Abstract
TGA transcription factors are implicated as regulators of pathogenesis-related (PR) genes because of their physical interaction with the known positive regulator, nonexpresser of PR gene1 (NPR1). A triple-knockout mutant tga2-1 tga5-1 tga6-1 was shown previously to be defective in the induction of PR genes and systemic acquired resistance, confirming their role in disease resistance. However, the contributions of individual TGA factors have been difficult to discern because of functional redundancy among these factors, as well as possible dual functions for some single factors. In this study, we characterized six TGA factors by reverse genetics. We show that TGA3 is required for both basal and 2,6-dichloroisonicotinic acid-induced transcription of PR genes. The tga3-1 mutants were found to be defective in basal pathogen resistance, whereas induced resistance was unaffected. TGA1 and TGA4 play partially redundant roles in regulation of basal resistance, having only moderate effects on PR gene expression. Additionally, an activation-tagged mutant of TGA6 was able to increase basal as well as induced expression of PR1, demonstrating a positive role for TGA6 on PR gene expression. In contrast, TGA2 has repressor activity on PR gene expression even though it can act as a positive regulator in the tga5-1 tga6-1 null mutant background. Finally, we examined the genetic interaction between tga2-2 and suppressor of npr1 inducible1 (sni1-1). TGA2's repressor activity overlaps with SNI1 because the tga2-2 sni1-1 double mutant shows a synergistic effect on PR gene expression.
Induction of plant systemic acquired resistance (SAR) correlates with the expression of pathogenesis-related (PR) genes (Métraux et al., 1991; Ryals et al., 1996; Durrant and Dong, 2004). These genes, some of which encode proteins with antimicrobial activity (Van Loon and Van Strien, 1999), are believed to be the effectors of SAR, conferring broad-spectrum resistance to pathogens. In some plants, such as tobacco (Nicotiana tabacum), cucumber (Cucumis sativus), and Arabidopsis (Arabidopsis thaliana), an increase in salicylic acid (SA) production is essential for establishment of SAR (Gaffney et al., 1993). Treatment of these plants with SA or its functional analogs 2,6-dichloroisonicotinic acid (INA) and benzothiodiazole is sufficient to induce SAR (Görlach et al., 1996; Lawton et al., 1996). In Arabidopsis, SA-mediated gene expression and disease resistance have been shown to require the function of nonexpresser of PR gene1 (NPR1; Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). The npr1 mutants are not only impaired in SAR, but also show reduced basal resistance to pathogen infection (Cao et al., 1997; Delaney, 1997).
The NPR1 protein contains a BTB/POZ domain and an ankyrin repeat domain, which are known to be involved in protein-protein interaction (Cao et al., 1997; Aravind and Koonin, 1999). It was discovered recently that activation of NPR1 involves reduction of the protein, releasing it from a large oligomeric complex to translocate into the nucleus (Kinkema et al., 2000; Mou et al., 2003). Given the absence of a canonical DNA-binding domain, NPR1 was proposed to regulate PR gene expression as a cofactor of the TGA transcription factors, which interact with NPR1 in the yeast (Saccharomyces cerevisiae) two-hybrid system (Zhang et al., 1999; Despres et al., 2000; Zhou et al., 2000) and in planta (Subramaniam et al., 2001; Fan and Dong, 2002).
There are 10 TGA transcription factors in Arabidopsis (Jakoby et al., 2002) of which seven (TGA1–TGA7) have been characterized with respect to their interaction with NPR1. These seven TGAs can be further divided into three subgroups based on sequence homology: TGA1 and TGA4 comprise group I; TGA2, TGA5, and TGA6 belong to group II; and TGA3 and TGA7 make up group III (Xiang et al., 1997). TGA2, TGA3, TGA5, TGA6, and TGA7 constitutively interact with NPR1 in yeast and in planta when transiently expressed. Interestingly, the two TGA factors (i.e. TGA1 and TGA4) that showed no interaction with NPR1 in yeast were found to bind NPR1 only in SA-induced leaves. Reduction of two Cys residues that are uniquely present in TGA1 and TGA4 are responsible for this SA-dependent interaction (Despres et al., 2003).
To add to the complexity of TGA-regulated gene expression, linker-scanning (LS) mutagenesis of the PR1 promoter revealed both positive and negative regulatory cis-elements (Lebel et al., 1998). Mutation in the LS7 element that contains a TGA-binding site resulted in complete loss of gene induction, whereas mutation in another TGA-binding element (LS5) augmented gene expression. A second negative element (LS4) was found to contain a WRKY transcription factor-binding site (W box). Consistent with this result, some WRKY factors have been shown to negatively regulate pathogen-induced PR gene expression (Journot-Catalino et al., 2006; Wang et al., 2006; Xu et al., 2006; Zheng et al., 2007).
Previous studies using gel mobility shift and chromatin immunoprecipitation (ChIP) assays have shown that TGA factors bind to the as-1 element (Lam and Lam, 1995; Miao and Lam, 1995; Jupin and Chua, 1996; Johnson et al., 2003; Rochon et al., 2006). Increased binding to the as-1 element was observed with SA-induced plant extracts. Additionally, binding of in vitro-synthesized TGA2 to the as-1 element was enhanced in the presence of NPR1 (Despres et al., 2000), suggesting that NPR1 enhances or stabilizes the binding of TGA factors to the promoter. To further prove that the TGA factors are present in the DNA-bound complex, a supershift in gel mobility was shown using an antibody against TGA2 (Lam and Lam, 1995; Pontier et al., 2002). Moreover, depletion of TGA2 and TGA3 from nuclear extracts resulted in reduced protein binding to the as-1 element (Jupin and Chua, 1996; Zhou et al., 2000; Johnson et al., 2003). ChIP assays in SA-induced plants showed that antibodies against TGA2 and TGA3 preferentially precipitated the PR1 promoter sequences in an NPR1-dependent manner, confirming that these factors were recruited to the PR1 promoter in vivo (Johnson et al., 2003).
Several molecular approaches used to study TGA factors in plant defense have produced conflicting results. Overexpression of a dominant-negative mutant of TGA2 that competes with the binding of all TGAs to the target promoter showed increased expression of PR genes (Pontier et al., 2001; Fitzgerald et al., 2005). On the other hand, overexpression of TGA5 in Arabidopsis protected the plants against Hyaloperonospora parasitica NOCO2 infection without affecting PR gene expression (Kim and Delaney, 2002). More direct genetic evidence linking TGA factors to PR gene induction and resistance was only obtained when a triple-deletion knockout mutant, tga2-1 tga5-1 tga6-1, was generated in Arabidopsis and found to be defective in SAR and PR1 induction (Zhang et al., 2003). However, unlike npr1-1, the tga2-1 tga5-1 tga6-1 mutant does not show a defect in basal resistance (Zhang et al., 2003). In fact, the triple mutant has elevated basal expression of PR genes. This result suggests that, besides TGA2, TGA5, and TGA6, other TGA factors may be transcriptional activators of PR gene expression. Furthermore, TGA2, TGA5, or TGA6 may have dual activity. This is consistent with the results of the PR1 promoter study showing that there are at least two TGA-binding elements in this promoter, which, when mutated, have the opposite effect on transcription (Lebel et al., 1998).
