Abstract
Homologous recombination between circular sister chromosomes during DNA replication in bacteria can generate chromosome dimers that must be resolved into monomers prior to cell division. In Escherichia coli, dimer resolution is achieved by site-specific recombination, Xer recombination, involving two paralogous tyrosine recombinases, XerC and XerD, and a 28-bp recombination site (dif) located at the junction of the two replication arms. Xer recombination is tightly controlled by the septal protein FtsK. XerCD recombinases and FtsK are found on most sequenced eubacterial genomes, suggesting that the Xer recombination system as described in E. coli is highly conserved among prokaryotes. We show here that Streptococci and Lactococci carry an alternative Xer recombination machinery, organized in a single recombination module. This corresponds to an atypical 31-bp recombination site (dif SL) associated with a dedicated tyrosine recombinase (XerS). In contrast to the E. coli Xer system, only a single recombinase is required to recombine dif SL, suggesting a different mechanism in the recombination process. Despite this important difference, XerS can only perform efficient recombination when dif SL sites are located on chromosome dimers. Moreover, the XerS/dif SL recombination requires the streptococcal protein FtsKSL, probably without the need for direct protein-protein interaction, which we demonstrated to be located at the division septum of Lactococcus lactis. Acquisition of the XerS recombination module can be considered as a landmark of the separation of Streptococci/Lactococci from other firmicutes and support the view that Xer recombination is a conserved cellular function in bacteria, but that can be achieved by functional analogs.
Author Summary
In bacteria, genetic information is mainly carried by a single circular chromosome. The replication of this circular molecule sometimes leads to the formation of a chromosome dimer unable to segregate in the daughter cells during the division process. In the bacterial model E. coli, chromosome dimers are resolved in monomers by site-specific recombination: two recombinases (XerC and XerD) cooperatively catalyze the recombination at a chromosomal site (dif), located at the junction of the two replication arms. This recombination is intimately coupled to cell division by the control of the septal protein FtsK. Xer recombination machinery as described in E. coli appears highly conserved among bacterial species. We show by comparative genomics and genetic studies that Streptococci use an alternative Xer recombination system, renamed XerS/dif SL, which is composed of a single recombinase phylogenetically unrelated to XerCD proteins and a noncanonical dif site. We also demonstrate that the streptococcal FtsK protein localizes at the division septum and operates the XerS/dif SL recombination. This is the first identification of an alternative Xer recombination system in prokaryotes to out knowledge, which might indicate that other unusual chromosome dimer resolution systems could exist in bacterial phyla where a canonical dif site is not detected.
Introduction
Chromosome replication is a key function in living cells, and any factor that impedes progression of replication forks can result in mutagenesis and genome instability. Several strategies have evolved to rescue replication forks stalled by DNA damage. Most of these depend on homologous recombination pathways but are not necessarily accompanied by strand exchange [1]. However, in cases where replication fork repair does lead to sister chromatid exchange, bacteria with circular chromosome(s) are faced with a potential topological problem because an odd number of crossovers between sister chromatids generates chromosome dimer, which must be converted back to monomers for a correct segregation to daughter cells. In E. coli, chromosome dimer formation occurs in 15% of the cell population [2,3], and conversion to monomers is carried out by the Xer site-specific recombination system (for recent reviews see [4,5]). This is composed of two paralogous tyrosine recombinases (integrases), XerC and XerD, which cooperatively catalyze strand exchanges at a 28-bp DNA sequence, the dif site, which must be located at the junction of the two replichores to be functional [3,6–8]. Xer recombination is intimately coupled to cell division [9] through the septal protein FtsK [10–12], a DNA translocase [8] with an essential N-terminal transmembrane domain involved in its localization at the septum [13], and a C-terminal DNA motor domain involved in positioning and synapsing the two dif sites of the chromosome dimer at the division septum [12,14–19] as well as in activating the strand exchange [8] by direct interaction with XerD [20,21].
Homologs of XerCD recombinases and FtsK are found in most eubacterial phyla and some archeal lineages [22] as well as the canonical dif site [23]. Moreover, interactions between the E. coli dif site and the XerCD recombinases of Haemophilus influenzae [24], Pseudomonas aeruginosa [25], Bacillus subtilis [26], Proteus mirabilis [27], and Caulobacter crescentus [28] have been experimentally demonstrated in vitro. These observations led to the general view that Xer recombination is a function conserved among bacteria harboring circular chromosome(s). However, regulation of strand exchange may differ, depending on the bacterial species: FtsK-mediated activation of Xer recombination in H. influenzae obeys the E. coli paradigm [21], whereas in B. subtilis, the model bacteria for firmicutes (formerly known as low GC-content Gram-positive bacteria), neither of the two FtsK analogs (SpoIIIE and YtpT) appears able to drive Xer recombination [26]. Several attempts have been made to identify the Xer recombination machinery in Streptococci, a taxonomic group belonging to firmicutes and comprising major pathogens [29] as well as innocuous food-grade species of major industrial importance [30,31]. These studies revealed putative XerCD recombinases but failed to identify a dif site [32,33].
We show here, by comparative genomics and functional analyses in L. lactis, S. pneumoniae, and E. coli, that Streptococci possess alternative Xer recombination machinery phylogenetically unrelated to the classical XerCD/dif system. This machinery involves a single tyrosine recombinase (XerS) and an atypical dif site (dif SL ), both located on a single genetic module. We also show that, in contrast to B. subtilis, the streptococcal FtsK protein localizes at the division septum and controls the XerS/dif SL recombination.
Results
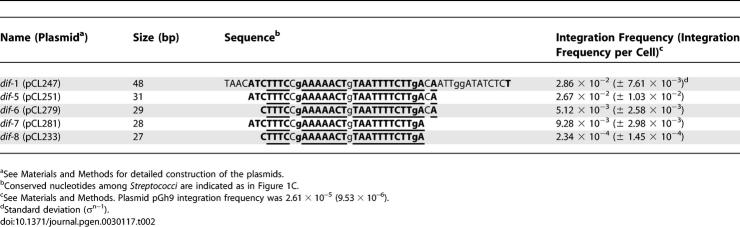
Identification of the Streptococcal dif Site by Comparative Genomics
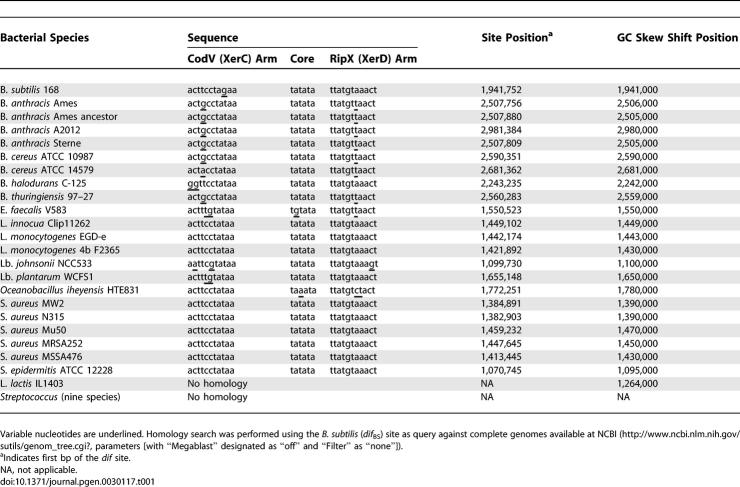
Assuming that Xer recombination is highly conserved in eubacteria with a significant homology of the dif sites even between distantly related species [26], we performed an in silico search for putative dif sites in several completely sequenced firmicutes genomes. Candidate dif sites should: (i) have a significant similarity with that of B. subtilis (dif BS ), (ii) occur only once per genome, and (iii) be localized in the replication terminus (terC), defined as the chromosomal region located opposite the replication origin (oriC) where compositional strand biases switch sign [34,35]. Using these rules, a canonical dif site could be identified in all species analyzed except for Streptococci and Lactococci (Table 1). We therefore used an alternative three-step approach based on comparative genomics to identify the streptococcal/lactococcal dif site (Figure 1). The terC region for three streptococcal genomes was localized (Figure 1A) using a cumulative GC skew diagram [34], and a comparison of the 10-kb region encompassing the GC skew shift was performed (Figure 1B). This analysis revealed a 2-kb segment that showed significant similarity within the three species (>70% identity at the DNA level) and included a 356-amino-acid tyrosine-recombinase–encoding gene (annotated ymfD on the L. lactis IL1403 genome [36] but hereafter renamed xerS) preceded by a ∼50-bp highly (>90%) conserved sequence. When used to scan 49 genomes of other streptococcal species (Figure 1C), this ∼50-bp fragment revealed a 31-bp consensus sequence (hereafter named dif SL) with weak homology to the B. subtilis or E. coli dif sites, but with an overall structure resembling the DNA targets for tyrosine recombinases (i.e., two imperfect inverted repeats separated by a 6–8-bp central sequence).
