Abstract
We report a chemical derivatization method that selects a class of metabolites from a complex mixture and enhances their detection by 13C NMR. Acetylation of amines directly in aqueous medium with 1,1′-13C2 acetic anhydride is a simple method that creates a high sensitivity and quantitative label in complex biofluids with minimal sample pretreatment. Detection using either 1D or 2D 13C NMR experiments produces highly resolved spectra with improved sensitivity. Experiments to identify and compare amino acids and related metabolites in normal human urine and serum samples as well as in urine from patients with the inborn errors of metabolism tyrosinemia type II, argininosuccinic aciduria, homocystinuria, and phenylketonuria demonstrate the method. The use of metabolite derivatization and 13C NMR spectroscopy produces data suitable for metabolite profiling analysis of biofluids on a time scale that allows routine use. Extension of this approach to enhance the NMR detection of other classes of metabolites has also been accomplished. The improved detection of low-concentration metabolites shown here creates opportunities to improve the understanding of the biological processes and develop improved disease detection methodologies.
Keywords: carbon-13, inborn errors of metabolism, metabolite profiling, metabolomics, metabonomics
The metabolic profile in biofluids represents a snapshot of ongoing biological processes in the human body. The presence of a particular metabolite, panel of metabolites, or a certain ratio of metabolites can indicate normal homeostasis, a response to biological stress, or even a specific disease state. Conventional medical diagnostic methods are typically based on the selective detection of a single or a few biochemical parameters that are associated with a given disease. A major challenge in the present practice of clinical medicine is the lack of suitable biomarkers and bioanalytical technologies for earlier detection of numerous diseases. Metabolic profiling or metabolomics, defined as the analysis of multiple biofluid metabolites in parallel, holds the promise of earlier disease detection and improved understanding of systems biology (1–3). Early indications of this potential have been reported for the detection of several diseases, including inborn errors of metabolism, cardiovascular diseases, and cancer (4–7).
NMR and mass spectrometry are the two most often used analytical methods for metabolite profiling because of their high resolution and rich data content (8–14). Although mass spectrometry is the more sensitive technique, NMR provides broad coverage of the metabolome by detecting all of the (hydrogen-containing) metabolites present in the biofluid simultaneously, with excellent reproducibility and only limited sample pretreatment. Thus, for example, many classic inborn errors of metabolism (IEM) can be diagnosed by the use of 1H NMR spectroscopy of body fluids (15–17). Several of the IEM are associated with the accumulation of amino acids as metabolites in serum and urine. 1H NMR studies on urinary excretion of diagnostic amino acids such as phenylalanine in phenylketonuria (PKU), branched chain amino acids (leucine, valine, isoleucine) in maple syrup urine disease, N-acetyl aspartic acid in Canavan disease, and tyrosine and N-acetyl tyrosine in tyrosinemia type I have been reported (18, 19). However, the diversity of the detectable metabolites found in intact biofluids and their highly variable concentrations often results in the severe overlap of amino acid signals in 1H NMR spectra with those from several other metabolites, thus alternatives are highly sought (20).
13C NMR can potentially serve as a useful alternative to 1H NMR for identifying and quantifying metabolites because of its larger chemical shift range and reduced spectral complexity (21). However, 13C NMR suffers from poor sensitivity because of the low natural abundance (≈1.1%) and low gyromagnetic ratio of 13C nuclei and therefore requires unacceptably long data acquisition times. Hence, the application of natural-abundance 13C NMR to metabolic profiling has been challenging (22, 23). A recent advance is the use of cryogenically cooled probes to collect improved 13C NMR data that may be potentially useful for metabolomic analysis on a time scale suitable for routine experiments (24). Despite this development, however, substantial gains in sensitivity are still needed for routine applications of 13C NMR to metabolomics-based biomarker discovery.
With these opportunities and challenges in mind, 13C isotope labeling provides a potentially useful strategy for improving sensitivity and resolution for NMR-based metabolomics. For more than two decades, 13C labeling has been used for a variety of applications, including tracing the metabolic pathways and fluxes in a variety of cells and tissues (25–28), for protein structure determination (29), and recently for the NMR structural elucidation of individual oligosaccharides using 13C-labeled peracetylation (30, 31). In the latter experiments, the samples were reacted under dry conditions in chloroform.
