Discovering the biological roles of a protein is best accomplished by observing the consequences of its removal. However, such loss-of-function studies are rarely straightforward. In the case of genetic experiments, including those dealing with knockout mice, gene functions in one tissue type are often disguised by deleterious phenotypes, including lethality, in another. Even when this problem is overcome, for example, by tissue-specific gene deletion using the Cre/loxP system, it can be difficult to dissect primary from secondary effects to determine the molecular basis of a phenotype. The difficulty lies in the speed at which the biological events being studied occur. For example, after transcription of the Cre recombinase is induced, considerable time will lapse before recombination of the targeted gene and dissipation of preexisting pools of the target gene's RNA and protein. During this period, the studied cells could have, for example, responded to extracellular signals, undergone cell divisions, changed position or shape, and even differentiated into a new cell type. Alternative methods, such as RNA interference or small-molecule inhibition, allow regulation of the protein of interest during tighter time windows. Unfortunately, these techniques have their own shortcomings. RNA interference suffers from nonspecific effects, unpredictable degrees of “knockdown,” and slow kinetics of onset and reversibility. Small-molecule regulation is generally very fast and usually reversible; however, identifying or developing a small molecule that is genuinely specific with reliable pharmacokinetics challenges even the largest pharmaceutical company. To this end, researchers have devoted considerable energy to develop new technologies that merge gene-based methods (to create impeccable specificity) with chemical-based strategies (to provide rapid on/off regulation). In a recent issue of PNAS, Pratt et al. (1) report a new approach that uses a generic drug to induce the recovery of a native target protein from a fusion protein that is otherwise destined for destruction (1). This method adds to the growing toolbox available to researchers interested in perturbing biological systems closer to physiologically relevant speeds.
Much of biology is regulated at the molecular level by changes in the proximity of molecules. For example, receptor dimerization is a common way that signals are transduced from the membrane into the cell. Similarly, protein phosphorylation requires recruitment of the substrate to its kinase, and transcriptional regulation depends on cooperative interactions between multiple transcription factors. Coopting this universal aspect of biological regulation by artificially inducing dimerization is an effective way to regulate and study cellular events (2). Small molecules that are able to simultaneously bind to two protein domains can be used for just this purpose. These protein domains can be individually fused to different proteins or protein moieties so that addition of the compound induces the association of the protein domains and triggers molecular responses, including receptor activation (2), nucleocytoplasmic transport (3), transcriptional activation (4), and the timing of mitotic chromosome separation (5). Although the original molecules were homodimerizers made by joining two molecules of FK506, one commonly used tripartite complex is the FKBP12–rapamycin–FRB system. Rapamycin is a macrolide antibiotic that is approved for pharmaceutical use as an immunosuppressant and shows considerable promise as an antitumor agent. Its suitability as a drug is based on its ability to inhibit mTor, a protein kinase that is involved in cell growth and proliferation. Rapamycin's ability to inhibit mTor depends on a prior high-affinity association with a cellular protein called FKBP12. Together, FKBP12 and rapamycin bind in a highly specific manner to the 89-aa FKBP12–rapamycin binding (FRB) domain of mTor (6).
