Abstract
CD6 is a lymphocyte receptor that belongs to the scavenger receptor cysteine-rich superfamily. Because some members of the scavenger receptor cysteine-rich superfamily act as pattern recognition receptors for microbial components, we studied whether CD6 shares this function. We produced a recombinant form of the ectodomain of CD6 (rsCD6), which was indistinguishable (in apparent molecular mass, antibody reactivity, and cell binding properties) from a circulating form of CD6 affinity-purified from human serum. rsCD6 bound to and aggregated several Gram-positive and -negative bacterial strains through the recognition of lipoteichoic acid and LPS, respectively. The Kd of the LPS–rsCD6 interaction was 2.69 ± 0.32 × 10−8 M, which is similar to that reported for the LPS–CD14 interaction. Further experiments showed that membrane CD6 also retains the LPS-binding ability, and it results in activation of the MAPK signaling cascade. In vivo experiments demonstrated that i.p. administration of rsCD6 before lethal LPS challenge significantly improved mice survival, and this was concomitant with reduced serum levels of the proinflammatory cytokines TNF-α, IL6, and IL-1β. In conclusion, our results illustrate the unprecedented bacterial binding properties of rsCD6 and support its therapeutic potential for the intervention of septic shock syndrome or other inflammatory diseases of infectious origin.
Keywords: bacterial cell component, innate immunity, lymphocyte surface receptor
The scavenger receptor cysteine-rich superfamily (SRCR-SF) is an ancient and highly conserved family of proteins, characterized by the presence of one or several repeats of a cysteine-rich extracellular domain named SRCR (1, 2). The SRCR-SF includes both cell-surface and secreted proteins that can be expressed on cells of hemopoietic origin such as macrophages (e.g., SR-AI/II, MARCO, CD163, Mac2-binding protein, and Spα) and lymphocytes (e.g., CD5, CD6, and T19/WT1), as well as on nonhematological cells such as those from of the digestive, respiratory, and urinary epithelial tracts (e.g., DMBT1, S4D-SRCRB, and SCARA5) (2). There is no unifying function for all of the members of the SRCR-SF, but some of them have been implicated in the development of the immune system and in the regulation of innate and adaptive immune responses (3). A few members of the SRCR-SF (i.e., SR-AI/II, MARCO, DMBT1, Spα, and SCARA5) are known to interact with bacteria and to bind to conserved pathogen-associated molecular patterns present on microbial surfaces, such as LPS, lipoteichoic acid (LTA), and peptidoglycan. These interactions were initially mapped outside the SRCR domains (4), but recent reports have demonstrated the direct involvement of the SRCR domains in such interaction (5–8). Given the conserved structure of SRCR domains throughout the evolution, it remains to be analyzed whether pathogen scavenging is a general property shared by all members of the SRCR-SF or only by a selected group of its members.
The CD5 and CD6 receptors are the two only members of the SRCR-SF that are expressed on human lymphocytes. Both are lymphoid-specific surface glycoproteins sharing important similarities in structure, function, and tissue expression (2). CD5 and CD6 are expressed on thymocytes, mature T cells, and the B1a B cell subset, although CD6 expression has also been reported on certain regions of the brain (9). They exhibit important differences in their cytoplasmic regions, but their extracellular regions are exclusively composed of three consecutive SRCR domains, which show extensive amino acid sequence identity (1, 10). Functionally, they are physically associated to the antigen-specific receptor complex present on T (TCR/CD3) and B (BCR) cells, where they contribute to either positive or negative modulation of the activation and differentiation signals delivered by that receptor complex (2, 11, 12). In the present study we have explored the bacterial binding capabilities of the ectodomains of the human lymphocyte receptors CD5 and CD6, both known to exist as membrane receptors, but also as soluble receptors circulating in serum (13, 14). Data are provided herein on the binding of soluble and membrane forms of CD6, but not CD5, to the surface of Gram-positive and Gram-negative bacteria through the recognition of pathogen-associated molecular patterns (namely, LPS and LTA). The relevance of such an interaction is illustrated by the beneficial effects of the infusion of a recombinant soluble form of CD6 (rsCD6) on the survival rate in a mouse experimental model of septic shock.
Results
rsCD6 Binds to Gram-Positive and Gram-Negative Bacteria.
To determine whether the ectodomain of human CD6 and CD5 could directly bind to the surface of whole bacteria, we used an approach that was previously used for the study of SR-AI binding to bacteria (15). Thus, biotin-labeled recombinant soluble proteins encompassing the ectodomains of human CD5, CD6, and Spα (rsCD5, rsCD6, and rSpα) (Fig. 1A) were incubated with bacterial suspensions, and their binding to bacterial pellets was further assayed by SDS/PAGE and Western blotting against streptavidin–HRP. Our results show that, as previously reported for rSpα (8), rsCD6 bound to Gram-positive and -negative bacteria (Fig. 1B), indicating that this protein also possesses bacterial binding activity. In contrast, neither rsCD5 nor the negative control BSA bound to bacterial suspensions. As illustrated by Fig. 2C, the presence of biotin-labeled rsCD6 was greatly reduced in Escherichia coli and Staphylococcus aureus bacterial cell pellets in the presence of EDTA. This indicates that, like rSpα (8) and CRP-ductin (16), rsCD6 recognition of cell wall components from Gram-positive and -negative bacteria is facilitated by Ca2+.
