Abstract
The enzyme hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase (HCT) is involved in the production of methoxylated monolignols that are precursors to guaiacyl and syringyl lignin in angiosperm species. We identified and cloned a putative HCT gene from Pinus radiata, a coniferous gymnosperm that does not produce syringyl lignin. This gene was up-regulated during tracheary element (TE) formation in P. radiata cell cultures and showed 72.6% identity to the amino acid sequence of the Nicotiana tabacum HCT isolated earlier. RNAi-mediated silencing of the putative HCT gene had a strong impact on lignin content, monolignol composition, and interunit linkage distribution. AcBr assays revealed an up to 42% reduction in lignin content in TEs. Pyrolysis-GC/MS, thioacidolysis, and NMR detected substantial changes in lignin composition. Most notable was the rise of p-hydroxyphenyl units released by thioacidolysis, which increased from trace amounts in WT controls to up to 31% in transgenics. Two-dimensional 13C-1H correlative NMR confirmed the increase in p-hydroxyphenyl units in the transgenics and revealed structural differences, including an increase in resinols, a reduction in dibenzodioxocins, and the presence of glycerol end groups. The observed modifications in silenced transgenics validate the targeted gene as being associated with lignin biosynthesis in P. radiata and thus likely to encode HCT. This enzyme therefore represents the metabolic entry point leading to the biosynthesis of methoxylated phenylpropanoids in angiosperm species and coniferous gymnosperms such as P. radiata.
Keywords: lignin, HCT, tracheary elements
The global trend toward a biomaterials-based economy makes plant cell walls increasingly important as renewable resources for the production of biofuels and biocomposites. Lignin is the second most abundant terrestrial biopolymer after cellulose and a major structural component of cell walls in wood-forming tissues (1). The content, composition, and structure of lignin all have considerable impact on the utilization of plant-derived materials and have therefore been the subject of intensive research (1, 2). Lignins are heterogeneous cell wall polymers derived primarily from hydroxycinnamyl alcohols via combinatorial radical coupling reactions (3). Typically, they make up 20–30% of the cell wall material in woody tissue of both angiosperm and gymnosperm species. Lignin in coniferous gymnosperms such as Pinus radiata does not contain syringyl (S) components, which makes it different from lignin of many other vascular plants including angiosperms (4).
We have developed a P. radiata callus culture system to better assign function to genes associated with cell wall-related processes such as lignification in conifers. These callus cultures can be transformed and subsequently induced to differentiate into tracheary elements (TEs), the main cellular components of wood in conifer species (5). The biochemical composition of cell wall polymers in differentiated TEs is similar to those produced in P. radiata wood (6). This culture system is also more rapid and efficient for the generation of transgenics than plant-forming somatic embryos (5). The activation of the phenylpropanoid pathway in those callus cultures via feeding experiments has already proven useful for the investigation of lignin-related genes (7). However, the usefulness of the TE differentiation process per se in identifying the function of cell wall-related genes still needs to be verified.
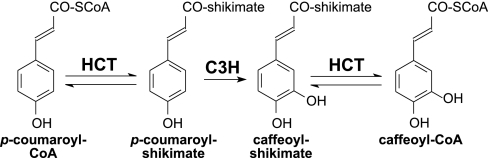
A key step in the biosynthesis of lignin in angiosperm species is catalyzed by the recently discovered enzyme hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase (HCT) (8). This enzyme plays a crucial role in the biosynthesis of monolignols by catalyzing the transfer of coumarate from p-coumaroyl-CoA to shikimate, resulting in the formation of the corresponding p-coumaroyl shikimate ester (Fig. 1). It is this ester rather than the previously accepted CoA-thioester that is used by p-coumarate 3-hydroxylase (C3H) to functionalize the aromatic ring toward guaiacyl (G)- and S-units (8). Silencing of HCT in Nicotiana benthamiana and Medicago sativa caused a reduction of both G- and S-type lignin subunits and a substantial increase in p-hydroxyphenyl (H) subunits, which confirmed its involvement in the production of methoxylated phenylpropanoids in angiosperms (9, 10). Here, we describe the isolation of a putative P. radiata HCT clone from a xylem-derived cDNA library and the effects of its silencing in TE-forming P. radiata callus cultures via RNAi. Our results demonstrate that this gene is required for the biosynthesis of G-type lignin in coniferous gymnosperms such as P. radiata.
Fig. 1.
Enzymatic function of HCT in the phenylpropanoid pathway in tobacco (8).
