Abstract
Glycosylation creates an intricate and complex code for biological information that plays a role in cell–cell communication, infection, and immunity among many biological events. Dynamic changes in the glycosylation status of cells have been observed in tumor cell metastasis and cell differentiation but have been difficult to analyze because of a lack of high-throughput and facile technologies. Here, we present a method for the rapid evaluation of differences in the glycosylation of heterogeneous mammalian samples using a ratiometric two-color lectin microarray approach. This work represents a significant improvement in glycomics technology and sets the stage for the systematic evaluation of how glycans encode biological information in complex systems.
Keywords: carbohydrate analysis, glycomics
Carbohydrates are intricate information-carrying biopolymers of rising interest in the postgenomic age. Dynamic changes in glycosylation are observed in a myriad of key biological events in mammalian systems, including embryogenesis, neuronal development, and tumor cell metastasis. Despite their importance, however, we have yet to unravel the complex mechanisms by which carbohydrates are modulated and how they in turn affect biological phenomena (1). The information carried by carbohydrates is often encoded in characteristic substructures rather than by precisely defined molecules, occurring in the context of both N- and O-linked glycoconjugates and glycolipids. Nature decodes glycosylation through the use of lectins, carbohydrate-binding proteins that can bind to multiple glycoproteins bearing the same carbohydrate motif and that, in mammalian systems, often mediate cell–cell interactions (2, 3). This argues that analysis of cellular patterns of sugar substructures, rather than of precisely defined glycans, is critical to understanding the carbohydrate code of an organism.
Recent efforts toward decoding the glycome have focused on the use of mass spectrometry (4–9). Although great strides have been made in the use of this technique, mass spectrometry has difficulty in discriminating between the many structural isomers presented by glycans. In addition, mass spectrometry requires expensive equipment and time-consuming separation and annotation techniques (4, 10). A typical glycomic analysis by mass spectrometry focuses on only a portion of the glycans (usually the N-linked fraction), and some critical substructures, such as sialic acids and sulfation of sugars, are often removed before analysis.
A complimentary method for the structural characterization of glycans is presented by lectin microarrays (11–17). These microarrays consist of a collection of, mostly plant-derived, carbohydrate-binding proteins immobilized onto a solid support at a high spatial density. Interrogation of these arrays with fluorescently labeled samples creates a pattern of binding that depends on the carbohydrate structures, providing a method for the rapid characterization of carbohydrates on glycoproteins, bacteria, or mammalian cells (11–16). Lectin microarrays require neither the separation of N- and O-linked glycans before analysis nor the removal of sialic acids or other substructural motifs that might impede analysis by mass spectrometry. Although initial experiments demonstrating the utility of lectin microarrays for the rough glycoprofiling of mammalian cells have been established, there are multiple flaws in the current protocols from the standpoint of collecting biologically relevant glycomic data sets (15). First, they rely on single-color analysis with no reliable quality control methodology to allow for fine comparisons between samples. Second, these studies have yet to examine issues such as the reproducibility of lectin microarray data. Third, they ignore cellular glycolipids, an important part of the glycome (15). The importance of quality control issues in microarray data collection has been recently highlighted by renewed efforts in the DNA microarray field to ensure accuracy in data collection (18). Here, we present a method for the differential comparison of whole mammalian glycomes using a ratiometric lectin microarray approach amenable to the adaptation of both established statistical tools and microarray quality control strategies. We demonstrate the accuracy and reproducibility of our lectin microarray system for both biological and technical replicates and apply this system to the examination of dynamic glycosylation changes upon cell differentiation. This work sets the stage for future work to systematically define the glycan code and to integrate glycomic and genomic information in an effort to understand how the code is controlled by the cell.
Results and Discussion
General Strategy.
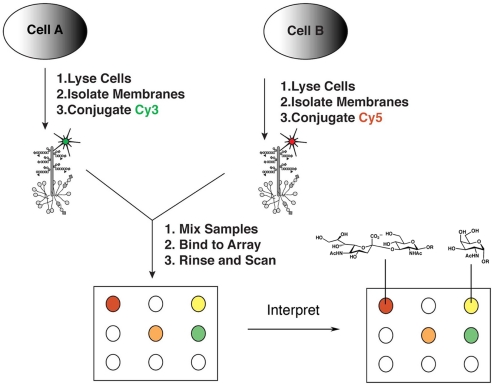
Our strategy was inspired by the two-color ratiometric approach used in DNA microarray analysis. Briefly, cells are harvested by scraping or centrifugation, avoiding the use of proteases, which can bias the glycoprotein pool (19), and detergents, which can solubilize glycolipids. Cell samples are then sonicated, causing the formation of micelle-like structures from the membranes (cellular micellae) (20). These cellular micellae are fluorescently labeled with orthogonal dyes (i.e., Cy3 or Cy5) via the lysines on incorporated proteins before analysis on the lectin microarray (Fig. 1). We anticipated that the vast majority of glycoproteins and glycolipids within our labeled cellular micellae would originate from the cell surface; however, we tested whether labeling of whole cells before cell lysis versus labeling of the preformed cellular micellae gives rise to altered patterns of glycan expression on our microarrays. No significant difference in the glycan patterns were observed (Pearson correlation coefficient R = 0.83, n = 21 lectins, P < 0.001), although labeling of the whole cells prelysis did result in lower levels of dye incorporation overall [supporting information (SI) Fig. 6]. We also verified the presence of glycolipid in our cellular micellae by TLC followed by resorcinol staining for sialic acids, which are found as major components in gangliosides (data not shown).
