Abstract
Lung cancer is the number one cause of cancer-related deaths in the world. Patients treated with current chemotherapies for non-small-cell lung cancers (NSCLCs) have a survival rate of ≈15% after 5 years. Novel approaches are needed to treat this disease. We show elevated NAD(P)H:quinone oxidoreductase-1 (NQO1) levels in tumors from NSCLC patients. β-Lapachone, an effective chemotherapeutic and radiosensitizing agent, selectively killed NSCLC cells that expressed high levels of NQO1. Isogenic H596 NSCLC cells that lacked or expressed NQO1 along with A549 NSCLC cells treated with or without dicoumarol, were used to elucidate the mechanism of action and optimal therapeutic window of β-lapachone. NSCLC cells were killed in an NQO1-dependent manner by β-lapachone (LD50, ≈4 μM) with a minimum 2-h exposure. Kinetically, β-lapachone-induced cell death was characterized by the following: (i) dramatic reactive oxygen species (ROS) formation, eliciting extensive DNA damage; (ii) hyperactivation of poly(ADP-ribose)polymerase-1 (PARP-1); (iii) depletion of NAD+/ATP levels; and (iv) proteolytic cleavage of p53/PARP-1, indicating μ-calpain activation and apoptosis. β-Lapachone-induced PARP-1 hyperactivation, nucleotide depletion, and apoptosis were blocked by 3-aminobenzamide, a PARP-1 inhibitor, and 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid acetoxymethyl ester (BAPTA-AM), a Ca2+ chelator. NQO1− cells (H596, IMR-90) or dicoumarol-exposed NQO1+ A549 cells were resistant (LD50, >40 μM) to ROS formation and all cytotoxic effects of β-lapachone. Our data indicate that the most efficacious strategy using β-lapachone in chemotherapy was to deliver the drug in short pulses, greatly reducing cytotoxicity to NQO1− “normal” cells. β-Lapachone killed cells in a tumorselective manner and is indicated for use against NQO1+ NSCLC cancers.
Keywords: DNA repair inhibitor, non-small-cell lung cancer, NQO1, apoptosis, μ-calpain cell death
Lung cancer is responsible for nearly one-third (29%) of all cancer-related deaths in the United States (1). Current therapies for non-small-cell lung cancer (NSCLC), which represents >80% of all lung cancers, use platinum-based drugs (e.g., cisplatin) in combination with ionizing radiation (2). Although progress has been made, the long-term survival of NSCLC patients remains bleak at ≈15% 5 yr after therapy (2). There is a great need to develop tumor-selective chemotherapeutic agents specific for NSCLCs.
Elucidation of novel cell death pathways for selective killing of chemoresistant NSCLC tumors may lead to more effective treatment strategies. Currently, there are no developed chemotherapeutic agents that exploit poly(ADP-ribose)polymerase-1 (PARP-1) hyperactivation for NSCLC therapy. PARP-1 facilitates DNA repair, specifically in resealing DNA single-strand breaks (3). PARP-1 binds and stabilizes DNA single-strand breaks, and then converts NAD+ (its substrate) into branched poly(ADP-ribose) (PAR) polymers that are used to posttranslationally modify itself and other specific protein substrates. When DNA damage overwhelms repair (e.g., after severe genotoxic stress), PARP-1 becomes hyperactivated. PARP-1 hyperactivation leads to rapid NAD+ depletion, dramatic ATP pool losses, and cell death referred to as “programmed necrosis” (4) or “necroptosis” (5, 6). Such cell death responses have been noted only after supralethal N-methyl-N′-nitro-N-nitrosoguanidine or hydrogen peroxide (H2O2) doses (7).
β-Lapachone (β-lap) (3,4-dihydro-2,2-dimethyl-2Hnaphtho[1,2-b]pyran-5–6-dione) is an antitumor agent with renewed interest because of its selective killing of human cancer cells that overexpress endogenous NAD(P)H:quinone oxidoreductase-1 (NQO1) (E.C. 1.6.99.2), a two-electron oxidoreductase (8). The development of novel delivery strategies that increase the bioavailability and tumor targeting of β-lap have further served to peak interest in its use as an antitumor agent (9–12). In fact, ARQ 501 (i.e., β-lap) is currently in phase II trials against pancreatic cancers.
Importantly, β-lap kills cancer cells regardless of cell cycle position (5, 6) or p53 status (6). It synergizes with ionizing radiation (13, 14) or selective chemotherapies (11), highlighting the compound's usefulness for cancer therapy. Here, we generated isogenically matched NQO1+ and NQO1− human H596 NSCLC cells and demonstrated that β-lap induces PARP-1-mediated cell death in an NQO1-dependent manner, an exploitable pathway for selective therapy of NSCLCs.
Results
Specific Elevation of NQO1 in NSCLC Determines β-Lap Lethality.
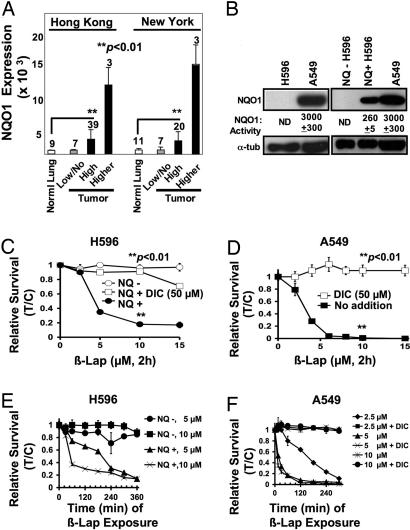
Prior data suggested that NQO1 expression was elevated in tumors of NSCLC patients (15); however, substantial variability was noted (15, 16). Analyses of microarray data from New York and Hong Kong databases showed that NQO1 mRNA levels were elevated 5- to 15-fold in NSCLC vs. associated normal lung tissues (Fig. 1A). The incidence of NQO1− tumors appears to match the reported frequency of individuals with known NQO1 polymorphisms that result in loss of enzyme activity (8).
