Abstract
The yeast Sch9 kinase has been implicated in the cellular adjustment to nutrient availability and in the regulation of aging. Here, we define a novel role for Sch9 in the transcriptional activation of osmostress inducible genes. Loss-of-function mutants sch9 are sensitive to hyperosmotic stress and show an impaired transcriptional response upon osmotic shock of several defense genes. We show that Sch9 is required for gene expression regulated by Sko1, a transcription factor, which is directly targeted by the Hog1 MAP kinase. Sch9 interacts in vitro with both Sko1 and Hog1. Additionally, Sch9 phosphorylates Sko1 in vitro. When artificially tethered to promoter DNA, Sch9 strongly activates transcription independently of osmotic stress. Using in vivo chromatin immunoprecipitation, we demonstrate that Sch9 is recruited to the GRE2 and CTT1 genes exclusively under osmostress conditions, and that this recruitment is dependent on Hog1 and Sko1. Furthermore, Sch9 is required for the proper recruitment of Hog1 at the same genes. Our data reveal the complexity of stress-induced transcription by the regulated association of signaling kinases to chromatin.
Keywords: Hog1, osmotic stress, Sch9, transcription
Introduction
All eukaryotic cells respond to environmental stresses by the activation of complex transcriptional programs, which affect the expression of many genes involved in the cellular adjustment to the adverse conditions. A great variety of stresses are sensed by specific signal transduction pathways, which very often culminate in the modulation of protein kinase activities. In yeast cells, several highly conserved signaling pathways regulate transcription via protein kinases like mitogen-activated protein (MAP) kinase (MAPK) (Gustin et al, 1998), protein kinase A (PKA) (Thevelein and de Winde, 1999), Snf1 (Carlson, 1999), TOR (Martin and Hall, 2005) and others, in response to nutrient, osmotic, oxidative and other stresses. Studies in the yeast model have revealed that signaling kinases involved in the stimulation of gene expression upon stress have very complex functions that go far beyond the simple modulation of specific transcription factor activities and involve the stable association of the active kinase with the chromatin of the regulated gene (Edmunds and Mahadevan, 2004, 2006; Chow and Davis, 2006). This much more direct function in transcriptional regulation is a general feature, as it has been reported for a variety of structurally unrelated protein kinases directly controlling gene activity such as MAPKs, PKA, Snf1 or Tor1 (Alepuz et al, 2001; Proft and Struhl, 2002; Lo et al, 2005; Li et al, 2006; Pokholok et al, 2006; Proft et al, 2006). The association of the signaling kinases with their target genes in the chromosome in vivo has opened up the possibility to identify stress genes regulated by a particular kinase by chromatin immunoprecipitation (ChIP) (Pascual-Ahuir et al, 2006; Pokholok et al, 2006; Proft et al, 2006).
In Saccharomyces cerevisiae, hyperosmotic stress rapidly activates the MAPK Hog1, which orchestrates cellular adaptations at the level of gene expression, ion homeostasis, translation and cell cycle progression (DeNadal et al, 2002). Although Hog1 is the key regulator of the yeast osmostress response, other signaling pathways also contribute to the osmostress resistance (Hohmann, 2002). The impact of hyperosmolarity on gene expression is considerable and involves the upregulation of a large number of stress-responsive genes, as revealed by genomic profiling (Posas et al, 2000; Rep et al, 2000). Upon activation, Hog1 rapidly associates with the chromatin of osmostress-responsive genes (Alepuz et al, 2001; Proft and Struhl, 2002; Pokholok et al, 2006; Proft et al, 2006) via the direct recruitment of the MAPK by various specific transcription factors, like the Sko1 repressor/activator and the Hot1 and Smp1 activators (Alepuz et al, 2001, 2003; Proft et al, 2001; DeNadal et al, 2003). Once tethered to the chromatin of inducible genes, activated Hog1 MAPK can stimulate transcription by surprisingly complex mechanisms. Hog1 can recruit chromatin modifying complexes in an indirect or direct manner (Proft and Struhl, 2002; DeNadal et al, 2004). Apart from chromatin modifications that favor transcriptional initiation upon stress, Hog1 seems to directly recruit the RNA polymerase II machinery to Hot1-dependent genes (Alepuz et al, 2003). Finally, the MAPK is a structural portion of the RNA pol II elongation complex selectively at osmostress-activated genes (Proft et al, 2006).
In mammalian cells, the stress-activated p38 MAPK (orthologue of yeast Hog1) activates transcription through various downstream effectors like transcription factors, protein kinases and other regulators. One of the direct targets of p38 are the Msk1/2 protein kinases (Deak et al, 1998), which link the MAPK activity to specific transcription factors and chromatin modifications (reviewed by Dunn et al (2005)). Msk1/2 activation is necessary to phosphorylate transcription factors like ATF1/2 and CREB (Wiggin et al, 2002; Arthur et al, 2004; Zhu et al, 2004). On the other hand, Msk1 can modify the chromatin structure of p38 target promoters by direct phosphorylation of histone H3 at serines 10 and 28 (Davie, 2003; Soloaga et al, 2003; Dyson et al, 2005). In yeast, the Hog1 MAPK targets the ATF/CREB protein Sko1 to modulate the expression of a subset of osmostress genes (Proft and Serrano, 1999; Proft et al, 2001). However, a downstream target similar to the MSK1/2 kinases of Hog1 has not been identified yet. Detailed studies on the Hog1–Sko1 interaction have revealed that a particular MAPK-regulated transcription factor is targeted by various kinases to modulate gene expression (Proft et al, 2001). Here, we report that the Sch9 protein kinase, which shares similarity with mammalian MSK1/2, regulates transcription through the Sko1 transcription factor.
The yeast Sch9 kinase was originally identified as being partially redundant to the catalytical subunits of cAMP-regulated PKA (Toda et al, 1988). Functional analyses characterized Sch9 as a regulator of cell size control and transcription in response to nutrient availability (Crauwels et al, 1997; Pedruzzi et al, 2003; Jorgensen et al, 2004). Furthermore, it has become clear that the Sch9-regulated transcriptional output is different from PKA and involves transcriptional activation (Denis and Audino, 1991; Roosen et al, 2005). Sch9, like other molecules involved in nutrient signaling, is also a determinant of cellular aging. Loss of Sch9 function increases survival in stationary, non-dividing yeast cultures (Fabrizio et al, 2001), and enhances replicative lifespan (Fabrizio et al, 2004; Kaeberlein et al, 2005). Here, we define a novel role for the Sch9 kinase: it is an important determinant for acute osmotic stress adaptation at the transcriptional level through the Sko1 transcription factor. Its Sko1- and Hog1-regulated recruitment to osmostress-responsive genes defines Sch9 as a direct transcriptional activator in the context of chromatin.