Fig. 1.
General schematic of the two-color approach. Cells are lysed and membranes are isolated after sonication as “cellular micellae.” These cellular micellae are then labeled by coupling of either Cy3-NHS or Cy5-NHS with the lysines on proteins. Equivalent amounts (10 μg of protein, ≈105 cells) of the appropriate Cy3- and Cy5-labeled samples are mixed and hybridized to each lectin microarray. After washing, the arrays are scanned by using a GenePix Pro 4100A scanner. The resultant data are extracted by using GenePix Pro 5.1, analyzed, and annotated by using known lectin specificities, deconvolving the array data into carbohydrate patterns.
The lectin microarrays used in this work consist of either 21 or 58 lectins (see SI Table 1 for the list of lectins used). All lectins were printed at concentrations optimized to give a minimum signal of 1,000 arbitrary fluorescent units (A.U.) under fixed scanning conditions with a range of labeled glycoproteins. Therefore, only signals above the 1,000-A.U. threshold were considered positive on our arrays. Each lectin microarray slide provides 14 replicate arrays of which two are used for quality control hybridizations with glycoprotein standards. This insures that all lectins on our array are appropriately active and allows us to discard data from inactive or misprinted lectins. In addition, multiple replicates of lectins on each array (5–10) provide yet another level of quality control.
To obtain ratiometric lectin microarray data, equal amounts of the two differentially labeled cellular micellae samples (≈10 μg of each labeled sample based on protein analysis, ≈105 cells) are mixed and the mixture is applied to the array. Competitive binding between the two samples results in the rapid evaluation of relative differences in the binding affinities to the probes via alterations in the ratio of Cy3:Cy5 signal per probe (Fig. 1). As in previous lectin microarray studies, the glycan specificity of lectin binding was confirmed by means of monosaccharide inhibition (SI Fig. 7) (11, 14). Data from the microarray are extracted by using standard image analysis software (GenePix Pro 5.1) to obtain a rough alignment of spots. This alignment is then adjusted to ensure proper segmentation. To minimize the effects of variant spot size and quality on data analysis, the median rather than mean signal and background is used and replicate data are subjected to the Grubbs outlier test at the 95% confidence interval (21). Hand annotation of the resultant data uses carbohydrate specificity profiles of the lectins obtained from both the publicly available glycan array screening by the Consortium for Functional Glycomics (www.functionalglycomics.org) and the literature to deconvolve the pattern into carbohydrate structural information.
Ratiometric Lectin Microarray Data Reflect Differences in Cellular Glycan Composition.
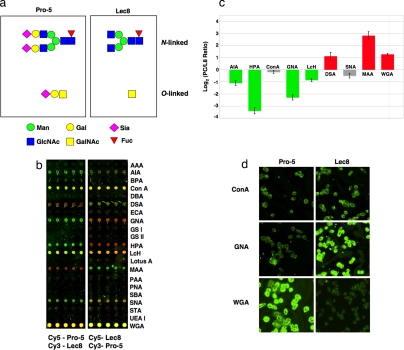
Our preliminary experiments focused on comparisons between Pro-5 Chinese hamster ovary (CHO) cells and Lec8, a mutant with decreased UDP-galactose transport derived as a wheat germ agglutinin (WGA)-resistant cell line (Fig. 2a) (22). Single-color analysis of these two cell lines revealed several differences but failed to reveal any loss of WGA binding by Lec8, a hallmark of these cells (SI Fig. 8). In contrast, the difference in WGA binding (2- to 4-fold) could be clearly observed by our ratiometric approach (Fig. 2c and SI Fig. 9). To control for any discrepancies due to dye labeling, we ran both sets of labeled samples (i.e., Cy5-Pro-5/Cy3-Lec8 and Cy5-Lec8/Cy3-Pro-5) in parallel microarrays on the same slide (Fig. 2b). We then used this data to compensate for dye bias by means of the method pioneered by Yang and colleagues (23) for gene arrays. In brief, the mean log2 (ratio of Pro-5 to Lec8) for each lectin was averaged over the two dye-swapped arrays (Fig. 2c). A positive log2 (ratio) indicates greater binding to the parent cell line Pro-5 (red in Fig. 2c); conversely, a negative signal indicates greater binding to the mutant Lec8 (green in Fig. 2c).
