Abstract
All eukaryotic cells contain the phospholipid phosphatidylinositol 4, 5-bisphosphate (PIP2) that serves multiple roles in signal transduction cascades. Type I phosphatidylinositol-4-phosphate 5-kinase (PIP5KI) catalyzes the synthesis of PIP2 by phosphorylating phosphatidylinositol 4 phosphate. Although the classical isoforms of PIP5KI (designated as α, β, and γ) all generate the same phospholipid product, they have significantly dissimilar primary structures and expression levels in different tissues, and they appear to localize within different compartments within the cell. Therefore, it appears likely that PIP5KI isoforms have overlapping, but not identical, functions. Here we show that targeted disruption of PIP5KIγ causes widespread developmental and cellular defects. PIP5KIγ-null embryos have myocardial developmental defects associated with impaired intracellular junctions that lead to heart failure and extensive prenatal lethality at embryonic day 11.5 of development. Loss of PIP5KIγ also results in neural tube closure defects that were associated with impaired PIP2 production, adhesion junction formation, and neuronal cell migration. These data, along with those of other PIP5KI isoforms, indicate that individual PIP5KI isoenzymes fulfill specific roles in embryonic development.
Keywords: phosphoinositide; phospholipid; phosphatidylinositol 4,5-bisphosphate
Over 40 years ago, Lowell and Mabel Hokin (1) showed that the head group of phosphatidylinositol can be transiently phosphorylated at the 3, 4, or 5 position to generate a family of phosphoinositides. This process is a key cellular event within all eukaryotic cells (2, 3). A major phosphoinositide found in these cells is phosphatidylinositol 4,5-bisphosphate (PIP2). PIP2 is widely known for the production of lipid second messengers from its hydrolysis by phospholipase C and phosphatidylinositol 3-kinase. PIP2 also functions to regulate vesicle secretion, GTP-binding proteins, actin-binding proteins, and PH domain-containing proteins, and serves as a cofactor for phospholipase D.
Three genes encode the three isoforms of type I phosphatidylinositol-4-phosphate 5-kinase (PIP5KI) known as PIP5KIα, PIP5KIβ, and PIP5KIγ (notably, PIP5KIγ also has two splice variants) (4–6). All three isoforms can be stimulated by small GTPases (Rho, Rac, Cdc42, and ARF), as well as by phosphatidic acid. Although PIP5KIα, PIP5KIβ, and PIP5KIγ are all capable of synthesizing PIP2, these isoenzymes have significantly dissimilar primary structures and different expression levels in different tissues, and they appear to localize within different compartments within some cells (7–12). For example, PIP5KIα localizes in membrane ruffles (8), PIP5KIβ localizes near endosomes (9), and PIP5KIγ is targeted to focal adhesions and nerve terminals (10–12).
Because most cells possess more than one isoform of PIP5KI, it appears likely that these isoforms have overlapping, but not identical, functions. Consistent with this hypothesis, a recently published murine line lacking PIP5KIα has a selective signaling defect in mast cells (13). PIP5KIγ is much larger than the other two isoforms and is the only isoform speculated to contribute to focal adhesion formation. To begin to understand the unique contribution of PIP5KIγ to developmental and cellular biology, we generated a murine line lacking this isoform of PIP5KI. Although PIP5KIγ is the predominate isoform within neurons, loss of PIP5KIγ induces developmental defects in a wide variety of tissues, indicating its unique and essential role in multiple cells types.
Results
Loss of PIP5KIγ Leads to Embryonic Lethality at Embryonic Day 11.5 (E11.5).
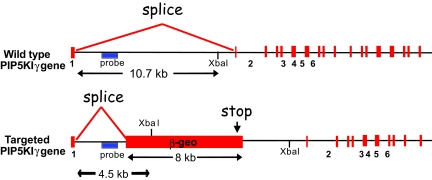
To elucidate the functions of PIP5KIγ, we used an embryonic stem (ES) cell line that contained a β-geo gene trap within the first intron of the PIP5KIγ gene (Table 1 and Fig. 1) (14). The gene trap strategy used to create this ES cell line was designed to create an abnormal mRNA transcript from the trapped allele that would produce a fusion protein corresponding to the first 32 amino acids of PIP5KIγ fused to β-gal. This ES cell line was used to create chimeric founders, which gave rise to germ-line heterozygotes harboring the targeted PIP5KIγ-null mutation.
Table 1.
Genotypes of embryos derived from matings of PIP5KIγ+/−
| Age | Wild type, % | Heterozygote, % | Null, % | Resorbed, % |
|---|---|---|---|---|
| E8.5 | 27 | 55 | 18 | 0 |
| E9.5 | 30 | 45 | 20 | 5 |
| E10.5 | 19 | 49 | 23 | 9 |
| E11.5 | 17 | 62 | 14 | 7 |
| E12.5 | 28 | 48 | 3 | 21 |
| Newborn | 36 | 64 | 0 |
Fig. 1.