Results
Clone Isolation and Characterization.
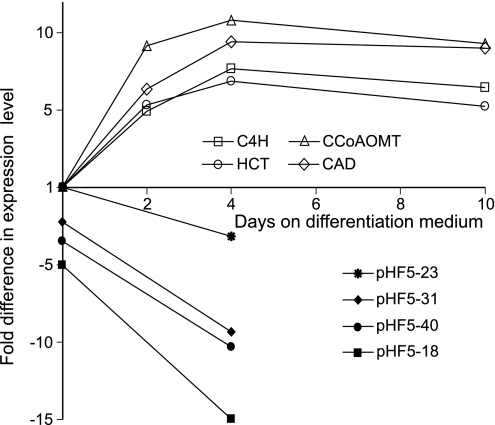
A 1,302-bp fragment of a putative HCT clone was isolated from a xylem-derived P. radiata cDNA library by using the PCR-based approach described in Materials and Methods. The deduced amino acid sequence of the coding region showed 72.6% identity to the amino acid sequence of the Nicotiana tabacum HCT clone NtHCT [supporting information (SI) Fig. 5]. The degree of amino acid similarity between the two protein sequences was 82.9%. All conserved motifs typical for plant acyltransferases (8) were present in the P. radiata clone. The expression level of the pine gene was also monitored during TE differentiation by using quantitative RT-PCR. The putative HCT gene was up-regulated during TE formation similar to other lignin-related genes such as cinnamate 4-hydroxylase (C4H), caffeoyl-CoA O-methyltransferase (CCoAOMT), and cinnamyl alcohol dehydrogenase (CAD) tested in this experiment (Fig. 2).
Fig. 2.
Steady-state mRNA pool of the lignin-related P. radiata genes C4H, HCT, CCoAOMT, and CAD during TE formation as assessed by quantitative RT-PCR at days 0, 2, 4, and 10 after initiation of the differentiation process compared with the HCT steady-state mRNA pool in transgenics pHF5-18, pHF5-23, pHF5-31, and pHF5-40 at days 0 and 4 after initiation of the differentiation process relative to WT controls.
Generation and Molecular Analysis of Transgenic Lines.
A total of 56 transgenic lines was generated through biolistic cobombardment using plasmids pAW16 and pHF5 (SI Fig. 6). Previous transformation experiments with pAW16 have established that the transformation process itself does not affect content or chemical composition of lignin in TEs (A.W., unpublished results). The presence of the HCT RNAi cassette in transgenic lines was tested via genomic PCR. Seventy-eight percent of all transgenics contained both PCR fragments of the HCT RNAi construct (data not shown). Pyrolysis-GC/MS was used to produce a chemical fingerprint of cell wall material generated after the differentiation process in transgenic and nontransgenic lines. Four transgenic lines, which exhibited substantial changes in lignin-derived signals compared with the WT control (data not shown), were chosen for further analysis. Quantitative RT-PCR experiments with those transgenic lines revealed that two of the four transgenics contained as little as 7–10% residual HCT message at day 4 on differentiation medium. The HCT steady-state mRNA levels in those transgenics were declining during early stages of TE formation, which contrasted with the situation in the WT control (Fig. 2).
Phenotypic Analysis of Transgenic Lines.
Analysis of purified TEs revealed reductions in AcBr-soluble lignin of up to 42% in silenced transgenics (Table 1). Pyrolysis-GC/MS with purified TEs indicated a strong increase in H lignin and a simultaneous reduction in G lignin in HCT-deficient lines (SI Fig. 7). Thioacidolysis, which selectively cleaves lignin β-ether linkages in lignin, released monomers, which were up to 31% of the H type (Table 1). This indicated a >50-fold increase in the H component compared with WT controls.
Table 1.
AcBr lignin and thioacidolysis yield of H/G monomers from WT and HCT-deficient TEs
| Sample | AcBr lignin, % wt/wt | H, μmol/g lignin | G, μmol/g lignin | %H |
|---|---|---|---|---|
| WT | 28.9 ± 0.8 | Trace | 1,886 | 0 |
| pHF5-18 | 22.9 ± 0.2 | 340 | 769 | 31 |
| pHF5-23 | 18.2 ± 0.1 | 189 | 1,149 | 14 |
| pHF5-40 | 16.7 ± 0.1 | 232 | 560 | 29 |
pHF5-18, -23, and -40 are the HCT-deficient transgenics.