Fig. 1.
Binding of rsCD6 to Gram-positive and Gram-negative bacteria. (A) Western blot analysis of the affinity-purified biotin-labeled proteins. (B) Protein binding to E. coli and S. aureus. (C) Calcium influence on the binding of rsCD6 and rSpα to E. coli and S. aureus. (D) Competition binding assays of rsCD6 to E. coli and S. aureus in the presence of increasing concentrations of LPS or LTA. For bacterial binding studies, biotin-labeled proteins were incubated with a suspension of 5 × 107 bacteria. Unbound protein was washed off, and then bacteria and bound protein were solubilized with SDS/PAGE loading buffer and electrophoresed. Detection of biotin-labeled proteins was performed by Western blot using HRP–streptavidin.
Fig. 2.
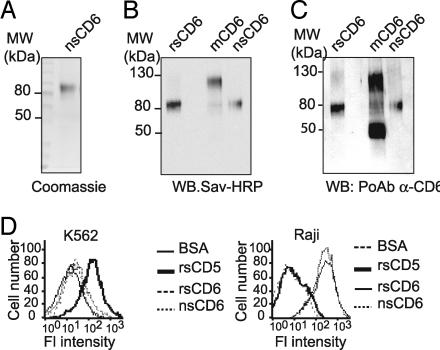
Characterization of affinity-purified circulating CD6 from human serum. (A) Coomassie blue staining of affinity-purified nsCD6 from human serum. (B) Western blot analysis of biotin-labeled purified nsCD6 and rsCD6 proteins and membrane CD6 (mCD6) immunoprecipitated from surface biotinylated HUT-78 cells with streptavidin–HRP. (C) Membranes containing the same proteins as in B, Western blotted with a rabbit polyclonal antiserum specific for the extracellular region of CD6. (D) Flow cytometry analysis of the reactivity of biotinylated rsCD5, rsCD6, nsCD6, or BSA (used as a negative control) with the K652 and Raji cells. Bound protein was detected with streptavidin–Tricolor.
We next sought to determine whether the observed binding of rsCD6 to bacteria was specific and to identify which bacterial cell-surface structures were being recognized. To answer these questions, competition experiments were designed in which biotin-labeled rsCD6 was incubated with increasing concentrations of purified LPS or LTA before the addition of a suspension of either E. coli or S. aureus (5 × 107 cells). LPS and LTA were assayed because they are ubiquitous cell-surface components of these microorganisms. As illustrated by Fig. 2D, binding of biotin-labeled rsCD6 to E. coli was competed in a dose-dependent manner by LPS (from E. coli), but not by LTA (from S. aureus). On the contrary, when the binding of rsCD6 to S. aureus was studied, LPS did not affect such interaction. Interestingly, this binding was competed in a dose-dependent manner by LTA from S. aureus.
Purification of nsCD6 from Human Serum.
Purification yielded 6 μg of a single protein with a molecular mass of 80 kDa as deduced from SDS/PAGE analysis and Coomassie blue staining (Fig. 2A). The observed molecular mass closely resembles that of recombinant soluble CD6 (rsCD6) (17), which is exclusively composed of the three extracellular SRCR domains of CD6, and is in contrast to that of the membrane form of CD6 (mCD6), which ranges from 105 to 130 kDa depending on its degree of phosphorylation (18). The observed molecular mass of the three different CD6 forms, i.e., rsCD6, nsCD6, and mCD6, immunoprecipitated from HUT-78 T cells is shown in Fig. 2B. The purified nsCD6 protein was identified as CD6 by Western blotting assays with a polyclonal antiserum raised against the extracellular region of human CD6 (Fig. 2C). Interestingly, mCD6, but not rsCD6 or nsCD6, was reactive with a polyclonal antiserum raised against the intracytoplasmic region of human CD6 (12) (data not shown). In cell binding experiments, both biotin-labeled rsCD6 and nsCD6 bound to Raji B cells but not to K562 erythroleukemic cells, in accordance with the differential expression of the CD6 ligand (ALCAM/CD166) (17, 19) (Fig. 2D).
Binding of rsCD6 to LPS and Kinetics of the rsCD6–LPS Interaction.
The results presented in Fig. 3A show that, in accordance with the bacterial binding experiments in Fig. 1, both natural and recombinant soluble CD6 forms bound to plastic-coated LPS in a dose-dependent fashion. No BSA–LPS interaction could be observed.
Fig. 3.
