Abstract
NF-κB and STATs regulate multiple cellular processes through the transcriptional activation of genes with diversified functions. Although the molecular mechanisms that can turn on/off the overall NF-κB/STAT signaling have been extensively studied, how NF-κB/STAT-target genes can be differentially regulated is poorly understood. Here we report that PIASy, a member of the PIAS (for protein inhibitor of activated STAT) protein family, is a physiologically important transcriptional repressor of NF-κB and STAT1. Piasy deletion in dendritic cells resulted in enhanced expression of a subset of NF-κB and STAT1-dependent genes in response to LPS or IFN-γ treatment, respectively. Consistently, Piasy null mice are hypersensitive to the LPS-induced endotoxic shock. Furthermore, PIASy and PIAS1 display specific as well as redundant effects on the regulation of NF-κB/STAT1 signaling. Pias1−/−Piasy−/− embryos died before day 11.5. The disruption of one allele of Pias1 in the Piasy−/− background significantly enhanced the effect of Piasy deletion on the transcriptional induction of NF-κB/STAT1-dependent genes, and vice versa. Our results demonstrate that PIASy cooperates with PIAS1 to regulate the specificity and magnitude of NF-κB/STAT1-mediated gene activation.
Keywords: transcriptional regulation
NF-κB and STATs are two important families of transcription factors that are activated in response to a variety of stimuli to regulate multiple cellular processes such as immune responses (1–5). NF-κB and STATs have distinct as well as synergistic effects on gene induction. The activity of NF-κB/STAT is tightly regulated. Understanding the regulation of NF-κB/STAT pathways is important because aberrant NF-κB/STAT signaling is associated with human cancers and immune disorders (2, 5–7). Extensive research efforts have been made in the identification and characterization of signaling molecules that can either positively or negatively regulate the NF-κB/STAT-mediated gene activation pathways. Among genes that are regulated by NF-κB/STAT, some of them show similar cellular functions, whereas others have different or even opposite biological effects. However, most regulators of the NF-κB/STAT signaling pathways identified so far have been shown to affect the overall NF-κB/STAT responses. How NF-κB/STAT-target genes can be differentially regulated is poorly understood. The identification of signaling molecules that control the specificity of NF-κB/STAT-mediated gene activation should enhance our ability to design specific therapeutic strategies for the treatment of human diseases.
The mammalian PIAS (protein inhibitor of activated STAT) protein family contains four members: PIAS1, PIAS3, PIASx, and PIASy (8). PIAS proteins were originally isolated in studies aimed at understanding the regulation of STAT signaling (9, 10). Among the PIAS family, both PIAS1 and PIASy have been shown to inhibit STAT1-mediated gene activation (10, 11). PIAS proteins possess SUMO E3 ligase activities (8, 12, 13). Subsequent biochemical studies suggest that members of the PIAS protein family are involved in the regulation of a growing list of >70 proteins, most of which are transcription factors, including NF-κB, SMAD, and p53 (8).
To understand the physiological functions of mammalian PIAS proteins, gene-targeting studies have been carried out. Characterization of Pias1 null mice demonstrates a role of PIAS1 as a physiologically important negative regulator of STAT1 and NF-κB (14, 15). Most interestingly, Pias1 disruption reveals an unexpected specificity of PIAS1 in gene regulation: PIAS1 selectively regulates a subset of STAT1 or NF-κB-dependent genes, with a notable preference for cytokines and chemokines. Piasy null mice have been generated by two independent groups (16, 17). However, initial studies using mouse embryonic fibroblasts (MEFs) isolated from Piasy null mice did not reveal a significant effect of Piasy disruption on the transcriptional induction of endogenous STAT1-target genes (16, 17). The physiological role of PIASy in the regulation of cytokine signaling has not been established.
In this work, we provide strong evidence to demonstrate that PIASy is a physiologically important negative regulator of STAT1 and NF-κB, and PIASy cooperates with PIAS1 to control the specificity and magnitude of STAT1 and NF-κB-mediated gene expression.
Results
PIASy As a Transcriptional Repressor of NF-κB.