In this study, we present genetic analysis of six TGA genes belonging to all three subgroups of this transcription factor gene family. We found that disruption of individual TGA4 and TGA7 did not significantly alter PR gene expression. Although the tga1-1 mutant showed a moderate decrease in INA-induced PR gene expression, it plays a significant role in regulating basal defense with partial dependence on TGA4. The tga3-1 single-knockout mutations significantly compromised expression of PR genes and displayed enhanced disease susceptibility (EDS). We also describe a novel function for TGA2 as a repressor of PR gene expression that appears to perform this function through genetic interaction with SNI1, another negative regulator of PR gene expression.
RESULTS
Isolation of T-DNA Insertion in Different TGA Family Members
To perform a comprehensive genetic study of TGA transcription factors, we screened for T-DNA insertion mutants in the NPR1-interacting TGA genes. To complement the work by Zhang et al. (2003) on tga2-1 tga5-1 (a single deletion of these adjacent genes) and tga2-1 tga5-1 tga6-1, we characterized a new allele of tga2 (tga2-2), which is wild type for TGA5, and an activation-tagged TGA6ACT allele that overexpresses the endogenous TGA6 gene. In addition, we found previously uncharacterized insertion mutants tga1-1, tga3-1, tga4-1, and tga7-1, which belong to the other two subgroups of the TGA gene family.
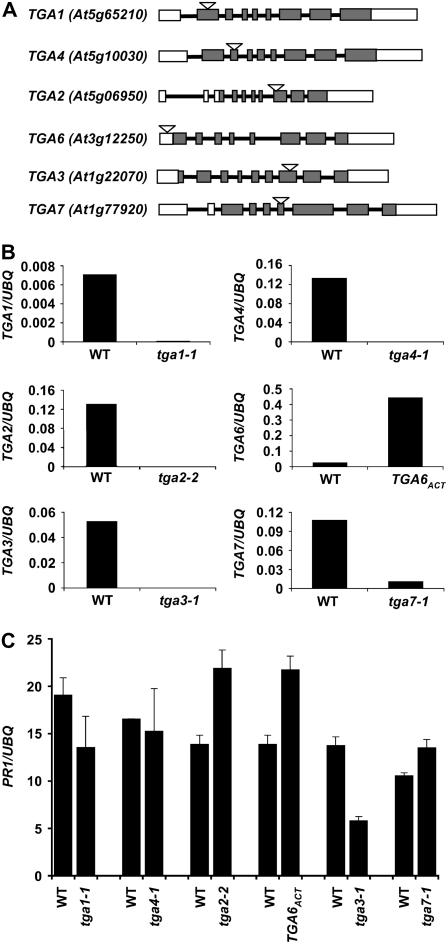
The tga1-1 and tga4-1 mutants were isolated from the SALK collection with T-DNA insertions in the first and second exons, respectively (Fig. 1A), causing complete loss of the respective transcripts (Fig. 1B). The tga2-2 and TGA6ACT homozygous lines were screened from the T-DNA-tagged population generated at the University of Wisconsin. As shown in Figure 1A, the T-DNA insertion in tga2-2 was found to be 1,000 nucleotides downstream of the start codon. Although no full-length cDNA could be amplified from the homozygous line (Fig. 1B), the region before the insertion site could be detected by reverse transcription (RT)-PCR (data not shown). Western blotting was then performed using a polyclonal antibody against the N terminus of TGA2 (Johnson et al., 2003), which confirmed that tga2-2 is a knockout mutant, producing no detectable full-length or truncated protein (Supplemental Fig. S1). The insertion in TGA6ACT is 100 nucleotides upstream of the start codon (Fig. 1A) and RT-PCR showed a 20-fold increase in TGA6 expression compared to wild type (Fig. 1B). Both tga2-2 and TGA6ACT mutants were backcrossed once with wild type and the resulting homozygous mutants in the F2 generation were selected and propagated for further studies. The tga3-1 T-DNA insertion was isolated from Thomas Jack's enhancer trap population. The insertion in TGA3 was 1,341 nucleotides downstream of the start codon (Fig. 1A) and no cDNA could be amplified from tga3-1, suggesting that it is a knockout mutant (Fig. 1B). The tga7-1 mutant was obtained from the GABI-Kat population where the insertion was found to be within the fourth exon. All of these mutants bred true in multiple genetic crosses. No apparent morphological defect affecting any developmental stage was observed in any of the single mutants studied.
Figure 1.
Characterization of tga T-DNA insertion mutants. A, Schematic representations of different tga T-DNA insertion mutations. Arabidopsis Genome Initiative numbers of the genes are shown in parentheses. Rectangular boxes represent the exons, whereas the interconnecting lines represent the introns. White rectangles show the untranslated regions in the genes. The T-DNA insertion site is represented as a triangle above each gene. B, Real-time RT-PCR analysis of TGA gene expression in the mutants. RNA was extracted from 2-week-old seedlings grown on a Murashige and Skoog plate. The individual bar diagrams shows the relative amount of the transcript of the corresponding TGA gene in the wild type and mutant normalized to the levels of a UBQ gene. The normalized values were multiplied by 1,000 for TGA2, TGA3, TGA4, and TGA7 and by 10 for TGA4. C, Expression of PR1 in each tga mutant was determined from 2-week-old seedlings harvested from Murashige and Skoog plates containing 20 μm INA using real-time RT-PCR. The y axis values are normalized to UBQ5 expression. Error bars represent se. The experiments were repeated at least three times with similar results. WT, Wild type; UBQ, ubiquitin.
PR Gene Expression in tga Mutants
To evaluate the role of individual TGA factors in PR gene expression, we tested a range of INA concentrations (2–50 μm) for induction of these genes. Plants with the Wassilewskija (Ws-0) genotype showed stress symptoms at INA concentrations higher than 20 μm. At 20 μm INA, significant differences in PR1 gene expression were detected in tga2-2 and TGA6ACT (both are in Ws-0) using real-time RT-PCR (Fig. 1C). We observed significant differences in PR gene expression in tga3-1 mutants at all the concentrations tested (Fig. 1C; Supplemental Fig. S2A). The tga1-1 mutant showed a small decrease in PR1 expression. However, no clear-cut differences were seen in tga7-1 and tga4-1 in any of the INA concentrations tested. Contrary to previous reports (Fan and Dong, 2002; Zhang et al., 2003; Kang and Klessig, 2005), which suggest that TGA2 is a transcriptional activator of PR genes, the tga2-2 knockout mutant showed higher expression of PR1 in response to induction (Fig. 1C). This result suggests that, in the presence of other TGA factors, TGA2 is a transcriptional repressor of PR genes. The TGA6ACT plants showed an increase in PR1 expression as compared to wild type, indicating that TGA6 is a transcriptional activator of PR1 as previously reported (Zhang et al., 2003). Disruption of TGA3 in the tga3-1 mutant showed >50% decrease in the expression of PR1 as compared to wild type. Similar results were obtained with SA induction of tga3-1 plants (Supplemental Fig. S2B). A prominent role of TGA3 as a transcription activator of PR genes is consistent with the fact that TGA3 is the strongest interactor of NPR1 (Zhou et al., 2000) and is the most abundantly expressed TGA gene in leaves (Zimmermann et al., 2004), where PR genes are expressed. INA responsiveness of other PR genes (PR2 and PR5) was also examined in tga2-2, tga3-1, and TGA6ACT, and they were found to have an expression pattern similar to PR1 (Supplemental Figs. S2C and S3).