Fig. 1.
NQO1 overexpression in NSCLC confers susceptibility to β-lap. (A) DNA microarray array data of NSCLC patient samples from New York and Hong Kong tissue banks were analyzed for NQO1 mRNA levels as described in Materials and Methods. Numbers above bars indicate the number of patients analyzed in each category. Associated normal tissue from patient samples was also evaluated but was not always available. (B) NQO1 protein levels and enzyme activities (nanomoles per minute per microgram) in H596 and A549 NSCLC cells, vector-alone H596 (NQ− H596) cells, and in H596 cells stably expressing NQO1 (NQ+ H596). Equivalent loading was monitored by using α-tubulin (α-tub). NQO1 activity <1.0 nmol·min−1·μg−1 was nondetectable (ND). (C) Long-term relative survival assays of NQ− H596 (○) and NQ+ H596 (●) cells treated for 2 h with β-lap or β-lap plus DIC (□) at the indicated doses. Shown are means ± SE for three independent experiments performed in triplicate. (D) A549 cells were treated as described for C, with β-lap (■) with or without 50 μM DIC (□). (E) NQ+ H596 and NQ− H596 cells were exposed to doses of β-lap, and drugs were removed at various times up to 360 min to determine minimum exposure times required for lethality. (F) A549 cells were treated with various β-lap doses, with or without 50 μM DIC, and long-term relative survival assays were performed. Results (means ± SE) in C–F are representative of experiments performed at least three times. In C and D, Student's t tests were performed. ∗∗, P < 0.01 vs. NQ− cells treated with β-lap alone (C) and A549 cells exposed to β-lap plus DIC (D).
To elucidate the mechanism of action of β-lap in NSCLC cells, we generated isogenically matched H596 cells expressing or lacking NQO1 at levels consistent with tumor tissues from NSCLC patients (16) (Fig. 1B). NQO1-containing H596 (NQ+ H596) cells were susceptible to β-lap at ≥5 μM for 2 h (Fig. 1C). NQO1-deficient H596 (NQ− H596) cells were resistant to the drug, even after 15 μM, 2 h (Fig. 1C). We then examined the responses of human A549 NSCLC cells to β-lap, because these cells express endogenously elevated NQO1 levels (Fig. 1B). NQO1 activity in A549 cells was ≈10-fold higher than in NQ+ H596 cells (Fig. 1B). Interestingly, the LD50 of A549 cells exposed to β-lap was ≈2.5 μM, 2 h, only slightly lower than the ≈4 μM LD50 value calculated for NQ+ H596 cells (Fig. 1 C and D). The similar cytotoxicity of NQO1+ A549 and H596 cells, despite greatly differing enzyme levels, indicates a required threshold level of NQO1 activity for β-lap-mediated lethality. Consistently, 5–10 μM β-lap gave similar lethality responses when given for 2 h, a minimum exposure time to kill NQO1+ NSCLC cells (Fig. 1 E and F). NQO1 inhibition by dicoumarol (DIC), or lack of enzyme expression, conferred dramatically increased resistance to β-lap (Fig. 1 E and F).
NQO1-Dependent Oxidative Stress in NSCLC Cells.
β-Lap is metabolized by NQO1 in a futile manner, cycling between the parent quinone form and an unstable hydroquinone that spontaneously reverts back to the parent compound (17). Therefore, we monitored reactive oxygen species (ROS) formation using the following: (i) 5- (and 6)-carboxy-2′,7′-dichlorodihydrofluorescein (DCF) staining and flow cytometry; and (ii) glutathione-recycling assays [supporting information (SI) Fig. 6]. Time course data indicated ROS formation within minutes after β-lap exposure in NQ+ H596 and A549 cells (Fig. 2 A and B). At 10 μM, β-lap induced ROS with saturation in 60 min (Fig. 2 A and B). Similar responses were noted in β-lap-exposed A549 cells, except that saturating ROS levels were not noted until 120 min (Fig. 2A Right). DIC dramatically prevented ROS in A549 cells (Fig. 2 A Right and B Right). In contrast, β-lap-exposed NQ− H596 cells showed little to no ROS formation at all doses examined (Fig. 2 A Left and B Left). Exposure of H596 or A549 cells with 2 mM H2O2 resulted in elevated ROS levels independent of NQO1 expression.
Fig. 2.
