Abstract
Mitochondrial injury, characterized by outer membrane permeabilization and consequent release of apoptogenic factors, is a key to apoptosis of mammalian cells. Bax and Bak, two multidomain Bcl-2 family proteins, provide a requisite gateway to mitochondrial injury. However it is unclear how Bax and Bak cooperate to provoke mitochondrial injury and whether their roles are redundant. Here, we have identified a unique role of Bak in mitochondrial fragmentation, a seemingly morphological event that contributes to mitochondrial injury during apoptosis. We show that mitochondrial fragmentation is attenuated in Bak-deficient mouse embryonic fibroblasts, baby mouse kidney cells, and, importantly, also in primary neurons isolated from brain cortex of Bak-deficient mice. In sharp contrast, Bax deficiency does not prevent mitochondrial fragmentation during apoptosis. Bcl-2 and Bcl-XL inhibit mitochondrial fragmentation, and their inhibitory effects depend on the presence of Bak. Reconstitution of Bak into Bax/Bak double-knockout cells restores mitochondrial fragmentation, whereas reconstitution of Bax is much less effective. Bak interacts with Mfn1 and Mfn2, two mitochondrial fusion proteins. During apoptosis, Bak dissociates from Mfn2 and enhances the association with Mfn1. Mutation of Bak in the BH3 domain prevents its dissociation from Mfn2 and diminishes its mitochondrial fragmentation activity. This study has uncovered a previously unrecognized function of Bak in the regulation of mitochondrial morphological dynamics during apoptosis. By this function, Bak may collaborate with Bax to permeabilize the outer membrane of mitochondria, unleashing the apoptotic cascade.
Keywords: Bax, mitochondria, Bcl-2, cytochrome c
Mitochondrial injury is central to apoptosis (1–3). The permeabilization of mitochondrial outer membrane leads to the release of apoptogenic factors such as cytochrome c (cyt.c), Smac/Diablo, Omi/HtrA, endonuclease G, and apoptosis-inducing factor (1–3). Critical regulators of mitochondrial integrity during apoptosis include Bcl-2 family proteins (4–8). In particular, Bax and Bak, two proapoptotic multidomain Bcl-2 proteins, are essential to the permeabilization of mitochondrial outer membrane (9, 10). Despite these findings, it remains unclear how Bax and Bak cooperate to provoke the membrane permeabilization and whether their roles are overlapping or redundant.
Recent studies have revealed a striking morphological change of mitochondria during apoptosis (11–14). Upon apoptotic stimulation, mitochondria collapse from a filamentous network into punctate fragments. Importantly, the morphological change seems to contribute to mitochondrial injury and consequent release of apoptogenic factors including cyt.c (15–24). Mitochondrial morphological dynamics is determined by a balance between two opposing processes, fission and fusion (25, 26). Thus, mitochondrial fragmentation during apoptosis may be a result of increased fission and/or decreased fusion. However, it is unknown how the morphological dynamics is regulated and shifted during apoptosis.
Whether Bcl-2 family proteins regulate mitochondrial morphology during apoptosis is unclear. Sugioka et al. (18) showed that overexpression of Bcl-2 did not affect mitochondrial fragmentation during apoptosis. Similar results were suggested for Bcl-XL, the antiapoptotic Bcl-2 homolog (16). In sharp contrast, Kong et al. (24) demonstrated the inhibitory effects of Bcl-2 on mitochondrial fragmentation during apoptosis of MCF-7 cells. In yeast, Bcl-2 and Bcl-XL expression blocked mitochondrial fission and cell death (19). In Caenorhabditis elegans, mitochondrial fragmentation during developmental cell death was affected by mutations of CED-9, a Bcl-2 ortholog in the worm (20). Moreover, CED-9, when transfected into mammalian cells, could antagonize mitochondrial fragmentation during apoptosis (27). Recent work by Karbowski et al. (28) further suggested a role of Bax and Bak in mitochondrial morphogenesis in normal healthy cells. Nevertheless, whether and how they are involved in the regulation of mitochondrial fragmentation during apoptosis is unknown.
Using loss- and gain-of-function approaches, we now show that Bak, but not Bax, has a critical role in mitochondrial fragmentation during apoptosis. Mechanistically, Bak interacts with Mfn1 and Mfn2, two mitochondrial fusion proteins. During apoptosis, Bak dissociates from Mfn2 and associates with Mfn1. Mutation of Bak in the BH3 domain prevents its dissociation from Mfn2, which is accompanied by the loss of mitochondrial fragmentation activity. Thus, Bak may regulate mitochondrial morphology and pathology during apoptosis by interacting with mitofusins.
Results
Mitochondrial Fragmentation Occurs Early During Apoptosis and Is Inhibited by Dominant-Negative Drp1 and Bcl-2 but Not by Caspase Inhibitors.