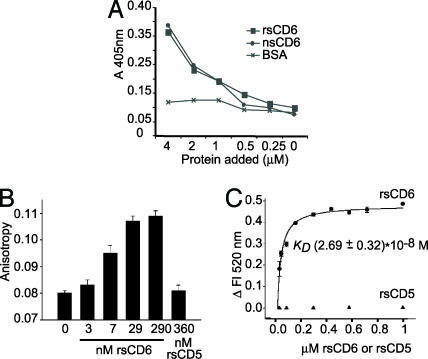
Binding of rsCD6 to LPS. (A) ELISA showing direct binding of nsCD6 and rsCD6 to LPS. Several concentrations of biotinylated rsCD6, nsCD6, or BSA (as negative control) were added to the LPS-coated wells, and bound protein was detected with streptavidin–HRP. (B and C) Binding of rsCD6 or rsCD5 to Re-LPS was monitored by changes in FITC–Re-LPS fluorescent properties. (B) rsCD6, but not rsCD5, induces a significant increase in fluorescence anisotropy upon binding to FITC–Re-LPS, which increases with increasing rsCD6 concentration. (C) Net change in fluorescence emission intensity of FITC–Re-LPS at 520 nm upon addition of increasing amounts of rsCD6 or rsCD5. The apparent Kd for FITC–Re-LPS/sCD6 complexes, calculated from the saturation curve fitted to a rectangular hyperbola, was 2.69 ± 0.32 × 10−8 M.
The binding of rsCD6 and rsCD5 to a rough mutant (Re595) of LPS (Re-LPS) in solution was studied next by analyzing the changes in fluorescent properties of FITC–Re-LPS such as anisotropy and intensity. Fig. 3B shows the binding of rsCD6 and rsCD5 to FITC–Re-LPS by measuring fluorescence anisotropy of the labeled LPS molecule. Fluorescence anisotropy measurements depend on the rate and extent of the rotational motion of the fluorophore during the lifetime of the excited state. Addition of different amounts of rsCD6 to FITC–Re-LPS caused a protein concentration-dependent increase of the anisotropy values of FITC–Re-LPS, indicating that the binding of rsCD6 to Re-LPS caused mechanical restrictions of the rotational mobility of the dye. Control experiments were done with free fluorescein to demonstrate that all of these changes did not result from the interaction of rsCD6 with fluorescein, but with the LPS molecule; the fluorescence emission anisotropy of free fluorescein was very low and was not affected by addition of 3-fold excess of rsCD6 (data not shown). On the other hand, rsCD5 did not cause any change in FITC–Re-LPS fluorescence anisotropy, indicating that this protein does not bind to Re-LPS.
Addition of rsCD6, but not rsCD5, to FITC–Re-LPS in solution also produced an increase of total fluorescence emission intensity of fluorescent LPS. Fig. 3C shows that the magnitude of the fluorescence intensity change at 520 nm increased as a function of rsCD6 concentration, but not rsCD5 concentration, and was saturable. These results allowed us to determine the affinity of rsCD6 binding to LPS. The apparent Kd for FITC–Re-LPS/rsCD6 complexes, calculated from the saturation curve fitted to a rectangular hyperbola, was 2.69 ± 0.32 × 10−8 M.
Binding of LPS to Cell-Surface CD6.
We next questioned whether the LPS–CD6 interaction occurs as well with the receptor expressed on the cell surface. These studies were performed by staining with FITC-labeled LPS of 2G5 cells, a Jurkat cell derivative selected for deficient CD5 and CD6 expression (20). As shown in Fig. 4 A and B, fluorescence intensity was higher in 2G5 cells stably expressing wild-type CD6 (2G5-CD6.wt) (12) compared with parental untransfected 2G5 cells. Further confirmation of our results was obtained from competition binding experiments. In these experiments, binding of FITC–LPS to 2G5-CD6.wt cells was inhibited in a dose-dependent manner by rsCD6, but not with rsCD5 or BSA (data not shown), used as a negative control (Fig. 4C), indicating that the inhibition was specific. 2G5 and 2G5-CD6.wt cells were negative for CD14 expression but expressed equivalent amounts of Toll-like receptor 4 (TLR4) on their surface (data not shown), in accordance with a recent report (21). Therefore, differences in LPS binding cannot be attributed to differential expression of TLR4. From these data we conclude that LPS is able to interact with CD6 on the cell surface.
Fig. 4.
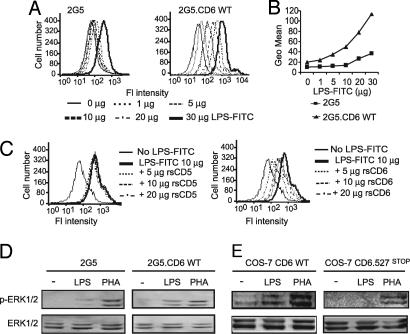
LPS from E. coli binds to cell-surface CD6 and activates ERK1/2. (A) Flow cytometry analysis showing direct binding of increasing amounts of LPS–FITC to parental and CD6.wt-transfected 2G5 cells. (B) To ease comparison, mean fluorescence intensities of A were plotted against the amount of LPS–FITC added to each cell line. (C) Competition studies of LPS–FITC binding to the 2G5-CD6.wt transfectants. Cells were incubated with 10 μg of LPS–FITC in the presence of increasing amounts of rsCD6 or rsCD5. (D and E) Analysis of ERK1/2 phosphorylation after LPS or PHA stimulation of parental and CD6.wt-transfected 2G5 cells (D) and COS-7 cells transiently expressing wild-type (CD6.wt) or cytoplasmic tail-truncated CD6 (CD6.P527stop) molecules (E). In both cases, serum-starved cells were stimulated for 40 min with 100 μg/ml LPS or 100 ng/ml PHA at 37°C. Cell lysates were resolved by SDS/PAGE, transferred to nitrocellulose, and subjected to immunoblotting with anti-phospho ERK1/2 (p-ERK1/2) antiserum. Further reprobing with anti-ERK1/2 antiserum was used as loading control.
