Abstract
Filopodia are thin, spike-like cell surface protrusions containing bundles of parallel actin filaments. So far, filopodial dynamics has mainly been studied in the context of cell motility on coverslip-adherent filopodia by using fluorescence and differential interference contrast (DIC) microscopy. In this study, we used an optical trap and interferometric particle tracking with nanometer precision to measure the three-dimensional dynamics of macrophage filopodia, which were not attached to flat surfaces. We found that filopodia act as cellular tentacles: a few seconds after binding to a particle, filopodia retract and pull the bound particle toward the cell. We observed F-actin-dependent stepwise retraction of filopodia with a mean step size of 36 nm, suggesting molecular motor activity during filopodial pulling. Remarkably, this intracellular stepping motion, which was measured at counteracting forces of up to 19 pN, was transmitted to the extracellular tracked particle via the filopodial F-actin bundle and the cell membrane. The pulling velocity depended strongly on the counteracting force and ranged between 600 nm/s at forces <1 pN and ≈40 nm/s at forces >15 pN. This result provides an explanation of the significant differences in filopodial retraction velocities previously reported in the literature. The measured filopodial retraction force–velocity relationship is in agreement with a model for force-dependent multiple motor kinetics.
Keywords: actin filaments, interferometric three-dimensional particle tracking, molecular motors, nanomechanics, optical trapping
Filopodia are found in various cell types. They function as sensors of the local environment and take part in cell migration (1, 2). It is widely accepted that filopodial extension and retraction are regulated by filament elongation and retrograde actin flow (1, 3–5). There is increasing evidence that, in addition to actin polymerization and depolymerization, myosin motors play an important role for filopodial dynamics. In the case of filopodial extension, roles have been identified for myosin-V (6), myosin-VII (7), and myosin-X (8). The first evidence for myosin involvement in the retrograde flow of actin has been shown in neuronal growth cones (9). More recent studies identified myosin-II as playing an important role in the retrograde flow (10, 11). However, other pieces of evidence indicate that myosin-II is not directly essential for filopodial dynamics (12). So the molecular machinery driving filopodial extension and retraction is still not yet revealed and unraveled.
It has been shown that filopodia are able to pull on objects (13–15) and are able to exert forces when they retract (15, 16). In the latter two studies, it was estimated that these forces can be as high as several hundreds of piconewtons (pN) (16) or even larger (15). However a precise direct measurement of filopodial pulling forces in combination with a precise measurement of the filopodial three-dimensional (3D) dynamics is still missing.
Although filopodia are abundant in macrophages, little is known about their role during phagocytosis. It has been shown that Dictyostelium mutants lacking myosin-VII have almost no filopodia, which was associated with a decreased phagocytosis rate (7). Earlier analyses by light and electron microscopy have shown macrophage filopodia attached to pathogens before their engulfment into the cell (17, 18). Recently, it has been shown (15) that phagocytosis can be initiated by filopodia, which pull invasin-coated beads toward the cell body. However, very little is known about the precise mechanical properties of filopodial retraction during phagocytosis.
Here, we studied the dynamic behavior of macrophage filopodia and membrane ruffles (2) upon binding to antibody-coated particles. We used optically trapped IgG-coated beads as a phagocytic model system (19) in mouse J774 and RAW macrophage cell lines as well as in primary bone marrow macrophages (PBMM). Bacteria could also be trapped and used instead of beads in such an assay. However, because most bacteria have an elongated shape, 3D tracking of bacteria is much more difficult compared with 3D tracking of spherical beads because of the bacteria's additional rotational degrees of freedom (20). Furthermore, it has been shown in many studies (21) that beads can be an excellent phagocytic model system.
Up to now, filopodial dynamics was mainly studied in two dimensions by using fluorescence and differential interference contrast (DIC) microscopy on filopodia adherent to coverslips. In this study, we used an optical trap and interferometric particle tracking to measure the 3D retraction behavior of filopodia, which were not attached to coverslips. The optically trapped beads were moved toward the cell and bound to filopodia or ruffles. The beads were tracked in 3D with an interferometric laser-tracking system equipped with a quadrant-photodiode (QPD) (22). We tracked the beads in 3D with nanometer precision at sampling rates between 10 and 100 kHz. Trap and position detector were calibrated by using the Langevin method (22). In this way, we recently measured the thermal fluctuations of a trapped bead during the binding to the cell membrane, which provided information about the dynamics of the binding process with an unequaled spatiotemporal precision (19).
In the experiments described here, the 3D tracking of the filopodia- and ruffle-bound beads allowed us to quantify the filopodial and ruffle motion in response to particle binding with nanometer precision. Additionally, the optical trap served both as a mechanical force transducer and as an instrument to measure the cellular forces on the piconewton scale.
Results
Retraction upon Bead Binding.