TGA3 Is Required for Basal Resistance and PR Gene Expression
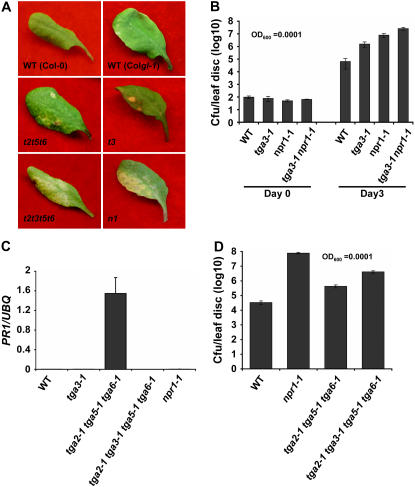
To examine the effect of the tga3-1 mutation on disease resistance, plants were infected with a low inoculum (OD600 = 0.0001) of Pseudomonas syringae pv maculicola ES4326 (Psm4326). At this level of inoculum, wild-type plants and mutants with enhanced (EDS) can be easily distinguished. In the EDS test shown in Figure 2A, wild-type plants did not show any disease symptoms, whereas npr1-1 plants showed clear chlorosis. The tga3-1 plants sporadically displayed disease symptoms. The tga3-1 mutants were then analyzed for growth of Psm4326 in leaves. As shown in Figure 2B, wild-type plants showed limited growth of Psm4326, whereas the npr1-1 mutant had substantial growth of this pathogen 3 d after inoculation. The tga3-1 mutant, in comparison, had a bacterial titer significantly higher than wild type, but lower than the npr1-1 mutant. Interestingly, the tga3-1 npr1-1 double mutant had even higher pathogen growth compared to either npr1-1 or tga3-1 alone. These results clearly demonstrate that TGA3 is required for basal defense and that the function of TGA3 is not completely dependent on NPR1.
Figure 2.
TGA3 is a positive regulator of PR1 expression and basal resistance. A, Disease symptoms from representative leaves of 4-week-old plants. Infections were carried out using P. syringae pv maculicola ES4326 OD600 = 0.0001. Photographs were taken 3 d postinfection. The experiment was repeated three times with similar results. WT, Wild type; t3, tga3-1; n1, npr1-1; t2t5t6, tga2-1 tga5-1 tga6-1; t2t3t5t6, tga2-1 tga3-1 tga5-1 tga6-1. B, Bacterial titers in the leaves of 4-week-old plants infiltrated with Pseudomonas as described in A. Samples were collected at 0 and 3 d postinfection. Values were averaged from eight samples per genotype and error bars represent se. The experiment was repeated at least three times with similar results. WT, Wild type. C, Basal level expression of PR1 in wild type, tga3-1, tga2-1 tga5-1 tga-6-1, tga2-1 tga3-1 tga5-1 tga6-1, and npr1-1 in the absence of INA. RNA was extracted from 2-week-old seedlings grown on Murashige and Skoog plates. Transcript levels were quantified by real-time RT-PCR and y axis values were normalized to UBQ5 expression. Error bars represent se from three PCR reactions. The experiment was repeated at least three times with similar results. UBQ, Ubiquitin. D, Bacterial titers in the leaves of 4-week-old plants infiltrated with P. syringae pv maculicola ES4326 OD600 = 0.0001. Samples were collected 3 d postinfection. Values were averaged from 16 samples per genotype and the error bars represent se. The experiment was repeated at least three times with similar results.
It has been shown previously that the tga2-1 tga5-1 tga6-1 triple mutant fails to establish SAR similar to npr1 (Zhang et al., 2003). However, unlike npr1, the triple mutant has elevated background expression of PR genes, accompanied by intact basal resistance to Psm4326. We hypothesize that this is due to the activity of TGA3. To test this hypothesis, the tga2-1 tga5-1 tga6-1 triple mutant was crossed with tga3-1 to generate a quadruple-knockout mutant. As shown in Figure 2C, the tga3-1 mutation in the quadruple mutant completely blocked the basal expression of PR1 observed in the tga2-1 tga5-1 tga6-1 triple mutant. Furthermore, the EDS test showed that the quadruple mutant developed symptoms similar to those observed in npr1, whereas wild type and tga2-1 tga5-1 tga6-1 did not show any symptoms (Fig. 2A). However, when in planta bacterial growth was quantified, the quadruple mutant showed a bacterial titer higher than those in tga3-1 and the triple mutant, but lower than the titer observed in npr1-1 (Fig. 2D). These results further establish the importance of TGA3 in the regulation of basal PR gene expression and resistance. Moreover, TGA3 also seems to be involved in development of disease symptoms.
The tga1-1 tga4-1 Double Mutant Is Compromised in Resistance to Pseudomonas Infection
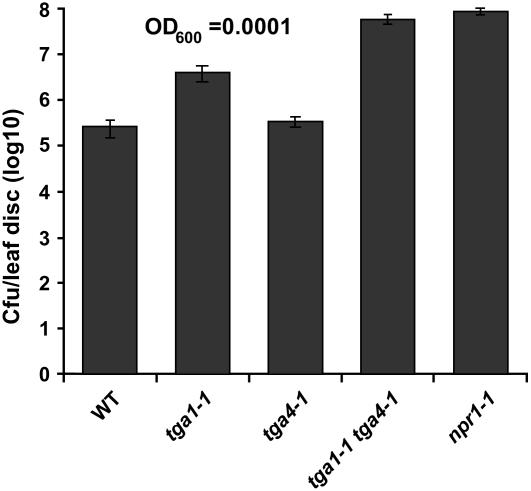
It has been shown previously that the reduced forms of TGA1 and TGA4 can interact with NPR1 (Despres et al., 2003), suggesting an important role for this subgroup of TGA factors in SAR. However, we only observed a small decrease in PR1 expression in the tga1-1 mutant in response to INA induction (Fig. 1C). Expecting that this was due to functional redundancy, we generated the tga1-1 tga4-1 double mutant through a genetic cross. Still, PR1 expression in response to INA in the double mutant was comparable to the tga1-1 single mutant. Similarly, the double mutant showed only a marginal effect on induced resistance (data not shown). To further assess the susceptibility of the tga1-1 tga4-1 mutant, we carried out an EDS test using Psm4326 as described in the previous section. As shown in Figure 3, the tga1-1 mutant supported significantly higher pathogen growth than the wild type, whereas the tga4-1 mutant seemed to be similar to the wild type. Even greater susceptibility was seen in the tga1-1 tga4-1 double mutant, which is similar to npr1-1 (Fig. 3). These results demonstrate that TGA1 and TGA4 positively regulate basal resistance. Additionally, TGA1 and TGA4 are partially redundant, with TGA1 having a greater effect than TGA4.
