Abstract
As indicated by various and some overlapped phenotypes of the dominant mutants, the Aux/IAA genes of Arabidopsis (Arabidopsis thaliana) concomitantly exhibit a functional similarity and differentiation. To evaluate the contributions of their expression patterns determined by promoter activity and molecular properties of their gene products to Aux/IAA function, we examined phenotypes of transgenic plants expressing the green fluorescent protein (GFP)-tagged msg2-1/iaa19, axr2-1/iaa7, or slr-1/iaa14 cDNA by the MSG2 or AXR2 promoter. When driven by the MSG2 promoter (pMSG2), each GFP-tagged cDNA caused the msg2-1 phenotype, that is, the wild-type stature in the mature-plant stage, long and straight hypocotyls in the dark, reduced lateral root formation, relatively mild agravitropic traits in hypocotyls, and a normal gravitropic response in roots. However, development of one or two cotyledonary primordia was often arrested in embryogenesis of the pMSG2∷axr2-1∷GFP and pMSG2∷slr-1∷GFP plants, resulting in monocotyledonary or no cotyledonary seedlings. Such defects in embryogenesis were never seen in pMSG2∷msg2-1∷GFP or the msg2-1, axr2-1, or slr-1 mutant. The MSG2 promoter-GUS staining showed that expression of MSG2 started specifically in cotyledonary primordia of the triangular-stage embryos. When driven by the AXR2 promoter (pAXR2), each GFP-tagged mutant cDNA caused, in principle, aberrant aboveground phenotypes of the corresponding dominant mutant. However, either the axr2-1∷GFP or slr-1∷GFP cDNA brought about dwarf, agravitropic stems almost identical to those of axr2-1, and the pAXR2∷msg2-1∷GFP and pAXR2∷slr-1∷GFP hypocotyls exhibited complete loss of gravitropism as did axr2-1. These results showed functional differences among the msg2-1, axr2-1, and slr-1 proteins, though some phenotypes were determined by the promoter activity.
Auxin acts as a signaling molecule in many aspects of plant growth and development, including embryogenesis, root and shoot patterning, apical dominance, and tropic responses. These processes include the regulation of gene expression by two protein families, auxin response factors (ARFs) and the Aux/IAA proteins (Leyser, 2002; Hagen et al., 2004), that function directly downstream of the auxin F box receptors (AFBs; Dharmasiri et al., 2005; Kepinski and Leyser, 2005).
ARFs were initially identified by their ability to bind to auxin-responsive elements via an amino-terminal DNA-binding domain, and they regulate the expression of genes containing such promoter elements in an auxin-dependent manner (Ulmasov et al., 1997; Hagen et al., 2004). The Aux/IAA genes were first identified as genes whose transcripts were rapidly induced by auxin (Abel and Theologis, 1996). They encode short-lived nuclear proteins, and most of them contain four highly conserved domains (I–IV) that contribute to their functional properties. Domains I and II are unique to the Aux/IAA proteins, while domains III and IV are also conserved in the carboxyl-terminal domain (CTD) of ARFs and serve as protein-protein interaction domains that promote both homo- and heterodimerization between members of the Aux/IAA and ARF families (Kim et al., 1997; Ulmasov et al., 1997; Hagen et al., 2004). Domain I is a repressor domain that is dominant over the activation function of ARF (Tiwari et al., 2004). Domain II confers instability to the protein (Worley et al., 2000; Ouellet et al., 2001). Recent studies indicate that an AFB, TIR1, interacts with the Aux/IAA proteins and stimulates their degradation in an auxin-dependent manner (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). Both the ARF and Aux/IAA proteins consist of large families with 23 and 29 members, respectively, in Arabidopsis (Arabidopsis thaliana).
Genetic studies of loss-of-function mutants of the ARF genes have revealed both distinct and redundant roles of ARFs in plant growth and development. For example, NPH4/ARF7 plays a central role in tropistic responses in hypocotyls and lateral root formation (Liscum and Briggs, 1995; Watahiki and Yamamoto, 1997; Harper et al., 2000; Tatematsu et al., 2004), and so does MP/ARF5 in embryo patterning and vascular tissue formation (Mattsson et al., 2003). However, ARF19 is also involved in tropic responses and lateral root formation because the arf19 mutation, which does not affect these processes alone, enhances the nph4 defects significantly (Okushima et al., 2005; Wilmoth et al., 2005). Similarly, the nph4 mp double mutants exhibit more severe defects than the corresponding single mutants, indicating that MP and NPH4 act redundantly (Hardtke et al., 2004). In the Aux/IAA genes, several dominant mutations in domain II, which increase the stability of the gene products, have been identified, such as iaa1/axr5 (Park et al., 2002; Yang et al., 2004), iaa3/shy2 (Tian and Reed, 1999), iaa6/shy1 (Reed, 2001), iaa7/axr2 (Nagpal et al., 2000), iaa12/bdl (Hamann et al., 2002), iaa14/slr (Fukaki et al., 2002), iaa17/axr3 (Rouse et al., 1998), iaa18 (Reed, 2001), iaa19/msg2 (Tatematsu et al., 2004), and iaa28 (Rogg et al., 2001). Various aberrant phenotypes involved in auxin-mediated growth and development, as well as altered gene expression in response to auxin, are found in these mutants. Each of the altered phenotypes is distinct in a different subset of the aux/iaa mutants, which indicates functional similarity and differentiation of the Aux/IAA family.
The similarity and differentiation of the Aux/IAA genes may result from their expression patterns produced by their promoter activity and/or molecular properties of their gene products. Knox et al. (2003) and Weijers et al. (2005) evaluated the contributions of the two factors to function of the Aux/IAA genes in transgenic plants expressing the mutant Aux/IAA (mAux/IAA) cDNA by the same promoter. Their results show that the specificity of the Aux/IAA action is primarily regulated at the level of gene transcription, while there exist some functional differences between the aux/iaa proteins. The msg2 mutants provide a rare opportunity to address this question because the msg2 defects appear to be very specific compared to the other dominant Aux/IAA mutants. msg2 is defective only in tropic responses of hypocotyls, formation of lateral roots and fecundity, and looks almost normal at the mature stage in contrast to the pleiotropic defects displayed by most of the aux/iaa mutants. By taking advantage of the unique msg2 phenotype, we also investigated the molecular basis of the similarity and differentiation among the msg2-1, axr2-1, and slr-1 genes.
