Abstract
Telomerase is a cellular reverse transcriptase that extends one strand (the G-strand) of the telomere terminal repeats. Aside from this role in telomere length maintenance, telomerase has been proposed to serve a protective function at chromosome ends, although this is not well understood mechanistically. Earlier analysis suggests that, in the pathogenic yeast Candida albicans, the catalytic reverse transcriptase subunit of telomerase (TERT/EST2) can protect telomeres against nucleolytic degradation. In this report we demonstrate that the RNA component (TER1) has a similar function; in addition to complete loss of telomerase activity and progressive telomere attrition, the ter1-ΔΔ strains manifested a dramatic increase in the amount of G-strand overhangs, consistent with aberrant degradation of the complementary C-strand. We also demonstrate that a catalytically incompetent EST2 protein can suppress such overhang accumulation in the est2-ΔΔ mutant to the same extent as the wild-type protein. Altogether, our data support the notion that the Candida telomerase core components mediate a protective function through a mechanism that is independent of its catalytic activity.
Keywords: G-strand, telomerase RNA, telomerase reverse transcriptase
Telomeres are specialized nucleoprotein structures that maintain the integrity of eukaryotic chromosomal termini by protecting them from fusion and recombination and promoting their replication (for reviews see refs. 1–4). In most organisms, telomeric DNA consists of short repetitive sequences that are rich in G-residues on the 3′ end-containing strand. These repeats are maintained by a ribonucleoprotein known as telomerase, which acts as an unusual reverse transcriptase (for reviews see refs. 3 and 5–7). The regulation of telomere length and telomerase activity has been shown to be pivotal in the control of genome stability and cellular lifespan.
Components of the telomerase enzyme complex have been analyzed in a variety of organisms including ciliated protozoa, yeasts, and mammals. Two components are essential for the polymerization activity: an RNA that provides the template [telomerase RNA (TER)] and a protein subunit that catalyzes nucleotide addition [telomerase reverse transcriptase (TERT)/EST2]. In addition to TER and TERT, telomerases from different organisms have been shown to possess a number of accessory or regulatory subunits that promote telomerase ribonucleoprotein biogenesis, assembly, and function.
Aside from telomere extension, telomerase has been proposed to serve a protective function at telomeres (for reviews see refs. 7 and 8). This notion was based on observations in both yeast and mammalian systems, in which the effects of telomerase mutants on growth can be uncoupled from those on bulk telomere lengths (9–11). For example, a hypomorphic telomerase allele that cannot avert progressive telomere attrition can nevertheless prolong cellular lifespan (10). Conversely, overexpressing a catalytically inactive point mutant of TERT can cause premature senescence and apoptosis of primary human fibroblasts despite lack of obvious effect on bulk telomere lengths (9). These results can be explained by the ability of a low level of telomerase or defective telomerase to confer a protection function (possibly through direct binding). However, because all of the above positive effects reported to date required a catalytically competent TERT, the possibility that the apparently protective effects are due to preferential extension of short telomeres by the defective telomerase cannot be excluded. However, induced overexpression of TERT in the mouse can exert effects on proliferation of certain cell compartments even in mice that were deleted for TER (12), thus arguing for an activity of TERT that is independent of telomere extension.
We have earlier shown that, in the diploid pathogenic yeast Candida albicans, deletion of TERT/EST2 results in not only progressive telomere attrition, but also a large increase in the level of G-strand overhangs (13). In contrast, deletion of EST1 and EST3 (two regulatory subunits of the telomerase complex), although causing telomere loss, failed to elicit the same accumulation in G-strand overhangs. Notably, the overhang increase is reminiscent of the phenotypes of Saccharomyces cerevisiae mutants suffering from dysfunction of telomere binding proteins such as Cdc13p, Ku70, and Ku80 (14–17). Analysis of such mutants indicated that loss of telomere protection engenders preferential degradation of the telomere C-strand, leading to G-strand overhang accumulation (18–20). Thus the phenotypes of the est2-ΔΔ mutant suggest that, in C. albicans, the TERT/EST2 component may mediate a physically protective function akin to S. cerevisiae telomere binding proteins. To extend and further test this hypothesis, we cloned the Candida TER gene (TER1) and characterized the consequences of its loss. We also examined the ability of a catalytically incompetent TERT/EST2 to suppress G-strand accumulation. As reported below, our data provide strong support for the notion that telomerase can protect telomeres through a mechanism that is completely independent of its catalytic activity.
Results
Identification of a Candidate TER Gene (TER1).
Prior studies have revealed interesting phenotypic differences in Candida mutants missing different protein components of the telomerase complex (13, 21). In particular, there was substantial accumulation of G-strand overhang in the est2-ΔΔ mutant, but not the est1-ΔΔ or est3-ΔΔ mutant, suggesting the catalytic protein component may have an additional function in protecting telomeres against C-strand degradation. To gain further insights into the mechanisms of protection, we sought to determine the role of TER in telomere protection.