β-Lap induces oxidative stress and DNA damage in NQO1+ H596 and A549 cells. (A) NQ+ H596 (Left), NQ− H596 (Left), and A549 cells (Right) were treated with 10 μM β-lap with or without 50 μM DIC. ROS formation was monitored at the indicated times by DCF staining as described in Materials and Methods. Cells were also treated with 2 mM H2O2 or 10 μM antimycin A for 20 min as positive controls (data not shown). Results shown are means ± SE for three or more experiments. ∗∗, P < 0.01 vs. NQ− H596 (Left) and A549 cells treated with β-lap plus DIC (Right). (B) NQ+ H596, NQ− H596, and A549 cells were treated with the indicated doses of β-lap with or without 50 μM DIC for 20 min, and ROS formation was measured as in A. (C) NQ− H596, NQ+ H596, and A549 cells were exposed to β-lap with or without 50 μM DIC and assessed for DNA damage by using comet assays as described in Materials and Methods. Cells were also exposed to 2 mM H2O2 for 2 h as positive controls. Comet tail lengths of NQO1− and NQO1+ H596 cells were measured by using Image J software (a.u., arbitrary unit). Shown are representative micrographs of experiments performed at least three times. Graphed are means ± SE from three experiments. Student's t tests were performed. ∗∗, P < 0.001 vs. NQ− H596 treated with β-lap alone.
β-Lap Induces NQO1-Dependent DNA Damage.
Dramatic ROS induction suggested that β-lap exposure may cause damage and breaks in DNA. DNA damage, measured by comet tail formation, was detected in NQ+ H596 and A549 NSCLC cells after β-lap exposure (10 μM, 2 h) (Fig. 2C). DNA damage was not detected in NQ− H596 cells, nor in DIC-treated A549 or NQ+ H596 cells (Fig. 2C). Thus, NQO1-mediated ROS formation led to significant DNA breaks not observed in cells lacking this enzyme (Fig. 2C). Exposure of all cells to 2 mM H2O2 caused equivalent DNA damage (Fig. 2C).
β-Lap-Induced PARP-1 Hyperactivation Depletes Essential Nucleotides.
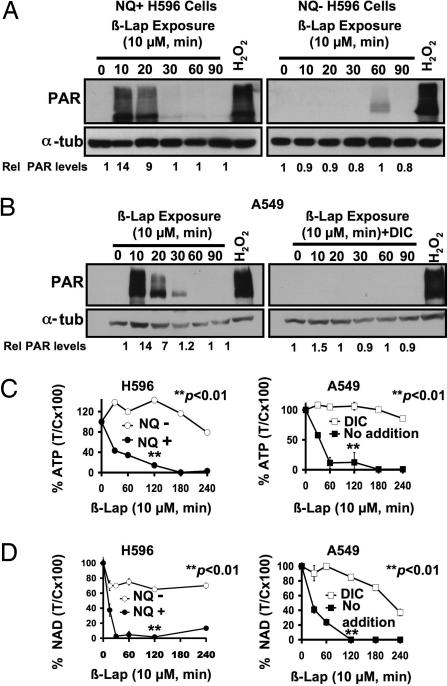
We previously noted a rapid loss of ATP during β-lap-stimulated cell death in a process leading to μ-calpain activation (18). Because the combination of DNA damage and nucleotide (ATP) loss was observed during high dose H2O2- or N-methyl-N′-nitro-N-nitrosoguanidine-induced cell death responses involving PARP-1 hyperactivation (19), we examined the role of this repair protein in β-lap-induced lethality. PAR accumulation, an indicator of PARP-1 hyperactivation, was detected in NQ+ H596 and A549 cells in 10 min of 10 μM β-lap exposure (Fig. 3A Left and B Left). DIC suppressed PAR formation in β-lap-exposed A549 cells (Fig. 3B Right), whereas NQ− H596 cells exposed to β-lap did not elicit PAR formation (Fig. 3A Right). In contrast, H2O2 (2 mM) exposure of all cells tested resulted in significant and comparable PAR formation (Fig. 3 A and B).
Fig. 3.
β-Lap-induced PARP-1 hyperactivation causes NQO1-dependent nucleotide depletion. (A and B) Western blot analyses of PAR formation in NQO1+ and NQO1− H596 cells treated with or without β-lap (10 μM) (A) or A549 cells exposed to β-lap (10 μM) (B Left) with or without 50 μM DIC (B Right) and harvested at the indicated times. PAR formation was detected as described in Materials and Methods. (C) NQ+ H596, NQ− H596, and A549 cells were mock-treated or exposed to β-lap (10 μM), harvested at the indicated times, and analyzed for ATP level changes as in Materials and Methods. For A549 cells, DIC (50 μM) was added with β-lap to examine the effects of NQO1 inhibition. (D) NQ+ H596, NQ− H596, and A549 cells were treated with β-lap (10 μM) with or without DIC (40 μM), and changes in intracellular NAD+ levels were analyzed at indicated times as described in Materials and Methods. For C and D, results shown are means ± SE for experiments performed three or more times. Student's t tests were performed. ∗∗, P < 0.01 vs. NQ− H596 cells treated with β-lap alone and A549 cells treated with β-lap plus DIC.
Rapid PAR-polymer formation correlated well with loss of essential metabolic nucleotides (i.e., ATP and NAD+) in both NQO1+ H596 and A549 cells (Fig. 3 C and D). Abrogation of NQO1 function, either by genetic polymorphism or by DIC (50 μM) addition, prevented β-lap-induced PAR formation and nucleotide loss (Fig. 3).
