Abstract
Proper metazoan mesoderm development requires the function of a basic helix-loop-helix (bHLH) transcription factor, Twist. Twist-containing dimers regulate the expression of target genes by binding to E box promoter elements containing the site CANNTG. In Caenorhabditis elegans, CeTwist functions in a subset of mesodermal cells. Our study focuses on how CeTwist controls the expression of its target gene, arg-1. We find that a 385 bp promoter region of arg-1, which contains three different E box elements, is sufficient for maintaining the full CeTwist-dependent expression pattern. Interestingly, the expression of arg-1 in different tissues is regulated distinctly, and each of the three E boxes plays a unique role in the regulation. The first and the third E boxes (E1 and E3) are required for expression in a distinct subset of the mesodermal tissues where arg-1 is normally expressed, and the second E box (E2) is required for expression in the full set of those tissues. The essential role of E2 in arg-1 regulation is correlated with the finding that E2 binds with greater affinity than E1 or E3 to CeTwist dimers. A potential role for additional transcription factors in mesodermal gene regulation is suggested by the discovery of a novel site that is also required for arg-1 expression in a subset of the tissues but is not bound in vitro by CeTwist. On the basis of these results, we propose a model of CeTwist gene regulation in which expression is controlled by tissue-specific binding of distinct sets of E boxes.
Keywords: C. elegans, Twist; bHLH, E box, hlh-2, hlh-8, arg-1, E/DA, mesoderm, DSL homolog
1. Introduction
Twist plays important roles in mesoderm development in many organisms including Drosophila melanogaster (Thisse et al., 1987), Caenorhabditis elegans (Harfe et al., 1998; Corsi et al., 2000) and humans (Wang et al., 1997). In Drosophila, where Twist was first identified, a null mutation in the gene causes embryonic lethality due to the complete absence of mesoderm (Simpson, 1983). In C. elegans, mutations in CeTwist disrupt normal development of a subset of muscles that are involved in egg laying and defecation, resulting in egg-laying defective and constipated phenotypes (Harfe et al., 1998; Corsi et al., 2000; Corsi et al., 2002). The homology among the downstream target genes of DTwist and CeTwist demonstrate a striking parallel in the pathways between the two organisms (Harfe et al., 1998; Krantz et al., 1998). In humans, mutations in the TWIST1 gene result in Saethre-Chotzen syndrome, one form of congenital craniosynostotic disease, characterized by the premature fusion of cranial sutures in the newborn (Wilkie, 1997). Interestingly, mutations in the human homologs of several CeTwist target genes cause various syndromes characterized by craniofacial defects (Jabs et al., 1993; Chieffo et al., 1997; Harfe et al., 1998; Krantz et al., 1998; Crosnier et al., 1999; Hehr and Muenke, 1999; Lajeunie et al., 1999). Moreover, FGFR2, the human homolog of the CeTwist target gene egl-15, has recently been demonstrated to be a target gene of human Twist1 (Connerney et al., 2006). These results demonstrate that Twist pathways in nematodes, insects and vertebrates are well conserved. Thus, the study of CeTwist and its target genes in C. elegans will help us to further understand how the Twist pathway functions in humans leading to a better understanding of the molecular basis for human congenital craniofacial diseases.
All bHLH family transcription factors function as either homodimers or heterodimers with another bHLH protein. Twist can homodimerize or heterodimerize with the bHLH proteins E in humans and Daughterless (DA) in Drosophila. Importantly, the two forms of dimers have distinct mesodermal functions during development (Castanon et al., 2001; Connerney et al., 2006). In C. elegans, the only identified partner for CeTwist (encoded by the hlh-8 gene) is CeE/DA (encoded by the hlh-2 gene), which is the homolog of E proteins and Daughterless (Krause et al., 1997; Harfe et al., 1998). CeTwist can form homodimers in vitro; however, whether CeTwist homodimers are present in vivo is not clear (Harfe et al., 1998). The CANNTG conserved core DNA binding sequence of bHLH dimers is called an E box. The middle two nucleotides of an E box have been postulated to provide binding specificity for certain bHLH dimers, with additional influence from flanking sequences. In vitro selection assays for binding sites indicate that different bHLH proteins prefer to bind different E boxes. For example, homodimers of MyoD, another bHLH family protein, prefer to bind to CAGCTG; while heterodimers of vertebrate Twist and E proteins prefer to bind to CATATG (Kophengnavong et al., 2000). How CeTwist regulates the expression of its target genes through specific binding sites in vivo is not well understood.
CeTwist is expressed in a subset of mesodermal tissues. These cells include the head mesodermal cell (hmc) whose function is unclear, the four enteric muscles which are required for defecation, the post-embryonically born mesodermal precursor cells that are derived from the M mesoblast, including precursors of the 16 sex muscles which are critical for proper egg laying, and eight of the 16 differentiated sex muscles (four vm1 and four vm2 vulval muscles (vm)) (Harfe et at., 1998; P. W. and A. K. C., unpublished; Fig. 1). Thirteen CeTwist target genes have been identified (Harfe and Fire, 1998; Harfe et al., 1998; Kostas and Fire, 2002; Wang et al., 2006). The expression pattern and promoter region of three of these genes, ceh-24, egl-15 and mls-1, have been studied (Harfe and Fire, 1998; Harfe et al., 1998; Kostas and Fire, 2002). These three target genes are expressed in a subset of the tissues where CeTwist is expressed and each gene shows a distinct expression pattern (Table 1). ceh-24 is expressed in the eight vulval muscles, egl-15 is expressed in a subset of the sex muscle precursors and the four vm1 vulval muscles, and mls-1 is expressed in a subset of the sex muscle precursors, the four vm2 vulval muscles, the uterine muscles (um), and three of the four enteric muscles (intestinal muscles and anal depressor, but not the anal sphincter). The differential expression patterns of CeTwist target genes suggest that CeTwist alone or in combination with other factors, regulates the expression of each of these genes in a distinct manner.
Figure 1. CeTwist is expressed in non-lineally related cells.
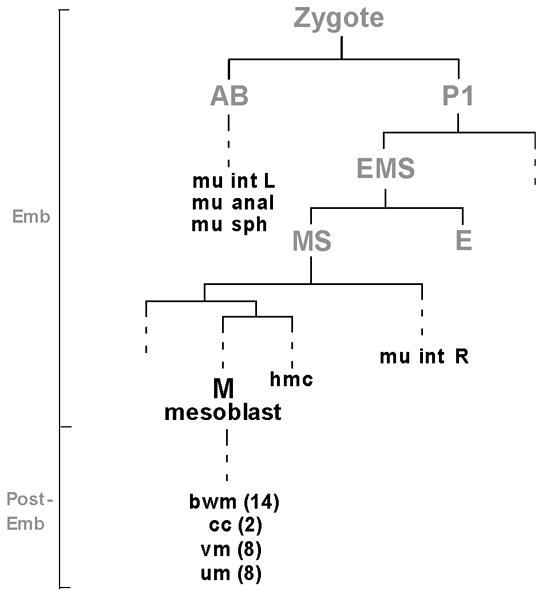
A partial lineage to illustrate the cells where hlh-8 is expressed (dark black letters). hlh-8 is expressed in the embryonically born hmc, four enteric muscles (mu int L and R, mu sph and mu anal), M mesoblast and post-embryonically born M descendants (cc and vm and the precursors to the um and bwm). Founder cells are labeled in gray. Vertical lines represent cells and horizontal lines represent cell divisions. Dashed lines are used where division events are not shown. The vertical line on the left is not drawn to scale but indicates the embryonic (Emb) and post-embryonic (Post-Emb) stages during development. The number of each type of cell is indicated in parentheses if the number is greater than one. Abbreviations used: mu int L and R, left and right intestinal muscles; mu sph, anal sphincter; mu anal, anal depressor; hmc, head mesodermal cell; bwm, body wall muscles; cc, coelomocytes; vm, vulval muscles; um, uterine muscles.
Table 1.