In contrast to the experiments described above, we have found that an isotope-labeled acetylation reaction can provide an approach for the analysis of complex mixtures. The reactions are facile and quantitative and can be carried out directly in aqueous solution at ambient temperature. In general, our approach consists of labeling specific classes of metabolites with easily observed, isotopically enriched reactant species under physiological pH. This procedure is especially attractive for complex mixtures such as urine, serum, or other biofluids when combined with sensitivity-improved 2D inverse detected (1H-13C) heteronuclear methods to yield spectra with good signal-to-noise ratios.
In this article, we perform chemical derivatization using 1,1′-13C2 acetic anhydride followed by 1D 13C NMR or inverse detected 1H-13C heteronuclear single quantum coherence (HSQC) (32) experiments to profile amino acids in human urine and serum with high resolution and sensitivity. We also apply the method to the analysis of urine from patients with the prior diagnosed metabolic disorders tyrosinemia type II (TYRO), argininosuccinic aciduria (ASA), homocystinuria (HCY), and PKU. Rapid detection and quantitation of amino acid metabolic profiles in human body fluids using the approach presented here could be useful for early diagnosis and prognosis of diseases associated with amino acid metabolism. The approach is extendable to detect other classes of complex metabolites and therefore other diseases in a similar fashion.
Results and Discussion
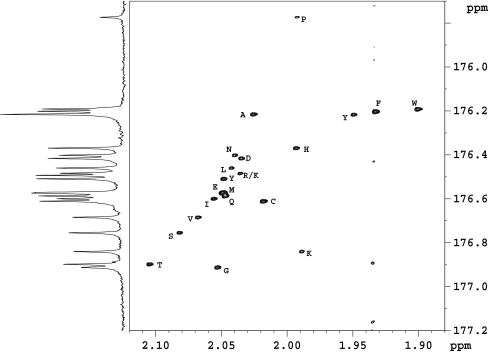
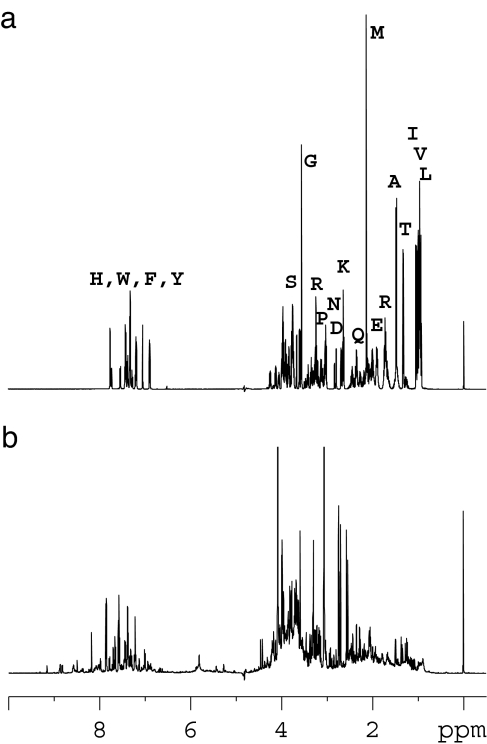
Derivatization of amino acids and amines takes place rapidly using acetic anhydride in aqueous solution at ambient temperature. The derivatization of a sample consisting of a mixture of 20 standard l-amino acids and detection by 1D and 2D NMR is shown in Fig. 1. The 13C NMR spectrum of the N-acetylated derivatives shows the presence of the isotopically labeled carbonyl carbons that appear in the 175.7- to 177-ppm region. It can be seen in Fig. 1 that the individual amino acids are well resolved, with a single peak for each amino acid. Lysine and tyrosine show additional peaks due to the derivatization of side-chain–NH2 (176.43 ppm) and phenolic hydroxyl (176.48 ppm) groups, respectively. The derivatization procedure results in a sensitivity gain of ≈100 for the 1D 13C NMR detection of the target amino acids. Additional enhancement of the sensitivity and resolution using 1H detection was obtained in the HSQC experiment. Chemical shifts for the amino acids in the mixture were identified from the 2D HSQC spectrum (Fig. 1) by comparing cross-peak positions with those from individual amino acid derivatives, determined separately. Compared with the normal 1D 1H spectrum for the same mixture (Fig. 2a) the derivatization approach results in a much simplified (13C) spectrum in 1D and an enhanced sensitivity for 2D when compared with the HSQC spectrum of the unmodified amino acids.