Schematic of PIP5KIγ gene targeting. Diagram shows the location of the β-geo within the first intron of the PIP5KIγ gene. Location of the Southern blot probe is shown in blue. The gene traps insert an XbaI site. The insertion leads to a read-through mutation within the first intron of the targeted gene and truncates PIP5KIγ after the 32nd amino acid.
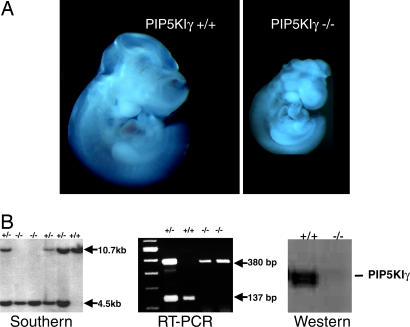
Heterozygous PIP5KIγ mice were intercrossed, but no viable PIP5KIγ−/− mice were identified (Table 1). Timed matings demonstrated that the majority of the PIP5KIγ-null embryo loss occurred between E11 and E12. Examination of viable E9.5 embryos revealed that the PIP5KIγ-null embryos were easily distinguished from their littermates by their smaller size and open anterior neural folds (Fig. 2A). Southern blot, PCR, and sequence analysis of genomic DNA from targeted mice revealed deletion of expression of exons 2 to 18, including the entire catalytic domain. Therefore, the expected fusion protein is predicted to lack phosphoinositol kinase activity. We confirmed that the gene trap insertion resulted in loss of wild-type PIP5KIγ expression by RT-PCR and by anti-PIP5KIγ immunoblotting (Fig. 2B).
Fig. 2.
Loss of PIP5KIγ induces lethality at E11.5. (A) E9.5 PIP5KIγ−/− embryos are smaller compared to wild-type littermates (Left vs. Right). (B) The Southern blot of XbaI-digested DNA shows a 10.7-kb (wild-type) band and 4.5-kb (PIP5KIγ-targeted) band. Shown is an RT-PCR using a sense primer from exon 1 and antisense primers from β-geo and exon 3. Anti-PIP5KIγ immunoblot (BD Biosciences, San Jose, CA) shows complete loss of protein in brain lysates of knockout embryos.
PIP5KIγ Is Critical for Cardiovascular Development.
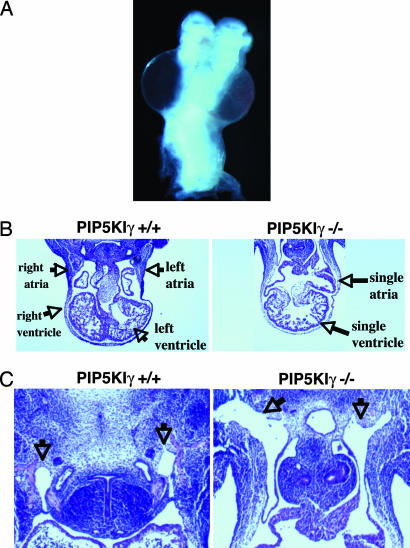
Possible explanations for lethality at this stage include defects in erythropoiesis and/or cardiovascular development. Histological examination of blood from E10.5 PIP5KIγ−/− embryos demonstrated normal morphology of RBCs (data not shown). However, close inspection of the homozygous embryos revealed massively dilated pericardial sacs (Fig. 3A). This result suggested that the embryos died of cardiac failure. Further analysis showed that these embryos had anomalous communication between the great vessels and the atria, engorged cardinal veins, and a lack of ventricular septation (Fig. 3 B and C).
Fig. 3.
PIP5KIγ-null embryos die of a cardiovascular defect. E11.5 PIP5KIγ-null embryos have a pericardial effusion (anterior view) (A), a single ventricle (B), and enlarged cardinal veins (C, arrows).
Our targeting strategy resulted in expression of β-geo under the control of the PIP5KIγ promoter. Therefore, staining embryos with lacZ revealed the cells that express β-geo, thereby indicating the expression pattern of PIP5KIγ. LacZ staining of PIP5KIγ+/− and PIP5KIγ−/− embryos revealed that normal myocardial cells are predicted to express PIP5KIγ (Fig. 4A). In situ staining with eHAND, the left ventricle-specific probe, demonstrated that PIP5KIγ−/− embryos had a markedly underdeveloped, but detectable right ventricle (data not shown). Together these results demonstrate that PIP5KIγ is critical for myocardial development, and that the lack of this enzyme leads to heart failure, resulting in lethality around E11.5.
Fig. 4.