Figure 3.
tga1-1 tga4-1 mutants are compromised in basal resistance. Bacterial titers in the leaves of 4-week-old plants infiltrated with P. syringae pv maculicola ES4326 OD600 = 0.0001. Samples were collected 3 d postinfection. Values were averaged from 16 samples per genotype and error bars represent se. The experiment was repeated at least three times with similar results. WT, Wild type.
TGA6 Can Activate PR Gene Expression in tga2-2
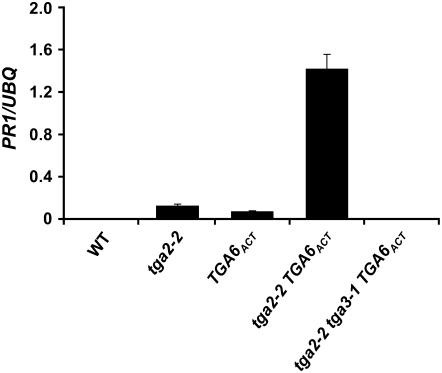
In the TGA6ACT mutant, the T-DNA insertion upstream of the regulatory region of TGA6 causes overexpression of this gene (Fig. 1B). Although this does not lead to constitutive expression of PR1, INA-induced PR1 transcription is significantly enhanced (Fig. 1C). This suggests that TGA6 activity may not be constitutive and may require an additional activation step. Alternatively, removal of a negative regulator may be required for TGA6 to perform its function as a constitutive transcriptional activator. To test this hypothesis, we crossed TGA6ACT into the tga2-2 background where the TGA2 function is knocked out. Indeed, the tga2-2 TGA6ACT double mutant showed dramatically increased basal PR1 expression compared to the TGA6ACT and the tga2-2 single mutants (Fig. 4). This result shows that TGA6 is a constitutive transcriptional activator of PR1 and its activity is repressed in the presence of TGA2. Because we found TGA3 to be the major transcriptional activator of PR genes, we tested whether the activator function of TGA6 requires TGA3. We generated the tga2-2 tga3-1 TGA6ACT triple mutant as described in the experimental procedures. Because tga2-2, TGA6ACT, and tga3-1 are in two different ecotypes, we analyzed multiple progeny from each of the crosses to ensure that the observed phenotypes were from these mutations, not from mixing the genetic backgrounds. As expected, in the absence of TGA3, basal PR1 expression observed in the tga2-2 TGA6ACT double mutant was completely abolished (Fig. 4). In the tga2-2 tga3-1 TGA6ACT triple mutant, INA-induced PR gene expression was also partially compromised compared to the double mutant, likely due to the absence of functional TGA3 (Supplemental Fig. S4).
Figure 4.
Overexpression of TGA6 leads to activation of PR1 expression in tga2-2. RNA was extracted from 2-week-old seedlings grown on Murashige and Skoog plates. Transcript levels were quantified by real-time RT-PCR and y axis values were normalized to UBQ5 expression. Error bars represent se from three PCR reactions. The experiment was repeated at least three times with similar results. WT, Wild type; UBQ, ubiquitin.
TGA2 Acts Synergistically with SNI1 as a Transcriptional Repressor of PR Genes
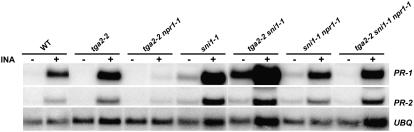
The enhanced PR gene expression observed in the tga2-2 knockout mutant suggests that TGA2 is a transcriptional repressor. In addition to TGA2, SNI1 is another negative regulator of PR genes (Li et al., 1999; Mosher et al., 2006). To determine whether there is any genetic interaction between these two negative regulators, we crossed tga2-2 with sni1-1. Interestingly, the tga2-2 sni1-1 double-homozygous plants were underrepresented in the F2 population (observed 1:100 rather than the expected 1:16 ratio; χ2 = 4.41; P > 0.5), suggesting that plant viability is compromised by the presence of both mutations. We analyzed the expression of PR1 and PR2 in the surviving tga2-2 sni1-1 double-homozygous plants by northern-blot analysis (Fig. 5). As reported previously, the sni1-1 single mutant had elevated background expression of PR1. Interestingly, the level of PR1 expression in the tga2-2 sni1-1 double mutant was dramatically elevated compared to the sni1-1 single mutant, resulting in expression levels that were similar to INA-induced wild-type plants. Expression of PR2 was similarly induced in the double mutant. To exclude the possibility that high PR gene expression is due to mixing of Ws and Columbia (Col) ecotypes, we analyzed multiple tga2-2 sni1-1 plants from independent crosses. All of the tga2-2 sni1-1 double-mutant progeny tested had dramatically increased PR gene expression (data not shown). The synergistic effect of these two mutations suggests that TGA2 and SNI1 have overlapping activities as negative regulators of PR genes. The PR1 transcript level in tga2-2 sni1-1 was induced further after INA treatment, suggesting that, although TGA2 and SNI1 are the predominant repressors for this gene, other factors may also be involved. In the absence of TGA2 and SNI1, transcription of PR1 seems to be partially dependent on NPR1 because in the tga2-2 sni1-1 npr1-1 triple mutant, the level of PR1 expression is significantly reduced in the presence or absence of INA (Fig. 5).
Figure 5.
TGA2 is a negative regulator of PR gene expression. Northern blot showing expression of PR1 and PR2 in tga2-2 sni1-1 and tga2-2 sni1-1 npr1-1 mutants. Total RNA was extracted from 2-week-old seedling grown on Murashige and Skoog agar plates with (+) or without (−) INA (20 μm). PR1 and PR2 expression levels were analyzed by hybridizing an RNA blot with respective gene-specific probes. A UBQ5-specific probe was used to hybridize the same blot as a loading control. WT, Wild type; UBQ, ubiquitin.
Surprisingly, the high levels of PR gene expression in the tga2-2 sni1-1 are not sufficient to confer resistance against Psm4326 (data not shown), indicating that activation of additional genes is required. Recently, we discovered that for the PR proteins to be effectively folded and secreted outside the cell, genes involved in protein folding, modification, and secretion have to be coordinately up-regulated during SAR (Wang et al., 2005). A mutation in one of these genes, BiP2, not only compromised SAR, but also caused massive cell death upon SAR induction due to accumulation of unfolded PR proteins. This study also showed that induction of the protein secretory pathway genes requires NPR1, but not TGA factors. We tested expression of two of the genes (BiP2 and GRP94) that are required for secretion of PR proteins in the tga2-2 sni1-1 double mutant. Real-time RT-PCR showed that the expression of BiP2 and GRP94 were not significantly changed in the tga2-2 sni1-1 mutant even though the PR gene levels were extremely high (Supplemental Fig. S5). This may explain the lack of enhanced resistance and low viability of the double mutant. In support of this hypothesis, viability was restored in the tga2-2 sni1-1 npr1-1 triple mutant because the triple mutant was obtained at the expected frequency (observed 1/72; expected 1/64).