Fig. 1.
Shown are 2D 1H-13C HSQC spectrum of a mixture of 20 standard amino acids derivatized with 1,1′-13C2 acetic anhydride. The 1D 13C NMR spectrum can be seen on the carbon axis and shows separate signals for each amino acid. Amino acids are labeled with the one-letter code. Some T1 noise is seen near 1.935 ppm.
Fig. 2.
Shown are 1D 1H NMR spectra of 20 standard amino acids (a) and normal human urine (b).
For assessing the reproducibility of the derivatization, reactions of individual amino acids and their mixtures using stock solutions were performed in triplicate under identical conditions. Both reproducibility and the reaction efficiency were assessed based on the integrals of the carbonyl carbon of the derivatized amino acids with reference to an internal standard [sodium 3-trimethylsilyl (2,2,3,3-2H4)-1-propionate]. The yield varied from 85% to 100% for most of the amino acids except for proline (60%) with excellent reproducibility (3–5%) as determined from triplicate experiments. Similar results were obtained for urine samples. The high yield results from the fact that the reaction of acetic anhydride with amines is much faster than its hydrolysis in water. Even better yield and reproducibility can be anticipated through the use of automated reaction procedures and a larger excess (>2-fold) of acetic anhydride. Although there are numerous methods that have been reported for obtaining isotopically 13C-labeled derivatives of amino acids, including carbamyl derivatives with KOCN (33), N-hydroxy-methyl derivatives with formaldehyde (34), and benzoyl derivatives with N-hydroxyscuccinamide sulfonyl benzoate (35), we found that 13C-labeled acetyl derivatization with acetic anhydride (36) was the superior method as it worked well at room temperature in aqueous solution, produced high yield, and is readily available commercially.
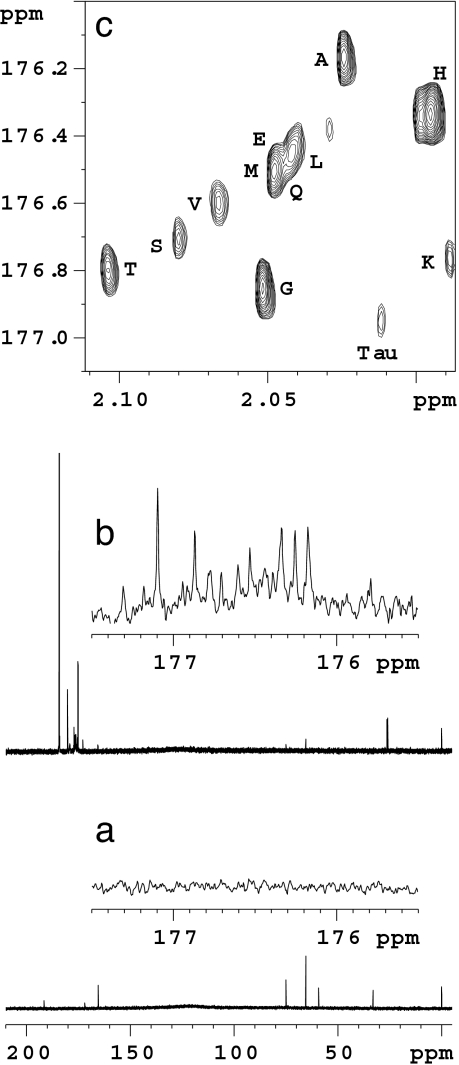
The difficulty of identifying amino acid peaks in a complex spectrum such as human urine (Fig. 2b) is compounded by the high degree of spectral overlap and their often low abundance. Chemical derivatization provides a method to simplify this analysis considerably. Fig. 3 compares 1D 13C spectra before (Fig. 3a) and after (Fig. 3b) derivatization of a normal urine sample. Whereas the 13C signals of amino acids were too weak to be seen in the spectrum before derivatization, labeled carbonyl carbons of the modified amino acids show up distinctly. The sensitivity and resolution of detection of amino acids could be further enhanced by using proton detected 2D experiments such as HSQC (Fig. 3c). Twelve amino acids were assigned in the 2D HSQC spectrum, while there were a few unassigned peaks of mainly low intensity. Assignments were made by using the results of the data generated from Fig. 1. Heteronuclear multiple bond correlation experiments were also performed, but produced inferior results to HSQC.