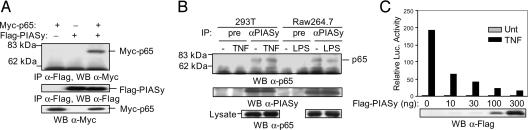
To explore a potential role of PIASy in the regulation of NF-κB, we first tested whether PIASy can interact with NF-κB in vivo by coimmunoprecipitation (co-IP) analysis. When both Myc-p65 and Flag-PIASy were coexpressed in 293T cells, Myc-p65 was coimmunoprecipitated by anti-Flag, indicating that PIASy and NF-κB p65 can interact in vivo (Fig. 1A). To address whether PIASy interacts with NF-κB-p65 endogenously, co-IP assays were performed with cell lysates from human 293T or murine macrophage-like cell line Raw264.7, with or without TNF or LPS treatment, respectively. As shown in Fig. 1B, p65 was immunoprecipitated by anti-PIASy, but not the preimmune serum, indicating that NF-κB p65 and PIASy interact endogenously. To examine the effect of PIASy on NF-κB-mediated gene activation, luciferase reporter assays were carried out in human 293T cells transiently transfected with an NF-κB reporter, together with increasing amounts of Flag-PIASy. In this experiment, the induction of luciferase activity was mediated by endogenous NF-κB proteins. TNF treatment induced an ≈200-fold increase of NF-κB-mediated gene activation, which was inhibited by PIASy in a dose-dependent manner (Fig. 1C). These results suggest that PIASy is an inhibitor of NF-κB.
Fig. 1.
PIASy interacts with NF-κB p65 and represses TNF-induced NF-κB-mediated gene activation. (A) 293T cells were transiently transfected with Myc-p65 and Flag-PIASy, either alone or together as indicated. Whole-cell lysates were used for IP with anti-Flag, followed by Western blot with anti-Myc or anti-Flag. The same extracts were also subjected to Western blot using anti-Myc. (B) 293T cells and Raw264.7 cells were either untreated or treated with TNF (10 ng/ml) or LPS (100 ng/ml), respectively. Whole-cell lysates were used for IP with anti-PIASy, followed by Western blot using anti-p65. The same filter was reprobed with anti-PIASy. (C) Luciferase reporter assays. 293T cells were transiently transfected with a luciferase reporter (2× NF-κB), together with increasing amounts of Flag-PIASy. At 24 h after transfection, cells were either untreated or treated with TNF (10 ng/ml) for 6 h, followed by luciferase assays. The same extracts were analyzed by Western blot using anti-Flag.
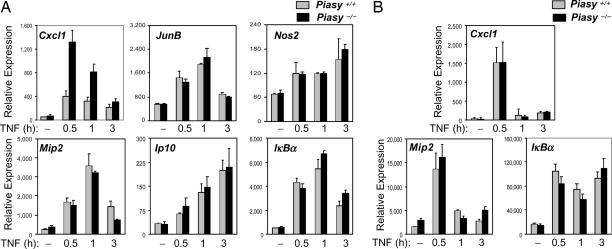
To validate a role of PIASy in the negative regulation of NF-κB, we examined whether NF-κB-mediated gene activation is altered in the absence of PIASy. Primary MEF cells prepared from wild-type (WT) and Piasy null mice (16) were untreated or treated with TNF for various time periods. Total RNA isolated from these cells was used for the analysis of transcriptional induction of NF-κB-dependent genes by quantitative real-time PCR (Q-PCR). The induction of Cxcl1 [Chemokine (C-X-C motif) ligand 1], an NF-κB target gene, was reproducibly enhanced in Piasy null cells compared with WT controls (Fig. 2A). However, the induction of many other NF-κB target genes, such as JunB, Nos2, Mip2, Ip10, and IκBα by TNF was not affected by Piasy deletion (Fig. 2A). In addition, Piasy disruption failed to show a significant effect on the induction of NF-κB-dependent genes, including Cxcl1, in bone marrow-derived macrophages (BMMs) (Fig. 2B). These results indicate that the removal of PIASy has a rather limited effect on the transcriptional activation of NF-κB-dependent genes in MEFs and BMMs.
Fig. 2.
Analysis of TNF-induced gene activation in Piasy−/− MEFs and BMMs. (A) Piasy+/+ and Piasy−/− primary MEF cells were either untreated or treated with TNF (20 ng/ml) for various time points. Total RNA was prepared and subjected to Q-PCR analyses. (B) Same as in A except that Piasy+/+ and Piasy−/− BMMs were used.