Table 1.
Localization of dif Sites in Some Firmicutes
Figure 1. Identification of the Streptococcal dif SL Site by Comparative Genomics.
(A) Cumulative GC skew diagrams of three streptococcal genomes. Lla, L. lactis IL1403; Spyo, S. pyogenes M1 GAS; Spn, S. pneumoniae R6.
(B) Multiple DNA comparison is presented for the 10-kb regions encompassing the region of GC skew shift. The result is scaled to the gene organization of the L. lactis IL1403 terC region, and the conserved ORF is indicated in black. The lactococcal ORFs A, B, and C, not found in other streptococcal genomes, showed significant homology to the YmfD protein when fused together, suggesting ancient duplication of the ymfD region in L. lactis. The conserved ∼50-bp sequence is indicated by an asterisk.
(C) Sequence similarity of putative dif SL sites from 49 publicly available streptococcal genomes (December 2006) and comparison with the E. coli and B. subtilis dif sites are presented. Nucleotides conserved in all except one species are indicated in bold, and those conserved in all species are indicated in bold and underlined. S. pyogenes M1 strains were M1 GAS, MGAS10750, MGAS5005, and MGAS10394. S. pyogenes M3 strains were MGAS315, MGAS2096, MGAS10270, MGAS9429, MGAS6180, M49–591, and SSI-1. S. agalactiae sequences of strains A909, H36B, COH1, CJB111, and 18RS21 were identical to strain 2603V/R. Asterisks indicate unfinished genomes.
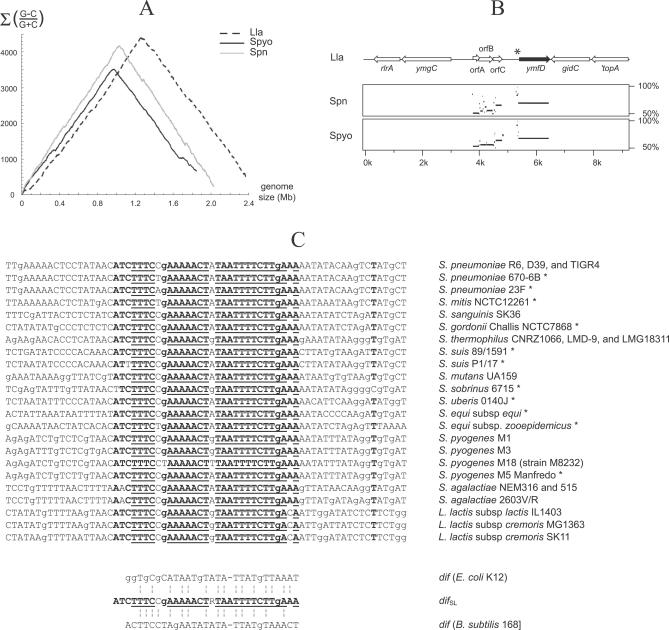
Comparative analysis of the genetic context in the 10-kb terC region of different streptococcal species revealed notable features strongly suggesting that streptococcal Xer recombination machinery is defined by one genetic module, corresponding to the dif SL site followed by one of its dedicated recombinases (Figure 2). The physical link between dif SL and xerS open reading frame (ORF) was found to be preserved among all Streptococci for which there is sequence data, and no genetic element other than dif SL-xerS was conserved in the 10-kb terC region. In addition, the genes surrounding dif SL-xerS did not show a preferred orientation that might indicate possible cotranscription with the xerS gene. Moreover, the xerS ORF often exhibits a putative ρ-independent transcription terminator at its end. These observations indicate that xerS is unlikely to be part of an operon and suggest that the dif SL-xerS pair behaves as an individual genetic module.
Figure 2. Gene Context Analysis of the 10-kb terC Region of Different Streptococcal Species.
Lla, L. lactis IL1403; Spn, S. pneumoniae R6; Spyo, S. pyogenes M1 GAS; Sagal, S. agalactiae NEM316; Sth, S. salivarius subsp. thermophilus CNRZ1066; Smut, S. mutans UA159. The ORF coding for the putative tyrosine recombinase is shown in black. Only putative ρ-independent transcription terminators with free energy (ΔG) <−12 kcal.mol−1 are indicated.
In Vivo Characterization of the dif SL Site
The candidate dif SL site was tested for its ability to support site-specific recombination in L. lactis and S. pneumoniae by intermolecular recombination assays. A 37-bp synthetic sequence encompassing the putative 31-bp lactococcal dif SL site was cloned in a plasmid that does not replicate in firmicutes (pCL52, Table S1). The resulting construction (pCL235, Table S1) was used to transform a wild-type (WT) strain of L. lactis (MG1363, Table S1). In contrast to pCL52, which did not yield transformant, pCL235 produced transformants at an efficiency representing 1% of the efficiency attained with a replicative plasmid (unpublished data). This demonstrates that the putative dif SL site was capable of rescuing pCL235, presumably by promoting integration of exogenous DNA into the lactococcal chromosome. When transformed into a recA strain (VEL1122, Table S1), plasmid pCL235 was also rescued with the same efficiency as in the WT strain (unpublished data), indicating that plasmid integration occurred in a RecA-independent manner. Moreover, as judged by Pulsed-Field gel Electrophoresis analysis (Figure S1, lanes 2 and 4), pCL235 integrated into the chromosome of both strains at the location predicted for the native dif SL site. Thus, the 37-bp sequence appeared to contain the DNA target of a site-specific recombination system. The dif SL-mediated site-specific integration was also demonstrated to be a general process in Streptococci, since plasmid pGh9, a temperature sensitive replication (repA ts ) mutant containing either the lactococcal 37-bp sequence described above (pCL231, Table S1) or its pneumococcal counterpart (pCL403, Table S1), integrated into the chromosomes of L. lactis and S. pneumoniae under nonpermissive conditions with comparable efficiencies (respectively 4.88 × 10−2 [± 2.33 × 10−2] cell−1 and 2.67 × 10−2 [± 1.55 × 10−2] cell−1). However, it should be mentioned that location of the insertion site of pCL403 has not been verified in S. pneumoniae.
