In this issue of PNAS, Duyn et al. (1) present a magnetic resonance imaging (MRI) technique that reveals astonishing and hitherto unseen details of the architecture of the cerebral cortex of living human beings. The cortex (Latin for “bark” or “outer rind”) is a thin layer of gray matter that is highly folded to attain a large surface area within the limited volume of the human skull, and it is thought to be the substrate for the vast majority of the cognitive skills that we possess. That the cortex has an interesting laminar structure, and that this structure may relate to its functional properties, has been known for more than 100 years (2, 3). Directly visualizing these laminar properties in living humans is one of the central goals of structural neuroimaging research. From a neuroscience standpoint, the importance of imaging cortical architecture stems from the fact that the human cortex can be parcellated into discrete regions based on changes in the laminar distributions of cell types and density, dating back to the classic work of Korbinian Brodmann a century ago, one of a number of famous cortical parcellations (3–7). These regions, frequently referred to as “cortical areas,” are strongly tied to the functional properties of the brain, and hence being able to robustly and routinely delineate them in vivo would be a fundamental tool in research aimed at deepening our understanding of the human brain.
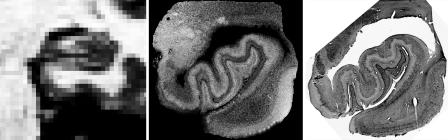
The challenge of visualization of cortical architecture is illustrated in Fig. 1, which shows a coronal section (a plane perpendicular to the line connecting the nose to the back of the head) through the human medial temporal lobe, one of the regions affected earliest by Alzheimer's disease (AD) (8). In MRI there is a direct relationship between resolution and signal-to-noise ratio (SNR). Specifically, the SNR varies with the third power of the linear dimension of a voxel (or linearly with the volume of a voxel). Fig. 1 Left shows a standard in vivo MRI scan obtained at ≈1-mm resolution, requiring 8.5 min to acquire. Fig. 1 Center is of a portion of a fixed ex vivo brain acquired at 100-μm isotropic resolution, and Fig. 1 Right is a standard Nissl stain, which marks cell bodies as dark spots. As can be seen, the ex vivo MRI scan in the center reveals much of the same laminar information as the histological image on the right, again showing that MRI has the potential to probe laminar architecture. The additional SNR required to obtain Fig. 1 Center relative to Left is approximately a factor of 1,000 or the ratio of the volume of the voxels in each image. This resolution is achievable in imaging ex vivo samples because of a number of factors that result in a dramatic increase in SNR, including scan times that would be prohibitively long in vivo (e.g., >12 h), the placement of small coils in close proximity to the sample with no intervening skull, and the absence of artifacts and image blurring due to respiratory and cardiac cycles. None of these factors are applicable in vivo, implying that the SNR needed to attain the image resolution to visualize these details in vivo must be obtained elsewhere.
Fig. 1.
Comparison of coronal sections through the medial temporal lobe in vivo (Left), ex vivo (Center), and histological images. (Left) Standard 1-mm in vivo image of a temporal lobe. (Center) A 100-μm isotropic ex vivo MRI image showing laminar structure in the cortex and the hippocampus. (Right) Nissl stain revealing distribution of cell bodies into layers. Neda Bernasconi and Jean Augustinack (Harvard Medical School, Boston, MA) provided these images.
The fundamental technique used by Duyn et al. (1), using phase changes in the MRI signal that arise from local variations in the magnetic susceptibility of brain tissue to reveal underlying anatomical structure, has been in use for some time (8, 9). The contributions of the present paper are in bringing this technique to the level where it can be used to study the internal structure of the cortex and the simply astonishing images that they have obtained. As always in MRI, the increase in resolution comes about because of an increase in SNR, which can then be traded off for smaller voxel sizes. The authors achieve the unprecedented SNR and resolution through advances on a number of fronts. First, they worked with state-of-the-art ultra-high-field magnetic resonance equipment and solved some of the confounds associated with imaging at this type of field strength, such as the modulations in the magnetic field induced by respiratory cycles. Second, they were quick to realize that by measuring phase changes rather than signal magnitude they were severalfold more sensitive to susceptibility-based contrast, a contrast that is enhanced at higher field. Finally, they realized that the internal structure of the cortex offers an attractive target for this contrast mechanism. The result is a convincing demonstration that these images can be useful for directly visualizing the underlying laminar architecture of the cortex.
The basic insight of using the signal phase to reveal anatomical and physiological details of the human brain was developed more than a decade ago (9) and has been dubbed susceptibility weighted imaging (SWI) (8). The technique allows the visualization of phase difference in the hydrogen protons precessing at a rate that is proportional to the local magnetic field. When brain structures magnetize to different degrees, the result is local variations in the magnetic fields in and around these structures, and changes in the phase thus reflect anatomical and physiological phenomena related to the source of the magnetization, such as the amount of iron in the tissue or blood. Originally this technique was applied to imaging of the vasculature by Haacke and colleagues (10), which resulted in significant clinical applications such as visualization of tumor neovasculature (11). More recently, it has been applied at ultra-high field by a group at Ohio State University to image brain structure with some success (12).
