Abstract
Tomato (Solanum lycopersicum) plants with the Cf-4 resistance gene recognize strains of the pathogenic fungus Cladosporium fulvum that secrete the avirulence protein Avr4. Transgenic tomato seedlings coexpressing Cf-4 and Avr4 mount a hypersensitive response (HR) at 20°C, which is suppressed at 33°C. Within 120 min after a shift from 33°C to 20°C, tomato mitogen-activated protein (MAP) kinase (LeMPK) activity increases in Cf-4/Avr4 seedlings. Searching tomato genome databases revealed at least 16 LeMPK sequences, including the sequence of LeMPK1, LeMPK2, and LeMPK3 that cluster with biotic stress-related MAP kinase orthologs from Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum). LeMPK1, LeMPK2, and LeMPK3 are simultaneously activated in Cf-4/Avr4 seedlings, and, to reveal whether they are functionally redundant or not, recombinant LeMPKs were incubated on PepChip Kinomics slides carrying peptides with potential phosphorylation sites. Phosphorylated peptides and motifs present in them discriminated between the phosphorylation specificities of LeMPK1, LeMPK2, and LeMPK3. LeMPK1, LeMPK2, or LeMPK3 activity was specifically suppressed in Cf-4-tomato by virus-induced gene silencing and leaflets were either injected with Avr4 or challenged with C. fulvum-secreting Avr4. The results of these experiments suggested that the LeMPKs have different but also overlapping roles with regard to HR and full resistance in tomato.
Plants are able to prevent or stop colonization by pathogens via highly effective defense systems. The constant battle between plants and pathogens can be described by a so-called zigzag model and consists of several layers of resistance responses of the plant, which are suppressed by the pathogen (Jones and Dangl, 2006). The primary resistance response is based on the recognition of common features of pathogens, also referred to as pathogen-associated molecular pattern-triggered innate immunity. Successful pathogens suppress this primary immune response with specific effectors. In turn, plants have developed resistance (R) proteins that mediate recognition of the pathogen via the secreted effector proteins, rendering them avirulence factors. As a result of this coevolution, resistance/avirulence-based host-pathogen interactions have evolved that follow the gene-for-gene model (Flor, 1942). When an R protein and its cognate avirulence (Avr) protein are present, a swift resistance response is initiated, which consists of localized cell death, the so-called hypersensitive response (HR), and associated defense responses (Chisholm et al., 2006; Jones and Dangl, 2006). The immediate response of the plant relies on rapid posttranslational modifications that alter the function of signaling proteins by changing their activity and/or localization.
Along this line, mitogen-activated protein (MAP) kinase (MAPK) cascades, some of the major signaling modules in eukaryotes, are rapidly activated by posttranslational modification upon recognition of pathogens by resistant plants (Pedley and Martin, 2005). MAPK cascades transfer signals from upstream receptors to downstream cellular effectors and rapid MAPK activation allows instantaneous modification of downstream signaling proteins (Krens et al., 2006; Zhang et al., 2006). In plants, these cascades have been implicated in typical defense responses, such as production of pathogenesis-related proteins, reactive oxygen species (ROS), ethylene, and cell death (Pedley and Martin, 2005). The phospho-relay system is based on specific activation of three types of kinases; MAPKK kinases (MAPKKKs), MAPK kinases (MAPKKs or MKKs), and MAPKs (also referred to as MPKs). Perception of external stimuli leads to MAPKKK activation, which subsequently phosphorylates the [Ser/Thr]-x(3,5)-[Ser/Thr] motif present in the target MAPKKs, thereby activating them. In their turn, MAPKKs phosphorylate the Thr (T) and Tyr (Y) residues in the TxY motif of the target MAPKs, which then become active and can phosphorylate downstream proteins that initiate the cellular response (Pedley and Martin, 2005). Plants trigger MAPK cascades upon biotic stress, but also when challenged by abiotic stresses, such as wounding, drought, ozone, and UV light (Nakagami et al., 2005; Mishra et al., 2006). Thus, signals from diverse stresses eventually converge into various overlapping, but also distinct, MAPK cascades (Zhang et al., 2006), which is reflected by the presence of, for example, 20 MAPK-, 10 MAPKK-, and 60 putative MAPKKK-encoding genes in Arabidopsis (Arabidopsis thaliana; Ichimura et al., 2002).