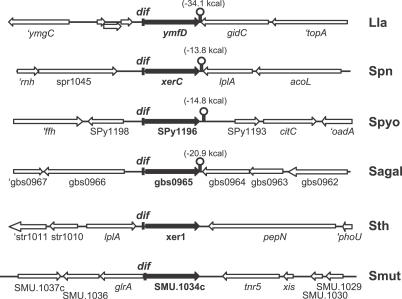
The minimal size of the dif SL site was determined in L. lactis by scoring the integration efficiency of pGh9 containing variants of the dif SL site (Table 2). Reducing the length of the dif SL region from 48 to 31 bp did not alter the plasmid integration efficiency, indicating that the strongly conserved T located 13 bp away from the 31-bp consensus sequence (Figure 1C) was not part of the dif SL site. However, removing the two external bp from both sides of the 31-bp consensus sequence (Table 2, dif-8) led to a 100-fold decrease in integration efficiency, though this sequence was still proficient in site-specific recombination at the native dif SL site (Figure S1, lanes 3 and 5). Finally, deleting two nucleotides at either side of the 31-bp consensus sequence led to a 2-fold (Table 2, compare dif-7 to dif-5; Wilcoxon test, p < 0.003) or 4-fold (Table 2, compare dif-6 to dif-5; Wilcoxon test, p < 0.01) reduction in integration frequency. Together, these results led us to propose that the 31-bp consensus sequence defines the authentic dif SL site.
Table 2.
Chromosomal Integration Frequencies of dif SL Variants in L. lactis
Recombination at dif SL Requires One Recombinase, XerS
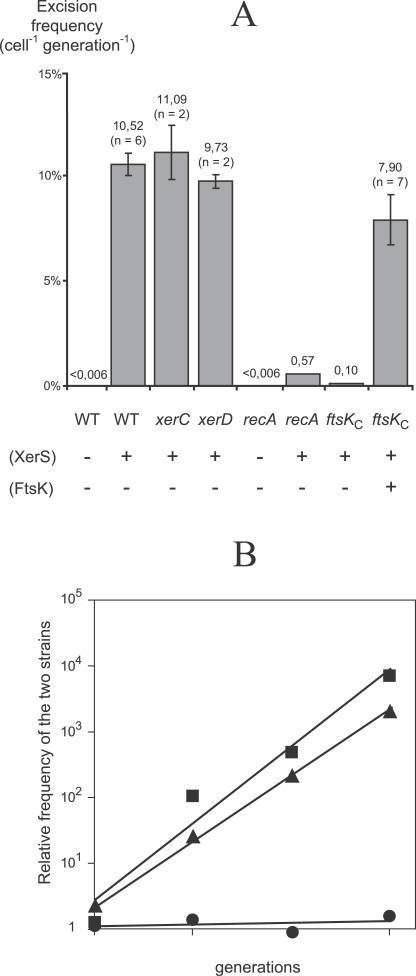
Given that predictive analyses revealed XerS as the prime candidate for the actual Xer recombinase, a recombination assay was performed in S. pneumoniae to test if XerS was needed for recombination at dif SL. This streptococcal species was selected mainly for its facility to construct mutants compared to L. lactis. The recombination at the dif SL site was totally abolished in a xerS mutant (strain S501, Table S1; Materials and Methods), with undetectable integration of pCL403, demonstrating that XerS was one catalytic recombinase of the XerS/dif SL system. To test whether XerS was the only recombinase involved in the Xer catalytic machinery, XerS/dif SL recombination was assayed in E. coli using an excision assay previously developed for the genetic analysis of the E. coli dif site activity [37]. Briefly, the native E. coli dif site was replaced by a cassette containing two directly repeated lactococcal dif SL sites flanking a kanamycin resistance (KmR) gene (strain E359, Table S1), and the excision frequency (cell−1 generation−1; Materials and Methods) was determined by counting the number of KmS recombinants at different generations during serial cultures (Figure 3A). In absence of the lactococcal XerS recombinase, almost no recombination was observed (<0.006% cell−1 generation−1) indicating that XerCD of E. coli do not recombine dif SL. In contrast, introduction of a plasmid expressing the lactococcal xerS gene (pCL297, Table S1) increased the excision frequency to 10% cell−1 generation−1 (Figure 3A), a value close to the excision frequency observed in E. coli when using the native XerCD/dif system [3]. In addition, the excision frequency was not significantly altered in E. coli xerC or xerD mutant (Figure 3A). This indicates that fortuitous interaction between XerS and E. coli XerC or XerD recombinases is unlikely to account for recombination at dif SL sites. However, as recombination assay has not been performed on a xerC xerD double mutant, this cannot be totally ruled out. XerS also promoted intermolecular recombination between one lactococcal dif SL site replacing the native E. coli dif site on the chromosome (strain E368, Table S1) and a second dif SL site located on a nonreplicative plasmid (unpublished data). Together, these data demonstrated that XerS is the only streptococcal tyrosine recombinase required to catalyze intra- and intermolecular recombination between dif SL sites.
Figure 3. XerS/dif SL Recombination in E. coli .
(A) KmR cassette excision mediated by XerS in E. coli in different genetic backgrounds is shown. The +/− signs indicated presence/absence of plasmid expressing the L. lactis xerS (pCL297) or the E. coli ftsK (pCL263) genes. When available the number of independent experiments (n) is indicated below each excision frequency mean value. Error bars correspond to the standard deviation (σn−1).
(B) Effect of the lactococcal XerS/dif SL system on chromosome dimer resolution in E. coli as measured by growth competition assays is presented. E368 (XerS + dif SL +) was mixed with E367 (XerS + dif SL −, squares), E378 (XerS+ dif SL + ftsK C −, triangles), or E375 (WT strain with XerS, circles) at a ratio of 1:1 and grown in serial culture for 60 generations. Values were calculated from two independent assays.
A phylogenetic analysis of all tyrosine recombinases present in the sequenced genome of five streptococcal species revealed another integrase conserved among Streptococci. This atypical recombinase, more related to phages' integrases (Figure S2) and previously identified as XerD in S. pneumoniae [33], lacks the extreme C-terminal region and two amino acids of the catalytic tetrad R-H-R-Y [38]. When tested alone in E. coli, YnbA (the lactococcal ortholog of S. pneumoniae XerD) showed no intra- or intermolecular recombination activity on dif SL and did not influence the recombination process when coexpressed with XerS. Moreover, it did not bind specifically to the lactococcal dif SL site in vitro (unpublished data). Therefore, YnbA is unlikely to belong to the streptococcal Xer system.
XerS/dif SL and Chromosome Dimer Resolution in E. coli
Although the XerS/dif SL system involves only one recombinase, as do the Cre/loxP and Flp/FRT systems, its location at the terC region of streptococcal chromosomes strongly suggests that it functions to resolve chromosome dimers. To examine whether XerS/dif SL can substitute the XerCD/dif system in E. coli, we used the growth competition assays (Figure 3B) previously developed to show that XerCD/dif resolved chromosome dimers in E. coli [3,39]. For that purpose, we constructed E. coli strains containing or not one lactococcal dif SL site replacing the native dif site. The strain harboring a complete streptococcal Xer system (E368, Table S1) showed a growth advantage of 10% generation−1 when competed with its isogenic strain missing the dif SL site (E367, Table S1). As found for XerCD in E. coli [37], this selective benefit matches the excision frequency of the KmR cassette in the excision assay described above. In addition, strain E368 showed no growth defect compared to a strain harboring a functional XerCD/dif system (E375, Table S1). These results were correlated with cell morphology changes: strain E367 retained the filamentation phenotype of an E. coli strain defective in Xer recombination, while the strain harboring the complete XerS/dif SL system displayed a WT cell morphology (unpublished data). Thus, the XerS/dif SL system can substitute XerCD/dif in E. coli to resolve chromosome dimers.