Several researchers have shown examples of being able to directly visualize laminar architecture both in vivo (13–16) and ex vivo (17–19), using signal magnitude. These studies have used exceedingly long scan sessions or extremely localized imaging to detect signatures of the cortical architecture, but none have been able to convincingly demonstrate the delineation of a entire cortical area in vivo. From a clinical standpoint, being able to detect the borders of cortical areas by using laminar properties would be a critical advance in our ability to diagnose disorders such as AD earlier in their course. In AD specifically, the earliest stage of the disease is thought to be confined to a small number of cortical areas (20, 21). Thus, detecting changes in brain structure in early AD can be decomposed into two separate problems: (i) localizing the regions where an effect is expected, and (ii) detecting the effect. The phase imaging technique has the potential to aid in each of these areas. Early detection would be vital because therapeutic intervention will likely be effective only before widespread cell death. The contribution of Duyn et al. (1) vividly demonstrates that signal phase has such significant advantages over signal magnitude in imaging cortical architecture, thus bringing neuroimaging one step closer to the in vivo delineation of cortical regions.
Signal phase has advantages over signal magnitude in imaging cortical architecture.
Acknowledgments
This work was supported in part by the National Center for Research Resources (NCRR) (P41-RR14075, R01 RR16594-01A1), the NCRR Biomedical Informatics Research Network Morphometric Project (BIRN002, U24 RR021382), the National Institute for Biomedical Imaging and Bioengineering (R01 EB001550), the National Institute for Neurological Disorders and Stroke (R01 NS052585-01), and the Mental Illness and Neuroscience Discovery (MIND) Institute, and is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health (NIH) through the NIH Map for Medical Research Grant U54 EB005149.
Footnotes
The authors declare no conflict of interest.
See companion article on page 11796.
References
- 1.Duyn JH, van Gelderen P, Li T-Q, de Zwart JA, Koretsky AP, Fukunaga M. Proc Natl Acad Sci USA. 2007;104:11796–11801. doi: 10.1073/pnas.0610821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogt O. J Psychol Neurol. 1910;15:221–232. [Google Scholar]
- 3.Brodmann K. Vergleichende Lokalisationslehre der Großhirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. Leipzig, Germany: J. A. Barth; 1909. [Google Scholar]
- 4.Vogt O. J Psychol Neurol. 1911;18:107–118. [Google Scholar]
- 5.von Economo C. The Cytoarchitectonics of the Human Cerebral Cortex. London: Oxford Univ Press; 1929. [Google Scholar]
- 6.Sarkissov S, Filimonoff IN, Kononova IP, Preobrazenskaja NS, Kukueva LA. Atlas of the Cytoarchitectonics of the Human Cerebral Cortex. Moscow: Medgiz; 1955. [Google Scholar]
- 7.Flechsig P. Anatomie des menschliches Gehirn Ruckenmark auf mielogenetische Grundlage. Leipzig, Germany: Thieme; 1920. [Google Scholar]
- 8.Haacke E, Xu Y, Cheng Y, Reichenbach J. Magn Reson Med. 2004;52:612–618. doi: 10.1002/mrm.20198. [DOI] [PubMed] [Google Scholar]
- 9.Yablonskiy D, Haacke E. Magn Reson Med. 1994;32:749–763. doi: 10.1002/mrm.1910320610. [DOI] [PubMed] [Google Scholar]
- 10.Reichenbach JR, Venkatesan R, Schillinger DJ, Kido DK, Haacke EM. Radiology. 1997;204:272–277. doi: 10.1148/radiology.204.1.9205259. [DOI] [PubMed] [Google Scholar]
- 11.Barth M, Nöbauer-Huhmann I, Reichenbach J, Mlynarik V, Schöggl A, Matula C, Trattnig S. Invest Radiol. 2003;38:409–414. doi: 10.1097/01.RLI.0000069790.89435.e7. [DOI] [PubMed] [Google Scholar]
- 12.Abduljalil AM, Schmalbrock P, Novak V, Chakeres DW. J Magn Reson Imaging. 2003;18:284–290. doi: 10.1002/jmri.10362. [DOI] [PubMed] [Google Scholar]
- 13.Walters NB, Egan GF, Kril JJ, Kean M, Waley P, Jenkinson M, Watson JGG. Proc Natl Acad Sci USA. 2003;100:2981–2986. doi: 10.1073/pnas.0437896100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark VP, Courchesne E, Grafe M. Cerebral Cortex. 1992;2:417–424. doi: 10.1093/cercor/2.5.417. [DOI] [PubMed] [Google Scholar]
- 15.Barbier EL, Marrett S, Danek A, Vortmeyer A, van Gelderen P, Duyn J, Bandettini P, Grafman J, Koretsky AP. Magn Reson Med. 2002;48:735–738. doi: 10.1002/mrm.10255. [DOI] [PubMed] [Google Scholar]
- 16.Bridge H, Clare S, Jenkinson M, Jezzard P, Parker AJ, Matthews PM. J Vis. 2005;5:93–102. doi: 10.1167/5.2.1. [DOI] [PubMed] [Google Scholar]
- 17.Augustinack JC, van der Kouwe AJW, Blackwell ML, Salat DH, Wiggins CJ, Frosch MP, Wiggins GC, Potthast A, Wald LL, Fischl BR. Ann Neurol. 2005;57:489–494. doi: 10.1002/ana.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fatterpekar GM, Naidich TP, Delman BN, Aguinaldo JG, Gultekin SH, Sherwood CC, Hof PR, Drayer BP, Fayad ZA. AJNR Am J Neuroradiol. 2002;23:1313–1321. [PMC free article] [PubMed] [Google Scholar]
- 19.Pfefferbaum A, Sullivan E, Adalsteinsson E, Garrick T, Harper C. Neuroimage. 2004;21:1585–1595. doi: 10.1016/j.neuroimage.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 20.Gomez-Isla T, Price JL, McKeel DW, Jr, Morris JC, Growdon JH, Hyman BT. J Neurosci. 1996;16:4491–4500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braak H, Braak E. Acta Neuropathol (Berlin) 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]