Binding of LPS to Membrane CD6 Induces ERK1/2 Activation.
Further evidence of LPS binding to cell-surface CD6 was obtained from activation of the MAPK signaling cascade in transient and stable transfectants expressing membrane CD6. As shown in Fig. 4D, LPS stimulation of 2G5-CD6.wt transfectants resulted in marked ERK1/2 phosphorylation responses compared with parental 2G5 cells. However, the two transfectants showed similar responses after addition of PHA, the latter used as a positive control and to show the integrity of the MAPK signaling cascade. Similar results were obtained with transient transfection of CD6.wt in the heterologous COS-7 cell system (Fig. 4E). Fig. 4E shows that COS-7 cells transiently expressing a cytoplasmatic tail-truncated molecule (CD6.P527stop) (22) failed to respond to LPS but not to PHA, demonstrating that integrity of the cytoplasmic region of CD6 is also required for proper LPS-induced ERK1/2 phosphorylation. The differences in ERK1/2 phosphorylation are not due to differential CD6 surface expression, as assessed by FACS analysis and Western blot experiments (data not shown).
Binding of rsCD6 Leads to both Bacteria and LPS Aggregation.
We hypothesized that the existence of multiple bacterial binding domains on the rsCD6 molecule would lead to bacterial aggregation phenomena. Fig. 5A shows that presence of rsCD6 induced aggregation of Gram-negative (E. coli) as well as Gram-positive (S. aureus) bacteria, to a similar extent as the positive control rSpα (8). In accordance with its inability to bind bacteria, rsCD5 was also unable to induce their aggregation, and this also was the case of the negative control HSA.
Fig. 5.
rsCD6 induces bacterial aggregation. (A) FITC-labeled E. coli and S. aureus bacterial suspensions were incubated overnight at room temperature with rsCD6 or rsCD5 (2 μM) in the presence of 5 mM Ca2+. Equimolar concentrations of rSpα and HSA were used as positive and negative control, respectively. Aggregation was observed by direct examination on a fluorescence microscope. (B) Kinetics of Ca2+-dependent Re-LPS aggregation in the absence (filled circles) and presence of increasing concentrations of rsCD6, as described in Materials and Methods. The final concentrations of Re-LPS, Ca2+, and EDTA were 100 μg/ml, 2.5 mM, and 5 mM, respectively. One representative experiment of two performed is shown.
We further explored the process of Re-LPS aggregation induced by Ca2+ in the presence of rsCD6. This was analyzed by measuring changes in light absorbance at 400 nm (Fig. 5B). These experiments were carried out under the same ionic conditions as binding studies with fluorescent LPS, except that Ca2+ as well as concentrations of Re-LPS 200 times higher were needed to produce detectable light absorption at 400 nm. Fig. 5B shows that LPS molecules were able to aggregate in buffers containing Ca2+ and that low concentrations of rsCD6 induced a further aggregation of LPS. Control assays showed that addition of Ca2+ (2.5 mM) does not affect the aggregation status of rsCD6 as measured by fluorescence emission spectra of rsCD6 at 275 nm (data not shown). Together, these data suggest that, in the presence of Ca2+, rsCD6 may contribute to increase the size of bacterial aggregates as well as of LPS aggregates.
rsCD6 Prevents LPS-Induced Septic Shock in Mice.
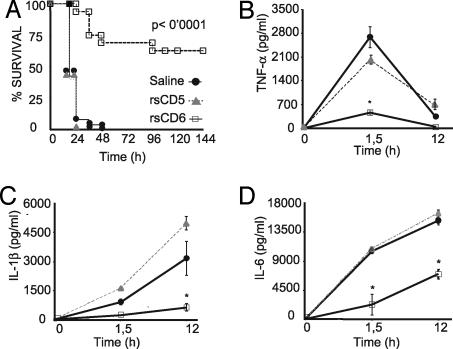
We assessed whether administration of rsCD6 into mice would improve their survival in front of LPS-induced septic shock. As observed in Fig. 6A, administration of a single i.p. dose of 25 μg of rsCD6, but not rsCD5, in mice 1 h before i.p. LPS challenge significantly enhanced their survival rate as compared with the saline control (up to 70%). In accordance with these data, administration of rsCD6 induced a significant reduction on the levels of plasma proinflammatory cytokines TNF-α, IL-1β, and IL-6 in these mice (Fig. 6 B, C, and D, respectively).
Fig. 6.
