAACA-3′). The full-length coding sequence of YAB1 was amplified from the full-length cDNA clone using the primer pair YAB1CDS-F (5′-GAATTCGAAATGTCGGTCCAGTTTACATC-3′) and YAB1CDS-R (5′- GGATCCTATATCGTCGTCGTCCCTCC-3′). The PCR product was inserted into pGADT7-Rec2 to fuse with the GAL4 activation domain. The constructs were sequenced and used to transform the yeast strain Y187 for one-hybrid analysis.
Gel Shift Assays
To produce the YAB1 protein, the full length of the indica YAB1 cDNA was inserted into and translationally fused with the pET-32a expression vector (Novagen) and expressed in Rosetta-gami (DE3) Escherichia coli cells (Novagen). The target protein was purified with B-PER 6×His Spin Purification kit (Pierce). The 0.8-kb GA3ox2 promoter fragment was cut into two segments by restriction digestion and used as probes in gel shift assays. Oligonucleotides corresponding to the five overlapping regions of the upstream segment of the promoter or to the GARE elements were synthesized and annealed into double strands. The sequence of wild-type GARE oligonucleotide was 5′-TATAACAGATTTATAACAGATT and that of the mutant GARE sites 5′-TATATGAGATTTATATGAGATT. The probes were end-labeled by filling with 32P-dCTP using the Klenow fragment. DNA-binding reactions were performed at room temperature for 20 min in 10 mm Tris, pH 7.5, 50 mm NaCl, 1 mm DTT, 1 mm EDTA, 1 mm MgCl2, 5% glycerol, and 50 mg L−1 poly(dI-dC)·poly(dI-dC) (Amersham Pharmacia Biotech) and separated on 6% polyacrylamide gels in Tris-glycin (0.3% Tris, 1.88% glycin) buffer.
Growth Regulator Treatments
Transgenic and wild-type seedlings were grown on hydroponic culture media to the four-leaf stage before treatment. Paclobutrazol (at the final concentration of 10 μm) was added to the hydroponic culture 1 d before adding GA3 at the concentrations of 0.01, 0.05, 0.1, 0.5, 1, 10, and 20 μm. The length of the second leaf sheath was measured 10 d after the treatment. The hydroponic culture media and treatments were repeated every 2 d.
For YAB1 expression analysis, wild-type seedlings at the four-leaf stage were sprayed with 50 μm GA3 or 10 μm paclobutrazol and harvested at different time points for RNA extraction and quantitative RT-PCR analysis. For expression analysis of GAox genes, transgenic and wild-type plants were sprayed with 50 μm GA3 and harvested at different time points for RNA extraction and quantitative RT-PCR analysis. The expression levels of these genes in these samples were determined relative to time zero, normalized against actin1, and averaged over three biological replicates.
Light Microscopy
Stems and leaves were harvested from the mature plants of wild type and transgenic ones. Stems were treated with 15% of hydrofluoric acid before dehydration. The procedures of dehydration, clearing, infiltration, and embedding were carried out as mentioned above. The microtome sections (15 μm) were mounted on glass slides for imaging.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF098752 and BAF12697.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Rice YAB1 sequences from indica and japonica subspecies.
Supplemental Figure S2. Southern-blot analysis of YAB1 transgene copy numbers.
Supplemental Figure S3. Cosuppression of YAB1 expression induced GA3ox2.
Supplemental Figure S4. GA transiently induced expression of YAB1.
Supplementary Material
Acknowledgments
We thank Li Xianhua for technical assistance, Xu Chaiguo for rice field management, Dr. M. Matsuoka for providing slr and gid1 mutants, and Dr. Z. He for sending eui mutants.