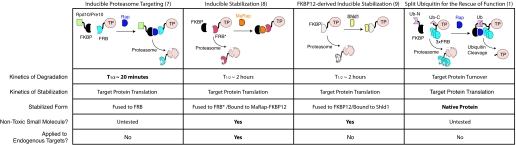
Unlike mTor, most proteins lack a specific inhibitor. Instead of devoting the immense time and monetary expense to develop one inhibitor for each target, would it be possible to engineer any target protein in a reproducible manner to make it sensitive to existing small molecules? One attractive means to accomplish this would be to use cellular degradation machinery to regulate the destruction of a tagged protein in the presence or absence of a small molecule. Several groups, including Pratt et al. (1), have established systems that provide this ability. One method uses the FKBP12–rapamycin–FRB system to allow loss-of-function studies in a “drug-off” manner by inducing the degradation of a target protein through recruitment to the Rpd10 or Pre10 subunits of the yeast proteasome (Fig. 1) (7). This method can induce degradation of target proteins in <1 h. However, it has not been applied to metazoans or been used with nontoxic rapamycin analogues (rapalogues). A converse approach called inducible stabilization works in a “drug-on” method (Fig. 1) (8). Here, mutated forms of FRB (one is a triple mutant, FRB*, that contains a crucial T2098L substitution) act as “degrons” to cause degradation of fusion proteins. Upon recruitment of FKBP12 using rapamycin or rapalogues, the fusion protein is thermodynamically stabilized, and activity of the target protein is recovered. Inducible stabilization has been expanded to a nondimerizing method during which a mutated form of FKBP12 destabilizes fusion proteins until a small molecule called Shld1 is added (Fig. 1) (9). This approach has the advantage of minimizing interference of the target protein's functions in its stabilized form. Both of the inducible stabilization methods are rapidly reversible, producing a fusion protein half-life of ≈2 h after drug withdrawal.
Fig. 1.
A comparison of four methods for the conditional control of proteins using inducible protein degradation. Inducible proteasome targeting uses the FKBP12–rapamycin–FRB system to direct a target protein (TP) for proteasome-mediated degradation upon recruitment of a proteasome subunit (Rpd10 or Pre10). This approach has the fastest kinetics of degradation of the four methods. Inducible stabilization stabilizes target proteins that are otherwise destabilized by fusion to a triple-mutant form of FRB (FRB*) upon binding of FKBP12–rapamycin. This method uses a nontoxic rapamycin analogue called MaRap and has been used for endogenous gene regulation in mice. FKBP12-derived inducible stabilization stabilizes target proteins fused to a mutant form of FKBP12 (FKBP′) with addition of a nontoxic small molecule called Shld1 alone, minimizing disruption of the target protein's activity. SURF restores the native target protein by cleavage of a reconstituted ubiquitin from the target protein upon drug-induced stabilization. FKBP, FKBP12; FRB, FKBP12–rapamycin binding domain; TP, target protein; MaRap, methallyl rapamycin; Ub-N, N-terminal split ubiquitin; Ub-C, C-terminal split ubiquitin.
The method developed by Pratt et al. (1) is a clever modification of inducible stabilization that may have utility in certain settings for drug-dependent protein regulation (Fig. 1). Their study combines the use of the FKBP12–rapamycin–FRB method and some unique features of ubiquitin biology. When ubiquitin chains are added to proteins, they are targeted for proteasome-mediated destruction. However, when a single ubiquitin is fused to the N termini of proteins, it is cleaved by cellular ubiquitin proteases, releasing the rest of the polypeptide. This ability of ubiquitin is disrupted when it is “split” into two halves. When these halves are reconstituted by induced dimerization, the refolded ubiquitin once again creates a substrate for cleavage (10). Pratt et al. create a fusion protein that includes a multimerized FRB domain (which acts as a degron), half of the ubiquitin molecule, and a target protein. Dimerization of the FRB domains to FKBP12 molecules fused to the other half of ubiquitin reconstitutes a complete ubiquitin structure, which is subsequently cleaved. This cleavage releases the target protein from the FRB-based degron, restoring a functional and native target protein. Pratt et al. call this method “split ubiquitin for the rescue of function” (SURF). They demonstrate SURF using several targets, including Luciferase; caspase-8, a protease; v-Src, a cellular tyrosine kinase; and Smad6, a transcription factor. Importantly, these proteins are of different classes, supporting general applicability for this strategy.
