Abstract
The recognition of carrier proteins by multiple catalytic partners occurs in every cycle of chain elongation in the biosynthesis of fatty acids and of the pharmacologically important polyketide and nonribosomal peptide natural products. To dissect the features of carrier proteins that determine specific recognition at distinct points in assembly lines, we have used the two-module Escherichia coli enterobactin synthetase as a model system. Using an entB knockout strain, we developed a selection for growth on iron-limiting medium to evolve aryl carrier protein domains. The aryl carrier proteins from VibB of Vibrio cholerae vibriobactin and HMWP2 of Yersinia pestis yersiniabactin assembly lines were evolved by random mutagenesis to support growth under selection conditions, yielding a convergent set of mutations. Subsequent in vitro biochemical characterizations with partner enzymes EntE, EntF, and Sfp on the evolved VibB aryl carrier protein revealed a ≈500-fold improvement in reconstituted enterobactin production activity. Mechanistic characterization identified three distinct specific recognition surfaces of VibBArCP for three catalytic partners in enterobactin biosynthesis. Our results suggest that heterologous carrier protein interactions can be engineered with a small number of mutations given a suitable selection scheme and provide insights for reprogramming nonribosomal peptide biosynthesis.
Keywords: nonribosomal peptide synthetase, recognition, siderophores
A large number of medicinally important natural products are polyketides (PKs) and nonribosomal peptides (NRPs) (1). Members of these natural products include the antibiotics vancomycin (NRP) and erythromycin (PK), the immunosuppressant FK506 (hybrid PK/NRP), and the anticancer agents epothilone and bleomycin (both are hybrid PK/NRP) (2). PKs, NRPs, and fatty acid scaffolds are assembled with comparable biosynthetic logic and by similar protein machinery. The monomers to be elongated (malonyl and methylmalonyl groups in FA and PK, and amino acids in NRP frameworks), as well as the resultant growing acyl/peptide chains, are covalently tethered to carrier proteins (CPs) as protein phosphopantetheinyl thioesters at each stage of chain initiation, elongation, and termination. The phosphopantetheinyl (Ppant) arm is introduced posttranslationally by dedicated Ppant transferases (PPTases) onto a serine side chain in the 80- to 100-residue CPs (3). The terminal thiol of the Ppant arm provides the nucleophile for thioester formation, affording the thermodynamic activation for the C–C bond-forming thioclaisen condensation/chain elongation reactions in FA and PK assembly and for the C-N amide formations in NRP chain growth (4, 5).
Thus, fatty acid synthases (FASs), polyketide synthases (PKSs), and nonribosomal peptide synthetases (NRPSs) are composed of modules. Each module contains multiple catalytic domains and one carrier protein domain that bears the acyl/aminoacyl/peptidyl chain on which the catalytic domains act. Different CPs are serviced by subsets of specific catalytic domains. Three variants of carrier proteins have been defined based on the type of assembly line involved and on the type of acyl group tethered to the Ppant arm: acyl carrier proteins (ACPs) in FAS and PKS; peptidyl carrier proteins (PCPs) in NRPS; and aryl carrier proteins (ArCPs) in the biosynthesis of bacterial virulence-conferring nonribosomal peptide siderophores (iron chelators) and other aryl-capped natural products (6).
Despite the fact that ACPs, PCPs, and ArCPs serve cognate functions in their natural product assembly lines, and all carry acyl-thioesters on their Ppant prosthetic groups, they are clearly differentially recognized by catalytic partners. We have been interested in defining the surface features of the 80- to 100-residue CP domains that are recognized by partner enzymes. To that end, we have previously dissected the two-module E. coli enterobactin synthetase (EntBDEF) assembly line to evaluate the role and recognition of the two carrier proteins: (i) an ArCP, in EntB carrying 2,3-dihydroxybenzoyl; and (ii) a PCP in EntF carrying an l-seryl group during chain assembly and cyclotrimerization (2, 7, 8).
We performed those studies by using a combinatorial mutagenesis and selection scheme for functional EntB ArCP or EntF homologs by growth of Escherichia coli under iron-limiting conditions, in which production of enterobactin is required. To further reveal rules for carrier protein recognition, assess the portability of carrier proteins between assembly lines, and reprogram their recognition, we now report that noncognate aryl carrier proteins, VibB ArCP from the Vibrio cholerae vibriobactin siderophore pathway (9) and the HMWP2 ArCP domain from the Yersinia pestis siderophore yersiniabactin pathway (10), can be evolved to replace the EntB ArCP with a convergent set of mutations. The evolved ArCP domains are recognized by the three enzymes Sfp, EntE, and EntF to produce functional enterobactin.
Results
Domain Swapping and Initial Characterization of Isochorismate Lyase (ICL)-ArCP Chimeric Proteins.