RESULTS
Growth and Development of Shoots
Adult plants of msg2-1/iaa19 look similar to the wild type except for having a reduced fecundity (Fig. 1B; Tatematsu et al., 2004). slr-1/iaa14 has small leaves and short and thin inflorescence stems. The number of inflorescence stems is also reduced (Fig. 1C; Fukaki et al., 2002). The axr2-1/iaa7 is dwarf and has agravitropic stem (Fig. 1D; Nagpal et al., 2000). In an attempt to investigate whether the phenotype of each mutant was determined by the specific pattern of expression of each gene, we compared phenotype of transgenic Arabidopsis plants that expressed the msg2-1, axr2-1, or slr-1 cDNA under the control of a 2-kb portion of the MSG2 or AXR2 promoter (pMSG2 and pAXR2, respectively). The mAux/IAA cDNAs were also fused translationally with GFP to detect their expression.
Figure 1.
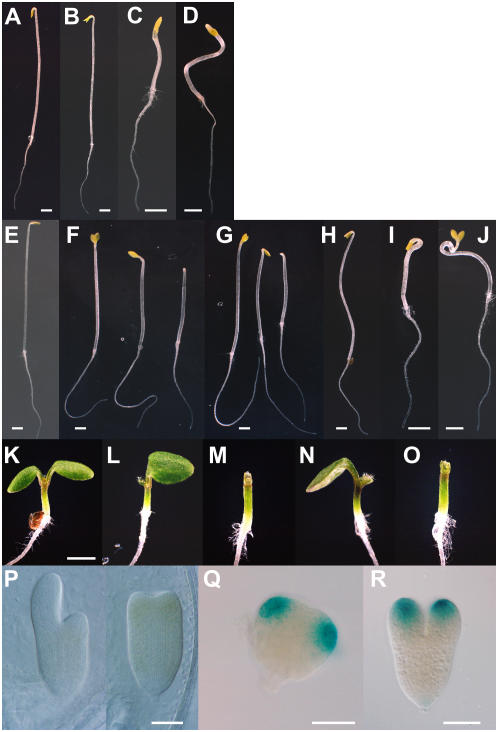
Phenotypes of 4-week-old transgenic plants that express mutant Aux/IAA genes tagged with GFP. A, Wild type; B, msg2-1/iaa19; C, slr-1/iaa14; D, axr2-1/iaa7; E, pMSG2∷msg2-1∷GFP; F, pAXR2∷msg2-1∷GFP; G, pMSG2∷axr2-1∷GFP; H, pAXR2∷axr2-1∷GFP; I, pMSG2∷slr-1∷GFP; and J, pAXR2∷slr-1∷GFP. All plants were sown in 55-mm-diameter pots. [See online article for color version of this figure.]
Transgenic plants harboring the pMSG2∷msg2-1∷GFP transgene showed a normal morphology (Fig. 1E). The reduced fecundity as observed in msg2-1 was found in five out of 50 independent T1 plants. On the other hand, seedlings of multiple pMSG2∷axr2-1∷GFP and pMSG2∷slr-1∷GFP lines often lacked one or both cotyledons (Fig. 2, F, L, and M, and G, N, and O, respectively). In one pMSG2∷axr2-1∷GFP line, one and no cotyledon phenotypes appeared in 59% and 25% of the seedlings (n = 334); 44% and 27% were observed for a pMSG2∷slr-1∷GFP line, respectively (n = 126). The single cotyledon of defective seedlings was normal in shape and normally expanded after germination (Fig. 2, L and N). Plants with the defective cotyledons were smaller than wild type, but they produced true leaves with normal phyllotaxis except for the first leaf, which was sometimes fused (data not shown). The pMSG2∷axr2-1∷GFP and pMSG2∷slr-1∷GFP plants with two cotyledons looked almost normal, though they showed reduced fecundity that was more severe than that of msg2-1 (Fig. 1, G and I).
Figure 2.
Phenotypes of transgenic plants in the seedling and embryo stages. A to J, Dark-grown seedlings; K to O, light-grown seedlings. A and K, Wild type; B, msg2-1; C, axr2-1; D, slr-1; E, pMSG2∷msg2-1∷GFP; F, L, and M, pMSG2∷axr2-1∷GFP; G, N, and O, pMSG2∷slr-1∷GFP; H, pAXR2∷msg2-1∷GFP; I, pAXR2∷axr2-1∷GFP; and J, pAXR2∷slr-1∷GFP. The seedlings of pMSG2∷axr2-1∷GFP and pMSG2∷slr-1∷GFP sometimes lacked one cotyledon (F [middle], G [middle], L, and N) or had no cotyledons (F [right], G [right], M, and O). All seedlings were grown for 3 d. P, Aberrant development of torpedo-stage embryos of pMSG2∷axr2-1∷GFP that lacked one (left) or two (right) cotyledon primordia. Q and R, Expression of the MSG2 promoter-GUS at the triangular (Q) and early torpedo stages (R). Scale bar, 1 mm (A–K) and 100 μm (P–R). Scale in L to O is same as that in K.
To our knowledge, the aberrant development of cotyledons had never been seen in msg2-1, slr-1, axr2-1, or the pMSG2∷msg2-1∷GFP plants. The abnormality became evident from the torpedo-stage embryo, in which development of primordia of one or both cotyledons was arrested (Fig. 2P). Examination of the MSG2 promoter-GUS line showed MSG2 expression in primordia of the cotyledon in as early as the triangular stage of embryo (Fig. 2, Q and R). This suggests that ectopic expression of axr2-1 or slr-1 in cotyledon primordia tends to repress development of the cotyledon, but that native expression of msg2-1 does not.