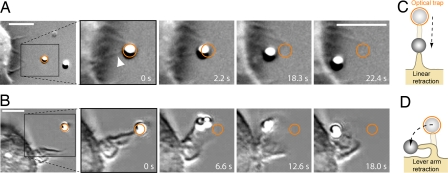
A few seconds after binding to IgG-coated beads, the filopodia and ruffles on J774, RAW, and PBMM cells retracted and pulled the beads toward the cell to initiate phagocytic uptake of the beads. The retraction occurred either linearly [Fig. 1 A and C and supporting information (SI) Movie 1] or in a lever arm manner (Fig. 1 B and D and SI Movie 2). Filopodia mainly retracted in a linear way whereas ruffles retracted either linearly or in a lever arm like.
Fig. 1.
Filopodial and ruffle retraction preceding phagocytosis. (A) An IgG-coated bead in the optical trap (orange circle) is moved toward a filopodium of a J774 macrophage. Upon binding, the filopodium (arrowhead) retracts and pulls the bead toward the cell to initiate phagocytic uptake. (B) Two trapped beads are moved toward a membrane ruffle. Upon binding (second picture), the ruffle retracts in a lever arm manner and pulls the beads toward the cell. (C and D) Sketch of the linear and the lever arm retraction respectively. (Scale bars: 5 μm.)
To test whether the retraction behavior of filopodia and ruffles is ligand-specific, we used control beads coated with avidin and control beads coated with bacterial LPS (lipopolysaccharide). In both cases, filopodia and ruffle retraction showed the same behavior as in the case of IgG-coated beads.
To measure the unobstructed retraction dynamics of filopodia and ruffles, we probed only membrane protrusions that were not attached to the coverslip (“free protrusions”). Linear filopodial elongation and retraction is in principle a one-dimensional behavior that can be studied on a flat surface like a coverslip. However, an attachment of the filopodium to a surface results in adhesion forces that affect the retraction speed (we show that counteracting forces exerted by the optical trap slow down the retraction speed). Therefore, strong variations of the (typically unknown) adhesion forces in different experimental setups will result in strong variations of the filopodial retraction speeds, which hamper the analysis and interpretation of experimental results. Furthermore, if meaningful force–velocity relations shall be measured, it is necessary to prevent any uncontrolled adhesion forces between the filopodium and a substrate.
Conventional tracking techniques such as 2D video-microscopy enable the unambiguous classification of any motion only if it is restricted to the x–y plane of the setup (e.g., if the investigated parts of the cell are very thin and attached to the coverslip). In the case of free protrusions, linear retraction could then only be discriminated against any other type of motion (e.g., lever arm motion) if, by chance, it occurs in the focal plane of the microscope.
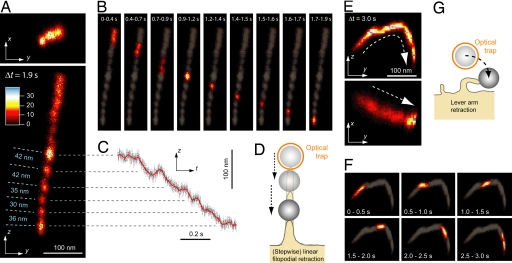
To address and solve this restriction, we used a 3D interferometric laser-tracking system to create bead position histograms in 3D. From these 3D histograms, we derived 2D projections to classify the protrusion retraction behavior (Fig. 2). Fig. 2 A and B shows x–y and y–z projections of a linear retraction event, and Fig. 2 E and F shows x–y and y–z projections of a lever arm retraction. Fig. 2 D and G shows sketches of these two retraction types. The dynamics of the linear and the lever arm retraction event are shown in Fig. 2 B and F, respectively, by displaying position histograms over varying time intervals. In Fig. 2 A and B, it can be seen that part of the retraction occurs stepwise. Fig. 2C shows a trace of the bead's z position as a function of time. The transition times of the steps range from 30 ms to 120 ms.
Fig. 2.
Nanometer-precise 3D tracking of linear and lever arm retraction. 2D projections (x–y and y–z plane) of 3D bead position histograms reveal whether a filopodial retraction is purely linear (A and B) or lever arm-like (E and F). The sampling rate of all of the 3D position measurements shown in this figure was 10 kHz. (A) The x–y (Upper) and y–z (Lower) histogram of linear retraction over a time interval of Δt = 1.9 s (the number of data points in each histogram is 1.9 s × 10 kHz = 19,000). The bin size of the position histograms is 2 nm in the x, y, and z direction. (B) The dynamics of the retraction process shown in A is visualized by displaying the y–z histogram for short time intervals: t = 0 to 0.4 s, 0.4 to 0.7 s, 0.7 to 0.9 s, etc. (in the background, the histogram shape is outlined for the whole time interval t = 0 to 1.9 s). (A and B) Part of the retraction occurs stepwise. The step sizes in this figures range from 30 to 42 nm. The counteracting optical force was <3 pN. (C) The z position vs. time traces of the retraction shown in A. The step transition times are between 30 ms and 120 ms. (D) Sketch of the linear, stepwise filopodial retraction. (E) The x–y (Upper) and y–z (Lower) histogram of lever arm retraction over a time interval of Δt = 3.0 s (the number of data points in each histogram is 3.0 s × 10 kHz = 30,000). The bin size of the position histograms is 2 nm in the x, y, and z direction. (F) The dynamics of the retraction process shown in E is visualized by displaying the y–z histogram for short time intervals: t = 0 to 0.5 s, 0.5 to 1.0 s, 1.0 to 1.5 s, etc. (in the background, the histogram is shown for the whole time interval t = 0 to 3.0 s). (G) Sketch of the lever arm retraction process.