Fig. 2.
Two-color evaluation of Pro-5 vs. Lec8. (a) Examples of N- and O-linked oligosaccharides for Pro-5 and Lec8 cells (31, 32). Carbohydrates are shown in the representational code of the Consortium for Functional Glycomics (www.functionalglycomics.org/static/consortium). (b) Representative microarray data for Pro-5/Lec8 dye-swapped pairs. The two subarrays shown are from the same slide. (c) Plot of the dye-bias-compensated log2 (Pro-5/Lec8 ratios) for each lectin. Signals in the Cy3 or Cy5 channel were converted to log2 and then averaged over 10 microarray spots. The mean log2 (Pro-5 signal) was divided by the mean log2 (Lec8 signal) for each of the dye pairs [log2 (Cy5-Pro-5/Cy3-Lec8) and log2 (Cy3-Pro-5/Cy5-Lec8)] and averaged to remove dye bias (23). Error was propagated by using standard formulas from the standard deviations. Only positive lectin signals are shown (see Materials and Methods). Signals colored in green show ≥2-fold increase in binding to Lec8, whereas signals in red show ≥2-fold better binding to the parent Pro-5. Signals marked in gray display a <2-fold difference in the (Pro-5/Lec8) ratio. (d) Validation of microarray results for Pro-5/Lec8 by means of histology. Cells were fixed in methanol and stained with biotinylated lectins (Con A, GNA, WGA, 80 μg/ml, followed by staining with FITC-labeled avidin). The images were acquired at the same exposure time and are shown on the same scale for every lectin. For comparison, the log2 (Pro-5:Lec8 ratio) from the histology images for ConA = 0.09 ± 0.27, GNA = −1.64 ± 0.28, and WGA = 3.05 ± 0.41. Ratios were calculated from the images as described in Materials and Methods.
The relative glycan composition afforded by our ratiometric microarray method accurately reflected the expected glycan compositions from the predicted biological expression patterns (22), flow cytometry data from the literature (15), and the analysis of fixed cells by means of lectin histology (Fig. 2d). Major differences in glycan expression between Lec8 and Pro-5 included the loss of α-2,3 sialosides as reflected by MAA binding and increased exposure of core epitopes from both N-linked (mannose or fucosylated mannose core, reflected by GNA and LcH, respectively) and O-linked (core GalNAc residues, reflected in HPA and AIA binding) glycans in Lec8 cells.
Lectin Microarray Data Are Reproducible.
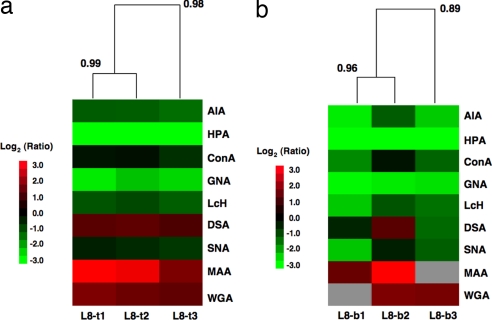
A general concern about microarrays is the reproducibility of the data across microarray slides. We tested the reproducibility of our array data by using both biological and technical replicate data sets. We again chose the comparison between Lec8 and Pro-5 as our model system. For technical replicates, identical mixtures of Lec8 and Pro-5 samples were hybridized to three different lectin microarray slides on two different dates. Hierarchical clustering of the resultant Yang correlation data sets by using the Pearson correlation coefficient with average linkage analysis revealed excellent correlation between the three sample sets (R = 0.98, n = 9, P < 0.0001; Fig. 3a). Biological replicates, in which distinct sample mixtures of Lec8 and Pro-5 derived from different passages of cells were prepared and analyzed on different dates, also gave excellent correlation between the samples, although more variation in the signals was observed (R = 0.89, n = 9, P = 0.001; Fig. 3b). Overall, this work demonstrates that the ratiometric lectin microarray method yields highly reproducible glycomic data sets.
Fig. 3.
Heat maps with dendrograms of hierarchical clustering for technical and biological replicates. All heat maps were generated by using Cluster 3.0 (30) and Java TreeView with the Pearson correlation as the distance metric for the arrays with average linkage analysis. The Pearson correlation coefficients are indicated at each branching point. Only the Yang correlations for positive lectins were used in the correlation. Gray boxes represent misprinted lectins that were excluded from the analysis. Misprints were evaluated by examination of the glycoprotein quality control standards. (a) Technical replicates of Pro-5/Lec8. (b) Biological replicates of Pro-5/Lec8.