Differential expression patterns and important promoter regions of CeTwist and target genes.
| expression sites
|
minimal promoter region b (bp) | E boxes in minimal promoter region (bp) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| hmc a | M lineage | vm1 | vm2 | um | mu ints | mu sph | mu anal | |||
| hlh-8 c | + d | + | + | + | − | + | + | + | ------ | ------ |
| egl-15 | − | + e | + | − | − | − | − | − | −701 to −1 | CATATGs (−602, −371, & −239), CAGATC (−572) & CATCTG (−471), |
| mls-1 | − | + f | − | + | + | + | − | + | −798 to −1 | CACGTG (−270), CAGATG (−179) & CATATG (−123) |
| ceh-24 | − | − | + | + | − | − | − | − | −2066 to −2019 | CATATGs (−2037 & −2028) |
| arg-1 | + | − | + | + | + | + | + | + | ? | ? |
Abbreviations used: hmc: head mesodermal cell; M lineage: descendants of M mesoblast before differentiation occurs; vm1 & vm2: type 1 & type 2 vulval muscles; um: uterine muscles; mu ints: left and right intestinal muscles; mu sph: anal sphincter; mu anal: anal depressor.
Minimal promoter region: the smallest promoter region for maintaning the full expression pattern of a gene.
References for the data. hlh-8: Harfe et al., 1998; P. W. and A. K. C., unpublished. egl-15: Harfe et al., 1998. mls-1: Kostas and Fire, 2002; S. Kostas and A. Fire, personal communication. ceh-24: Harfe and Fire, 1998. arg-1: Kostas and Fire, 2002; this paper.
Symbols used: +, positive expression; - , no expression.
egl-15 is expressed in the two sex myoblasts (SM) in M lineage.
mls-1 is expressed in a subset of SM descendants which give rise to the vm2 and um.
In several CeTwist target genes, the potential roles of the E boxes within the minimal promoter region (the smallest promoter region sufficient to drive reporter gene expression in a wild-type pattern) of CeTwist target genes have been previously investigated (Table 1). For mls-1, mutational analysis of one of the three E boxes (CAGATG, -179 bp) within the 798 bp minimal promoter region (−798 to −1 bp) revealed its important role for the expression and function of this gene (Kostas and Fire, 2002; S. Kostas and A. Fire, personal communication). For ceh-24, the two CATATGs (called NdE boxes) in the 48 bp minimal upstream sequence (−2066 to −2019 bp) are required for its expression (Harfe and Fire, 1998). And for egl-15, deletion mutations suggest that the five E boxes within the 701 bp minimal promoter region (−701 to −1 bp), which consist of three different E box types, are important for the expression of this gene (Harfe et al., 1998) (Table 1). These studies indicate that distinct sets of E boxes might be involved in the differential regulation of each gene. However, the CeTwist-dependent role that each E box plays within the minimal promoter region of any of these genes is not known.
In order to understand how CeTwist can differentially regulate its target genes, we wanted to examine a target with the broadest expression pattern. The target gene, arg-1, was chosen because it is expressed in multiple, lineally-unrelated mesodermal cell types (Table 1, Fig. 1). arg-1 is a member of the Delta/Serrate/LAG-2 (DSL) family of Notch signaling ligands (Mello et al., 1994). Although the function of the ARG-1 protein has not been defined, ARG-1 can substitute for LAG-2 in vivo when expressed from the lag-2 promoter, indicating that it can act as a Notch ligand (Fitzgerald and Greenwald, 1995). Additionally, it is not clear how arg-1 functions as a CeTwist target during mesoderm development since a deletion mutation in the gene and arg-1 RNAi experiments did not result in an obvious mesodermal phenotype (J. Z. and A. K. C., unpublished). This lack of phenotype may be due to overlapping functions with either the DSL homolog, apx-1, as the two genes seem to have arisen from a gene duplication event during evolution (www.treefam.org/cgi-bin/TFinfo.pl?ac=TF316224) or with one of the other eight C. elegans DSL homologs (Chen and Greenwald, 2004). An arg-1::gfp reporter is expressed in multiple mesodermal tissues including the hmc, vm1 and vm2 vulval muscles, and the four enteric muscles (Kostas and Fire, 2002) (Fig. 2). This expression pattern overlaps only partially with the cells where CeTwist and its other known target genes are found (Table 1).
Figure 2. A transcriptional arg-1::gfp reporter is expressed in the head mesodermal cell, the vulval muscles, and the enteric muscles.
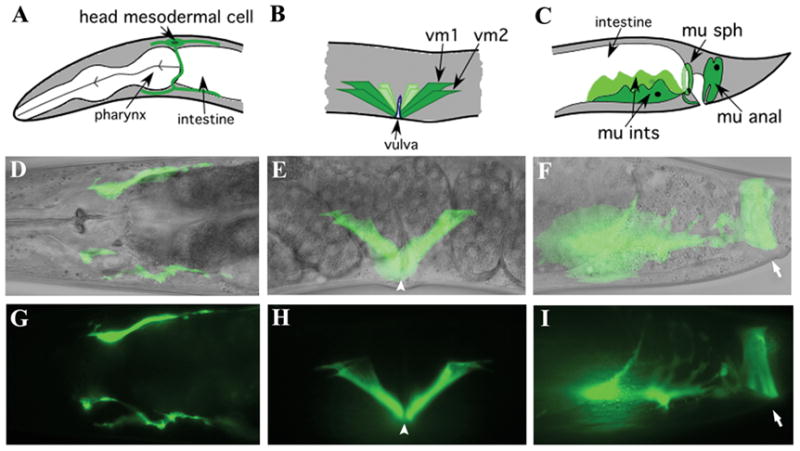
The upper panels show line drawings, middle panels show merged micrographs of Nomarski and GFP images, and lower panels show GFP images alone. (A, D, G) the head mesodermal cell, (B, E, H) vm1 and vm2 vulval muscles, and (C, F, I) the four enteric muscles. (F, I) The left intestinal muscle, anal sphincter and anal depressor are visible and the right intestinal muscle is in a different focal plane. The position of the vulval opening is marked by arrowheads and the anal opening is marked by arrows.
Our studies focus on understanding the mechanism of tissue-specific regulation of arg-1 expression by CeTwist. We performed a detailed analysis of the upstream sequence of arg-1 to identify the specific sequences in the promoter region that are required for its expression. Here, we report that a minimal 385 bp upstream sequence of arg-1 is sufficient to maintain its full expression pattern. Moreover, the tissue-specific expression of this gene is regulated differentially. There are three E boxes within the minimal promoter region that play unique roles, with each one required for expression in a distinct set of tissues. This type of regulation has not been identified in previously studied CeTwist target genes. In addition, we identified a six-nucleotide sequence that is also involved in the regulation of arg-1 expression in the vm1 vulval muscles. Altogether, our results provide insights into how a single transcription factor can mediate differential temporal and spatial gene expression.
2. Results
2.1. A 436 bp promoter region is sufficient to drive arg-1::gfp expression in the hmc, vulval muscles, and enteric muscles
An arg-1::gfp integrated line (PD4444) has been reported to express GFP in the hmc, the eight vms, and the four enteric muscles (Kostas and Fire, 2002) (Fig. 2). In this line, we found that arg-1::gfp was also expressed in the uterine muscles, although the GFP intensity and frequency was relatively low (data not shown). We also detected GFP expression in a few unidentified neurons in the nerve ring region late in embryogenesis, which gradually disappeared at the L1 larval stage. The PD4444 arg-1::gfp reporter was generated by inserting the 2824 bp upstream sequence of arg-1 into a gfp reporter vector (J. K. Liu., personal communication). We reconstructed the original arg-1::gfp reporter with a larger upstream region in order to include the full complement of potential 5’ regulatory sites. We generated a transcriptional arg-1::gfp reporter, pZJ02 [−6240 to −1], by inserting a 6240 bp upstream fragment (without the ~150 bp sequence immediately downstream of F31A9.4, Fig. 3A) into a gfp vector, and we examined the GFP expression pattern. Because we intended to make both 5’ as well as 3’ deletions of the arg-1 upstream region, we used a vector that contains an egl-18 basal promoter. This basal promoter vector has been used previously to identify potential enhancer elements of other genes (Wagmaister et al., 2006).