Step Sizes.
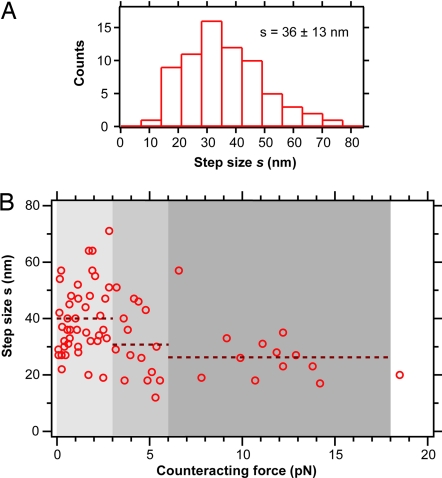
We analyzed 70 steps from 17 filopodial retraction events (steps were found in about half of the investigated retraction events) and found that the step size distribution had a mean value of 36 ± 2 nm (mean ± error of mean) and a standard deviation of 13 nm (Fig. 3A). Recently, intracellular stepwise molecular motion during organelle transport was observed in 2D by using fluorescence microscopy-tracking techniques (23, 24). However, no study so far reported intracellular stepwise motion in 3D. As shown in Fig. 3B, most of the steps (42 of 70) were found at low counteracting forces ranging from 0 to 3 pN. However, some steps were also found at higher forces: 15 steps were found at forces from 3 to 6 pN, 12 steps at forces from 6 to 18 pN, and 1 step at a force >18 pN. The chosen force intervals are integer multiples of 3 pN, which is approximately the stall force of a molecular motor with a step size of 36 nm (see Discussion). Fig. 3B shows that the mean step size decreases with higher forces. In the range from 0 to 3 pN, the step size was s = 40 ± 12 nm, in the range from 3 to 6 pN, the step size was s = 31 ± 12 nm and in the range from 6 to 18 pN, the step size was s = 27 ± 11 nm. We also found that steps occur at filopodial retraction velocities ranging from 10 nm/s to 430 nm/s (SI Fig. 6).
Fig. 3.
Filopodial retraction step size. (A) Histogram (n = 70) of step sizes of linear filopodial retraction at counteracting forces ranging from 0 to 19 pN. The mean step size is s = 36 ± 13 nm (mean ± standard deviation). (B) Step size of linear filopodial retraction as a function of the counteracting force applied by the optical tweezers. In the force range from 0 pN to 3 pN, the step size was s = 40 ± 12 nm (n = 42); from 3 pN to 6 pN, the step size was s = 31 ± 12 nm (n = 15); and from 6 pN to 18 pN, the step size was s = 27 ± 11 nm (n = 12).
Cytoskeleton and Motors.
To determine the cytoskeletal components involved, we investigated the role of F-actin and microtubules in the protrusion retraction behavior. After depolymerization of the actin-filaments by Latrunculin A (2 μM), no filopodial retraction upon bead binding was observed. Depolymerization of microtubules by Nocodazole (20 μM) had no effect on the retraction behavior.
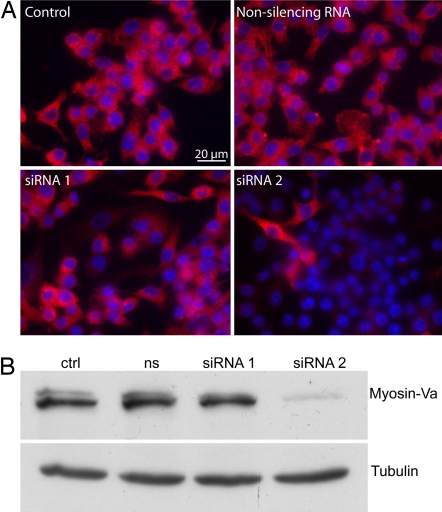
The filopodial retraction step size of 36 ± 13 nm (mean ± standard deviation) is within the range (30–38 nm) of the known step sizes of the processive motors myosin-V (25) and myosin-VI (26) measured in vitro. Therefore, we tested the role of those two molecular motors in the retraction. Myosins of class VI are unique because they are the only motors that are established to move toward the minus end of actin filaments (27). Furthermore, myosin-VI is present in membrane ruffles and filopodia (28). To test for a role of myosin-VI, we used PBMM isolated from myosin-VI knockout mice (MyoVI−/−). No strong difference was observed between the retraction behavior of wt and MyoVI−/− macrophages. The role of myosin-Va was tested by using RNA silencing in RAW macrophages by using RNAi. The quality of the knockdown was checked by immunofluorescence and only those experiments, where myosin-Va was knocked down in >90% of the cells (Fig. 4A) were used for further analysis. The knockdown was also confirmed by Western blot analysis (Fig. 4B). There was no strong difference between the retraction behavior of the control and the myosin-Va knockdown cells.