Upstream signaling components that activate MAPK cascades remain largely unknown, although ROS, auxin and abscisic acid, and phosphatidic acid have been reported to be involved (Lee et al., 2001; Mishra et al., 2006). Signaling events downstream of activated MAPK cascades also remain a black box because hardly any MAPK substrates have been identified. So far, 1-aminocyclopropane-1-carboxylic acid synthase (ACS), the rate-limiting enzyme in ethylene biosynthesis (Liu and Zhang, 2004), and MKS1, which is required for full salicylic acid-dependent resistance (Andreasson et al., 2005), have been reported to be phosphorylated by MAPKs in Arabidopsis. Furthermore, plant-specific WRKY transcription factors that contain the WRKYGQK core sequence followed by a zinc-finger motif are phosphorylated by MAPKs (Menke et al., 2005). Many putative MAPK substrates were identified by employing a high-throughput proteomic screen in Arabidopsis (Feilner et al., 2005).
In tomato (Solanum lycopersicum; former name Lycopersicon esculentum [Le]), several components of MAPK signaling cascades have been identified. The tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 are activated upon stress responses caused by the wound-signaling peptide systemin, oligosaccharide elicitors, UV-B radiation, and the fungal toxin fusicoccin (Holley et al., 2003; Higgins et al., 2006). Furthermore, LeMPK2 and LeMPK3 are activated in a Pto-specific manner upon expression of AvrPto and AvrPtoB, and upon expression of LeMAPKKKα (Pedley and Martin, 2004). The authors also identified four MAPKKs, of which LeMKK2 and LeMKK4 activate LeMPK2 and LeMPK3 in vivo. In vitro experiments revealed that both LeMKKs are able to phosphorylate LeMPK1, LeMPK2, and LeMPK3. In addition to its activation by phosphorylation, LeMPK3 mRNAs are specifically induced in resistant tomato upon inoculation with bacterial strains Xanthomonas campestris pv vesicatoria and Pseudomonas syringae pv tomato, and upon treatment with a fungal ethylene-inducing xylanase (Mayrose et al., 2004). Finally, virus-induced gene silencing (VIGS) of both LeMPK1 and LeMPK2, LeMPK3, or LeMKK2 revealed a role for these kinases in Mi-1-mediated aphid resistance (Li et al., 2006).
We study the resistance response of tomato to the fungal pathogen Cladosporium fulvum. Several C. fulvum (Cf) R genes of tomato and their cognate Avrs from C. fulvum have been identified (Thomma et al., 2005), including the gene pairs Cf-4/Avr4 and Cf-9/Avr9 (Rivas and Thomas, 2005). To study specific activation of kinases in typical defense responses leading to cell death, tobacco (Nicotiana tabacum) cell suspensions expressing Cf genes were elicited with its cognate Avr protein. In accordance with these studies, Cf-9-expressing cells were reported to activate calcium-dependent protein kinases and the MAPKs salicylic acid-induced protein kinase and wound-induced protein kinase (SIPK and WIPK, respectively; Romeis et al., 1999, 2000). Activation of the latter two kinases was confirmed in Cf-9 transgenic tobacco leaves. Furthermore, LeMAPKKKα has been shown to be a positive regulator of the Cf-9-mediated HR and overexpression of the encoding gene causes MAPK activation and cell death (Del Pozo et al., 2004). Interestingly, Cf-9-expressing cells also showed an increase in MAPK activity after treatment with Avr9 (De Jong et al., 2000).