The pAXR2∷axr2-1∷GFP plants reproduced the axr2-1 phenotype in respect of their dwarfism and agravitropism in stems (Fig. 1H). Out of 34 T1 plants harboring the pAXR2∷axr2-1∷GFP transgene, 17 plants showed the axr2-like phenotype, nine plants were normal in morphology, and the others exhibited an intermediate phenotype with semi-dwarf traits. The pAXR2∷slr-1∷GFP plants also showed the axr2 phenotype (Fig. 1J). In contrast, the pAXR2∷msg2-1∷GFP plants displayed normal morphology even though we examined 48 T1 lines (Fig. 1F). These results suggest again that axr2-1 and slr-1 perform very similar functions, and that their functions differ from that of msg2-1 under the control of the AXR2 promoter. This observation is consistent with phylogenetic evidence that AXR2 and SLR are most closely related in the Aux/IAA family (Remington et al., 2004).
Growth of Dark-Grown Hypocotyls
Dominant mutations in Aux/IAA genes result in varied phenotypes in hypocotyls grown in the dark. axr2-1 exhibits a de-etiolated phenotype (Fig. 2C; Nagpal et al., 2000), but msg2-1 and slr-1 do not, except that they have a partially open hook structure. In addition, hypocotyls of msg2-1 grow straight, whereas those of slr-1 display curly growth (Fig. 2, B and D; Fukaki et al., 2002; Tatematsu et al., 2004). Under the control of the MSG2 promoter, none of the three mAux/IAA∷GFPs caused either de-etiolated phenotype or curly growth of hypocotyls. Their hypocotyls looked like those of msg2-1 (Fig. 2, E–G). Interestingly, hypocotyls of etiolated seedlings with one or no cotyledon were shorter than normal ones by up to 30%, possibly due to a shortage of auxin or nutrients. On the other hand, growth in the dark of hypocotyls of the three transgenic plants expressing mAux/IAA∷GFPs by the AXR2 promoter were different. Seedlings of pAXR2∷axr2-1∷GFP, like those of axr2-1, were de-etiolated (Fig. 2I). Seedlings of pAXR2∷msg2-1∷GFP, like those of msg2-1, had straight hypocotyls (Fig. 2H). Seedlings of pAXR2∷slr-1∷GFP, like those of slr-1, had curly hypocotyls (Fig. 2J). It is thus concluded that when driven by the AXR2 promoter, each mAux/IAA protein fused with GFP reproduced the phenotype of the corresponding dominant mutant from which it was derived. These results suggest that the three mAux/IAAs had different effects on growth regulation in hypocotyls.
Growth and Development of Roots
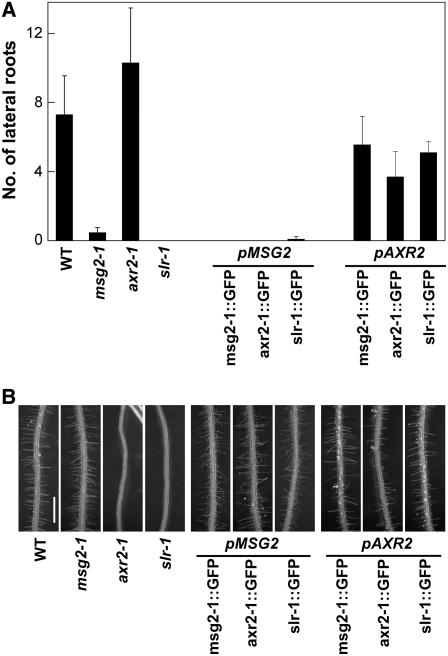
msg2-1 and slr-1 are defective in formation of lateral roots, but msg2-1, unlike slr-1, exhibits normal root hairs (Fig. 3; Fukaki et al., 2002; Tatematsu et al., 2004). On the other hand, axr2-1 has more abundant lateral roots than wild type but has no root hairs (Fig. 3; Wilson et al., 1990; Nagpal et al., 2000). Under the control of the MSG2 promoter, all three mAux/IAA∷GFPs decreased the number of lateral roots, but had no effect on the abundance of root hairs. Thus, roots of the pMSG2∷mAux/IAA∷GFP plants looked similar to those of the msg2 mutants (Fig. 3), suggesting that the msg2 phenotype in roots is dependent on the activity of the MSG2 promoter. On the other hand, the axr2-1∷GFP cDNA driven by the AXR2 promoter did not show significant effects on the formation of lateral roots (P = 0.081 in t test) or root hairs, which was distorted in axr2-1. Therefore, other cis-acting elements outside the 2-kb promoter used in this study must be required to control the AXR2 gene expression in roots. The pAXR2∷msg2-1∷GFP and pAXR2∷slr-1∷GFP plants also showed a normal root phenotype (0.18 < P <0.34 in t test for lateral root formation), which might be due to insufficient activity of the AXR2 promoter that we used in this study.
Figure 3.
Formation of lateral root (A) and root hair (B) in transgenic plants expressing mutant Aux/IAA genes tagged with GFP. Seedlings were grown on vertically oriented plates for a week under continuous white light. A, Number of lateral roots; B, images of the primary roots of the seedlings. Scale bar, 1 mm.
Tropic Responses
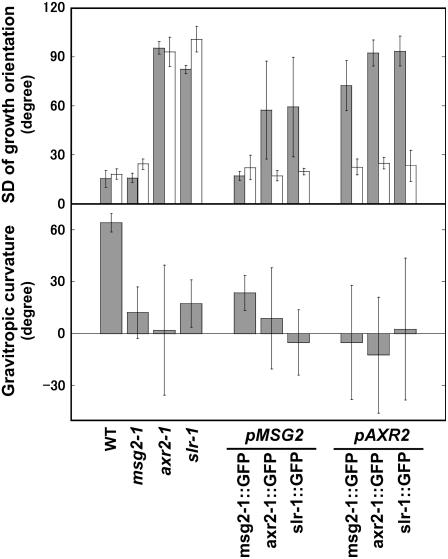
The gravitropic response is reduced in most aux/iaa mutants, including msg2, axr2, and slr, but in a distinct manner. For example, although axr2 and slr exhibit agravitropism in both roots and hypocotyls, msg2 is defective only in hypocotyls (Tatematsu et al., 2004). We first examined growth orientation of hypocotyls and roots of seedlings, which reflects their ability to respond to gravity. Seedlings were grown on vertically held agar plates. Growth orientation of hypocotyls and roots was randomized in axr2-1 and slr-1 as revealed by their much greater sd than that of wild type, whereas it was normal in msg2-1 (Fig. 4, top). However, when turned 90°, msg2-1 hypocotyls did not respond to gravity as readily as wild type (Fig. 4, bottom), showing that msg2-1 hypocotyls were also defective in gravitropism. The large se of bending curvature of axr2-1 hypocotyls indicated almost complete loss of gravitropism in axr2-1, while the smaller se in msg2-1 and slr-1 showed that they retained some gravitropic sensitivity.