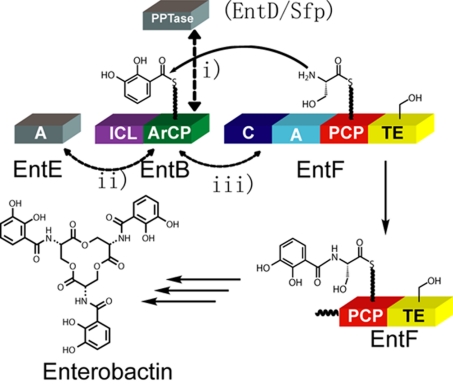
Our selection for enterobactin production was based on the previous discovery that the expression of plasmid-encoded EntB is able to complement the chromosomal deletion of the entB gene of the enterobactin synthetase, which is essential for E. coli under iron-limited conditions (2, 11). The iron in selection media was sequestered by the addition of the iron chelator 2,2′-dipyridyl. Cells producing enterobactin, which has an estimated Kd of 10−35 M for ferric iron at physiological pH (12), are able to recruit the iron from 2,2′-dipyridyl for growth on the iron-depleted media. The enterobactin biosynthetic scheme is shown in Fig. 1. The synthetase consists of four proteins [E. coli enterobactin synthetase (EntBDEF)] (13). EntE is a free-standing adenylation (A) domain that activates 2,3-dihydroxybenzoate (DHB). EntD is a dedicated phosphopantetheinyl transferase (PPTase). EntB is a two-domain protein that contains an N-terminal ICL domain (involved in production of DHB), and a C-terminal ArCP onto which DHB is loaded. The four-domain (C-A-PCP-TE) protein EntF is a combination elongation/termination module protein that is responsible for the activation/loading of serine (by the A domain), condensation of DHB (presented on EntBArCP) and the loaded serine [by the conensation (C) domain], and production of the macrolactone [by the thioesterase (TE) domain].
Fig. 1.
Enterobactin biosynthesis scheme. The endogenous PPTase in the enterobactin synthetase is EntD; the broad-substrate PPTase Sfp was used in our selection. Protein–protein interactions between EntBArCP or hererologous ArCPs during enterobactin are indicated by double-headed, dashed arrows. The ArCP must be recognized by PPTase (EntD or Sfp) (i), EntE (ii), and EntF C domain (iii).
The crystal structure of EntB (14) revealed a proline-rich linker (amino acid sequence: PAPIP) between the ICL and ArCP domains [supporting information (SI) Fig. 5]. The two domains do not appear to have extensive interdomain contacts, and we have previously shown in vitro that the ICL domain is not required for the function of the NRPS (13). These observations suggest that the two domains function independently and that incorporation of exogenous ArCPs downstream of the proline-rich linker in an ICL-ArCP hybrid (SI Fig. 5) might provide a reasonable starting point for directed evolution of ArCP interactions in the enterobactin synthetase.
The DNA encoding ArCPs of VibB from Vibrio cholerae for vibriobactin biosynthesis, and of HMWP2 from Yersinia pestis for yersiniabactin biosynthesis were fused to the ICL domain of EntB and cloned into pET22b vector to create the chimeric constructs pET22b-EntBICL-VibBArCP and pET22b-EntBICL-HMWP2ArCP. These chimeric constructs were introduced into E. coli cells containing a chromosomal replacement of the entB gene for a kanamycin resistance gene (entB::kanR) (2), and the growth of the resulting clones was tested. The pET22b-EntBICL-VibBArCP and pET22b-EntBICL-HMWP2ArCP clones grew quickly on iron-rich medium, yielding colonies of comparable sizes to entB::kanR cells complemented with WT entB after 16 h of growth (data not shown). However, the growth of entB::kanR cells harboring pET22b-EntBICL-VibBArCP and pET22b-EntBICL-HMWP2ArCP was not observed under iron-limiting conditions after 4 days (data not shown). When these chimeric genes were cloned into a polycistronic vector (pET-DUET), along with the gene for the broad-substrate PPTase Sfp from B. subtilis, slow growth of entB::kanR cells was observed when transformed with the chimeric constructs and grown for 2 days (data not shown). The observed lack of growth for pET22b-EntBICL-VibBArCP and pET22b-EntBICL-HMWP2ArCP was likely due to lack of recognition of VibBArCP and HMWP2 ArCP by the PPTase EntD, which has a slow kcat and high substrate specificity (15). (The only functional CP substrates that have been demonstrated for EntD are EntBArCP and EntF PCP.) Inclusion of the sfp gene (16) on the cloning vector likely improved growth of entB::kanR cells harboring the chimeric genes on iron-deficient media by improving the degree of phosphopantetheinylation of the exogenous ArCP. Despite this improvement, cells harboring these chimeric genes were still much slower in growth on selection medium than cells complemented with WT entB. We reasoned that this disparity was due to deficiencies in recognition between the ArCPs and their catalytic partners. Therefore, we used the polycistronic constructs containing both sfp and the chimeric genes as a starting point for directed evolution to improve the ArCP interactions.
ArCP Library Production and Selection for Functional Enterobactin Synthetases.