Results
Sch9 is important for osmotic and oxidative stress resistance
Yeast cells respond to hyperosmotic stress by activation of the HOG (High-Osmolarity Glycerol) MAPK cascade. Several other kinases of the AGC family of protein kinases like PKA or PKC have been implicated in the adaptation to changes in osmolarity. Here, we investigate the function in osmostress adaptation of three structurally related AGC kinases, Sch9, Ypk1 and Ypk2. We directly tested whether loss of any of these kinases affected the resistance to salt and osmotic stress. We compared the growth of mutant strains ypk1Δ, ypk2Δ and sch9Δ under various stress conditions with the wild type and the hog1Δ mutant (Figure 1). Deletion of SCH9 resulted in a strong hypersensitivity to salt (NaCl plates) and hyperosmotic stress (KCl plates). A modest sensitivity was observed for Li+ stress. The phenotype for sch9Δ mutants was almost as pronounced as the sensitivity of the hog1Δ MAPK deletion strain. ypk1Δ mutants showed a moderate sensitivity to NaCl and KCl, while loss of YPK2 did not produce any observable growth defect under the conditions tested. Analysis of the expression of typical osmostress inducible defense genes revealed that only sch9Δ mutants showed a defect in the transcriptional response (see below); therefore, we focused on identifying the function of the Sch9 kinase in the adaptation to hyperosmolarity. We next tested whether the osmosensitivity of sch9Δ mutants was due to the lack of the Sch9 kinase function. We retransformed the mutant with wild-type SCH9 or a point mutated allele in the Sch9 kinase domain (K441A; Morano and Thiele, 1999). These complementation assays demonstrated that the observed osmosensitivity phenotype is directly caused by the lack of the Sch9 kinase function (Figure 1B and C).
Figure 1.
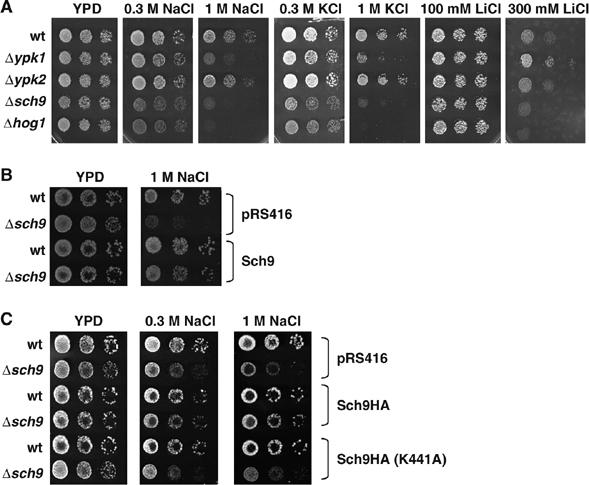
Osmotic and salt stress sensitivities of yeast mutants ypk1Δ, ypk2Δ and sch9Δ. (A) Isogenic yeast strains BY4741 (wild type) and deletion mutants ypk1Δ, ypk2Δ, sch9Δ and hog1Δ were spotted onto YPD plates with the indicated concentrations of salt. (B) BY4741 (wild type) and sch9Δ mutants were transformed with pRS416 (vector control) or pRS416-SCH9 and assayed for NaCl resistance. (C) HA-tagged fusion proteins (wild type or point mutated K441A Sch9) were expressed from pRS416 in BY4741 (wild type) and sch9Δ mutants. Transformed strains were assayed for NaCl resistance.
Both kinases, Sch9 and the MAPK Hog1, are important for survival under osmostress (Figure 1). We wanted to further investigate the epistatic relation of both molecules in the stress response. Therefore, we used hog1Δ and sch9Δ single mutants and hog1Δsch9Δ double mutants, to assay for salt stress survival. As depicted in Figure 2A, the phenotype of the highly osmosensitive hog1Δ strain was slightly exacerbated by the additional deletion of SCH9 (0.3 M NaCl and KCl plates). Since the HOG pathway is also necessary to properly adapt to oxidative stress (Rep et al, 2001; Bilsland et al, 2004), and sch9 mutants have been characterized as hyperresistant to oxidative stress in stationary phase (Fabrizio et al, 2001), we included H2O2 treatment in the phenotypic analysis of the kinase mutants. As shown in Figure 2B, sch9Δ and hog1Δ mutants were hypersensitive to oxidative stress, while the hog1Δsch9Δ double deletion showed a slightly more pronounced growth defect. Interestingly, H2O2 sensitivity of sch9Δ cells was observed exclusively in exponentially growing cultures, whereas we confirmed H2O2 resistance in stationary cultures (Figure 2C). We conclude that Sch9 kinase is an important determinant for osmotic and oxidative stress adaptation in actively dividing cells.
Figure 2.
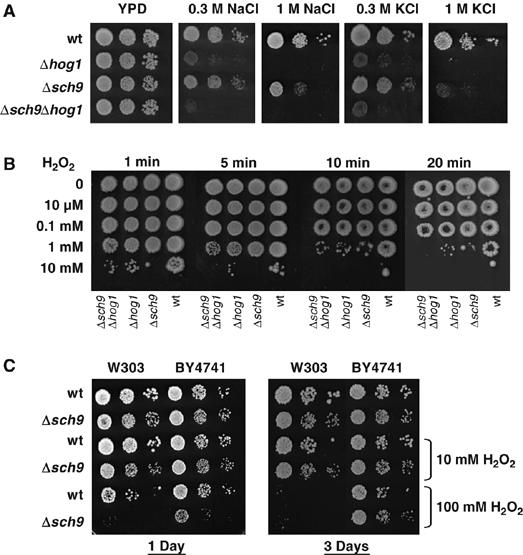
Osmotic and oxidative stress phenotypes of sch9Δ and hog1Δ mutants. (A) Isogenic yeast strains W303-1A (wild type) and deletion mutants sch9Δ (MAP85), hog1Δ(MAP32), and sch9Δhog1Δ (MAP84) were assayed for growth on YPD plates with or without the indicated salt concentrations. (B) The same strains were incubated with different H2O2 concentrations for the indicated times and then assayed for survival on YPD plates. (C) Comparison of hydrogen peroxide sensitivities of exponentially growing (1 day, OD600=0.8) and PDS shift (3 days) cultures of wild type and sch9Δ mutants in the indicated genetic backgrounds.
Sch9 function is required for osmostress-induced gene expression
Adaptation of yeast cells to hyperosmolarity involves the transient upregulation of many defense genes at the transcriptional level, dependent of the Hog1 MAPK (Posas et al, 2000; Rep et al, 2000). We addressed the question of whether the observed osmosensitivity of sch9Δ mutants could be linked to an impaired transcriptional response. We therefore measured the mRNA levels of several highly inducible defense genes before and during osmotic shock, in wild type, sch9Δ and hog1Δ mutants. We assayed for GRE2, CTT1 and STL1 expression, three genes whose activation is driven by distinct subsets of specific transcription factors, Sko1 [GRE2], Msn2,4 [CTT1] and Hot1 [STL1]. As depicted in Figure 3, expression of all three genes was robustly induced in yeast wild-type cells, whereas induction was completely abolished in hog1Δ mutant cells. Loss of Sch9 function significantly affected the stress-induced mRNA levels of all three genes tested, with the CTT1 gene affected the most. The impaired expression of the defense genes strongly suggests that the Sch9 kinase is directly or indirectly involved in the transcriptional control upon osmostress.
Figure 3.