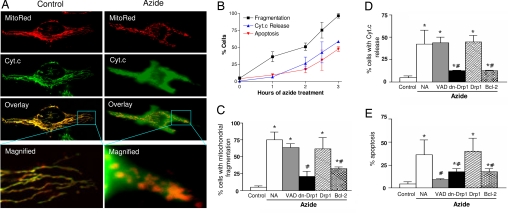
To study the mitochondrial morphological dynamics during apoptosis, HeLa cells were transfected with Mito-DsRed2 (MitoRed) to fluorescently label mitochondria (Fig. 1A) and then subjected to three types of apoptotic treatment including azide, staurosporine (STS), and cisplatin (29, 30). The treatments led to mitochondrial fragmentation. As shown in Fig. 1A, control cells had filamentous mitochondria (Fig. 1A, Control: MitoRed) and maintained cyt.c in the organelles (Fig. 1A, Control: Cyt.c). After azide incubation, mitochondria became fragmented (Fig. 1A, Azide: MitoRed) and lost cyt.c into cytosol (Fig. 1A, Azide: Cyt.c). These changes were clearly shown by superimposing the MitoRed and Cyt.c images and by higher magnification (Fig. 1A). Cell counting showed that <5% of control cells had mitochondrial fragmentation (Fig. 1B). Azide treatment for 1 h induced mitochondrial fragmentation in 36% of cells, and mitochondrial fragmentation increased progressively thereafter, reaching maximal levels in 3 h (Fig. 1B). The percentages of cells showing cyt.c release and apoptosis were always lower than that of mitochondrial fragmentation. Thus, 1 h of azide treatment did not induce cyt.c release or apoptosis, but did induce mitochondrial fragmentation (Fig. 1B). Similar results were shown for STS and cisplatin treatments (data not shown). The results suggest that mitochondrial fragmentation occurs early during apoptosis, before mitochondrial membrane permeabilization and cyt.c release.
Fig. 1.
Mitochondrial fragmentation precedes cytochrome c release during apoptosis and is inhibited by dominant-negative Drp1 mutant and Bcl-2. (A) Representative cell images showing mitochondrial fragmentation and cyt.c release after azide treatment. HeLa cells were transfected with MitoRed to fluorescently label mitochondria in red. The transfected cells were subjected to control incubation (Control) or 3 h of 10 mM azide treatment (Azide). Cyt.c was stained in green by indirect immunofluorescence. Images of MitoRed-labeled mitochondria (MitoRed) and cyt.c immunofluorescence (Cyt.c) were collected by confocal microscopy. (B) Time courses of mitochondrial fragmentation, cyt.c release, and apoptosis. HeLa cells transfected with MitoRed were treated with 10 mM azide for indicated time and stained for cyt.c immunofluorescence. Percentages of cells showing mitochondrial fragmentation, cyt.c release, and apoptotic morphology were evaluated by cell counting. Data are means ± SD of three separate experiments. (C–E) Effects of VAD, Drp1, dn-Drp1, and Bcl-2 on mitochondrial fragmentation, cyt.c release, and apoptosis. HeLa cells were cotransfected with MitoRed and dn-Drp1, Drp1, or Bcl-2. The cells were then treated with 10 mM azide for 3 h in the presence or absence of 100 μM VAD. Percentages of mitochondrial fragmentation, cyt.c release, and apoptosis in MitoRed-labeled cells were quantified by cell counting. Data are means ± SD of three separate experiments. *, significantly different from the untreated (Control) group; #, significantly different from the treated no-addition (NA) group.
VAD, a general peptide inhibitor of caspases, did not suppress mitochondrial fragmentation during apoptotic treatment (Fig. 1C). VAD did not attenuate cyt.c release either (Fig. 1D), although it diminished apoptosis as expected (Fig. 1E). Thus, mitochondrial fragmentation is not secondary to caspase activation. Mitochondrial fragmentation during apoptosis might be a result of changes in mitochondrial morphological dynamics dictated by fission and fusion. We tested this possibility by targeting Drp1, a critical mitochondrial fission protein (31). HeLa cells were transfected with wild-type Drp1 or its dominant-negative mutant [dn-Drp1 (31)] and were then subjected to apoptotic treatment. As shown in Fig. 1C, dn-Drp1 (and not wild-type Drp1) suppressed mitochondrial fragmentation during apoptosis. Notably dn-Drp1 also ameliorated cyt.c release and apoptosis [Fig. 1 D and E and supporting information (SI) Fig. 6]. The results support earlier studies (15–23) for a role of mitochondrial fission or fragmentation in mitochondrial injury during apoptosis.