Redundant Role of PIAS1 and PIASy in Animal Development.
Although PIASy originally was identified as an inhibitor of STAT1, initial studies using Piasy null MEFs failed to show any significant effect of Piasy deletion on the induction of endogenous STAT1-dependent genes (16, 17). In addition, no significant difference in STAT1-induced genes was observed in WT and Piasy null BMMs (data not shown).
To test a possible functional redundancy of PIASy and PIAS1, we performed experiments to generate PIAS1 and PIASy double-knockout mice. Pias1−/− and Piasy−/− mice were crossed to generate Pias1+/−Piasy+/− mice. However, the subsequent compound cross (Pias1+/−Piasy+/− × Pias1+/−Piasy+/−) produced no Pias1 and Piasy double-null mice [see supporting information (SI) Table 1]. To determine whether Pias1−/−Piasy−/−double knockouts are embryonic lethal, we carried out embryo dissection experiments at day 11.5 (SI Table 2). No Pias1−/−Piasy−/− embryos were found, suggesting that these embryos died before day 11.5. In addition, Pias1−/−Piasy+/− embryos also showed reduced survival (7% compared with the theoretical value 12.5%), whereas Pias1−/+Piasy−/− embryos at day 11.5 were detected at the expected frequency. These results suggest that PIAS1 and PIASy have functional redundancy during mouse embryonic development, with a more profound effect associated with Pias1 disruption.
Cooperative Effect of PIAS1 and PIASy on the Regulation of NF-κB and STAT1.
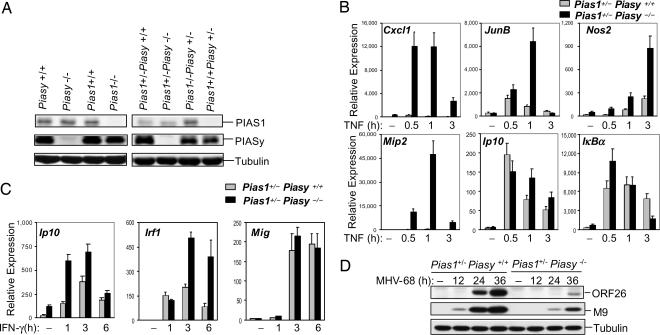
It was not possible to obtain Pias1 and Piasy double-knockout MEFs because Pias1−/−Piasy−/− embryos died before day 11.5. We tested the hypothesis that the removal of one allele of Piasy or Pias1 in the Pias1−/− or Piasy−/− background, respectively, may affect the ability of PIASy or PIAS1 to regulate NF-κB or STAT1 responses. Pias1−/−Piasy+/− and Pias1+/−Piasy−/− MEFs, as well as the matched control cells, were prepared from embryos. Western blot analysis indicated that the removal of one allele of Piasy or Pias1 showed gene dosage effect as the level of PIASy or PIAS1 protein expression was correspondingly reduced to ≈50% (Fig. 3A Right). However, Piasy disruption had no significant effect on PIAS1 expression, and vice versa (Fig. 3A Left).
Fig. 3.
Analysis of TNF and IFN-γ-induced gene transcription in Piasy−/−Pias1+/− and Piasy+/+Pias1+/− MEFs. (A) Western blot analyses were performed with whole-cell lysates prepared from various cell types as indicated, by using Abs against PIAS1, PIASy, and tubulin. (B) Piasy+/+Pias1+/− and Piasy−/−Pias1+/− MEFs were untreated or treated with TNF (20 ng/ml) for various time points. Total RNA was prepared and subjected to Q-PCR analyses. (C) Same as in A except cells were treated with IFN-γ. (D) Piasy+/+Pias1+/− and Piasy−/−Pias1+/− MEFs were infected with MHV-68, followed by Western blot analyses with anti-ORF-26, anti-M9, or anti-tubulin.