The DNA of the exogenous ArCPs (full-length) were subjected to random mutagenesis by error-prone PCR with two types of mutagenic dNTPs: 8-oxo-dGTP and dPTP (2′-deoxy-p-nucleoside-5′-triphosphate) (17). Mutation rates were adjusted by diluting the mutagenic dNTPs with normal nucleotides; we used libraries containing an average of 3- to 4-aa mutations within the ArCP regions. The ArCP mutant library DNA was then fused with DNA encoding the EntBICL, ligated into the polycistronic vector containing sfp, and introduced into entB::kanR cells by electroporation. This procedure yielded library sizes of 105-106 unique clones. Sequencing of unselected clones from the mutagenic libraries demonstrated that mutations were almost randomly dispersed throughout the ArCP region (data not shown). The libraries were then subjected to selection conditions for a functional enterobactin synthetase by plating onto the iron-deficient medium, and those colonies that displayed improvement in supporting the entB::kanR growth under iron-limiting condition were defined as “hits” (SI Fig. 6). These hits were isolated and the corresponding DNA was sequenced. SI Table 3 shows a summary of the mutations from the pool of hits (25–50 for each library). Mutations with observed occurrences no less than three times were considered significant and are indicated in SI Table 3. The highest-occurring convergent mutations were accepted and introduced into the corresponding exogenous ArCP scaffold, yielding a mutant clone that served as the template for the next round of evolution. Similar mutagenesis, selection, and mutation strategies were used in subsequent rounds. Table 1 contains a summary of the high-occurrence mutations after each round of selection for both clones; a sequence alignment displaying this information is also shown in SI Fig. 7a. The VibBArCP was evolved for three rounds, yielding a total of six high-occurrence mutations. The HMWP2ArCP was evolved for two rounds, yielding a total of six high-occurrence mutations; in this case, the third round did not generate high-occurrence mutations. Two types of numbering schemes were used to describe the position of each residue/mutation on the carrier proteins: (i) the absolute position (AP); (ii) the position relative to the phosphopantetheinylated serine [relative position (RP)]. The numbering for the AP of each ArCP begins at the junction with the ICL domain. However, we predict that the position of the residues relative to the Ppant serine is more relevant to function. Therefore, RP numbering is used in this paper: residues C-terminal to the Ppant serine are indicated with positive numbers, and residues N-terminal to the ArCP active site serine are indicated with negative numbers. To distinguish the two types of numbering, a “+” or “−” symbol is included before the RP, but not before the AP.
Table 1.
Summaries for highest-occurring convergent mutations during three rounds of ArCP evolution
| Carrier protein | Round | Absolute position | Relative position | Mutations | Mutation group |
|---|---|---|---|---|---|
| VibBArCP | I | 70 | +24 | E → K | I |
| 54 | +8 | E → G | II | ||
| 47 | +1 | I → V | I | ||
| II | 89 | +43 | C → R | I | |
| 38 | −8 | N → D | I | ||
| III | 41 | −5 | F → L | II | |
| HMWP2ArCP | I | 10 | −23 | I → M | II |
| 75 | +42 | N → D | I | ||
| 25 | −8 | N → D | I | ||
| II | 57 | +23 | Y → C | I | |
| 38 | +5 | R → G | II | ||
| 33 | +1 | I → V | I |
Analysis of Convergent Mutations and Structure-Function Analysis.
We grouped the high-occurrence mutations into two categories (Table 1): convergent mutations (group I) and nonconvergent mutations (group II). The mutations at −8, +1, +24, and +43 positions in VibBArCP and the mutations at −8, +1, +23, and +42 in HMWP2ArCP fall in the convergent mutation category because mutations at these positions were observed in the evolved clones of both libraries (one-residue position perturbations were tolerated); the other mutations were nonconvergent mutations. Here, we focus on group I, because these mutations are likely to reflect the common features and rules in ArCP recognition and evolution. The aligning residues in EntBArCP for + 1, +23, +24, and −8 positions are V, A, K, and N, respectively (SI Fig. 7b).
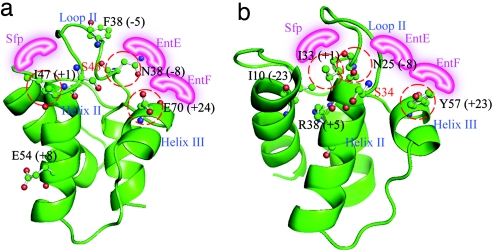
To better understand the information from the group of convergent mutations, we mapped them onto structural homology models of the ArCPs (Fig. 2 a and b). The C-terminal mutation (+42/43) could not be mapped onto the model structures because these positions contain no aligning residues in EntBArCP. For the other group I mutations, (i) the +1 position is located in helix II immediately C-terminal to the phosphopantetheinylated serine; (ii) the +24/23 positions are located in helix III region; and (iii) the −8 positions located in the loop II regions. The positions of group II mutations are also indicated in the models.
Fig. 2.
Structural models. The convergent mutations were mapped onto structural models of VibBArCP (a) and HMWP2ArCP (b). The proposed differential recognition surfaces are shown in pink. The numbers in parentheses indicates the relative position (RP) of each residue. The position of group I mutations are circled in red.