Fig. 4.
Myosin-Va RNA silencing: immunofluorescence and Western blot analysis. (A) Immunofluorescence analysis: myosin-Va antibody (DIL-2) labeling (red) and a DAPI staining (blue) of RAW cells. The siRNA 2 had a knockdown efficiency of ≈90%. (B) Western blot analysis: DIL-2 antibody against myosin-Va and the anti-α tubulin (Sigma) for the control (ctrl), the nonsilencing RNA (ns), and the silencing RNA (siRNA 1 and siRNA 2). The siRNA 2 against myosin-Va had a knockout-efficiency of ≈90%.
We also tested the role of myosin-II, because this molecular motor is known to play an important role in cellular tail retraction events during cell motility (29). We used blebbistatin (50 μM) to inhibit myosin-II in J774 cells. Although some of the filopodial retraction traces were very noisy in the case of the blebbistatin-treated cells, there was no strong affect on the dynamics of the retraction behavior. Furthermore, we still observed discrete steps in the filopodial retraction after blebbistatin treatment.
Force–Velocity Relationship.
To characterize the mechanical properties of linear filopodial retraction, we next asked how filopodia bound to beads respond to external forces. We measured the filopodial retraction velocity v as a function of the counteracting force F applied by the optical tweezers. We identified linear retraction events and determined the retraction velocity and the counteracting optical force as described in Materials and Methods. The linear retraction events took between 0.3 s and 10 s, with an average value of 1.8 s. Because a sampling rate of 10–100 kHz was used, each linear retraction event therefore consisted of several thousands to several hundred thousands of tracked bead position data points. Because drifts in the setup (e.g., a sample stage drift or a laser intensity drift) would cause systematic errors in the force and velocity measurements, we measured the setup drift by tracking beads which were attached to the coverslip. These measurements showed that the setup drift is on the order of ≈1 nm/s.
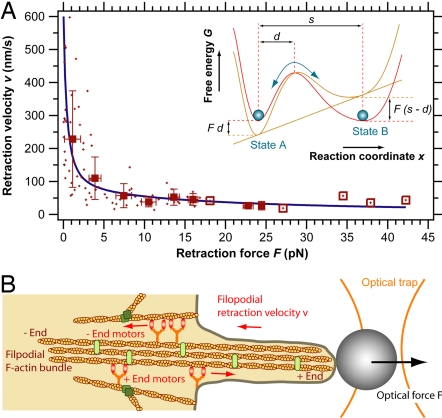
The filopodial retraction force–velocity data points are shown in Fig. 5A. The large range of forces was achieved by using different laser powers. The retraction velocities were strongly dependent on the counteracting force and ranged between 600 nm/s at forces <1 pN and 40 nm/s at forces >15 pN (Fig. 5A).
Fig. 5.
(A) Force–velocity curve for linear filopodial retraction: speed of linear filopodial retraction as a function of the counteracting force applied by the optical tweezers. Shown are the raw data points (crosses) and the pooled data points (squares) as well as the theoretical curve (blue line) according to Eq. 2. The data pooling was done over 3-pN force intervals. The open squares (which have no error bars) indicate data points where pooling was done over a single raw data point. (Inset) Two-state model for motor kinetics. Shown is the force-dependent free energy G of a two-state molecular motor system. State A and state B denote sequential motor positions along its track (reaction coordinate x). An external force F alters the free energy of the two states and thereby the transition rates. (B) Model for filopodial retraction. The data presented here are in agreement with the hypothesis that multiple myosin motors with a step size ≈36 nm are involved in filopodial retraction.
Discussion
Retraction upon Bead Binding.
Macrophage filopodia and ruffles are very dynamic cellular structures that actively probe their environment. Thereby they increase the effective surface area of the cell. Their tentacle-like retraction upon particle binding could be a general mechanism to increase phagocytic uptake rates.
3D Position Histograms.
We found that 3D bead-tracking histograms are highly suited to discriminate linear retraction behavior from other retraction behaviors (e.g., lever arm). In contrast to 3D-tracking techniques, conventional 2D video microscopy would not be able to discriminate different retraction behaviors as shown in Fig. 2. Furthermore, Fig. 2A shows that the filopodial retraction velocity would be strongly underestimated if only the x–y projection were used to determine the retraction speed.