Effect of rsCD6 and rsCD5 on survival rate and cytokine serum levels after LPS-induced septic shock. (A) Survival graph. C57BL/6J mice (8 weeks old) were injected i.p. with a lethal dose of LPS (30 mg/kg) 1 h after i.p. administration of sterile saline solution (n = 26), rsCD5 (n = 10), or rsCD6 (n = 16) (25 μg each). The percentage of survival mice was analyzed, and the log-rank t test P values were calculated. (B–D) Circulating levels of cytokines in LPS-challenged mice. Plasma levels of TNF-α (B), IL-1β (C), and IL-6 (D) were quantified by ELISA at different times after LPS injection. Data are expressed as mean ± SEM. Statistical differences in the results were evaluated by the two-tailed Student t test. ∗, Statistically significant difference (P < 0.05).
Discussion
In the present study it is shown that human CD6, a cell-surface receptor mainly expressed by cells of the lymphoid lineage, is able to bind to conserved bacterial structures such as LPS and LTA. Interestingly, the Kd of the LPS–CD6 interaction appeared to be of relative high affinity, similar in magnitude to that reported for the interaction of LPS with CD14, the most widely accepted LPS receptor in mammalian cells. These data are of relevance by themselves to the knowledge of the biological role of the CD6 receptor, but also because they led to the finding that i.p. administration of a recombinant form of CD6 abolished the lethal effects of LPS-induced septic shock in mice.
Innate immune responses rely on the ability of multiple nonpolymorphic germ-line-encoded receptors to recognize the so-called pathogen-associated molecular patterns, which are conserved products of microbial pathogens not shared by the host and essential for their survival (23). Pattern-recognition receptors are mainly expressed by phagocytic cells (granulocytes, macrophages, and dendritic cells) and cells of epithelial barriers (23). However, recent studies report the expression of pattern-recognition receptors, namely TLR, in cells central to adaptive immune responses such as T and B lymphocytes (21, 24, 25). Among the receptors involved in pattern recognition, there are several members of the SRCR-SF expressed on macrophages and mucosal surfaces (5, 8, 16, 26). As far as we know, no information is currently available on that regard for the two only members of the SRCR-SF expressed on human lymphocytes, CD5 and CD6. In a series of in vitro studies we have shown that CD6, but not CD5, can bind to Gram-positive and -negative bacteria (Fig. 1) and that this binding was facilitated by the presence of Ca2+. CD6 is not a type C lectin, and it does not aggregate in the presence of Ca2+. On the contrary, presence of Ca2+ is known to affect the structure of the bacterial external membrane and the aggregation status of LPS (27). Therefore, Ca2+ may facilitate the rsCD6–LPS interaction through its effect on bacteria and LPS. Competition binding experiments showed that the observed interaction of rsCD6 with the bacterial surfaces is specific. They also indicate that, like Spα (8), rsCD6 binds to LTA and LPS through independent and nonoverlapping sites of the molecule.
Low levels of soluble CD6 have been detected in normal human sera (14), but its biochemical characterization has not been achieved until present. We have purified from pooled human sera a natural soluble CD6 (nsCD6) protein with molecular mass, antibody reactivity, and cell binding characteristics similar to rsCD6 (17) (Fig. 2). These data, together with the shared ability to bind to LPS in ELISAs (Fig. 3), suggest that rsCD6 retains the biological activities of the circulating form of CD6 (nsCD6). These results validate the use of rsCD6 in the ensuing studies given the low availability of nsCD6.
The interaction of rsCD6 with LPS has an apparent Kd of 2.69 ± 0.32 × 10−8 M. Using the same methodology and the same ionic conditions, Kd values for FITC–Re-LPS/LBP, FITC–Re-LPS/sCD14, FITC–Re-LPS/SP-A, and FITC–Re-LPS/rSpα complexes are 3.5 × 10−9 M, 2.9 × 10−8 M, 2.8 × 10−8 M, and 1.83 × 10−7 M, respectively (8, 28, 29). These data indicate that rsCD6 binds to Re-LPS ≈10-fold more tightly than SRCR-containing rSpα but with affinity similar to CD14 (28).
In contrast to rsCD6, the recombinant form encompassing the ectodomain of CD5 (rsCD5) did not bind to bacteria (Fig. 1) or LPS (Fig. 3). It seems, therefore, that the SRCR domains of CD6 still retain the bacterial binding properties shared by some members of the SRCR-SF, whereas those of CD5 do not. This suggests that CD5 and CD6 may have followed a divergent evolution, which is of relevance for the function of these receptors and the SRCR structure.
Although CD6 is found circulating in serum, this receptor is mainly found expressed on the cell surface. Enhanced binding of LPS–FITC to the surface of 2G5 cell transfectants expressing CD6 versus the parental cells suggested that membrane-bound CD6 also retains the ability to bind to LPS (Fig. 4 A and B). Moreover, the ability of LPS to induce ERK1/2 phosphorylation on 2G5 and COS-7 cells is in agreement with previous results showing that CD6 ligation induces MAPK activation (22) and suggest that membrane CD6 is able to sense the presence of LPS in the extracellular milieu and to deliver intracellular signals. The involvement of additional intracellular signaling pathways and its functional consequences will require future investigation.