In Vivo Characterization of Evolved ArCPs.
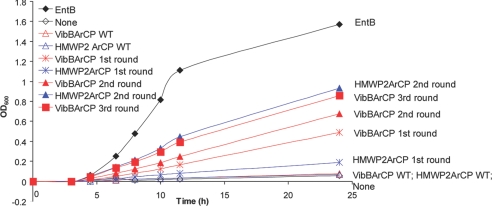
To confirm the significant improvement of growth under iron-limiting conditions for clones containing the high-occurrence mutations, we developed a liquid culture assay in iron-limiting medium. The entB::kanR strains coexpressing Sfp and chimeric proteins VibBArCP (WT and first, second, and third round) and HMWP2 ArCP (WT and first and second round) were synchronized and inoculated into liquid minimal media containing 200 μM 2,2′-dipyridyl. The time course for growth, measured by OD600, was recorded (Fig. 3). The clones containing WT VibBArCP and HMWP2ArCP displayed almost no growth (OD600 < 0.1) after 24 h, whereas the third-round-evolved VibBArCP and second-round-evolved HMWP2ArCP constructs grew to an OD600 of ≈0.9 in 24 h. Cells complemented with EntBArCP grew to OD600 1.6 in 24 h. The resulting mutant from each round showed better growth than the corresponding ArCP mutant from the previous round. We observed no significant difference in growth rates in iron-rich medium (data not shown). Similar growth behavior was observed in a spot-test assay on solid media (SI Fig. 8). These growth patterns indicate that the mutations from the selection provide a significant gain of function for supporting entB::kanR growth in iron-limiting conditions, for which the production of enterobactin is required.
Fig. 3.
Liquid culture assays for evolved ArCPs. The evolved ArCPs (VibBArCP and HMWP2ArCP) showed significant growth curve improvement compared with the WT ArCP starting points.
In Vitro Enterobactin Reconstitution Assay for VibB ArCPs.
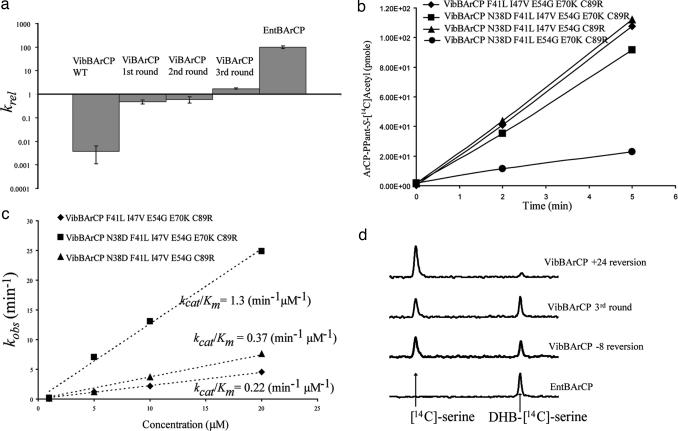
To confirm that the gain of function for the evolved ArCPs was indeed due to its improved activity in enterobactin biosynthesis and to further probe how mutations in ArCP improve enterobactin biosynthesis, we used the series of VibBArCP mutants as a model for in vitro characterizations. ViBArCP (WT, first, second, and third rounds) were His-tagged at the N terminus, overexpressed in E. coli, and purified. An enterobactin production reconstitution assay was performed in the presence of DHB, l-Ser, ATP, CoASH, and the enzymes EntE, EntF, Sfp. The products were separated and quantified by HPLC. Initial rates were calculated relative to assays with EntBArCP (Table 2 and Fig. 4a). Overall, the three rounds of evolution improved the enterobactin in vitro reconstitution rate for VibBArCP by ≈500-fold. The first-round evolution of VibBArCP provided the largest improvement for in vitro enterobactin production (>130-fold) relative to that WT starting point. This result is consistent with the extremely high convergent occurrences observed (27 of 37) of E-to-K mutations at the +24 position during the first round evolution of VibBArCP. Although the activity of the third round VibBArCP is still 50-fold lower than that of WT EntBArCP, a 500-fold improvement with only three rounds of directed evolution is very significant. The important residues and their positions revealed by our directed evolution scheme would provide a good starting point for future engineering of carrier proteins by saturation mutagenesis and selection for further optimization.
Table 2.
Initial rates of in vitro Enterobactin reconstitution
| Construct | krel |
|---|---|
| EntBArCP | 100 ± 13 |
| VibBArCP WT | (3.6 ± 2.5) × 10−3 |
| VibBArCP first round | 0.48 ± 0.10 |
| VibBArCP second round | 0.59 ± 0.17 |
| VibBArCp third round | 1.7 ± 0.1 |
All the rates were scaled relative to EntBArCP (defined as 100) and expressed as krel.
Fig. 4.