Fig. 3.
13C NMR spectrum of normal human urine (a), normal human urine after derivatization with 1,1′-13C2 acetic anhydride (b), and 2D HSQC spectrum of derivatized normal urine showing distinct amino acid signals with increased sensitivity (c). Tau, taurine.
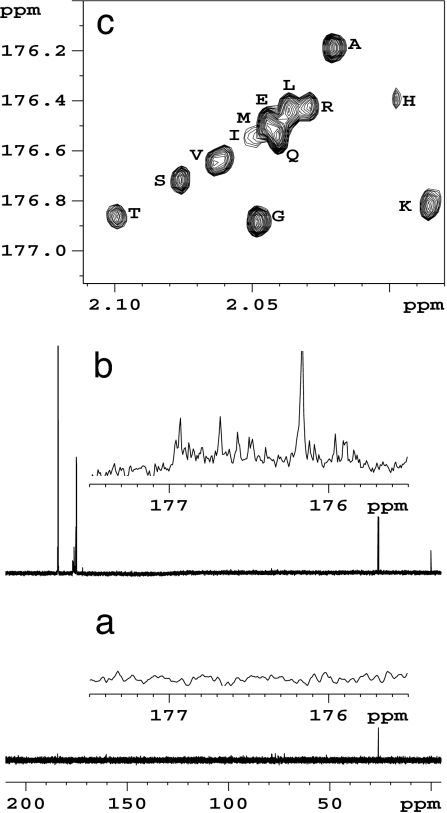
The method can also be applied to serum samples. To detect amino acids and other amines selectively in serum, we extended the derivatization approach by first precipitating the interfering proteins normally present in high concentrations. The protein-depleted serum was then treated with 13C-labeled reagent. As evidenced in the 1D 13C NMR spectra of Fig. 4, amino acids that are not detected before derivatization (Fig. 4a) are clearly observed after derivatization (Fig. 4b). As expected, the sensitivity and resolution is further enhanced by using the HSQC experiment on the derivatized serum, and the various amino acids identified are shown with assignments (Fig. 4c).
Fig. 4.
13C NMR spectrum of normal human serum after protein precipitation (a), normal serum after protein precipitation and derivatization with 1,1′-13C2 acetic anhydride (b), and 2D HSQC spectrum of derivatized human serum showing distinct amino acid signals with increased sensitivity (c).
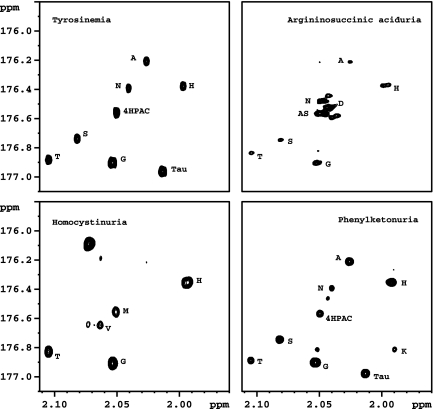
To demonstrate the application of this approach to disease detection, we have analyzed the urine from patients previously diagnosed with TYRO, ASA, HCY, and PKU. TYRO is an autosomal recessive condition caused by a defect in the hepatic tyrosine aminotransferase gene responsible for converting tyrosine to 4-hydroxyphenylpyruvate. Presymptomatic identification and treatment of patients with TYRO is reported to prevent any long-term problems. Elevated levels of tyrosine in blood and increased levels of 4-hydroxyphenylacetate (4HPAC), 4-hydroxyphenyllactate, and 4-hydroxyphenyl-pyruvate in urine, in addition to several amino acids, are the typical indications of the disease. The 2D HSQC spectrum of derivatized urine from a TYRO patient shows high levels of 4HPAC, in addition to several amino acids, including tyrosine (Fig. 5), thus uniquely confirming the findings. The signals from acetylation of phenolic–OH groups of 4HPAC and tyrosine overlap because of similarities in their structure. However, the identification of these markers in the urine can also be supported by 1H NMR. ASA is caused by the deficiency of argininosuccinate (AS) lyase. This deficiency results in the increased levels of AS, which is excreted in urine. In the derivatized urine 2D HSQC spectrum of ASA, the specific biomarker AS was distinctly visible (Fig. 5). HCY is caused by the deficiency of the enzyme cystathionine β-synthase, which converts the potentially toxic amino acid homocysteine to cysteine. This disease is characterized by elevated levels of methionine and homocysteine in plasma and urine. An increased level of methionine in urine is clearly seen from the 2D HSQC spectrum of derivatized urine of HCY. The metabolic disorder PKU is characterized by the deficiency of phenylalanine hydroxylase. Urinary excretion of phenylalanine and different forms of hydroxyphenylacetic acids are characteristic of this disease. The specific biomarkers phenylalanine and 4HPAC were clearly observed in the 2D spectrum of the derivatized urine of the PKU patient. These results clearly demonstrate the utility of the proposed approach for the detection of the amino acid class of metabolites with increased sensitivity and improved resolution. In NMR spectra of all of these biofluids, the chemical shifts of the derivatized amino acids were found to be highly reproducible (<0.005 ppm variation) except that of amino acids such as histidine (0.01 to 0.02 ppm variation), which is known to be more sensitive to small changes in ion concentration or pH. An additional unknown biomarker near histidine is also seen in Fig. 3 and in the ASA spectrum shown in Fig. 5.