It has been suggested that blockage of the main receptors that mediate recognition of bacteria and their products may be a suitable strategy for treating sepsis shock syndrome, which, at present, has a high mortality rate (up to 70% in septic patients) (30, 31). Accordingly, strategies targeting or using several innate immune receptors, such as activated protein C, TLR2, or CD14, are being currently tested in animal models of septic shock and also in clinical trials (32–34). Administration of a single dose (25 μg) of rsCD6 into mice 1 h before LPS challenge significantly enhanced their survival rate (up to 70%) as compared with rsCD5 or saline treatment (Fig. 6A) and concomitantly induced a significant reduction in the levels of proinflammatory cytokines TNF-α, IL-1β, and IL-6 in these mice (Fig. 6 B–D). The bacterial aggregation data (Fig. 5A), together with the increase of Ca2+-induced LPS aggregation in the presence of rsCD6 (Fig. 5B), suggest that rsCD6 may contribute to increase the size of particles. This would facilitate particle clearance from the circulation (by, for instance, facilitating phagocyte engulfment) and reduce subsequent inflammatory processes, which in cases such as sepsis may even result in death.
It cannot be ruled out, however, that the antiinflammatory effects of rsCD6 on LPS-induced septic shock may also result from its inhibitory effects on different cell subsets involved in the outcome of sepsis such as CD8 T cells and natural killer cells (35). In fact, rsCD6 has been shown to inhibit human antigen-presenting cell–T cell interactions (17, 36), and CD6 is known to be expressed on natural killer cells (36). In any case, the septic shock data are of great relevance because they constitute a therapeutic approach in the prevention of LPS-induced septic shock.
In conclusion, the results of the present report illustrate the unprecedented bacterial binding properties of the ectodomain of CD6 and support its therapeutic potential for the intervention of septic shock syndrome or other inflammatory diseases of infectious origin. They also suggest that not only cells of the innate immune system, but also T lymphocytes, may sense the presence of bacterial components through CD6 as well as other well known pattern-recognition receptors (e.g., TLRs), although the functional consequences of such a recognition are yet to be analyzed in depth. It can be hypothesized that, even if its main role were the modulation of T cell activation and differentiation signals, CD6 may have retained the ability to recognize microbial components as an accessory property, which is shared with other members of the ancient and highly conserved SRCR-SF. This adds further evidence to the notion that SRCR domains may have emerged as protein modules of the innate immune system for recognition of pathogen-associated molecular patterns.
Materials and Methods
Cells, Antibodies, and Reagents.
The cell lines COS-7, Raji, K562, and HUT-78 were from the American Type Culture Collection (Manassas, VA). The CD5- and CD6-negative 2G5 cells (20) were stably transfected with the pHβ-CD6.P527stop and pHβ-CD6.wt constructs (12, 22). Cell growth conditions are detailed in supporting information (SI) Methods. A full list of providers of antibodies and reagents can be found in SI Methods.
Expression, Affinity Purification, and Biotin Labeling of Recombinant Proteins.
The recombinant proteins were expressed in HEK 293-EBNA and affinity-purified as reported (12, 37). Their purity was assessed by SDS/PAGE and staining with Coomassie blue. Protein biotinylation was performed with EZ-Link PEO-maleimide-activated biotin (Pierce/Perbio Science, Cheshire, U.K.) following the manufacturer's instructions (8) and monitored by Western blotting.
Bacterial Strains and Bacterial Binding Studies.
The bacterial strains used in this study are clinical isolates characterized by the Department of Microbiology of the Hospital Clinic of Barcelona using standard biochemical procedures. Bacterial growth conditions are detailed in SI Methods. Binding of rsCD6 to bacteria was studied following a method described previously (15), with slight modifications (8).
Purification of Soluble CD6 from Human Serum.
Soluble CD6 was affinity-purified from 1 liter of human plasma pooled from healthy blood donors obtained from the Blood Bank of the Hospital Clinic de Barcelona as detailed in SI Methods.
LPS-Binding ELISAs.
LPS purified from E. coli O55:B5, O111:B4, or O26:B6 (Sigma, St. Louis, MO) was used to coat 96-well microtiter plates (Nunc, Roskilde, Denmark) and assayed for BSA, rsCD6, or nsCD6 binding as detailed in SI Methods.
Binding Assays of Soluble Proteins to FITC–Re-LPS.
A fluorescent Re-LPS derivative (FITC–Re-LPS) in which the phosphoethanolamine group of Re-LPS was bound to FITC by a previously described method (38). Fluorescence measurements were carried out as previously described (8, 28, 29) and as described in SI Methods.
Bacteria and LPS Aggregation Assays.
Bacteria aggregation assays were performed as described (8). LPS aggregation induced by rsCD6 was studied as before (29). For further details see SI Methods.
Flow Cytometry Assays.
Cell-binding properties of soluble proteins were assessed as described (13). Binding of LPS to cell-surface CD6 was assessed by incubating cells with different amounts of LPS–FITC from E. coli 0111:B4 (Sigma) in the presence or absence of rsCD5, rsCD6, or BSA as detailed in SI Methods.
ERK1/2 Phosphorylation Assays.
ERK1/2 phosphorylation was analyzed by Western blot as described (22) (see SI Methods).
CD6 Immunoprecipitation.
Immunoprecipitation of membrane CD6 (mCD6) from HUT-78 T cells was performed and analyzed by Western blotting as described (12).