Bcl-2 family proteins are critical regulators of mitochondrial injury during apoptosis (4–8). Whether these proteins regulate mitochondrial morphological dynamics under the pathological condition is unclear (16, 18–20, 24, 27, 32). We demonstrated the inhibitory effects of Bcl-2 on mitochondrial fragmentation during apoptosis (Fig. 1C). For example, mitochondrial fragmentation during azide treatment was reduced from 75% in untransfected cells to 33% in Bcl-2 transiently transfected cells. Bcl-2 also inhibited cyt.c release and apoptosis (Fig. 1 D and E). Mitochondrial fragmentation during apoptosis was also suppressed in cells stably transfected with Bcl-2 and by transient transfection of Bcl-XL (SI Fig. 7). Bcl-2 has dual subcellular localizations, mitochondria and endoplasmic reticulum (ER). Using Bcl-2 constructs of targeted expression (33, 34), we showed that mitochondrial Bcl-2 was more effective than ER-Bcl-2 in suppressing mitochondrial fragmentation; nevertheless, mitochondrial Bcl-2 was not as effective as the wild-type gene (SI Fig. 8). Together, the results suggest that Bcl-2 family proteins may regulate mitochondrial morphological dynamics during apoptosis.
Mitochondrial Fragmentation During Apoptosis Is Inhibited in Bak-Deficient Cells but Not in Bax-Deficient Cells.
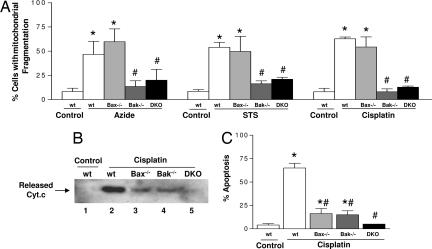
To pursue the mitochondrial morphological regulation by Bcl-2 family proteins, we focused on Bax and Bak, two crucial proapoptotic Bcl-2 proteins (4–10). We first examined Bax and Bak single- or double-knockout mouse embryonic fibroblasts (MEF) (9, 10); deficiency of Bax or Bak in these cells was confirmed by immunoblot analysis (SI Fig. 9). Regardless of their genotypes, all cells showed low mitochondrial fragmentation under control condition (SI Fig. 9 B–D). After apoptotic treatment, mitochondrial fragmentation was induced in wild-type cells and Bax-knockout cells (Fig. 2A). In sharp contrast, mitochondrial fragmentation was drastically reduced in Bak-knockout and Bax/Bak double-knockout cells (Fig. 2A). Consistent with previous observations (10), single knockout of Bax or Bak reduced, and double knockout of Bax/Bak completely blocked, cyt.c release and apoptosis (Fig. 2 B and C). The results suggest a unique role of Bak in the regulation of mitochondrial fragmentation during apoptosis. This finding was validated by using transformed baby mouse kidney cells (35) (SI Fig. 10). We further confirmed the unique role of Bak in mitochondrial fragmentation during apoptosis in primary cultures of brain cortical neurons (SI Fig. 11) isolated from wild-type, Bax-knockout, and Bak-knockout mice. Of note, in Bak-knockout cells, the limited cyt.c release was shown in the low percentage of cells that had fragmented mitochondria (SI Fig. 12). These results suggest that both Bax and Bak contribute to mitochondrial injury; however, their roles may not be completely overlapping. By inducing mitochondrial fragmentation, Bak may collaborate with Bax to ensure an efficient outer membrane permeabilization and complete cyt.c release.
Fig. 2.
Bak (but not Bax) deficiency blocks mitochondrial fragmentation during apoptosis. Wild-type (wt), Bax-knockout (Bax−/−), Bak-knockout (Bak−/−), and Bax/Bak double-knockout (DKO) MEFs were subjected to apoptotic treatments with 10 mM azide for 3 h, 1 μM STS for 4 h, or 20 μM cisplatin for 16 h. To evaluate mitochondrial fragmentation, the MEFs were transfected with MitoRed before apoptotic treatments. (A) Cells with mitochondrial fragmentation were examined by fluorescence microscopy and quantified by cell counting. (B) To analyze cyt.c release, MEFs after cisplatin treatment were extracted to collect the cytosolic fraction for immunoblot analysis of cyt.c. (C) To analyze apoptosis, MEFs after cisplatin treatment were stained with Hoechst 33342. Apoptosis was evaluated by counting of the cells with typical apoptotic morphology including cellular and nuclear condensation and fragmentation. Data are presented as means ± SD of four separate experiments. *, significantly different from the untreated control group; #, significantly different from the treated wild-type group.
Inhibitory Effects of Bcl-2 on Mitochondrial Fragmentation Depend on the Presence of Bak and Not Bax.
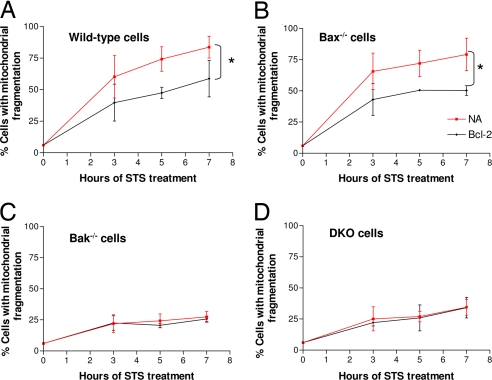
We determined whether Bak was required for the inhibitory effects of Bcl-2 on mitochondrial fragmentation (Fig. 3). Consistent with our earlier results shown in Fig. 2, mitochondrial fragmentation during apoptotic treatment in Bak-knockout and Bak/Bax double-knockout MEF was significantly lower than that of wild-type and Bax-knockout cells (Fig. 3 C and D versus Fig. 3 A and B). Importantly, whereas Bcl-2 suppressed mitochondrial fragmentation in both wild-type and Bax-knockout cells (Fig. 3 A and B), it did not inhibit mitochondrial fragmentation in Bak-knockout or Bak/Bax double-knockout cells (Fig. 3 C and D). Thus, Bak seems to be an essential regulator of mitochondrial fragmentation whereby Bcl-2 exerts its inhibitory effects.