The 3D histogram method also facilitates the discovery of stepping events in livings cells. Live cell measurements are in general noisier than in vitro measurements with single molecular motors where stepping events are typically observed. In addition, cells present a mechanical buffer that blurs out discrete molecular motor stepping events. Therefore the discovery of discrete molecular motor steps is in general more difficult in live cells than in vitro. However, a comparison of Fig. 2 A and C shows that steps are more easily detected if the histogram method is applied instead of the position versus time (r–t) graph method, which is usually used to identify molecular motor steps. The histograms are projections along the time-axis and require less abstraction work from the viewer than the integration by eye along the time-axis in an r–t graph.
The step transition times in Fig. 2C range from 30 ms to 120 ms, which is much longer than the typical rise times of molecular motors measured in vitro, which are on the order of several tens of microseconds (30). However, if the motors are coupled to the tracked bead via actin-fibers and the cell membrane (Fig. 5B), fast motor steps will be damped viscoelastically, which can account for the relatively long transition times observed here.
Cytoskeleton and Motors.
Our experiments with drugs such as Latrunculin A and Nocodazole, which disrupt the cytoskeleton, indicate that the filopodial retraction depends on F-actin, but not on microtubules. The measured step size of 36 ± 13 nm together with the dependency of the retraction on F-actin suggests that an F-actin-based molecular motor with a step size of ≈36 nm is involved in filopodial retraction. We tested the role of the molecular motors myosin-Va and -VI using RNA silencing and knockout mice, respectively. We did not find a strong difference between the retraction behavior of the control and the myosin-Va knockdown cells or between the control and the myosin-VI knockdown cells. Plausible explanations for these results are that another (myosin) motor is responsible for the observed steps in filopodial retraction or that multiple motors (possibly including myosin-Va or myosin-VI) work together in filopodial retraction.
In principle, it is also possible that the observed steps are not directly due to molecular motors, but rather due to actin depolymerization. Actin filaments form a long-pitch helix with a pseudo repeat of 36 nm (25, 26). Therefore it is imaginable that the actin depolymerization rate is modulated by the helical pitch e.g., through actin binding proteins. Furthermore, the filaments might not always disassemble monomer by monomer, but rather in oligomeric chunks of the size of one helical pitch. Although an oligomeric polymerization was recently observed in microtubules (31), neither an oligomeric polymerization nor an oligomeric depolymerization was so far observed in actin filaments. Therefore, we believe that the observed step size is rather caused by molecular motor stepping than by actin depolymerization.
Step Sizes.
Most of the steps during filopodial retraction were found at counteracting forces between 0 and 3 pN. However some steps were also observed at higher forces of up to 19 pN. This observation is remarkable because the maximal stall force of a molecular motor with a step size of 36 nm is ≈3 pN if the motor hydrolyzes one single adenosine 5′-triphosphate (ATP) molecule per step. An upper limit for the stall force Fs of a motor with a step size s can be calculated from the free energy GATP [≈100 pN·nm in live cells (32)] available from ATP hydrolysis: Fs ≤ GATP/s ≈ 3 pN. The assumption that a single ATP molecule is hydrolyzed per motor step is based on experiments with the motor proteins kinesin-1 (33, 34) and myosin-V (35), which were shown to hydrolyze one single ATP molecule per step.
If a molecular motor with a given stall force Fs is responsible for the filopodial retraction, then multiple motors N sharing the load F (Fig. 5B) must be active at counteracting forces F > Fs. In the case of 3 pN stall force, at least two motors must contribute to the filopodial retraction in the force regime between 3 pN and 6 pN. In the range from 6 pN to 18 pN, the minimum number of contributing motors is 3–6.
If multiple motors pull on a common cargo the observed step size depends on the number of motors and on the degree of synchronization of the motor stepping. If the motors are considered as linear springs and if they are not synchronized (on a time scale smaller than the temporal detection resolution), the observed step size s of the cargo after a single motor step s0 decreases strongly with the number of contributing motors: it decreases by a factor of 1/2 for two motors, by a factor of 1/3 for three motors, by a factor of 1/4 for four motors, etc. However, such a strong decrease of the step size as a function of the counteracting force is not observed experimentally (Fig. 3B).
An explanation for the observation of relatively high step sizes at loads above the stall force of a single motor is that the kinetics of each motor is not independent from the other motors. If multiple motors pull together on a common load, a partial synchronization of the motor stepping can be achieved if each motor has a load-dependent stepping rate. If one of N pulling motors is stepping forward, the load onto this particular motor is increased and the load onto the other N − 1 motors is reduced. Because the velocity of molecular motors typically decreases with increasing counteracting force (25, 32, 36, 37), the reduced load of the N − 1 motors will increase their probability for a stepping event. If such a mechanism synchronizes the motion of a subset of n motors fast enough compared with the time resolution of the detector, the individual n sub steps will not be resolved and the observed net step size will be s = s0 n/N.