Taking advantage of the large (23-bp) and homogeneous telomeric repeat unit sequence in C. albicans, we directly probed a phage λ library of the C. albicans genome with the telomeric repeat sequence. From this library we identified three candidate TER gene clones. Each clone contained a 28-bp sequence that matched one complete repeat unit, plus a portion of an adjacent repeat unit, of the C. albicans telomere sequence (Fig. 1A). Such a telomere-complementary element that contains a terminally reiterated sequence of a few nucleotides is characteristic of all TER templating domains identified thus far. Northern blot analysis revealed a single large transcript >1,500 nt long that hybridized to the cloned sequence [supporting information (SI) Fig. 6]. Mapping of the 5′ and 3′ ends revealed a length of 1,540–1,544 nucleotides for this RNA (henceforth referred to as Ter1). Whereas the 5′ end of the RNA could be established unambiguously, some heterogeneity within a range of 5 nt was observed for the 3′ end. Interestingly, both the 3′ end of Tetrahymena thermophila and S. cerevisiae TER are also variable and are thought to be subjected to posttranscriptional processing (22–24).
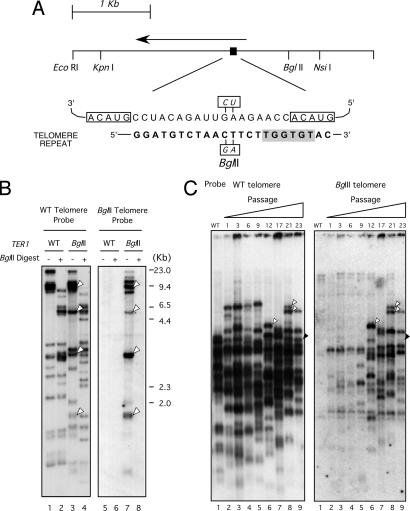
Fig. 1.
The identification of TER1 and the demonstration that it encodes a TER. (A Upper) Schematic diagram of the genomic fragment containing the TER1 gene. The RNA transcript (arrow) is estimated to be 1,544 bp in length. The template region is indicated by the black rectangle. (Lower) The RNA template encoded by the 28 bp of the telomere homology region within the TER1 gene and a copy of the telomere repeat are illustrated. The duplicated parts of the template sequence are indicated by boxes. Shown in a shaded background is the conserved core of the RAP1 binding site. The base changes made to create the BglII mutation within the template region are also illustrated. (B) DNAs from the CAI4 strain and a CAI4 derivative containing an introduced TER1-BglII allele were analyzed by Southern blotting. The same filter was probed with labeled Ca7-3 (specific for wild-type telomere repeat) in Left and with labeled CaBgl14 (specific for the TER1-BglII mutant telomeric repeat) in Right. Arrowheads indicate bands that hybridized to both the wild-type and TER1-BglII telomere probes. Each DNA sample was digested with ClaI or ClaI plus BglII as indicated. (C) Genomic DNAs from mutant cells at different passages as indicated at the top were digested with HindIII and subjected to telomere Southern blot using the wild-type telomeric probe (Left) and a BglII-specific telomeric probe (Right). The white arrowheads in lanes 6 and 8 point to examples of well resolved new telomeric bands that appeared during passage and that have gained BglII repeats. The black arrowhead in lane 9 points to a telomeric fragment that remained free of BglII repeats after >20 passages. Each serial passage corresponds to 20–25 generations. The 30th passage of the wild-type strain is shown on the far left in each panel.
The Putative TER1 Template Sequence Could Be Incorporated into Candida Telomeres.
We used two strategies to confirm the authenticity of TER1. First, we altered the putative template sequence and monitored telomeres for incorporation of the correspondingly altered sequence in vivo (25). A two-base mutation was engineered into the putative template sequence to yield a BglII restriction site (named the TER1-BglII allele) (Fig. 1A). Telomeric repeats specified by this allele were predicted to contain this two-base change. To minimize the possibility of perturbing telomere length regulation, the mutated positions were away from the predicted Rap1 protein binding site (26). Rap1 is the major double-stranded telomere binding protein in other budding yeasts (27); C. albicans has a known Rap1 homolog, although its function at telomeres has not been established (28).
We transformed C. albicans strain CAI4 with an integrating plasmid, pSR3-U-B4D, containing the TER1-BglII allele and the C. albicans URA3 gene as a selectable marker. Transformants with the plasmid integrated by homologous recombination at the native TER1 locus were identified by Southern blot analysis. Because C. albicans is diploid, the correct transformant (referred to as the Bgl strain) contained three copies of the TER1 gene: two wild-type copies and a TER1-BglII mutant copy. To monitor the incorporation of mutant repeats into telomeres, genomic DNA was isolated from both wild-type and Bgl strains, digested with ClaI alone or ClaI plus BglII, and subjected to Southern blot analysis using a wild-type telomere probe or a BglII telomere probe (Fig. 1B). Two features of the banding patterns strongly support the notion that BglII-containing repeats were incorporated into the telomeres of the Bgl strain. First, the BglII probe hybridized only to DNA fragments derived from the Bgl but not the wild-type strain. Most of these DNA fragments also bound wild-type probe (indicated by white arrowheads in Fig. 1B, lanes 3 and 7) and therefore likely represent telomere restriction fragments. Second, as expected, digestion of these putative BglII-containing telomere fragments with BglII resulted in complete loss of BglII-specific hybridization signal (Fig. 1B, compare lanes 7 and 8). To gain additional support for the incorporation of mutant repeats, we performed a separate analysis of the samples using the HindIII and BglII restriction enzymes (SI Fig. 7). Again, the BglII probe detected fragments only in the Bgl strain, and some of these fragments evidently contained wild-type telomere repeats. These results confirm that the TER1 gene locus encodes a Candida TER.