Figure 3. Identification of the arg-1 promoter region that retains full expression activity.
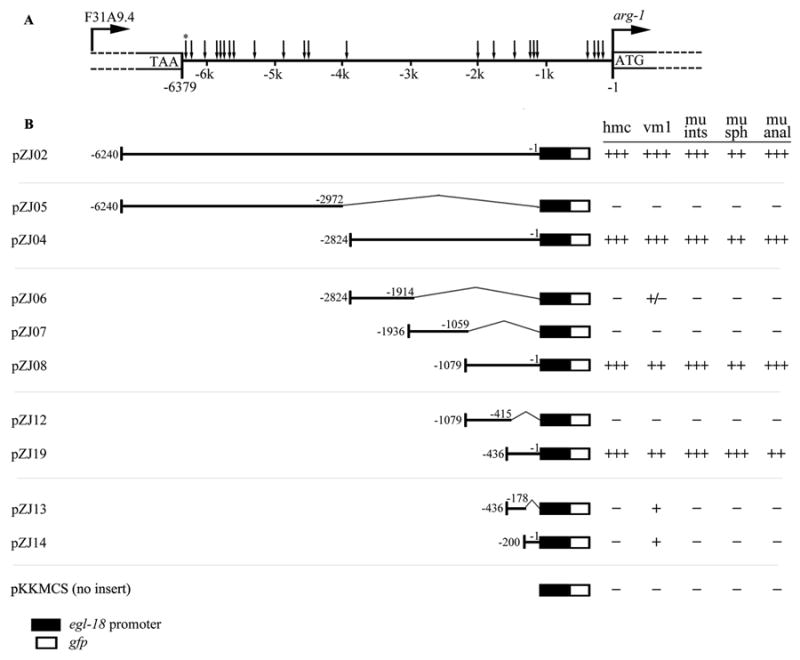
(A) The 23 E boxes present in the upstream sequence of arg-1 are indicated by arrows located between the stop site of F31A9.4 and the start site of arg-1. The asterisk (*) indicates the E box at position −6359 which was not included in the pZJ02 [−6240 to −1] gfp construct. (B) Four sets of successively smaller bi-directional deletion constructs derived from pZJ02 [−6240 to −1] in addition to a construct with no insert (pKKMCS) are shown. The smallest construct that retained full expression activity was pZJ19 [−436 to −1]. The gfp vector used for each construct was pKKMCS which contains an egl-18 basal promoter (black box) in front of the gfp cDNA (empty box). Each construct was tested for activity in at least two independent transgenic lines. For each line, at least 20 adult transgenic animals were observed for expression of GFP in the head mesodermal cell (hmc), vm1 vulval muscles (vm1), intestinal muscles (mu ints), anal sphincter (mu sph) and anal depressor (mu anal). Activities of each DNA insert were assigned based on percentage of animals observed with GFP expression according to the following criteria: +++ (90–100%), ++ (60–89%), + (20–59%), ± (11–19%), and − (≤ 7%).
The transgenic animals harboring the pZJ02 [−6240 to −1] reporter showed a broader GFP expression pattern than the PD4444 animals with the previously integrated arg-1::gfp that includes a shorter arg-1 upstream region. In addition to the hmc, the sex muscles, and the four enteric muscles, GFP was also observed in the anterior and posterior gonadal sheath cells in four independent lines (Table 2). The entire intergenic region was then subdivided into four sets of increasingly smaller fragments to assay for gfp expression (Fig. 3B). Only the hmc, sex muscle, and enteric muscle expression depended on CeTwist (see later section) so we focused our expression analysis on these tissues. As expected, the 2.8 kb region directly upstream of the ATG drove expression in all of these tissues. After subdividing the 2.8 kb region, we determined that the proximal 436 bp upstream of the arg-1 coding sequence was sufficient to drive GFP expression in all of the tissues (Fig. 3B, pZJ19 [−436 to −1]). When this fragment was further divided into two segments, with 22 bp overlapping at the junction, in pZJ13 [−436 to −178] and pZJ14 [−200 to −1], GFP expression was lost in the majority of the tissues (Fig. 3B), suggesting that the proximal 436 bp upstream sequence is necessary and sufficient for arg-1::gfp expression in the hmc, vm1, and four enteric muscles.
Table 2.
Expression pattern of arg-1 promoter segments in wild-type, hlh-8 (-) and hlh-2 RNAi animals
| arg-1 promoter region (bp) | genotypes/treatment | GFP expression pattern
|
|||
|---|---|---|---|---|---|
| vm1 a | hmc & enteric muscles | gonadal sheath cells | pm6 & pm7 | ||
| pZJ02 [−6240 to −1] | WT | + | + | + | − |
| hlh-8 (-) b | − | − | − | − | |
| hlh-2 RNAi c | −/+d | + e | + f | − | |
| pZJ05 [−6240 to −2972] | WT | − | − | + | + |
| hlh-8 (-) | − | − | + | + | |
| hlh-2 RNAi | − | − | + f | + | |
| pZJ04 [−2824 to −1] | WT | + | + | − | − |
| hlh-8 (-) | − | − | − | − | |
| hlh-2 RNAi | −/+ | + | − | − | |
| pZJ19 [−436 to −1] | WT | + | + | − | − |
| hlh-8 (-) | − | − | − | − | |
| hlh-2 RNAi | −/+ | + | − | − | |
Abbreviations used: hmc, head mesodermal cell; vm1, type 1 vulval muscles; enteric muscles, left and right intestinal muscles, anal sphincter and anal depressor; pm6 and pm7, pharyngeal muscles 6 and 7; hlh-8 (-), hlh-8 (nr2061).
hlh-8 (-) mutation is a predicted null allele (Corsi et al., 2000).
L1 larvae were fed bacteria harboring an hlh-2 RNAi vector or an L4440 empty vector (data not shown), and the GFP expression pattern was observed in at least 30 adult worms for each RNAi treatment.
Residual GFP expression in the vm1 was observed in < 25% of the hlh-2 RNAi treated animals; in contrast, GFP expression in the vm1 was observed in at least 91% of the animals in RNAi control group (not shown).
GFP expression in the hmc and enteric muscles was unaffected possibly because these cells are already formed during embryogenesis prior to the RNAi treatment.
The amount of GFP expression in the gonadal sheath cells was essentially unchanged after hlh-2 RNAi treatment compared to the control group. However, the morphology of the cells was abnormal because the hlh-2 RNAi treatment disrupted their development. Sterility and protruding vulva phenotypes were also observed in the hlh-2 RNAi-treated animals.
2.2. The proximal 436 bp promoter region of arg-1 is regulated by CeTwist and CeE/DA
In order to test the dependence of arg-1::gfp expression on CeTwist function, some of our gfp reporters were crossed into a predicted null allele, hlh-8 (nr2061), referred to as hlh-8 (-) in this paper. Previous work demonstrated that most aspects of PD4444 arg-1::gfp expression are abolished in homozygous hlh-8 (-) null mutant animals (Wang et al., 2006). In order to test whether the full length upstream sequence is regulated by CeTwist, we crossed one transgenic line derived from each of the three constructs pZJ02 [−6240 to −1], pZJ04 [−2824 to −1] and pZJ05 [−6240 to −2972] into the hlh-8 (-) background and examined whether the expression persisted or not (Table 2). The GFP expression in the hmc, vm1, and four enteric muscles in animals containing constructs of either the ~6.2kb insert (pZJ02) or the ~2.8kb insert (pZJ04) was completely abolished.