Chromosome dimers in E. coli are mostly formed by homologous recombination [2]. As a recA mutation also drastically reduces the XerCD-mediated recombination at dif [2,37], this argued toward the fact that chromosome dimer is mandatory for creating the conditions necessary for a recombination between two directly repeated dif sites. Such dependence was investigated for the XerS system using the same E. coli excision assay [37]. The frequency of the KmR cassette excision by the XerS system fell from 10% to less than 0.6% cell−1 generation−1 (Figure 3A) in a recA derivative of the E359 strain (strain E379, Table S1). As this 20-fold reduction of recombination efficiency was similar to that observed in E. coli [37], this strongly suggests that, as for the XerCD/dif system, XerS can perform efficient recombination only when dif SL sites are located on chromosome dimers.
XerS/dif SL Recombination Depends on the Septal Protein FtsK
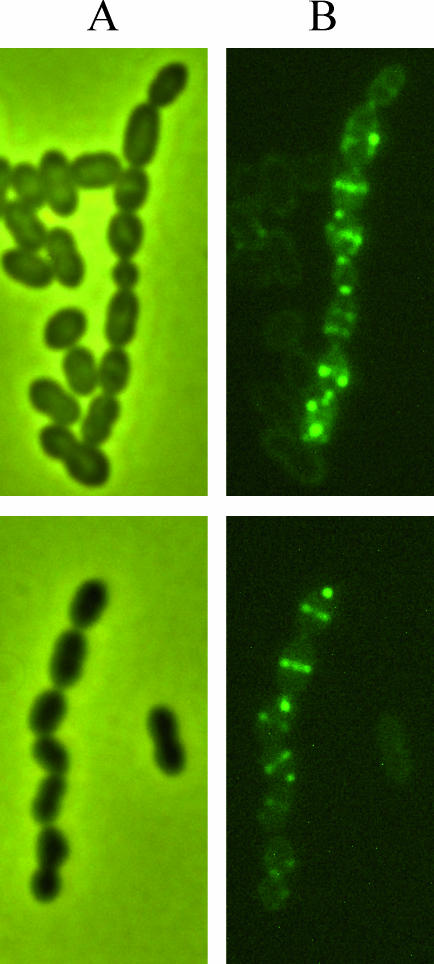
All streptococcal genomes sequenced so far contain one ORF encoding a protein homologous to the 787-amino-acid B. subtilis protein SpoIIIE. These SpoIIIE-like proteins (hereafter named FtsKSL) range from 758 (S. mutans) to 816 (S. agalactiae) amino acids in length and retain the structural signatures of proteins from the FtsK-HerA superfamily [40]: they contain a ∼180-amino-acid N-terminal region of weak similarity that includes four predicted transmembrane domains and a strongly conserved ∼500-amino-acid C-terminal region corresponding to the DNA translocase domain (unpublished data). The cellular localization of FtsKSL was determined in L. lactis using GFP fusions, corresponding to the GFP protein fused to the C-terminal of the full-length (FtsK1 − 763 − GFP) or N-terminal region (FtsK1 − 181 − GFP) of lactococcal FtsKSL. Both GFP fusions clearly localized at the septum of L. lactis (Figure 4), indicating that as expected, FtsKSL localizes at the lactococcal division septum and its 181 amino acids containing the four transmembrane domains were sufficient to drive this localization.
Figure 4. Subcellular Localization of FtsKSL-GFP Proteins in L. lactis .
Phase-contrast (A) and fluorescence (B) microscopy of L. lactis NZ900 overexpressing full-length lactococcal FtsK-GFP (upper) or N-ter Ftsk-GFP (lower) are presented. Cells were analyzed by microscopy on midexponential growth phase.
The control of XerS-mediated recombination by FtsK was examined in S. pneumoniae and E. coli. For that purpose, strains expressing FtsK proteins deleted of their C-terminal part were constructed (ftsK C mutants, only the first 405 amino acids and the first 316 amino acids of FtsK are synthesized in S. pneumoniae and E. coli respectively; Materials and Methods). Surprisingly, during the construction of the ftsKC mutants in S. pneumoniae, insertions of the Mariner minitransposon were obtained into the C-terminal or the N-terminal domain of FtsKSL. This suggests that neither the C- nor the N terminus is essential for the growth of this bacterium, though all pneumococcal ftsK mutants were severely impaired in growth rate and cell viability as xerS mutants (unpublished data). In S. pneumoniae, XerS/dif SL recombination depended on the C-terminal part of FtsKSL, because integration of the repA ts plasmid containing the pneumococcal dif SL site (pCL403), though not totally abolished as for the xerS mutant, became severely impaired in the ftsK C mutant (S502, Table S1), with an efficiency of 1.52 × 10−4 (± 3.5 × 10−4) cell−1 corresponding to less than 1% of the integration efficiency of the WT strain. Similar observations were made in the ftsK C mutant of E. coli (E372, Table S1), using the excision assay. The excision frequency of the dif SL-KmR- dif SL cassette dropped from 10% in WT strain to 0.1% cell−1 generation−1 in the ftsK C strain (Figure 3A). This decrease was unambiguously associated to the lack of the C-terminal part of FtsK, since expression of the full-length E. coli ftsK gene carried on a pBAD-derivative plasmid (pCL263, Table S1) restored KmR cassette excision to nearly the WT frequency (Figure 3A). In addition, results from growth competition assays (Figure 3B) or cell morphology observations (unpublished data) also showed that the XerS system was unable to resolve chromosome dimers in E. coli in absence of the C-terminal domain of FtsK. Together our data demonstrated that XerS/dif SL recombination in S. pneumoniae and E. coli, as well as dimer resolution in E. coli, depends on FtsK.
Discussion
In this work, we provide experimental evidence that Streptococci possess an unconventional Xer recombination machinery that requires only one tyrosine recombinase, XerS, to catalyze the site-specific recombination at a 31-bp sequence dif SL. This raises an important question as to whether this system is orthologous to the “classical” E. coli XerCD system found in most bacterial species, including many other firmicutes. Not only does XerS differ significantly in length and primary sequence from members of the XerCD recombinases family (unpublished data), but dif SL also differs in length and shows a weak similarity with the E. coli or B. subtilis dif sites (Figure 1C). Moreover, dif SL is located immediately upstream the xerS coding sequence in all streptococcal and lactococcal species analyzed. Such genetic organization contrasts with that of classical XerCD systems, with the two recombinases encoded by genes located far from each other and distant from dif, and is more comparable to the integration modules of mobile elements such as integrons [41], bacteriophages such as P1 [42] or mycobacteriophage L5 [43], and some ICEs such as the clc element from Pseudomonas [44] or CTnDOT from Bacteroides [45]. As Streptococci and Lactococci (together defining the taxonomic family of Streptococcaceae) represent a rather homogeneous phylogenetic group among firmicutes when compared to other genera such as Clostridium or Lactobacillus [46,47], we speculate that acquisition of the XerS system might have replaced the “classical” XerCD system at the time of or soon after their split from other firmicutes, with this event representing one landmark of this separation.
As demonstrated in this study, the cis-organization of the dif SL-xerS module is not mandatory for efficient recombination, but this probably reveals a selective pressure to maintain that arrangement. Although at present the xerS transcription start point location is unknown, we speculate that dif SL either lies between the xerS ORF and its promoter or is part of the xerS promoter. If this is true, this unusual arrangement might reflect a regulatory mechanism in which binding of XerS to dif SL might autoregulate xerS expression. Alternatively, as it has been recently observed that some filamentous phages [48] or genetic islands [49] can divert the XerCD recombination system to integrate themselves at the chromosomal dif site of several bacteria, another hypothesis could be that the dif SL-xerS arrangement might serve to prevent insertion of additional genetic material at dif SL, because such event would separate the xerS ORF from its promoter and lead to inactivation of the chromosome dimer resolution system.