Because there were two wild-type copies of the TER1 gene and just one BglII mutant allele, the Bgl cells were predicted to contain a mixture of telomerases with wild-type and mutant RNAs. Therefore, mixed wild-type and BglII repeat sequences were expected to be added to telomeres. In addition, all telomeres in Bgl cells were expected to retain wild-type repeats internally, regardless of whether the BglII repeats have been added (29). Interestingly, it appeared that during early passages relatively few chromosome ends had incorporated BglII repeats, because the pattern of BglII-positive telomeric bands was much less complex than that detected with the wild-type probe (Fig. 1C). After more prolonged propagation, a greater proportion of telomere fragments in the Bgl strain became detectable with the BglII telomere probe. The slow average kinetics by which telomeres acquired BglII repeats suggests that telomerase acts rather infrequently in Candida to maintain telomere length homeostasis. Alternatively, the mutated template or the mutated telomere repeat might be less functional than the wild-type sequence, thus causing the underrepresentation of the mutated sequence.
Deletion of the TER1 Gene Results in Complete Loss of Telomerase Activity.
To rule out the possible existence of other TER genes and to characterize further the function of TER1, we generated homozygous deletion strains by replacing both copies of TER1 with HisG using the URA-Blaster cassette (30). To facilitate comparison with other telomerase deletion mutants, we generated the ter1-ΔΔ strains in the BWP17 strain background (13, 21). In addition, to rule out unintended effects of strain manipulation, we also constructed multiple reconstituted clones by reintegrating a wild-type TER1 gene into ter1-ΔΔ strains. The genotypes of the deletion and reconstituted strains were all confirmed by Southern blot analysis (data not shown).
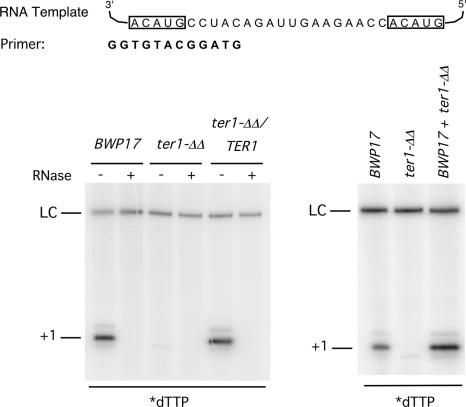
The strains were first examined for the presence or absence of telomerase activity. Cell extracts and telomerase-enriched fractions were prepared as previously described and subjected to primer extension analysis. As shown in Fig. 2, the parental BWP17 and the reconstituted strain contained an RNase-sensitive primer extension activity that supported the addition of dTMP to the primer, whereas the ter1-ΔΔ strain lacked this activity completely. Mixing the BWP17 and ter1-ΔΔ extract did not abolish the activity, thus ruling out the presence of inhibitors in the mutant extracts. Extracts from multiple deletion and reconstituted strains were analyzed with similar results. We conclude that the TER1 gene is essential for telomerase activity in C. albicans and that there is no alternative TER in this genome.
Fig. 2.
Loss of telomerase activity in the ter1-ΔΔ strain. (Left) Extracts and DEAE fractions were prepared from the BWP17, ter1-ΔΔ, and ter1-ΔΔ/TER1 strains and subjected to primer extension assays using the indicated primer and 32P-labeled dTTP. As controls, fractions were also pretreated with RNase A before the assays. (Right) Telomerase fractions from the BWP17 and ter1-ΔΔ strains were tested in primer extension assays either separately or after mixing as indicated. A 46-nt labeled oligonucleotide was added to each reaction mixture before ethanol precipitation as a control for sample recovery (LC).
Deletion of TER1 Gene Results in Progressive Telomere Attrition.
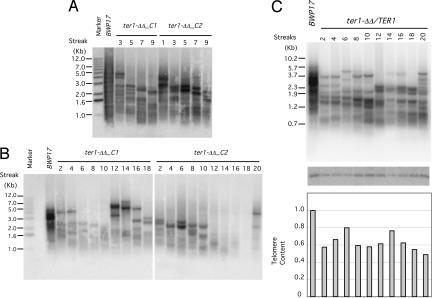
We then analyzed the telomere dynamics of several deletion strains and a reconstituted strain. As shown in Fig. 3A, two ter1-ΔΔ strains exhibited progressive telomere attrition for at least 10 streaks (≈200 generations), consistent with loss of TER. Interestingly, both clones exhibited sudden gain of telomere contents during passage, but with very different kinetics. One clone manifested substantial telomere elongation at the 12th streak, and the other at the 20th streak (Fig. 3B). Both events were likely to reflect the emergence of “survivors,” and the distinct kinetics testifies to the stochastic nature of this recombinational process (31, 32). In addition, we observed relatively stable telomere maintenance in a ter1-ΔΔ/TER1 reconstituted strain over 20 streaks, confirming that telomere attrition in the deletion strains was due to loss of the TER1 gene (Fig. 3C). Notably, the overall telomere content of the reconstituted strain was found to be ≈60% of BWP17. A single copy of the TER1 gene may therefore be insufficient for normal telomere maintenance. Alternatively, the reintegrated copy of TER1 may not be expressed properly despite its native chromosome location. Further studies will be necessary to address this issue. Interestingly, recent studies indicate that TER is haploinsufficient in many organisms including S. cerevisiae, mice, and humans (33–36).