In our initial observation of pZJ05 [−6240 to −2972], we saw GFP expression not only in the gonadal sheath cells, but also in the pharyngeal muscles pm6 and pm7 (Table 2). The pharyngeal muscle expression was suppressed by the proximal ~2.8 kb upstream sequence since the whole 6240 bp upstream sequence did not show GFP expression in these cells. However, in the hlh-8 (-) mutants harboring pZJ05 [−6240 to −2972], the expression in the pharyngeal muscles persisted, indicating that GFP expression in this tissue was independent of CeTwist function. Additionally, in the hlh-8 (-) background, GFP expression of the pZJ02 [−6240 to −1] construct in the gonadal sheath cells disappeared; however, the pZJ05 [−6240 to −2972] reporter was still expressed in this tissue. One possible explanation is that the GFP expression in the gonadal sheath cells was dependent on CeTwist protein only in the presence of the proximal 2.8 kb promoter region. The explanation for this phenomenon will require further investigation.
To confirm that expression of arg-1 is activated by CeTwist through the proximal 436 bp promoter region, two independent transgenic lines derived from pZJ19 [−436 to −1] were crossed into the hlh-8 (-) mutant background. The GFP expression in the mutants was completely abolished (Table 2) indicating that CeTwist is required for the expression.
In order to investigate whether the CeTwist heterodimeric partner CeE/DA is also important to activate arg-1::gfp expression through this promoter region, we took two approaches: one to remove CeE/DA function and the other to overexpress the protein along with CeTwist. In the first approach, RNAi experiments were performed to knock down hlh-2 expression, because, in contrast to hlh-8, there are no null alleles of hlh-2 currently available. Additionally, treating adults with RNAi leads to embryonic lethality in the offspring (Krause et al., 1997) so L1 animals were fed dsRNA to degrade the hlh-2 mRNA during larval development. This procedure allowed us to examine the expression of arg-1::gfp in the vulval muscles since they are born later in the L4 stage. Compared to control animals that were not fed the hlh-2 dsRNA, the treated animals had a 66% decrease in the number of animals that expressed arg-1::gfp in the vm1 muscles indicating that CeE/DA plays an important role for arg-1 expression in these cells (Table 2). The expression of arg-1::gfp in the hmc and enteric muscles was unchanged when the treated and control group of animals were compared (Table 2). These cells are all born and expressing the gfp reporter during embryogenesis prior to the RNAi treatment and would not be expected to be affected by the treatment. However, if newly expressed CeE/DA was required to maintain the arg-1 expression in these cells during larval development we might expect to see a decrease in GFP expression, which was not observed in the experiment. Therefore, the requirement for CeE/DA function in these particular cells will require further investigation.
If CeTwist/CeE/DA heterodimers are required to activate arg-1 expression through the proximal 436 bp sequences, this promoter region of arg-1 should also respond to the ectopic expression of CeTwist and CeE/DA. Therefore a second strategy of overexpressing the bHLH proteins was employed. The pZJ19 [−436 to −1] was injected into a strain (AK103) which contains integrated phs::hlh-8 and phs::hlh-2 transgenes in which CeTwist and CeE/DA can be overexpressed upon heat shock. Ectopic arg-1::gfp expression in this strain, especially in embryos and young larvae, was observed after heat shock treatment (Table 3). Strong GFP expression was observed in the intestine cells, hypodermis, and neurons where the heat shock promoter is most active, as well as in the tissues where arg-1 is normally expressed. In addition, the pZJ19 construct was also injected into a strain (AK100) containing only an integrated phs::hlh-8 plasmid, in which CeTwist but not CeE/DA can be overexpressed upon heat shock. Interestingly, ectopic GFP expression was also observed in the transgenic animals at various stages, especially in embryos and young larvae (Table 3). The onset of the ectopic GFP expression was delayed and the intensity was less strong when compared to overexpression of both bHLH proteins (compare AK105 to AK104 in Table 3). The arg-1::gfp reporter pZJ19 [−436 to −1] alone did not respond to heat shock (AK107 in Table 3). Altogether these results suggested that arg-1::gfp expression was activated through the 436 bp promoter region by CeTwist/CeE/DA heterodimers and, possibly, CeTwist homodimers. It is also formally possible that CeE/DA homodimers could be activating arg-1, however this scenario is less likely since CeE/DA homodimers do not bind the E boxes in the 436 bp region (see later section describing Fig. 6).
Table 3.
Ectopic arg-1::gfp expression upon overexpression of CeTwist alone or with CeE/DA.
| Strain name | heat shock plasmidsa | arg-1::gfp reporter | heat shock 20 minb | heat shock 2 hrb | ||
|---|---|---|---|---|---|---|
| recover 2 hr | recover 20 hr | recover 2 hr | recover 20 hr | |||
| AK100c | phs::hlh-8 | none | −d | − | −e | − |
| AK107 | none | pZJ19 [−436 to −1] | − | − | − | − |
| AK105c | phs::hlh-8 | pZJ19 [−436 to −1] | +/− | + | + | +++ |
| AK104 |
phs::hlh-8
phs::hlh-2 |
pZJ19 [−436 to −1] | ++ | ++++ | ++ | ++++ |
Integrated plasmids containing cDNA under the control of the hsp16.41 promoter.
Populations of mixed stage animals of each strain were subjected to heat shock treatment at 33°C for 20 min or 2 hrs followed by recovery at 20°C. The GFP expression pattern of the animals was examined under the fluorescent stereomicroscope during recovery.
Strains AK100 and AK105 also contain hlh-8::gfp, which is a transcriptional reporter that does not contain any coding sequence. This gfp construct was linked to the integrated phs::hlh-8 and could not be crossed out of the strain. However, the difference between the response to heat shock in AK100 compared to AK105 is specific to arg-1::gfp since AK100 has essentially no ectopic expression from hlh-8::gfp.
-:no ectopic GFP expression compared to animals without heat shock treatment; +/−: low level of ectopic GFP expression; +: modest level of ectopic GFP expression; each additional + indicates a significant increase in GFP expression.
Sporadic, low level of ectopic hlh-8::gfp expression was observed in embryos but not larvae after heat shock treatment for 2 hrs and recovery for 4 hrs; the GFP expression disappeared after recovery for 8 hrs.
Figure 6. The three arg-1 E boxes are bound by CeTwist homodimers and CeTwist heterodimers but not by CeE/DA homodimers.
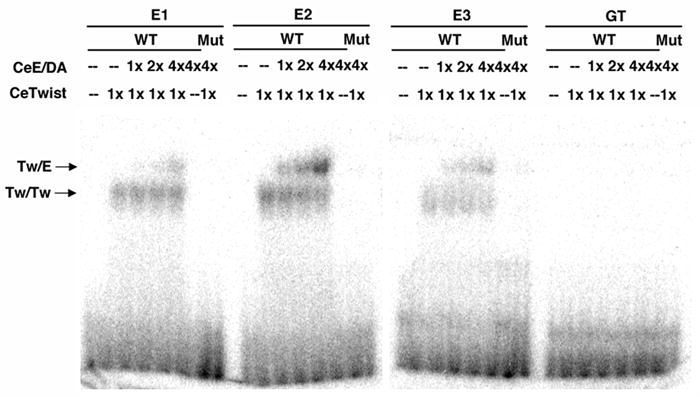
An Electrophorectic Mobility Shift Assay (EMSA) was performed using CeTwist and CeE/DA purified from a bacterial expression system. The proteins were incubated with 20mer probes made using the sequences of the wild-type (WT) and mutated (Mut) promoter regions (E1, E2, E3) identified in Fig. 5. The protein-DNA mixture was separated on native gels followed by autoradiography. The relative amount of each protein (1X, 2X, 4X) incubated with the probes was determined by SDS-PAGE analysis. CeTwist/CeE/DA heterodimers (Tw/E) and CeTwist homodimers (Tw/Tw) bound with high affinity to E2 and with relatively lower affinity to E1 and E3. Using a phosphorimager to quantify the binding, Tw/E binds with 4-fold greater affinity to E2 than to E1 or E3 and Tw/Tw binds with nearly 2-fold greater affinity to E2 than to E1 or E3. The binding was abolished with the mutated probes. The GT box was not bound by any of these bHLH proteins, suggesting that it is bound by another factor in vivo.