Fig. 5.
2D HSQC spectra of 13C-derivatized urine samples from patients with four metabolic disorders as indicated. Each sample shows altered metabolite signal intensities, typical for the disease. Labeled amino acids and other identified metabolites are described in Results and Discussion.
Although we have focused on the derivatization of amino acids in this study, the method used here is not restricted to N-acetylation of amino acids alone; several other metabolites will also be acetylated. For example, the observation of labeled 4HPAC, a biochemical marker of TYRO (Fig. 5), is caused by the acetylation of its phenolic hydroxyl group; even the hydroxyl group of the amino acid, tyrosine, becomes acetylated. Therefore, in addition to identifying all amino acids, this approach would also enable detection of several other amines and phenolic metabolites in body fluids. Extension to another class of metabolites using this approach opens up additional possibilities for the analysis of complex samples and potentially for improved disease detection. For example, we have also had success in derivatizing carboxylic acid groups of metabolites in urine by using excess methyl iodide (37), which further expands the types of metabolite profiles accessible with the method. Combinations of derivatization, sensitivity-enhanced NMR experiments such as 2D HSQC, and pattern recognition methods such as principle component analysis could be potentially useful in biomarker discovery for a number of diseases.
Conclusion
Chemical derivatization of human body fluids, such as urine and serum, with 13C-labeled substrates combined with sensitivity-enhanced 2D HSQC experiments provide sufficient sensitivity and spectral resolution to acquire well resolved spectra of biofluids. This approach offers the possibility of identifying and quantifying amino acids and other metabolite target groups in biological samples without separation. This approach is consistent with the requirements for a fast screening method for body fluids, such as urine and serum, for medical diagnoses. 2D HSQC spectra of derivatized urine/serum can be acquired in <10 min, making possible the efficient use of 13C NMR spectroscopy alongside 1H NMR spectroscopy in metabolomic studies.
Materials and Methods
Chemicals and Samples.
l-amino acids, sodium 3-trimethylsilyl (2,2,3,3-2H4)-1-propionate, and human serum (all from Sigma–Aldrich, St. Louis, MO), and 1,1′-13C2 acetic anhydride (Cambridge Isotope Laboratories, Andover, MA) were used without further purification. Normal urine samples were obtained from healthy volunteers, and previously banked and deidentified urine samples from patients diagnosed with TYRO, ASA, HCY and PKU were obtained from the Section of Pediatric Metabolism and Genetics, Department of Pediatrics, Indiana University School of Medicine. Sodium azide (0.1% wt/vol) was added to the urine samples to prevent bacterial growth. The samples were then filtered with a centrifugal filter device Centriprep YM-10 (10,000 molecular weight cutoff), aliquoted, and stored at −70°C until analyses were performed. All human samples collection procedures were approved by the Institutional Review Board at Purdue University.