To study HR-related signaling processes in intact tomato plants, transgenic tomato lines lacking a functional Cf gene (‘Money Maker’-Cf-0) and expressing Avr4 were crossed to ‘Money Maker’-Cf-4 tomato, resulting in Cf-4/Avr4 offspring that displays lethality at the seedling stage (Thomas et al., 1997; Cai et al., 2001). Because specific Avr perception appeared to be temperature sensitive, Cf-4/Avr4 seedlings can be rescued upon incubation at 33°C (De Jong et al., 2002). When the seedlings are subsequently transferred to 20°C, a synchronous systemic HR-related cell death program is initiated and this biological system has successfully been employed to study early transcriptional changes by cDNA-AFLP analysis (Gabriëls et al., 2006). This study revealed that, in these plants, the typical defense-related genes are up-regulated and novel genes were identified that play a role in plant defense.
Here, we report on the specific Cf-4/Avr4-mediated activation of the tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 upon initiation of the HR. Interestingly, analysis of the phosphorylation specificity of these LeMPKs using PepChip Kinomics slides (hereafter referred to as PepChips) revealed overlapping, but also different, phosphorylation preferences for each kinase, indicating different downstream roles for the LeMPKs. VIGS of the genes encoding the individual kinases suggested that LeMPK1, LeMPK2, and LeMPK3 play different but overlapping roles in the establishment of the HR and resistance of tomato to C. fulvum.
RESULTS
Kinase Activation upon Cf-4/Avr4-Induced Defense Signaling
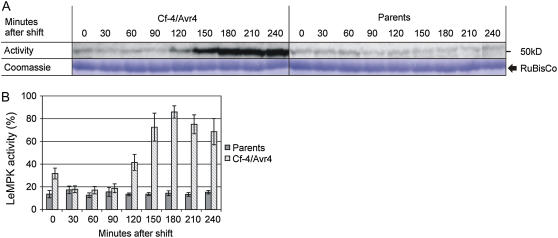
Tomato seedlings expressing both Cf-4 and Avr4 develop systemic necrosis at 20°C, but can be rescued from lethality at 33°C. When incubated at 33°C and subsequently transferred to 20°C, defense signaling leading to systemic HR is induced in Cf-4/Avr4 seedlings, but not in seedlings from the parental lines (De Jong et al., 2002). A video covering a 5-d period shows seedlings from the moment of the temperature shift until the Cf-4/Avr4 seedlings had become completely necrotic (Supplemental Movie S1). To study the activation of kinases during Cf-4/Avr4-induced defense signaling, the HR was induced in 1-week-old Cf-4/Avr4 seedlings and seedlings from the parents by a temperature shift to 20°C. Cotyledons were sampled at intervals of 30 min, starting from the actual temperature shift, and were analyzed for kinase activity using myelin basic protein (MBP) as a substrate in in-gel kinase assays. MBP kinase activity in the parents, representing a mixture of the Cf-4 and transgenic Avr4-expressing lines, remained at basal levels throughout the experiment (Fig. 1A). However, in Cf-4/Avr4 seedlings, MBP kinase activity had significantly increased at 120 min after the temperature shift, reached its maximum at 180 min, and subsequently stabilized over the next 60 min (Fig. 1A). The activity was quantified and expressed as a percentage of the maximal activity per experiment (Fig. 1B). Statistical analysis revealed that the MBP kinase activity in Cf-4/Avr4 seedlings is significantly increased compared to the parental lines at t = 0 and from 120 min onward (P ≤ 0.05). MBP kinase activity was present in a band with a molecular mass of about 50 kD, suggesting that this is a reflection of MAPK activity. Samples taken from primary leaves of older Cf-4/Avr4 seedlings that were subjected to the same treatment showed a similar MBP kinase activation pattern (data not shown), indicating that the response in cotyledons is representative of the response in true leaves.
Figure 1.