2.3. The expression of arg-1 is differentially regulated
Considering that arg-1 is expressed in multiple tissues, we next investigated whether the expression of this gene in different tissues is regulated by distinct promoter elements. We made a series of 5’ deletions of the 436 bp promoter region and asked if any of the deletions resulted in loss of GFP in some or all of the cells that normally express arg-1::gfp (Fig. 4). These deletions are intermediate-scale deletions used to define small regions with critical tissue-specific functions that may point to specific E boxes for further study. There are four E boxes in the 436 bp promoter region of arg-1 (Fig. 4). Our series of constructs successively removed one E box at a time in addition to other surrounding promoter sequences (E boxes CAATTG (−391), CATATG (−266), CATCTG (−215), and CACATG (−155), the last three were referred to as E1, E2, and E3, respectively) (Fig. 4). Removal of the first 51 bp (pZJ23 [−385 to −1]) from the 5’ end did not result in any obvious change in the GFP expression pattern. However, removal of the next 125 bp (pZJ24 [−260 to −1]) caused GFP expression to be lost in the hmc, greatly diminished in the vm1 muscles, and unchanged in the enteric muscles, indicating that this piece of DNA (containing E box E1) was required for arg-1::gfp to be expressed in the hmc, and was also important for its expression in vm1 muscles. Upon deleting the next 51 bp (pZJ26 [−209 to −1]), the GFP expression was completely abolished, indicating that the region containing E2 was critical for arg-1::gfp to be expressed in the four enteric muscles and was also important for its expression in the vm1 vulval muscles. Since the enteric muscles are born during embryogenesis and the vm1 vulval muscles are born post-embryogenesis, it is possible that arg-1 expression in the two groups of tissues is controlled separately (Fig. 1). To investigate this possibility, another deletion construct was generated in which only 20 bp instead of 51 bp was removed from construct pZJ24 [−260 to −1] creating pZJ25 [−240 to −1] (Fig. 4). Interestingly, removal of this region, which does not contain an E box, abolished the residual GFP expression in the vm1 but did not affect expression in the enteric muscles. Therefore, this small region was potentially involved in regulating arg-1::gfp expression in the vm1 muscles, and the 31 bp removed in pZJ26 [−209 to −1], including one E box, was likely required for the enteric muscle expression. These results suggested that the 385 bp promoter region of arg-1 was sufficient to maintain the full expression pattern. This region can be divided into four segments, from −385 to −261 bp, −260 to 241 bp, −240 to −210 bp and −209 to −1 bp. Each of the first three regions was involved in the differential regulation of arg-1 expression and played a distinct role in this process. The fourth segment (−209 to −1 bp) could not be assessed in these experiments, yet its importance was indicated by construct pZJ13 [−436 to −178] (Fig. 3B), in which the proximal 177 bp was removed and the GFP expression in the hmc and enteric muscles was completely lost. Next, we focused on the E boxes present in these regions of the arg-1 promoter to determine their contribution to the differential expression.
Figure 4. The expression of arg-1 is regulated in a tissue-specific manner.
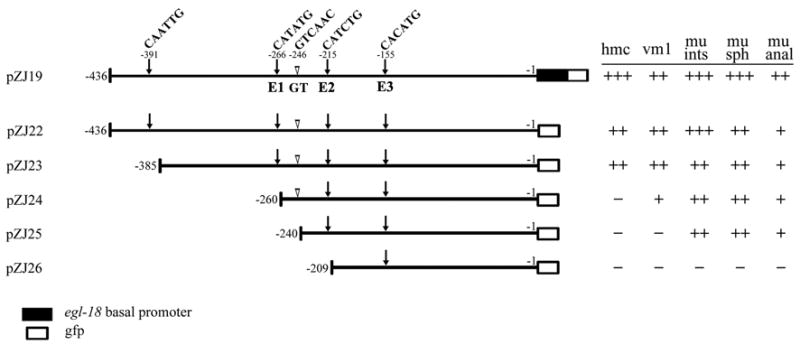
A series of 5’ unidirectional deletion constructs derived from promoter construct pZJ19 [−436 to −1] are diagrammed. The 436 bp promoter region contains four E boxes (arrows): −391: CAATTG, −266: CATATG (E1), −215: CATCTG (E2), and −155: CACATG (E3). The GT box sequence GTCAAC (−246 bp) is indicated with open arrowheads. Constructs pZJ22 [−436 to −1] through pZJ26 [−209 to −1] were made using pPD95.67 which does not contain any basal promoter in front of the gfp cDNA (empty box only). Examination of the transgenic animals derived from each construct was conducted as described in the Fig. 3 legend.
2.4. Individual E boxes are required for the differential regulation of arg-1::gfp expression
Three different E boxes are present in the proximal 385 bp upstream of arg-1. Each E box was included in one of the four promoter segments described above (Fig. 4). Since CeTwist is predicted to activate target gene expression upon binding an E box, it is possible CeTwist binds each E box under certain conditions leading to the differential expression of arg-1. We next investigated the roles of the E boxes by mutating the consensus sequence from CANNTG to AANNAG. This sequence alteration can eliminate the function of an E box (Karp and Greenwald, 2003). The mutations were all made to the proximal 385 bp promoter region of arg-1 (pZJ23 [−385 to −1]) (Fig. 5). Since each construct was identical except for one E box mutation, we could query the function of each E box individually.
Figure 5. Differential regulation of the arg-1 expression pattern requires distinct E boxes.
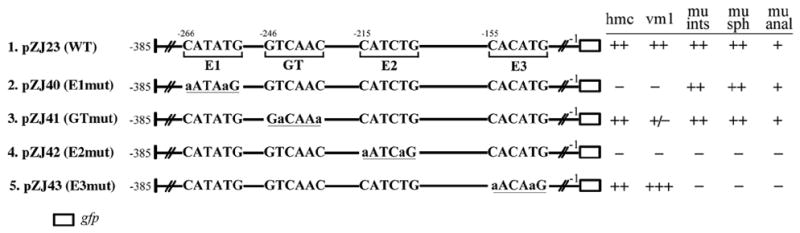
The E boxes (E1, E2, E3) in the minimal promoter region, pZJ23 [−385 to −1] (line 1), were examined using site-directed mutagenesis. GFP reporters containing the mutated sites revealed tissue-specific requirements for each E box (lines 2,4,5). A GFP reporter made with a mutated GT box, which has the same base composition as an E box, leads to a decrease in expression in vulval muscles (line3). The mutated sequence in each construct is underlined and the specific mutated nucleotides are in lower case. Black bars indicate the sequences that are not changed in the mutagenesis. Refer to Fig. 3 for details on examining the GFP expression pattern.
When E1 was mutated, the expression of arg-1::gfp in both the hmc and vm1 was abolished, yet expression in the enteric muscles was comparable to pZJ23 [−385 to −1] (Fig. 4, 5). Interestingly, when E2 was mutated, the expression in all the tissues was completely abolished. When E3 was mutated, the expression of arg-1::gfp in the four enteric muscles disappeared, whereas the hmc and the vm1 expression was unchanged (Fig. 5). Altogether, our mutant analysis indicated that E1 and E3 were individually required for arg-1 expression in a subset of tissues, whereas E2 was required in all tissues.
2.5. A unique six-nucleotide sequence is involved in regulating arg-1 expression in the vm1 muscles
Within the 385 bp minimal promoter region, the sequence from −260 to −241 bp, which is important for arg-1 expression in the vm1 muscles, does not contain an E box (Fig. 4). It does contain a six-nucleotide sequence GTCAAC at −246 bp that we refer to as a GT box with the same composition as an E box (CANNTG) but in an opposite orientation. The potential function of this sequence was also investigated due to its interesting composition and location. This sequence was mutated to GACAAA (pZJ41 GTmut), following the same strategy as mutating an E box (Fig. 5). The mutation resulted in a loss of the GFP expression in the vm1 muscles in 81% of the transgenic animals (n = 53), while expression in all other tissues was unaffected. This result was consistent with the data obtained from the unidirectional deletion constructs (pZJ24 [−260 to −1] and pZJ25 [−240 to −1]; Fig. 4), suggesting that the GT box was also involved in regulating the expression of arg-1 in the vm1 muscles.