We performed experiments to analyze the effect of Pias1 single allele disruption on the ability of PIASy to regulate NF-κB-mediated gene expression. Pias1+/−Piasy+/+ and Pias1+/−Piasy−/− MEFs were challenged with TNF for various time periods, followed by Q-PCR analysis. Interestingly, the induction of genes that are not affected by Piasy deletion alone, such as JunB, Mip2, and Nos2 (Fig. 2), was significantly enhanced in the absence of PIASy in the Pias1−/+ background (Fig. 3B). In addition, Piasy deletion had a more profound effect on the induction of PIASy-sensitive gene Cxcl1 in the Pias1+/− background as compared with the Pias1+/+ background (Figs. 2A and 3B). However, the induction of Ip10 and IκBα was not significantly affected by Piasy disruption in the Pias1+/− background. These results demonstrate that the level of PIAS1 protein affects the ability of PIASy to regulate the specificity and magnitude of NF-κB-mediated gene activation.
Although PIASy initially was identified as an inhibitor of STAT1, earlier studies with Piasy null MEFs failed to show a significant effect of PIASy on the induction of endogenous STAT1-target genes (16, 17). To test whether the lack of PIASy effect on STAT1-mediated gene expression in Piasy null MEFs is due to a redundant function of PIAS1, we examined the IFN-induced activation of STAT1-target genes in primary Pias1+/−Piasy−/− and Pias1+/−Piasy+/+ MEFs (Fig. 3C). The induction of STAT1-target genes Ip10 and Irf1, but not Mig, was significantly enhanced in Pias1+/−PIasy−/− cells as compared with the Pias1+/−Piasy+/+ controls. Thus, the removal of one allele of Pias1 reveals a role of PIASy in the regulation of STAT1-dependent gene activation. These results support the hypothesis that PIAS1 and PIASy have a redundant role in the regulation of STAT1 signaling.
To examine the functional significance of PIASy in STAT1 signaling, we examined the effect of Piasy disruption on STAT1-dependent antiviral responses in the Pias1+/− background. The IFN-activated STAT1 signaling pathway is critical for the antiviral response against MHV-68 (mouse γ-herpes virus-68) infection (14). Pias1+/−PIasy−/− and Pias1+/−Piasy+/+ MEFs were infected with MHV-68 for various time periods. The viral replication was examined by Western blot analysis with antibodies (Abs) to the viral capsid proteins M9 or ORF26 (14). The expression of viral proteins M9 and ORF26 was significantly inhibited in Pias1+/−PIasy−/− cells as compared with the Pias1+/−Piasy+/+ control. These findings support a role of PIASy in the negative regulation of STAT1 signaling.
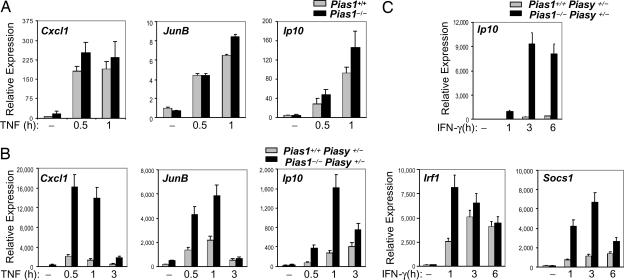
PIAS1 selectively regulates a subset of IFN and TNF-responsive genes (14, 15). We next performed reciprocal experiments to test whether PIASy affects the ability of PIAS1 to regulate NF-κB and STAT1 signaling. Pias1+/+Piasy+/− and Pias1−/−Piasy+/− MEFs were treated with TNF-α for various time periods, followed by Q-PCR analyses. The transcriptional activation of NF-κB-target genes such as Cxcl1, JunB, and Ip10 was significantly up-regulated in Pias1−/−Piasy+/− MEFs as compared with the matched Pias1+/+Piasy+/− controls (Fig. 4B). In contrast, in the Piasy WT background, the induction of Cxcl1, Junb, and Ip10 was not significantly affected by Pias1 disruption (Fig. 4A). These results indicate that PIAS1 has a distinct specificity in the regulation of NF-κB-mediated gene activation as compared with PIASy (e.g., on the induction of Cxcl1 gene), and the PIAS1 specificity is affected by the level of PIASy expression.
Fig. 4.
Analysis of the effect of single allele disruption of Piasy on PIAS1-mediated inhibition of TNF and IFN-γ-induced gene transcription. (A) Pias1+/+ and Pias1−/− MEFs were untreated or treated with TNF (20 ng/ml) for various time points. Total RNA was prepared and subjected to Q-PCR analyses. (B) Same as in A except Piasy+/−Pias1+/+ and Piasy+/−Pias1−/− MEF cells were used. (C) Same as in B except cells were treated with IFN-γ.