Fig. 3.
Progressive telomere attrition in the ter1-ΔΔ strain. (A) Two independently derived ter1-ΔΔ strains were passaged by repeatedly streaking for single colonies on plates. Chromosomal DNAs were isolated and subjected to telomere Southern blot analysis. (B) Same as A except that DNAs from different streaks were used for the telomere analysis. (C Top) Chromosomal DNAs were isolated from successive streaks of a ter1-ΔΔ/TER1 strain and subjected to telomere length analysis. (C Middle) The same blot was hybridized to an EST2 gene probe. (C Bottom) The telomere signals were divided by the EST2 signal, normalized against the BWP17 sample, and plotted.
Deletion of TER1 Gene Caused a Substantial Increase in G-Strand Overhangs.
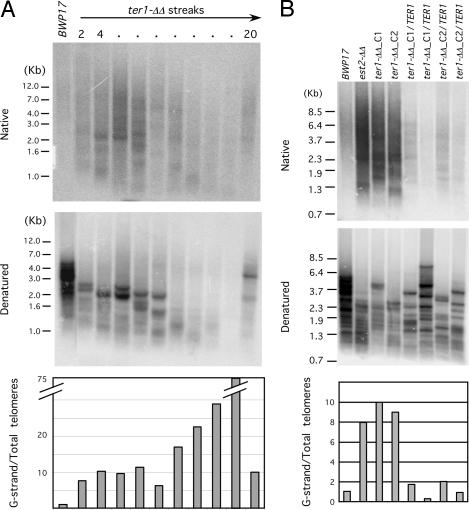
We next used the in-gel hybridization assay to assess the levels of unpaired G-strand overhang in the TER1 deletion and reconstituted strain (Fig. 4). As observed earlier, the parental BWP17 strain exhibited a very low level of unpaired G-strand. Interestingly, during early passages (the first 10 streaks) of a ter1-ΔΔ clone, the level of unpaired G-strand increased by ≈10-fold. Thereafter, as telomeres continued to shorten, the relative intensity of the G-strand signal became even greater compared with total telomeric DNA signal, peaking at the point of minimal telomere content. This increase in relative G-strand signal was due primarily to a more precipitous decline in total telomere signal, rather than an increase in the absolute level of G-strand overhangs (Fig. 4A, compare Top and Middle). After the emergence of putative survivors, the overhang levels declined to ≈10 times the wild-type level. Prior treatment of the DNA with Escherichia coli Exo I (a 3′ to 5′ single-stranded exonuclease) abolished the majority of G-strand signals in the ter1-ΔΔ samples, indicating that such signals were mostly due to terminal overhangs (SI Fig. 8). Analysis of the TER1-reconstituted strains confirmed that the overhang accumulation in deletion strains was due to the loss of the TER1 gene; both reconstituted strains manifested near wild-type levels of unpaired G-strand as judged by in-gel hybridization (Fig. 4B). Notably, G-strand accumulation is not simply a secondary consequence of telomere attrition, because the deletion and reconstituted strains in this experiment had comparable amounts of total telomeric DNA, but the former manifested much higher levels of overhangs. Altogether, our results indicate that, like Candida EST2, the TER1 gene is also required to suppress abnormal G-strand overhang accumulation in Candida.
Fig. 4.
Substantial accumulation of unpaired G-strand overhangs in the ter1-ΔΔ strains. (A Top) Chromosomal DNAs were isolated from successive streaks of a ter1-ΔΔ strain, digested with AluI and NlaIII, and subjected to electrophoresis and in-gel hybridization using a G-strand specific probe. (A Middle) The gel was treated with an alkaline solution to denature the DNA and then subjected to a second round of hybridization using the same probe. (A Bottom) The ratios of the G-strand to total telomere signals were calculated and plotted. (B) Same as A except that the DNAs were derived from early passages of BWP17, est2-ΔΔ, and multiple ter1-ΔΔ and ter1-ΔΔ/TER1 clones.
A Catalytically Incompetent EST2 Gene Can Suppress the Accumulation of G-Strand Overhang in est2-ΔΔ Strains.