With only one catalytic recombinase involved in the recombination reaction, the XerS system is more similar to Cre/loxP and Flp/FRT than to XerCD/dif. However, XerS retains particular features that could indicate alternative mechanism in the recombination process. For instance, in vivo characterization of the dif SL site in L. lactis revealed an asymmetry in its organization, with left and right arms differing in length (the left arm being 2-bp longer than the right one) as well as in nucleotide sequence (Table 2). This differs from loxP and FRT sites, which contain two perfectly symmetrical 13-bp arms surrounding the core region [50,51]. How a single recombinase can accommodate dissimilar binding sites to perform the DNA strand exchange reaction without accessory factor has to be analyzed, but we speculate that asymmetry of the dif SL site might have a role in the control of this strand exchange order.
Though we did not provide direct experimental data demonstrating that XerS/dif SL is involved in chromosome dimer resolution in Streptococci, several lines of evidence strongly suggest that dimer resolution is the primary task of this site-specific recombination system. First, classical XerCD recombinases and canonical dif site are not present in streptococcal/lactococcal genomes but substituted by the XerS/dif SL recombination module at the chromosomal location predicted for a site-specific recombination system acting on chromosome dimer resolution. Second, to catalyze the strand exchange reaction XerS seems to require at least one of the two dif SL sites located on the chromosome, because recombination between two dif SL sites contained within a multicopy plasmid with theta replication (pSC101 derivative in E. coli, and pAMB1 in L. lactis, unpublished data) could be detected neither in E. coli nor in L. lactis. At last, not only is XerS/dif SL able to resolve chromosome dimers in E. coli as efficiently as the native XerCD/dif system (Figure 3B), but XerS/dif SL recombination also hinged on formation of chromosome dimers, as revealed by the RecA-dependency of the KmR cassette excision (Figure 3A), with the excision efficiency exactly matching the frequency of chromosome dimers resolution.
We also demonstrated that, in contrast to SpoIIIE from B. subtilis that only infrequently (∼6%) concentrates at the vegetative septum [52] and is not involved in the Xer recombination [26], the streptococcal FtsKSL protein localizes at the division septum and still directs the XerS/dif SL recombination, as dimer resolution and intra- or intermolecular recombination were drastically inhibited in E. coli and S. pneumoniae cells lacking the C-terminal part of FtsK. Although our preliminary analyses of the pneumococcal ftsK mutants need to be confirmed, the ability to obtain viable cells depleted of FtsK suggests that neither the N- nor the C terminus of the protein is essential in Streptococci. As essentiality of FtsK seems to be species dependent, with only the N-terminal part in E. coli and the C terminus in C. crescentus being essential [53,54], we hypothesize that activity of FtsK, though still involved in cell division and DNA translocation, could slightly differ among the different bacteria. However, it appears that no correlation can be done between essentiality and localization of FtsK to the division septum. In E. coli and some other γ-proteobacteria, the C-terminal part of FtsK drives the XerCD recombination reaction in two ways: by participating to the formation of the recombination synapse through its DNA translocase activity [14–16] and activating the recombination reaction by direct interaction with XerD [8]. Some of our data strongly indicated that such interaction between FtsKSL and XerS is unnecessary to activate the XerS/dif SL recombination in Streptococci, though this cannot be totally ruled out. First, the XerS-mediated intramolecular recombination frequency at dif SL in E. coli (Figure 3A) was close to that measured with XerCD/dif [3], suggesting no species specificity for FtsK requirement. This observation contrasts to that made in E. coli where the H. influenzae FtsK was inefficient in activation of the E. coli XerD and vice versa, implying that the FtsK-XerD interaction is highly species specific in these bacteria [21]. In addition, both pneumococcal and lactococcal XerS protein sequences do not contain the amino acids motif (RQ–QQ) found in E. coli XerD and involved in its specific interaction between with FtsK [20]. At last, the cassette excision by recombination at dif SL in E. coli, as well as plasmid integration in S. pneumoniae, was not totally abolished in ftsK C mutants but dropped to 1% of the recombination activity of WT strains, suggesting that some productive recombination synapses might form independently of FtsK, most probably by the random collision of two dif sites. This observation also contrasts the results obtained with the cassette excision assay performed with the E. coli XerCD/dif system, wherein no recombination was detected in an ftsK C mutant [55], suggestive of the FtsK-mediated activation of the recombination. Our data are more easily accommodated to a model where XerS is unable to form a productive synapse and requires the DNA translocase activity of FtsKSL to bring the two dif SL sites of a chromosome dimer close to each other and in an active geometry before performing the strand exchange. However, the recombination would not need direct activation by protein interaction between FtsKSL and XerS. However, as for the XerCD model [11], our model cannot provide satisfactory explanation to how FtsK is involved in the intermolecular recombination between one dif SL site located on a suicide (or repA ts) plasmid and the chromosomal dif SL site, and the mechanism of the FtsKSL-mediated control has to be analyzed further.
In conclusion, the discovery of a Xer recombination system phylogenetically unrelated to the classical XerCD system reinforces the idea that chromosome dimer resolution can be viewed as a housekeeping function conserved among bacteria with circular chromosome(s), but that some species can use functional analogs to perform this task. We expect that other bacterial species among those whose genome(s) are missing a canonical dif site also contain alternative chromosome dimer resolution systems. Finally, we note that the particularity of the XerS system makes it a valuable candidate for the development of new antibacterial drugs specifically directed against the pathogenic Streptococci.
Materials and Methods
Plasmids, bacterial strains, and growth conditions.
The plasmids and bacterial strains used in this study are listed in Table S1. E. coli strains and plasmids containing synthetic lactococcal or pneumococcal variants of dif SL sites were constructed using the procedure provided in Text S1. E. coli strains were grown at 30 °C in LB medium. Antibiotics were used at the following concentrations: erythromycin (Em) 150 μg ml−1, chloramphenicol (Cm) 20 μg ml−1, spectinomycin (Spc) 100 μg ml−1, kanamycin (Km) 50 μg ml−1, and ampicillin (Ap) 25 μg ml−1. L. lactis strains were grown semi-anaerobically at 30 °C in M17 broth (Merck KGaA, http://www.merck.de) supplemented with 0.5% (w/v) glucose (GM17) and transformed as previously described [56]. Antibiotics used for selection of lactococcal transformants were: Em 1 μg ml−1, Cm 5 μg ml−1, and Spc 200 μg ml−1. S. pneumoniae strains were grown in Todd-Hewitt broth (Difco/BD Biosciences, http://www.bdbiosciences.com) supplemented with 0.5% yeast extract (THY) and transformed using synthetic competence-stimulating peptide (CSP) as described [57]. Antibiotic concentrations used for selection of pneumococcal transformants were: Em 0.2 μg ml−1 and Km 500 μg ml−1.
DNA manipulation.
Restriction and modification enzymes were purchased from New England Biolabs (http://www.neb.com) and used as recommended by the supplier. Plasmid DNA from E. coli was isolated with the Qiaprep spin kit according to the manufacturer's instructions (Qiagen, http://www.qiagen.com). Chromosomal DNA from E. coli, L. lactis, and S. pneumoniae was isolated with the DNeasy tissue kit according to the manufacturer's instructions (Qiagen). Preparation of lactococcal genomic DNA embedded in agarose matrix, Pulsed-Field gel Electrophoresis, and Southern blot with dried agarose gels were performed as previously described [58]. Hybridization signals were detected with a Bioimaging BAS1000 analyzer system (FUJI Photo Film, http://www.fujifilm.com) and analyzed with TINA version 2.07c software (Raytest Isotopenβgeräte GmBH, http://www.raytest.de).