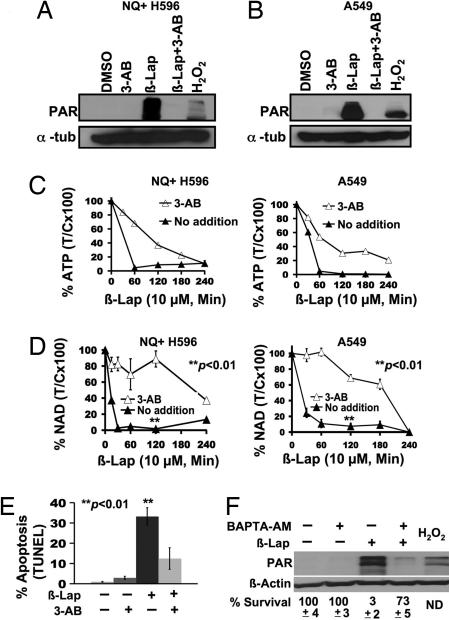
Inhibition of PARP-1 activity using 3-aminobenzamide (3-AB), an NAD+ analog, spared NQ+ H596 and A549 cells from β-lap-induced PAR formation (i.e., PARP-1 hyperactivation) (Fig. 4 A and B) and attenuated nucleotide depletion (Fig. 4 C and D). Thus, PARP-1 inhibition protected cells from β-lap-induced PARP-1 hyperactivation and short-term (48-h TUNEL assays) downstream cell death responses (Fig. 4E). Because 3-AB inhibits PARP-1-dependent DNA repair, long-term survival is prevented. By using NQO1+ breast cancer cells (19), we showed that 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid acetoxymethyl ester (BAPTA-AM), a Ca2+ chelator, spared cells from β-lap-induced PARP-1 hyperactivation, but allowed repair, enhancing long-term survival (19). Similarly, BAPTA-AM prevented PARP-1 hyperactivation, spared NAD+ loss (Fig. 4F and SI Fig. 7), and greatly increased long-term survival in β-lap-treated NQO1+ NSCLC cells (Fig. 4F and SI Fig. 7), highlighting the important role of Ca2+-mediated PARP-1 hyperactivation in β-lap-induced cell death.
Fig. 4.
Inhibition of PARP-1 hyperactivation delays NQO1-dependent nucleotide pool depletion in NSCLC cells after β-lap. (A and B) NQ+ H596 or A549 cells were pretreated with or without 25 mM 3-AB or mock-treated for 2 h and then exposed to β-lap with or without 3-AB for 20 min. (C and D) NQ+ H596 and A549 cells were treated with β-lap with or without 3-AB as in A, harvested at the indicated times, and analyzed for changes in nucleotide (NAD+ and ATP) pool levels as described in Materials and Methods. Results were graphed as means ± SE of experiments performed three times. Student's t tests were performed. ∗∗, P < 0.01 vs. NQ− H596 cells treated with β-lap alone; A549 cells treated with β-lap plus DIC. (E) A549 cells were mock-treated or pretreated with 3-AB as described (19) and then exposed to β-lap (10 μM, 2 h) and harvested 48 h later. Cells were assessed for apoptosis by TUNEL assays. Student's t tests were performed. ∗∗, P < 0.01 vs. A549 cells treated with β-lap plus 3-AB. (F) Western blot of A549 cells pretreated with 5 μM BAPTA-AM as described in Materials and Methods or mock-treated and then exposed to β-lap (6 μM, 10 min). Samples were then harvested for PAR formation. Survival data are expressed as %T/C. The percentage of NAD+ loss was also monitored as a surrogate for PARP-1 hyperactivation in cells pretreated with 5 μM BAPTA-AM or mock-treated and then exposed to β-lap (6 μM, 1 h). BAPTA-AM-pretreated, β-lap-exposed cells retained NAD+ (87% control), whereas β-lap-treated NQO1+ A549 cells lost significant NAD+ levels (43%) measured at 1 h after treatment (see SI Fig. 7B). ND, not determined.
NQO1 Expression Provides a Unique Therapeutic Window for β-Lap Efficacy in NSCLC Cells.
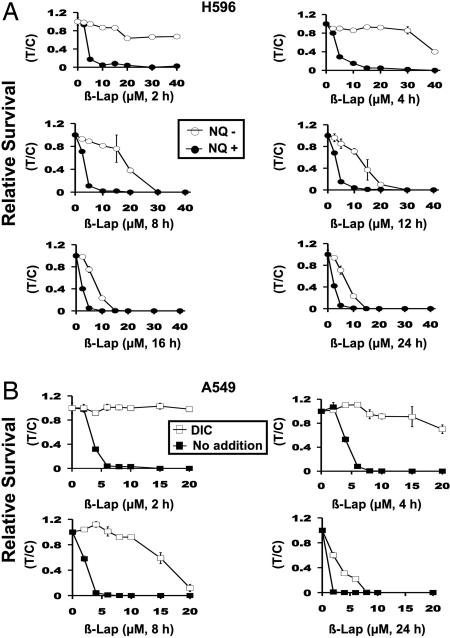
NQO1 is endogenously overexpressed in tumors from NSCLC patients vs. associated normal tissue (Fig. 1A) (15), providing a potential therapeutic window for the use of “bioactivated” compounds, such as β-lap, in the treatment of this disease. Because 2-h β-lap exposures of NQO1+ NSCLC cells were minimally required to irreversibly commit cells to cell death (Fig. 1 D and E), we optimized the therapeutic window of this drug. Reports by others (20) suggested that β-lap cytotoxicity in A549 cells was cell cycle and caspase-3 dependent. However, these studies were performed by using continuous administration of β-lap in vitro, a regimen that seemed counterindicated based on our data.