Derivatization of Standard Amino Acids.
Stock solutions of 20 naturally occurring amino acids (0.1 M each) in 20 mM phosphate buffer (pH 8.0) were prepared in separate tubes. For the derivatization of individual amino acids, 100 μl (10 μmol) of each amino acid diluted with 650 μl of 20 mM phosphate buffer (pH 8.0) was reacted with 1,1′-13C2 acetic anhydride (2 μl, 20 μmol) at room temperature. For the derivatization of amino acid mixtures, 50 μl of each amino acid solution was mixed together in a reaction container (producing 100 μmol of total amino acids). The 1,1′-13C2 acetic anhydride (20 μl, 200 μmol) was then added manually in steps of 2 μl to the well stirred solution of amino acids at room temperature. The pH of the reaction media was maintained constant at 8.0 by the addition of 1 M NaOH solution at regular intervals using 10-μl aliquots from a micropipette. For individual amino acids such as lysine, tyrosine, and cysteine, in which cases the side-chain functional groups also reacts, four-fold excess of acetic anhydride was used. All reactions of individual and mixture of amino acids were performed in triplicate.
Derivatization of Metabolites in Human Urine.
Frozen human urine samples were thawed and concentrated ≈2-fold in vacuo. Eight hundred microliters of each concentrated urine sample was transferred to a separate tube, and the pH was adjusted to 8.0 by the addition of 1 M NaOH solution. The 1,1′-13C2 acetic anhydride (12 μl, 120 μmol) was then added manually in steps of 2 μl to the well stirred solution of urine at room temperature, while the pH was maintained at 8.0 by the addition of 10-μl aliquots of 1 M NaOH solution. All reactions of normal urine were performed in triplicate.
Derivatization of Metabolites in Human Serum.
Two serum samples (1 ml each) were mixed with methanol in the ratio 1:2 (vol/vol) in separate tubes. The mixtures were vortexed and incubated for 20 min at −20°C to precipitate the majority of serum proteins (38). The deproteinized supernatant was then decanted after centrifugation at 13,200 × g for 10 min. The solutions were dried and reconstituted in 600 μl of phosphate buffer (pH 8). One sample was transferred to a reaction container where 1,1′-13C2 acetic anhydride (4 μl, 40 μmol) was added in steps of 1 μl while stirring at room temperature. The pH of the reaction medium was maintained at 8.0 by the addition of 10-μl aliquots of a 1-M NaOH solution at regular intervals using a micropipette. The second sample served as a control.
NMR Spectroscopy.
Measured volumes (530 μl) of control and derivatized solutions were mixed with 70 μl of sodium 3-trimethylsilyl (2,2,3,3-(2)H4)-1-propionate/D2O (0.5% wt/vol) solution separately, pH adjusted to 7.05 ± 0.03, and placed in separate 5-mm NMR tubes. All NMR experiments were carried out at 25°C on a DRX 500-MHz spectrometer (Bruker, Billerica, MA) equipped with 1H inverse detection and triple-axis field gradients. 1D 13C NMR spectra were recorded with inverse-gated proton decoupling by using the WALTZ-16 sequence. A total of 64 or 128 transients were averaged, and 64,000 data points were acquired for each sample. Line broadening of 1.0 Hz was applied before Fourier transformation. A long recycle delay of 30 s was used for 13C NMR experiments for derivatized amino acids to enable quantitation and to assess reproducibility of the reactions. 2D HSQC experiments were performed for all samples by using an insensitive nuclei enhanced by polarization transfer delay of 100 ms, corresponding to 3JCH = 5.0 Hz, which represents an average value determined for standard derivatized amino acids. Spectral widths of 500 and 625 Hz were used in the 1H and 13C dimensions, respectively. Sixty-four free induction decays were collected along t1 using four transients per increment. Phase-sensitive data were obtained by using echo-antiecho mode with GARP (Globally Optimized Alternating-Phase Rectangular Pulses) carbon decoupling during acquisition. The resulting 2D data were zero-filled to 1,024 points in the t1 dimension after forward linear prediction to 128 points and were Fourier-transformed after multiplying by a squared sine-bell window function shifted by π/4 along t1 and π/2 along the t2 dimension. Chemical shifts were referenced to the glycine peak in the HSQC spectra.