Retraction Velocities.
The filopodial retraction velocities reported here range between 600 nm/s and 40 nm/s (Fig. 5A). These velocities are high compared with previously reported retraction values, which ranged between 160 nm/s and 25 nm/s (see also SI Table 2) (4, 6, 12, 38).
We can exclude a drift of the setup as a significant source of error for our velocity measurements because the setup drift of ≈1 nm/s is negligible compared with the filopodial retraction velocities. We can also exclude instantaneous jumps in the system as a source of the high velocities as they would have been easily detected in the retraction traces because the retraction events sustained for a long time (0.3 s to 10 s) compared with the time sampling (0.01 ms to 0.1 ms).
The large variance of the previously reported filopodial retraction velocities (4, 6, 12, 38), as well as our relatively high velocities measured at low load can be explained as follows. First, we measured velocities on a short timescale of ≈1 s (time sampling: 0.01 to 0.1 ms) compared with ≈1–5 min (time sampling on the order of several seconds) in the previous studies. Secondly, the previously reported retraction velocities were determined on filopodia, which were attached to coverslips. Because we show that counteracting forces slow down the retraction speed (Fig. 5A), we assume that varying unknown adhesion forces between filopodia and coverslips are one reason for these relatively low velocities and their large variance.
Force–Velocity Relationship.
From our measurements of the filopodial retraction velocity v as a function of the counteracting force F (Fig. 5A), various energetic values were calculated (Table 1). The values for the velocity and the force are the pooled data points shown in Fig. 5A. The mechanical power of the filopodial retraction is P = Fv. By using the measured average step size of s = 36 nm, the stepping rate (number of steps per second) rs = v/s and the mechanical work per motor step Ws = P/rs = Fs were calculated. If one motor hydrolyses one ATP molecule per step, then a value Ws > GATP shows that more than one motor is involved in the filopodial retraction. For a given motor efficiency η, the stall force of the motor is Fs = ηGATP/s. The minimal number of active motors Nm = ceil[Ws/(ηGATP)] = ceil(F/Fs) was calculated and also listed in Table 1. The value for the motor efficiency (η = 90%) was estimated from previous measurements (see Model).
Table 1.
Energetic and mechanical parameters of filopodial retraction
| Measured values |
Values derived from measurements |
||||
|---|---|---|---|---|---|
| Force, pN (F) | Velocity, nm/s (v) | Motor steps per sec, 1/s (rs = v/s) | Mechanical power of retraction, GATP/s* (P = Fv) | Mechanical work per motor step, GATP* (Ws = P/rs) | Minimal no. of motors at work† (Nm) |
| 1.1 | 228 | 6.3 | 2.5 | 0.4 | 1 |
| 3.9 | 110 | 3.1 | 4.3 | 1.4 | 2 |
| 7.4 | 57 | 1.6 | 4.2 | 2.7 | 3 |
| 10.5 | 38 | 1.1 | 4.0 | 3.8 | 5 |
| 13.6 | 52 | 1.4 | 7.1 | 4.9 | 6 |
| 16.0 | 45 | 1.3 | 7.2 | 5.8 | 7 |
From the measured force F, velocity v, and step size s, several energetic and mechanical parameters of the filopodial retraction were derived: the number of motor steps per second, the mechanical power applied during the retraction, the work done per motor step, and the minimal number of motors at work.
*GATP = 100 pN·nm (32).
†For a motor efficiency of η = 90%.
If N motors pull on a common cargo and if each of these motors makes one step, the free energy required is G = NGATP. Therefore, the number of pulling motors should be close to the minimal number of motors N ≈ Nm if the cell minimizes the ATP hydrolysis to work efficiently. To test whether the data shown in Fig. 5A are in agreement with this hypothesis, we chose the simplest dynamic model for the reaction kinetics of the motors.
Model.