Hierarchical Clustering of Lec Mutants Reveals Subtle Differences in Carbohydrate Composition.
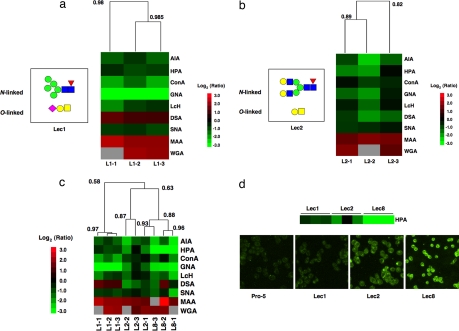
To further validate our system, we acquired the comparative lectin binding profiles of two additional Lec mutants: Lec1, which is deficient in GlcNAcT1, the enzyme necessary for hybrid and complex N-linked glycans; and Lec2, a mutant lacking CMP-sialic acid transport (24, 25). Biological replicates of Lec1 clustered very tightly (R = 0.98, n = 9, P < 0.001) and displayed a marked increase in exposed mannose residues when compared with Pro-5 as reflected by differences in ConA and GNA binding (Fig. 4a). In addition, a loss of MAA binding was observed, indicating that the majority of the α-2,3-sialic acid recognized by this lectin on Pro-5 cells exists in N-linked glycans. Biological replicates of Lec2 also clustered well (R = 0.82, n = 9, P = 0.007) and demonstrated the expected loss of sialic acid epitopes (MAA, WGA; Fig. 4b).
Fig. 4.
Comparison of all three Lec mutant cell lines. (a) Hierarchical cluster map of ratiometric data for dye-bias-corrected Lec1 hybridized against the common biological reference Pro-5. The heat map was generated and annotated as explained in Fig. 3. Example N- and O-linked glycans for Lec1 are indicated on the left side of the heat map (33). (b) Hierarchical cluster map of ratiometric data for dye-bias-corrected Lec2 hybridized against the common biological reference Pro-5. The heat map was generated as explained previously. Example N- and O-linked glycans for Lec2 are indicated on the left side of the heat map. (c) Hierarchical cluster map of ratiometric data for all three Lec mutants. (d) Comparison between ratiometric data sets and lectin histology for relative HPA binding of Pro-5, Lec1, Lec2, and Lec8 cells. The color bar shows ratiometric data from the microarrays. Cells were stained with biotinylated HPA, followed by FITC-avidin. All fluorescence microscopy images were obtained and displayed by using identical settings.
Because Pro-5 was used as a common biological reference for all hybridizations, we could directly compare the ratiometric data between all three Lec mutants (Fig. 4c). The samples grouped into two sets representing Lec1 (R = 0.97, n = 9, P < 0.001; Fig. 4c) and a mixture of Lec2 and Lec8 (R = 0.63, P = 0.06; all Lec8 and a single Lec2 sample clustered at R = 0.88, P = 0.002). The grouping of Lec2 and Lec8 may reflect both the general similarities in the glycan composition between the cell lines and the fact that the clustering is hierarchical. Interestingly, a closer examination of HPA binding (α-GalNAc) across all cell samples revealed subtle but consistent differences in the levels of ratiometric change for each cell line. To validate that the observed discrete differences in HPA levels were indicative of real differences in the cell-surface carbohydrate composition, we obtained the accompanying histology (Fig. 4d). As shown, our ratiometric microarray data closely reflect HPA binding to the mutant cells as observed by conventional fluorescence microscopy. This demonstrates that our system can be used to elucidate not only the rough presence or absence of an epitope but also subtle variations in the quantitative levels of exposed carbohydrate residues.
Glycomic Analysis of HL-60 Differentiation into Neutrophils.
We next applied our system to the rapid evaluation of glycosylation changes during blood cell differentiation. For a model system, we chose the differentiation of the promyelocytic leukemia cell line HL-60 into neutrophils upon treatment with DMSO. In brief, we differentiated HL-60 cells by treatment with DMSO (1.25%) over 7 days (26). We prepared five biological replicates of the differentiated HL-60 cells. Labeled cellular micellae derived from these samples were prepared for lectin microarray analysis as previously described. An orthogonally labeled biological reference sample from the parent undifferentiated HL-60 cells was also prepared. Single-color lectin microarray analysis of the parent HL-60 cell line revealed a mixture of glycan epitopes expressed on the cells including high mannose N-linked oligosaccharides, α-2,6-linked sialic acid, and terminal α- and β-GalNAc residues (SI Fig. 10). These data are in line with recent glycan profiling data for HL-60 cells obtained by means of mass spectrometry analysis by the Consortium for Functional Glycomics, once again confirming the validity of our system.