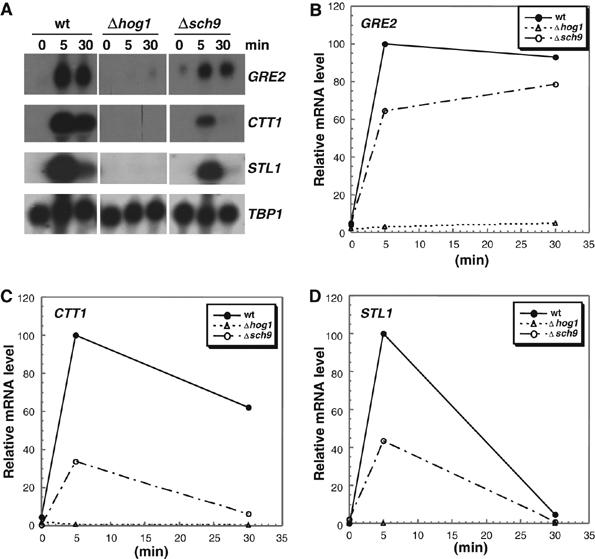
Roles of Sch9 and Hog1 kinases in the osmotic stress-induced transcription of GRE2, CTT1 and STL1. (A) Transcript levels of the indicated genes were monitored by Northern analysis before and during acute osmotic stress (0.4 M NaCl). (B–D) The mRNA levels for all three stress genes were quantified, normalized for the TBP1 internal loading control and depicted in the graphs. The highest signal for each gene was arbitrarily set to 100%. The isogenic yeast strains BY4741 (wild type) and mutants sch9Δ and hog1Δ were used. Results shown come from the same original blot which was probed with the indicated genes.
Sch9 affects Sko1-regulated gene expression
The Sko1 repressor/activator is one of several transcription factors, which regulates osmostress-induced gene expression under the direct control of the Hog1 MAPK (Proft et al, 2001, 2005; Proft and Struhl, 2002). Sko1 binds to cAMP-responsive elements (CRE) sequences. Expression analysis of CRE-driven reporter genes can be used to quantitate the osmostress-activated expression, which depends exclusively on the Sko1 and Hog1 proteins (Proft and Serrano, 1999; Pascual-Ahuir et al, 2001; Proft et al, 2001). We used a 2xCRE-lacZ reporter to test whether the Sch9 kinase acts through the Sko1 protein to induce transcription upon osmotic stress. As shown in Figure 4, the reporter gene is highly inducible by NaCl treatment in wild-type cells, which is completely dependent on the Sko1 and Hog1 proteins (Proft et al, 2001). In sch9Δ mutant cells, we observed a dramatic reduction in the activation of the reporter fusion upon stress. This lack of activation is not due to reduced Sko1 or Hog1 protein levels, because epitope-tagged Sko1 or Hog1 proteins expressed from their chromosomal locus were equally abundant in wild-type and sch9Δ mutant cells (data not shown). We conclude that the Sch9 kinase has a function in the osmostress-regulated transcriptional response via the Sko1 transcription factor.
Figure 4.
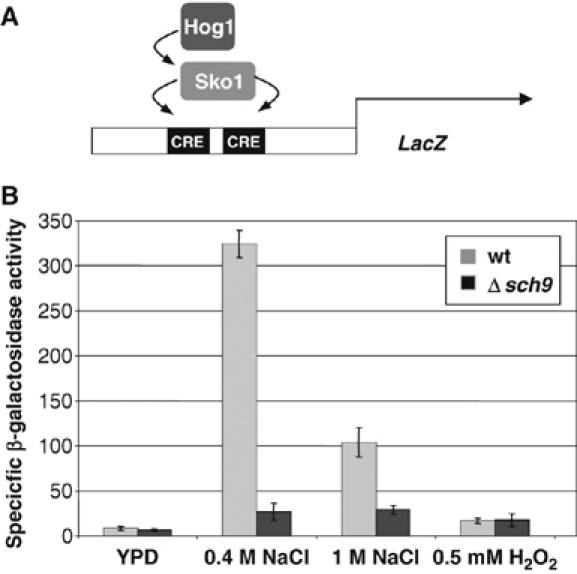
Sch9 function is necessary for Sko1-dependent transcriptional activation. (A) An Sko1- and Hog1-dependent CRE-lacZ reporter gene (CYC1-(2xCRE)-lacZ; pMP253) was used. (B) Yeast strains W303-1A (wild type) and deletion mutant sch9Δ (MAP85) were transformed with pMP253 and subjected or not to the indicated salt and oxidative stress treatments. Specific β-galactosidase activities (nmol/min/mg) are the result of measurements in duplicate of three independent transformants.
Sch9 interacts with Sko1 and Hog1, and phosphorylates the Sko1 transcription factor in vitro
We have characterized Sch9 as a positive modulator of osmostress-induced gene expression, which acts, at least in part, through the Sko1/Hog1 transcription factor/MAPK complex. Next we wanted to test whether the Sch9 kinase formed a complex with either Sko1 or Hog1. We performed co-immunoprecipitation experiments using HA-tagged Sch9 from osmostressed or unstressed yeast cells, with bacterial GST fusions of Hog1, Sko1 and GST alone as a control. The results of these in vitro interaction experiments are shown in Figure 5. Sch9 robustly copurified with Sko1-GST and weakly with Hog1-GST. Poor coprecipitation of GST alone and no enrichment of the GST proteins in the absence of the HA antigen demonstrated that the observed interactions were specific. While the amount of coprecipitated Sko1 was the same with Sch9 from unstressed or stressed cells, the interaction with Hog1 increased slightly when Sch9 was purified after osmotic shock. Taken together, we show that Sch9 forms a complex with Sko1 and Hog1 in vitro. We next addressed the question of whether the Sch9 kinase phosphorylated one or both of its interaction partners. Therefore, we performed in vitro phosphorylation studies with Sch9 and Sko1 or Hog1. We immunopurified HA-tagged Sch9 from yeast, before and after a brief hyperosmotic shock, and tested for phosphorylation of purified GST-Sko1 or GST-Hog1. As depicted in Figure 5B, Sch9 readily phosphorylated Sko1-GST in vitro, independently of the stress conditions. Equal amounts of Hog1-GST were not detectably phosphorylated by Sch9 in the same assay. Thus, Sch9 specifically phosphorylates the transcription factor Sko1 but not Hog1. These results correlate well with the ability of Sch9 to retain Sko1 in the coprecipitation experiments.
Figure 5.
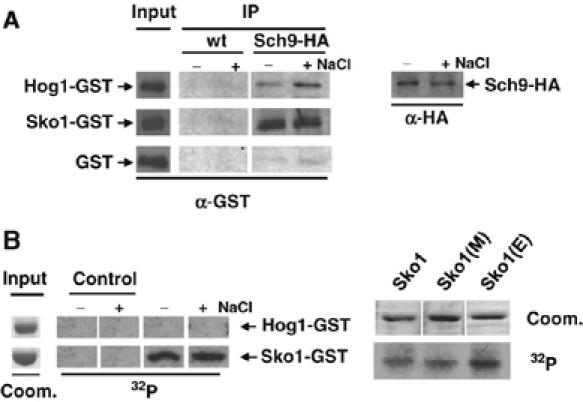
Sch9 interaction and phosphorylation with Sko1 and Hog1 in vitro. (A) HA epitope-tagged Sch9 was immunopurified from yeast strain MAP85 before and after osmotic shock (5 min, 0.4 M NaCl), in the presence of affinity purified GST (control), Hog1-GST and Sko1-GST proteins. As a control, the untagged wild-type strain W303-1A (wt) was used in the same copurification assay. Protein input (10% of total protein) was checked by anti-HA or anti-GST Western blot. Copurified GST proteins (IP) were visualized by anti-GST Western blot. For each interaction, the amounts of the input and IP samples were determined at the same time by anti-GST Western blot to make the IP efficiencies comparable between the different GST fusions. (B) Sch9 phosphorylates Sko1 in vitro. Affinity purified Hog1-GST and Sch9-GST full-length fusion proteins were incubated in the presence of [32P]ATP with immunopurified Sch9-HA kinase obtained from strain MAP85 before and after osmotic shock (A). Control reactions were performed in the absence of GST proteins. Phosphorylated proteins were resolved by SDS–PAGE, transferred to PVDF membranes and detected by autoradiography. Point mutated versions of Sko1-GST were: Sko1(E)=Sko1S108A,T113A,S126A; Sko1(M)=Sko1S380A,S393A,S399A. Input lanes represent approximately 10% of the total protein visualized by Coomassie staining of the same original gel. The different small blots come from the same original autoradiography for each experiment.