DISCUSSION
PR1 expression is a paradigm for the coregulation of PR genes during SAR (Eulgem, 2005); therefore, understanding the regulation of this gene can help us understand SAR. The PR1 promoter contains both negative and positive regulatory elements, the importance of which has been shown through LS mutagenesis (Lebel et al., 1998). Molecular studies of TGA factors also strongly suggest differential effects of TGAs in regulating PR gene expression. However, activator or repressor functions are difficult to assign to individual TGA factors due to functional redundancy and lack of knockout mutants in all of the TGA genes. Through our comprehensive characterization of single, double, triple, and quadruple mutants of TGA genes, we clearly revealed the functions of TGA1, TGA2, TGA3, TGA4, and TGA6.
Characterization of the tga3-1 knockout mutant showed that this transcription factor is an important activator for both basal and induced expression of PR genes (Figs. 1C, 2C, and 4). This is consistent with the fact that TGA3 is one of the most highly expressed TGA genes in rosette leaves (Zimmermann et al., 2004) and is the strongest interactor of NPR1 (Zhou et al., 2000). Although the tga1-1 mutation only had a moderate effect on INA-induced PR1 expression (Fig. 1C), it significantly impaired the plant's basal resistance to Pseudomonas infection (Fig. 3). TGA1 and TGA4 may have functional redundancy because the double mutant showed an additive effect on bacterial growth. These data indicate that the TGA1 and TGA4 subgroup of TGA factors is critical to basal resistance, with TGA1 playing a more dominant role. We were unable to find a consistent phenotype for the tga7-1 mutant under the conditions tested. It is possible that TGA7 plays a redundant role with TGA3 because it belongs to the same subgroup.
Our study of the TGA6ACT mutant is consistent with that of Zhang et al. (2003), where TGA6 is a transcription activator of PR genes. Interestingly, we found that overexpression of TGA6 in TGA6ACT is not sufficient to cause a significant increase in PR gene expression in the absence of an SAR inducer (Fig. 4). Rather, TGA6 activity requires removal of TGA2 because only in the tga2-2 knockout background did overexpression of TGA6 result in a significant increase in PR gene expression. Additionally, in the absence of TGA2, TGA6 may also be able to replace the activator function of TGA2. Moreover, this activity is dependent on TGA3 because in the tga2-2 tga3-1 TGA6ACT mutant, basal PR gene expression is abolished.
Studies carried out by Fan and Dong (2002) using a chimeric TGA2-GAL4 fusion protein showed that TGA2 can act as an activator of a GAL4-mediated reporter gene in an NPR1-dependent manner. The role of TGA2 as a transcription activator was also shown by the loss of PR1 induction in tga2-1 tga5-1 tga6-1. When the wild-type TGA2 gene was transformed back into this triple mutant, the inducibility of PR genes was restored (Zhang et al., 2003). This indicated that, in the absence of TGA5 and TGA6, TGA2 can be a transcription activator. In contrast, studies with dominant-negative mutants of the TGA2 homologs in tobacco and rice (Oryza sativa) showed a negative role for TGA2 in PR gene expression and disease resistance (Pontier et al., 2001; Fitzgerald et al., 2005; Thurow et al., 2005). However, the role of Arabidopsis TGA2 alone was previously unknown because the tga2 single-knockout mutant was not available. Interestingly, we observed that, in the tga2-2 single mutant, both the basal and the induced levels of PR1 expression were moderately increased (Figs. 1C and 4). The phenotype of tga2-2 was synergistically enhanced in the sni1-1 mutant background (Fig. 5). To reconcile the negative role of TGA2, we hypothesize that TGA2 may function both as a transcription activator and as a repressor. The dual function of TGA2 could be achieved by any of several mechanisms. First, it may function as an activator or a repressor depending on which of the two as-1-type elements it binds in the PR1 promoter. TGA2 has been shown to bind to both the negative (LS5) as well as positive (LS7) TGA-binding element in the PR1 promoter (Despres et al., 2000). Contrary to the previous report, a recent publication (Rochon et al., 2006) showed constitutive recruitment of TGA2 to the PR1 promoter by ChIP. This result supports our hypothesis that TGA2 may be present at the promoter to repress transcription before induction. However, the low resolution of the ChIP experiments did not distinguish TGA2 binding between the two TGA-binding elements in the PR1 promoter. Some transcription factors have been known to have dual activities. One such example is the tumor suppressor protein p53 whose activation and repression activities largely depend on the specific sequence it binds (Marks et al., 2003). The second possible mechanism to explain the dual functions of TGA2 is through association with other trans-acting factors or the basal transcription machinery. A third mechanism for the dual functions is through posttranslational modification. A good example is the SP3 transcription factor, which acts as a repressor of the transforming growth factor-β receptor type II gene when unmodified whereas acetylated SP3 acts as a transcriptional activator (Ammanamanchi et al., 2003). Phosphorylation has also been shown as a repressor-to-activator switch for c-jun, a bZIP transcription factor (Lamph et al., 1990). The unphosphorylated c-jun is part of the histone deacetylase inhibitory complex and, upon phosphorylation, c-jun dissociates from the histone deacetylase repressor complex and acts as an activator (Weiss et al., 2003). The role of phosphorylation was also proposed in inhibiting the binding of TAG1a to the as-1 element in tobacco (Jupin and Chua, 1996). A recent study showed that TGA2 is phosphorylated upon SA activation (Kang and Klessig, 2005). Even though mutating the putative phosphorylation site in TGA2 did not affect its binding activity to the LS7 element, it remains to be tested whether it affects binding to the negative (LS5) element.
The synergistic effect of tga2-2 and sni1-1 mutations and the involvement of WRKY factors on PR gene expression suggest the presence of multiple negative regulators with overlapping functions. This effect of the tga2-2 sni1-1 double mutant on PR genes could not be explained by replacement of TGA2 function by TGA6 because the TGA6ACT sni1-1 mutant did not show the similar effect as tga2-2 sni1-1 on the expression of PR genes (data not shown). Although we have not detected any direct physical interaction between TGA2 and SNI1 (data not shown), the genetic interaction of these two factors could be at the level of chromatin modification. The SNI1 has been shown to be a nuclear protein with significant structural similarities to armadillo repeats that are present in many regulatory proteins, such as armadillo and β-catenin. Moreover, loss of SNI1 function leads to chromatin modifications at the PR1 promoter that mimic SAR induction (Mosher et al., 2006). Because it is known that NPR1 resides predominantly in the cytosol before induction, it is intriguing and interesting that NPR1 is required for the high level of PR1 expression seen in tga2-2 sni1-1. To understand this phenomenon, we first measured the SA levels in the tga2-2 sni1-1 plants and found them to be similar to wild type. We then measured reduced glutathione levels in the sni1-1 and tga2 sni1-1 double mutant because reducing conditions cause monomerization and nuclear localization of NPR1. We detected high levels of reduced glutathione and constitutive nuclear localization of NPR1 in the sni1-1 mutant (Z. Mou and X. Dong, unpublished data). The levels of reduced glutathione remained high in the tga2-2 sni1-1 double mutant, which can explain the requirement of NPR1 for the high levels of PR gene expression observed in this mutant. The nuclear NPR1 may recruit positive TGA factors, such as TGA3 and TGA1, to the PR1 promoter in the absence of the negative regulators. Interestingly, the sni-1 tga2-2 double mutant is further inducible by INA, which is probably because nuclear-localized NPR1 needs further activation to be fully functional. This is consistent with the recent study by Rochon et al. (2006).