For NMR analysis, so-called enzyme lignins enriched in lignin by removing a substantial proportion of the polysaccharides by using polysaccharidases were generated (11). These lignins were then dissolved in a cell wall dissolution system (12), acetylated, and subsequently used for structural analysis. Two-dimensional NMR dispersion is sufficient in that remaining polysaccharides interfere only minimally in the lignin analysis (12).
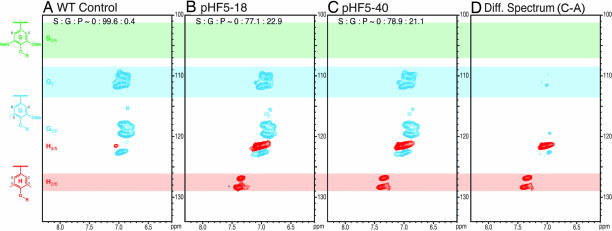
Aromatic region: H/G distribution.
Changes in the H/G distribution in the lignins were visualized from the aromatic region of NMR spectra, particularly the 2D 13C–1H correlation [heteronuclear single-quantum coherence (HSQC)] spectra correlating protons with their attached carbons. H and G aromatic resonances were well separated at 500 MHz. Substantial differences in the aromatic nature of the lignins in the WT (Fig. 3A) and the HCT-deficient lines (Fig. 3 B–D) were revealed. Wild-type TE lignins were G-rich with H components derived from p-coumaryl alcohol visible only at low-level contours (H3/5 contour in Fig. 3A). The lignin from the HCT-deficient transgenics had strikingly higher H levels, as seen in Fig. 3 B–D. Volume integration (Table 2) allowed reasonable quantification of the differences. The integrals suggested that the H-unit content was elevated by >50-fold, consistent with the thioacidolysis data.
Fig. 3.
Partial short-range 13C–1H (HSQC) correlation spectra (aromatic regions only) of acetylated enzyme lignins isolated from the WT control (A), the HCT-deficient line pHF5-18 (B), the HCT-deficient line pHF5-40 (C), and a difference spectrum revealing that the major relative difference is in enhanced H-units (D). Traces of H-units are seen in the typically G lignin in the WT tracheids, whereas H-units are strikingly more prevalent in the spectra of the transgenics.
Table 2.
NMR-derived H/G and interunit linkage data for WT and HCT-deficient P. radiata TE-acetylated enzyme lignins
| Sample | %H | %G | H/G | %A | %B | %C | %D | %X1 | %X7 | Σ β–O–4 |
|---|---|---|---|---|---|---|---|---|---|---|
| WT | 0.4 | 99.6 | 0.4 | 58.9 | 17.5 | 8.7 | 1.4 | 13.6 | 0 | 60.3 |
| pHF5-18 | 22.9 | 77.1 | 29.8 | 61.3 | 15.8 | 11.1 | 0.5 | 8.9 | 2.4 | 61.8 |
| pHF5-40 | 21.1 | 78.9 | 26.8 | 55.3 | 15.9 | 11.5 | 0 | 16.3 | 1.0 | 55.3 |
pHF5-18 and pHF5-40 are the HCT-deficient transgenics. A, β–O–4 (β-aryl ether); B, β–5 (phenylcoumaran); C, β–β (resinol); D, β–O-4/5–5 (dibenzodioxocin); X1, cinnamyl alcohol endgroup; X7, arylglycerol end group (see Fig. 4 for structures).
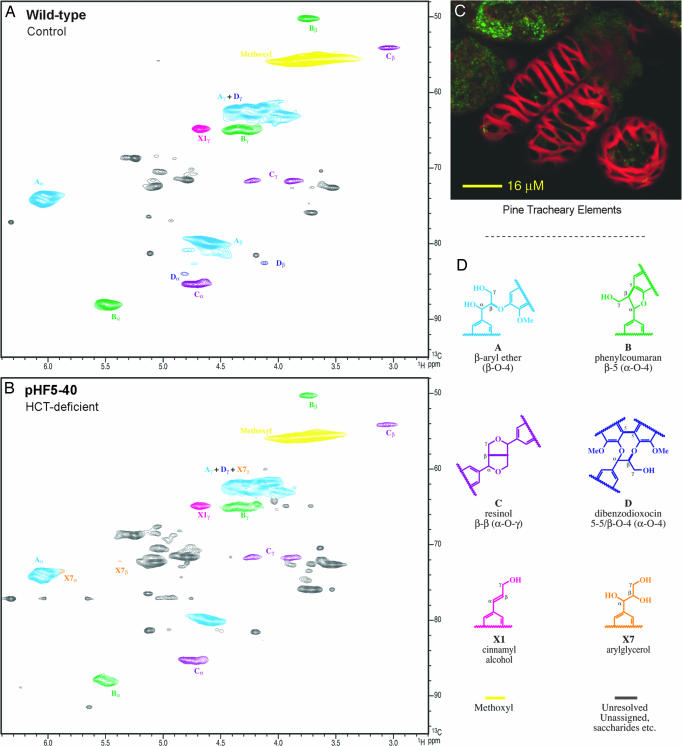
Side-chain region: Interunit linkage and end-group distribution.