MBP kinase activity upon initiation of Cf-4/Avr4-induced defense signaling. Cf-4/Avr4 tomato seedlings were grown at 33°C, after which defense signaling was induced by a shift to 20°C. Seedlings were subsequently sampled at intervals of 30 min over a period of 4 h (sample at t = 0 represents the harvest just before the temperature shift). Cf-4- and transgenic Avr4-expressing seedlings were combined (parents) and treated similarly. A, Protein extracts from the seedlings were analyzed using in-gel kinase assays with MBP as a substrate. In each lane, similar amounts of protein were loaded as shown by Coomassie staining of the Rubisco large subunit. B, MBP kinase activity represented by the 50-kD band was quantified by phosphor imaging and expressed as the percentage of the maximal activity determined per experiment, which was set to 100%. For each time point, the average activity (bars) of five independent experiments is presented and ses of mean (sem) are shown by error bars. [See online article for color version of this figure.]
LeMPK Family Analysis
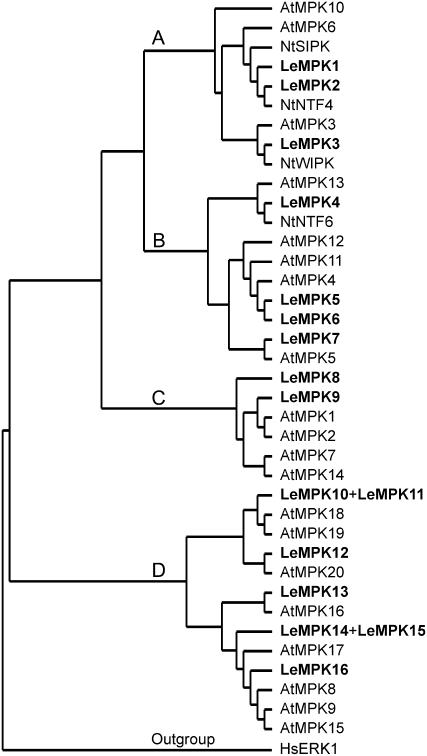
In most cases, MBP kinase activity reflects MAPK activity. For Arabidopsis, 20 AtMPKs were described to cluster in groups A to D (Ichimura et al., 2002), and MAPKs that cluster in group A have been described to be positive regulators of defense signaling (Mishra et al., 2006). Some members of group B are negative regulators of defense signaling, whereas only in rice (Oryza sativa) two MAPKs of group D were activated by pathogens (Song et al., 2006; Zhang et al., 2006). Because information on the tomato MAPK gene family is limited, the size of the LeMPK family and the variation between the family members was studied to identify potential additional homologs of the already described LeMPK1, LeMPK2, and LeMPK3 (Holley et al., 2003) that cluster in group A. The open reading frames (ORFs) of these LeMPKs were used in BLAST queries on The Institute for Genomic Research (TIGR), the National Center for Biotechnology Information (NCBI), and the SOL Genomics Network (SGN) databases and additional LeMPKs were identified, which, to identify all homologs, were in turn also used in BLAST queries on the same databases. Thirteen additional sequences that putatively encode a LeMPK were identified (Table I). The LeMPK sequences were translated into protein sequences and the ORFs were aligned with those of LeMPK1, LeMPK2, and LeMPK3, the ORFs from AtMPK1 to AtMPK20, the tobacco ORFs from NtWIPK, NtSIPK, NtNTF4, and NtNTF6, and the ORF of Homo sapiens HsERK1 that was assigned as an outgroup. As presented in Figure 2, the 13 additional LeMPKs cluster over groups B to D. LeMPKs present in groups A, B, and C all have a Thr-Glu-Tyr (TEY) activation domain, whereas those of group D have a Thr-Asp-Tyr (TDY) activation domain, except for two incomplete sequences lacking this part of the sequence. LeMPK6 might represent a nonfunctional homolog because the Met-Glu-Tyr (MEY) sequence that replaces the TEY domain in this MAPK probably renders the protein inactive. Because the tomato genome has not been fully sequenced yet, additional LeMPKs might be found in the near future. However, it is unlikely that large numbers of novel LeMPKs will be identified because all groups presented in the cladogram contain LeMPK sequences. Furthermore, other higher plant species of which the genome has been fully sequenced, such as poplar (Populus trichocarpa) and rice, contain comparable numbers of MAPKs (21 and 15, respectively; Hamel et al., 2006). The results mentioned above suggest that group A is complete, and, therefore, to identify the MAPKs responsible for the MBP phosphorylation shown in Figure 1, we focused on LeMPK1, LeMPK2, and LeMPK3.