Figure 4.
Growth orientation (top) and gravitropic responses (bottom) of transgenic plants that express the mutant Aux/IAA genes tagged with GFP. Top, Seedlings were grown on vertically oriented plates for 3 d in the dark, and growth angle was measured. Variation of growth orientation of hypocotyl (gray bars) or root tip (white bars) was represented by sd of growth angles. Values indicate mean and sd of three independent experiments, each using eight to 13 seedlings. Bottom, Curvature of hypocotyls of seedlings that were grown on vertically oriented plates for 3 d in the dark and then reoriented by 90°. Curvature was measured 15 h after the turn. Values indicate mean and se of three independent experiments, each using seven to 13 seedlings.
Growth orientation of hypocotyls with each mAux/IAA∷GFP under the control of the MSG2 or AXR2 promoter tended to be random except for pMSG2∷msg2-1∷GFP. However, the randomness of hypocotyl orientation with pMSG2∷axr2-1∷GFP and pMSG2∷slr-1∷GFP was smaller than that of hypocotyls with mAux/IAA∷GFPs driven by the AXR2 promoter, which were almost perfectly agravitropic as judged from their sd values (Yamamoto and Yamamoto, 1998). The sd of growth orientation of pMSG2∷axr2-1∷GFP and pMSG2∷slr-1∷GFP was variable because hypocotyls with one or two cotyledons and with no cotyledons responded to gravity differently. The sd of the former hypocotyls was approximately 40°, while that of the latter was approximately 95°. Therefore, if all the pMSG2∷axr2-1∷GFP and pMSG2∷slr-1∷GFP seedlings have a pair of cotyledons, their gravitropic defects would be certainly weaker than those of the pAXR2∷mAux/IAA∷GFP plants. This clearly shows that the gravitropic responses in the transgenic plants were primarily determined by their promoter activities. However, even when driven by the same MSG2 promoter, gravitropic defects were not the same between the mAux/IAA∷GFP proteins: Defects of hypocotyls with axr2-1∷GFP and slr-1∷GFP proteins were larger than those observed in the pMSG2∷msg2-1∷GFP hypocotyls, which were normal with respect to growth orientation on the vertically oriented agar plates. This result indicates that the axr2-1 and slr-1 proteins have more deleterious effects on gravitropic responses than msg2-1.
Roots were normally orientated in all the transgenic plants tested, like those of msg2-1 (Fig. 4, top, white bars). The gravitropic response also was not affected in roots (data not shown). MSG2 has been shown to be expressed in the elongation zone and pericycle of roots (Tatematsu et al., 2004), indicating that expression of neither msg2-1, axr2-1, nor slr-1 there affects root gravitropism. Expression of axr2-1∷GFP under the control of the AXR2 promoter also did not affect the gravitropic response in roots, while in the axr2-1 mutant the root was agravitropic. Since lateral roots and root hairs in pAXR2∷axr2-1∷GFP formed normally as described above, the axr2 phenotype was produced only in aerial parts in this transgenic line, probably because the AXR2 promoter that we used had insufficient activity. Consequently, the function of msg2-1 and slr-1 could not be determined in root when they were driven by the AXR2 promoter.
Expression of Transgenes
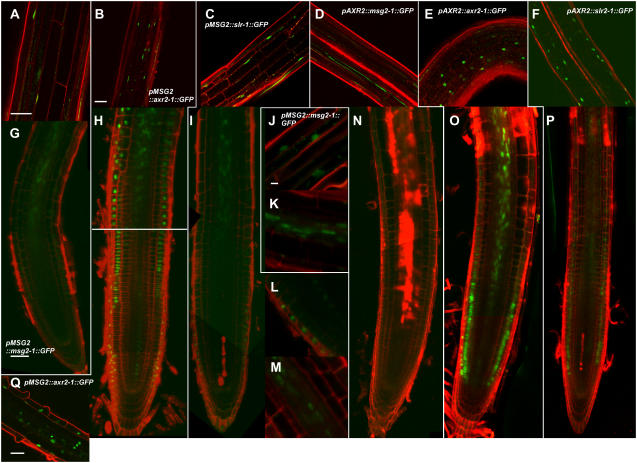
Expression of the mAux/IAA∷GFP transgenes was measured with fluorescent confocal microscopy (Fig. 5) and quantitative reverse transcription (RT)-PCR (Fig. 6). GFP fluorescence was detected in nuclei in all the transgenic plants (Fig. 5), except for pAXR2∷msg2-1∷GFP (Fig. 5, D and N). However, quantitative RT-PCR analysis revealed that all transgenic plants, including pAXR2∷msg2-1∷GFP, expressed the transgenes (Fig. 6). GFP fusion proteins whose expressions were driven by the MSG2 promoter were found in hypocotyls (Fig. 5, A–C) and roots except for their meristematic zone (Fig. 5, G–M), which is consistent with the staining pattern reported for the pMSG2-GUS gene (Tatematsu et al., 2004). Under the control of the AXR2 promoter, the expression of axr2-1∷GFP and slr-1∷GFP was observed in hypocotyls (Fig. 5, E and F) and the meristematic and elongation zones of roots (Fig. 5, O and P). In the case of the pAXR2∷axr2-1∷GFP roots, GFP signals were also obvious in the central stele of the apical region of the maturation zone (Fig. 5O). The expression of AXR2 has been investigated by the use of the GUS fusion gene with the 2.3-kb AXR2 promoter. GUS activity was found in shoot and root apical meristems of seedlings, but not in either hypocotyls or dark-grown roots except for the vasculature of hypocotyl/root junctions (Tian et al., 2002). Therefore, we observed a wider distribution of the AXR2 promoter activity in this study. The use of the GFP-tagged axr2-1 or slr-1, both of which are stabilized by a mutation in domain II, may make it possible to detect a lower activity of the AXR2 promoter.
Figure 5.