The side-chain region peripherally reflected the changes in the H/G distribution, and was rich in detail regarding the types and distribution of interunit bonding patterns present in the lignin fraction. The control lignin spectrum (Fig. 4A) was typical of a G lignin containing residual polysaccharides (13, 14). The HSQC spectrum resolved most of the correlations for the various linkage types in the polymer, the exception being in the complex γ-region, where only the correlations from the phenylcoumarans B and the cinnamyl alcohol end groups X1 were fully resolved. The lignin was seen as being rich in β-aryl ether units A, with substantial amounts of phenylcoumarans B, resinols C, and cinnamyl alcohol end groups X1.
Fig. 4.
Partial short-range 13C–1H (HSQC) spectra (side-chain regions) of acetylated enzyme lignins isolated from P. radiata TEs. (A) WT control. (B) HCT-deficient line pHF5-40. HCT deficiency and the incorporation of higher levels of p-coumaryl alcohol into the lignin produce changes in the distribution of interunit linkage types (see text). Note that the contour levels used to display the two spectra were chosen to highlight the structural similarities and differences. With no internally invariant peaks, interpretation of apparent visual quantitative differences needs to be cautious. Volume integrals and semiquantitative data are given in Table 2. The analogous spectrum for line pHF5-18 is provided in SI Fig. 8. (C) Photograph shows the appearance of P. radiata TEs. (D) Interunit type designations A–D, X1, and X7 follow conventions established previously (1, 3, 15).
Lignins from HCT-deficient transgenics had spectra with several differences (Fig. 4B, Table 2, and SI Fig. 8). Glycerol end groups X7 were apparent and the dibenzodioxocin level was reduced. Relative levels of the other units changed marginally as well, there being fewer phenylcoumarans B but more resinols C in the transgenics.
Discussion
This study has demonstrated that HCT has a crucial role in determining lignin content, monolignol composition, and interunit linkage distribution in TE-forming P. radiata callus cultures.
HCT silencing caused a reduction in lignin content in all species tested so far (refs. 9 and 10; this study). Whereas lignin was reduced by 15% in N. benthamiana (9), both M. sativa and P. radiata showed more substantial lignin reductions of ≈50 and 42%, respectively (ref. 10; this study). These quantitative differences might reflect the different silencing technologies used in the individual studies. Virus-induced gene silencing was used to silence HCT in N. benthamiana, whereas transgenic antisense or RNAi were used in the other two studies. Virus-induced gene silencing is unable to alter lignin deposited before virus infection and is therefore more limited in elucidating quantitative variation. Nonetheless, HCT silencing is likely to have a substantial impact on lignin quantity in both angiosperm and pine species. Silencing of HCT also had considerable impact on the monolignol distribution in deposited lignins. Most notable was the rise in H-type lignin, which was observed in both angiosperms tested (9, 10). Here, a >50-fold increase in H-units was identified by using both thioacidolysis and 2D NMR. NMR-derived H/G ratios (Table 2) were consistent with the ratios derived from thioacidolysis (Table 1), despite the fact that thioacidolysis measures only the distribution in the monomers that can be released by cleaving β-ether bonds. The increase in H-units was at the expense of G-units in P. radiata (Table 1). This relationship was also observed in M. sativa, whereas HCT silencing in N. benthamiana preferentially depleted S-type lignin (9). The depletion of G-type lignin in M. sativa and P. radiata establish that HCT is essential for the production of coniferyl alcohol in both angiosperm and coniferous gymnosperm species. The results in tobacco might again represent a somewhat distorted picture based on virus-induced gene silencing, and stable HCT silencing should be used to verify the original phenotype.