Table I.
Tomato MAPKs
The Unigene Identifier, type of activation domain (TxY), the number of residues of the protein, and the clustering in the groups presented in Figure 2 are indicated for the 16 LeMPKs.
| Name | Unigene Identifier | TxY | Residues | Group |
|---|---|---|---|---|
| LeMPK1 | SGN-U316697 | TEY | 397 | A |
| LeMPK2 | SGN-U316695 | TEY | 395 | A |
| LeMPK3 | SGN-U313928 | TEY | 374 | A |
| LeMPK4 | SGN-U323634 | TEY | 373 | B |
| LeMPK5 | SGN-U313996 | TEY | 281 | B |
| LeMPK6 | SGN-U313995 | MEY | 377 | B |
| LeMPK7 | SGN-U323219 | TEY | 380 | B |
| LeMPK8 | SGN-U318773 | TEY | 371 | C |
| LeMPK9 | SGN-U316113 | TEY | 373 | C |
| LeMPK10 | SGN-U317229 | TDY | 576 | D |
| LeMPK11a | SGN-U322516 and TC168576 | – | 395 | D |
| LeMPK12 | SGN-U318438 | TDY | 622 | D |
| LeMPK13 | SGN-U316366 and SGN-316367 | TDY | 596 | D |
| LeMPK14 | SGN-U318361 | TDY | 496 | D |
| LeMPK15a | SGN-U332259 | – | 207 | D |
| LeMPK16 | SGN-U318101 | TDY | 576 | D |
These sequences lack the N-terminal region and therefore do not contain the TDY domain.
Figure 2.
Relationships among the MAPKs of tomato (Le), Arabidopsis (At), and Nicotiana tabacum (Nt). Thirteen sequences homologous to the ORF of LeMPK1, LeMPK2, or LeMPK3 were obtained from the tomato TIGR, NCBI, and SGN databases and translated. All 16 LeMPK protein sequences were aligned with the known AtMPK and NtMPK sequences and a cladogram showing four distinct groups was generated in which LeMPK4 to LeMPK16 are numbered from top to bottom according to their position in the cladogram. LeMPK11 and LeMPK13 each represent a fusion of two database entries with an identical overlapping part. LeMPK11 and LeMPK15 are positioned manually next to their closest homolog because their sequence is not complete, and, in this way, misclustering in the ClustalX alignment is avoided.
Cf-4/Avr4-Induced Defense Signaling Causes Activation of LeMPK1, LeMPK2, and LeMPK3
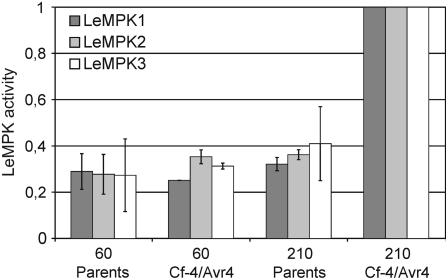
To identify which LeMPK is activated upon Cf-4/Avr4-induced defense signaling, LeMPK1, LeMPK2, and LeMPK3 were immunoprecipitated from protein extracts of Cf-4/Avr4 and the parents, of which the MBP kinase activity is shown in Figure 1, using antiserum raised against either LeMPK1, LeMPK2, or LeMPK3 (Holley et al., 2003). Subsequently, the precipitated kinases were incubated with MBP to reveal whether they had been activated in the responding plants. Interestingly, the activity of all three LeMPKs had increased upon the initiation of Cf-4/Avr4-induced defense signaling when compared to the LeMPK activity in the parents (Fig. 3). Although the MAPK protein levels were unaltered (data not shown), LeMPK3 transcription was significantly up-regulated at 180 min after the temperature shift in the Cf-4/Avr4 seedlings (I.J.E. Stulemeijer and M.H.A.J. Joosten, unpublished data). This observation matches with the earlier described transcriptional regulation of LeMPK3 upon recognition of a bacterial avirulence factor (Mayrose et al., 2004). Furthermore, the transcript levels of LeMPK1 and LeMPK2 were not altered.