Expression of mAux/IAA∷GFP transgenes observed with confocal fluorescent microscopy. Longitudinal images of GFP were observed in hypocotyls of 3-d-old etiolated seedlings (A–F) and roots of 1-week-old light-grown seedlings (G–Q) counterstained with 10 μg mL−1 propidium iodide. A, G, J, and K, pMSG2∷msg2-1∷GFP; B, H, and Q, pMSG2∷axr2-1∷GFP; C, I, L, and M, pMSG2∷slr-1∷GFP; D and N, pAXR2∷msg2-1∷GFP; E and O, pAXR2∷axr2-1∷GFP; F and P, pAXR2∷slr-1∷GFP. Images from A to C, E, J, L, and Q are lateral optical sections; the other images are medial sections. J to M and Q, Enlarged images of the central stele in the elongation zone (K and M), and epidermis in the elongation zone (J), the meristematic zone (L), and the root hair initiation zone (Q). Scale bar, 50 μm (A, B, G, and Q) and 10 μm (J). Scales in B to F are the same; scales in G to I and N to P are the same; and scales in J to M are the same.
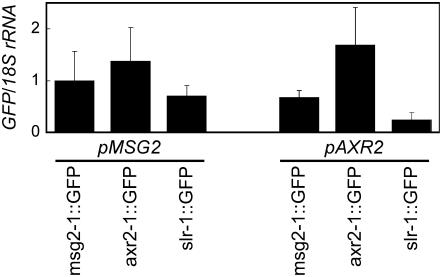
Figure 6.
Expression of mAux/IAA∷GFP transgenes determined with quantitative RT-PCR. RT-PCR was done for the sequence encoding GFP in transgenic seedlings grown for 1 week under white-light condition. Expression of the 18S ribosomal RNA was determined as a control. Transcript levels are shown as values relative to those in pMSG2∷msg2-1∷GFP after normalization to the 18S ribosomal RNA levels. Values indicate mean and sd of three independent experiments.
Expression of pMSG2∷axr2-1∷GFP was seen in all the epidermal cells of the root hair initiation zone (Fig. 5Q). Root hair formation is regulated through interaction between two types of epidermal cells, trichoblasts and atrichoblasts (Kurata et al., 2005). The fact that the presence of the axr2-1 protein in all the epidermal cells of the root hair initiation zone of the transgenic plants did not inhibit root hair formation suggests that inhibitory effects of the axr2-1 mutation on the root hair formation may not be epidermis autonomous, but may be brought about by global defects caused by the mutation. We also carried out confocal microscopic measurements of the pMSG2∷msg2-1∷GFP and pMSG2∷axr2-1∷GFP embryos. However, we have failed to detect their GFP fluorescence so far (data not shown).
Transcription level of the mAux/IAA∷GFP transgene, determined with quantitative RT-PCR, was relatively similar when driven by the MSG2 promoter. However, fluorescence of the axr2-1∷GFP protein (Fig. 5, B and H) was stronger than that of msg2-1∷GFP (Fig. 5, A, G, J, and K) or slr-1∷GFP (Fig. 5, C, I, L, and M). When driven by the AXR2 promoter, mRNA level varied 7-fold among the three transgenic plants (Fig. 6). The varied level was not proportional to level of the mAux/IAA proteins estimated from fluorescent intensity of GFP (Fig. 5) and severity of abnormal phenotypes of the transgenic plants as mentioned above. In particular, no fluorescence of the GFP-tagged msg2-1 protein was observed in pAXR2∷msg2-1∷GFP (Fig. 5, D and N), while the pAXR2∷slr-1∷GFP plants fluoresced significantly (Fig. 5, F and P). But, mRNA level of the latter transgene was lower than that of the former transgene (Fig. 6). These results suggest that the stability of the mAux/IAA proteins might differ, possibly with the msg2-1 protein being the least stable of the three.
DISCUSSION
Promoter Activity Often Determines Phenotype of Transgenic Plants
In this study, we created transgenic Arabidopsis plants harboring dominantly mutated MSG2, AXR2, or SLR cDNA driven by the MSG2 or AXR2 promoter. All the cDNAs were translationally fused with GFP. Their phenotypes, summarized in Table I, indicate that promoter activity often specifies the phenotype of transgenic plants. This promoter-dependent phenotype must result from the different expression patterns of MSG2 and AXR2. Promoter-GUS analyses show that the expression patterns of these genes are rather complementary to each other in etiolated hypocotyls: AXR2 is expressed only in the apical meristem (Tian et al., 2002), while MSG2 is expressed in the epidermis, the cortex, and especially the central stele of hypocotyls. In particular, no obvious staining is observed in the apical meristem except for the procambium (Tatematsu et al., 2004; Saito et al., 2007; our unpublished data). These results suggest that the extent of gravitropic defects in hypocotyls may be determined by the expression patterns of the mAux/IAA proteins, and that each of the three proteins may be able to repress the activity of NPH4 and ARF19 (Okushima et al., 2005; Wilmoth et al., 2005) to a similar extent once expressed in the same region. Similar scenarios are conceivable for lateral root formation since all the pMSG2∷mAux/IAA∷GFP plants exhibit similar defects and since MSG2 is expressed in root pericycles (Tatematsu et al., 2004) from which the primordia of lateral roots are formed. Similarly, normal root gravitropism in the pMSG2∷mAux/IAA∷GFP plants may be explained if we simply assume that root meristem is a critical tissue for root gravitropic response because MSG2 does not appear to express there. The wild-type stature of the pMSG2∷mAux/IAA∷GFP plants may also be explained in a similar manner.
Table I.