The Sko1 repressor/activator is phosphorylated by the MAPK Hog1 and PKA at multiple sites (Proft et al, 2001). We made use of Sko1-GST mutant proteins, which are no longer phosphorylated by Hog1 (Sko1S108A,T113A,S126A=Sko1[E]) or PKA (Sko1S380A, S393A, S399A)=Sko1[M]), to determine whether Sch9 phosphorylated Sko1 at the identified residues. As shown in Figure 5B, both mutant versions of Sko1 ([E] and [M]) were as efficiently phosphorylated by Sch9 as the wild-type protein, in vitro. Taken together, we show that the Sch9 kinase forms a complex with Sko1 and Hog1 in vitro, but only the transcription factor Sko1 is a target of the Sch9 kinase. However, phosphorylation of Sko1 is independent of the previously described residues targeted by Hog1 and PKA.
Sch9 activates transcription independently of stress when recruited artificially to a promoter
We have shown that Sch9 modulates stress-regulated expression via the Sko1 transcription factor, and that both molecules can form a complex in vitro. These data, together with the nuclear localization of Sko1 under the conditions tested, suggested that the Sch9 function in transcriptional activation was closely linked to molecular events on the responsive gene promoters. We therefore tested whether artificial recruitment of Sch9 to a promoter was sufficient to stimulate transcription. We employed fusions of Sch9 with the DNA binding domain of Gal4 (Gal4DBD) and measured the expression of the Gal4-dependent GAL1-lacZ gene. We compared the transcriptional activator capacity of Sch9-Gal4DBD with the Hog1-Gal4DBD hybrid protein. Hog1 has been described to readily activate transcription in monohybrid experiments strictly under hyperosmotic stress conditions (Alepuz et al, 2003). As shown in Figure 6, Hog1-Gal4DBD activated transcription from the GAL1 promoter exclusively after a brief hyperosmotic shock, and was inactive under normal or oxidative stress conditions. The Sch9-Gal4DBD hybrid strongly activated lacZ expression constitutively and independently of the growth conditions (Figure 6). The activation capacity of Sch9 was comparable to that of the stress-activated Hog1 MAPK. We conclude that artificial recruitment of the Sch9 kinase is sufficient to activate transcription independently of stress.
Figure 6.
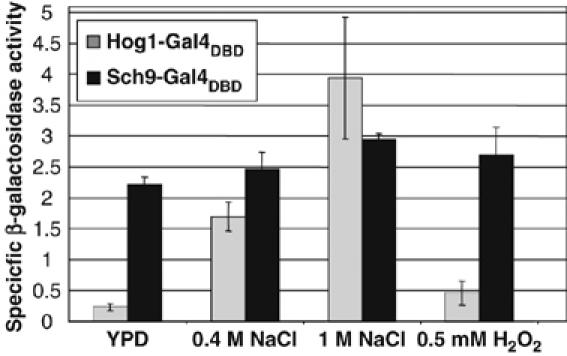
Sch9-Gal4DBD activates transcription independently on stress. Full-length Hog1-Gal4DBD and Sch9-Gal4DBD fusion proteins were expressed in yeast strain YULH. The expression of the chromosomal GAL1-lacZ reporter was monitored in the absence of stress (YPD) or after the indicated osmotic and oxidative stress. Specific β-galactosidase activities are give in (nmol/min/mg). Measurements were obtained in duplicate of three independent transformants.
Sch9 is recruited to the chromatin of osmostress- responsive genes in a HOG-dependent manner in vivo
We have found that Sch9 acts as a constitutive transcriptional activator when artificially tethered to a promoter in vivo. However, the phenotype of sch9Δ mutants indicated that the kinase mainly played a role in the osmostress-induced gene expression. We therefore investigated whether Sch9 physically associates with natural stress-responsive genes in vivo, and whether this recruitment was regulated by osmotic stress. A strain expressing HA-tagged Sch9 from its chromosomal locus was used to determine the association of the kinase with the osmostress-regulated genes GRE2 and CTT1 by ChIP in vivo. The transcriptional activation of both defense genes was significantly affected by the loss of Sch9 function (Figure 3). As shown in Figure 7A, Sch9 does not associate with the GRE2 and CTT1 genes under normal conditions. Upon brief hyperosmotic shock, however, we detected Sch9 at the GRE2 and CTT1 promoters, and at the 5′ regions of the respective ORFs. Occupancy of Sch9 was the highest at the promoter regions, but extended significantly to the 5′ portion of the adjacent transcribed regions. At the end of the ORF regions, we measured much less (GRE2) or no occupancy (CTT1) of Sch9. We tested whether the observed stress-regulated recruitment of Sch9 depended on the Hog1 MAPK. When HA-Sch9 was expressed in a hog1Δ mutant, its association with GRE2 and CTT1 chromatin was completely abolished, both at the promoter and transcribed regions (Figure 7A).
Figure 7.
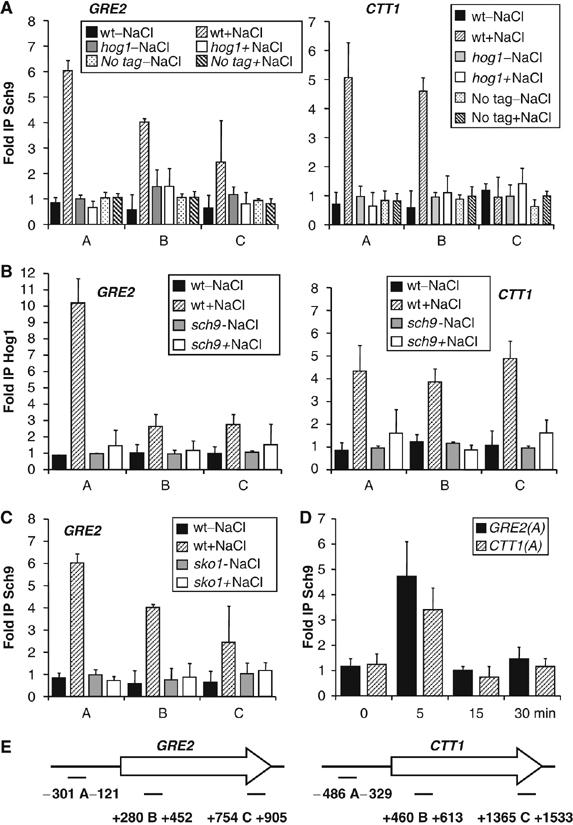
Interdependent Sch9 and Hog1 recruitment to the osmostress-responsive GRE2 and CTT1 genes in vivo. (A) Sch9 association with the indicated GRE2 and CTT1 promoter and ORF regions. Yeast wild type (MAP85) or hog1Δ mutants (MAP90) expressing Sch9-(HA)3 or the no tag control strain W303-1A were used. ChIP was performed with unstressed cells (−NaCl) or after brief osmostress (5 min, 0.4 M NaCl). (B) Hog1 association with the same chromosomal regions. Hog1-HA was expressed in wild type (MAP51) or in sch9Δ mutants (MAP83), before and after osmotic shock. (C) Sch9 association with the GRE2 promoter and ORF region. Yeast wild type (MAP85) or sko1Δ mutants (MAP93) expressing Sch9-(HA)3 were used before or after brief hyperosmotic shock. (D) Kinetic ChIP analysis of Sch9-(HA)3 association at the GRE2 and CTT1 promoters. (E) A schematic representation of the amplified regions in the ChIP experiments. Immunoprecipitation (IP) efficiencies are presented as the fold over the POL1 coding sequence control. Expression of HA-tagged Sch9 and Hog1 was not affected in the mutant strains used, as detected by Western blot.