2.6. The three E boxes but not the GT box are bound by CeTwist-containing dimmers
Since the E boxes in the minimal arg-1 promoter were playing unique roles in the gene’s expression, we tested whether they could each be bound by CeTwist and/or CeE/DA. We also wanted to test whether CeTwist and/or CeE/DA could bind to the GT box in the arg-1 promoter as well. An electrophoretic mobility shift assay (EMSA) was performed using bacterially-expressed recombinant proteins that were incubated with 20mer probes containing individual E or GT boxes and their surrounding sequences. Each of the three E boxes was bound by both CeTwist homodimers and heterodimers but not by CeE/DA homodimers (Fig. 6), suggesting that CeTwist-containing dimers regulate arg-1 expression by directly binding to the E boxes in vivo. Interestingly, E2 bound more CeTwist/CeE/DA heterodimers than the other two E boxes, suggesting this site might have greater affinity for the dimers in vivo. The bHLH binding activity for the three E boxes was specific since it was lost when mutated probes were used in the assay. Additionally, the other site we found to be important for arg-1 expression, the GT box, was not bound by either CeTwist or CeE/DA-containing dimers, suggesting the presence of another factor which binds to this sequence and contributes to the regulation of arg-1 expression.
3. Discussion
3.1. Selective E boxes are required for arg-1 expression
There are 22 canonical E boxes in the 6240 bp upstream sequence of arg-1. Each E box is in a distinct context of flanking sequence. Our studies demonstrated that only three of those E boxes are necessary for the expression of arg-1. Our results also indicated that each of the three E boxes plays a distinct role in the regulation of arg-1 expression. Three lines of evidence indicated that all three E boxes in arg-1 are dependent on CeTwist function. First, a null mutation of hlh-8 completely abolished the expression of the gfp reporter, pZJ19 [−436 to −1], in which all three E boxes were present (Table 2). Second, pZJ19 [−436 to −1] was ectopically expressed in response to ectopic overexpression of CeTwist with or without overexpression of CeE/DA. Third, CeTwist homodimers and CeTwist/CeE/DA heterodimers bind to each of the three E boxes in vitro.
3.2. Different sets of E boxes are important for the expression of different CeTwist target genes
Previous studies on three CeTwist target genes, ceh-24, egl-15 and mls-1, together with our study on arg-1, indicated that distinct sets of E boxes, at different positions relative to the initiator ATG, are required for the expression of each gene (Harfe and Fire, 1998; Harfe et al., 1998; Kostas and Fire, 2002). Interestingly, a common E box, the NdE box (CATATG), is involved in vulval muscle expression of three CeTwist target genes (egl-15, ceh-24 and arg-1), suggesting a unique relationship between a specific type of E box and tissue-specific expression pattern (Harfe and Fire, 1998; Harfe et al., 1998). The importance of an NdE box was also revealed by in vitro selection assays, in which vertebrate Twist-E12 heterodimers prefer to bind to sites containing a CATATG (Kophengnavong et al., 2000). Upon examination of the region near the CATATG in egl-15 and ceh-24, a GT box can be found within 20 bp from the CATATG sites in ceh-24 but not in the minimal promoter of egl-15. The GT box, therefore, may be required for strong expression in the vms of both ceh-24 and arg-1. CeTwist target gene expression in the enteric muscles does not seem to be related to a particular E box. For mls-1, the E box CAGATG is required for most of the activity, and for arg-1, both CATCTG and CACATG are necessary for its expression.
The presence of a specific E box alone may not be sufficient to determine its importance in CeTwist target gene expression. It is possible that the flanking sequences influence the binding activity of CeTwist dimers to specific E boxes. In vitro binding site selection assays identified specific flanking regions of an NdE box, GC/AA CATATG TT/GC, that were preferred for binding to vertebrate Twist-E12 heterodimers (Kophengnavong et al., 2000). Mutations made with the minimal promoter of ceh-24 also indicated that the immediate flanking sequences of the two NdE boxes are essential for expression (Harfe et al., 1998). The importance of flanking sequences is further supported by the position of the GT box in the arg-1 promoter. This site was not bound by CeTwist-containing dimers, yet it is important for arg-1 expression in the vm1 muscles. The GT box is positioned halfway between E1 and E2, only 20–30 nucleotides away from each E box. The GT box could be bound directly by another factor that influences the binding or transcriptional activity of CeTwist at E1 or E2 (see model below). Additionally, the distance of an E box to the ATG and to other E boxes could be a factor as well. In egl-15, mls-1, and arg-1, the minimal promoter regions are all in close proximity to the ATG and the E boxes within those regions are all closely positioned to one another. The combinatorial effect of all of these factors may account for the distinct expression patterns of the four CeTwist target genes (Table 1).
3.3. A working model for the differential regulation of arg-1 expression
It is intriguing that in the promoter region of the same gene, two different E boxes are used in regulating its expression in two distinct subsets of tissues, while another E box is essential for expression in all the tissues. Combining both in vivo and in vitro data from the arg-1 promoter leads us to a working model for how the regulation of this gene can be accomplished through CeTwist (Fig. 7). In our model, all three E boxes can be bound by either CeTwist heterodimers or homodimers. The main expression sites of arg-1 are the hmc, vm1, and enteric muscles. Both our site-directed mutagenesis and EMSA experiments indicate that E2 plays a critical role in directing arg-1 expression and is, therefore, likely to be bound in all arg-1-expressing tissues. This site may have preference for CeTwist/CeE/DA heterodimers as these molecules had relatively higher affinity when binding to E2 rather than the other two E boxes. In the hmc and vm1, arg-1 expression requires functional E1 as well as E2. The lower affinity binding of CeTwist dimers to E1 suggests that an additional factor X could be required in vivo to increase the binding affinity specifically in these tissues. Similarly, in the enteric muscles, arg-1 expression requires both E2 and E3. The weaker affinity for site E3 suggests an additional factor Y could facilitate binding to E3 and drive expression in these muscles (Fig. 7).
Figure 7. A model for the tissue-specific regulation of arg-1 expression by CeTwist.
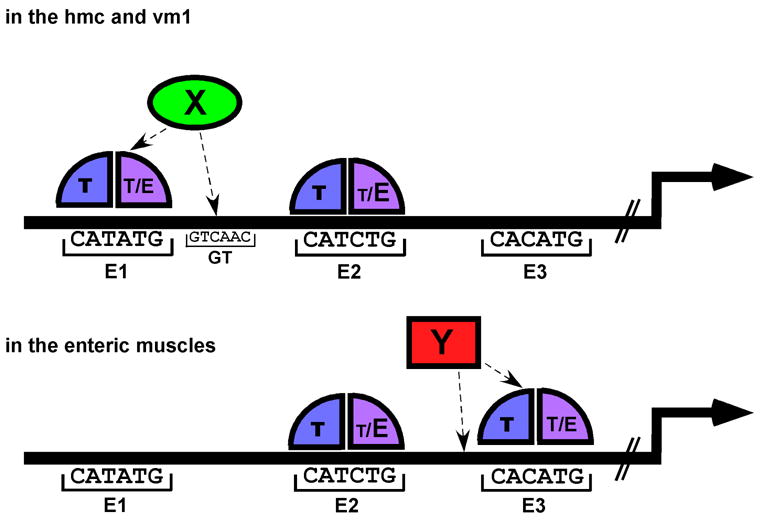
Each of the three E boxes in the 385 bp minimal promoter region could be bound by either CeTwist/CeE/DA heterodimers (T/E) or CeTwist homodimers (T/T). E2 is required for arg-1 expression in all of the tissues and is bound by T/E with high affinity in vitro (shown by the larger E vs. the smaller T bound at E2). In contrast, E1 and E3 are bound by T/E or T/T with lower affinity in vitro. The expression of arg-1 in the hmc and vm1 requires T/E or T/T to bind to E1 and to E2. It is proposed in this model that an ancillary factor (X) is present in the hmc and vm1 to facilitate the binding of CeTwist dimers to E1, allowing arg-1 expression to occur in these tissues. Similarly, in the enteric muscles, CeTwist dimers bind to both E2 and E3. A proposed ancillary factor (Y) in the enteric muscles may increase the binding affinity of CeTwist dimers to E3. Factors X and Y would be present in a subset of the tissues, thereby conferring tissue-specific regulation of arg-1 expression through the E boxes. An alternative possibility (not shown) could exist where T/E or T/T predominate in the individual tissues and have more affinity for either E1 or E3 whereas E2 can bind either dimer. In such a model, the presence of the specific CeTwist containing dimer would lead to tissue-specific regulation through the E boxes.