In vitro characterization of evolved the VibBArCP mutants. (a) Enterobactin reconstitution assay. Initial rates of enterobactin formation relative to a control experiment using EntBArCP are plotted. After three rounds of evolution, the activity of VibBArCP has been improved by ≈500-fold. (b) Phosphopantetheinylation of VibBArCP (WT and mutants) by Sfp. The ArCPs were incubated with the PPTase Sfp and [1-14C]-acetyl CoA, and the resulting time course for [1-14C]-acetyl-S-ArCP formation was plotted. (c) Acylation of holo-VibBArCPs monitored by incorporation of [14C]salicylate by EntE. The kinetics for different VibBArCP mutants, under kcat/Km conditions, are shown. (d) Single-turnover condensation assay to check interactions between the ArCP domains and EntF. The third-round VibBArCP, and its −8 and +24 reversion mutants were preloaded with DHB, and an EntF tridomain construct (C-A-PCP) lacking the thioesterase domain was loaded with [14C]serine. The HPLC traces show the result of condensation under single-turnover conditions.
Mechanistic Dissection of Group I Mutations.
We focused our attention on the group I mutations, because these selected mutations are most likely to reflect the rules for carrier protein recognition required for enterobactin biosynthesis. (Group II mutations were not characterized in this study.) The alignment of EntBArCP, VibBArCP, and HMWP2ArCP are shown in SI Fig. 7b. The +1 position residue in EntBArCP is valine (SI Fig. 7b); during the rounds of evolution, the +1 position residue converged to valine in VibBArCP and HMWP2ArCP (Fig. 2). In the cocrystal structure of the PPTase AcpS in complex with ACP from B. subtilis, the +1 position residue on ACP has direct contacts with AcpS (18). Therefore, we predicted that the +1 position mutation might contribute to PPTase recognition and that the enhancement obtained by mutation of this residue during the rounds of directed evolution is likely due to improvement of Sfp recognition. The +23/24 positions on helix III have previously been characterized as the interaction surface for carrier proteins with their immediate downstream catalytic partner (2, 7). Elements of helix III are involved in the recognition between EntB-ArCP and EntF, and between the PCP and TE domains of EntF (2, 7). This region has also been reported to be conformationally dynamic in several carrier proteins characterized by NMR (19), further suggesting a role in mediating interdomain interactions. Therefore, we postulated that the +23/24 position mutation might have a significant contribution to improvement of carrier protein recognition during the condensation step and possibly other steps as well. We predicted that mutations at the −8 position would be involved in EntE recognition, because previously reported site-directed mutagenesis at the −9 position on HMWP2ArCP improved the recognition between this ArCP and the DHB-activating adenylation domain from vibriobactin biosynthesis, VibE (20). The C-terminal +42/43 mutations do not have aligning residues on EntBArCP (SI Fig. 7b) and therefore were not characterized further.
To test experimentally the mechanisms of each group I convergent mutation, we made individual reversion mutants of third-round-evolved VibBArCP at positions −8, +1, and +24. The third-round VibBArCP contains the following mutations: VibBArCP N38D F41L I47V, E54G E70K C89R. Reversion of the mutations in the third-round-evolved VibB-ArCP resulted in the following clones: VibBArCP N38D F41L E54G E70K C89R (+1 reversion mutant), VibBArCP F41L I47V E54G E70K C89R (−8 reversion mutant), and VibBArCP N38D F41L I47V E54G C89R (+24 reversion mutant). The evolved VibBArCP mutant (from the third round) and the reversion mutants listed above were expressed in E. coli, purified, and assayed for individual steps in enterobactin production. The premise for these studies was that we could evaluate the effect of the single mutation in the context of the other mutations by this single-residue reversion comparison.
Phosphopantetheinylation Assay for Reversion Mutants.
To examine our hypothesis that the +1 position mutations from Ile to Val might contribute to improving PPTase recognition, we tested the recognition of ArCPs by the PPTase Sfp by using an assay that measures the Sfp-catalyzed incorporation of [14C]acetyl-CoA into the ArCP over time (Fig. 4b). The initial rates of these reversion mutants were compared with third-round-evolved VibBArCP (Fig. 4b). The rates for loading of [1-14C]acetyl-Ppant were similar for the third-round-evolved VibBArCP, and its +24 and −8 reversion mutants. However, the +1 reversion mutant displayed a 3-fold decrease of initial rates for Sfp recognition. The decrease in phosphopantetheinylation associated with the +1 reversion confirmed our prediction that the +1 isoleucine to valine mutation improves the recognition of VibB-ArCP by the PPTase Sfp.
EntE Recognition Assay for Reversion Mutants.
Acylation with [14C]salicylate (a radioactive analog for the EntE substrate DHB) was used to evaluate the recognition of −8 and +24 reversion mutants by EntE in comparison with the evolved third-round VibBArCP (Fig. 4c). Treating the holo-ArCP as the substrate to obtain kinetics for insight into the ArCP-EntE interaction, saturation was not observed for VibBArCPs at 20 μM ArCPs (thus, Michaelis–Menten kinetics could not be obtained). However, we were able to compare among mutants under kcat/Km conditions, which reflects the efficiency of the ArCPs as the substrate of EntE (SI Table 4). The reversion mutation at the −8 position decreases the kcat/Km by 6-fold and the reversion mutation at the +24 position decreases the kcat/Km by 3.5-fold relative to the evolved third-round VibB-ArCP. These results imply that both the +8 and +24 mutations during the evolution process improved recognition of the ArCP by EntE.