The GRE2 and CTT1 genes are also targeted by Hog1 during transcriptional activation, and the MAPK associates with both promoters and the whole ORF regions upon stress (Proft et al, 2006). We next tested whether the recruitment of Hog1 was affected by the Sch9 kinase. We performed ChIP experiments with wild-type yeast or sch9Δ mutants expressing HA-tagged Hog1 from its chromosomal locus. As expected, we found Hog1 to be present at the GRE2 and CTT1 genes (promoter and transcribed regions), exclusively upon osmotic stress (Figure 7B). In the absence of Sch9, the recruitment of Hog1 to both stress genes was severely diminished and reduced to near background levels. Thus, both the Sch9 and Hog1 kinases modulate osmoshock inducible transcription directly at the chromatin of some responsive genes, and they are mutually dependent on each other with respect to their chromatin recruitment.
We have shown that Sch9 acts through the Sko1 transcription factor to stimulate transcription and that both proteins form a complex in vitro. We analyzed whether Sko1 was responsible for the recruitment of Sch9 to stress-activated genes in vivo. We expressed HA-tagged Sch9 in a sko1Δ mutant strain and used ChIP to quantify the recruitment of the kinase to the GRE2 gene, which is a typical Sko1 target (Proft et al, 2001; Rep et al, 2001; Proft and Struhl, 2002). As depicted in Figure 7C, the recruitment of Sch9 to the GRE2 gene depended completely on the function of Sko1. These data indicate that the Sko1 repressor/activator recruits Sch9 to the stress-activated GRE2 gene.
We finally quantified Sch9 recruitment to the GRE2 and CTT1 promoters by ChIP in a kinetic manner. We found that similar to Hog1, Sch9 kinase very transiently associates with both regions in the early stage (5 min) of the transcriptional osmostress response (Figure 7D).
Discussion
Here, we establish a novel function for the Sch9 protein kinase in yeast and show that the kinase is necessary to properly adapt to acute hyperosmotic stress at the level of the expression of defense genes. We present several independent lines of evidence showing that Sch9 activates transcription of stress-responsive genes directly at the chromatin of the regulated genes. Although Sch9 was originally characterized as being partially redundant to PKA, based mainly on its overexpression phenotype or genetic interactions (Toda et al, 1988), recent work has shown that Sch9 has PKA-independent or -opposing functions (Roosen et al, 2005). Genomic transcript profiling experiments identified a great number of genes, which are positively regulated by Sch9 and very interestingly revealed that many stress genes, while repressed by PKA, are activated by Sch9 (Roosen et al, 2005). Among those genes are typical hyperosmotic stress marker genes like GRE2 or CTT1, whose expression is highly inducible in a Hog1-dependent manner. We show here that Sch9 is required for the transcriptional activation of such genes under hyperosmotic stress conditions (Figure 3). Consequently, the loss of Sch9 function renders yeast cells hypersensitive to osmotic and salt stress (Figures 1 and 2). The contribution of each signaling kinase, Sch9 and Hog1, to osmostress resistance is different. While the Hog1 MAPK is absolutely required for induced transcript levels and resistance, sch9Δ mutants display a milder phenotype. This difference might be explained by the differential regulation of distinct subsets of stress genes regulated by each kinase. Indeed, we have detected genes (i.e., GPD1) whose expression during osmoshock was Sch9 independent, but strongly dependent on Hog1 (data not shown). Defense genes with the opposite pattern of regulation (Sch9 dependent, Hog1 independent) also exist (i.e., SSA3/4; data not shown). Taken together, both regulatory kinases do not necessarily coincide at the same target genes, which might explain how each regulator contributes, at least in part, additively to osmostress adaptation.
Here, we focus at the transcriptional regulation of Sch9 and Hog1-targeted defense genes. We show that the Sch9 kinase acts through the Sko1 transcription factor, which regulates gene expression bound at CRE in osmostress-responsive promoters. Several independent experimental approaches shown here support a model in which Sch9 phosphorylates the Sko1 bZIP protein and forms a stable complex with the transcription factor, to stimulate transcription from Sko1-bound promoters. These data are important, because although the impact of Sch9 on stress-regulated transcription was known (Pedruzzi et al, 2003; Roosen et al, 2005), its role in the regulation of gene expression remained speculative, mainly because of the lack of an unambiguous downstream target. Sch9 specifically copurifies with Sko1 and phosphorylates the transcription factor in vitro (Figure 5). Additionally, Sko1-regulated transcriptional activation (CRE-lacZ) almost completely depends on the Sch9 protein (Figure 4). Therefore, we conclude that the Sch9 kinase directly targets Sko1 to activate a subset of osmostress-responsive genes. The Sko1 protein is one of the direct phosphorylation targets of the activated Hog1 MAPK (Proft and Serrano, 1999; Proft et al, 2001), which operates a transcriptional switch from repressor to activator upon stress (Proft and Struhl, 2002). The DNA-bound Sko1 might therefore integrate inputs from several different regulatory kinases. Interestingly, the existence of other kinases targeting Sko1 under the control of the HOG pathway was suggested, because the transcription factor was still phosphorylated in vivo in a stress- and Hog1-dependent manner, after the removal of all Hog1 phosphorylation sites (Proft et al, 2001). One of the contributing kinases identified here is Sch9. The multitude of regulatory inputs at the Sko1 molecule might be even more complex, since PKA phosphorylations have also been identified in Sko1 (Proft et al, 2001), and Sko1-dependent reporter genes are regulated by PKA activity (Pascual-Ahuir et al, 2001). However, the function of PKA in the regulation of Sko1 activity remains to be elucidated. In the case of Sch9, the phosphorylation sites are different from the previously identified Hog1- and PKA-targeted residues in the Sko1 protein (Figure 5). Sch9 most likely acts through other specific transcription factors. A good candidate is the zinc finger protein Gis1, which activates gene expression from post-diauxic shift (PDS) promoter elements upon nutrient starvation (Pedruzzi et al, 2000). PDS containing promoters are enriched among the Sch9-controlled genes and some Gis1-regulated stress genes (SSA3, GRE1) depend on Sch9 with respect to their activation during the diauxic shift (Roosen et al, 2005). It remains to be seen, however, whether Gis1 or other transcription factors are directly targeted by Sch9.