To optimize the therapeutic window of β-lap, we exposed NQ+ vs. NQ− NSCLC cells to various doses of β-lap. NQO1-deficient cells (by polymorphism in H596 or in DIC-treated A549 cells) may represent responses of enzyme-deficient cells (e.g., normal tissue) (21). Exposure times of β-lap from 2–8 h were optimal to exploit NQO1+ H596 or A459 NSCLC cells, while minimally affecting NQO1− cells (Fig. 5). In contrast, longer prolonged exposures (12–24 h) of cells to β-lap abrogated the therapeutic window of this drug in that NQO1− cells were significantly killed by β-lap, presumably through two one-electron oxidoreductions. Responses in A549 cells cotreated with DIC plus β-lap mimicked responses of β-lap-exposed NQ− H596 NSCLC cells. These data strongly suggest that bolus and not continuous doses of this drug should be efficacious.
Fig. 5.
Defining the therapeutic window of β-lap. (A) Long-term relative survival assays of NQ+ and NQ− H596 cells treated with β-lap at the doses (micromolar) and times (hours) indicated. After various exposures, relative survival was assessed. (B) Relative survival assays of A549 cells treated with various concentrations of β-lap with or without 50 μM DIC for 2–24 h. Data are graphed as in A, except that β-lap alone vs. β-lap plus DIC were graphed. ○, NQO1− cells treated with or without various doses and times of β-lap. ● or ■, NQO1+ cells treated in the same manner. □, NQO1+ cells treated with β-lap plus 50 μM DIC to inhibit NQO1. Addition of 50 μM DIC to β-lap-treated NQO1− cells had no additional effect. The graphs show the means ± SE of three independent experiments performed in triplicate.
Discussion
In numerous animal and cell culture models, NQO1 is elevated/induced in normal tissues/cells in response to low-level carcinogen (22) or ionizing radiation (23) exposures. Importantly, levels of this two-electron oxidoreductase are commonly endogenously elevated 2- to 30-fold in many tumors, including NSCLCs, vs. most human normal tissues (8, 16, 24). Elevated NQO1 levels in NSCLCs were confirmed in our tumor vs. normal tissue survey (Fig. 1A). Elevated NQO1 levels in tumor vs. normal lung tissue represent a therapeutically exploitable target for compounds bioactivated by NQO1.
An important consideration in the development of any “NQO1 bioactivatable” compound for NSCLC therapy is that a considerable fraction (10–15%) of tumors from NSCLC patients may contain NQO1 polymorphisms that confer expression loss and drug resistance. Two well characterized NQO1 polymorphisms (NQO1*2, NQO1*3) that inactivate gene expression (25) occur in the human population at reported incidences of 10 and 1%, respectively, (8). Tumors with low or no NQO1 levels in our screen of New York and Hong Kong patient samples (Fig. 1A) are predicted to contain NQO1 polymorphisms.
To examine the mechanism of cell death caused by β-lap in NSCLC, we developed an isogenically matched human NSCLC model system lacking or expressing NQO1 at levels consistent with tumors from NSCLC patients. H596 cells are homozygous for the NQO1*2 polymorphism (8) and were resistant to β-lap. Expression of NQO1 in these cells greatly increased their sensitivity to β-lap, which was inhibited by DIC. By using NQO1+ A549 or NQ+ H596 cells, we showed that β-lap-induced cell death was optimal when short-term drug exposures were administered (2–8 h) in survival assays. Cells with low levels of NQO1 or those treated with DIC, as would be the case for normal tissue, were spared. Indeed, normal lung IMR-90 fibroblasts (SI Fig. 8) that expressed low NQO1 levels were resistant to β-lap, consistent with prior normal cell responses to this drug (21). Importantly, the therapeutic window was eliminated with long-term (>12-h) β-lap exposures. These data suggested that continuous exposure of patients to β-lap should be avoided, and preclinical screening for patients with NQO1 polymorphisms before therapy should be conducted.
The mechanism of cell death induced by β-lap in NSCLC cells (Figs. 1–4) serves to explain and define the compound's optimal dose-dependent therapeutic window. The metabolism of β-lap by NQO1 results in dramatic ROS formation, extensive DNA damage, and PARP-1 hyperactivation, corroborating our findings in NQO1+ breast cancer cells (19). Excessive DNA damage associated with PARP-1 hyperactivation was noted in ischemia/reperfusion responses in tissues and after conditions (e.g., supralethal doses of H2O2), where extensive DNA single-strand breaks occurred (26, 27). We demonstrated that PARP-1 hyperactivation was a major determinant in NAD+ and ATP loss (Fig. 3), based on kinetics of nucleotide pool loss (i.e., NAD+), and the fact that PARP-1 inhibition by 3-AB (an NAD+ analog) strongly blocked both NAD+ and ATP losses, as well as β-lap-induced lethality. Loss of ATP resulted in Ca2+ influx that ultimately led to μ-calpain activation and atypical cleavage of PARP-1 and p53 (18, 28) (SI Fig. 9). A prior report suggested that caspase activation and cell cycle status were key players in β-lap-induced toxicity in A549 cells (20). However, our data contradict these findings and clearly show that ROS formation, DNA damage, and PARP-1 hyperactivation are critical factors in cell death in NQO1+ NSCLC cells exposed to β-lap regardless of cell cycle status.