Perturbing the monolignol composition also perturbs lignin structure, producing lignins with a different distribution of unit types. Two-dimensional 13C–1H NMR proved to be particularly useful to reveal those changes in HCT-deficient transgenics. Lignins are characterized by various types of interunit bonds, the most prominent being labeled A–D, and side-chain end units X1 and X7 (Fig. 4). This linkage-type distribution differed between the WT and the HCT-deficient P. radiata lignins. In particular, G dibenzodioxocins D appeared to be depleted. In previously examined lignins from C3H-deficient M. sativa plants, which contained up to ≈65% H-units, new dibenzodioxocins involving 5-coupled H-units were discovered at enhanced levels (15). Such dibenzodioxocins were not observed here, presumably because p-coumaryl alcohol is at more modest levels, coupling mainly to G-units in an H/G-copolymer. β-Aryl ether A levels were similar, whereas phenylcoumarans B were slightly depleted and resinols C were enhanced. Quantities of cinnamyl alcohol end groups X1 were the most variable, and no conclusions are drawn here. Finally, the glycerol structures X7 were present at substantially higher levels. Such structures have also been found to be more prevalent in the H-rich lignins from C3H-deficient M. sativa plants (15). Oxidative coupling reactions using p-coumaryl alcohol produce substantial levels of glycerols in synthetic polymers that have not been ball milled (15), so we suspect that they may be, at least in part, authentic units in the native lignins.
Some features of the lignins in P. radiata TEs deserve further comment. In pine lignins, cinnamyl end groups X1 and resinols C each typically comprise only ≈2% of the units (16). Both types of units are signatures of monomer dehydrodimerization and are consequently minor in softwood lignification. The main lignin polymerization pathway in softwoods is the so-called “end-wise” cross-coupling (17) of coniferyl alcohol (at its β-position) with the phenolic end of the growing polymer (at its 5- or 4-O-positions). Lignification in trees appears to occur under conditions where the monolignol radical concentrations are limiting such that cross-coupling with the growing polymer is more prevalent (3). The levels of C and X1 in these spectra suggest that the lignins in TEs are formed by a more “bulk” type polymerization in which monomer dehydrodimerization is more frequent than “end-wise” polymerization. S resinols C are seen at similarly high levels in angiosperms (14). This might also suggest that radical concentrations are not as limiting in the TE system as they are in developing wood.
Conclusions
This first study on HCT manipulation in softwoods establishes this enzyme as the metabolic entry point leading to the biosynthesis of G-lignin in coniferous gymnosperms such as P. radiata. HCT silencing significantly affected lignin content and monomer distribution, which in turn perturbed lignin structure. Such structural changes are consistent with the metabolic plasticity inherent in the existing theory of lignification in which monolignols are cross-coupled onto the growing polymer in a chemically controlled fashion, independent of proteinaceous control (3). The metabolic plasticity of the lignification process allows the lignin component to be widely varied in composition and structure, providing potential avenues toward improved feedstocks for biomaterials and biomass conversion processes. Finally, this study provides additional validation for the P. radiata TE system as a model to obtain relevant information on the biological role of cell wall-related genes.
Materials and Methods
Clone Isolation, Construct Design, and Transformation of Callus Cultures.
The coding region of a putative P. radiata HCT cDNA clone was isolated from a xylem-derived cDNA library either by using the primer pairs HCTfwd1, 5′-GC-GGATCC-ATGCTTATAACAGTGAAGAATTCGC, and HCTrev1, 5′-CT-GCGGCCGCC-TAGATTTCATACAACATTTTGGCA; or HCTfwd2, 5′-CG-CCCGGG-ATGCTTATAACAGTGAAGAATTCGC, and HCTrev2, 5′-CG-AGATCT-CTAGATTTCATACAACATTTTGGCA, respectively. The amplified 1,302-bp PCR fragments of the putative HCT cDNA clone were sequenced and cloned SmaI, BglII (antisense orientation) and BamHI, NotI (sense orientation) into a derivative of pAHC25 (18) by using the restriction sites contained in the primers (above in bold). The resulting plasmid containing the HCT RNAi construct was sequenced and named pHF5 (SI Fig. 6). P. radiata callus cultures were cotransformed with pHF5 and pAW16. Plasmid pAW16 contains an NPT II selection cassette and a GUS reporter gene cassette (SI Fig. 6). Plasmid pAW16 was generated by replacing the 35S promoter of the NPT II cassette in pCW122 (19) with the Zea mays ubi1 promoter from pAHC25. All transformation procedures were performed as described earlier (5).
Tissue Culture Procedures.