Phenotypes of the transgenic lines examined in this study
| Expressed Protein | Driving Promoter
|
|
|---|---|---|
| pMSG2 | pAXR2 | |
| msg2-1∷GFP | msg2 phenotype | msg2 phenotype except for root and severer agravitropism in hypocotyl |
| axr2-1∷GFP | Largely msg2 phenotype with abnormal cotyledon and more reduced fecundity | axr2 phenotype except for root |
| slr-1∷GFP | Largely msg2 phenotype with abnormal cotyledon and more reduced fecundity | axr2 phenotype in shoot morphology and hypocotyl gravitropism; slr phenotype in hypocotyl; wild-type phenotype in root |
| Phenotypes of Dominant Aux/IAA Mutants
| ||
| msg2 | Agravitropic hypocotyl, fewer lateral roots, reduced fecundity | |
| axr2 | Dwarf morphology, agravitropic hypocotyl and root, de-etiolation in the dark, increased number of lateral roots, no root hair | |
| slr | Agravitropic hypocotyl and root, no lateral root, no root hair, curly growth of etiolated hypocotyl | |
When expressed by the AXR2 promoter, the axr2-1∷GFP and slr-1∷GFP proteins induced almost identical morphologies in mature plants. This is another example of promoter-dependent phenotypes. Because the two proteins are most closely related in the Aux/IAA family, the distinct shapes of mature plants and the opposite phenotypes in lateral root formation observed in the axr2 and slr mutants (Wilson et al., 1990; Nagpal et al., 2000; Fukaki et al., 2002) must be determined by different activities of the AXR2 and SLR promoters. Essentially the same promoter-dependent phenotypes have been reported in promoter-exchange experiments between SHY2/IAA3 and BDL/IAA12 (Weijers et al., 2005). Whereas shy2-2 mutants have no embryonic phenotypes (Tian and Reed, 1999), pBDL∷shy2-2 plants showed a rootless phenotype similar to that of bdl. pBDL∷shy2-2 plants also showed bdl-like postembryonic growth abnormalities. Furthermore, pSHY2∷bdl plants resemble shy2-2 mutants at both the seedling and mature stages.
Different Functions Are Also Observed among Mutated Aux/IAA Proteins
These results also indicate functional differentiation among the three mAux/IAA proteins when driven by the AXR2 promoter. Growth fashion of etiolated hypocotyls of each pAXR2∷mAux/IAA∷GFP was distinct and was similar to that of the original dominant aux/iaa mutant. The msg2-1 and pAXR2∷msg2-1∷GFP hypocotyls were long and straight, and looked like those of nph4 (Tatematsu et al., 2004). The axr2-1 and pAXR2∷axr2-1∷GFP hypocotyls were short and similar to the arf6 arf8 double mutants (Nagpal et al., 2005). The curly slr-1 and pAXR2∷slr-1∷GFP hypocotyls were distinct from those of other arf and aux/iaa mutants that have been characterized (Fukaki et al., 2002). The distinct effects of the mAux/IAA∷GFPs should be posttranslational because each transgene was driven by the same promoter with the same 5′ and 3′ untranslated regions. Therefore, the msg2-1 proteins may specifically interfere with the activities of NPH4 and the axr2-1 proteins may specifically interfere with the activities of ARF6 and ARF8, in the AXR2 expression domain. At present, we cannot identify any ARFs that work with slr-1. These speculations about specific interference between Aux/IAA and ARF were also suggested by a comparison of the shape of transgenic plants at the mature stage when mAux/IAA∷GFP was driven by the AXR2 promoter. The specificity of mAux/IAA proteins could also arise from a possible difference in stability of the proteins as mentioned above. This suggests the difference of mAux/IAA proteins in AFB-dependent degradation pathway. In this assumption, msg2-1 proteins may have a higher affinity to AFBs than axr2-1 and slr-1 proteins.
Interestingly, expressions of axr2-1∷GFP and slr-1∷GFP under the control of the MSG2 promoter resulted in aberrant cotyledon development, which had never been observed in pMSG2∷msg2-1∷GFP plants or the msg2-1 mutant so far. Taken together, these results clearly show that msg2-1 functions differently from axr2-1 and slr-1, and there appear to be some differences between functions of axr2-1 and slr-1 even though they are most closely related with respect to their primary sequences (Remington et al., 2004).
Similar conclusions have been reached for the relationship between shy2/iaa3 and axr3/iaa17 (Knox et al., 2003) or bdl/iaa12 (Weijers et al., 2005). Transient expression of the shy2-6 and axr3-1 cDNAs by the same soybean (Glycine max) heat shock promoter reproduces the different root hair phenotype of the original mutant (Knox et al., 2003). On the other hand, expression of shy2-2 protein under the control of the SHY2 promoter inhibits gravitropism, auxin sensitivity, and auxin-induced gene expression in roots. However, expression of bdl protein under the control of the SHY2 promoter does not have these effects. Although embryonic defects are qualitatively determined by transcriptional regulation, there are clear quantitative differences in embryonic root initiation between shy2-2 and bdl action when driven by the BDL promoter: The shy2-2 protein is less effective than the bdl protein (Weijers et al., 2005). Although different properties are observed between mAux/IAA proteins that are stabilized by dominant mutations in domain II, these results suggest that the wild-type Aux/IAA proteins also have distinct functions as well as shared functions.
Weijers et al. (2005) found the promoter-dependent phenotype in embryo, hypocotyl, and shoot, and the protein-dependent phenotype in root development. In this study, we observed both the promoter-dependent and the expressed protein-dependent phenotypes in one organ, the hypocotyl. However, our finding is not surprising considering that hypocotyls consist of a few cell layers and a dozen cell types. Recently, Fukaki et al. (2005) investigated the slr defects in roots using tissue-specific promoters and succeeded in identifying the cell types in which slr functioned. Their results highlight the importance of knowledge on cells crucial for each auxin phenotype. In conclusion, it is necessary to know the sites of Aux/IAA action at the cell level, as well as the strength of the molecular interaction between Aux/IAAs and target ARFs or AFBs to further understand the molecular basis for specificity and similarity of the function of Aux/IAA proteins.
Cotyledons Develop Aberrantly When axr2-1∷GFP or slr-1∷GFP Is Expressed by the MSG2 Promoter
The pMSG2∷axr2-1∷GFP or pMSG2∷slr-1∷GFP plants with no cotyledons look very similar to the pin-formed1 (pin1) pinoid (pid) double mutants (Furutani et al., 2004), which develop pin-like inflorescences. However, vegetative and reproductive development of the transgenic plants is relatively normal. The transgenic plants with only one cotyledon are somewhat similar to the single cotyledonous phenotype sometimes observed in the mp single mutants or the nph4 mp double mutants (Hardtke et al., 2004), although these mutations also affect embryonic development of axes and roots. During the transition from the globular stage to the heart stage of the embryo, cotyledonary primordia are formed in places where auxin accumulates, owing to the function of the auxin efflux facilitator PIN1 (Friml et al., 2004) and PID kinase that appears to determine subcellular positioning of PIN1 (Benková et al., 2003; Jenik and Barton, 2005). Our present results suggest that ectopic expression of axr2-1 or slr-1 in the cotyledonary primordia interferes with auxin signaling and arrests growth and development of the cotyledon. The molecular targets of both mutated proteins might be NPH4 and MP.