Two-color analysis of the differentiated cells versus the parent HL-60 reference revealed a number of distinct differences between the two cell types. As expected, the biological replicate data sets showed a high degree of correlation (R = 0.81, n = 32, P < 0.001). Differentiated neutrophils displayed an increase in the levels of high mannose N-linked oligosaccharides as evidenced by ratiometric changes in GNA, NPA, PEA, LcH, and ConA (Fig. 5). These data are in line with previous mass spectrometry data showing a similar result for N-linked analysis (27). In addition, increases in N-linked structures containing β-1,6-GlcNAc branches (PHA-L) and structures containing both α- and β-terminal GalNAc residues (CSA, AIA, VVA, HPA) are observed. In contrast, neutrophils display fewer α-2,6-sialic acid epitopes on their N-linked oligosaccharides than do undifferentiated HL-60 cells, as evidenced by ratiometric changes in binding of the highly specific α-2,6-sialic acid lectin PSL and the general α-sialic acid lectin LPA (28, 29). These data conflict with data from SNA, a less specific lectin that binds α-2,6-sialic acid on both N- and O-linked glycoproteins but that has also been found to bind nonsialylated epitopes as evidenced by both its binding to the sialylation-deficient Lec2 cells and work by others (28). One of the advantages of the lectin microarray system over traditional lectin analysis is that cross-correlating lectins of related but distinct specificities leads to more accurate annotation of the carbohydrate composition.
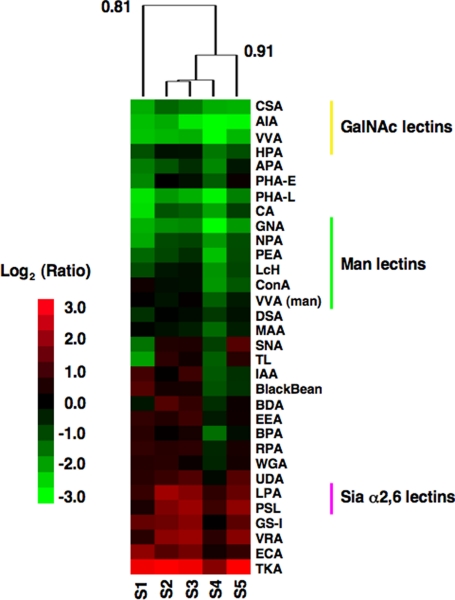
Fig. 5.
Differentiation of HL-60 cells into granulocytes causes distinct changes in the glycosylation pattern. Hierarchical cluster map of ratiometric data for differentiated HL-60 cells (Cy3) biological replicates hybridized against a pooled common undifferentiated HL-60 reference (Cy5). Only lectins giving positive signals were used in the cluster analysis (see Materials and Methods). As described previously, the Pearson correlation was used as the distance metric for the arrays with average linkage analysis. Correlation coefficients are shown at select nodes of the dendrogram. In this heat map, green indicates stronger binding to a lectin by the differentiated cells, whereas red indicates stronger binding by the undifferentiated HL-60 cells.
Conclusions
Here, we present a method for rapid and sensitive differential mammalian glycomic analysis. Using a biological referencing system and a ratiometric approach, we gain both sensitivity and a means to directly compare samples across multiple slides. This enables us to determine subtle differences between samples. At the present time, single-color analysis does not allow for close comparisons because there are no clear internal standards that are appropriate for calibration of the data. Calibration to the highest signal is arbitrary and no cell-surface protein is known that is consistently expressed across all cell lines. We have demonstrated that our system provides reproducible semiquantitative lectin-binding analysis of complex cell-surface glycosylation and have applied it to the analysis of blood cell differentiation. This technology fills a current gap in glycomic analysis by providing a simple high-throughput method to obtain an overall view of both N- and O-linked glycosylation and potentially glycolipids at an intermediate level of substructural resolution. One caveat to any microarray approach is that it cannot distinguish between increased expression of a strongly binding epitope, which may be in lower overall abundance, and higher amounts of a lower affinity ligand. For gene microarrays this problem has led to the design of increasingly specific oligonucleotide probes and the verification of microarray results with other techniques such as RT-PCR (34). A similar approach for lectin microarrays can be envisioned in which cross-correlations with increasingly selective recombinant lectins in combination with validation of interesting lectin microarray data by means of lower throughput mass spectrometry or chromatography based approaches are used to address this issue. In the future, expansion of the array to include antibodies, new more highly selective lectins, and other glycan binding proteins will allow us to gather even more detailed information about the glycome, including the ability to analyze structures such as sulfation and sialyl Lewis x that are difficult to observe in the mass spectrometer. This technology will enable us to study alterations in glycans associated with dynamic biological states, including differentiation and the development of disease states such as cancer, in a systematic manner. In addition, it opens the door to the correlation of glycomic and genomic changes, allowing us to gain insight into the genetic underpinnings of the carbohydrate code.
Materials and Methods
CHO Cell Culture.