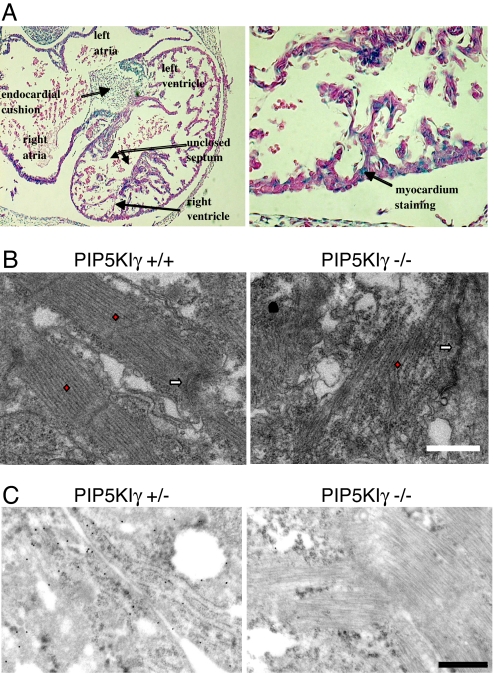
PIP5KIγ is required for actin organization and fascia adhesion formation in myocardiocytes. (A) E10.5 PIP5KIγ knockout embryo has an extremely atrophic right ventricle and a ventricular septum that has failed to close (Left). As seen by distribution of β-gal in the PIP5KIγ+/− heart shown, only myocardial cells are predicted to express PIP5KIγ mRNA (Right). (B) Electron micrographs of wild-type and PIP5KIγ-null cardiocytes. The red diamonds overlay the actin-rich sarcomeres, and the white arrows indicate the location of the fascia adherens. Loss of PIP5KIγ leads to actin disorganization, and the loss of the normal association of actin cables with the fascia adherens. (Scale bar: 1.0 μm.) (C) Immunogold-coupled anti-N-cadherin staining of PIP5KIγ+/− and PIP5KIγ−/− cardiocytes demonstrates that cells lacking PIP5KIγ do not have N-cadherin at the fascia adherens. (Scale bar: 500 nm.)
Anderson and colleagues (15) observed that PIP5KIγ contributes to adherens junction formation. Embryonic cardiomyocytes do form cadherin-rich structures, and they form fascia adherens at cellular interfaces. We analyzed whether loss of PIP5KIγ affected the organization of these structure. As shown in Fig. 4B Left, cardiomyocytes obtained from E9.5 wild-type embryos displayed normal development of actin-rich sarcomeres. The actin cables were orderly and terminated as expected at the fascia adherens between adjacent cardiomyocytes. In contrast, the actin cables within the sarcomeres of PIP5KIγ−/− myocytes were disorganized and failed to associate with the fascia adherens. Furthermore, N-cadheren was absent from this cellular region in PIP5KIγ-null cardiomyocytes (Fig. 4C). These data suggest that cardiac developmental abnormality in the PIP5KIγ-null embryos may be due, at least in part, to an underlying cytoskeletal defect.
Embryos Lacking PIP5KIγ Exhibit Exencelphaly.
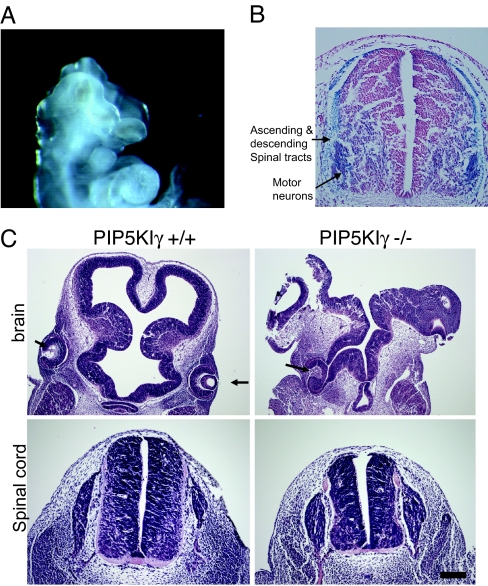
In addition to their overall small size, the PIP5KIγ-null embryos exhibited rostral neural tube closure defects (excencephaly; Figs. 2A and 5A and C). Staining for β-gal predicts that PIP5KIγ is expressed most prominently in the motor column (Fig. 5B) and in the developing ascending and descending spinal tracts. Analysis of the PIP5KIγ-null embryos revealed that the telancephalic neural folds failed to close, giving rise to a disorganized neuroepithelium. This finding was also noted in the rare PIP5KIγ−/− embryos viable at E12.5 (data not shown). The disorganized overgrowth of the neuroepithelium (Fig. 5C Right Upper) is characteristic of neural tube closure defects and not attributable to a delay in development (16). The neural tube closure defect was restricted to the head region; closure of the caudal neural tube over the spinal cord was complete (Fig. 5C Right Lower). Therefore, PIP5KIγ is only necessary for rostral neural tube closure, similar to many other mouse mutants showing selective neural tube closure defects caudally or rostrally (17). Although caudal neural tube closure appears to occur normally, the spinal cord showed a reduction in volume in PIP5KIγ−/− embryos (Fig. 5C Lower Left and Lower Right).
Fig. 5.