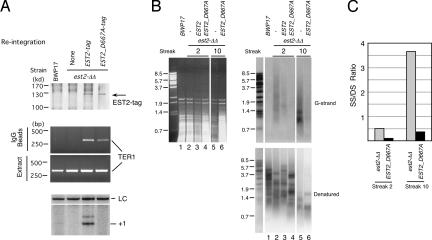
That Candida EST1 and EST3 (in contrast to EST2 and TER1) are not required to suppress G-strand overhang accumulation indicates that the protective effect of telomerase core components can be uncoupled from the in vivo telomere extension activity of the complex (13). However, the question remains as to whether the catalytic activity of the core complex plays a role in protection. To address this question, we tested the ability of a catalytically inactive allele of EST2 (D667A) to suppress overhang accumulation. To facilitate analysis, both the wild-type and D667A EST2 genes used for transformation were tagged at their C termini with a FLAG3 and a TAP tag (37). Each gene was reintegrated back into a disrupted locus of the est2-ΔΔ strain to generate the respective reconstituted strain. The reconstituted EST2 genes were thus expressed from endogenous chromosomal EST2 promoters. The expression of the wild-type and mutant Est2 proteins was first assessed by IP Western blot and was found to be comparable (Fig. 5A Top). Both proteins also associated with Ter1 RNA at similar levels (Fig. 5A Middle). However, the IP sample obtained from the mutant extract was completely devoid of primer extension activity (Fig. 5A Bottom). As predicted, the est2-ΔΔ/EST2 strain maintained stable telomeres whereas the est2-ΔΔ/EST2_D667A strain manifested progressive telomere shortening (data not shown). Remarkably, we found that both reconstituted strains exhibited substantially reduced G-strand overhang in comparison with the est2-ΔΔ strain during passaging. Interestingly, suppression of G-strand overhang by the D667A allele occurred despite continued telomere loss in this strain. As observed for the ter1-ΔΔ strain, the relative abundance of G-strand (in relation to total telomeres) in the est2-ΔΔ strain became even more elevated in late passages (Fig. 5B, compare lanes 2 and 5). However, the magnitude of suppression by the D667A allele (fold reduction of relative G-strand signal) was comparable in early and late passages (Fig. 5C). Analysis of three other independent est2-ΔΔ/EST2 and two other independent est2-ΔΔ/EST2_D667A strains yielded similar results (SI Fig. 9). We conclude that a catalytically incompetent Est2 protein in Candida is fully capable of protecting telomeres against abnormal accumulation of unpaired G-strand.
Fig. 5.
A catalytically incompetent EST2 can protect telomeres against G-strand overhang accumulation. (A Top) Extracts were prepared from the wild-type, est2-ΔΔ, est2-ΔΔ/EST2-tag, and est2-ΔΔ/EST2_D667A-tag strains and subjected to IgG-Sepharose pull-down followed by Western blot using antibodies directed against protein A. The position of the tagged Est2p is indicated by an arrow. (A Middle) RNAs were prepared from the crude extracts and IgG-Sepharose pull-down samples and analyzed by RT-PCR to detect Ter1. (A Bottom) The extracts were subjected to IgG-Sepharose pull-down followed by primer extension assay to detect telomerase enzyme activity. A 12-nt primer (5′-GGTGTACGGATG-3′) and labeled dTTP were included in the reactions. A 46-nt labeled oligonucleotide was added to each reaction mixture before ethanol precipitation as a control for sample recovery (LC). (B) DNAs were isolated from different streaks of BWP17, est2-ΔΔ, est2-ΔΔ/EST2-tag, and est2-ΔΔ/EST2_D667A-tag strains, digested with AluI and NlaIII, and subjected successively to agarose gel electrophoresis and ethidium bromide staining (Left), in-gel hybridization to detect G-strand overhangs (Upper Right), and hybridization to detect all telomeric DNA (Lower Right). The identity of the strain and the streak from which the DNA was derived are indicated at the top. (C) The ratio of G-strand overhang to total telomeric DNA was determined for samples 2, 4, 5, and 6 in B and plotted. The unit is arbitrary.
Discussion
Candida TER.
That TER1 encodes a unique TER in C. albicans is supported by incorporation of mutant repeats as well as the phenotypes of the deletion and reconstituted strains. A remarkable feature of TER1 is that it represents the largest TER gene yet identified. At ≈1,554 nt, it is significantly longer than TERs from Kluyveromyces lactis (≈1,259 nt) and S. cerevisiae (≈1,157 nt) (24, 38, 39). Unfortunately, the overall sequence conservation between Candida Ter1 and other yeast RNAs is too low to allow for alignment and prediction of structural elements. The template region of Candida TER1 is located much closer to the 5′ end (≈250 nt) than other yeast RNAs, but the significance of this finding is unclear. Further analysis of TERs from other Candida spp. will likely be informative with regard to the structure and function of this unusual RNA.
Distinct Functions of Candida Telomerase Components in Telomere Maintenance and Protection.