P. radiata callus cultures were maintained and induced to differentiate TEs as described earlier (20). For TE purification, differentiated callus cultures (≈3 g) were resuspended in Mops-KOH buffer (20 mM; pH 6.8; 10 ml), containing 20 mM sodium metabisulfite and homogenized by using a Retsch (Newtown, PA) PM200 ball mill for 20 min (vibration frequency, 1,800 min−1) in Teflon vials (25 ml) containing Teflon beads (2 × 8 mm). The homogenate was transferred into a sterile 50-ml tube, washed with 40 ml of Mops-KOH buffer five times, and filtered through a 40-μm nylon mesh. The cells were collected from the filter and resuspended in 25 ml of buffer, and TEs were separated from residual nondifferentiated cells by differential sedimentation. The proportion of TEs was determined by counting the TEs and the total number of cells on 10 samples prepared after the enrichment procedure.
Molecular Analysis of TE Cultures.
The integration of the HCT RNAi construct into the genome was monitored by genomic PCR using the primer pair M13fwd, 5′-CCCAGTCACGACGTTGTAAAACG, and HCTfwd1 (as above), as well as the primer pair HCTfwd1 (as above) and NOS-terrev2, 5′-ATTGCCAAATGTTTGAACGA. The expression of the lignin-related genes C4H, CCoAOMT, CAD, and the putative HCT gene were assessed by quantitative RT-PCR as described earlier (21). The following primers were used for quantitative RT-PCR experiments: for C4H: C4Hfwd, 5′-GCTTCAGGCTATTGGTTTGG, and C4Hrev, 5′-AGGAGCTGCCCTGGAATTAT; for HCT: HCTfwd, 5′-AGCAGTCCTCAGTCTCATCATAA, and HCTrev, 5′-TAATGACTACGCCCAATTACAGG; for CCoAOMT: CCoAOMTfwd, 5′-TTGCAGGCGTGTCTATTGAAAACAATC, and CCoAOMTrev, 5′-CAAATGGCTTCAACCCCATA; and for CAD: CADfwd, 5′-GTCCGTTACAGATTTGTGGTGGATGTT, and CADrev, 5′-GGCAGAAGACATTGGATTATTATTT.
Pyrolysis-GC/MS.
Pyrolysis-GC/MS was carried out on powdered, freeze-dried purified TEs as described in ref. 5. Thermal breakdown products were identified by using mass spectra of lignin and polysaccharide-derived pyrolysis products (22–25).
Thioacidolysis.
Analytical thioacidolysis of purified TEs from WT controls and silenced HCT transgenics was carried out by using the standard method (26), except reactions were performed at 100°C for 2 h as described by Pasco and Suckling (27).
AcBr Lignin Assay.
TEs were analyzed for AcBr-soluble lignin according to the procedure of Fukushima and Hatfield (28) with the following modifications. Powdered, freeze-dried TEs (2–4 mg) were dissolved in 25% AcBr in glacial acetic acid (0.5 ml) by warming to 50°C for 2 h. After cooling, 2 ml of 2 M NaOH solution, 2.4 ml of glacial acetic acid, and 0.35 ml of 0.5 M hydroxylamine hydrochloride solution were added. The contents of each tube were transferred quantitatively to a 25-ml volumetric flask, and the volume was adjusted to 25 ml with acetic acid. Absorbance was measured at 280 nm on a Varian (Palo Alto, CA) Cary 300 Bio UV-visible spectrophotometer 20 min after final dilution. The concentration of AcBr-soluble lignin was calculated by using an extinction coefficient of 20.0 liter·g−1·cm−1 according to Iiyama and Wallis (29).
NMR Spectroscopy.
Cell wall preparation and ball milling.
Isolated TEs were extracted with methanol (sonnication, 20 min, three times). Isolated cell walls (160, 285, and 112 mg for WT, HCT-40, and HCT-18) were ball milled for 20 min by using a Retsch PM100 ball mill vibrating at 600 min−1 with zirconium dioxide vessels (50 ml) containing ZrO2 ball bearings (3 × 30, 7 × 10 mm).
Crude cellulases digestion.
The ball-milled walls were transferred to centrifuge tubes (50 ml) and digested at 30°C with crude cellulases (Cellulysin; Calbiochem, San Diego, CA; lot no. B23109; 30 mg/g of sample, in pH 5.0 acetate buffer; three times 2 days; fresh buffer and enzyme each time) leaving all of the lignin and residual polysaccharides totaling 44 mg (28% of the original cell wall, WT), 90 mg (32%, HCT-40), and 36 mg (32%, HCT-18) (11, 30).