Similar experiments were performed to test whether the disruption of one Piasy allele affects the inhibitory function of PIAS1 in STAT1 signaling (Fig. 4C). The IFN-induced activation of Ip10, a previously known PIAS1-sensitive gene in the IFN pathway (14), was dramatically up-regulated in Pias1−/−Piasy+/− cells as compared with that in Pias1+/+Pias1y+/− control cells. Most importantly, the induction of genes that were previously identified as PIAS1-insensitive genes, such as Irf1 and Socs1 (14), was significantly enhanced in Pias1−/−Piasy+/− cells, as compared with the Pias1+/+Piasy+/− control cells. These data strongly suggest that the level of PIASy expression affects the specificity and magnitude of PIAS1-mediated inhibitory effect on NF-κB and STAT1 signaling.
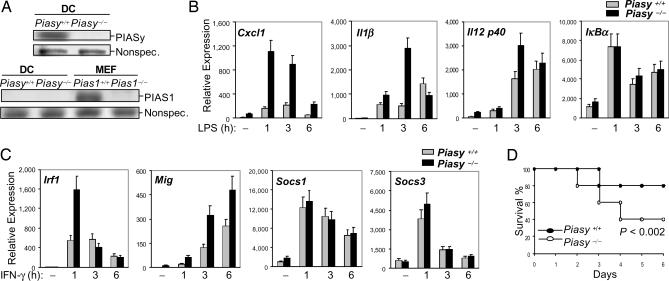
Enhanced NF-κB and STAT1-Dependent Gene Activation in Piasy-Null Dendritic Cells (DCs).
Based on our studies in MEFs described above, we predicted that PIASy should play a more critical role in the regulation of NF-κB and STAT1 signaling in certain types of cells where no/low PIAS1 is expressed. To test this hypothesis, we searched for the types of cells with no/low PIAS1 expression. As shown in Fig. 5A, PIASy is expressed in bone marrow-derived DCs (BMDCs), whereas PIAS1 protein is undetectable. To examine the role of PIASy in the regulation of NF-κB and STAT1 signaling in DCs, WT and Piasy−/− BMDCs, untreated or treated with LPS or IFN-γ for various time periods, were subjected to Q-PCR analyses. The transcriptional activation of NF-κB-target genes such as Cxcl1 and IL1β in response to LPS stimulation was dramatically enhanced in Piasy−/− DCs as compared with the WT controls (Fig. 5B). The transcriptional activation of IL-12 p40 was slightly increased, whereas the activation of IκBα was not altered in Piasy−/− DCs. Similarly, the transcriptional activation of STAT1-dependent genes such as Irf1 and Mig, but not Socs1 or Socs3, in response to IFN-γ was enhanced in Piasy−/− DCs as compared with the WT controls (Fig. 5C). These results further support the conclusion that PIASy is a physiologically important negative regulator of NF-κB and STAT1.
Fig. 5.
PIASy has an indispensable role in LPS and IFN signaling. (A) Cell extracts from Piasy+/+ and Piasy−/− DCs, or Pias1+/+ and Pias1−/− MEFs were subjected to Western blot analyses by using anti-PIASy (Left) or anti-PIAS1 (Right). (B) Piasy+/+ and Piasy−/− DCs were either untreated or treated with LPS (100 ng/ml) for various time points. Total RNA was subjected to Q-PCR analyses. (C) Same as in A except that cells were treated with IFN-γ (10 ng/ml). (D) Piasy−/− mice are hypersensitive to LPS-induced endotoxic shock. Piasy+/+ (filled circles) and Piasy−/− (open circles) littermates (n = 5) were i.p. injected with 20 μg per g of body weight LPS from E. coli serotype O55:B5 and monitored for survival. P was determined by nonpaired t test.
Piasy Null Mice Are Hypersensitive to LPS-Induced Endotoxic Shock.
Both NF-κB and STAT1 are known to play an important role in LPS signaling. To validate the physiological role of PIASy in the negative regulation of NF-κB and STAT1 signaling, Piasy null mice were subjected to LPS-induced endotoxic shock analysis. Piasy−/− mice, together with the matched WT controls, were challenged with a sublethal dose of LPS. At day 6 after LPS injection, whereas 80% of WT mice survived the LPS challenge, only 40% of Piasy null mice remained viable (Fig. 5D). These results are consistent with a role of PIASy in the negative regulation of LPS signaling.