Neuronal defects in PIP5KIγ−/− embryos. (A) PIP5Kγ-null embryos fail to close their neuroepithelium. (B) Distribution of β-gal expression within the spinal cord of an E10.5 PIP5Kγ+/− embryo. This staining pattern predicts that PIP5Kγ is expressed most prominently in the motor column and the developing ascending and descending spinal tracts. (C) Cranial sections through telencephalon (Upper) and diencephalon (Lower) of E10.5 embryos at the level of the eyes (arrows.) The wild-type embryo displays normal development (Left). The PIP5KIγ−/− embryo (Right) has an unclosed neural tube and an abnormally organized neuroepithelium. Although the PIP5KIγ−/− cord has a normal general morphology, it is reduced in its dorsoventral axis and medial-lateral thickness. (Scale bar: 25 μm.)
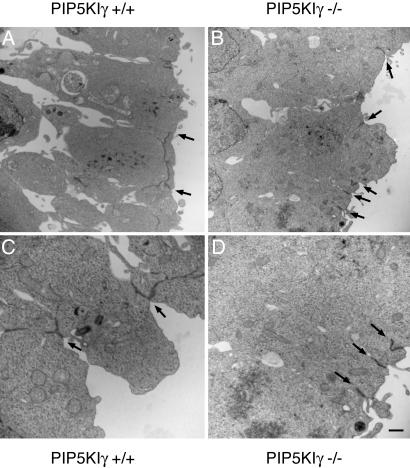
Given the adherens junction defects observed in cardiomyocytes and that cell structure/polarity in the neuroepithelium is essential for normal neural tube closure (34), we sought to determine whether adherens junctions were also disrupted in the CNS. Ultrastructural examination of the neuroepithelium in E8.5 embryos demonstrated normal, long, and branched adherens junctions PIP5KIγ+/+ mice (Fig. 6 A and C). In contrast, neuroepithelial cells exhibited short and uniformly simple adherens junctions in PIP5KIγ−/− mice. Although the significance of these data remains uncertain, the observed structural defect indicates that PIP5KIγ is necessary to establish normal junctions in the embryonic nervous system. Future studies will be directed at understanding the mechanisms and role of PIP5KIγ in these developmental processes.
Fig. 6.
Loss of PIP5Kγ impairs adherens junction formation in the brain. Electron micrographs from neuroepithelium of E8.5 PIP5KIγ+/+ (A and C) and PIP5KIγ−/− (B and D) embryos. Adherens junctions (arrows) are found between cells at the apical border. PIP5KIγ+/+ neuroepithelial cells exhibit longer and more complex branched junctions compared to PIP5KIγ−/− cells. (Scale bar: 2 μm.)
Neuronal Cells Lacking PIP5KIγ Have Defective PIP2 Production and a Migration Defect.
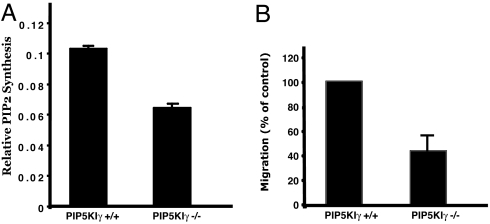
Given the abundance of PIP5KIγ in neuronal tissue, we analyzed whether brain PIP2 production was impaired in knockout embryos. As shown in Fig. 7A, PIP5KI activity was markedly decreased in brains derived from PIP5KIγ-null embryos compared to brains derived from wild-type littermates. We next analyzed whether this biochemical defect induced abnormalities on cytoskeletal-dependent processes.
Fig. 7.
PIP5KIγ−/− neuronal cells exhibit defective PIP2 synthesis and cell migration. (A) Lysates of embryonic brains were analyzed for in vitro kinase activity by using PI4P as the exogenous substrate. (B) PIP5KIγ-null neuronal precursor cells have impaired migration in a transwell assay. Shown is mean ± SEM for three experiments (migration is normalized for control cells).
Neural tube closure requires numerous cellular events, including extensive cell movement. Given the speculated role of PIP5KIγ in actin dynamics and focal adhesion formation (10, 11, 18), we analyzed whether neuronal cells derived from knockout embryos exhibited normal ex vivo migration. Using a modified Boyden chamber, we found a 50% reduction in PIP5KIγ-null neuronal precursor cell migration across a matrigel-coated filter after 24 h, compared to cells derived from wild-type littermates (Fig. 7). Although these data do not directly define a role for a cell migration defect contributing to the failure of neural tube closure, this hypothesis is reasonable and requires further exploration.
Discussion
Anderson and colleagues (19) were able to identify two separate PIP kinase (PIPK) families known as type I PIPK and type II PIPK from erythrocytes. These kinases were later found to have differing substrate specificity. Type I phosphorylated the d-5 position of the inositol ring on phosphatidylinositol 4-phosphate (PI4P), and type II PIPK preferred to phosphorylate the d-4 position of the inositol ring of phosphatidylinositol 5-phosphate (4–6, 20). Both types yield PIP2. The type I PIPK became known as type I PIP5KI, and the type II PIPK became known as type II PIP4K.