This work was supported by the National Special Key Program of Rice Functional Genomics, by the National Natural Science Foundation of China, and by the Biotechnology and Biological Sciences Research Council of the United Kingdom (grants to Rothamsted Research).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Dao-Xiu Zhou (dao-xiu.zhou@u-psud.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Amador V, Monte E, Garcia-Martinez JL, Prat S (2001) Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106 343–354 [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11 605–612 [DOI] [PubMed] [Google Scholar]
- Bowman JL (2000) The YABBY gene family and abaxial cell fate. Curr Opin Plant Biol 3 17–22 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR (1999) CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126 2387–2396 [DOI] [PubMed] [Google Scholar]
- Cercós M, Gómez-Cadenas A, Ho T-HD (1999) Hormonalregulation of a cysteine proteinase gene, EPB-1, in barley aleurone layers: cis- and trans-acting elements involved in the coordinated gene expression regulated by gibberellins and abscisic acid. Plant J 19 107–118 [DOI] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, Garcia-Lepe R, Lewis MJ, Hedden P (1999) Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J 17 547–556 [DOI] [PubMed] [Google Scholar]
- Cowling RJ, Kamiya Y, Seto H, Harberd NP (1998) Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis Plant Physiol 117 1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker SJ, Hedden P, Lenton JR, Stoddart JL (1990) Comparison of gibberellins in normal and slender barley seedlings. Plant Physiol 94 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis Plant Physiol 136 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Hu Y, Zhao Y, Liu H, Zhou D-X (2007) A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol 144 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Sun T (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209 [DOI] [PubMed] [Google Scholar]
- Fleet CM, Sun TP (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8 77–85 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Yamane H, Spray CR, Gaskin P, MacMillan J, Phinney BO, Takahashi N (1988) Qualitative and quantitative analyses of gibberellins in vegetative shoots of normal, dwarf-1, dwarf-2, dwarf-3, and dwarf-5 seedlings of Zea mays L. Plant Physiol 88 1367–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y (2000) Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7 373–385 [DOI] [PubMed] [Google Scholar]
- Golz JF, Hudson A (1999) Plant development: YABBYs claw to the fore. Curr Biol 9 R861–R863 [DOI] [PubMed] [Google Scholar]
- Gubler F, Jacobsen JV (1992) Gibberellin-responsive elements in the promoter of a barley high-pI a-amylase gene. Plant Cell 4 1435–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV (1995) Gibberellin-regulated expression of a myb gene in barley aleuronecells: evidence for Myb transactivation of a high-pI a-amylase gene promoter. Plant Cell 7 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV (1999) Target genes and regulatory domains of the GAMyB transcriptional activator in cereal aleurone. Plant J 17 1–9 [DOI] [PubMed] [Google Scholar]
- Hay A, Craft J, Tsiantis M (2004) Plant hormones and homeoboxes: bridging the gap? Bioessays 26 395–404 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5 523–530 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Hur J, Kim SJ, Han MJ, Kim SR, An G (2004) Ectopic expression of OsYAB1 causes extra stamens and carpels in rice. Plant Mol Biol 56 133–143 [DOI] [PubMed] [Google Scholar]
- Kaneko M, Itoh H, Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Ashikari M, Matsuoka M (2003) Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J 35 104–115 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Yamaguchi I, Murofushi N, Ota Y, Takahashi N (1988) Fluctuation and localization of endogenous gibberellins in rice. Agric Biol Chem 52 1189–1194 [Google Scholar]
- Martin DN, Proebsting WM, Parks TD, Dougherty WG, Lange T, Lewis MJ, Gaskin P, Hedden P (1996) Feed-back regulation of gibberellin biosynthesis and gene expression in Pisum sativum L. Planta 200 159–166 [DOI] [PubMed] [Google Scholar]
- Mason HS, Lam DM, Arntzen CJ (1992) Expression of hepatitis B surface antigen in transgenic plants. Proc Natl Acad Sci USA 89 11745–11749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell (Suppl) 14 S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raventós D, Skriver K, Schlein M, Karnahl K, Rogers SW, Rogers JC, Mundy J (1998) HRT, a novel zinc finger, transcriptional repressor from barley. J Biol Chem 273 23313–23320 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Ito T, Shimura Y, Okada K (1999) FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 11 69–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Lechtenberg B, Forsbach A, Gils M, Bahadur S, Schmidt R (2004) Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16 2561–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutoh K, Yamauchi D (2003) Two cis-acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant J 34 635–645 [DOI] [PubMed] [Google Scholar]
- Talon M, Koornneef M, Zeevaart JA (1990) Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc Natl Acad Sci USA 87 7983–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M (2000) Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA 97 11638–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Li L, Wu K, Peeters AJ, Gage DA, Zeevaart JA (1995) The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA 92 6640–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY (2004) The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Feng Q, Jin C, Qiu D, Zhang L, Xie K, Yuan D, Han B, Zhang Q, Wang S (2005) Features of the expressed sequences revealed by a large-scale analysis of ESTs from a normalized cDNA library of the elite indica rice cultivar Minghui 63. Plant J 42 772–780 [DOI] [PubMed] [Google Scholar]
- Zhu YY, Nomura T, Xu YH, Zhang YY, Peng Y, Mao BZ, Hanada A, Zhou HC, Wang RX, Li PJ, et al (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.