PIASy Regulates the Promoter Recruitment of NF-κB-p65.
To understand the molecular mechanism of PIASy-mediated gene regulation, the effect of PIASy on various steps of NF-κB or STAT1 signaling was examined. Protein extracts were prepared from Pias1+/−Piasy+/+ and Pias1+/−Piasy−/− MEFs, untreated or treated with TNF or IFN-γ for various time periods. Western blot analyses with several signaling proteins in the NF-κB or STAT1 pathways indicated that PIASy does not affect the cytoplasmic signaling events of TNF or IFN (SI Fig. 7A). Similar experiments were performed with Pias1+/+Piasy+/− and Pias1−/−Piasy+/− MEFs after TNF or IFN stimulation. The results showed that Pias1 disruption had no effect on the ligand-induced nuclear translocation of NF-κB or STAT1 in the Piasy heterozygous background (SI Fig. 7B).
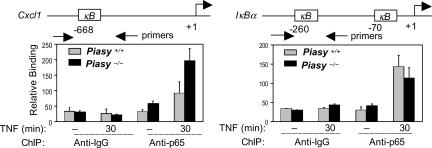
Next, we performed experiments to test whether PIASy regulates the ability of NF-κB-p65 to bind to the promoters of endogenous genes. Chromatin IP (ChIP) assays were performed with WT and Piasy null MEFs using anti-p65 Ab. The recruitment of p65 to the promoter of Cxcl1 (a PIASy-sensitive gene) after TNF stimulation was examined by Q-PCR analysis of the p65 immunoprecipitates by using primers that amplify the NF-κB binding region on the Cxcl1 promoter (Fig. 6). The binding of p65 to the Cxcl1 promoter was significantly enhanced in Piasy null cells as compared with the WT control. In contrast, the recruitment of p65 to the IκBα gene (a PIASy insensitive gene) promoter was not affected in the absence of PIASy. These findings are consistent the enhanced transcriptional induction of Cxcl1, but not IκBα, in Piasy null MEFs (Fig. 2). Thus, PIASy functions by regulating the recruitment of p65 to the endogenous gene promoters.
Fig. 6.
PIASy regulates the promoter recruitment of p65. ChIP assays were performed with WT and Piasy−/− MEFs, untreated or treated with TNF, followed by IP with anti-p65. The p65-immunoprecipitates were then analyzed by Q-PCR with specific primers as indicated.
Discussion
Studies described in this paper demonstrate that PIASy is a physiologically important negative regulator of NF-κB and STAT1. In addition, we show that the relative levels of PIAS1 and PIASy expression in a given cell type can affect the specificity and magnitude of NF-κB/STAT-mediated gene activation. Our studies support a hypothesis that NF-κB- and STAT-responsive genes can be regulated in subgroups by specific transcriptional regulators such as PIAS1 and PIASy. Understanding the specificity involved in the regulation of NF-κB/STAT-mediated gene activation is highly important because many NF-κB or STAT-induced genes have distinct or even opposite biological effects. Therefore, novel therapeutic strategies that target gene-specific regulators of NF-κB/STAT may prove to be more efficient for the treatment of human disorders.
PIASy originally was identified as a transcriptional repressor of STAT1 (11). However, initial characterization of Piasy null MEFs failed to show a significant effect of Piasy disruption on the induction of endogenous STAT1-dependent genes (16, 17). Studies described in this paper have now solved this puzzle: the lack of PIASy effect on STAT1-dependent genes in MEFs is due to a redundant role of PIAS1. Our studies indicate that PIASy has an indispensable role in the regulation of NF-κB/STAT1-mediated gene activation in cells that express low/no PIAS1, such as DCs. It remains to be determined whether additional PIAS1 low/no-expressing cell types may exist where PIASy plays an indispensable role. We noticed that even in DCs where PIAS1 expression is low, the induction of some NF-κB or STAT1-dependent genes was not significantly affected by Piasy disruption (Fig. 5). It is possible that the presence of additional regulators, including other members of the PIAS family, also may influence the activity of PIASy in DCs. Nevertheless, Piasy null mice are hypersensitive to the LPS-induced septic shock, consistent with an important role of PIASy as a negative regulator of NF-κB and STAT1.