Because PI4P, the substrate of PIP5KI, is ≈50-fold more abundant than phosphatidylinositol 5-phosphate, the substrate of PIP4K, it appeared that phosphorylation of PI4P by PIP5KI is the predominant pathway of PIP2 synthesis in most cells. This hypothesis was confirmed by radiolabeled phosphate pulse–chase experiments that analyzed the relative labeling rate of the d-4 and d-5 positions of the inositol ring (7, 21). Therefore, it stands to reason that the bulk of mammalian PIP2 synthesis is by and large mediated by PIP5KI. Our data demonstrate that the PIP5KIγ isoform synthesizes a pool of PIP2 that is critical for complete cardiovascular and neuronal development.
Cell-Matrix Adhesions.
Reports by Di Paolo et al. (11) and Ling et al. (10) have shown that, under certain circumstances, PIP5KIγ can associate with talin (22–25). The solution structure of a fragment of mouse talin-1 (amino acids 306–429) with a peptide corresponding to PIP5KIγ amino acids 641–648 has recently been published (26). Furthermore, these groups have shown that only the γ isoform of PIP5KI localizes within and contributes to the formation of focal adhesions. Early cardiac development involves proper migration of the bilateral precardiac mesoderm and subsequent complex bending and looping of the primitive cardiac tube. Given such a complex morphological event, it is not surprising that cell-matrix interactions, including cell migration, play an important role in this process. Knockouts of p130Cas, vinculin, and α4-integrins, which also contribute to focal adhesion formation, die of a cardiovascular developmental defect at this same stage of development (27–29). Although it is tempting to speculate that the cardiovascular developmental defect is because of impaired focal adhesion formation, at this point, we have not detected absent focal adhesion formation in any PIP5KIγ-null cells studied to date (data not shown). However, given the importance of cell migration in cardiac looping morphogenesis, such a mechanism potentially contributes to the defects observed in PIP5KIγ mutants.
Intercellular Adhesions.
Recent work by Anderson and coworkers (15) demonstrated that PIP5KIγ directly associates with N-cadherin and regulates its trafficking within epithelial cells. This interaction is critical for the formation of adherens junctions within these cells. In the intact heart, myocardiocytes associate with each other via dimers of the transmembrane adhesion molecule, N-cadherin. In these cells, N-cadherin localizes in a counterpart of the zona adherens called the fascia adherens. It is at this structure that the actin cytoskeleton found within the sarcomeres associates with the cell membrane. Our studies demonstrate that loss of PIP5KIγ leads to a markedly disordered organization of the actin cytoskeleton in myocardiocytes. In these cells, actin does not associate at all with the fascia adherens. These results provide in vivo confirmation that PIP5KIγ is essential for cell junction formation and that this mechanism may explain some of the cardiac developmental defect.
This study demonstrates that PIP5KIγ-null myocardiocytes lack N-cadherin at the fascia adherens. N-cadherin plays an important role in cardiac development, and N-cadherin-null embryos die by E10 because of cardiovascular failure (30). Similar to embryos lacking PIP5KIγ, N-cadherin−/− cardiomyocytes lack normal assembly of adherens junctions (31). However, in contrast to PIP5KIγ-null cells, myocytes lacking N-cadherin have sarcomeres containing ordered arrays of actin bundles that associate with the fascia adherens. Thus, PIP5KIγ contributes to sarcomere actin organization via mechanisms that are also independent of N-cadherin.
Neurologic Development.
Neural tube closure requires cytoskeletal rearrangements, cell migration, fusion of separate epithelial layers, and programmed cell death (reviewed in ref. 32). Furthermore, these processes, which occur along the entire neural axis, are under regional control. As a result, neural tube closure defects are often restricted to the cranial region, the caudal end of the neural tube, or may involve the entire neural axis (33). PIP5KIγ−/− mice exhibit neural tube closure defects restricted to the cranial region, adding to a list of candidate genes involved in the pathogenesis of cranial neural tube closure genes (34). Given the recognized role for PIP5KIγ in cell migration and the requirement of cell migration in neural tube closure, we predicted that PIP5KIγ−/− neuronal cells would have a defective migration phenotype. Using a modified Boyden chamber assay, we found a significant defect in neural precursor cell migration. These data suggest that the cell migration required for cranial neural tube closure depends on PIP5KIγ, although we cannot exclude the role of other cellular processes, including adherens junction formation, that are also dependent on PIP5KIγ for cranial neural tube closure. These possibilities will be investigated in future studies.
Lethality.