Condensation Assay for Reversion Mutants.
We predicted that the +24 position mutation on VibBArCP during first round evolution might dramatically improve the condensation step between the DHB on VibBArCP and the serine on EntFPCP. To test this hypothesis, we monitored the build-up of condensation product DHB-serine by using a radioactivity assay under single-turnover conditions (7). The ratio of uncondensed [14C]serine to DHB-[14C]serine reflects the efficiency of ArCPs as the substrate for EntF C domain. As shown in Fig. 4d, after 1 min reaction the ratio of [14C]serine and DHB-[14C]serine on EntF PCP for third-round VibBArCP and its −8 position mutation is ≈1:1. However, for the +24 position reversion mutant, the major species on EntFPCP is uncondensed [14C]serine, and DHB-[14C]serine is barely detectable. This result confirmed our prediction that the highly convergent mutation at the +24 position indeed dramatically improved the recognition of ArCPs by the EntF C domain.
Discussion
Carrier protein domains serve as central architectural elements in natural product assembly lines for polyketides and nonribosomal peptides. These domains are way stations where specific catalytic partner proteins interact, with the timing and specificity of the carrier protein interactions being crucial for production of the natural product in high fidelity. Basic domains/enzymes with which the carrier protein must interact in each elongation cycle of PK/NRP assembly include: PPTases, acyltransferase or adenylation domains, and condensing enzymes (ketosynthases in PKS and condensation domains in NRPS). Additional enzymes can interact with the CP domains harboring the growing chain in a given elongation cycle (e.g., tailoring domains). In PKS assemblies, these can include ketoreductase, dehydratase, and enoyl reductase enzymes, as well as C-methyltransferases. In NRPS assemblies, tailoring domains can include epimerase, oxidase, and N-methyltransferase domains. Finally, at the end of most PKS and NRPS assembly lines, a thioesterase domain is frequently located immediately downstream of the last carrier protein for the purpose of releasing the full-length natural product chain from the assembly line.
Rules for recognition of one carrier protein but not another one must exist, especially in such contexts as the andrimid PKS/NRPS assembly line, in which six carrier proteins are in separate subunits and require precisely timed recognition choreography for proper chain growth (21). Although both x-ray and NMR structures have been reported for a few ACPs and PCPs (14, 18, 19, 22), these structures are clearly dynamic and do not immediately reveal how many surfaces would be recognized by distinct partner enzymes. Successful reprogramming of PKS and NRPS machinery will therefore require detailed knowledge of carrier protein recognition by catalytic domains.
Our studies on the enterobactin synthetase, the V. cholerae vibriobactin synthetase, and the Y. pestis yersiniabactin synthetase have been a prototype to dissect assembly line organization, the role of the Ppant arm, the timing of apo-to-holo carrier protein conversion, the nature of the covalent intermediates during siderophore chain elongation and maturation, and the interfaces between PKS and NRPS modules (9, 10, 13, 23–26). To assess portability of carrier protein domains between assembly lines and thereby to delineate residues and surfaces of carrier proteins that might be recognition determinants, we have turned to the aryl carrier proteins in these three assembly lines. Siderophore NRPSs provide suitable system for evolution and analysis of carrier protein function in an assembly line for several reasons. First, the growth requirement for the pathway under iron starvation allows a selection for restoration of function. Second, we have previously devised assays for interaction of all three enzymes Sfp, EntE, and EntF with the ArCP domain of EntB. Thus, phenotypic assays can be followed up with biochemical assays to assess which step(s) have been affected, allowing molecular interpretation.
We replaced the EntB ArCP domain from the two-domain ICL-ArCP protein of E. coli with ArCP domains from either V. cholerae or Y. pestis. We used random mutagenesis coupled with in vivo selection for siderophore production to evolve the heterologous ArCPs to be functional in the enterobactin synthetase. From the series of clones isolated during the evolution process, we were able to identify several convergent mutations. These mutations were mapped onto the structural models of VibBArCP and HMWP2ArCP: (i) the +1 position mutations immediately followed the Ppant serine on helix II; (ii) the −8 position mutations were in the loop II region; (iii) and the + 23/24 position mutations were in the dynamic helix III region; (iv) the C terminus had no aligning residues in EntBArCP and may be in an unstructured region.