Fig. 3.
Bcl-2 inhibits mitochondrial fragmentation in Bax-knockout cells but not in Bak-knockout cells. MEF cells of different genotypes wild-type (A), Bax-knockout (B), Bak-knockout (C), and Bax/Bak double knockout (D) were transfected with MitoRed alone (NA) or cotransfected with MitoRed and Bcl-2 (Bcl-2). The cells were then subjected to apoptotic treatment with 1 μM STS. Mitochondrial fragmentation was examined by fluorescence and confocal microscopy and quantified by cell counting. Data are means ± SD of three separate experiments. ∗, significant difference between the nontransfected NA group and the Bcl-2-transfected group.
Bak Is More Effective Than Bax in Restoring Mitochondrial Fragmentation in Bax/Bak Double-Knockout Cells.
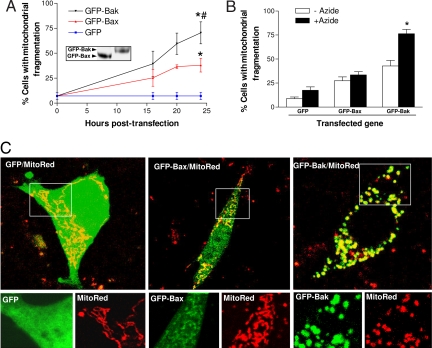
If Bak is indeed a key to mitochondrial fragmentation during apoptosis, reconstitution of Bak into Bak-knockout cells should restore mitochondrial fragmentation. To test this, we transfected Bax/Bak double-knockout MEF cells with GFP-Bax, GFP-Bak, or GFP. MitoRed was cotransfected to reveal mitochondrial morphology. As shown in Fig. 4A, both GFP-Bax and GFP-Bak induced a transfection time-dependent increase of mitochondrial fragmentation, but GFP-Bak was significantly more effective in this function (Fig. 4A). The efficacy of GFP-Bak in mitochondrial fragmentation was not due to higher expression of this protein (Fig. 4A Inset). We further determined the effects of Bax or Bak reconstitution on mitochondrial fragmentation during apoptotic treatment. To this end, Bax/Bak double-knockout cells were cotransfected for 16 h with MitoRed and either GFP-Bax or GFP-Bak and then subjected to azide treatment. As shown in Fig. 4B, azide did not induce significant mitochondrial fragmentation in cells reconstituted with GFP or GFP-Bax, but it did increase mitochondrial fragmentation from 43% to 76% in Bak-reconstituted cells (Fig. 4B, GFP-Bak, open versus filled columns). Representative images of GFP-, GFP-Bax-, or GFP-Bak-transfected cells after azide treatment are shown in Fig. 4C. As expected, GFP was expressed all over the cell, whereas Bax showed some accumulation in mitochondria and Bak had a restrictive mitochondrial localization. Notably, the cells reconstituted with Bak, but not GFP or Bax, fragmented their mitochondria [Fig. 4C, magnified (Lower)]. Similarly, Bak-reconstituted cells showed higher mitochondrial fragmentation during STS treatment than the cells reconstituted with Bax (SI Fig. 13). This gain-of-function study further supports a role of Bak in mitochondrial fragmentation during apoptosis.
Fig. 4.
Bak is more effective than Bax in restoring mitochondrial fragmentation in Bax/Bak double-knockout cells. (A) Induction of mitochondrial fragmentation by Bax or Bak transfection in Bax/Bak double-knockout (DKO) MEF cells. DKO cells were cotransfected with MitoRed and GFP, GFP-Bax, or GFP-Bak. At indicated time points, the cotransfected cells were examined by fluorescence and confocal microscopy to evaluate mitochondrial fragmentation. Data are means ± SD of five separate experiments. ∗, significantly different from GFP-transfected group; #, significantly different from GFP-Bax-transfected group. (Inset) Immunoblot analysis of GFP-Bax and GFP-Bak expression by using an anti-GFP antibody. (B) Restoration of azide-induced mitochondrial fragmentation in DKO cells by Bak but not by Bax. DKO cells were cotransfected with MitoRed and GFP, GFP-Bax, or GFP-Bak for 16 h. The cells were then incubated for another 3 h with control medium or 10 mM azide. Mitochondrial fragmentation in transfected cells was examined by fluorescence and confocal microscopy. Data are means ± SD of four separate experiments. ∗, significant difference between the treated (+Azide) and untreated (−Azide) group. (C) Representative images of MitoRed/GFP, MitoRed/GFP-Bax, and MitoRed/GFP-Bak cotransfected cells after azide treatment. Shown are transfected cells with superimposed MitoRed and GFP signals (Upper) and magnified areas with separated MitoRed and GFP signals (Lower).