The effect on lethality that we observed in PIP5KIγ−/− mice is earlier than that reported by Di Paolo et al. (35), who used an independently derived murine line. Their mutant mice are viable until a few hours after birth. Although the phenotype of both murine lines is lethality, the developmental abnormalities do vary in their degree. Several possible explanations exist to account for the differences between the two mouse lines. First, our mutant may represent a complete null mutation that causes lethality at midgestation, whereas a hypomorphic allele might develop until the first day of life. Our analysis using RT-PCR indicates a complete loss of PIP5KIγ transcripts beyond the first exon. Our anti-PIP5KIγ immunoblots are consistent with this conclusion. A second possible explanation for the divergent result is that our gene trap may be generating a dominant negative protein. The gene trap used for our murine line is situated in the first intron. This strategy predicts that any protein generated from this message would be truncated after the 32nd amino acid. Because we cannot detect a phenotype in the PIP5KIγ+/− mice, this finding demonstrates that a putative truncated protein does not have a dominant negative effect. Together these results give us confidence that the murine line we have described contains a complete null mutation within the PIP5KIγ gene. The final, and perhaps most probable, alternative explanation for the apparent discrepancy between our results and those of Di Paolo et al. is that modifier genes in the genetic background of the two murine lines influence the severity of the phenotypes.
Conclusion
In summary, our findings demonstrate that loss of PIP5KIγ leads to pleotrophic developmental defects and embryonic lethality at E11.5. Embryos lacking PIP5KIγ fail to undergo proper cardiac chamber septation and exhibit neural tube closure defects. Furthermore, cells lacking this enzyme fail to adequately synthesize PIP2, form normal adherens junctions, or migrate in transwell assays. Given the widespread distribution of the three PIP5KI isoforms, these data suggest that individual PIP5KI isoforms have overlapping, but not completely redundant, functions within cells.
Materials and Methods
Targeting of PIP5KIγ Genes.
Berkeley Bay Genomics Group provided ES cell lines (XD096) containing disruption of one allele of the PIP5KIγ genes by β-geo random insertion mutagenesis (14). Using an RT-PCR, Southern blotting, and a sequencing-based strategy, we identified the specific site of insertion of the β-geo cassette within the first intron of the gene. Location of the Southern blot probe is shown in Fig. 1. The sense PCR primer corresponding to the first exon was 5′-TGGTCTGCGGAGAGTGGG-3′, the antisense primer corresponding to exon 3 was 5′-CTCTCCCGACGCATCCAC-3′, and the antisense primer corresponding to β-geo was 5′-TTGAGGGGACGACGACAGTATC-3′. Generation of chimeric mice was performed at the Transgenic Core Facility at the University of Pennsylvania.
Morphologic Examination of Embryos.
Embryos were collected on the indicated days postconception and fixed in 4% paraformaldehyde for 24 to 48 h. Embryos were dehydrated through a series of ethanol solutions and embedded in paraffin. In situ hybridization with the eHAND probe was performed as previously described (36, 37). In situ hybridization slides were photographed on a Nikon E600 microscope with fluorescent lighting using darkfield imaging with a red filter (Nikon, Tokyo, Japan). Further details on histological procedures can be found at the University of Pennsylvania Molecular Cardiology Research Center web site (www.med.upenn.edu/mcrc/).
EM.
The embryonic hearts were processed as previously described (38, 39). Briefly, after careful dissection, the tissue was immediately fixed with prewarmed 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer for 1 h. The samples were then rinsed with sodium cacodylate buffer and postfixed with 1% osmium tetroxide. They were dehydrated with ascending grade of ethanol, stained en bloc with uranyl acetate, embedded in epoxy medium, and polymerized at 68°C for 72 h. Ultrathin sections (≈80 nm) were cut with a diamond knife, mounted on single-slot copper grids, stained with uranyl acetate and lead citrate, and examined with an FEI Tecnai T12 electron microscope operated at 80-kV accelerating voltage.
Immunostaining was done by using an N-cadherin antibody from BD Biosciences Transduction Laboratories (San Jose, CA). For those studies, tissue was fixed in 0.1 M sodium cacodylate buffer containing 4% paraformaldehyde and 0.1% glutaraldehyde. After dehydration in graded alcohol, they were infiltrated and embedded in the LR White resin. Blocks were cured under a UV lamp at −20°C for 72 h. Then 80-nm-thick sections were picked up on formvar-coated slotted nickel grids treated with blocking buffer and then incubated in a 1:20 dilution of antibody overnight at 4°C. The next day, the grids were washed in saline-based Tris buffer and then incubated with 10-nm gold anti-mouse for 1 h at room temperature. Unbound gold was washed first with Tris buffer followed by deionized water and then stained with saturated aqueous uranyl acetate. Images were captured by using JEOL (Tokyo, Japan) JEM 1010 equipped with AMT 12-HR and a CCD camera (Hamamatsu Corporation, Bridgewater, NJ).
In Vitro Kinase Assay.