Our mechanistic dissection revealed that: the +1 position mutation improved the recognition of VibBArCPs by the PPTase Sfp by 3-fold; the −8 position mutation on VibBArCP improved its EntE recognition by 6-fold. Chemical shift changes were observed for NMR titration-experiments of 15N-holo EntBArCP at the −8 position residue asparagine and nearby residues when titrated with unlabeled EntE (A. Koglin, J.R.L., and C.T.W., unpublished results), suggesting specific interactions between EntE and EntBArCP at −8 position and nearby residues. Interestingly, we also observed a 3.5-fold improvement of EntE recognition by a +24 position residue mutation from E to K. The corresponding helix/loop III region in EntBArCP has been revealed as important recognition surface by the downstream C domain (2). The NMR structures of TycC3PCP and other carrier proteins (19) have suggested helix III region is a conformationally dynamic region. Therefore, a possible mechanism for improving EntE recognition at the +24 position might be through tuning the dynamics and equilibrium of carrier protein conformations. In the heteronuclear single quantum correlation (HSQC) spectra of 15N-holo-EntBArCP, the aligning +24 position residue lysine demonstrated chemical exchange, implying two dynamic states, and one state was selected during EntE titration (A. Koglin, J.R.L., and C.T.W., unpublished results), consistent with our mechanistic interpretation. Furthermore, by condensation assay, we found that the +24 position mutation dramatically improved the condensation efficiency. It is interesting that a single mutation at the +24 position could improve two distinct enzymatic steps in enterobactin biosynthesis, suggesting the dynamics and complexity of carrier proteins at this region, as well as the potential power of directed evolution approaches in understanding and reprogramming protein–protein recognitions. In sum, we have placed three different regions on EntBArCPs for different protein–protein recognitions.
The results in this study complement our previous experiments that defined regions on E. coli EntBArCP that interact with the PPTase EntD (8) and the C domain of EntF (2). In regard to EntBArCP shotgun alanine scanning and enterobactin production screening, an EntE recognition surface was not identified, probably because EntE catalyzed acylation is not rate-limiting. In this study, the noncognate pair of VibBArCP with EntE allowed us to identify the −8 position in the loop II region as an EntE recognition surface. The +24 position mutation dramatically improved the condensation step, which is consistent with our previous understanding of this position as the recognition surface of the immediate downstream partner. This residue could also tune EntE recognition dynamically, raising the acylation rate by 3.5-fold, extending our understanding about the roles of this region.
This approach reveals residues and regions of the surface of the carrier proteins that interact with specific partner proteins. Clearly, the 80- to 100-residue carrier protein fold is an information-rich surface to be read by potential partner proteins as one layer of selectivity in assembly line function. This couples with the recent inputs from NMR and x-ray investigations that carrier proteins can populate different conformations as potential switches. Carrier proteins also are highly flexible in terms of their position within a module, befitting the required visits to multiple active sites of catalytic partners. The work described here is a start to understand how one carrier protein can be morphed into another, to function effectively in a heterologous assembly line or perhaps to redirect chain elongation flux in a system with in trans components. A future challenge will be coordinating the evolution of heterologous carrier protein recognition with alterations in the chemical composition of the natural product. Such evolution of natural product assembly lines will be required for reprogramming of biosynthetic machinery to produce new variants in high efficiency.
Materials and Methods
Mutagenesis and Library Construction.
Randomly mutagenized libraries of VibArCP and HMWP2ArCP were produced by error-prone PCR using chimeric EntBICL-VibBArCP and EntBICL-HMWP2ArCP constructs as the templates. Mutations were introduced by mutagenic dNTPs: 8-oxo-dGTP and dPTP (17). Detailed procedures are provided in SI Materials and Methods. For subsequent rounds of mutagenesis and evolution, EntBICL-VibArCP (first and second rounds) and HMWP2ArCP (first and second rounds) chimeric genes served as templates.
Silent mutations (residues 204–205) were introduced into entB with primer 5′-CGGGTGGTGATGACCGAAGAGCTCCTGCCAGCACCTATCCCC-3′, resulting in the installation of a SacI site between the DNA encoding the ICL domain and ArCP domain of EntB. An inactive template was then produced by cloning an unrelated “stuffer” DNA sequence in place of the DNA for the ArCP region by using the newly stalled SacI site. DNA libraries from the mutagenic PCR were then cloned into this inactive template, replacing the stuffer sequence with the library DNA encoding the mutant ArCP.
Convergent mutations from directed evolution were introduced into the VibArCP and HMWP2ArCP by splice overlap extension, and the resulting ArCPs were cloned into pET-DUET-Sfp backbones, generating templates for next round mutagenesis and evolution.
Enterobactin Production Selection.
VibBArCP and HMWP2ArCP library DNA were electroporated into electrocompetent entB::kanR cells and plated onto minimal media in which iron was sequestered by the addition of 100 μM 2,2′-dipyridyl. The transformants were allowed to grow for 24–60 h and the largest colonies were isolated and sequenced.
Liquid Culture Assay for Evolved ArCPs.
pET-DUET vector containing the sfp gene and DNA encoding the chimeric EntBICL-VibBArCP (WT, first, second, and third round) or EntBICL-HMWP2ArCP (WT, first, second, and third round) were electroporated into entB::kanR cells. The resulting clones were grown in LB culture to reach saturation overnight. Then, 5 μl of the saturation cultures was inoculated into 5 ml of minimal media containing 200 μM 2,2′-dipyridyl and grown at 37°C, with shaking at 200 rpm in an Innova 2300 Platform Shaker (New Brunswick Scientific, Edison, NJ). The growth was monitored by OD600. A control experiment was performed in rich media to ensure that no intrinsic growth differences between clones existed. Assays were performed in duplicate.