When confronted with manageable levels of DNA damage, PARP-1 uses NAD+ as a substrate to add branched and linear poly(ADP-ribose) polymers (PARs) to itself (PAR-PARP-1), as well as other nuclear acceptor proteins, to facilitate DNA break repair. Under normal repair conditions, branched polymers are rapidly degraded by poly(ADP-ribose)glycohydrolase, an exo- and endoglycohydrolase that does not directly inhibit PARP-1 activity (3, 29, 30). Poly(ADP-ribose)glycohydrolase exerts its affect on PARP-1-mediated toxicity by decreasing PAR turnover, slowing the rate of NAD+ depletion. Using 3-AB, our data suggested that NAD+ depletion was a critical factor in β-lap-induced lethality, significantly delaying ATP loss. Preventing NQO1 activity (Fig. 3) or scavenging ROS upstream (K. F. Reinicke, M.S.B., E.A.B., Y.D., W.G.B., D. R. Spitz, and D.A.B., unpublished data) was far more efficient and significantly inhibited PARP-1 hyperactivation and β-lap lethality. Thus, β-lap exposure causes DNA damage (Fig. 2), and simultaneously inhibits DNA repair by dramatic ATP and NAD+ pool losses. This is a major advantage for β-lap in NSCLC therapy, because very few compounds are efficacious after a short “bolus” administration, and few induce irreversible, cancer-selective lethality regardless of cell cycle status, loss of p53 function, or loss of proapoptotic factors (E.A.B., J. Meng, and D.A.B., unpublished data).
We theorize that β-lap lethality can be further enhanced by using improved drug delivery strategies in combination with a detailed understanding of the drug's mechanism of action. Our ongoing studies include developing efficacious delivery vehicles [e.g., functionalized micelles, millirod implantable devices (10, 31)], for efficacious delivery of this water-insoluble drug and its potentially more potent prodrugs (32). Future β-lap and NQO1 bioactivated chemotherapies for NSCLC have the potential to play a major role in treating devastating drug resistant cancers, such as NSCLC.
Materials and Methods
Chemicals and Reagents.
β-Lap was synthesized by us and dissolved in DMSO (33). 3-AB, DIC, hydrogen peroxide (H2O2), staurosporine, Hoechst dye 53326, BSA, cytochrome c (practical grade), and propidium iodide were purchased from Sigma–Aldrich (St. Louis, MO). BAPTA-AM was purchased from Calbiochem (La Jolla, CA) and dissolved in DMSO. 5-(and-6)-Chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate was purchased from Invitrogen Life Technologies (Eugene, OR).
NQO1 Gene Expression Profiling on Human Lung Cancers by DNA Microarray Analyses.
Clinical specimens were obtained from New York and Hong Kong tissue banks containing 30 adenocarcinoma, 11 normal lung tissues (New York), 49 adenocarcinoma, and 9 normal samples (Hong Kong), respectively (SI Tables 1 and 2). mRNA preparation, Affymetrix (Santa Clara, CA) microarray (human U133 plus 2) hybridization, and probe set analyses were performed by using Affymetrix MAS 5.0. The genomic profiling database was maintained by the Lung Cancer SPORE Program at University of Texas Southwestern. NQO1 expression was detected by using two probes (201468_s_at; 210519_s_at), and expression was graphed by histogram (bin size, 1,000 units) using raw data without log transformation (SI Figs. 10 A and C and 11 A and C). Expression was classified as low, high, or higher (SI Tables 1 and 2). Analyses of data using the 201468_s_at probe set was graphed (Fig. 1A).
Tissue Culture.
NQO1-deficient H596 NSCLC cells were provided by Su-Shu Pan (University of Pittsburgh, Pittsburgh, PA). A549 cells were generously donated by Steven Dubinett (University of California, Los Angeles, CA). IMR-90 lung fibroblasts were obtained from the American Tissue Type Culture Collection (Manassas, VA). Cells were grown in DMEM with 10% FBS, 2 mM l-glutamine, 100 units/ml penicillin, and 100 mg/ml streptomycin at 37°C in a humidified incubator with a 5% CO2–95% air atmosphere. All cells were mycoplasma free (17, 18).
Generation of Isogenically Matched NQO1-Expressing H596 NSCLC Cells.
H596 NSCLC cells are homozygous for the NQO1*2 polymorphism (8), resulting in lack of NQO1 expression/activity (Fig. 1). Log-phase H596 cells were infected with retroviral vectors expressing NQO1 or empty vector, and stable pooled populations, LPC-NQO1 (NQ+ H596) or LPCX (NQ− H596), were selected in 1.0 μg/ml puromycin.
NQO1 Assays.
Cell lysates were prepared and NQO1 enzyme assays performed with or without DIC (17). Activities were calculated as DIC-inhibited nanomoles of cytochrome c reduced per minute per microgram of protein based on initial rates of OD change at 550 nm and an extinction coefficient for cytochrome c of 21.1 mM/cm (17). Nondetectable NQO1 levels were <1 nmol·min−·μg−.
Western Blot Analyses.
Whole-cell lysates were prepared and proteins were separated by 10% SDS/PAGE (33). α-PARP-1 (sc-8007) and α-p53 (DO-1) antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), as well as α-PAR (BD Pharmingen, San Jose, CA), were used at 1:2,000.
Relative Survival Assays.
Long-term, relative survival assays based on DNA content after 7 days of growth posttreatment were performed in 48-well dishes (17). NQ− and NQ+ H596, and A549 cells were treated with or without β-lap at the indicated doses/times, in the presence or absence of 40 or 50 μM DIC. Cells were also pretreated with 5 μM BAPTA-AM (19) before β-lap addition. Results were confirmed by colony-forming ability assays (33) and graphed as means ± SE from at least three experiments performed in triplicate.