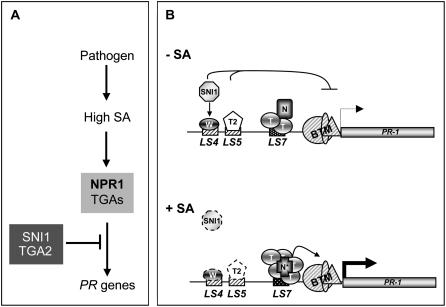
This study has defined the transcriptional activities of five TGA transcription factors in PR gene expression and disease resistance. It also revealed genetic interactions between these factors and other known regulators of PR genes. TGA4 and TGA7 may still be important in the regulation of PR gene expression and disease resistance under conditions not tested here. Based on the information generated in this study and previous work, we propose the following model (Fig. 6A): NPR1, in association with TGA factors, is required for basal and induced PR gene expression. In the absence of an inducer, TGA2 and SNI1 are responsible for repression of PR genes. As illustrated in Figure 6B, the PR1 promoter contains both positive (LS7) and negative (LS4 and LS5) cis-elements. Before induction, TGA2 and SNI1 repress basal expression through direct and indirect interactions, respectively, with the negative elements. We do not rule out constitutive binding of transcription activators such as TGA1, TGA3, and TGA6 to the positive element in the uninduced state. But this does not lead to gene expression in the presence of TGA2 and SNI1. Upon induction, TGA2 and SNI1 repression is removed through an unknown mechanism and transcription is activated by NPR1 in association with the positive TGA transcription factors, including TGA1, TGA2, TGA3, TGA5, and TGA6. Further biochemical studies are required to understand the mechanism by which TGA2 represses transcription.
Figure 6.
Proposed model for TGA transcription factors in the regulation of PR1. A, Flow chart showing the genetic interactions between key regulatory components in the regulation of PR gene expression. Arrows represent activation and blocked arrows indicate repression. B, Mechanistic model showing recruitment of different factors under uninduced (−SA) and induced (+SA) conditions on the PR1 promoter. T, TGA transcription factors (TGA1–TGA6); T2, TGA2; N, NPR1; N*, activated NPR1; W, WRKY transcription factor; BTM, basal transcription machinery. The negative cis-regulatory elements, LS5 and LS4, are shown as hatched boxes and the positive element, LS7, is shown by a dotted rectangle. The dotted trans-acting factors represent an inactive form. Thickness of the arrow represents the level of expression and the blocked arrow represents repression.
MATERIALS AND METHODS
Plant Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown either on soil (Metro-Mix 200; Grace Sierra) or on Murashige and Skoog medium (Murashige and Skoog, 1962) containing 2% Suc and 0.8% agar under conditions described in Clarke et al. (1998).
Isolation of TGA T-DNA Insertion Mutants
The tga2-2 and TGA6ACT T-DNA insertion alleles were isolated from the Ws-0 ecotype of Arabidopsis. The insertion was identified by PCR screening of pooled genomic DNA using the protocol provided by the Arabidopsis gene knockout research facility at the University of Wisconsin (Krysan et al., 1999). The tga2-2 and TGA6ACT homozygous plants were obtained by PCR screening the progeny using two gene-specific primers flanking the T-DNA insertion and a primer complementary to the T-DNA left-border sequence. Sequencing of the PCR product from the left border revealed the exact insertion site of the T-DNA. The primer pairs used for PCR are shown in Table I. The sequences of the primers are described in Table II. Plants in which a PCR product was detected using one gene-specific primer and the T-DNA primer were identified and homozygosity for the insertion mutation was confirmed by the lack of segregation in the next generation. The tga3-1 mutant was identified in Thomas Jack's T-DNA-tagged population (http://www.arabidopsis.org/abrc/jack.jsp) made in the Col-gl1 ecotype by performing three rounds of PCR with pooled genomic DNA. The exact insertion site was determined by sequencing the region flanking the T-DNA. Homozygous tga3-1 plants were obtained by genomic Southern blotting. The T-DNA insertion was detected by the presence of an extra HindIII site in the T-DNA. Southern blotting was carried out using a standard protocol and the full-length TGA3 coding region was used to make the probe. The tga7-1 T-DNA insertion mutant (no. 434F04) was obtained from German plant genomic research plan Kolner Arabidopsis (GABI-Kat; Rosso et al., 2003). The homozygosity of the insert was confirmed by the absence of the wild-type PCR product using gene-specific primers (Table I). The tga1-1 and tga4-1 mutants were identified in the T-DNA insertion collection from the SALK Institute Genomic Laboratory (http://signal.salk.edu). T3 progeny (SALK_082821 and SALK_127923 for tga1-1 and tga4-1, respectively) were analyzed individually by PCR. Insertion was identified as described above for tga2-2 using the T-DNA left-border primer and gene-specific primers (Table I).
Table I.
Summary of primer pairs used for genotyping and RT-PCR
Primer pairs listed in the first column were used to amplify either the genomic DNA or the cDNA from wild-type and mutant plants. The gene names are given before the kind of template used for amplification.
| Primer Pair | Amplification |
|---|---|
| TGAKOF + TGAKOR | TGA1 wild type genomic |
| LBb1 + TGA1KOF | TGA1 T-DNA insertion |
| KOTGA2 + TGA2RKO | TGA2 wild type genomic |
| KOTGA2 + JL202 | TGA2 T-DNA insertion |
| TGA4KOF + TGA4KOR | TGA4 wild type genomic |
| LBb1 + TGA4KOF | TGA4 T-DNA insertion |
| KOT6 + TGA6RKO | TGA6 wild type genomic |
| JL202 + TGA6RKO | TGA6 T-DNA insertion |
| TGA7F + TGA7LCR | TGA7 wild type genomic |
| TGA1LCF + TGA1LCR | TGA1 cDNA |
| TGA2LCF + TGA2LCR | TGA2 cDNA |
| TGA3LCF + TGA3LCR | TGA3 cDNA |
| TGA4LCF + TGA4LCR | TGA4 cDNA |
| TGA6LCF + TGA6LCR | TGA6 cDNA |
| TGA7LCF + TGA7LCR | TGA7 cDNA |
Table II.