Bak Interaction with Mitofusins: Changes During Apoptotic Treatment and Effects of BH3 Mutation.
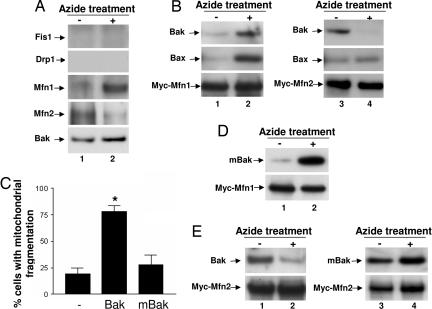
To gain mechanistic insights into mitochondrial morphological regulation by Bak, we determined Bak interaction with mitochondrial fission–fusion proteins. We first analyzed endogenous protein interactions by coimmunoprecipitation (IP). In this assay, Bak did not interact with Fis1 or Drp1, two mitochondrial fission proteins, under either control or apoptotic conditions (Fig. 5A). On the contrary, Bak showed co-IP with two fusion proteins Mfn1 and Mfn2 in control HeLa cells (Fig. 5A, lane 1). Notably, upon apoptotic induction by azide, Bak dissociated from Mfn2 and increased its interaction with Mfn1 (Fig. 5A, lane 2). Similar interactions between Bak and Mfn1 and 2 were demonstrated during cisplatin treatment (SI Fig. 14). The specificity of the co-IP assay was supported by control experiments by using Mfn1- or 2-deficient cells and nonimmune serum (SI Fig. 15). To further determine the interaction between Bak and Mfn1 and 2, we transfected HeLa cells with Myc-tagged Mfn1 or -2 to collect lysates for IP using an anti-Myc antibody. As shown in Fig. 5B, small yet consistently detectable amounts of Bak and Bax coimmunoprecipitated with Myc-Mfn1 under control condition (Fig. 5B, lane 1); the molecular interactions increased markedly during apoptosis (Fig. 5B, lane 2). Bak and Bax also interacted with Myc-Mfn2 under control condition (Fig. 5B, lane 3). After apoptotic treatment, Bak, but not Bax, dissociated from Myc-Mfn2 (Fig. 5B, lane 4). Thus, Bak showed a unique interaction pattern with both endogenous (Fig. 5A) and transfected (Fig. 5B) mitofusins; remarkably the molecular interactions changed upon apoptotic induction. The molecular interaction between Bak and mitofusins was further confirmed by FRET assay (SI Fig. 16). In addition, transfected Bak and mitofusins showed colocalization (SI Fig. 17). Collectively, the results suggest that Bak may regulate mitochondrial morphological dynamics by interacting with mitofusins. To further investigate this possibility, we used a loss-of-function mutant of Bak that had a point (L75E) mutation in the BH3 domain (36). When transfected into Bax/Bak double-knockout MEF cells, the mutant Bak (mBak) did not induce apoptosis (data not shown), and, importantly, unlike wild-type Bak, mBak could not induce mitochondrial fragmentation either (Fig. 5C). We then determined the interaction of mBak with Mfn1 and Mfn2. Similar to wild-type Bak (Fig. 5 A and B), mBak interacted with Mfn1 weakly under control condition (Fig. 5D, lane 1), and, upon apoptotic stimulation, the molecular interaction was markedly enhanced (Fig. 5D, lane 2). The most striking differences between Bak and mBak were shown in their interactions with Mfn2. As shown in Fig. 5E, wild-type Bak interacted with Mfn2 in control cells (Fig. 5E, lane 1), and the interaction was disrupted during apoptosis (Fig. 5E, lane 2). In sharp contrast, mBak did not dissociate from Mfn2 during apoptotic treatment (Fig. 5E, lane 4).
Fig. 5.
Bak interaction with mitofusins: changes during apoptotic treatment and effects of BH3 mutation. (A) HeLa cells were untreated or treated with 10 mM azide for 3 h. Whole lysates were collected with CHAPS buffer and subjected to immunoprecipitation using an anti-Bak antibody. The resultant immunoprecipitates were analyzed for Fis1, Drp1, Mfn1, Mfn2, and Bak by immunoblotting. (B) HeLa cells were transfected with Myc-Mfn1 or Myc-Mfn2. The cells were untreated or treated with azide to collect whole-cell lysates for immunoprecipitation using an anti-Myc antibody. The resultant immunoprecipitates were analyzed for Bak, Bax, Myc-Mfn1, or Myc-Mfn2 by immunoblotting. (C) Bax/Bak double-knockout MEFs were cotransfected with Mito-Red and Bak, mBak, or empty vector. Cells with mitochondrial fragmentation were examined and counted by fluorescence microscopy. (D and E) Bax/Bak double-knockout MEFs were cotransfected with Bak or mBak and Myc-Mfn1 or Myc-Mfn2. The cells were then untreated or treated with azide to collect whole-cell lysates for immunoprecipitation using an anti-Myc antibody. The resultant immunoprecipitates were analyzed for Bak, mBak, and Myc-Mfn1 or Myc-Mfn2 by immunoblotting. Results in A, B, D, and E are representatives of at least three separate experiments. Data in C are presented as means ± SD of three separate experiments. ∗, significantly different from the empty vector transfection group.