We assume that the kinetics of a single motor can be described by a reversible two-state model. Fig. 5A Inset shows the free energy G of the rate-limiting step within the motor's work cycle. State A and state B denote sequential motor positions along its track (reaction coordinate x), s denotes the motor step size, and d is the transition distance between state A and the transition point. An external force F changes the free energy of the system and thereby the transition rates k1 = k10e−Fd/kBT(A→B) and k2 = k20eF(s−d)kBT(B→A) between the two states (32, 39), where k10 and k20 are the transition rates without external load. At the stall force Fs both transition rates are equal k10 e−Fsd/kBT = k20 eFs(s−d)kBT, which results in zero net motor motion. The previous three equations can be used to directly calculate the single motor force–velocity relationship
If N motors pull on a given load F in a coordinated way, the force is shared between these motors, yielding an effective load of F/N for each motor. If the motors do not hinder each other and if N ≈ Nm = ceil(F/Fs) ≈ F/Fs + ½ is used, the force–velocity relationship is
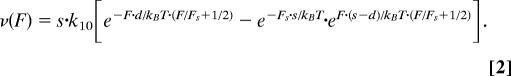 |
Except from the stall force, which depends on the unknown motor efficiency, all parameters in Eq. 2 were derived from our experimental data. The step size s = 36 ± 13 nm (mean value ± standard deviation) was directly measured (Fig. 3A). Previous studies on the stall forces of processive myosin motors have reported Fs ≈ 1.7 − 3 pN for myosin-V (25, 37, 40) and Fs ≈ 2.8 pN for myosin-VI (26). As an estimate for our analysis, we chose the average of the abovementioned values Fs ≈ 2.6 pN (which requires a motor efficiency of η = Fss/GATP = 90% for GATP = 100 pN·nm). The transition rate k10 ≈ vmax/s0 ≈ 17 1/s was estimated from the highest measured velocities (vmax ≈ 600 nm/s). The remaining parameter d = 4.6 ± 0.4 nm was derived by a fit of Eq. 2 to the raw data shown in Fig. 5A. The transition rate d determines the force-dependence of the rate-limiting step within the motor cycle. The values for s, k10, and d measured here are all within the range of the mechanical parameters determined so far for processive myosin motors in vitro (25, 26, 37). Our findings are therefore in agreement with the hypothesis that an F-actin plus end or minus end directed processive myosin motor is involved in filopodial retraction (Fig. 5B).
Summary and Conclusion.
In this study, we have reported that filopodia of macrophages act as cellular tentacles possibly to increase the efficiency of the uptake of pathogens. A few seconds after binding to optically trapped beads, filopodia retracted and pulled the beads toward the cell to initiate phagocytic uptake of the beads. We measured the retraction of the filopodia by tracking the attached beads with an interferometric 3D-tracking system. We found that the filopodial retraction velocity depends strongly on the counteracting force. Furthermore, we observed stepwise F-actin-dependent retraction of the filopodia with an average step size of 36 nm, suggesting that a myosin motor with a step size of ≈36 nm is involved in filopodial retraction. Although damped by cellular structures like the filopodial F-actin bundle and the plasma membrane, steps could be resolved in about half of the investigated retraction events. Steps are also observed at counteracting forces above the stall force of a single motor supporting the idea that multiple synchronized motors are involved. Although we show that a basic model for force-dependent multiple motor kinetics is in agreement with the measured filopodial retraction force–velocity relationship, more light needs to be shed on the cellular mechanisms that potentially steer the synchronization or the number of active motors enabling variable retraction forces and velocities. Further insights into these cellular mechanisms not only would provide information about the underlying molecular machinery but also may bear fascinating novel concepts for biology-based nano-technological actuators.
Materials and Methods
Cell Culturing and Primary Cell Isolation.
J774A.1 and RAW 264.7 mouse macrophages were cultured as described in SI Materials and Methods. Primary murine bone marrow macrophages were isolated and cultured as described in SI Materials and Methods.
Beads.
Polystyrene beads with a diameter of 1 μm were coated with IgG as described in SI Materials and Methods.
Myosin-Va RNA Silencing, Immunofluorescence, and Western Blotting.
Myosin-Va silencing with siRNA, immunofluorescence, and Western blotting was performed as described in SI Materials and Methods.
Optical Trapping, Interferometric 3D Tracking, and Force–Velocity Measurements.
The calibration of the optical trap and the quadrant–photodiode detection system as well as the force–velocity measurements were done as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Eric Dufresne, Paul Forscher, Jonathon Howard, Laurent Vonna, Thomas Surrey, and Philipp Keller for helpful discussions; Christian Klasen for mouse colony management; John A. Hammer III (National Institutes of Health, Bethesda, MD) for providing the DIL-2 antibody; and Eik Hoffmann for Western blotting and helpful discussions. H.K. was supported by a grant from the German National Academic Foundation (Studienstiftung des deutschen Volkes).
Abbreviations
- PBMM
primary bone marrow macrophages
- MyoVI−/−
myosin-VI knockout mice.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702449104/DC1.