It is interesting that neither pMSG2∷msg2-1∷GFP nor msg2-1 exhibits any defects in embryogenesis. msg2-1 might not be able to interact with MP as readily as axr2-1 or slr-1. However, the interaction between MSG2 and CTD of MP (MP-CTD) has been shown to be as strong as that between MSG2 and NPH4-CTD by yeast two-hybrid assay (Tatematsu et al., 2004) and fluorescence cross-correlation spectroscopy in HeLa cells (Muto et al., 2006). The interaction between MSG2 and NPH4-CTD appears strong enough to bring about tropistic defects in msg2-1 hypocotyls (Tatematsu et al., 2004). Apparently, full-length MP and NPH4 proteins must be used in further analyses of molecular interaction with Aux/IAA proteins.
We found that MSG2 was expressed in primordia of the cotyledon during the transition stage from globular to heart stage. The restricted expression of MSG2 in the primordia is in sharp contrast to the largely uniform, low-level expression of NPH4 and MP in the embryo (Hamann et al., 2002; Hardtke et al., 2004; Weijers et al., 2006). NPH4 and MP are expressed uniformly in subepidermal tissues of early embryos, being gradually confined to the vascular precursor cells. MSG2 expression in the embryo is also distinct from that of other Aux/IAA genes, BDL/IAA12, SHY2/IAA3 (Hamann et al., 2002; Weijers et al., 2006), and IAA13 (Weijers et al., 2005). The expression pattern of BDL is very similar to that of MP, while SHY2 is not expressed until the mid-torpedo stage. SHY2 is subsequently expressed in provascular cells. The distinct pattern of MSG2 expression in embryogenesis suggests that MSG2, in addition to functioning in tropistic responses in hypocotyls and lateral root formation (Tatematsu et al., 2004; Saito et al., 2007), also functions in the formation of cotyledons.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana) were first imbibed in water in the dark at 4°C for 3 d. They were surface sterilized with 1% hypochlorite, and sown on nutrient agar plates that contained half-strength Murashige and Skoog salts (Murashige and Skoog, 1962), 1% (w/v) Suc, and 1% (w/v) agar. Plants were grown at 23°C under continuous illumination at a fluence rate of 14 W m−2, obtained from three 40-W white fluorescent tubes (FL40SW; NEC). In some experiments, plants were grown on a 1:1 (v/v) vermiculite:Metromix 350 (Scotts-Sierra).
Gravitropism of hypocotyls was examined according to Nakamoto et al. (2006). In brief, seedlings were grown on vertically oriented agar plates for 3 d in the dark and then turned 90° to a horizontal position. An image of the seedlings was taken 15 h after the turn with a digital camera (C-4040 Zoom; Olympus), and hypocotyl curvature was measured with Image Pro-Plus (Media Cybernetics).
Transgenic Plants
For promoter-swapping experiments, 2-kb promoter regions of MSG2 and AXR2, which contained 5′ untranslated region, were amplified by PCR from DNA of wild-type Columbia using a pair of oligonucleotide primers: 5′-CACCTAAGAAACATGAGACATGTCACAA-3′ and 5′-GCATGCATATATATAGTCGACTTCTTGAACTTCTTTTTTTCCTCT-3′ for MSG2, and 5′-CACCGATCAAAACGGATCACAAAATTAA-3′ and 5′-GCATGCATATATATAGTCGACGTTACTTGTAATAGATTAGAAATA-3′ for AXR2. All the forward primers contained CACC sequence at the 5′ ends for directional TOPO-cloning (Invitrogen), and all the reverse primers contained the SalI and SphI sites at the 5′ ends. The PCR products were subcloned into pENTR/D-TOPO (Invitrogen). The sGFP gene (Niwa, 2003) was amplified from pGWB4 (Huang et al., 2006) by PCR using forward primers containing the SalI and SmaI sites at the 5′ end and reverse primers with the SphI site at the 5′ end. The PCR product was inserted between the SalI and SphI sites at the 3′ end of each promoter. The Aux/IAA cDNA that has a dominant mutation was amplified from total RNA prepared from msg2-1, axr2-1, and slr-1 by RT-PCR using a pair of oligonucleotide primers: 5′-GTCGACATGGAGAAGGAAGGACTCGGGCTT-3′ and 5′-GCATGCCTCGTCTACTCCTCTAGGCTGCAG-3′ for msg2-1, 5′-GTCGACATGATCGGCCAACTTATGAACCTC-3′ and 5′-GCATGCAGATCTGTTCTTGCAGTACTTCTC-3′ for axr2-1, and 5′-GTCGACATGAACCTTAAGGAGACGGAGCTT-3′ and 5′-GCATGCTGATCTGTTCTTGAACTTCTCCAT-3′ for slr-1. All the forward and reverse primers contained the SalI and SmaI sites at the 5′ ends, respectively. The PCR product was inserted between promoter and sGFP using the SalI and SmaI sites. The DNA fragment, promoter∷mAux/IAA cDNA∷GFP, was inserted in T-DNA of the gateway binary vector pGWB1 (Huang et al., 2006) using LR clonase (Invitrogen). The constructs were introduced into Agrobacterium tumefaciens strain pGV3101 by electroporation, which was then used to transform Arabidopsis Columbia ecotype by flower dip method (Clough and Bent, 1998).
Microscopy
For GFP analysis, hypocotyls and roots were counterstained with 10 μg mL−1 propidium iodide (Dojindo) and placed on slides in a drop of water. Fluorescence was imaged by a confocal microscope (LSM510; Zeiss). GFP was excited at the 488 nm laser line of a CW Ar+ laser, and propidium iodide was excited at the 543 nm laser line of a CW He-Ne laser through a water immersion objective (C-Apochromat, 40×, 1.2NA; Zeiss). Emission signals were detected at 500 to 530 nm for GFP and >560 nm for propidium iodide by sequential scanning.