LPS-Induced Endotoxic Shock.
C57BL/6J mice (8 weeks old) were injected i.p. with 25 μg of saline solution, rsCD5, or rsCD6 1 h before injection of an i.p. lethal dose of LPS from E. coli. For further details see SI Methods.
Determination of Cytokine Serum Levels.
The systemic release of TNF-α, IL-1β, and IL-6 cytokines was determined by ELISA in pooled serum samples as described in SI Methods.
Supplementary Material
Acknowledgments
We thank Dr. Jordi Vila for assistance with bacterial strains and Belén Suárez for technical assistance. This work was supported by Ministerio de Educación y Ciencia Grants SAF2004-03251 (to F.L.), BIO-2005-01393 (to J.Y.), and SAF2006-04434 (to C.C.) and Instituto de Salud Carlos III Grants PI05/1013 (to F.L.) and CB06/06/0002 (to C.C.). M.-R.S. is supported by Fondo de Investigación Sanitaria Grant FIS CP05/100. M.F., A.I., and I.G. are recipients of fellowships from Institut d'Investigacions Biomèdiques August Pi i Sunyer, Departament d'Universitats, Recerca i Societat de la Informació, and Ministerio de Sanidad y Consumo–Institut d'Investigacions Biomèdiques August Pi i Sunyer, respectively.
Abbreviations
- SRCR-SF
scavenger receptor cysteine-rich superfamily
- LTA
lipoteichoic acid
- TLR
Toll-like receptor.
Footnotes
Conflict of interest statement: This work is the subject of patent application ES200700893 submitted by the University of Barcelona.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702815104/DC1.
References
- 1.Freeman M, Ashkenas J, Rees DJ, Kingsley DM, Copeland NG, Jenkins NA, Krieger M. Proc Natl Acad Sci USA. 1990;87:8810–8814. doi: 10.1073/pnas.87.22.8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarrias MR, Gronlund J, Padilla O, Madsen J, Holmskov U, Lozano F. Crit Rev Immunol. 2004;24:1–37. doi: 10.1615/critrevimmunol.v24.i1.10. [DOI] [PubMed] [Google Scholar]
- 3.Aruffo A, Bowen MA, Patel DD, Haynes BF, Starling GC, Gebe JA, Bajorath J. Immunol Today. 1997;18:498–504. doi: 10.1016/s0167-5699(97)01130-4. [DOI] [PubMed] [Google Scholar]
- 4.Doi T, Higashino K, Kurihara Y, Wada Y, Miyazaki T, Nakamura H, Uesugi S, Imanishi T, Kawabe Y, Itakura H, et al. J Biol Chem. 1993;268:2126–2133. [PubMed] [Google Scholar]
- 5.Elomaa O, Kangas M, Sahlberg C, Tuukkanen J, Sormunen R, Liakka A, Thesleff I, Kraal G, Tryggvason K. Cell. 1995;80:603–609. doi: 10.1016/0092-8674(95)90514-6. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y, Oliver P, Davies KE, Platt N. J Biol Chem. 2006;281:11834–11845. doi: 10.1074/jbc.M507599200. [DOI] [PubMed] [Google Scholar]
- 7.Peiser L, Gough PJ, Kodama T, Gordon S. Infect Immun. 2000;68:1953–1963. doi: 10.1128/iai.68.4.1953-1963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarrias MR, Rosello S, Sanchez-Barbero F, Sierra JM, Vila J, Yelamos J, Vives J, Casals C, Lozano F. J Biol Chem. 2005;280:35391–35398. doi: 10.1074/jbc.M505042200. [DOI] [PubMed] [Google Scholar]
- 9.Mayer B, Funke I, Seed B, Riethmuller G, Weiss E. J Neuroimmunol. 1990;29:193–202. doi: 10.1016/0165-5728(90)90162-g. [DOI] [PubMed] [Google Scholar]
- 10.Brannstrom A, Sankala M, Tryggvason K, Pikkarainen T. Biochem Biophys Res Commun. 2002;290:1462–1469. doi: 10.1006/bbrc.2002.6378. [DOI] [PubMed] [Google Scholar]
- 11.Lozano F, Simarro M, Calvo J, Vila JM, Padilla O, Bowen MA, Campbell KS. Crit Rev Immunol. 2000;20:347–358. [PubMed] [Google Scholar]
- 12.Gimferrer I, Farnos M, Calvo M, Mittelbrunn M, Enrich C, Sanchez-Madrid F, Vives J, Lozano F. J Biol Chem. 2003;278:8564–8571. doi: 10.1074/jbc.M209591200. [DOI] [PubMed] [Google Scholar]
- 13.Calvo J, Places L, Espinosa G, Padilla O, Vila JM, Villamor N, Ingelmo M, Gallart T, Vives J, Font J, et al. Tissue Antigens. 1999;54:128–137. doi: 10.1034/j.1399-0039.1999.540203.x. [DOI] [PubMed] [Google Scholar]
- 14.Ramos-Casals M, Font J, Garcia-Carrasco M, Calvo J, Places L, Padilla O, Cervera R, Bowen MA, Lozano F, Ingelmo M. Rheumatology. 2001;40:1056–1059. doi: 10.1093/rheumatology/40.9.1056. [DOI] [PubMed] [Google Scholar]