The major advantage of SURF is that, after drug addition, the recovered target protein is not fused to any additional domains that may interfere with its activity. The researchers demonstrate how this can be problematic because stabilization of the triple-FRB fusion proteins of both v-Src and Smad6 fails to restore the function of those proteins when cleavage of the degron/ubiquitin is prevented. In these experiments, nine additional protein domains (three FRB domains, three FKBP12 domains, and three split ubiquitins) are simultaneously assembled on the target protein, so it is not surprising that this bulk inactivates the protein. In contrast, most small protein tags are not deleterious and are often used for assessing protein function. Indeed, many proteins have been shown to retain activity when fused to FRB and recruited to FKBP12 (11). Furthermore, WT FRB is a far less potent degron than FRB* in most cases (8, 12), and many proteins are unaffected by fusion to WT FRB. Likewise, FKBP12 fusion proteins also are generally active, including in the setting of FKBP-derived inducible stabilization (9). Nevertheless, it is clearly important to restrict tag size and to fully vet the activity of fused target proteins before creating conditional alleles.
The disadvantage of SURF compared with other inducible stabilization methods is that SURF would not be rapidly reversible upon drug removal because degradation of the target protein would depend on the normal kinetics of protein turnover. In addition, all inducible stabilization methods depend on protein translation before the restoration of protein function after drug addition [for example, it takes 24 h to fully recover luciferase levels after rapamycin addition in the assays of Pratt et al. (1)]. For dissections of molecular pathways, conditional regulation would be ideally accomplished in <15 min after drug addition (the minimum time for protein accumulation after changes in transcription), which clearly is not provided by SURF. Applying SURF to proteins with intrinsic rapid turnover may improve the kinetics, as would exploring ubiquitin mutants, which may mediate faster cleavage. It also will be important to confirm that SURF works using nontoxic rapalogues to avoid inhibiting endogenous mTor, because its signaling pathway is active in many settings (reviewed in ref. 13).
The ultimate test of the promising “inducible proteolytic shunt” method of Pratt et al. (1) and other inducible regulation systems is to apply them to endogenous genes to make new biological insights. In this light, conditional control of genes at the protein level in mice is a particularly salient goal because mouse genetics are increasingly prevalent in studies of development and disease, but sensitive and reproducible conditional systems are wanting. Although inducible stabilization has been used to define developmental time windows for GSK-3β function in palate and skeletal development (14), further pioneering experiments are required to validate the conditional protein degradation approaches before their use becomes widespread. Nevertheless, the overriding need for methods that can be used to create conditional protein alleles means that we can certainly look forward to exciting applications of SURF and related techniques in mice and other organisms.
Footnotes
The authors declare no conflict of interest.
See companion article on page 11209 in issue 27 of volume 104.
References
- 1.Pratt MR, Schwartz EC, Muir TW. Proc Natl Acad Sci USA. 2007;104:11209–11214. doi: 10.1073/pnas.0700816104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 3.Klemm JD, Beals CR, Crabtree GR. Curr Biol. 1997;7:638–644. doi: 10.1016/s0960-9822(06)00290-9. [DOI] [PubMed] [Google Scholar]
- 4.Ho SN, Biggar SR, Spencer DM, Schreiber SL, Crabtree GR. Nature. 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 5.Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 6.Choi J, Chen J, Schreiber SL, Clardy J. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 7.Janse DM, Crosas B, Finley D, Church GM. J Biol Chem. 2004;279:21415–21420. doi: 10.1074/jbc.M402954200. [DOI] [PubMed] [Google Scholar]
- 8.Stankunas K, Bayle JH, Gestwicki JE, Lin YM, Wandless TJ, Crabtree GR. Mol Cell. 2003;12:1615–1624. doi: 10.1016/s1097-2765(03)00491-x. [DOI] [PubMed] [Google Scholar]
- 9.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnsson N, Varshavsky A. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clackson T. Chem Biol Drug Design. 2006;6:440–442. doi: 10.1111/j.1747-0285.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 12.Stankunas K, Bayle JH, Havranek JJ, Wandless TJ, Baker D, Crabtree GR, Gestwicki JE. ChemBioChem. 2007 May 24; doi: 10.1002/cbic.200700087. [DOI] [PubMed] [Google Scholar]
- 13.Wullschleger S, Loewith R, Hall MN. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Liu KJ, Arron JR, Stankunas K, Crabtree GR, Longaker MT. Nature. 2007;446:79–82. doi: 10.1038/nature05557. [DOI] [PubMed] [Google Scholar]