Genome sequences analyses.
Nucleotide sequences were obtained from NCBI (http://www.ncbi.nlm.nih.gov/genomes/static/eub_g.html), JGI (http://genome.jgi-psf.org/tre_home.html), and the Sanger Institute (http://www.sanger.ac.uk/Projects/Microbes). Cumulative GC skews were obtained from an in-house build program (Laurent Lestrade, Laboratoire de Biologie Moléculaire des Eucaryotes, Toulouse, France). Multiple DNA comparison was performed using MultiPipMaker program [59].
Intermolecular recombination assays in L. lactis and S. pneumoniae.
Chromosomal integration assays of repA ts plasmid pGh9 or dif SL-containing derivatives in L. lactis were performed according to [60]. For S. pneumoniae, frozen strains containing the pGh9 or derivatives were grown at 39 °C (water bath) to an OD600 = 0.3 in THY without antibiotics. Appropriate dilutions were plated on 10 ml of D medium [57] containing 2% of defibrinated horse blood (bioMérieux, http://www.biomerieux.com) and supplemented when appropriate with Em, and plates were incubated at 39 °C. Integration of the pGh9 plasmid was undetectable in S. pneumoniae (no colonies were observed when plating 0.1 ml of the undiluted bacterial culture). The integration frequency per cell (ipc) was calculated as the geometric mean of the ratio of colonies on selective versus nonselective plates obtained from five to 19 independent experiments.
In vitro mariner mutagenesis in S. pneumoniae.
Mutagenesis was carried out as described [61]. The target DNA for in vitro transposition of the KmR mariner minitransposon pR410 [61] were obtained by PCR reactions using R800 chromosomal DNA as template. The sizes of PCR fragments were: 2.012 bp for xerS gene (primers: forward, 5′-TAgAAAACCgATTCTCAgAAATgAgATC-3′; reverse, 5′-gAAgAAgAATTggCCgA AATCAA-3′) and 4.053 bp for ftsK SL gene (primers: forward, 5′-AAAACAAAgCCTTggTggTgC CT-3′; reverse, 5′-CTTgCgACAAgAAAgggAAA-TTT-3′). The mutagenized PCR fragments were then transformed into strain R800. For each mutagenesis, ten KmR transformants were checked by PCR and shown to carry a mariner insertion. The accurate insertion position of the transposon, as well as its orientation, was determined by PCR and DNA sequencing as described [61]. The resulting chromosome structures of the selected mutants were: R800 xerS, insertion of mariner 167 bp downstream the ATG (insertion allows the synthesis of only the first 55 amino acids of XerS); R800 ftsK C, insertion of mariner 1217 bp downstream the ATG (insertion allows the synthesis of only the first 405 amino acids of FtsK).
dif SL-KmR-dif SL cassette excision in E. coli.
XerS-mediated excision of the dif SL-KmR-dif SL was performed according to [37]. Briefly, E. coli strains transformed with the XerS expression plasmid pCL297 (or cotransformed with pCL297 and pCL263) were growth in serial cultures at 30 °C (because of the thermosensitive replication of plasmid pCL297) for 60 generations in nonselective LB medium. The ratio of KmR/total colonies was measured each 20 generations, and excision frequencies (cell−1 generation−1) deduced according to Perals et al. [37]. For low excision frequencies (<1% cell−1 generation−1), such as those obtained in the absence of XerS in recA or ftsK C single mutants and recA ftsK C double mutants, 200 colonies picked from nonselective plates after 85 generations of serial growth were replica plated on Km-containing LB plates, the ratio of KmR/total colonies was measured, and used to estimate the excision frequencies per generation.
Growth competition assays in E. coli.
Coculture experiments were performed as described by Perals et al. [3], except that the growth temperature was 30 °C (because of the thermosensitive replication of plasmid pCL297). The growth advantage per generation, corresponding to the frequency of chromosome dimer resolution, was calculated from the slope of each competition curve as according to [3]. The strains containing one dif SL site located at the native position of the E. coli dif site correspond to KmS strains obtained from replica plating from nonselective plates in the dif SL-KmR-dif SL excision experiments described above.
Subcellular localization of FtsK-GFP fusions in L. lactis.
The lactococcal ftsK gene was amplified by PCR from chromosomal DNA of L. lactis MG1363. The 557-bp PCR fragment (encoding the first 181 amino acids of FtsK; primers: forward, 5′-CATgCCATgggTgAAAgTAAAAAAATgCCT-3′; reverse 5′-CCATCgATTTTAgggAAAAgTCC TggAA-3′) and the 2303-bp PCR fragment (full-length ftsK gene; primers: forward, 5′-CATgCCATgggTgAAAgTAAAAAAATgCCT-3′; reverse 5′-CCATCgATCTCTTCTACTCCTC CAATA-3′) were cleaved with NcoI (bold) and EcoRV (underlined) and cloned into corresponding sites of pNG8048e [62], generating respectively pFtsKNter and pFtsKFL. The ClaI-XbaI fragment containing the gfpmut1 gene of pSG1154 [63] was cloned into the corresponding sites of pFtsKNter and pFtsKFL yielding plasmids pKNtergfp and pKFLgfp respectively. For fluorescence microscopy, cells from an overnight culture were diluted (1:100) into fresh medium and grown at 30 °C with agitation. At OD600 = 0.5, supernatant of the nisin-producing strain NZ9700 [64] was added at dilution 1:1,000. At OD600 = 1, 2 μl of culture was directly applied to a polysine microscope slide (Omnilabo, http://www1.omnilabo.nl) and covered with a cover glass. Cells were examined using a 100× oil immersion objective on a Zeiss microscope (Carl Zeiss, http://www.zeiss.com), and the fluorescent signal of GFP was detected using filter set 09 (excitation, 450–490 nm and emission, > 520 nm) from Zeiss. Images were captured with an Axion Vision camera (Axion Technologies, http://www.axiontech.com), and assembly of the final figures used Adobe Photoshop version 6.0.
Supporting Information
Shown is Pulsed-Field gel Electrophoresis of SmaI-digested chromosomes (A) and corresponding Southern hybridization (B) of WT strain (MG1363) and its recA-derivative (VEL1122) after dif-mediated integration of plasmids pCL235 or pCL237. Lanes: 1, MG1363; 2, MG1363::pCL235; 3, MG1363::pCL237; 4, VEL1122::pCL235; and 5, VEL1122::pCL237. (Electrophoresis conditions: 10 V/cm−1/13 h/15 s pulse time in Tris/borate/EDTA 0.05M). As predicted from the physical map of MG1363 chromosome [58] and genome sequence, insertion of integrative plasmids at the dif SL site into the 610-kb SmaI fragment generated two new SmaI fragments of 350- and 260-kb in size. These corresponding restriction fragments were visualized in Southern hybridization using pCL235 and ilvD gene as probes in the presence of 32P-labeled λ DNA. A part of the chromosome population displayed a WT structure (i.e., with the plasmids pCL235 or pCL237 excised from the chromosome), an instability also observed when transforming E. coli strain with a nonreplicative plasmid containing the E. coli dif sequence (F. Cornet, personal communication).
(918 KB PDF)
Colors: red, integrases conserved in each streptococcal species; green, phage-related integrases; blue, transposon-related integrases; black, uncharacterized integrases. Bootstrap values are indicated for each branch.
(228 KB PDF)
(28 KB PDF)
(16 KB PDF)
Accession Numbers
The National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) accession number for L. lactis is NC_009004.