The hypothesis that the presence of tissue-specific additional factors (X and Y) are also necessary for complete arg-1 expression is supported by the observation that the GT box in between E1 and E2 was important for arg-1 to be expressed in the vm1 but could not be bound by either of the CeTwist dimers (Fig. 6), suggesting another factor is involved. However, this regulation must also depend on normal CeTwist function because the expression of the arg-1::gfp in the vm1 muscles, as well as in the hmc and enteric muscles, was completely lost in the hlh-8 (-) mutant animals (Table 2). There are several possibilities for how factor X could be working at the arg-1 promoter. It is possible that in the vm1 muscles, factor X both interacts with CeTwist dimers and is able to bind to the GT box. The proximity of the GT box to E1 allows factor X to effectively recruit the CeTwist dimer to E1 for tighter binding. Alternatively, factor X may indirectly enhance the binding affinity of CeTwist dimers to E1 by binding to the GT box and altering the DNA conformation in that region to allow CeTwist dimers to bind to E1 with higher affinity in the vm1 muscles. The expression of arg-1 in the hmc also requires functional E1 and E2. It is possible that another site is fulfilling this function in the hmc, and we have not located the DNA element yet. By analogy, the lower affinity binding of CeTwist dimers to E3 and its requirement for expression in the enteric muscles suggests a factor Y could be fulfilling the same function as factor X in these cells. Additional examination of intervening sequences in the arg-1 promoter will provide support for this hypothetical factor.
3.4. CeTwist dimerization status could play a role in transcriptional regulation
Although in humans and Drosophila Twist has been found to form both homodimers and heterodimers in vivo (Castanon et al., 2001; Connerney et al., 2006), there has been no direct evidence supporting the presence of functional CeTwist homodimers in C. elegans. However, our EMSA experiments demonstrated that CeTwist homodimers were able to bind to all three E boxes in the arg-1 promoter. In addition, the pZJ19 [−436 to −1] arg-1::gfp reporter was expressed ectopically in response to overexpression of CeTwist alone (Table 3). It is unlikely that the ectopic expression of arg-1 was due exclusively to heterodimer formation when only CeTwist was overexpressed; the reporter was activated even in tissues where no evidence exists that high levels of endogenous CeE/DA detectable by immunofluorescence is present (e.g. in the intestine, Krause et al., 1997). The overexpression experiment supports the possibility that CeTwist homodimers could turn on arg-1 expression in vivo. Recently, a model was proposed for the regulation of human craniosuture development by hTwist1 (Connerney et al., 2006). In this model, Twist1/E2A heterodimers and Twist1 homodimers have distinct functions in adjacent cells. Homodimer formation is favored in certain cells by an inhibitory HLH protein with no basic domain, which sequesters the E protein from dimerizing with Twist1 in this region. Similar regulation could exist in C. elegans where a tissue-specific inhibitory molecule could lead to a bias for homodimers in the hmc and vm1 and heterodimers in the enteric muscles or vice versa. Alternatively, partner choice for CeTwist dimerization could also be influenced by phosphorylation of the partner proteins as it is for the vertebrate homolog (Cai and Jabs, 2005; Firulli et al., 2005). In this alternative model, the specific dimers would have greater affinity for either E1 or E3 and lead to tissue-specific regulation through the E boxes. Such a model presupposes the presence and regulation of CeTwist homodimers, which are a subject of ongoing study in our laboratory.
3.5. Future analysis
Overall, our detailed analysis of the promoter region of arg-1 revealed a unique aspect of CeTwist function in regulating the tissue-specific expression pattern of one of its target genes. We will proceed to test our model to investigate the mechanism by which different E boxes are utilized. In addition, our data suggest that additional enhancer elements in the promoter of arg-1 are involved in the regulation. Nine other CeTwist target genes have recently been identified (Wang et al., 2006) and our knowledge of arg-1 regulation together with previous data on the other three target genes will greatly facilitate our future analysis of the promoter regions of these new target genes. Finally, careful analysis of the CeTwist pathway in C. elegans could potentially help us to better understand how TWIST functions in humans at a molecular level.
4. Experimental Procedures
4.1. C. elegans strains
PD4444 ccIs4444[arg-1::gfp; dpy-20(+)] II (Kostas and Fire, 2002) is expressed in the head mesodermal cell, vm1 and vm2 vulval muscles, uterine muscles and four enteric muscles.
To assess whether different promoter fragments of arg-1 still respond to CeTwist protein expression, gfp reporter constructs containing various promoter regions of arg-1 were either injected or crossed into the following strains:
AK103 thIs1[phs::hlh-8; phs::hlh-2; pRF4] (Wang et al., 2006) contains heat-shock (hs) inducible constructs phs::hlh-8 (pAC17) (Corsi et al., 2002) and phs::hlh-2 (pKM1035) (Harfe et al., 1998) which overexpress CeTwist and CeE/DA upon heat shock treatment.
AK100 thIs5[phs::hlh-8; pRF4]; ayIs6[hlh-8::gfp] X overexpresses CeTwist alone under heat shock treatment.
KM20 hlh-8 (nr2061), is a predicted null mutant of hlh-8 in which 1267 bp is deleted from the locus including the sequence encoding the HLH domain (Corsi et al., 2000).
4.2. Construction of gfp reporters and transgenic lines
Various promoter regions of arg-1 were amplified by PCR and inserted into the multiple cloning sites of the gfp vector pKKMCS (egl-18::gfp) (kindly provided by J. Wagmaister and D. Eisenmann) and/or pPD95.67 (gift from A. Fire). The gfp reporter constructs (100 μg/ml) were then injected into wild-type N2 animals together with the pRF4 plasmid (50 μg/ml), which contains the dominant rol-6(su1006) allele and serves as a transformation marker (Mello et al., 1991). The construct pZJ19 [−436 to −1] was also injected into strains AK103 and AK100. In addition, constructs pZJ02 [−6240 to −1], pZJ04 [−2824 to −1], pZJ05 [−6240 to −2972] and pZJ19 [−436 to −1] were crossed into the hlh-8(-) mutant background.
4.3. hlh-2 RNAi
hlh-2 dsRNA was introduced into C. elegans strains harboring the arg-1::gfp reporters by feeding them E. coli producing dsRNA from an L4440-based vector as described by Kamath and colleagues with minor modifications (Kamath et al., 2001). Briefly, overnight cultures of E. coli strain HT115 transformed with either an hlh-2 RNAi vector or an empty L4440 vector (gifts from A. Golden) were transferred to NGM plates containing 100 μg/ml ampicillin in addition to 0.35 mM IPTG to induce dsRNA production and incubated at room temperature for 24 hr before transferring L1 animals to them. After 24 hr, rolling transgenic animals were transferred to a plate with fresh RNAi bacteria and incubated at 20°C for an additional 24 hr. GFP expression was then observed in >30 young adults from the RNAi treatment.