Sequences of primers used in this study
| Gene | Primer Name | Primer Sequence (5′-3′) |
|---|---|---|
| TGA1 | TGA1LCF | GGATTTCGACCCTCCG |
| TGA1 | TGA1LCR | GGCTCTAAGCCGTTGA |
| TGA1 | TGA1TF | TCTTCGAAGAATTTGGCGAAGA |
| TGA1 | TGA1TR | TTCCTGCTGTTCCATGGGAAGTAT |
| TGA2 | TGA2LCF | GACGACACAGATCATCCT |
| TGA2 | TGA2LCR | TCAAGCGGCTGTTCTC |
| TGA2 | TGA2RKO | CGGATGAACGAAATCCACCGA |
| TGA2 | KOTGA2 | ATCAAGCCCTTTACTTGTGCACCTTCAAG |
| TGA3 | TGA3LCF | GGGAATGTGGCGAACT |
| TGA3 | TGA3LCR | CTTTCGACCAAACCCTG |
| TGA4 | TGA4KOF | GTTCCACCGAGAAGGTTTG |
| TGA4 | TGA4KOR2 | TATCACTAACTTGTCCATGT |
| TGA4 | TGA4LCF | ATGCGTTATCCCAAGG- |
| TGA4 | TGA4LCR | TGTCGCGTGGTTAAGAT |
| TGA6 | TGA6LCF | AGTTGGATCAAAAGACCCT |
| TGA6 | TGA6LCR | CCATTGCCACCAGTAG |
| TGA6 | TGA6RKO | GACTATTCTCCAGCTGCTGAACA |
| TGA6 | KOT6 | GGACTGATGTCTCAACTGATGGTGACACAG |
| TGA7 | TGA7F | GTCCCAATACTGCTACTTCCTC |
| TGA7 | TGA7LCF | CGTTTAGCGCAGAAC |
| TGA7 | TGA7LCR | CTTGTAGCCAGTGTGAAT |
| TGA7 | TGA7LCR | CTTGTAGCCAGTGTGAAT |
| NPR1 | NPR1F | GATCATAAGGCACTTGACTCG |
| NPR1 | NPR1R | ATGAGTGCGGTTCTACCTTC |
| PR1 | PR-1LCF | CTCATACACTCTGGTGGG |
| PR1 | PR-1LCR | TTGGCACATCCGAGTC |
| SNI1 | sni1F | TGGTTTTGTTTTGCAGGCTTGGTCACCATT |
| SNI1 | sni1R | TCTTGCTCATGTTCTTAAGCTAGATTTCAC |
| UBQ5 | UBQLCF | GACGCTTCATCTCGTCC |
| UBQ5 | UBQLCR | GTAAACGTAGGTGAGTCCA |
Generation of Double, Triple, and Quadruple Mutants
Because tga3-1 was generated in the Col ecotype containing the recessive glabrous mutation (gl1), it was used as female in all of the crosses, from which the F1 progeny would be distinguishable from the tga3-1 parent by the presence of trichomes. To obtain tga3-1 npr1-1 and tga2-2 tga3-1, tga3-1 was crossed with npr1-1 and tga2-2, respectively. F1 plants (with trichomes) were allowed to self-fertilize and double mutants were screened from the F2 population by identifying tga2-2 homozygous T-DNA insertion using PCR as described above. The npr1-1 allele was also confirmed by PCR using the cleaved-amplified polymorphic sequence marker primers described in Table II, followed by digestion with NlaIII. The tga3-1 mutation was screened using Southern hybridization as described in the previous section. To create the tga1-1 tga4-1 double mutant, tga4-1 plants were used as pollen donors and F1 plants were analyzed for the presence of the TGA4 T-DNA allele; the successful F1 plants were self-fertilized to produce F2 seeds. These seeds were planted and screened for the homozygous tga1-1 and the tga4-1 allele by PCR using the T-DNA left-border primer and gene-specific primers to TGA1 and TGA4. Multiple homozygous plants were recovered and used for further analysis. To make the tga2-2 TGA6ACT double mutant, tga2-2 was used as the pollen donor and the F1 plants were identified by the presence of a band using the TGA2 gene-specific primer and the T-DNA left-border primer. The F2 progeny were then generated through self-fertilization and screened for T-DNA insertion at both TGA2 and TGA6 loci as described above. To obtain tga2-2 tga3-1 TGA6ACT, TGA6ACT was crossed with the tga2-2 tga3-1 double mutant and the F2 progeny containing homozygous mutation at TGA2, TGA3, and TGA6 loci were screened as described above. To generate the tga2-1 tga3-1 tga5-1 tga6-1 quadruple mutant, tga3-1 was crossed with the tga2-1 tga5-1 tga6-1 triple mutant (Zhang et al., 2003). The tga2-1 tga5-1 deletion mutation in the progeny was obtained by lack of the PCR product using gene-specific primers for TGA2. Similarly, the tga6-1 deletion was confirmed by lack of the PCR product using the TGA6 gene-specific primer. To obtain tga2-2 sni1-1 and tga2-2 sni1-1 npr1-1, tga2-2 pollen was used to fertilize sni1-1 and sni1-1 npr1-1 plants, respectively. The recessive sni1-1 mutation has a pleiotropic effect on plant morphology; therefore, wild-type-looking F1 plants were progeny of successful crosses and were allowed to self-fertilize to produce F2 plants. These plants were first screened for the sni1-1 locus using cleaved-amplified polymorphic sequence markers (Table II) followed by digestion with Tsp509I. The npr1-1 and tga2-2 loci were identified as described above. To make tga2-2 npr1-1, tga2-2 pollens were used to fertilize npr1-1 plants. The F1 progeny that expressed the PR gene were chosen and selfed to produce F2 plants. Genotyping of tga2-2 and the npr1-1 allele was carried out as described above. Homozygosity for all of the mutants identified in the F2 generation was further confirmed in the F3 by the lack of segregation.
RNA Extraction and Detection
Seedlings were grown on plates containing Murashige and Skoog medium or on Murashige and Skoog medium supplemented with different concentrations of INA for 2 weeks. RNA was extracted from seedlings by using the method described previously (Bowling et al., 1994). To synthesize cDNA, RNA (10 μg) was treated with 1 unit of DNaseI (Ambion) for 30 min at 37°C prior to RT. First-strand cDNA synthesis was carried out with the oligo(dT) primer using the first-strand cDNA synthesis kit (Invitrogen) following the manufacturer's protocol. Gene-specific primers were designed using LightCycler probe design software (Roche). Real-time PCR was carried out on a 10× diluted reverse-transcribed template using a Quantitect SYBR green PCR kit (Qiagen) in LightCycler (Roche) following the manufacturer's protocol. Primers used for real-time PCR are described in Tables I and II. Northern-blot analysis was carried out as described previously in Clarke et al. (1998).