ROS Analyses.
Disulfide glutathione and total glutathione levels were assessed (32, 34). Data were expressed as percent disulfide glutathione normalized to protein content measured by Lowry (35). Shown were means ± SE for experiments performed at least three times (SI Fig. 6). ROS formation was confirmed by conversion of nonfluorescent 5-(and 6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate to its fluorescent derivative by flow cytometry (FC-500 flow cytometer; Beckman Coulter Electronics, Miami, FL) (36). Data were graphed as means ± SE for experiments performed three times in duplicate.
Alkaline Comet Assays.
DNA lesions, including DNA single- and double-strand breaks, were assessed in single cells by using alkaline comet assays from TREVIGEN (Gaithersburg, MD) (19, 37). Slides were stained with SYBR Green and visualized by using a Nikon (Melville, NY) Eclipse TE2000-E fluorescence microscope. Digital photomicrographs were taken and comet tail lengths were quantified (19).
Nucleotide Analyses.
Changes in intracellular NAD+ levels were measured and intracellular NAD+ levels expressed as percent treated divided by control (%T/C) ± SE from at least three individual experiments, each in duplicate (19). In some experiments, cells were pretreated with 5 μM BAPTA-AM or 25 mM 3-AB as described (19). ATP levels were analyzed from whole-cell extracts by using luciferase-based bioluminescence assays (18). Data were graphed as means ± SE of experiments performed three or more times in triplicate for ATP, or three or more independent experiments for NAD.
Apoptotic Assays.
Apoptosis was quantified by using Apo Direct Apoptotic assays (TUNEL) from BD Pharmingen as described (18). Samples were analyzed by using an EPICS Elite ESP or FC-500 flow cytometer (Beckman Coulter Electronics) and Elite acquisition software. Data were expressed as means ± SE from three independent experiments performed in triplicate.
Statistical Analyses.
Statistical differences were determined by using Student's t tests, and P values were reported.
Supplementary Material
Acknowledgments
We thank members of the D.A.B. laboratory for their helpful comments and Drs. Chi-Leung Lam, Maria P. Wong (both at University of Hong Kong, Hong Kong, China), and William Gerald (Memorial Sloan–Kettering Cancer Center, New York) for assistance with the Hong Kong and New York databases. This work was supported by National Institutes of Health (NIH)/National Cancer Institute (NCI) Grant CA102792 (to D.A.B.) and University of Texas Southwestern M. D. Anderson Lung Cancer Specialized Programs of Research Excellence Grant NCI/NIH P50 CA70907 (to J.D.M.). Stipends for K.E.R. and M.S.B. were provided by U.S. Army Department of Defense Breast Cancer Predoctoral Fellowships W81XWH-05-1-0248 and W81XWH-04-1-0301, respectively. This paper is document no. CSCN 007 of the Cell Stress and Cancer Nanomedicine Program at the University of Texas Southwestern Medical Center at Dallas.
Abbreviations
- 3-AB
3-aminobenzamide
- BAPTA-AM
1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid acetoxymethyl ester
- DCF
5- (and 6)-carboxy-2′,7′-dichlorodihydrofluorescein
- DIC
dicoumarol
- β-lap
β-lapachone (3,4-dihydro-2,2-dimethyl-2H-naphtho[1,2-b]pyran-5–6-dione)
- NQO1
NAD(P)H:quinone oxidoreductase-1
- NQO1+
NQO1-proficient cell
- NQ+ H596
NQO1-proficient H596 cell
- NQO1−
NQO1-deficient cell
- NQ− H596
NQO1-deficient H596 cell
- NSCLC
non-small-cell lung cancer
- PAR
poly(ADP-ribose) modified protein
- PARP-1
poly(ADP-ribose)polymerase-1
- ROS
reactive oxygen species
- T/C
treated/control.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702176104/DC1.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Spira A, Ettinger DS. N Engl J Med. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 3.Kim MY, Zhang T, Kraus WL. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 4.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pink JJ, Wuerzberger-Davis S, Tagliarino C, Planchon SM, Yang X, Froelich CJ, Boothman DA. Exp Cell Res. 2000;255:144–155. doi: 10.1006/excr.1999.4790. [DOI] [PubMed] [Google Scholar]
- 6.Bentle MS, Bey EA, Dong Y, Reinicke KE, Boothman DA. J Mol Histol. 2006;37:203–218. doi: 10.1007/s10735-006-9043-8. [DOI] [PubMed] [Google Scholar]
- 7.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 8.Ross D, Siegel D. Methods Enzymol. 2004;382:115–144. doi: 10.1016/S0076-6879(04)82008-1. [DOI] [PubMed] [Google Scholar]