Acknowledgments
This work was supported by the National Institutes of Health Roadmap Initiative on Metabolomics Technology, National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grant 3 R33 DK070290, and a Collaborative Biomedical Research Grant from Purdue University/Discovery Park and the Indiana University School of Medicine.
Abbreviations
- HSQC
heteronuclear single quantum coherence
- TYRO
tyrosinemia type II
- ASA
argininosuccinic aciduria
- HCY
homocystinuria
- PKU
phenylketonuria
- 4HPAC
4-hydroxyphenylacetate.
Footnotes
The authors declare no conflict of interest.
References
- 1.Nicholson JK, Lindon JC, Holmes E. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 2.Saxena V, Gupta A, Nagana Gowda GA, Saxena R, Yachha SK, Khetrapal CL. NMR Biomed. 2006;19:521–526. doi: 10.1002/nbm.1034. [DOI] [PubMed] [Google Scholar]
- 3.Bala L, Ghoshal UC, Ghoshal U, Tripathi P, Misra A, Nagana Gowda GA, Khetrapal CL. Magn Reson Med. 2006;56:738–744. doi: 10.1002/mrm.21041. [DOI] [PubMed] [Google Scholar]
- 4.Moolenaar SH, Gohlich-Ratmann G, Engelke UF, Spraul M, Humpfer E, Dvortsak P, Voit T, Hoffmann GF, Brautigam C, van Kuilenburg AB, et al. Magn Reson Med. 2001;46:1014–1017. doi: 10.1002/mrm.1289. [DOI] [PubMed] [Google Scholar]
- 5.Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, Bethell HWL, Clarke S, Schofield PM, Mckilligin E, Mosedale DE, Grainger DJ. Nat Med. 2002;8:1439–1444. doi: 10.1038/nm1202-802. [DOI] [PubMed] [Google Scholar]
- 6.Odunsi K, Wollman RM, Ambrosone CB, Hutson A, McCann SE, Tammela J, Geisler JP, Miller G, Sellers T, Cliby W, et al. Int J Cancer. 2005;113:782–788. doi: 10.1002/ijc.20651. [DOI] [PubMed] [Google Scholar]
- 7.Chen HW, Pan Z, Talaty N, Cooks RG, Raftery D. Rapid Commun Mass Spectrom. 2006;20:1577–1584. doi: 10.1002/rcm.2474. [DOI] [PubMed] [Google Scholar]
- 8.Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 9.Lindon JC, Holmes E, Nicholson JK. Prog Nucl Mag Reson Spectrosc. 2004;45:109–143. [Google Scholar]
- 10.van der Greef J, Smilde AK. J Chemom. 2005;19:376–386. [Google Scholar]
- 11.Wagner S, Scholz K, Donegan M, Burton L, Wingate J, Völkel W. Anal Chem. 2006;78:1296–1305. doi: 10.1021/ac051705s. [DOI] [PubMed] [Google Scholar]
- 12.Crockford DJ, Holmes E, Lindon JC, Plumb RS, Zirah S, Bruce SJ, Rainville P, Stumpf CL, Nicholson JK. Anal Chem. 2006;78:363–371. doi: 10.1021/ac051444m. [DOI] [PubMed] [Google Scholar]
- 13.Pan Z, Raftery D. Anal Bioanal Chem. 2007;387:525–527. doi: 10.1007/s00216-006-0687-8. [DOI] [PubMed] [Google Scholar]
- 14.Pan Z, Gu H, Talaty N, Chen HW, Shanaiah N, Hainline BE, Cooks RG, Raftery D. Anal Bioanal Chem. 2007;387:539–549. doi: 10.1007/s00216-006-0546-7. [DOI] [PubMed] [Google Scholar]
- 15.Iles RA, Hind AJ, Chalmers RA. Clin Chem. 1985;31:1795–1801. [PubMed] [Google Scholar]