Figure 3.
Initiation of Cf-4/Avr4-induced defense signaling causes activation of LeMPK1, LeMPK2, and LeMPK3. Cf-4/Avr4 tomato seedlings were grown at 33°C, after which defense signaling was induced by a temperature shift to 20°C. Seedlings were subsequently sampled at 60 and 210 min after the temperature shift. ‘Money Maker’-Cf-4 and transgenic Avr4-expressing seedlings were combined (parents) and treated similarly. LeMPK1, LeMPK2, or LeMPK3 was immunoprecipitated from total protein extracts using specific antisera and incubated with MBP and radiolabeled ATP and protein bands were quantified. Bars represent the average activity of two independent experiments and ses of mean (sem) are shown by error bars. LeMPK1, LeMPK2, and LeMPK3 activities in the Cf-4/Avr4 seedlings after 210 min were set to 1 and the remaining bars were related to these. Note that the bars do not represent absolute levels of LeMPK activity.
LeMPK1, LeMPK2, and LeMPK3 Have Overlapping and Distinct Specificities Based on PepChip Analysis
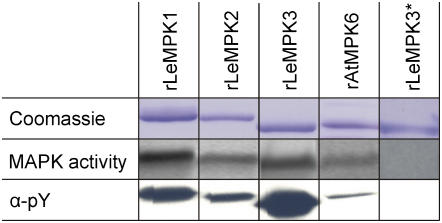
The experiments described above show that LeMPK1, LeMPK2, and LeMPK3 are specifically activated after triggering Cf-4/Avr4-induced defense signaling. To investigate whether the different LeMPKs have overlapping and/or distinct phosphorylation specificities, recombinant (r) LeMPKs were produced in Escherichia coli and their peptide phosphorylation specificity was tested. We included rAtMPK6 as a control in this assay because, for this MAPK, a substrate has been identified (Liu and Zhang, 2004), and AtMPK6 clusters with LeMPK1, LeMPK2, and LeMPK3 in group A (Fig. 2). The four rMAPKs were expressed as His-tagged proteins, purified and visualized by Coomassie staining, and kinase assays were performed to confirm basal MAPK activity (Fig. 4, top and middle). MAPKs produced in E. coli autophosphorylate the Tyr residue in the TEY motif (Crews et al., 1991; Mayrose et al., 2004), thereby gaining activity that is probably controlled by specific MAPK phosphatases in vivo. Monoclonal anti-phospho-Tyr (α-pY) antiserum recognizes the rMAPKs, indicating that the rMAPKs are indeed phosphorylated on a Tyr residue (Fig. 4, bottom). To show that loss of activity is coupled to a loss of phosphorylation, rLeMPK3 was stored in a solution without kinase storage buffer, which results in an inactive enzyme (rLeMPK3*; Fig. 4). Probing with the α-pY antiserum revealed that this inactive rMAPK is indeed no longer phosphorylated (Fig. 4). Thus, MAPKs that are produced in E. coli have basal activity. This conclusion is supported by the observations of Feilner et al. (2005), who produced AtMPK3 and AtMPK6 in E. coli and used the active MAPKs to perform protein microarray-based kinase assays.
Figure 4.
rMAPKs of group A have basal kinase activity and are phosphorylated on a Tyr residue. rMAPKs stored in kinase storage buffer (rLeMPKs and rAtMPK6; see “Materials and Methods” for details) were run on SDS-PAGE gels and stained with Coomassie Brilliant Blue. Basal kinase activity was determined by incubation of the rMAPKs with MBP and radiolabeled ATP (middle panel) and a blot carrying the rMAPK proteins was incubated with antiserum specific for pY. rLeMPK3* represents an inactive form of the LeMPK3 enzyme obtained by storage in a solution without kinase storage buffer. [See online article for color version of this figure.]