The most notable outcome of the current analysis is the establishment of a special role for the core telomerase components (Est2p and Ter1) in suppressing aberrant accumulation of G-strand overhangs in C. albicans. Interestingly, accumulation of G-strand overhangs has been observed in S. cerevisiae mutants with abnormal telomere binding proteins such as Cdc13p or the Ku complex (14, 15). Analysis of such mutants indicated that G-strand accumulation was due to aberrant degradation of the complementary C-strand (18). Thus, it is tempting to ascribe G-strand accumulation in Candida ter1-ΔΔ and est2-ΔΔ strains to aberrant C-strand degradation as well. The overhang accumulation in the Candida mutants could be due to either limited C-strand degradation at the majority of telomeres or more severe C-strand resection at a minority of telomeres (or both). Curiously, in some ter1-ΔΔ and est2-ΔΔ samples, almost all telomere restriction fragments have shortened to <3 kb (as judged by probing of denatured DNA), yet a large proportion of G-strand signal resided in longer DNA fragments (e.g., see Fig. 4B). This discrepancy may be due to very extensive C-strand resection of some telomeres such that subtelomeric restriction enzyme sites became single-stranded and resistant to cutting. Because the relative abundance of such fragments was evidently low, our results support the notion that in a ter1-ΔΔ or est2-ΔΔ mutant cell population only a small fraction of the total telomeres have suffered drastic C-strand degradation.
G-strand accumulation has not been reported for telomerase mutants in S. cerevisiae or other yeasts. However, unlike C. albicans, S. cerevisiae wild-type cells have relatively short telomeres and senesce rapidly upon the loss of telomerase components, potentially making it difficult to detect excess G-strand (especially if overhang accumulation occurred at only a small fraction of telomeres). Alternatively, telomerase in S. cerevisiae and other systems may be functionally redundant in telomere protection and may be required in this role only in the absence of other protective factors.
Strikingly, the protective function of Candida telomerase can be fully mediated by a catalytically inactive allele of EST2. Thus, protection in this case cannot be accounted for by any mechanism that invokes directly or indirectly the nucleotide addition activity of telomerase. Although the precise mechanism awaits further analysis, we speculate that the core complex may bind directly to telomere ends during parts of the cell cycle to block the access of degrading enzymes. Recent analysis of telomere protection by end-binding proteins in S. cerevisiae suggests that C-strand degradation occurs only during parts of the cell cycle, specifically after completion of S-phase (40). It should be interesting to determine in the future the cell cycle dependence of Candida telomerase localization and its functional requirements.
A Multiplicity of Telomeric and Nontelomeric Functions for TERT.
It should be stressed that the protective function that we uncovered for Candida telomerase is mechanistically distinct from previous reports of protection by telomerase, which all seem to require a catalytically competent enzyme. For example, S. cerevisiae telomerase has been shown to suppress aberrant telomere fusion, especially in combination with checkpoint proteins such as Tel1p (41, 42). This ability is probably attributable to telomerase's ability to extend short telomeres, which are prone to fusion. Also as mentioned earlier, hypomorphic alleles of human TERT can extend cellular lifespan without causing bulk telomere elongation. Again this activity may be due to the ability of the mutant human TERT to extend critically short telomeres.
Telomerase even appears to have functions off telomeres; overexpression of human TERT in mammalian cells has been shown to cause global chromatin structure change and alter DNA repair efficiency, activities that nevertheless depend on a catalytically competent TERT (43). In addition, ectopic expression of human TERT has been shown to alter the expression of genes away from telomeres, including growth-promoting genes (44). Expression of mouse TERT, even in the absence of TER, leads to alterations in mouse stem cell properties (12). More recently, TER was reported to regulate ATR (ATM- and RAD3-related) and DNA damage response irrespective of the telomerase status of the cells (45). Thus, while we have apparently uncovered a mechanism of telomere protection by telomerase, our results also reinforce the notion that telomerase is a functionally versatile complex that can mediate multiple activities/processes on and possibly off telomeres.
Materials and Methods
Strains and Plasmids.
The C. albicans strains CAI4 (ura3Δ::imm434/ura3Δ::imm434) and BWP17 (ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG) were used as the parental strains (30, 46). The derivations of mutant strains are described below.
Cloning of the C. albicans TER1 Gene and Mapping of the 5′ and 3′ Ends of Mature RNA.
Details of the cloning and characterization of TER1 are provided in SI Materials and Methods.
Construction of Mutant Candida Strains.
An ≈3.9-kb EcoRI-SacI fragment containing the TER1 gene was cloned into EcoRI- and SacI-digested pBluescript SK− to generate pSR3. The EcoRI site of the insert was derived from C. albicans sequences, and the SacI site was derived from polylinker bordering the phage insert. The C. albicans URA3 gene, isolated as a 1566 HpaII fragment from pMB7 (30), was introduced into pSR3 at the ClaI site to create plasmid pSR3-U. Two base substitutions were then made in the putative template region of pSR3-U to create a BglII site, resulting in plasmid pSR3-U-B4D (47). We then transformed CAI4 with pSR3-U-B4D and identified correct integrants by Southern blotting. The integrants contained two copies of wild-type TER1 and a single copy of TER1-BglII.