- 9.Nasongkla N, Wiedmann AF, Bruening A, Beman M, Ray D, Bornmann WG, Boothman DA, Gao J. Pharm Res. 2003;20:1626–1633. doi: 10.1023/a:1026143519395. [DOI] [PubMed] [Google Scholar]
- 10.Wang F, Blanco E, Ai H, Boothman DA, Gao J. J Pharm Sci. 2006;95:2309–2319. doi: 10.1002/jps.20721. [DOI] [PubMed] [Google Scholar]
- 11.Li CJ, Li YZ, Pinto AV, Pardee AB. Proc Natl Acad Sci USA. 1999;96:13369–13374. doi: 10.1073/pnas.96.23.13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro GI, Supko JG, Ryan DP, Appelman L, Berkenbilt A, Craig AR, Jones S, Yagovane D, Li C, Elder J. J Clin Oncol. 2005;23:3042–3043. [Google Scholar]
- 13.Park HJ, Ahn KJ, Ahn SD, Choi E, Lee SW, Williams B, Kim EJ, Griffin R, Bey EA, Bornmann WG, et al. Int J Radiat Oncol Biol Phys. 2005;61:212–219. doi: 10.1016/j.ijrobp.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 14.Boothman DA, Trask DK, Pardee AB. Cancer Res. 1989;49:605–612. [PubMed] [Google Scholar]
- 15.Siegel D, Franklin WA, Ross D. Clin Cancer Res. 1998;4:2065–2070. [PubMed] [Google Scholar]
- 16.Marin A, Lopez de Cerain A, Hamilton E, Lewis AD, Martinez-Penuela JM, Idoate MA, Bello J. Br J Cancer. 1997;76:923–929. doi: 10.1038/bjc.1997.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pink JJ, Planchon SM, Tagliarino C, Varnes ME, Siegel D, Boothman DA. J Biol Chem. 2000;275:5416–5424. doi: 10.1074/jbc.275.8.5416. [DOI] [PubMed] [Google Scholar]
- 18.Tagliarino C, Pink JJ, Dubyak GR, Nieminen AL, Boothman DA. J Biol Chem. 2001;276:19150–19159. doi: 10.1074/jbc.M100730200. [DOI] [PubMed] [Google Scholar]
- 19.Bentle MS, Reinicke KE, Bey EA, Spitz DR, Boothman DA. J Biol Chem. 2006;281:33684–33696. doi: 10.1074/jbc.M603678200. [DOI] [PubMed] [Google Scholar]
- 20.Woo HJ, Choi YH. Int J Oncol. 2005;26:1017–1023. [PubMed] [Google Scholar]
- 21.Li Y, Sun X, LaMont JT, Pardee AB, Li CJ. Proc Natl Acad Sci USA. 2003;100:2674–2678. doi: 10.1073/pnas.0538044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Begleiter A, Fourie J. Methods Enzymol. 2004;382:320–351. doi: 10.1016/S0076-6879(04)82018-4. [DOI] [PubMed] [Google Scholar]
- 23.Boothman DA, Meyers M, Fukunaga N, Lee SW. Proc Natl Acad Sci USA. 1993;90:7200–7204. doi: 10.1073/pnas.90.15.7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malkinson AM, Siegel D, Forrest GL, Gazdar AF, Oie HK, Chan DC, Bunn PA, Mabry M, Dykes DJ, Harrison SD, et al. Cancer Res. 1992;52:4752–4757. [PubMed] [Google Scholar]
- 25.Phillips RM, Basu S, Brown JE, Flannigan GM, Loadman PM, Martin SW, Naylor B, Puri R, Shah T. Int J Oncol. 2004;24:1005–1010. [PubMed] [Google Scholar]
- 26.Lam TT. Res Commun Mol Pathol Pharmacol. 1997;95:241–252. [PubMed] [Google Scholar]
- 27.Halmosi R, Berente Z, Osz E, Toth K, Literati-Nagy P, Sumegi B. Mol Pharmacol. 2001;59:1497–1505. doi: 10.1124/mol.59.6.1497. [DOI] [PubMed] [Google Scholar]
- 28.Tagliarino C, Pink JJ, Reinicke KE, Simmers SM, Wuerzberger-Davis SM, Boothman DA. Cancer Biol Ther. 2003;2:141–152. doi: 10.4161/cbt.2.2.237. [DOI] [PubMed] [Google Scholar]
- 29.Koh DW, Dawson VL, Dawson TM. Cell Cycle. 2005;4:397–399. doi: 10.4161/cc.4.3.1559. [DOI] [PubMed] [Google Scholar]
- 30.Ying W, Sevigny MB, Chen Y, Swanson RA. Proc Natl Acad Sci USA. 2001;98:12227–12232. doi: 10.1073/pnas.211202598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nasongkla N, Shuai X, Ai H, Weinberg BD, Pink J, Boothman DA, Gao J. Angew Chem Int Ed Engl. 2004;43:6323–6327. doi: 10.1002/anie.200460800. [DOI] [PubMed] [Google Scholar]
- 32.Reinicke KE, Bey EA, Bentle MS, Pink JJ, Ingalls ST, Hoppel CL, Misico RI, Arzac GM, Burton G, Bornmann WG, et al. Clin Cancer Res. 2005;11:3055–3064. doi: 10.1158/1078-0432.CCR-04-2185. [DOI] [PubMed] [Google Scholar]
- 33.Wuerzberger SM, Pink JJ, Planchon SM, Byers KL, Bornmann WG, Boothman DA. Cancer Res. 1998;58:1876–1885. [PubMed] [Google Scholar]
- 34.Lee YJ, Galoforo SS, Berns CM, Chen JC, Davis BH, Sim JE, Corry PM, Spitz DR. J Biol Chem. 1998;273:5294–5299. doi: 10.1074/jbc.273.9.5294. [DOI] [PubMed] [Google Scholar]
- 35.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 36.Panduri V, Weitzman SA, Chandel NS, Kamp DW. Am J Physiol. 2004;286:L1220–L1227. doi: 10.1152/ajplung.00371.2003. [DOI] [PubMed] [Google Scholar]
- 37.Olive PL, Banath JP, Durand RE. Radiat Res. 1990;122:86–94. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.