Acknowledgments
The authors thank François-Xavier Barre, Sarah Bigot, Chantal Granadel, Bernard Martin, Jean-Pierre Claverys, and François Cornet for helpful discussions and the gift of plasmids and/or protocols. PLB particularly thanks Claude Bruand and Romain Mercier for useful suggestions in the E. coli experiments, Jonathan Filée for help in the phylogenetic analysis of streptococcal tyrosine recombinases, and François Cornet and Michael Chandler for insightful comments on the manuscript.
Abbreviations
- KmR
kanamycin resistance
- ORF
open reading frame
- WT
wild type
Footnotes
† Dedicated to my colleague and friend Frédérique Messud (1962–2005)
Author contributions. PLB and PR conceived and designed the experiments. PLB, MB, NC, MLDM, JL, DL, and TL performed the experiments. PLB, NC, and PR analyzed the data. PLB, MB, NC, MLDM, JL, DL, TL, and CP contributed reagents/materials/analysis tools. PLB, NC, MLDM, and PR wrote the paper.
Funding. All work in our laboratory is supported by the CNRS.
Competing interests. The authors have declared that no competing interests exist.
References
- Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc Natl Acad Sci U S A. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W, Kuempel PL. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J Bacteriol. 1998;180:6269–6275. doi: 10.1128/jb.180.23.6269-6275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perals K, Cornet F, Merlet Y, Delon I, Louarn JM. Functional polarization of the Escherichia coli chromosome terminus: The dif site acts in chromosome dimer resolution only when located between long stretches of opposite polarity. Mol Microbiol. 2000;36:33–43. doi: 10.1046/j.1365-2958.2000.01847.x. [DOI] [PubMed] [Google Scholar]
- Sherratt DJ, Soballe B, Barre FX, Filipe S, Lau I, et al. Recombination and chromosome segregation. Philos Trans R Soc Lond B Biol Sci. 2004;359:61–69. doi: 10.1098/rstb.2003.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesterlin C, Barre FX, Cornet F. Genetic recombination and the cell cycle: What we have learned from chromosome dimers. Mol Microbiol. 2004;54:1151–1160. doi: 10.1111/j.1365-2958.2004.04356.x. [DOI] [PubMed] [Google Scholar]
- Blakely GW, May G, McCulloch R, Arciszewska LK, Burke M, et al. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993;75:351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- Hallet B, Arciszewska LK, Sherratt DJ. Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: An enzymatic switch in site-specific recombination. Mol Cell. 1999;4:949–959. doi: 10.1016/s1097-2765(00)80224-5. [DOI] [PubMed] [Google Scholar]
- Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, et al. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- Steiner W, Kuempel PL. Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli . Mol Microbiol. 1998;27:257–268. doi: 10.1046/j.1365-2958.1998.00651.x. [DOI] [PubMed] [Google Scholar]
- Steiner W, Liu G, Donachie WD, Kuempel PL. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- Recchia GD, Aroyo M, Wolf D, Blakely GW, Sherratt DJ. FtsK-dependent and -independent pathways of Xer site-specific recombination. EMBO J. 1999;18:5724–5734. doi: 10.1093/emboj/18.20.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre FX, Aroyo M, Colloms SD, Helfrich A, Cornet F, et al. FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 2000;14:2976–2988. doi: 10.1101/gad.188700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- Corre J, Louarn JM. Evidence from terminal recombination gradients that FtsK uses replichore polarity to control chromosome terminus positioning at division in Escherichia coli . J Bacteriol. 2002;184:3801–3807. doi: 10.1128/JB.184.14.3801-3807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capiaux H, Lesterlin C, Perals K, Louarn JM, Cornet F. A dual role for the FtsK protein in Escherichia coli chromosome segregation. EMBO Rep. 2002;3:532–536. doi: 10.1093/embo-reports/kvf116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip SC, Bregu M, Barre FX, Sherratt DJ. Decatenation of DNA circles by FtsK-dependent Xer site-specific recombination. EMBO J. 2003;22:6399–6407. doi: 10.1093/emboj/cdg589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O, Ptacin JL, Pease PJ, Gore J, Eisen MB, et al. Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc Natl Acad Sci U S A. 2005;102:17618–17623. doi: 10.1073/pnas.0508932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Saleh OA, Lesterlin C, Pages C, El Karoui M, et al. KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J. 2005;24:3770–3780. doi: 10.1038/sj.emboj.7600835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Saleh OA, Cornet F, Allemand JF, Barre FX. Oriented loading of FtsK on KOPS. Nat Struct Mol Biol. 2006;13:1026–1028. doi: 10.1038/nsmb1159. [DOI] [PubMed] [Google Scholar]
- Yates J, Zhekov I, Baker R, Eklund B, Sherratt DJ, et al. Dissection of a functional interaction between the DNA translocase, FtsK, and the XerD recombinase. Mol Microbiol. 2006;59:1754–1766. doi: 10.1111/j.1365-2958.2005.05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J, Aroyo M, Sherratt DJ, Barre FX. Species specificity in the activation of Xer recombination at dif by FtsK. Mol Microbiol. 2003;49:241–249. doi: 10.1046/j.1365-2958.2003.03574.x. [DOI] [PubMed] [Google Scholar]
- Recchia GD, Sherratt DJ. Conservation of xer site-specific recombination genes in bacteria. Mol Microbiol. 1999;34:1146–1148. doi: 10.1046/j.1365-2958.1999.01668.x. [DOI] [PubMed] [Google Scholar]
- Hendrickson H, Lawrence JG. Mutational bias suggests that replication termination occurs near the dif site, not at Ter sites. Mol Microbiol. 2007;64:42–56. doi: 10.1111/j.1365-2958.2007.05596.x. [DOI] [PubMed] [Google Scholar]
- Neilson L, Blakely GW, Sherratt DJ. Site-specific recombination at dif by Haemophilus influenzae XerC. Mol Microbiol. 1999;31:915–926. doi: 10.1046/j.1365-2958.1999.01231.x. [DOI] [PubMed] [Google Scholar]
- Blakely GW, Davidson AO, Sherratt DJ. Sequential strand exchange by XerC and XerD during site-specific recombination at dif . J Biol Chem. 2000;275:9930–9936. doi: 10.1074/jbc.275.14.9930. [DOI] [PubMed] [Google Scholar]
- Sciochetti SA, Piggot PJ, Blakely GW. Identification and characterization of the dif Site from Bacillus subtilis . J Bacteriol. 2001;183:1058–1068. doi: 10.1128/JB.183.3.1058-1068.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villion M, Szatmari G. The XerC recombinase of Proteus mirabilis: Characterization and interaction with other tyrosine recombinases. FEMS Microbiol Lett. 2003;226:65–71. doi: 10.1016/S0378-1097(03)00577-9. [DOI] [PubMed] [Google Scholar]
- Jouan L, Szatmari G. Interactions of the Caulobacter crescentus XerC and XerD recombinases with the E. coli dif site. FEMS Microbiol Lett. 2003;222:257–262. doi: 10.1016/S0378-1097(03)00311-2. [DOI] [PubMed] [Google Scholar]
- Mitchell TJ. The pathogenesis of streptococcal infections: From tooth decay to meningitis. Nat Rev Microbiol. 2003;1:219–230. doi: 10.1038/nrmicro771. [DOI] [PubMed] [Google Scholar]
- Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, et al. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol Rev. 2005;29:435–463. doi: 10.1016/j.femsre.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Hugenholtz J, Smid EJ. Nutraceutical production with food-grade microorganisms. Curr Opin Biotechnol. 2002;13:497–507. doi: 10.1016/s0958-1669(02)00367-1. [DOI] [PubMed] [Google Scholar]
- Chalker AF, Lupas A, Ingraham K, So CY, Lunsford RD, et al. Genetic characterization of gram-positive homologs of the XerCD site-specific recombinases. J Mol Microbiol Biotechnol. 2000;2:225–233. [PubMed] [Google Scholar]