4.4. Heat shock experiments
Mixed populations of strains AK100, AK104 thIs1[phs::hlh-8; phs::hlh-2; pRF4]; thEx6[pZJ19; pRF4], AK105 thIs5[phs::hlh-8; pRF4]; ayIs6[hlh-8::gfp] X; thEx6[pZJ19; pRF4], and AK107 thEx6[pZJ19; pRF4] were grown at 20°C. Two groups of each population were subjected to heat-shock treatment at 33°C for 20 min or 2 hr, followed by recovery at 20°C. The arg-1::gfp expression was observed at 2 hr and 20 hr after recovery.
4.5. Site-directed mutagenesis
Site-directed mutagenesis was performed using pZJ23 [−436 to −1] as the plasmid backbone and mutant primers with Quick Change Site-Directed Mutagenesis Kit (Stratagene). Each resulting construct was sequenced to confirm that only the desired mutation was obtained. The constructs were then injected into wild-type animals to make transgenic lines.
4.6. Electrophoretic mobility shift assay
EMSA with bacterially-expressed 6-His+-tagged CeTwist and CeE/DA was performed as described in Harfe et al. (1998) and Krause et al. (1997). Labeled 20mer probes were made using the sequence of the wild-type or mutated promoter regions identified in Fig. 5 (probe sequences available upon request). The probes were incubated with the purified proteins, run on native gels and subjected to autoradiography. The relative amount of each protein (1X, 2X, 4X) was determined by prior SDS-PAGE analysis.
Acknowledgments
We thank Mike Krause, Andy Golden, Jun Kelly Liu, David Eisenmann, Javier Wagmaister and Andy Fire for providing reagents, protocols and useful suggestions, Bonnie Draper, Kiran Kondabagilu, Zhihong Zhang, and Yingzi Huang for help with the EMSA experiments, Rob Donnelly for help with crosses, other members in the Corsi lab, Joe Campbell, and Baltimore-Washington Worm club members for insightful discussions, and Mike Krause, Andy Golden, Jun Kelly Liu and Jane Hubbard for critical reading of the manuscript. This work was supported by NIH grant K22DE14541 to A. K. C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cai J, Jabs EW. A twisted hand: bHLH protein phosphorylation and dimerization regulate limb development. Bioessays. 2005;27:1102–6. doi: 10.1002/bies.20313. [DOI] [PubMed] [Google Scholar]
- Castanon I, Von Stetina S, Kass J, Baylies MK. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development. 2001;128:3145–59. doi: 10.1242/dev.128.16.3145. [DOI] [PubMed] [Google Scholar]
- Chen N, Greenwald I. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev Cell. 2004;6:183–92. doi: 10.1016/s1534-5807(04)00021-8. [DOI] [PubMed] [Google Scholar]
- Chieffo C, Garvey N, Gong W, Roe B, Zhang G, Silver L, Emanuel BS, Budarf ML. Isolation and characterization of a gene from the DiGeorge chromosomal region homologous to the mouse Tbx1 gene. Genomics. 1997;43:267–77. doi: 10.1006/geno.1997.4829. [DOI] [PubMed] [Google Scholar]
- Connerney J, Andreeva V, Leshem Y, Muentener C, Mercado MA, Spicer DB. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev Dyn. 2006;235:1345–57. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- Corsi AK, Brodigan TM, Jorgensen EM, Krause M. Characterization of a dominant negative C. elegans Twist mutant protein with implications for human Saethre-Chotzen syndrome. Development. 2002;129:2761–72. doi: 10.1242/dev.129.11.2761. [DOI] [PubMed] [Google Scholar]
- Corsi AK, Kostas SA, Fire A, Krause M. Caenorhabditis elegans twist plays an essential role in non-striated muscle development. Development. 2000;127:2041–51. doi: 10.1242/dev.127.10.2041. [DOI] [PubMed] [Google Scholar]
- Crosnier C, Driancourt C, Raynaud N, Dhorne-Pollet S, Pollet N, Bernard O, Hadchouel M, Meunier-Rotival M. Mutations in JAGGED1 gene are predominantly sporadic in Alagille syndrome. Gastroenterology. 1999;116:1141–8. doi: 10.1016/s0016-5085(99)70017-x. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Krawchuk D, Centonze VE, Vargesson N, Virshup DM, Conway SJ, Cserjesi P, Laufer E, Firulli AB. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet. 2005;37:373–81. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K, Greenwald I. Interchangeability of Caenorhabditis elegans DSL proteins and intrinsic signalling activity of their extracellular domains in vivo. Development. 1995;121:4275–82. doi: 10.1242/dev.121.12.4275. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Fire A. Muscle and nerve-specific regulation of a novel NK-2 class homeodomain factor in Caenorhabditis elegans. Development. 1998;125:421–9. doi: 10.1242/dev.125.3.421. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Vaz Gomes A, Kenyon C, Liu J, Krause M, Fire A. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 1998;12:2623–35. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehr U, Muenke M. Craniosynostosis syndromes: from genes to premature fusion of skull bones. Mol Genet Metab. 1999;68:139–51. doi: 10.1006/mgme.1999.2915. [DOI] [PubMed] [Google Scholar]
- Jabs EW, Muller U, Li X, Ma L, Luo W, Haworth IS, Klisak I, Sparkes R, Warman ML, Mulliken JB, et al. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75:443–50. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. Epub 2000 Dec 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X, Greenwald I. Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. 2003;17:3100–11. doi: 10.1101/gad.1160803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kophengnavong T, Michnowicz JE, Blackwell TK. Establishment of distinct MyoD, E2A, and twist DNA binding specificities by different basic region-DNA conformations. Mol Cell Biol. 2000;20:261–72. doi: 10.1128/mcb.20.1.261-272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostas SA, Fire A. The T-box factor MLS-1 acts as a molecular switch during specification of nonstriated muscle in C. elegans. Genes Dev. 2002;16:257–69. doi: 10.1101/gad.923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, Colliton RP, Genin A, Rand EB, Li L, Piccoli DA, Spinner NB. Spectrum and frequency of jagged1 (JAG1) mutations in Alagille syndrome patients and their families. Am J Hum Genet. 1998;62:1361–9. doi: 10.1086/301875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Park M, Zhang JM, Yuan J, Harfe B, Xu SQ, Greenwald I, Cole M, Paterson B, Fire A. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development. 1997;124:2179–89. doi: 10.1242/dev.124.11.2179. [DOI] [PubMed] [Google Scholar]
- Lajeunie E, Catala M, Renier D. Craniosynostosis: from a clinical description to an understanding of bone formation of the skull. Childs Nerv Syst. 1999;15:676–80. doi: 10.1007/s003810050457. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Priess JR. The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell. 1994;77:95–106. doi: 10.1016/0092-8674(94)90238-0. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P. Maternal-zygotic gene interactions during formation of the dorsoventral pattern in Drosophila embyros. Genetics. 1983;105:615–632. doi: 10.1093/genetics/105.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, el Messal M, Perrin-Schmitt F. The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 1987;15:3439–53. doi: 10.1093/nar/15.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagmaister JA, Miley GR, Morris CA, Gleason JE, Miller LM, Kornfie lK, Eisenmann DM. Identification of cis-regulatory elements from the C. elegans Hox gene lin-39 required for embryonic expression and for regulation by the transcription factors LIN-1, LIN-31 and LIN-39. Dev Biol. 2006 2006 May 19; doi: 10.1016/j.ydbio.2006.05.008. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Wang P, Zhao J, Corsi AK. Identification of novel target genes of CeTwist and CeE/DA. Dev Biol. 2006;293:486–98. doi: 10.1016/j.ydbio.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Wang SM, Coljee VW, Pignolo RJ, Rotenberg MO, Cristofalo VJ, Sierra F. Cloning of the human twist gene: its expression is retained in adult mesodermally-derived tissues. Gene. 1997;187:83–92. [PubMed] [Google Scholar]
- Wilkie AO. Craniosynostosis: genes and mechanisms. Hum Mol Genet. 1997;6:1647–56. doi: 10.1093/hmg/6.10.1647. [DOI] [PubMed] [Google Scholar]