- 15.Dunne DW, Resnick D, Greenberg J, Krieger M, Joiner KA. Proc Natl Acad Sci USA. 1994;91:1863–1867. doi: 10.1073/pnas.91.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madsen J, Tornoe I, Nielsen O, Lausen M, Krebs I, Mollenhauer J, Kollender G, Poustka A, Skjodt K, Holmskov U. Eur J Immunol. 2003;33:2327–2336. doi: 10.1002/eji.200323972. [DOI] [PubMed] [Google Scholar]
- 17.Gimferrer I, Calvo M, Mittelbrunn M, Farnos M, Sarrias MR, Enrich C, Vives J, Sanchez-Madrid F, Lozano F. J Immunol. 2004;173:2262–2270. doi: 10.4049/jimmunol.173.4.2262. [DOI] [PubMed] [Google Scholar]
- 18.Cardenas L, Carrera AC, Yague E, Pulido R, Sanchezmadrid F, Delandazuri MO. J Immunol. 1990;145:1450–1455. [PubMed] [Google Scholar]
- 19.Bowen MA, Patel DD, Li X, Modrell B, Malacko AR, Wang WC, Marquardt H, Neubauer M, Pesando JM, Francke U, et al. J Exp Med. 1995;181:2213–2220. doi: 10.1084/jem.181.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simarro M, Pelassy C, Calvo J, Places L, Aussel C, Lozano F. J Immunol. 1997;159:4307–4315. [PubMed] [Google Scholar]
- 21.Caron G, Duluc D, Fremaux I, Jeannin P, David C, Gascan H, Delneste Y. J Immunol. 2005;175:1551–1557. doi: 10.4049/jimmunol.175.3.1551. [DOI] [PubMed] [Google Scholar]
- 22.Ibanez A, Sarrias MR, Farnos M, Gimferrer I, Serra-Pages C, Vives J, Lozano F. J Immunol. 2006;177:1152–1159. doi: 10.4049/jimmunol.177.2.1152. [DOI] [PubMed] [Google Scholar]
- 23.Medzhitov R, Janeway C., Jr Immunol Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- 24.Gelman AE, Zhang J, Choi Y, Turka LA. J Immunol. 2004;172:6065–6073. doi: 10.4049/jimmunol.172.10.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarember KA, Godowski PJ. J Immunol. 2002;168:554–561. doi: 10.4049/jimmunol.168.2.554. [DOI] [PubMed] [Google Scholar]
- 26.Gordon S. Cell. 2002;111:927–930. doi: 10.1016/s0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]
- 27.Snyder S, Kim D, McIntosh TJ. Biochemistry. 1999;38:10758–10767. doi: 10.1021/bi990867d. [DOI] [PubMed] [Google Scholar]
- 28.Tobias PS, Soldau K, Gegner JA, Mintz D, Ulevitch RJ. J Biol Chem. 1995;270:10482–10488. doi: 10.1074/jbc.270.18.10482. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Verdugo I, Sanchez-Barbero F, Soldau K, Tobias PS, Casals S. Biochem J. 2005;391:115–124. doi: 10.1042/BJ20050529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Crit Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen HB, Rivers EP, Abrahamian FM, Moran GJ, Abraham E, Trzeciak S, Huang DT, Osborn T, Stevens D, Talan DA. Ann Emerg Med. 2006;48:28–54. doi: 10.1016/j.annemergmed.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 32.Meng G, Rutz M, Schiemann M, Metzger J, Grabiec A, Schwandner R, Luppa PB, Ebel F, Busch DH, Bauer S, et al. J Clin Invest. 2004;113:1473–1481. doi: 10.1172/JCI20762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schimke J, Mathison J, Morgiewicz J, Ulevitch RJ. Proc Natl Acad Sci USA. 1998;95:13875–13880. doi: 10.1073/pnas.95.23.13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Amersfoort ES, van Berkel TJ, Kuiper J. Clin Microbiol Rev. 2003;16:379–414. doi: 10.1128/CMR.16.3.379-414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enoh VT, Lin SH, Lin CY, Toliver-Kinsky T, Murphey ED, Varma TK, Sherwood ER. Shock. 2007;27:507–519. doi: 10.1097/SHK.0b013e31802b5d9f. [DOI] [PubMed] [Google Scholar]
- 36.Zimmerman AW, Joosten B, Torensma R, Parnes JR, van Leeuwen FN, Figdor CG. Blood. 2006;107:3212–3220. doi: 10.1182/blood-2005-09-3881. [DOI] [PubMed] [Google Scholar]
- 37.Sarrias MR, Padilla O, Monreal Y, Carrascal M, Abian J, Vives J, Yelamos J, Lozano F. Tissue Antigens. 2004;63:335–344. doi: 10.1111/j.0001-2815.2004.00193.x. [DOI] [PubMed] [Google Scholar]
- 38.Skelly RR, Munkenbeck P, Morrison DC. Infect Immun. 1979;23:287–293. doi: 10.1128/iai.23.2.287-293.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.