Specific transcription factors can serve to recruit signaling kinases to promoters for their activation upon specific stress conditions. This paradigm has been shown for the activated Hog1 MAPK, which is recruited to the chromatin of osmostress-responsive promoters through various transcription factors, including Sko1 and Hot1 (Alepuz et al, 2001; Proft and Struhl, 2002). Here, we show that Sch9 associates with defense genes like GRE2 or CTT1 upon osmostress in the living cell (Figure 7). Our ChIP data demonstrate that transcriptional control of osmostress-responsive genes involves the association of at least two signaling kinases, Sch9 and Hog1, at some activated genes. At the GRE2 gene, the recruitment of Sch9 completely depends on the Sko1 protein. This observation confirms the in vitro interaction and phosphorylation data and demonstrates that Sko1 is able to recruit Sch9 to chromatin under hyperosmotic stress conditions in vivo. Very recently, chromatin association of activated signaling kinases has been described in yeast for several structurally and functionally unrelated kinases, including Hog1 MAPK (Alepuz et al, 2001; Proft and Struhl, 2002; Proft et al, 2006), Snf1 (Lo et al, 2005), Tor1 (Li et al, 2006) or other MAP kinases and PKA subunits (Pokholok et al, 2006). These results indicate that kinases devoted to regulate transcription are much more intimately involved in gene expression than previously thought. Here, we describe that, in the case of osmostress, the transcriptional output depends on the interdependent association of two different kinases, Sch9 and Hog1, with activated loci on the chromosome. Both kinases are recruited by the Sko1 transcription factor to osmolarity-regulated promoters. Although the mechanism of the mutual interdependence of both signaling molecules is unknown, the stable interaction of both kinases with DNA-bound Sko1 might depend on the function of each kinase through the non-overlapping phosphorylation events on Sko1. In a more indirect model, Sch9 and Hog1 could modulate each other's activity. However, we were not able to detect an interaction between both molecules in vivo, or phosphorylation of the two kinases in vitro (data not shown). It is important to note that, although Hog1 recruitment to the GRE2 or CTT1 promoters is greatly affected in sch9 mutants, a considerable residual induction can still be observed (especially for GRE2). This might reflect the fact that Hog1 recruitment at promoters has a different impact on transcription, depending on the inducible locus.
Here, we show that recruitment of Sch9 to a promoter is sufficient to activate gene expression. When artificially tethered to the GAL1 promoter by fusion with the Gal4 DNA binding domain, Sch9 constitutively activated transcription (Figure 6). On the other hand, our ChIP experiments demonstrate that Sch9 interacts physically with target genes in vivo, exclusively after osmotic shock and dependent on Hog1. We therefore believe that regulated association of the kinase is the essential step during osmostress stimulated gene expression, which can be bypassed by artificial recruitment. Direct transcriptional activation in monohybrid assays has been reported for several stress-regulated kinases, like Hog1 or Snf1 (Kuchin et al, 2000; Alepuz et al, 2003), and Sch9 has been identified among the most potent transcriptional activators in an unbiased genomic screen using Gal4DBD-fusions (Titz et al, 2006). However, in contrast to Hog1 or Snf1, which retained most of their inducibility by environmental stress, even when constitutively bound to promoters, Sch9 behaved as a constitutive activator irrespective of the growth conditions. Selective recruitment of Sch9 to stress promoters might therefore be the main regulatory step upon osmostress. Once physically present at regulated genes, the precise function of Sch9 in transcriptional activation remains to be determined. Although direct functions for signaling kinases at the chromatin of activated genes have been generally postulated based on their regulated recruitment, little is known about the molecular mechanisms employed to stimulate transcription (Edmunds and Mahadevan, 2006). It can be expected that those functions are complex and not uniform among the kinases, because no uniform pattern of recruitment has been observed. While some kinases (Tpk2, Tor1) exclusively occupy promoter regions, others (Tpk1) associate with ORF regions, while MAP kinases seem to generally target both promoters and the whole transcribed region (Li et al, 2006; Pokholok et al, 2006; Proft et al, 2006). Here, we show that Sch9 crosslinks preferentially to promoter regions. However, the kinase is also detectable at the 5′ region within the ORF regions. Insights into the possible functions of chromatin-recruited signaling kinases come from studies on the Hog1 and Snf1 molecules. Hog1 can indirectly or directly recruit chromatin modifying complexes like the SAGA histone acetylase, the SWI/SNF chromatin remodeling complex (Proft and Struhl, 2002), or the histone deacetylase Rpd3 (DeNadal et al, 2004) to osmostress-activated promoters. Additionally, Hog1 and Snf1 seem to recruit the RNA pol II transcription machinery directly (Kuchin et al, 2000; Alepuz et al, 2003). Hog1 also stably associates with the RNA pol II elongation complex, which indicates a possible function of the MAPK in transcription elongation during stress (Proft et al, 2006). Also direct modification of histones has been implied as a mechanism of activation for the chromatin-associated Snf1 kinase (Lo et al, 2001, 2005). While the molecular function of Sch9 at chromosomal loci is yet to be determined, we speculate that the catalytic activity of Sch9 is essential, because a kinase dead mutant for Sch9 is osmosensitive. Furthermore, mechanisms like the phosphorylation of the Sko1 transcription factor described here might favor transcriptional initiation directly or indirectly through the modulation of the promoter chromatin.
In mammalian cells, stress-activated p38 MAPK modulates transcription through another kinase pair, Msk1/2. Msk1 is responsible for the stress-induced phosphorylation of histone H3 at serines 10 and 28 at p38-dependent promoters (Davie, 2003; Soloaga et al, 2003). The dual control of transcription by a MAPK and a downstream kinase directly at the chromatin of the regulated gene is similar to what we report here in the yeast model. It is important to note that Sch9, in addition to the previously reported homology to mammalian Akt/PKB, shares structural similarity to mammalian Msk1 (31% identity, 47% homology) and Msk2 (32% identity, 49% homology). It is therefore likely that yeast Sch9 might recapitulate the functions of different mammalian kinases. Here, we show that both kinases, Hog1 MAPK and Sch9, activate osmostress-regulated gene expression at the chromatin structure of defense genes. Our results demonstrate previously unanticipated complexity of transcriptional activation upon stress by multiple inputs from chromatin-recruited signaling kinases.