- Reichmann P, Hakenbeck R. A XerD recombinase with unusual active site motifs in Streptococcus pneumoniae . J Mol Microbiol Biotechnol. 2002;4:101–110. [PubMed] [Google Scholar]
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004;150:1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, et al. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001;11:731–753. doi: 10.1101/gr.169701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perals K, Capiaux H, Vincourt JB, Louarn JM, Sherratt DJ, et al. Interplay between recombination, cell division and chromosome structure during chromosome dimer resolution in Escherichia coli . Mol Microbiol. 2001;39:904–913. doi: 10.1046/j.1365-2958.2001.02277.x. [DOI] [PubMed] [Google Scholar]
- Ferreira H, Butler-Cole B, Burgin A, Baker R, Sherratt DJ, et al. Functional analysis of the C-terminal domains of the site-specific recombinases XerC and XerD. J Mol Biol. 2003;330:15–27. doi: 10.1016/s0022-2836(03)00558-8. [DOI] [PubMed] [Google Scholar]
- Leslie NR, Sherratt DJ. Site-specific recombination in the replication terminus region of Escherichia coli: Functional replacement of dif . EMBO J. 1995;14:1561–1570. doi: 10.1002/j.1460-2075.1995.tb07142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: Implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D. Integrons: Agents of bacterial evolution. Nat Rev Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- Sternberg N, Sauer B, Hoess R, Abremski K. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by DNA methylation. J Mol Biol. 1986;187:197–212. doi: 10.1016/0022-2836(86)90228-7. [DOI] [PubMed] [Google Scholar]
- Lee MH, Pascopella L, Jacobs WR, Jr, Hatfull GF. Site-specific integration of mycobacteriophage L5: Integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci U S A. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravatn R, Studer S, Zehnder AJ, van der Meer JR. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. Strain B13. J Bacteriol. 1998;180:5505–5514. doi: 10.1128/jb.180.21.5505-5514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Paszkiet BJ, Shoemaker NB, Gardner JF, Salyers AA. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J Bacteriol. 2000;182:4035–4043. doi: 10.1128/jb.182.14.4035-4043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV. Evolutionary genomics of lactic acid bacteria. J Bacteriol. 2007;189:1199–1208. doi: 10.1128/JB.01351-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henz SR, Huson DH, Auch AF, Nieselt-Struwe K, Schuster SC. Whole-genome prokaryotic phylogeny. Bioinformatics. 2005;21:2329–2335. doi: 10.1093/bioinformatics/bth324. [DOI] [PubMed] [Google Scholar]
- Huber KE, Waldor MK. Filamentous phage integration requires the host recombinases XerC and XerD. Nature. 2002;417:656–659. doi: 10.1038/nature00782. [DOI] [PubMed] [Google Scholar]
- Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol. 2001;41:263–277. doi: 10.1046/j.1365-2958.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- Van Duyne GD. A structural view of Cre-loxP site-specific recombination. Annu Rev Biophys Biomol Struct. 2001;30:87–104. doi: 10.1146/annurev.biophys.30.1.87. [DOI] [PubMed] [Google Scholar]
- Chen Y, Rice PA. New insight into site-specific recombination from Flp recombinase-DNA structures. Annu Rev Biophys Biomol Struct. 2003;32:135–159. doi: 10.1146/annurev.biophys.32.110601.141732. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis . EMBO J. 1997;16:2161–2169. doi: 10.1093/emboj/16.8.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg KJ, Dewar SJ, Donachie WD. A new Escherichia coli cell division gene, ftsK . J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, West L, Shapiro L. The bifunctional FtsK protein mediates chromosome partitioning and cell division in Caulobacter . J Bacteriol. 2006;188:1497–1508. doi: 10.1128/JB.188.4.1497-1508.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Corre J, Louarn JM, Cornet F, Barre FX. FtsK activities in Xer recombination, DNA mobilization and cell division involve overlapping and separate domains of the protein. Mol Microbiol. 2004;54:876–886. doi: 10.1111/j.1365-2958.2004.04335.x. [DOI] [PubMed] [Google Scholar]
- Le Bourgeois P, Langella P, Ritzenthaler P. Electrotransformation of Lactococcus lactis . In: Teissié J, Eynard N, editors. Electrotransformation of bacteria, Springer Lab Manual. Heidelberg: Springer; 2000. pp. 56–65. [Google Scholar]
- Alloing G, Granadel C, Morrison DA, Claverys JP. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae . Mol Microbiol. 1996;21:471–478. doi: 10.1111/j.1365-2958.1996.tb02556.x. [DOI] [PubMed] [Google Scholar]
- Le Bourgeois P, Lautier M, van den Berghe L, Gasson MJ, Ritzenthaler P. Physical and genetic map of the Lactococcus lactis subsp. cremoris MG1363 chromosome: Comparison with that of Lactococcus lactis subsp. lactis IL1403 reveals a large genome inversion. J Bacteriol. 1995;177:2840–2850. doi: 10.1128/jb.177.10.2840-2850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, et al. MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguin E, Prévots H, Ehrlich SD, Gruss A. Efficient insertional mutagenesis in Lactococci and other Gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M, Camilli A, Claverys JP. In vitro mariner mutagenesis of Streptococcus pneumoniae: Tools and traps. In: Hakenbeck R, Chhatwal GS, editors. The molecular biology of Streptococci. Norwich (United Kingdom): Horizon Scientific Press; 2006. pp. 509–516. [Google Scholar]
- Gutierrez J, Larsen R, Cintas LM, Kok J, Hernandez PE. High-level heterologous production and functional expression of the sec-dependent enterocin P from Enterococcus faecium P13 in Lactococcus lactis . Appl Microbiol Biotechnol. 2006;72:41–51. doi: 10.1007/s00253-005-0233-1. [DOI] [PubMed] [Google Scholar]
- Lewis PJ, Marston AL. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis . Gene. 1999;227:101–110. doi: 10.1016/s0378-1119(98)00580-0. [DOI] [PubMed] [Google Scholar]
- Kuipers OP, Beerthuyzen MM, Siezen RJ, de Vos WM. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216:281–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shown is Pulsed-Field gel Electrophoresis of SmaI-digested chromosomes (A) and corresponding Southern hybridization (B) of WT strain (MG1363) and its recA-derivative (VEL1122) after dif-mediated integration of plasmids pCL235 or pCL237. Lanes: 1, MG1363; 2, MG1363::pCL235; 3, MG1363::pCL237; 4, VEL1122::pCL235; and 5, VEL1122::pCL237. (Electrophoresis conditions: 10 V/cm−1/13 h/15 s pulse time in Tris/borate/EDTA 0.05M). As predicted from the physical map of MG1363 chromosome [58] and genome sequence, insertion of integrative plasmids at the dif SL site into the 610-kb SmaI fragment generated two new SmaI fragments of 350- and 260-kb in size. These corresponding restriction fragments were visualized in Southern hybridization using pCL235 and ilvD gene as probes in the presence of 32P-labeled λ DNA. A part of the chromosome population displayed a WT structure (i.e., with the plasmids pCL235 or pCL237 excised from the chromosome), an instability also observed when transforming E. coli strain with a nonreplicative plasmid containing the E. coli dif sequence (F. Cornet, personal communication).
(918 KB PDF)
Colors: red, integrases conserved in each streptococcal species; green, phage-related integrases; blue, transposon-related integrases; black, uncharacterized integrases. Bootstrap values are indicated for each branch.
(228 KB PDF)
(28 KB PDF)
(16 KB PDF)