In unstimulated cells, endogenous p65 is localized in the cytoplasm, whereas PIASy is present in the nucleus. Although p65 and PIASy interaction was detected in co-IP assays in the absence of ligand stimulation (Fig. 1B), this was most likely due to the presence of both PIASy and p65 in the same protein exacts under cell-free conditions. In intact cells, p65 can only interact with PIASy after ligand-induced nuclear translocation of p65.
Our studies indicate that PIASy and PIAS1 have overlapping but specific effects on gene regulation. For example, PIASy, but not PIAS1, is indispensable for the induction of Cxcl1 by TNF in MEFs (Figs. 2 and 4), whereas the removal of PIAS1, but not PIASy, causes the increased expression of Mip2 (15). It is possible that the specificity of PIASy and PIAS1 may be affected by the promoter microenvironment of individual genes. In addition, the activity of PIASy and PIAS1 may be regulated differently in response to cytokine stimulation. Further studies are needed to understand the molecular basis that accounts for PIAS1 and PIASy specificity in gene regulation.
A notable difference between PIASy and PIAS1 is that PIAS1 appears to have a more profound effect on the regulation of NF-κB/STAT-mediated gene activation than PIASy as measured by Q-PCR analyses. Correspondingly, we observed a more severe effect of Pias1 disruption than Piasy disruption on animal development (SI Tables 1 and 2). It is possible that the altered NF-κB/STAT1 signaling resulting from Pias1 and Piasy double disruption may contribute in part to the lethality of Pias1−/−Piasy−/− embryos. PIASy has been reported to regulate the activity of several other proteins (8, 18–21). It will be interesting to determine whether PIAS1 and PIASy may also have functional redundancy in the regulation of other signaling events.
Materials and Methods
Reagents.
The following Abs were used in the co-IP or Western blot analyses: anti-p65 (C-20; Santa Cruz Biotechnology, Santa Cruz, CA), anti-Flag (M2; Sigma, St. Louis, MO), anti-Myc (Cell Signaling Technology, Beverly, MA), anti-tubulin (Sigma), anti-IκBα (SC-371; Santa Cruz Biotechnology), and anti-PIAS1 and anti-PIASy (11). The following reagents were purchased: murine recombinant GM-CSF, TNF-α, and IFN-γ (PeproTech, Rocky Hill, NJ), and LPS (from Escherichia coli serotype O55:B5; Sigma). Anti-M9, anti-ORF26, and MHV-68 were obtained from R. Sun (University of California, Los Angeles).
Mice and Cells.
Mixed 129 × C57 background Piasy−/− and Pias1−/− mice have been described (14, 16). Compound heterozygous mice (Piasy+/−Pias1+/−) were obtained from the crossing of Piasy−/− and Pias1−/− mice and were used for compound crossing.
MEFs were obtained from embryonic day (E)11.5 littermate embryos and grown in 1× DMEM containing 10% FBS and 1% penicillin–streptomycin. BMDCs were obtained from bone marrow suspensions prepared from femurs and tibias of mice as described (22). Briefly, the bone marrow cells were plated in 10-cm culture plates at a density of 106 cells per ml in RPMI medium 1640 (GIBCO-BRL, Carlsbad, CA) supplemented with 10% FBS, 1% penicillin–streptomycin, and 20 ng/ml GM-CSF. After 5 days of culture, DC clusters were replated into new culture plates and subjected to cytokine stimulations and subsequent Q-PCR analyses.
Co-IP Assays.
Co-IP assays were performed as described (9).
Transient Transfection and Luciferase Assays.
293T cells were transfected by a calcium-phosphate procedure as described (23), and cell lysates were collected for luciferase assays (Promega, Madison, WI) 30 h after transfection. The relative luciferase units were corrected for relative expression of β-galactosidase.
Q-PCR Analysis.
Q-PCR was performed as described (14). Actin was used to standardize the levels of cDNA. The specific primers used in Q-PCR analyses have been described (14, 15), except for the following genes: Cxcl1, 5′-GCAGACCATGGCTGGGATT, 3′-GTGTGGCTATGACTTCGGTTTG; Socs3, 5′-GCTCCAAAAGCGAGTACCAGC, 3′-AGTAGAATCCGCTCTCCTGCA; and IL-12 p40, 5′-CCCATTCCTACTTCTCCCTCAA, 3′-CCTTTCTGGTTACACCCCTCCT.