Discussion
In mammalian cells, Bax and Bak are essential to mitochondrial outer membrane permeabilization during apoptosis (9, 10). It is generally believed that the roles of Bax and Bak in mitochondrial injury and apoptosis are overlapping or redundant (4–8), despite the notable differences in their subcellular localizations and interacting proteins (36, 37), and the phenotypic differences between Bax- and Bak-deficient mice (9, 38). Here, we have identified a previously undescribed function of Bak: it regulates mitochondrial fragmentation. The role seems to be unique for Bak, because it cannot be effectively substituted by Bax.
Mitochondrial fragmentation, as a result of altered morphological dynamics, has been implicated in mitochondrial injury during apoptosis (11–14). Our results now further suggest that the mitochondrial fragmentation may involve Bak. On the other hand, mitochondrial fragmentation involving Bak is not sufficient to induce maximal cyt.c release, which apparently requires Bax (9, 10) (Fig. 2B). Thus, Bak and Bax need to collaborate to evoke a full mitochondrial pathology. It is also noteworthy that neither Bak nor Bax is absolutely required for cyt.c release. In Bax-deficient cells, Bax-mediated mitochondrial injury is attenuated, but Bak can induce a small amount of cyt.c release, probably via mitochondrial fragmentation. In Bak-deficient cells, Bak-mediated mitochondrial fragmentation is suppressed, but Bax can induce limited cyt.c release (Fig. 2B).
A recent study by Karbowski et al. (28) showed mitochondrial fragmentation in Bax/Bak double-knockout cells under control culture condition. However, in our experiments, mitochondria in these cells were mainly filamentous (SI Fig. 9 B–D). Filamentous mitochondria were also shown in Bax/Bak double-knockout cells by other groups (39, 40). The exact cause of the discrepancy between these observations is unclear. Nevertheless the mitochondrial morphological dynamics seems to be quite sensitive to culture conditions, and subtle stress may induce fragmentation. In addition, the morphological dynamics is regulated during cell cycle, where mitotic cells tend to have fragmented mitochondria (41).
It is unclear how Bak regulates mitochondrial morphological dynamics during apoptosis. Nevertheless, CED-9, a Bcl-2 protein in C. elegans, interacts with Mfn2 (27). More recent work further suggests a functional interaction between Bax and Mfn2 in nonapoptotic cells (28). We now show that Bak interacts with Mfn-1 and -2 but not with mitochondrial fission proteins. Upon apoptosis induction, Bak dissociates from Mfn2 and associates with Mfn1. Moreover, mutation of Bak in the BH3 domain prevents Bak dissociation from Mfn2 during apoptosis, which is accompanied by the loss of mitochondrial fragmentation activity (Fig. 5). It is unclear why the mutant Bak cannot dissociate from Mfn2. One possibility is that, upon apoptotic stimulation, wild-type Bak can change its conformation to induce its dissociation from Mfn2, whereas the mutant Bak does not undergo the conformational change. Regardless of the underlying mechanisms, our results suggest a correlation of the mitochondrial fragmentation activity of Bak with its dissociation from Mfn2. It is plausible that the dissociation of Bak from Mfn2 may decrease the mitochondrial fusion activity of Mfn2 and, as a result, lead to cessation of fusion, inducing mitochondrial fragmentation. Further investigation is needed to test this possibility and gain mechanistic insights into the regulation and functionality of Bak–mitofusin interaction.
Our observation of the partial inhibitory effects of Bcl-2 on mitochondrial fragmentation is at odds with Sugioka et al. (18) but is consistent with Kong et al. (24) and Fannjiang et al. (19). We further show that mitochondrially targeted Bcl-2 is more effective than ER Bcl-2, but less effective than wild-type Bcl-2, in suppressing mitochondrial fragmentation. Bcl-2 may not directly interact with Bak (37), but it may regulate mitochondrial morphological dynamics by sequestering BH3-only proteins, regulating Ca2+ homeostasis, and/or interacting with mitochondrial fission–fusion proteins.
Knockout of Mfn1 or 2 leads to embryonic lethality in mice. MEFs derived from these gene knockout animals show very high levels of mitochondrial fragmentation, yet do not show much spontaneous apoptosis (42), suggesting that the lack of mitofusins and consequent mitochondrial fragmentation does not necessarily induce cell death. On the other hand, a regulatory role of mitofusins in apoptosis has been suggested by gene overexpression and siRNA knockdown studies (18, 43, 44). Despite these findings, it is unclear how mitofusins are regulated during apoptosis. Our current results suggest that their regulation may involve Bak. Identification of a function of Bak in mitochondrial morphological dynamics should have significant implications in our understanding of Bcl-2 family proteins, apoptosis, and mitochondrial pathology.