The deletion strain ter1-ΔΔ was generated by subjecting BWP17 to two rounds of transformation and 5-FOA selection using a ter1::hisG-URA3-hisG cassette (containing ≈700 bp of TER1 upstream and ≈600 bp of downstream sequence). The reconstituted strain ter1-ΔΔ/TER1 was then obtained by transforming the deletion strain with the pGEM-URA3-TER1 integrating plasmid, which contained a 2.6-kb fragment spanning the TER1 gene cloned into the PstI and SacI site of pGEM-URA3. C. albicans transformations and 5-FOA selections were carried out as previously described (13).
Analysis of Telomeres and G-Strand Overhangs.
Chromosomal DNAs were isolated by Smash and Grab as previously described (48) except that for some preparations the initial aqueous phase was subjected to two additional rounds of phenol/chloroform/isoamyl alcohol (25:24:1) extraction to minimize nuclease contamination. Analysis of the CAI4 and Bgl strains used a wild-type telomere probe named Ca7-3 (5′-TGTCTAACTTCTTGGTGTAC-3′) and two mutant probes named CaBgl14 (5′-GTCTAAGATCTTGG-3′) and CaBgl16b (5′-GTCTTAGATCTTGGTG-3′). The blots were hybridized and washed at different temperatures depending on the probe: 48°C for Ca7-3, 32°C for CaBgl14, and 45°C for CaBgl16b. Hybridization was performed in the Church and Gilbert mix containing 7% SDS (49), and washing was done with 2% SDS and 200 mM Na2HPO4.
Analysis of ter1-ΔΔ and reconstituted strains used a 23-nt telomere probe named CaC1 (5′-ACACCAAGAAGTTAGACATCCGT-3′). Standard telomere Southern blot analysis followed established protocols (21). For in-gel hybridization, the gel was first soaked in 2× SSC for 30 min and then dried at room temperature on a gel dryer for 30 min. A bleed valve was used to maintain the pressure during drying at 550 mmHg. The gel was then transferred to a bag and subjected to prehybridization at 40–42°C for 20 min (Church and Gilbert mix containing 100 μg/ml salmon sperm DNA) and hybridization at 40–42°C for 16–20 h (the prehybridization solution supplemented with 2 × 106 cpm of CaC1 probe). The gel was washed three times with 4× SSC at 22°C for 20 min each and twice with 4× SSC/0.1% SDS at 52°C for 20 min each, and then subjected to PhosphorImager analysis. The procedure for denaturation of the DNA and rehybridization has been described (13).
Purification of and Primer Extension Assay for C. albicans Telomerase.
Whole-cell extracts of C. albicans and active telomerase fractions from DEAE columns were prepared as previously described (21, 50).
IP Western Blot and RT-PCR.
TMG(n) buffer (10 mM Tris·HCl, pH 8.0/1.2 mM MgCl2/0.1 mM EDTA/0.1 mM EGTA/10% glycerol; n refers to NaOAc concentration in millimolar) was used for the experiment. IgG-Sepharose beads (45 μl) were pretreated with 400 μg of tRNA in 1 ml of TMG(0) at 4°C for 30 min to minimize nonspecific binding. After one wash in TMG(0), the beads were incubated with extracts from est2-ΔΔ or reconstituted strains containing tagged Est2p (4.8 mg) in 1.2 ml of TMG (400) and subjected to gentle rotation at 4°C for 2 h. The beads were then washed five times with TMG(800) and twice with TMG(0) and divided into two equal aliquots. One aliquot was subjected to Western blot analysis using antibodies directed against protein A as previously described (24, 51). The second aliquot was treated with proteinase K and extracted with phenol/chloroform/isoamyl alcohol, and the RNA was isolated by ethanol precipitation. The level of Ter1 was then quantified by semiquantitative RT-PCR (2× RT-PCR Master Mix; USB) using primer pairs designed to amplify a 350-bp fragment (forward, 5′-CCCATATTCAATGCTCTTGGAGTGTG-3′; reverse, 5′-CTCCACAAGGTATCATACAAATTATGG-3′). The linearity of the assay was confirmed by titrating the samples.
Supplementary Material
Acknowledgments
We thank C. Strahl for technical help and E. Nock and I. K. Reichardt for assistance in making the Bgl mutant. This work was supported by National Institutes of Health Grants GM069507 (to N.F.L.), GM 26259 (to E.H.B.), and DE11356 (to E.H.B. and M.J.M.) and Human Frontier Science Program Fellowship LT-415/96 (to Y.T.).
Abbreviations
- TERT
telomerase reverse transcriptase
- TER1
telomerase RNA 1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700327104/DC1.