For GUS staining, young siliques were fixed with ice-cold 50% acetone for 15 min. After rinse with 50 mm sodium phosphate, pH 7.0, thrice, ovules were taken off from siliques and stained for GUS activity by incubation in 50 mm sodium phosphate, pH 7.0, containing 2 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 10 mm K3Fe(CN)6, 10 mm K4Fe(CN)6, 1 mm EDTA, and 0.1% Triton X-100, at 37°C for 16 h. Then, they were rinsed with the above phosphate buffer and examined with a light microscope (Zeiss Axioplan) equipped with a digital camera (DXM1200; Nikon).
RT-PCR
Total RNA was prepared from 1-week-old seedlings grown under continuous white light using RNeasy Plant Mini kit (QIAGEN). After DNase treatment (RQ1 RNase-free DNase; Promega), RT was carried out using Moloney murine leukemia virus reverse transcriptase RNase H− (ReverTra Ace; Toyobo) with random primers. Quantitative PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) and a real-time PCR system (model 7300; Applied Biosystems). The PCR primers 5′-AAGCAGAAGAACGGCATCAAG-3′ and 5′-GGACTGGGTGCTCAGGTAGTG-3′ were used to amplify the transgene. ACTIN2 and 18S rRNA genes were used as internal controls with primers 5′-CGCTCTTTCTTTCCAAGCTCATA-3′ and 5′-CCATACCGGTACCATTGTCACA-3′, and 5′-ACGCGCGCTACACTGATGTA-3′ and 5′-TGATGACTCGCGCTTACTAGGA-3′, respectively. Essentially the same results were obtained by use of either control gene. Experiments were carried out for three independently prepared total RNA samples.
Acknowledgments
We thank Dr. T. Nakagawa (Shimane University) for pGWB1 and 4, and Ms. J. Iwasaki and M. Sasaki for technical assistance.
This work was supported in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (grant no. 17770026 to H.M. and grant no. 15370062 to M.K.) and from the Ministry of Education, Culture, Sports, Science and Technology (grant no. 17050001 to M.K. and grant no. 14036201 to K.T.Y.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hideki Muto (h-muto@imd.es.hokudai.ac.jp).
Some figures in this article are displayed in color online but in black and white in print.
Open Access articles can be viewed online without a subscription.
References
- Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435 441–445 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PBF, Ljung K, Sandberg G, et al (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M (2005) Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44 382–395 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29 153–168 [DOI] [PubMed] [Google Scholar]
- Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M (2004) PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131 5021–5030 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TG, Gray WM (2004) Auxin signal transduction. In PJ Davies, ed, Plant Hormones. Biosynthesis, Signal Transduction, Action! Kluwer, Dordrecht, The Netherlands, pp 282–303
- Hamann T, Benkova E, Baurle I, Kientz M, Jürgens G (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16 1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen T, Guilfoyle TJ, Berleth T (2004) Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131 1089–1100 [DOI] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E (2000) The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen J-G, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik PD, Barton MK (2005) Surge and destroy: the role of auxin in plant embryogenesis. Development 132 3577–3585 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis TIR1 protein is an auxin receptor. Nature 435 446–451 [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O (2003) AXR3 and SHY2 interact to regulate root hair development. Development 130 5769–5777 [DOI] [PubMed] [Google Scholar]
- Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, et al (2005) Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132 5387–5398 [DOI] [PubMed] [Google Scholar]
- Leyser O (2002) Molecular genetics of auxin signaling. Annu Rev Plant Biol 53 377–398 [DOI] [PubMed] [Google Scholar]
- Liscum E, Briggs WR (1995) Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7 473–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J, Ckurshumova W, Berleth T (2003) Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol 131 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Muto H, Nagao I, Demura T, Fukuda H, Kinjo M, Yamamoto KT (2006) Fluorescence cross-correlation analyses of molecular interaction between Aux/IAA protein and protein-protein interaction domain of auxin response factors of Arabidopsis expressed in HeLa cells. Plant Cell Physiol 47 1095–1101 [DOI] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, et al (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132 4107–4118 [DOI] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto D, Ikeura A, Asami T, Yamamoto KT (2006) Inhibition of brassinosteroid biosynthesis by either a dwarf4 mutation or a brassinosteroid biosynthesis inhibitor rescues defects in tropic responses of hypocotyls in the Arabidopsis mutant, non-phototropic hypocotyl 4. Plant Physiol 141 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y (2003) A synthetic green fluorescent protein gene for plant biotechnology. Plant Biotechnol 20 1–11 [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A (2001) IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-Y, Kim H-J, Kim J (2002) Mutation in domain II of IAA1 confers diverse auxin-related phenotypes and represses auxin-activated expression of Aux/IAA genes in steroid regulator-inducible system. Plant J 32 669–683 [DOI] [PubMed] [Google Scholar]
- Reed J (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6 420–425 [DOI] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135 1738–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O (1998) Changes in auxin response from mutations in an AUX/IAA gene. Science 279 1371–1373 [DOI] [PubMed] [Google Scholar]
- Saito K, Watahiki MK, Yamamoto KT (2007) Differential expression of the auxin primary-response gene MASSUGU2/IAA19 during tropic responses of Arabidopsis hypocotyls. Physiol Plant 130 148–156 [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Reed JW (1999) Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126 711–721 [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed J (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276 1865–1868 [DOI] [PubMed] [Google Scholar]
- Watahiki MK, Yamamoto KT (1997) The massugu1 mutation of Arabidopsis identified with failure of auxin-induced growth curvature of hypocotyl confers auxin insensitivity to hypocotyl and leaf. Plant Physiol 115 419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G (2005) Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jürgens G (2006) Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10 265–270 [DOI] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW (2005) NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43 118–130 [DOI] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M (1990) A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol Gen Genet 222 377–383 [DOI] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 21 553–562 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Yamamoto KT (1998) Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin-resistant mutant of Arabidopsis, aux1. Plant Cell Physiol 39 660–664 [DOI] [PubMed] [Google Scholar]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J 40 772–782 [DOI] [PubMed] [Google Scholar]