Phosphoinositide kinase activity in total brain lysate was determined as described previously (40). Lipids were separated by TLC using 70 mM CHCl3, 100 mM MeOH, 25 mM H2O, and 15 mM NH4OH (vol/vol). Lanes containing commercial standards of PIP or PI4,5P2 were stained with iodine vapors. After overnight exposure of film to the plates, the radioactive spots were visualized and quantitated by a PhosphorImager STORM 820 (Molecular Dynamics, Sunnyvale, CA). The results were expressed as counts corresponding to PIP2 as a fraction of total radioactivity per lane.
Neuronal Cell Migration Assay.
Neuronal precursor cells were isolated from E9.5 to E11.5 embryos, and cell migration was quantitated essentially as described by Aarum et al. (41). Briefly, 4 × 104 neuronal cells derived from PIP5KIγ−/− or PIP5KIγ+/+ littermates were placed in the upper chamber of a transwell containing a matrigel-coated separation filter. Neurobasal media (Invitrogen, Carlsbad, CA) were added to the lower chamber, and the cells were incubated overnight at 37°C with 5% CO2. Cells were fixed, stained for neuronal and glial cell markers, and quantified. Migration is expressed as a percentage of migration of PIP5KIγ+/+ cells.
Acknowledgments
We thank Drs. Neelima Shah, Qian-Cun Yu, Min Min Lu, and Jean Richea for technical assistance. This work was supported in part by the National Institutes of Health.
Abbreviations
- En
embryonic day n
- PIP2
phosphatidylinositol 4,5-bisphosphate
- PIP5KI
phosphatidylinositol-4-phosphate 5-kinase
- PI4P
phosphatidylinositol 4-phosphate.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Hokin MR, Hokin LE. J Biol Chem. 1953;203:967–977. [PubMed] [Google Scholar]
- 2.Toker A. Curr Opin Cell Biol. 1998;10:254–261. doi: 10.1016/s0955-0674(98)80148-8. [DOI] [PubMed] [Google Scholar]
- 3.Doughman RL, Firestone AJ, Anderson RA. J Membr Biol. 2003;194:77–89. doi: 10.1007/s00232-003-2027-7. [DOI] [PubMed] [Google Scholar]
- 4.Ishihara H, Shibasaki Y, Kizuki N, Katagiri H, Yazaki Y, Asano T, Oka Y. J Biol Chem. 1996;271:23611–23614. doi: 10.1074/jbc.271.39.23611. [DOI] [PubMed] [Google Scholar]
- 5.Loijens JC, Anderson RA. J Biol Chem. 1996;271:32937–32943. doi: 10.1074/jbc.271.51.32937. [DOI] [PubMed] [Google Scholar]
- 6.Ishihara H, Shibasaki Y, Kizuki N, Wada T, Yazaki Y, Asano T, Oka Y. J Biol Chem. 1998;273:8741–8748. doi: 10.1074/jbc.273.15.8741. [DOI] [PubMed] [Google Scholar]
- 7.Stephens LR, Hughes KT, Irvine RF. Nature. 1991;351:33–39. doi: 10.1038/351033a0. [DOI] [PubMed] [Google Scholar]
- 8.Doughman RL, Firestone AJ, Wojtasiak ML, Bunce MW, Anderson RA. J Biol Chem. 2003;278:23036–23045. doi: 10.1074/jbc.M211397200. [DOI] [PubMed] [Google Scholar]
- 9.Padron D, Wang YJ, Yamamoto M, Yin H, Roth MG. J Cell Biol. 2003;162:693–701. doi: 10.1083/jcb.200302051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- 11.Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, Chang S, Guo J, Wenk MR, De Camilli P. Nature. 2002;420:85–89. doi: 10.1038/nature01147. [DOI] [PubMed] [Google Scholar]
- 12.Wenk MR, Pellegrini L, Klenchin VA, Di Paolo G, Chang S, Daniell L, Arioka M, Martin TF, De Camilli P. Neuron. 2001;32:79–88. doi: 10.1016/s0896-6273(01)00456-1. [DOI] [PubMed] [Google Scholar]
- 13.Sasaki J, Sasaki T, Yamazaki M, Matsuoka K, Taya C, Shitara H, Takasuga S, Nishio M, Mizuno K, Wada T, et al. J Exp Med. 2005;201:859–870. doi: 10.1084/jem.20041891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skarnes WC. Methods Enzymol. 2000;328:592–615. doi: 10.1016/s0076-6879(00)28420-6. [DOI] [PubMed] [Google Scholar]
- 15.Ling K, Bairstow SF, Carbonara C, Turbin DA, Huntsman DG, Anderson RA. J Cell Biol. 2007;176:343–353. doi: 10.1083/jcb.200606023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ybot-Gonzalez P, Cogram P, Gerrelli D, Copp AJ. Development (Cambridge, UK) 2002;129:2507–2517. doi: 10.1242/dev.129.10.2507. [DOI] [PubMed] [Google Scholar]