We purchased Pro-5, Lec1, Lec2, and Lec8 CHO cell lines from American Type Culture Collection (Manassas, VA). Cells were cultured in DMEM (alpha modification; HyClone, Logan, UT) supplemented with 10% fetal bovine serum (Media tech, Herndon, VA) at 37°C and 5% CO2.
HL-60 Cell Culture and Differentiation.
The HL-60 cells were purchased from American Type Culture Collection and cultured in RPMI medium 1640 supplemented with 20% fetal bovine serum at 37°C and 5% CO2. HL-60 cells were differentiated by addition of 1.25% DMSO for ≈7 days.
Manufacture of Lectin Microarrays.
All lectins were purchased from EY Laboratories (San Mateo, CA). The lectins were prepared in print buffer containing 1 mM appropriate monosaccharide to aid spot morphology (16). We printed the lectins on Nexterion H slides (SCHOTT, Elmsford, NY) using a SpotBot ArrayIt personal microarrayer maintained at ≈50% humidity and room temperature. SMP3 pins were used to print 14 identical arrays per slide with either 5 (58 lectin array) or 10 (21 lectin array) replicate spots per lectin (TeleChem, Sunnyvale, CA). We spaced the arrays to fit within the 16-well FAST-frame (Schleicher & Schuell, Keene, NH), which creates discrete wells when placed over the slide. Note that two wells are occupied by the slide's barcode so only 14 usable wells can be created with this system. We incubated the printed slides for 1 h at room temperature and ≈50% humidity to maximize the coupling efficiency of the lectins. We then immersed the slides in 50 mM ethanolamine (in 50 mM sodium borate buffer, pH 8) for 1 h to block any remaining activated coupling sites on the slide. After blocking, the slides were rinsed three times with PBST (0.1 M phosphate buffer/0.15 M NaCl, pH 7.2/0.05% Tween 20), followed by a final rinse in PBS (0.1 M phosphate buffer/0.15 M NaCl, pH 7.2). Excess buffer was removed from the slides by using a bench top slide spinner (Labnet, Edison, NJ) before their use.
Cellular Micellae Sample Preparation and Fluorescent Labeling.
We harvested cells using a cell scraper after a 10-min incubation in 0.5 M EDTA solution (ddI H2O, pH 8). We then pelleted the cells (500 × g), resuspended them in PBS, and sonicated them at 4°C [three times for 5 s, 70% power (280 W); Branson (Danbury, CT) 450 sonicator with 1/8-inch tapered probe tip]. After sonication, we isolated cellular micellae by means of ultracentrifugation (100,000 × g) and resuspended the pellet in Cy-labeling buffer (0.1 M NaCO3 in H2O, pH 9.3). At this point, we determined the protein concentration by using the DC protein assay (Bio-Rad). We then added at least 10 μg of Cy3- or Cy5- NHS (GE Life Sciences, Piscataway, NJ) to 1 mg of cell membrane (as determined by protein content) and incubated for 30 min at room temperature. The labeled cell membrane preps were then dialyzed against PBS overnight at 4°C, aliquoted, and snap frozen in liquid N2. Aliquots were stored at −80°C for future use.
For whole-cell labeling, cells were harvested by incubating with 0.5 M EDTA in PBS (pH 7.2) for 30 min at 4°C. At this point, the cells easily could be collected without lysis by gentle washing with PBS. The cells were pelleted by centrifugation (200 × g, 10 min). After removal of the EDTA/PBS buffer solution, cells were resuspended in PBS and labeled with NHS-Cy dye (2 h, 4°C). To remove excess dye and ensure that only membrane proteins were labeled, we centrifuged the labeled cells (200 × g, 5 min) and rinsed them with PBS three times. The cells were then sonicated and centrifuged as previously described for the formation of cellular micelle-like samples.
Microarray Hybridization.
We fit the lectin microarray slides within the 16-well FAST frame to create a separate well for each individual array. As a quality control for both the print and the lectins, we analyzed two previously characterized glycoproteins, ovalbumin and bovine mucin, on each slide. For single-color experiments, we added 10 μg of labeled cell sample (corresponds to ≈105 cells) in a final volume of 100 μl (in PBS) to each array. For two-color ratiometric analysis, we used 10 μg of each orthogonally labeled cell sample in a final total volume of 100 μl. We then allowed the samples to hybridize with the slides for 2 h at room temperature with gentle rocking. The individual wells were then rinsed with PBST (100 μl, three times for 3 min), the FAST frame was removed, and the slides were immersed in PBS (5 min). We then dried the slides by using a bench top slide spinner before analysis.
Analysis of Microarray Data.