References
- 1.Faix J, Rottner K. Curr Opin Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Small JV, Stradal T, Vignal E, Rottner K. Trends Cell Biol. 2002;12:112–120. doi: 10.1016/s0962-8924(01)02237-1. [DOI] [PubMed] [Google Scholar]
- 3.Welch MD, Mallavarapu A, Rosenblatt J, Mitchison TJ. Curr Opin Cell Biol. 1997;9:54–61. doi: 10.1016/s0955-0674(97)80152-4. [DOI] [PubMed] [Google Scholar]
- 4.Mallavarapu A, Mitchison T. J Cell Biol. 1999;146:1097–1106. doi: 10.1083/jcb.146.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mogilner A, Rubinstein B. Biophys J. 2005;89:782–795. doi: 10.1529/biophysj.104.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang FS, Wolenski JS, Cheney RE, Mooseker MS, Jay DG. Science. 1996;273:660–663. doi: 10.1126/science.273.5275.660. [DOI] [PubMed] [Google Scholar]
- 7.Tuxworth RI, Weber I, Wessels D, Addicks GC, Soll DR, Gerisch G, Titus MA. Curr Biol. 2001;11:318–329. doi: 10.1016/s0960-9822(01)00097-5. [DOI] [PubMed] [Google Scholar]
- 8.Berg JS, Cheney RE. Nat Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 9.Lin CH, Espreafico EM, Mooseker MS, Forscher P. Neuron. 1996;16:769–782. doi: 10.1016/s0896-6273(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 10.Medeiros NA, Burnette DT, Forscher P. Nat Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- 11.Cai Y, Biais N, Giannone G, Tanase M, Jiang G, Hofman JM, Wiggins CH, Silberzan P, Buguin A, Ladoux B, Sheetz MP. Biophys J. 2006;91:3907–3920. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diefenbach TJ, Latham VM, Yimlamai D, Liu CA, Herman IM, Jay DG. J Cell Biol. 2002;158:1207–1217. doi: 10.1083/jcb.200202028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapfhammer JP, Raper JA. J Neurosci. 1987;7:201–212. doi: 10.1523/JNEUROSCI.07-01-00201.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wessells NK, Letourneau PC, Nuttall RP, Luduena-Anderson M, Geiduschek JM. J Neurocytol. 1980;9:647–664. doi: 10.1007/BF01205031. [DOI] [PubMed] [Google Scholar]
- 15.Vonna L, Wiedemann A, Aepfelbacher M, Sackmann E. Eur Biophys J. 2007;36:145–151. doi: 10.1007/s00249-006-0118-y. [DOI] [PubMed] [Google Scholar]
- 16.Heidemann SR, Lamoureux P, Buxbaum RE. J Cell Biol. 1990;111:1949–1957. doi: 10.1083/jcb.111.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young VB, Falkow S, Schoolnik GK. J Cell Biol. 1992;116:197–207. doi: 10.1083/jcb.116.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koerten HK, Ploem JS, Daems WT. Exp Cell Res. 1980;128:470–475. doi: 10.1016/0014-4827(80)90084-1. [DOI] [PubMed] [Google Scholar]
- 19.Kress H, Stelzer EHK, Griffiths G, Rohrbach A. Phys Rev E. 2005;71 doi: 10.1103/PhysRevE.71.061927. 061927. [DOI] [PubMed] [Google Scholar]
- 20.Kress H, Stelzer EHK, Rohrbach A. Appl Phys Lett. 2004;84:4271–4273. [Google Scholar]
- 21.Desjardins M, Griffiths G. Curr Opin Cell Biol. 2003;15:498–503. doi: 10.1016/s0955-0674(03)00083-8. [DOI] [PubMed] [Google Scholar]
- 22.Rohrbach A, Tischer C, Neumayer D, Florin EL, Stelzer EHK. Rev Sci Instr. 2004;75:2197–2210. [Google Scholar]
- 23.Kural C, Kim H, Syed S, Goshima G, Gelfand VI, Selvin PR. Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe TM, Higuchi H. Biophys J. 2007;92:4109–4120. doi: 10.1529/biophysj.106.094649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 26.Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL. Proc Natl Acad Sci USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 28.Buss F, Kendrick-Jones J, Lionne C, Knight AE, Cote GP, Paul Luzio J. J Cell Biol. 1998;143:1536–1545. doi: 10.1083/jcb.143.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krendel M, Mooseker MS. Physiology. 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- 30.Carter NJ, Cross RA. Curr Opin Cell Biol. 2006;18:61–67. doi: 10.1016/j.ceb.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Kerssemakers JWJ, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 32.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- 33.Visscher K, Schnitzer MJ, Block SM. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 34.Coy DL, Wagenbach M, Howard J. J Biol Chem. 1999;274:3667–3671. doi: 10.1074/jbc.274.6.3667. [DOI] [PubMed] [Google Scholar]
- 35.Rief M, Rock RS, Mehta AD, Mooseker MS, Cheney RE, Spudich JA. Proc Natl Acad Sci USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svoboda K, Block SM. Cell. 1994;77:773–784. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 37.Clemen AE-M, Vilfan M, Jaud J, Zhang J, Bärmann M, Rief M. Biophys J. 2005;88:4402–4410. doi: 10.1529/biophysj.104.053504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu M, Witke W, Kwiatkowski DJ, Kosik KS. J Cell Biol. 1997;138:1279–1287. doi: 10.1083/jcb.138.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bustamante C, Chemla YR, Forde NR, Izhaky D. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 40.Uemura S, Higuchi H, Olivares AO, De La Cruz EM, Ishiwata S. Nat Struct Mol Biol. 2004;11:877–883. doi: 10.1038/nsmb806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.