Materials and Methods
Cells.
Transformed MEF and baby mouse kidney cells of various Bax/Bak genotypes were originally from Stanley Korsmeyer (Dana–Farber Cancer Institute, Harvard Medical School, Boston, MA) and Eileen White (Rutgers University, Piscataway, NJ). MEFs deficient in Mfn1 or Mfn2 were from David C. Chan (California Institute of Technology, Pasadena, CA) (42). Primary neurons were isolated and cultured by using a method modified from previous studies (45).
Plasmids.
The coding regions of Bax and Bak were amplified by RT-PCR from mRNA of kidney tubular epithelial cells and cloned into pEGFP-C3 (BD Clontech, Palo Alto, CA) by using HindIII/KpnI sites for Bax and HindIII/BamH I sites for Bak. Myc-Mfn1 and Myc-Mfn2 were kindly provided by David C. Chan (36, 42). Murine Bak and the Bak (L75E) mutant were from Emily H.-Y. Cheng (Washington University School of Medicine, St. Louis, MO) (36). Drp1 and dn-Drp1(K38A) were from Alexander van der Bliek (University of California, Los Angeles, CA) (46), and subcloned into pcDNA3.1 for this study. Bcl-2 and Bcl-XL plasmids were from Junying Yuan (Harvard Medical School, Boston, MA) and Xiao-Ming Yin (University of Pittsburgh, Pittsburgh, PA), respectively. Bcl-2 plasmids for targeted expression in mitochondria or ER were from David W. Andrews (McMaster University Health Sciences Centre, Ontario, Canada).
Apoptotic Treatment.
Cells were incubated with 10 mM azide in glucose-free buffer, 1 μM STS, or 20–50 μM cisplatin in culture medium. The treatments induced cellular stress (ATP-depletion by azide, protein kinase inhibition by STS, DNA damage by cisplatin), leading to apoptosis mediated by mitochondrial injury (29, 30).
Mitochondrial Morphology.
Cells transfected with MitoRed were identified by fluorescence microscopy. Mitochondrial morphology in individual cells was evaluated. Fragmented mitochondria were shortened, punctate, and sometimes rounded, whereas filamentous mitochondria showed a thread-like tubular structure. Consistent with earlier studies, the mitochondria within one cell were often either filamentous or fragmented. In rare cases of mixed morphologies, we classified the cells based on the majority (>70%) of mitochondria. In some experiments, apoptosis and cyt.c release after immunofluorescence staining were also evaluated in MitoRed-transfected cells. Apoptosis was indicated by cell morphology including cellular condensation, formation of apoptotic bodies, and nuclear condensation and fragmentation. Cyt.c release was indicated by the loss of mitochondrial cyt.c staining and the appearance of cyt.c in the cytosol. For each sample, several random fields of cells (≥100 cells per dish) were evaluated for mitochondrial morphology, apoptosis, and cyt.c release.
Additional detailed methods are available in SI Methods.
Supplementary Material
Acknowledgments
We thank Drs. David C. Chan, Stanley J. Korsmeyer, Eileen White, Alexander van der Bliek, Margaret T. Fuller (Stanford University School of Medicine, Stanford, CA), David W. Andrews, Roger Y. Tsien (University of California at San Diego, La Jolla, CA), Emily H.-Y. Cheng, Junying Yuan, Xiao-Ming Yin, Dean Tang (University of Texas M. D. Anderson Cancer Center, Houston, TX), and Shivendra Singh (University of Pittsburgh, Pittsburgh, PA) for research reagents including plasmids, cell lines, and antibodies. C.B. is supported in part by the Multidisciplinary Predoctoral Training Program in Integrative Cardiovascular Biology from the National Institutes of Health (NIH). This work was supported by grants from the NIH and the U.S. Department of Veterans Affairs.
Abbreviations
- cyt.c
cytochrome c
- ER
endoplasmic reticulum
- IP
coimmunoprecipitation
- MEF
mouse embryonic fibroblast
- STS
staurosporine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703976104/DC1.