Materials and methods
Yeast strains and plasmids
The following yeast strains were used in this study: BY4741 wild type (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0), BY4741 with Δypk1∷KAN, Δypk2∷KAN, or Δhog1∷KAN (EUROSCARF strain collection), BY4741 with Δsch9∷loxP-KAN-loxP (this study), W303-1A wild type (MATa; ura3; leu2; trp1; his3; ade2), MAP32 (W303-1A with Δhog1∷loxP-KAN-loxP), MAP91 (W303-1A with Δsch9∷loxP-KAN-loxP) (this study), MAP84 (W303-1A with Δhog1∷TRP1; Δsch9∷loxP-KAN-loxP) (this study), MAP85 (W303-1A with SCH9-3HA∷loxP-KAN-loxP) (this study), MAP51 (W303-1A with Δura3∷loxP; 3HA-HOG1), MAP90 (W303-1A with Δhog1∷TRP1; SCH9-3HA∷loxP-KAN-loxP) (this study), MAP93 (W303-1A with Δsko1∷loxP; SCH9-3HA∷loxP-KAN-loxP) (this study), MAP83 (W303-1A with Δura3∷loxP; 3HA-HOG1, Δsch9∷loxP-KAN-loxP) (this study), YULH (MATa, ura3, trp1, lys2, his3, leu2, gal4Δ, gal80Δ, GAL1-URA3, GAL1-lacZ). The chromosomal SCH9 locus was disrupted as described (Güldener et al, 1996). C-terminal fusions of the Sch9 protein with a triple HA epitope were obtained by chromosomal tagging of the SCH9 gene (De Antoni and Gallwitz, 2000). To assay for Sko1- and Hog1-dependent gene expression upon osmotic stress, we used the 2xCREENA1-lacZ reporter fusion pMP253 (2 μ, TRP1, CYC1prom-(2xCREENA1)-lacZ). Highly osmostress-regulated expression of this construct depends completely on the Sko1 and Hog1 functions (Proft et al, 2001). Full-length Hog1-GST and Sko1-GST fusion proteins were expressed from pGEX-4T bacterial expression vectors (Proft et al, 2001). Point mutated versions of Sko1-GST were: Sko1(E) with S108, T113, S126 triple amino-acid substitution to A; Sko1(M) with S380, S393, S399 triple amino-acid substitution to A (Proft et al, 2001). For monohybrid studies, full-length Hog1-Gal4DBD or Sch9-Gal4DBD fusion proteins were expressed from the ADH1 promoter in plasmid pOBD2 (CEN4, TRP1, ADH1 promoter) (kind gift from Peter Uetz). Complementation experiments were carried out with wild-type SCH9, 3xHA-SCH9 or 3xHA-SCH9(K441A) in pRS416 (CEN6, URA3) (kind gift from Kevin Morano).
Sensitivity assays
Growth of yeast strains was monitored on YPD agar plates containing or not the indicated concentrations of NaCl, KCl and LiCl. Serial dilutions of fresh overnight cultures were spotted directly onto the plates and growth was assayed after 2–5 days. For H2O2 resistance assays, exponentially growing (PDS) yeast cultures were diluted in 96-well plates in fresh YPD (YP) medium. Equal numbers of cells were then treated with different concentrations of H2O2, and spotted after the indicated times onto YPD plates to assay for surviving cells.
Northern blot analysis
Northern analysis was carried out as described previously (Proft et al, 2001).
β-Galactosidase assays
For analyzing LacZ expression driven by the Sko1–Hog1-regulated CRE element (pMP253 plasmid), we quantified β-galactosidase activity in yeast wild-type (W303-1A) and sch9Δ (MAP91) cells. Transcriptional activation capacity of Hog1-Gal4DBD and Sch9-Gal4DBD was assayed in yeast strain YULH. β-Galactosidase activity was measured as described previously (Gaxiola et al, 1992).
Coprecipitation and in vitro phosphorylation assays
MAP85 cells expressing 3HA-tagged Sch9 were grown in YPD and harvested before or after a brief osmotic shock (0.4 M NaCl). Total extracts (∼1 mg) were obtained by glass bead disruption in buffer A (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 15 mM EDTA, 2 mM DTT, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), supplemented with protease inhibitors (Complete Mini, Roche)). Samples were incubated with 10 μl of anti-HA monoclonal antibody (12CA5, Roche) and protein A Sepharose beads (Amersham), overnight at 4°C. Beads were washed extensively in buffer A and used for coprecipitation and/or in vitro kinase assays. Hog1-GST and all Sko1-GST fusion proteins were expressed from pGEX-4T expression vectors in bacterial strain DH5α. Bacteria were lysed in buffer B (50 mM Tris–HCl pH 8, 150 mM NaCl, 1 mM EDTA, 2 mM DTT, 1% Triton X-100, 1 mM PMSF, supplemented with protease inhibitors). Fusion proteins were purified by incubation with glutathione Sepharose beads (Amersham) at 4°C. Beads were thoroughly washed and GST fusion proteins were eluted in in vitro kinase buffer (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 2 mM DTT) supplemented with 10 mM glutathione. Protein quantity and quality were confirmed by SDS–PAGE and Coomassie staining, or anti-GST Western blot. For coprecipitation experiments, Sch9-HA was immunoprecipitated in the presence of equal amounts of affinity purified GST (negative control), Hog1-GST or Sko1-GST. Control precipitations were performed in the absence of the HA-antigen using extracts from untagged wild-type cells. The amount of copurified GST proteins was determined by Western blotting with a polyclonal anti-GST anibody (Z-5, Santa Cruz Biotechnology). For in vitro kinase assays, immunopurified Sch9-HA bound to protein A Sepharose beads was obtained in in vitro kinase buffer from MAP85 cells, before and after brief osmotic shock. As a substrate, affinity purified Hog1-GST or Sko1-GST proteins were added. Kinase reactions were incubated for 10 min at 30°C in the presence of [γ-32P]ATP (0.2 μCi/μl) and stopped by addition of SDS–PAGE loading buffer. Phosphorylated proteins were detected by autoradiography.
ChIP assays
ChIP was performed as described previously (Kuras and Struhl, 1999). Quantitative PCR analyses at the indicated chromosomal loci were performed in real-time using an Applied Biosystems 7000 sequence detector, using the POL1 (+1796/+1996) coding sequence as a negative control. Each immunoprecipitation was performed twice with different chromatin samples. All data are presented as fold IP efficiency over the POL1 control sequence.
Acknowledgments
We thank Peter Uetz and Takashi Ito for the generous gift of yeast strain YULH and Sch9-Gal4DBD and Hog1-Gal4DBD expression plasmids, Kevin Morano and Joris Winderickx for SCH9 expression plasmids, and Lynne Yenush for critical reading of the manuscript. This work was supported by the ‘Ramón y Cajal' program and grants from Ministerio de Educación y Ciencia (BFU2005-01714, partially founded by FEDER) and Generalitat de Valencia (GV06/041) to MP.