We scanned the slides by using a GenePix 4100A fluorescent slide scanner (gain = 500, 5-μm scan; Molecular Devices, Sunnyvale, CA) and extracted the data with GenePix Pro 5.1 software (Molecular Devices). As previously mentioned the background-subtracted median values for each spot were used. Replicate data were statistically tested by using the Grubbs' outlier method (21). Microsoft Excel was used for statistical analysis and graphing. P value calculations were done by using the Student t determination at http://statpages.org/pdfs.html and with n-2 for the degrees of freedom. All errors were propagated from the standard deviations by using standard propagation of error formulas. For arrays printed and hybridized on different days (Pro-5 vs. Lec mutants), we used the Yang method (as described in the main text) to calculate the dye-bias-corrected ratios. For the HL-60 data, which were hybridized simultaneously to the same array against a common biological reference, the ratios were calculated without a dye-bias correction. To create the hierarchical clustering map, we used Cluster 3.0 with Java TreeView (http://rana.lbl.gov/EisenSoftware.htm) (30). We counted as positive lectins that gave a majority of signals above the 1,000 A.U. threshold for either the reference samples (Pro-5, undifferentiated HL-60) or the alternate samples (Lec mutants, differentiated HL-60). For the CHO samples, we used the lectins that gave positive signals in four of the six arrays used in the original biological replicate data set for either Lec8 or Pro-5. For the HL-60 samples, lectins that gave positive signals in either HL-60 or differentiated HL-60 for four of five biological replicates were counted as positive. Only positive lectins were used in the hierarchical clustering analysis for the two-colored data sets.
Histology.
We cultured Pro-5 and Lec8 cell in tissue culture-treated, black-wall, clear-bottom 96-well plates (Greiner Bio-One). We selected wells at 60–80% confluency. The cells were fixed with ice-cold methanol for 20 min and rinsed three times with PBS. We stained the cells with 4′,6′-diamidino-2-phenylindole (DAPI) (300 nM in PBS; Molecular Probes). The cells were incubated with the DAPI stain (40 μl) for 5 min at room temperature and rinsed three times with PBS. We then incubated the cells with biotinylated ConA, GNA, or WGA (80 μg/ml in PBS-T with 5% BSA; EY Laboratories) for 1 h at room temperature while rocking. After rinsing three times with PBS, we incubated the cells with FITC-avidin (1:1,000 dilution in PBS-T with 5% BSA) for 30 min at room temperature with gentle rocking. The cells were then rinsed three times with PBS before obtaining images. The images were taken at ×10 magnification (PlanFluor objective, numerical aperture = 0.3; Nikon) by using an inverted microscope (Nikon Eclipse TE 2000-U; Photometrics CoolSNAP ES monochrome camera) and MetaMorph image analysis software (Version 6.2r6; Molecular Devices). For each lectin, the exposure times were optimized to give the half maximal signal (≈2,000 A.U.) for the more positive cell line (either Pro-5 or Lec8, depending on the lectin). Images for both cell lines (Pro-5 and Lec8) were then obtained by using these conditions. Each pair of images (per lectin) is shown at the same scale, allowing us to directly compare the fluorescence levels visually. DAPI images were also obtained for each cell sample (data not shown). For HPA staining, an identical protocol was used. For image analysis, fluorescent images were background-subtracted and the total fluorescence intensity of the image was obtained. This value was normalized to the cell count, and the values over several fields of view were averaged. Outlier data were excluded by statistical testing.
Supplementary Material
Acknowledgments
We thank Dr. David Graham (University of Texas) for the use of his ultracentrifuge, The Arnold and Mabel Beckman Foundation for financial support, and the Consortium for Functional Glycomics (www.functionalglycomics.org; Grant GM62116) for data on the mass spectrometry of HL-60 glycans and detailed analysis of lectin specificities.
Abbreviation
- WGA
wheat germ agglutinin.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704954104/DC1.