- 16.Wevers RA, Engelke U, Heerschap A. Clin Chem. 1994;40:1245–1250. [PubMed] [Google Scholar]
- 17.Wevers RA, Engelke U, Wendel U, de Jong JG, Gabreels FJ, Heerschap A. Clin Chem. 1995;41:744–751. [PubMed] [Google Scholar]
- 18.Constantinou MA, Papakonstantinou E, Spraul M, Sevastiadou S, Costalos C, Koupparis MA, Shulpis K, Tsantili-Kakoulidou A, Mikros E. Anal Chim Acta. 2005;542:169–177. [Google Scholar]
- 19.Engelke UFH, Liebrand-van Sambeek MLF, de Jong JGN, Leroy JG, Morava E, Smeitink JAM, Wevers RA. Clin Chem. 2004;50:58–66. doi: 10.1373/clinchem.2003.020214. [DOI] [PubMed] [Google Scholar]
- 20.Sandusky P, Raftery D. Anal Chem. 2005;77:7717–7723. doi: 10.1021/ac0510890. [DOI] [PubMed] [Google Scholar]
- 21.Fan TW-M. Prog Nucl Mag Reson Spectrosc. 1996;28:161–219. [Google Scholar]
- 22.Moolenaar SH, Poggi-Bach J, Engelke UFH, Corstiaensen JMB, Heerschap A, de Jong JGN, Binzak BA, Vockley J, Wevers RA. Clin Chem. 1999;45:459–464. [PubMed] [Google Scholar]
- 23.Krawczyk H, Gryff-Keller A, Gradowska W, Duran M, Pronicka E. J Pharmaceut Biomed Anal. 2001;26:401–408. doi: 10.1016/s0731-7085(01)00420-4. [DOI] [PubMed] [Google Scholar]
- 24.Keun HC, Beckonert O, Griffin JL, Richter C, Moskau D, Lindon JC, Nicholson JK. Anal Chem. 2002;74:4588–4593. doi: 10.1021/ac025691r. [DOI] [PubMed] [Google Scholar]
- 25.Cohen SM, Ogawa S, Shulman RG. Proc Natl Acad Sci USA. 1979;76:1603–1607. doi: 10.1073/pnas.76.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen SM, Rognstad R, Shulman RG, Katz J. J Biol Chem. 1981;256:3428–3432. [PubMed] [Google Scholar]
- 27.Cohen SM. Biochemistry. 1987;26:573–580. doi: 10.1021/bi00376a032. [DOI] [PubMed] [Google Scholar]
- 28.Cohen SM. Biochemistry. 1987;26:581–589. doi: 10.1021/bi00376a033. [DOI] [PubMed] [Google Scholar]
- 29.Wüthrich K. NMR of Proteins and Nucleic Acid. New York: Wiley; 1986. [Google Scholar]
- 30.Bendiak B. Carbohydr Res. 1999;315:206–221. doi: 10.1016/s0008-6215(98)00311-5. [DOI] [PubMed] [Google Scholar]
- 31.Armstrong GS, Cano KE, Mandelshtam VA, Bendiak B, Shaka AJ. J Magn Reson. 2004;170:156–163. doi: 10.1016/j.jmr.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Bodenhausen G, Ruben DJ. Chem Phys Lett. 1980;69:185–189. [Google Scholar]
- 33.Stark GR, Smyth DG. J Biol Chem. 1963;238:214–222. [PubMed] [Google Scholar]
- 34.Kitamoto Y, Maeda H. J Biochem (Tokyo) 1980;87:1519–1530. doi: 10.1093/oxfordjournals.jbchem.a132893. [DOI] [PubMed] [Google Scholar]
- 35.Julka S, Regnier FE. Anal Chem. 2004;76:5799–5806. doi: 10.1021/ac049688e. [DOI] [PubMed] [Google Scholar]
- 36.Smyth DG. J Biol Chem. 1967;242:1592–1598. [PubMed] [Google Scholar]
- 37.Avila-Zárraga JG, Martínez R. Synth Commun. 2001;31:2177–2183. [Google Scholar]
- 38.Want EJ, O'Maille G, Smith CA, Brandon TR, Uritboonthai W, Qin C, Trauger SA, Siuzdak G. Anal Chem. 2006;78:743–752. doi: 10.1021/ac051312t. [DOI] [PubMed] [Google Scholar]