Pathogen Infection
Fully expanded leaves of 3- to 4-week-old soil-grown plants were infiltrated with Psm4326 suspension in 10 mm MgCl2 at OD600 = 0.0001. Disease symptoms were recorded 4 d postinfection. To determine bacterial growth, infected leaves were collected at 0 and 3 d after inoculation. Leaf discs of the same size were made using a hole puncher and bacterial titers from those leaf discs were determined.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Western blot of TGA2 in tga2-2 mutant.
Supplemental Figure S2. Expression of PR genes in tga3-1.
Supplemental Figure S3. Expression of PR genes in tga2-2 and TGA6ACT.
Supplemental Figure S4. Relative expression of PR-1 in tga2-2 TGA6ACT and tga2-2 tga3-1 TGA6ACT.
Supplemental Figure S5. Expression of BiP2 and GRP94 in tga2-2 sni1-1 mutant.
Supplementary Material
Acknowledgments
We thank Jonathan Arias for the gift of TGA2 antibody, Xin Li for providing the tga2-1 tga5-1 tga6-1 triple mutant, and James Siedow, Aparna Sharma, Lisa Anderson, Steven Spoel, and Natalie Spivey for critical comments on the manuscript.
This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (grant no. 2003–00981); by the National Science Foundation (grant no. 0090887); and by Monsanto (grant to X.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Xinnian Dong (xdong@duke.edu).
The online version of this article contains Web-only data.
References
- Ammanamanchi S, Freeman JW, Brattain MG (2003) Acetylated sp3 is a transcriptional activator. J Biol Chem 278 35775–35780 [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV (1999) Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J Mol Biol 285 1353–1361 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Liu Y, Klessig DF, Dong X (1998) Uncoupling PR gene expression from NPR1 and bacterial resistance: characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP (1997) Genetic dissection of acquired resistance to disease. Plant Physiol 113 5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Friedrich L, Ryals JA (1995) Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA 92 6602–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, Fobert PR (2003) The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres C, DeLong C, Glaze S, Liu E, Fobert PR (2000) The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12 279–290 [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X (2004) Systemic acquired resistance. Annu Rev Phytopathol 42 185–209 [DOI] [PubMed] [Google Scholar]
- Eulgem T (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10 71–78 [DOI] [PubMed] [Google Scholar]
- Fan W, Dong X (2002) In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald HA, Canlas PE, Chern MS, Ronald PC (2005) Alteration of TGA factor activity in rice results in enhanced tolerance to Xanthomonas oryzae pv oryzae. Plant J 43 335–347 [DOI] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261 754–756 [DOI] [PubMed] [Google Scholar]
- Görlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel K-H, Oostendorp M, Staub T, Ward E, Kessman H, et al (1996) Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7 106–111 [DOI] [PubMed] [Google Scholar]
- Johnson C, Boden E, Arias J (2003) Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15 1846–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T (2006) The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18 3289–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupin I, Chua N-H (1996) Activation of the CaMV as-1 cis-element by salicylic acid: differential DNA-binding of a factor related to TGA1a. EMBO J 15 5679–5689 [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Klessig DF (2005) Salicylic acid-inducible Arabidopsis CK2-like activity phosphorylates TGA2. Plant Mol Biol 57 541–557 [DOI] [PubMed] [Google Scholar]
- Kim HS, Delaney TP (2002) Over-expression of TGA5, which encodes a bZIP transcription factor that interacts with NIM1/NPR1, confers SAR-independent resistance in Arabidopsis thaliana to Peronospora parasitica. Plant J 32 151–163 [DOI] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X (2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12 2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Lam YK (1995) Binding site requirements and differential representation of TGF factors in nuclear ASF-1 activity. Nucleic Acids Res 23 3778–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamph WW, Dwarki VJ, Ofir R, Montminy M, Verma IM (1990) Negative and positive regulation by transcription factor cAMP response element-binding protein is modulated by phosphorylation. Proc Natl Acad Sci USA 87 4320–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 10 71–82 [DOI] [PubMed] [Google Scholar]
- Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E (1998) Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J 16 223–233 [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y, Clarke JD, Li Y, Dong X (1999) Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98 329–339 [DOI] [PubMed] [Google Scholar]
- Marks J, Saifudeen Z, Dipp S, El-Dahr SS (2003) Two functionally divergent p53-responsive elements in the rat bradykinin B2 receptor promoter. J Biol Chem 278 34158–34166 [DOI] [PubMed] [Google Scholar]
- Métraux J-P, Ahl-Goy P, Staub T, Speich J, Steinemann A, Ryals J, Ward E (1991) Induced resistance in cucumber in response to 2,6-dichloroisonicotinic acid and pathogens. In H Hennecke, DPS Verma, eds, Advances in Molecular Genetics of Plant-Microbe Interactions, Vol 1. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 432–439
- Miao ZH, Lam E (1995) Construction of a trans-dominant inhibitor for members of the TGA family of transcription factors conserved in higher plants. Plant J 7 887–896 [DOI] [PubMed] [Google Scholar]
- Mosher RA, Durrant WE, Wang D, Song J, Dong X (2006) A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18 1750–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X (2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Pontier D, Miao ZH, Lam E (2001) Trans-dominant suppression of plant TGA factors reveals their negative and positive roles in plant defense responses. Plant J 27 529–538 [DOI] [PubMed] [Google Scholar]
- Pontier D, Privat I, Trifa Y, Zhou JM, Klessig DF, Lam E (2002) Differential regulation of TGA transcription factors by post-transcriptional control. Plant J 32 641–653 [DOI] [PubMed] [Google Scholar]
- Rochon A, Boyle P, Wignes T, Fobert PR, Despres C (2006) The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 18 3670–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53 247–259 [DOI] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner H-Y, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Tsui F, Klessig DF (1997) Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant Microbe Interact 10 69–78 [DOI] [PubMed] [Google Scholar]
- Subramaniam R, Desveaux D, Spickler C, Michnick SW, Brisson N (2001) Direct visualization of protein interactions in plant cells. Nat Biotechnol 19 769–772 [DOI] [PubMed] [Google Scholar]
- Thurow C, Schiermeyer A, Krawczyk S, Butterbrodt T, Nickolov K, Gatz C (2005) Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J 44 100–113 [DOI] [PubMed] [Google Scholar]
- Van Loon LC, Van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55 85–97 [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2 1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X (2005) Induction of protein secretory pathway is required for systemic acquired resistance. Science 308 1036–1040 [DOI] [PubMed] [Google Scholar]
- Weiss C, Schneider S, Wagner EF, Zhang X, Seto E, Bohmann D (2003) JNK phosphorylation relieves HDAC3-dependent suppression of the transcriptional activity of c-jun. EMBO J 22 3686–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Miao Z, Lam E (1997) DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol Biol 34 403–415 [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fan W, Kinkema M, Li X, Dong X (1999) Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci USA 96 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tessaro MJ, Lassner M, Li X (2003) Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Mosher SL, Fan B, Klessig DF, Chen Z (2007) Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol 7 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF (2000) NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant Microbe Interact 13 191–202 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.