References
- 1.Green DR, Kroemer G. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 2.Martinou JC, Green DR. Nat Rev Mol Cell Biol. 2001;2:63–67. doi: 10.1038/35048069. [DOI] [PubMed] [Google Scholar]
- 3.Wang X. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 4.Cory S, Huang DC, Adams JM. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 5.Danial NN, Korsmeyer SJ. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 6.Harris MH, Thompson CB. Cell Death Differ. 2000;7:1182–1191. doi: 10.1038/sj.cdd.4400781. [DOI] [PubMed] [Google Scholar]
- 7.Tsujimoto Y. J Cell Physiol. 2003;195:158–167. doi: 10.1002/jcp.10254. [DOI] [PubMed] [Google Scholar]
- 8.Kuwana T, Newmeyer DD. Curr Opin Cell Biol. 2003;15:691–699. doi: 10.1016/j.ceb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Lindsten T, Ross AJ, King A, Zong WX, Rathmell JC, Shiels HA, Ulrich E, Waymire KG, Mahar P, Frauwirth K, et al. Mol Cell. 2000;6:1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cereghetti GM, Scorrano L. Oncogene. 2006;25:4717–4724. doi: 10.1038/sj.onc.1209605. [DOI] [PubMed] [Google Scholar]
- 12.Heath-Engel HM, Shore GC. Biochim Biophys Acta. 2006;763:549–560. doi: 10.1016/j.bbamcr.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Parone PA, Martinou JC. Biochim Biophys Acta. 2006;1763:522–530. doi: 10.1016/j.bbamcr.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Youle RJ, Karbowski M. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 15.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 16.James DI, Parone PA, Mattenberger Y, Martinou JC. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 17.Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Mol Biol Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugioka R, Shimizu S, Tsujimoto Y. J Biol Chem. 2004;279:52726–52734. doi: 10.1074/jbc.M408910200. [DOI] [PubMed] [Google Scholar]
- 19.Fannjiang Y, Cheng WC, Lee SJ, Qi B, Pevsner J, McCaffery JM, Hill RB, Basanez G, Hardwick JM. Genes Dev. 2004;18:2785–2797. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jagasia R, Grote P, Westermann B, Conradt B. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- 21.Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ. J Cell Biol. 2004;164:493–499. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breckenridge DG, Stojanovic M, Marcellus RC, Shore GC. J Cell Biol. 2003;160:1115–1127. doi: 10.1083/jcb.200212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Graber S, Kovacs I, Lee WD, Waggoner J, Cui J, et al. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong D, Xu L, Yu Y, Zhu W, Andrews DW, Yoon Y, Kuo TH. Mol Cell Biochem. 2005;272:187–199. doi: 10.1007/s11010-005-7323-3. [DOI] [PubMed] [Google Scholar]
- 25.Chan DC. Annu Rev Cell Dev Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 26.Okamoto K, Shaw JM. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 27.Delivani P, Adrain C, Taylor RC, Duriez PJ, Martin SJ. Mol Cell. 2006;21:761–773. doi: 10.1016/j.molcel.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 28.Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 29.Dong Z, Wang J. J Biol Chem. 2004;279:9215–9221. doi: 10.1074/jbc.M312225200. [DOI] [PubMed] [Google Scholar]
- 30.Jiang M, Wei Q, Wang J, Du Q, Yu J, Zhang L, Dong Z. Oncogene. 2006;25:4056–4066. doi: 10.1038/sj.onc.1209440. [DOI] [PubMed] [Google Scholar]
- 31.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnoult D, Rismanchi N, Grodet A, Roberts RG, Seeburg DP, Estaquier J, Sheng M, Blackstone C. Curr Biol. 2005;15:2112–2118. doi: 10.1016/j.cub.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 33.Zhu W, Cowie A, Wasfy GW, Penn LZ, Leber B, Andrews DW. EMBO J. 1996;15:4130–4141. [PMC free article] [PubMed] [Google Scholar]
- 34.Fiebig AA, Zhu W, Hollerbach C, Leber B, Andrews DW. BMC Cancer. 2006;6:213. doi: 10.1186/1471-2407-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Degenhardt K, Sundararajan R, Lindsten T, Thompson C, White E. J Biol Chem. 2002;277:14127–14134. doi: 10.1074/jbc.M109939200. [DOI] [PubMed] [Google Scholar]
- 36.Cheng EH, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ. Science. 2003;301:513–517. doi: 10.1126/science.1083995. [DOI] [PubMed] [Google Scholar]
- 37.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DC. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fannjiang Y, Kim CH, Huganir RL, Zou S, Lindsten T, Thompson CB, Mito T, Traystman RJ, Larsen T, Griffin DE, et al. Dev Cell. 2003;4:575–585. doi: 10.1016/s1534-5807(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 39.Alirol E, James D, Huber D, Marchetto A, Vergani L, Martinou JC, Scorrano L. Mol Biol Cell. 2006;17:4593–4605. doi: 10.1091/mbc.E06-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wasiak S, Zunino R, McBride HM. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neuspiel M, Zunino R, Gangaraju S, Rippstein P, McBride H. J Biol Chem. 2005;280:25060–25070. doi: 10.1074/jbc.M501599200. [DOI] [PubMed] [Google Scholar]
- 44.Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ. J Cell Biol. 2002;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu XJ, Wang CZ, Dai PG, Xie Y, Song NN, Liu Y, Du QS, Mei L, Ding YQ, Xiong WC. Nat Cell Biol. 2007;9:184–192. doi: 10.1038/ncb1535. [DOI] [PubMed] [Google Scholar]
- 46.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.