References
- 1.Varki A, Cummings R, Esko JD, Freeze H, Hart GW, Marth J, editors. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1999. [PubMed] [Google Scholar]
- 2.Gorelik E, Galili U, Raz A. Cancer Metastasis Rev. 2001;20:245–277. doi: 10.1023/a:1015535427597. [DOI] [PubMed] [Google Scholar]
- 3.Sharon N, Lis H. Science. 1989;246:227–234. doi: 10.1126/science.2552581. [DOI] [PubMed] [Google Scholar]
- 4.Zaia J. Mass Spectrom Rev. 2004;23:161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- 5.Uematsu R, Furukawa JI, Nakagawa H, Shinohara Y, Deguchi K, Monde K, Nishimura SI. Mol Cell Proteomics. 2005;4:1977–1989. doi: 10.1074/mcp.M500203-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Naka R, Kamoda S, Ishizuka A, Kinoshita M, Kakehi K. J Proteome Res. 2006;5:88–97. doi: 10.1021/pr0502976. [DOI] [PubMed] [Google Scholar]
- 7.Guerardel Y, Chang LY, Maes E, Huang CJ, Khoo KH. Glycobiology. 2006;16:244–257. doi: 10.1093/glycob/cwj062. [DOI] [PubMed] [Google Scholar]
- 8.Mahal LK. Anticancer Agents Med Chem. 2007 in press. [Google Scholar]
- 9.An HJ, Ninonuevo M, Aguilan J, Liu H, Lebrilla CB, Alvarenga LS, Mannis MJ. J Proteome Res. 2005;4:1981–1987. doi: 10.1021/pr0501620. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg D, Sutton-Smith M, Paulson J, Dell A. Proteomics. 2005;5:865–875. doi: 10.1002/pmic.200401071. [DOI] [PubMed] [Google Scholar]
- 11.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. ChemBioChem. 2005;6:985–989. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 12.Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Nat Methods. 2005;2:851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 13.Angeloni S, Ridet JL, Kusy N, Gao H, Crevoisier F, Guinchard S, Kochhar S, Sigrist H, Sprenger N. Glycobiology. 2005;15:31–41. doi: 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- 14.Hsu KL, Pilobello KT, Mahal LK. Nat Chem Biol. 2006;2:153–157. doi: 10.1038/nchembio767. [DOI] [PubMed] [Google Scholar]
- 15.Ebe Y, Kuno A, Uchiyama N, Koseki-Kuno S, Yamada M, Sato T, Narimatsu H, Hirabayashi J. J Biochem (Tokyo) 2006;139:323–327. doi: 10.1093/jb/mvj070. [DOI] [PubMed] [Google Scholar]
- 16.Hsu KL, Mahal LK. Nat Protocols. 2006;1:543–549. doi: 10.1038/nprot.2006.76. [DOI] [PubMed] [Google Scholar]
- 17.Koshi Y, Nakata E, Yamane H, Hamachi I. J Am Chem Soc. 2006;128:10413–10422. doi: 10.1021/ja0613963. [DOI] [PubMed] [Google Scholar]
- 18.Ji H, Davis RW. Nat Biotechnol. 2006;24:1112–1113. doi: 10.1038/nbt0906-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takekawa H, Ina C, Sato R, Toma K, Ogawa H. J Biol Chem. 2006;281:8528–8538. doi: 10.1074/jbc.M513773200. [DOI] [PubMed] [Google Scholar]
- 20.Marikar Y, Zachariah B, Basu D. Anal Biochem. 1992;201:306–310. doi: 10.1016/0003-2697(92)90343-6. [DOI] [PubMed] [Google Scholar]
- 21.Grubbs F. Technometrics. 1969;11:1–21. [Google Scholar]
- 22.Deutscher SL, Hirschberg CB. J Biol Chem. 1986;261:96–100. [PubMed] [Google Scholar]
- 23.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanley P, Caillibot V, Siminovitch L. Cell. 1975;6:121–128. doi: 10.1016/0092-8674(75)90002-1. [DOI] [PubMed] [Google Scholar]
- 25.Deutscher SL, Nuwayhid N, Stanley P, Briles EI, Hirschberg CB. Cell. 1984;39:295–299. doi: 10.1016/0092-8674(84)90007-2. [DOI] [PubMed] [Google Scholar]
- 26.Chang HH, Oh PY, Ingber DE, Huang S. BMC Cell Biol. 2006;7:11. doi: 10.1186/1471-2121-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizoguchi A, Takasaki S, Maeda S, Kobata A. J Biol Chem. 1984;259:11943–11948. [PubMed] [Google Scholar]
- 28.Toma V, Zuber C, Winter HC, Goldstein IJ, Roth J. Histochem Cell Biol. 2001;116:183–193. doi: 10.1007/s004180100304. [DOI] [PubMed] [Google Scholar]
- 29.Wu AM, Wu JH, Tsai MS, Herp A. Life Sci. 2000;66:2571–2581. doi: 10.1016/s0024-3205(00)00591-9. [DOI] [PubMed] [Google Scholar]
- 30.Eisen MB, Spellman PT, Brown PO, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Sundaram S, Shaper NL, Raju TS, Stanley P. J Biol Chem. 2001;276:13924–13934. doi: 10.1074/jbc.M010046200. [DOI] [PubMed] [Google Scholar]
- 32.Itoh S, Kawasaki N, Ohta M, Hayakawa T. J Chromatogr A. 2002;978:141–152. doi: 10.1016/s0021-9673(02)01423-1. [DOI] [PubMed] [Google Scholar]
- 33.Lin AI, Philipsberg GA, Haltiwanger RS. Glycobiology. 1994;4:895–901. doi: 10.1093/glycob/4.6.895. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Shmulevich I, Astola J. Microarray Quality Control. Hoboken, NJ: Wiley; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.