Biochemistry Assays.
Methods for enterobactin reconstitution, phosphopantetheinylation, acylation, and condensations assays are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Michael Fischbach and Alex Koglin for helpful discussions. This research was supported by National Institutes of Health Grants AI042738, GM020011, and GM047467. J.R.L. is supported by a Helen Hay Whitney Foundation Postdoctoral Fellowship.
Abbreviations
- ACP
acyl carrier protein
- ArCP
aryl carrier protein
- CP
carrier protein
- FA
fatty acid
- FAS
fatty acid synthase
- ICL
isochorismate lyase
- NRP
nonribosomal peptide
- NRPS
nonribosomal peptide synthetase
- PCP
peptidyl carrier protein
- PK
polyketide
- PKS
polyketide synthase
- PPTase
phosphopantetheinyl transferase.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705122104/DC1.
References
- 1.Cane DE, Walsh CT, Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 2.Lai JR, Fischbach MA, Liu DR, Walsh CT. Proc Natl Acad Sci USA. 2006;103:5314–5319. doi: 10.1073/pnas.0601038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh CT, Gehring AM, Weinreb PH, Quadri LE, Flugel RS. Curr Opin Chem Biol. 1997;1:309–315. doi: 10.1016/s1367-5931(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 4.Marahiel MA. Chem Biol. 1997;4:561–567. doi: 10.1016/s1074-5521(97)90242-8. [DOI] [PubMed] [Google Scholar]
- 5.Fischbach MA, Walsh CT. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 6.Lai JR, Koglin A, Walsh CT. Biochemistry. 2006;45:14869–14879. doi: 10.1021/bi061979p. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Z, Lai JR, Walsh CT. Chem Bio. 2006;13:869–879. doi: 10.1016/j.chembiol.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 8.Lai JR, Fischbach MA, Liu DR, Walsh CT. J Am Chem Soc. 2006;128:11002–11003. doi: 10.1021/ja063238h. [DOI] [PubMed] [Google Scholar]
- 9.Keating TA, Marshall CG, Walsh CT. Biochemistry. 2000;39:15522–15530. doi: 10.1021/bi0016523. [DOI] [PubMed] [Google Scholar]
- 10.Gehring AM, Mori I, Perry RD, Walsh CT. Biochemistry. 1998;37:11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- 11.Roche ED, Walsh CT. Biochemistry. 2003;42:1334–1344. doi: 10.1021/bi026867m. [DOI] [PubMed] [Google Scholar]
- 12.Crosa JH, Mey AR, Payne SM. Iron Transport in Bacteria. Washington, DC: Am Soc Microbiol; 2004. [Google Scholar]
- 13.Gehring AM, Mori I, Walsh CT. Biochemistry. 1998;37:2648–2659. doi: 10.1021/bi9726584. [DOI] [PubMed] [Google Scholar]
- 14.Drake EJ, Nicolai DA, Gulick AM. Chem Biol. 2006;13:409–419. doi: 10.1016/j.chembiol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 16.Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 17.Zaccolo M, Williams DM, Brown DM, Gherardi E. J Mol Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 18.Parris KD, Lin L, Tam A, Mathew R, Hixon J, Stahl M, Fritz CC, Seehra J, Somers WS. Structure (London) 2000;8:883–895. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- 19.Koglin A, Mofid MR, Lohr F, Schafer B, Rogov VV, Blum MM, Mittag T, Marahiel MA, Bernhard F, Dotsch V. Science. 2006;312:273–276. doi: 10.1126/science.1122928. [DOI] [PubMed] [Google Scholar]
- 20.Marshall CG, Burkart MD, Meray RK, Walsh CT. Biochemistry. 2002;41:8429–8437. doi: 10.1021/bi0202575. [DOI] [PubMed] [Google Scholar]
- 21.Jin M, Fischbach MA, Clardy J. J Am Chem Soc. 2006;128:10660–10661. doi: 10.1021/ja063194c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leibundgut M, Jenni S, Frick C, Ban N. Science. 2007;316:288–290. doi: 10.1126/science.1138249. [DOI] [PubMed] [Google Scholar]
- 23.Suo Z, Chen H, Walsh CT. Proc Natl Acad Sci USA. 2000;97:14188–14193. doi: 10.1073/pnas.260495697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller DA, Luo L, Hillson N, Keating TA, Walsh CT. Chem Biol. 2002;9:333–344. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 25.Miller DA, Walsh CT. Biochemistry. 2001;40:5313–5321. doi: 10.1021/bi002905v. [DOI] [PubMed] [Google Scholar]
- 26.Suo Z, Walsh CT, Miller DA. Biochemistry. 1999;38:17000. doi: 10.1021/bi9950950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.