References
- 1.de Lange T. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira M, Miller K, Cooper J. Mol Cell. 2004;13:7–18. doi: 10.1016/s1097-2765(03)00531-8. [DOI] [PubMed] [Google Scholar]
- 3.Cech T. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn E. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 5.Autexier C, Lue NF. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 6.Collins K. Nat Rev Mol Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackburn EH. FEBS Lett. 2005;579:859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 8.Chan S, Blackburn E. Philos Trans R Soc London B. 2004;359:109–121. doi: 10.1098/rstb.2003.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim M, Xu L, Blackburn E. Exp Cell Res. 2003;288:277–287. doi: 10.1016/s0014-4827(03)00217-9. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Wang H, Bishop JM, Blackburn EH. Proc Natl Acad Sci USA. 1999;96:3723–3728. doi: 10.1073/pnas.96.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prescott J, Blackburn EH. Genes Dev. 1997;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- 12.Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, Artandi SE. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinberg-Neifach O, Lue NF. Nucleic Acids Res. 2006;34:2710–2722. doi: 10.1093/nar/gkl345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 15.Garvik B, Carson M, Hartwell L. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandin N, Damon C, Charbonneau M. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grandin N, Damon C, Charbonneau M. EMBO J. 2001;20:6127–6139. doi: 10.1093/emboj/20.21.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maringele L, Lydall D. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zubko MK, Guillard S, Lydall D. Genetics. 2004;168:103–115. doi: 10.1534/genetics.104.027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Booth C, Griffith E, Brady G, Lydall D. Nucleic Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh S, Steinberg-Neifach O, Mian I, Lue N. Eukaryot Cell. 2002;1:967–977. doi: 10.1128/EC.1.6.967-977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greider CW, Blackburn EH. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 23.Chapon C, Cech TR, Zaug AJ. RNA. 1997;3:1337–1351. [PMC free article] [PubMed] [Google Scholar]
- 24.Bosoy D, Peng Y, Mian I, Lue N. J Biol Chem. 2003;278:3882–3890. doi: 10.1074/jbc.M210645200. [DOI] [PubMed] [Google Scholar]
- 25.Yu G-L, Bradley JD, Attardi LD, Blackburn EH. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 26.McEachern MJ, Blackburn EH. Proc Natl Acad Sci USA. 1994;91:3453–3457. doi: 10.1073/pnas.91.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McEachern MJ, Krauskopf A, Blackburn EH. Annu Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 28.Biswas K, Rieger KJ, Morschhauser J. Gene. 2003;307:151–158. doi: 10.1016/s0378-1119(03)00456-6. [DOI] [PubMed] [Google Scholar]
- 29.McEachern M, Underwood D, Blackburn E. Genetics. 2002;160:63–73. doi: 10.1093/genetics/160.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fonzi W, Irwin M. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McEachern MJ, Haber JE. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 32.Lundblad V. Oncogene. 2002;21:522–531. doi: 10.1038/sj.onc.1205079. [DOI] [PubMed] [Google Scholar]
- 33.Mozdy AD, Cech TR. RNA. 2006;12:1721–1737. doi: 10.1261/rna.134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiang YJ, Hemann MT, Hathcock KS, Tessarollo L, Feigenbaum L, Hahn WC, Hodes RJ. Mol Cell Biol. 2004;24:7024–7031. doi: 10.1128/MCB.24.16.7024-7031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hathcock KS, Hemann MT, Opperman KK, Strong MA, Greider CW, Hodes RJ. Proc Natl Acad Sci USA. 2002;99:3591–3596. doi: 10.1073/pnas.012549799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marrone A, Stevens D, Vulliamy T, Dokal I, Mason PJ. Blood. 2004;104:3936–3942. doi: 10.1182/blood-2004-05-1829. [DOI] [PubMed] [Google Scholar]
- 37.Hsu M, Yu EY, Singh S, Lue NF. Eukaryot Cell. 2007 Jun 1; doi: 10.1128/EC.0869-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McEachern MJ, Blackburn EH. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 39.Dandjinou A, Levesque N, Larose S, Lucier J, Abou Elela S, Wellinger R. Curr Biol. 2004;14:1148–1158. doi: 10.1016/j.cub.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 40.Vodenicharov MD, Wellinger RJ. Mol Cell. 2006;24:127–137. doi: 10.1016/j.molcel.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 41.Mieczkowski P, Mieczkowska J, Dominska M, Petes T. Proc Natl Acad Sci USA. 2003;100:10854–10859. doi: 10.1073/pnas.1934561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan S, Blackburn E. Mol Cell. 2003;11:1379–1387. doi: 10.1016/s1097-2765(03)00174-6. [DOI] [PubMed] [Google Scholar]
- 43.Masutomi K, Possemato R, Wong JM, Currier JL, Tothova Z, Manola JB, Ganesan S, Lansdorp PM, Collins K, Hahn WC. Proc Natl Acad Sci USA. 2005;102:8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith LL, Coller HA, Roberts JM. Nat Cell Biol. 2003;5:474–479. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
- 45.Kedde M, Sage CL, Duursma A, Zlotorynski E, Leeuwen BV, Nijkamp W, Beijersbergen R, Agami R. J Biol Chem. 2006;281:40503–40514. doi: 10.1074/jbc.M607676200. [DOI] [PubMed] [Google Scholar]
- 46.Wilson RB, Davis D, Mitchell AP. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunkel TA, Roberts JD, Zakour RA. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 48.Hoffman C, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 49.Church GM, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh S, Lue N. Proc Natl Acad Sci USA. 2003;100:5718–5723. doi: 10.1073/pnas.1036868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia J, Peng Y, Mian IS, Lue NF. Mol Cell Biol. 2000;20:5196–5207. doi: 10.1128/mcb.20.14.5196-5207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.