Enzyme lignin dissolution.
The polysaccharidase-digested cell wall fraction was subjected to solubilization in DMSO/N-methylimidazole (2/1 ml) (12). After acetylation (acetic anhydride, 0.5 ml, 1.5 h) and precipitation into water, the polysaccharidase-digested cell walls (38, 83, and 30 mg from WT, HCT-40, and HCT-18) yielded 45, 82, and 39 mg of acetylated product.
Spectroscopic procedures.
The NMR spectra presented here were acquired on a Bruker Biospin (Rheinstetten, Germany) DMX-500 instrument fitted with a sensitive cryogenically cooled 5-mm TXI 1H/13C/15N gradient probe with inverse geometry (proton coils closest to the sample). Acetylated lignin preparations (20–40 mg) were dissolved in 0.5 ml of CDCl3; the central chloroform solvent peak was used as internal reference (δC, 77.0; δH, 7.26 ppm). HSQC experiments (Figs. 3 and 4) had the following parameters: acquired from 8.6–2.4 ppm in F2 (1H) by using 1,236 data points (acquisition time, 200 ms), 158–40 ppm in F1 (13C) by using 512 increments (F1 “acquisition time” 17.2 ms) of 16 scans with a 1-s interscan delay, total acquisition time of 2 h and 55 min; the d24 delay was set to 1.72 ms (≈1/4J). Processing used typical matched Gaussian apodization in F2 and squared cosine-bell in F1. Volume integration of contours in HSQC plots was accomplished by using Bruker's TopSpin 1.3 software as described recently (31). For quantification of H/G distributions (Fig. 3), only the carbon-2 correlations from G-units, and the carbon-2/6 or 3/5 correlations from H-units were used, and the G integrals were doubled. No correction factors were used. We have been unable to develop satisfactory methods to establish any necessary “response factors” between H, G, and S correlation contour integrals. For pseudoquantification of the various interunit linkage types (Table 2), the following well-resolved contours (Fig. 4) were integrated: Aα, Bα, Cα, Dα, X7β, and X1γ. Integral correction factors determined previously (31) were used.
Supplementary Material
Acknowledgments
We thank Diane Steward, Lloyd Donaldson, Nigel Lee, and John Smith for technical assistance and Elspeth MacRae and Tim Strabala for critical reading of this manuscript. This work was partially supported by New Zealand Foundation for Research, Science, and Technology Contract C04X0207 and Department of Energy, Energy Biosciences Program Grant DE-AI02-00ER15067 (to J.R.). NMR experiments on the Bruker DMX-500 cryoprobe system made use of the National Magnetic Resonance Facility (Madison, WI).
Abbreviations
- TE
tracheary element
- HCT
hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyltransferase
- C3H
p-coumarate 3-hydroxylase
- G
guaiacyl
- S
syringyl
- H
p-hydroxyphenyl
- C4H
cinnamate 4-hydroxylase
- CCoAOMT
caffeoyl-CoA O-methyltransferase
- CAD
cinnamyl alcohol dehydrogenase
- HSQC
heteronuclear single-quantum coherence.
Footnotes
This article is a PNAS Direct Submission.
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. EF121452).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701428104/DC1.