- 17.Juriloff DM, Harris MJ. Hum Mol Genet. 2000;9:993–1000. doi: 10.1093/hmg/9.6.993. [DOI] [PubMed] [Google Scholar]
- 18.Shibasaki Y, Ishihara H, Kizuki N, Asano T, Oka Y, Yazaki Y. J Biol Chem. 1997;272:7578–7581. doi: 10.1074/jbc.272.12.7578. [DOI] [PubMed] [Google Scholar]
- 19.Bazenet CE, Ruano AR, Brockman JL, Anderson RA. J Biol Chem. 1990;265:18012–18022. [PubMed] [Google Scholar]
- 20.Rameh LE, Tolias KF, Duckworth BC, Cantley LC. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- 21.Whiteford CC, Brearley CA, Ulug ET. Biochem J. 1997;323:597–601. doi: 10.1042/bj3230597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ling K, Doughman RL, Iyer VV, Firestone AJ, Bairstow SF, Mosher DF, Schaller MD, Anderson RA. J Cell Biol. 2003;163:1339–1349. doi: 10.1083/jcb.200310067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SY, Voronov S, Letinic K, Nairn AC, Di Paolo G, De Camilli P. J Cell Biol. 2005;168:789–799. doi: 10.1083/jcb.200409028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bairstow SF, Ling K, Anderson RA. J Biol Chem. 2005;280:23884–23891. doi: 10.1074/jbc.M500576200. [DOI] [PubMed] [Google Scholar]
- 25.de Pereda JM, Wegener KL, Santelli E, Bate N, Ginsberg MH, Critchley DR, Campbell ID, Liddington RC. J Biol Chem. 2005;280:8381–8386. doi: 10.1074/jbc.M413180200. [DOI] [PubMed] [Google Scholar]
- 26.Kong X, Wang X, Misra S, Qin J. J Mol Biol. 2006;359:47–54. doi: 10.1016/j.jmb.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 27.Xu W, Baribault H, Adamson ED. Development (Cambridge, UK) 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- 28.Yang JT, Rayburn H, Hynes RO. Development (Cambridge, UK) 1995;121:549–560. doi: 10.1242/dev.121.2.549. [DOI] [PubMed] [Google Scholar]
- 29.Honda H, Oda H, Nakamoto T, Honda Z, Sakai R, Suzuki T, Saito T, Nakamura K, Nakao K, Ishikawa T, et al. Nat Genet. 1998;19:361–365. doi: 10.1038/1246. [DOI] [PubMed] [Google Scholar]
- 30.Radice GL, Rayburn H, Matsunami H, Knudsen KA, Takeichi M, Hynes RO. Dev Biol. 1997;181:64–78. doi: 10.1006/dbio.1996.8443. [DOI] [PubMed] [Google Scholar]
- 31.Kostetskii I, Li J, Xiong Y, Zhou R, Ferrari VA, Patel VV, Molkentin JD, Radice GL. Circ Res. 2005;96:346–354. doi: 10.1161/01.RES.0000156274.72390.2c. [DOI] [PubMed] [Google Scholar]
- 32.Zohn IE, Chesnutt CR, Niswander L. Trends Cell Biol. 2003;13:451–454. doi: 10.1016/s0962-8924(03)00173-9. [DOI] [PubMed] [Google Scholar]
- 33.Golden JA, Harding BN. Pathology and Genetics Series. Basel: ISN Neuropath Press; 2004. [Google Scholar]
- 34.Copp AJ, Greene ND, Murdoch JN. Nat Rev Genet. 2003;4:784–793. doi: 10.1038/nrg1181. [DOI] [PubMed] [Google Scholar]
- 35.Di Paolo G, Moskowitz HS, Gipson K, Wenk MR, Voronov S, Obayashi M, Flavell R, Fitzsimonds RM, Ryan TA, De Camilli P. Nature. 2004;431:415–422. doi: 10.1038/nature02896. [DOI] [PubMed] [Google Scholar]
- 36.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 37.Thomas T, Yamagishi H, Overbeek PA, Olson EN, Srivastava D. Dev Biol. 1998;196:228–236. doi: 10.1006/dbio.1998.8849. [DOI] [PubMed] [Google Scholar]
- 38.Allen E, Yu QC, Fuchs E. J Cell Biol. 1996;133:1367–1382. doi: 10.1083/jcb.133.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobinger GP, Weiner DJ, Yu QC, Wilson JM. Nat Biotechnol. 2001;19:225–230. doi: 10.1038/85664. [DOI] [PubMed] [Google Scholar]
- 40.Kinuta M, Yamada H, Abe T, Watanabe M, Li SA, Kamitani A, Yasuda T, Matsukawa T, Kumon H, Takei K. Proc Natl Acad Sci USA. 2002;99:2842–2847. doi: 10.1073/pnas.261715599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aarum J, Sandberg K, Haeberlein SL, Persson MA. Proc Natl Acad Sci USA. 2003;100:15983–15988. doi: 10.1073/pnas.2237050100. [DOI] [PMC free article] [PubMed] [Google Scholar]