ChIP Assay.
ChIP assays were performed by using the ChIP assay kit (Upstate Biotechnology, Lake Placid, NY) as instructed by the manufacturer. MEF cells (107) were either untreated or treated with TNF-α (20 ng/ml) for 30 min. Cell extracts were prepared, and chromatin was sheared by sonication (six 12-s pulses at 30% of the maximum strength). ChIP assays were performed with an anti-p65 Ab or rabbit IgG as a negative control. Bound DNA was quantified by Q-PCR and normalized with the input DNA. The sequences of the primers used are: Cxcl1 promoter, 5′-ATACAGCAGGGTAGGGATGC, 3′-TTGCCAACTGTTTTTGTGG; and IκBα promoter, 5′-GGACCCCAAACCAAAATC, 3′-GCCTGCACTCAGTAACCCCA.
Supplementary Material
Acknowledgments
We thank R. Sun for antibodies against MHV-68 viral proteins, S. Mink for discussion on MHV-68 infection, and C. Getman and N. Stein for technical assistance. This work was supported by National Institutes of Health grants (to K.S.). B.L. is supported by National Institutes of Health Research Scientist Development Award K01 AR52717-01.
Abbreviations
- co-IP
coimmunoprecipitation
- DC
dendritic cell
- BMDC
bone marrow-derived DC
- BMM
bone marrow-derived macrophage
- MEF
mouse embryonic fibroblast
- PIAS
protein inhibitor of activated STAT
- Q-PCR
quantitative real-time PCR.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701877104/DC1.
References
- 1.Ghosh S, Karin M. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 2.Li Q, Verma IM. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 3.Darnell JE, Jr, Kerr IM, Stark GR. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 4.Darnell JE., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 5.Shuai K, Liu B. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 6.Gilmore TD, Koedood M, Piffat KA, White DW. Oncogene. 1996;13:1367–1378. [PubMed] [Google Scholar]
- 7.Leonard WJ. J Invest Med. 1996;44:304–311. [PubMed] [Google Scholar]
- 8.Shuai K, Liu B. Nat Rev Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 9.Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, Shuai K. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 10.Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, Shuai K. Proc Natl Acad Sci USA. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Gross M, ten Hoeve J, Shuai K. Proc Natl Acad Sci USA. 2001;98:3203–3207. doi: 10.1073/pnas.051489598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson ES, Gupta AA. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 13.Sharrocks AD. Genes Dev. 2006;20:754–758. doi: 10.1101/gad.1421006. [DOI] [PubMed] [Google Scholar]
- 14.Liu B, Mink S, Wong KA, Stein N, Getman C, Dempsey PW, Wu H, Shuai K. Nat Immunol. 2004;5:891–898. doi: 10.1038/ni1104. [DOI] [PubMed] [Google Scholar]
- 15.Liu B, Yang R, Wong KA, Getman C, Stein N, Teitell MA, Cheng G, Wu H, Shuai K. Mol Cell Biol. 2005;25:1113–1123. doi: 10.1128/MCB.25.3.1113-1123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong KA, Kim R, Christofk H, Gao J, Lawson G, Wu H. Mol Cell Biol. 2004;24:5577–5586. doi: 10.1128/MCB.24.12.5577-5586.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth W, Sustmann C, Kieslinger M, Gilmozzi A, Irmer D, Kremmer E, Turck C, Grosschedl R. J Immunol. 2004;173:6189–6199. doi: 10.4049/jimmunol.173.10.6189. [DOI] [PubMed] [Google Scholar]
- 18.Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R. Genes Dev. 2001;15:3088–3103. doi: 10.1101/gad.944801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long J, Matsuura I, He D, Wang G, Shuai K, Liu F. Proc Natl Acad Sci USA. 2003;100:9791–9796. doi: 10.1073/pnas.1733973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bischof O, Schwamborn K, Martin N, Werner A, Sustmann C, Grosschedl R, Dejean A. Mol Cell. 2006;22:783–794. doi: 10.1016/j.molcel.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Mabb AM, Wuerzberger-Davis SM, Miyamoto S. Nat Cell Biol. 2006;8:986–993. doi: 10.1038/ncb1458. [DOI] [PubMed] [Google Scholar]
- 22.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.