References
- 1.Boerjan W, Ralph J, Baucher M. Ann Rev Plant Biol. 2003;54:519–549. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 2.Boudet AM, Kajita S, Grima-Pettenati J, Goffner D. Trends Plants Sci. 2003;8:576–581. doi: 10.1016/j.tplants.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH. Phytochem Rev. 2004;3:29–60. [Google Scholar]
- 4.Harris PJ. In: Plant Diversity and Evolution: Genotypic and Phenotypic Variation in Higher Plants. Henry RJ, editor. Wallingford, UK: CAB; 2005. pp. 201–227. [Google Scholar]
- 5.Möller R, McDonald AG, Walter C, Harris PJ. Planta. 2003;217:736–747. doi: 10.1007/s00425-003-1053-0. [DOI] [PubMed] [Google Scholar]
- 6.Möller R, Koch G, Nanayakkara B, Schmitt U. Tree Physiol. 2005;26:201–210. doi: 10.1093/treephys/26.2.201. [DOI] [PubMed] [Google Scholar]
- 7.Möller R, Steward D, Phillips L, Flint H, Wagner A. Plant Physiol Biochem. 2005;43:1061–1066. doi: 10.1016/j.plaphy.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M. J Biol Chem. 2003;278:95–103. doi: 10.1074/jbc.M209362200. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, Pollet B, Legrand M. Plant Cell. 2004;16:1446–1465. doi: 10.1105/tpc.020297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen F, Reddy MSS, Temple S, Jackson L, Shadle G, Dixon RA. Plant J. 2006;48:113–124. doi: 10.1111/j.1365-313X.2006.02857.x. [DOI] [PubMed] [Google Scholar]
- 11.Obst JR, Kirk TK. In: Methods in Enzymology. Biomass Part B: Lignin, Pectin, and Chitin. Wood WA, Kellogg ST, editors. London: Elsevier; 1988. pp. 3–12. [Google Scholar]
- 12.Lu F, Ralph J. Plant J. 2003;35:535–544. doi: 10.1046/j.1365-313x.2003.01817.x. [DOI] [PubMed] [Google Scholar]
- 13.Ralph J, Lu F. Org Biomol Chem. 2004;19:2714–2715. doi: 10.1039/B412633E. [DOI] [PubMed] [Google Scholar]
- 14.Ralph J, Marita JM, Ralph SA, Hatfield RD, Lu F, Ede RM, Peng J, Quideau S, Helm RF, Grabber JH. In: Advances in Lignocellulosics Characterization. Argyropoulos DS, Rials T, editors. Atlanta: TAPPI; 1999. pp. 55–108. [Google Scholar]
- 15.Ralph J, Akiyama T, Kim H, Lu F, Schatz PF, Marita JM, Ralph SA, Reddy MSS, Chen F, Dixon RA. J Biol Chem. 2006;281:8843–8853. doi: 10.1074/jbc.M511598200. [DOI] [PubMed] [Google Scholar]
- 16.Adler E. Wood Sci Technol. 1977;11:169–218. [Google Scholar]
- 17.Sarkanen KV, Hergert HL. In: Lignins. Occurrence, Formation, Structure, and Reactions. Sarkanen KV, Ludwig CH, editors. New York: Wiley–Interscience; 1971. pp. 43–94. [Google Scholar]
- 18.Christensen AH, Sharrock RA, Quail PH. Plant Mol Biol. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- 19.Walter C, Grace LJ, Wagner A, White DWR, Walden AR, Donaldson SS, Hinton H, Gardner RC, Smith DR. Plant Cell Rep. 1998;17:460–468. doi: 10.1007/s002990050426. [DOI] [PubMed] [Google Scholar]
- 20.Möller R, Ball RD, Henderson AR, Modzel G, Find J. Plant Cell Tissue Organ Cult. 2006;85:161–171. [Google Scholar]
- 21.Cato S, McMillan L, Donaldson L, Richardson T, Echt C, Gardner R. Plant Mol Biol. 2005;60:565–581. doi: 10.1007/s11103-005-5022-9. [DOI] [PubMed] [Google Scholar]
- 22.Faix O, Meier D, Fortmann I. Holz Roh Werkstoff. 1990;48:281–285. [Google Scholar]
- 23.Faix O, Fortmann I, Bremer J, Meier D. Holz Roh Werkstoff. 1991;49:213–219. [Google Scholar]
- 24.Faix O, Fortmann I, Bremer J, Meier D. Holz Roh Werkstoff. 1991;49:299–304. [Google Scholar]
- 25.Ralph J, Hatfield RD. J Agric Food Chem. 1991;39:1426–1437. [Google Scholar]
- 26.Rolando C, Monties B, Lapierre C. In: Methods in Lignin Chemistry. Dence CW, Lin SY, editors. Berlin: Springer; 1992. p. 335. [Google Scholar]
- 27.Pasco MF, Suckling ID. Holzforschung. 1994;48:504–508. [Google Scholar]
- 28.Fukushima RS, Hatfield RD. J Agric Food Chem. 2001;49:3133–3139. doi: 10.1021/jf010449r. [DOI] [PubMed] [Google Scholar]
- 29.Iiyama K, Wallis AFA. Wood Sci Technol. 1988;22:271–280. [Google Scholar]
- 30.Hu Z, Yeh T-F, Chang H-M, Matsumoto Y, Kadla JF. Holzforschung. 2006;60:389–397. [Google Scholar]
- 31.Ralph SA, Landucci LL, Ralph J. NMR Database of Lignin and Cell Wall Model Compounds. Washington, DC: Department of Agriculture; 2004. Available at http://ars.usda.gov/Services/docs.htm?docid=10491. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.