References
- Alepuz PM, de Nadal E, Zapater M, Ammerer G, Posas F (2003) Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA pol II. EMBO J 22: 2433–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alepuz PM, Jovanovic A, Reiser V, Ammerer G (2001) Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell 7: 767–777 [DOI] [PubMed] [Google Scholar]
- Arthur JS, Fong AL, Dwyer JM, Davare M, Reese E, Obrietan K, Impey S (2004) Mitogen- and stress-activated protein kinase 1 mediates cAMP response element-binding protein phosphorylation and activation by neurotrophins. J Neurosci 24: 4324–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsland E, Molin C, Swaminathan S, Ramne A, Sunnerhagen P (2004) Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol Microbiol 53: 1743–1756 [DOI] [PubMed] [Google Scholar]
- Carlson M (1999) Glucose repression in yeast. Curr Opin Microbiol 2: 202–207 [DOI] [PubMed] [Google Scholar]
- Chow CW, Davis RJ (2006) Protein kinases: chromatin-associated enzymes? Cell 127: 887–890 [DOI] [PubMed] [Google Scholar]
- Crauwels M, Donaton MC, Pernambuco MB, Winderickx J, de Winde JH, Thevelein JM (1997) The Sch9 protein kinase in the yeast Saccharomyces cerevisiae controls cAPK activity and is required for nitrogen activation of the fermentable-growth-medium-induced (FGM) pathway. Microbiology 143: 2627–2637 [DOI] [PubMed] [Google Scholar]
- Davie JR (2003) MSK1 and MSK2 mediate mitogen- and stress-induced phosphorylation of histone H3: a controversy resolved. Sci STKE 12: PE33. [DOI] [PubMed] [Google Scholar]
- De Antoni A, Gallwitz D (2000) A novel multi-purpose cassette for repeated integrative epitope tagging of genes in Saccharomyces cerevisiae. Gene 246: 179–185 [DOI] [PubMed] [Google Scholar]
- Deak M, Clifton AD, Lucocq LM, Alessi DR (1998) Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J 17: 4426–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNadal E, Alepuz PM, Posas F (2002) Dealing with osmostress through MAP kinase activation. EMBO Rep 3: 735–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNadal E, Casadome L, Posas F (2003) Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol Cell Biol 23: 229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNadal E, Zapater M, Alepuz PM, Sumoy L, Mas G, Posas F (2004) The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427: 370–374 [DOI] [PubMed] [Google Scholar]
- Denis CL, Audino DC (1991) The CCR1 (SNF1) and SCH9 protein kinases act independently of cAMP-dependent protein kinase and the transcriptional activator ADR1 in controlling yeast ADH2 expression. Mol Gen Genet 229: 395–399 [DOI] [PubMed] [Google Scholar]
- Dunn KL, Espino PS, Drobic B, He S, Davie JR (2005) The Ras-MAPK signal transduction pathway, cancer and chromatin remodeling. Biochem Cell Biol 83: 1–14 [DOI] [PubMed] [Google Scholar]
- Dyson MH, Thomson S, Inagaki M, Goto H, Arthur SJ, Nightingale K, Iborra FJ, Mahadevan LC (2005) MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J Cell Sci 118: 2247–2259 [DOI] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC (2004) MAP kinases as structural adaptors and enzymatic activators in transcription complexes. J Cell Sci 117: 3715–3723 [DOI] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC (2006) Cell signaling. Protein kinases seek close encounters with active genes. Science 313: 449–451 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pletcher SD, Minois N, Vaupel JW, Longo VD (2004) Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett 557: 136–142 [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD (2001) Regulation of longevity and stress resistance by Sch9 in yeast. Science 292: 288–290 [DOI] [PubMed] [Google Scholar]
- Gaxiola R, de Larriñoa IF, Villalba JM, Serrano R (1992) A novel and conserved salt-induced protein is an important determinant of salt tolerance in yeast. EMBO J 11: 3157–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24: 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin MC, Albertyn J, Alexander M, Davenport K (1998) MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62: 1264–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66: 300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M (2004) A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 18: 2491–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW III, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK (2005) Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196 [DOI] [PubMed] [Google Scholar]
- Kuchin S, Treich I, Carlson M (2000) A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc Natl Acad Sci USA 97: 7916–7920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Struhl K (1999) Binding of TBP to promoters in vivo is stimulated by activators and requires PolII holoenzyme. Nature 399: 609–612 [DOI] [PubMed] [Google Scholar]
- Li H, Tsang CK, Watkins M, Bertram PG, Zheng XF (2006) Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 442: 1058–1061 [DOI] [PubMed] [Google Scholar]
- Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhatter R, Berger SL (2001) Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293: 1142–1146 [DOI] [PubMed] [Google Scholar]
- Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL (2005) Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J 24: 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DE, Hall MN (2005) The expanding TOR signaling network. Curr Opin Cell Biol 17: 158–166 [DOI] [PubMed] [Google Scholar]
- Morano KA, Thiele DJ (1999) The Sch9 protein kinase regulates Hsp90 chaperone complex signal transduction activity in vivo. EMBO J 18: 5953–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Ahuir A, Posas F, Serrano R, Proft M (2001) Multiple levels of control regulate the yeast cAMP-response element-binding protein repressor Sko1p in response to stress. J Biol Chem 276: 37373–37378 [DOI] [PubMed] [Google Scholar]
- Pascual-Ahuir A, Posas F, Serrano R, Proft M (2006) Genome-wide location analysis of the stress-activated MAP kinase Hog1 in yeast. Methods 40: 272–278 [DOI] [PubMed] [Google Scholar]
- Pedruzzi I, Burckert N, Egger P, De Virgilio C (2000) Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J 19: 2569–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, Winderickx J, De Virgilio C (2003) TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell 12: 1607–1613 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA (2006) Activated signal transduction kinases frequently occupy target genes. Science 313: 533–536 [DOI] [PubMed] [Google Scholar]
- Posas F, Chambers JR, Heyman JA, Hoeffler JP, deNadal E, Arino J (2000) The transcriptional response of yeast to saline stress. J Biol Chem 275: 17249–17255 [DOI] [PubMed] [Google Scholar]
- Proft M, Serrano S (1999) Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol Cell Biol 19: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Struhl K (2002) Hog1 kinase converts the Sko1–Cyc8–Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9: 1307–1317 [DOI] [PubMed] [Google Scholar]
- Proft M, Gibbons FD, Copeland M, Roth FP, Struhl K (2005) Genomewide identification of Sko1 target promoters reveals a regulatory network that operates in response to osmotic stress in Saccharomyces cerevisiae. Eukaryot Cell 4: 1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, Posas F (2006) The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell 23: 241–250 [DOI] [PubMed] [Google Scholar]
- Proft M, Pascual-Ahuir A, de Nadal E, Ariño J, Serrano R, Posas F (2001) Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J 20: 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Krantz M, Thevelein JM, Hohmann S (2000) The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem 275: 8290–8300 [DOI] [PubMed] [Google Scholar]
- Rep M, Proft M, Remize F, Tamas M, Serrano R, Thevelein JM, Hohmann S (2001) The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol 40: 1067–1083 [DOI] [PubMed] [Google Scholar]
- Roosen J, Engelen K, Marchal K, Mathys J, Griffioen G, Cameroni E, Thevelein JM, DeVirgilio C, De Moor B, Winderickx J (2005) PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol Microbiol 55: 862–880 [DOI] [PubMed] [Google Scholar]
- Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS (2003) MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J 22: 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH (1999) Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol 33: 904–918 [DOI] [PubMed] [Google Scholar]
- Titz B, Thomas S, Rajagopala SV, Chiba T, Ito T, Uetz P (2006) Transcriptional activators in yeast. Nucleic Acids Res 34: 955–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Wigler M (1988) SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev 2: 517–527 [DOI] [PubMed] [Google Scholar]
- Wiggin GR, Soloaga A, Foster JM, Murray-Tait V, Cohen P, Arthur JS (2002) MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol Cell Biol 22: 2871–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Zhang Y, Bode AM, Dong Z (2004) Involvement of ERKs and mitogen- and stress-activated protein kinase in UVC-induced phosphorylation of ATF2 in JB6 cells. Carcinogenesis 25: 1847–1852 [DOI] [PubMed] [Google Scholar]