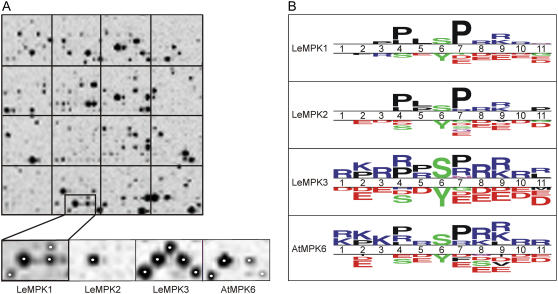
To investigate peptide phosphorylation specificities of the individual MAPKs, PepChips were incubated with the rMAPKs showing basal activity. PepChips carry a triplicate set of 976 peptides containing experimentally verified phosphorylation sites for different types of human kinases (see “Materials and Methods” for details). Each of the rMAPKs phosphorylated an overlapping, but also partially different, subset of the peptides (Fig. 5A). In the magnified region of the slide (Fig. 5A, small images), only one of the spotted peptides is phosphorylated by all three rLeMPKs and rAtMPK6, whereas two peptides are phosphorylated by both rLeMPK1 and rLeMPK3, as well as by rAtMPK6. Additionally, three peptides spotted in this area were only phosphorylated by rLeMPK1 or rLeMPK3. The peptide phosphorylation specificity of rAtMPK6 is most similar to that of rLeMPK1, although it has similarities with rLeMPK3 as well.
Figure 5.
PepChips reveal different phosphorylation patterns for LeMPK1, LeMPK2, and LeMPK3. PepChips were incubated with rMAPKs in the presence of radiolabeled ATP. A, Peptides phosphorylated on the PepChips were visualized with phosphor imaging. The top image shows one of the triplicate sets of peptides phosphorylated by rLeMPK1, whereas the bottom images show a subset of peptides differentially phosphorylated by rLeMPK1, rLeMPK2, rLeMPK3, and rAtMPK6, respectively. Spots marked by a white dot are represented in the selected subset of phosphorylated peptides presented in B. B, Selected phosphorylated and nonphosphorylated peptides were compared with TSL software. Putative phosphorylation sites (Ser [S], Thr [T], or Tyr [Y]) are aligned on position 6, and above the double line the TSL plots show for positions 1 to 11 whether a particular amino acid residue has an increased frequency in the phosphorylated peptides compared to the same position in the nonphosphorylated peptides. For the latter, the most frequently occurring residues are depicted below the double line (t test, P < 0.05). The size of the symbols is proportional to the relative frequencies of the residues in the phosphorylated and nonphosphorylated peptides. The largest (stack of) symbols in the TSL plots for rLeMPK1, rLeMPK2, rLeMPK3, and rAtMPK6 have a frequency of 62%, 58%, 34%, and 34%, respectively. L, Leu; R, Arg; K, Lys; D, Asp; E, Glu; F, Phe; V, Val; P, Pro.
The phosphorylation intensity of most of the peptides did not exceed background levels and these peptides probably do not contain motifs that could be present in putative in vivo substrates. For further analysis, only peptides showing phosphorylation intensity above the average peptide phosphorylation intensity of a complete PepChip and peptides with a phosphorylation intensity of zero were selected and will be referred to as phosphorylated and nonphosphorylated peptides, respectively (Supplemental Fig. S1; see “Materials and Methods” for details). Comparison of the phosphorylated peptides for LeMPK1, LeMPK2, or LeMPK3 revealed that 30% of the peptides were phosphorylated by LeMPK1, LeMPK2, and LeMPK3, 19% by LeMPK1 and LeMPK2, 5% by LeMPK1 and LeMPK3, and 4% by LeMPK2 and LeMPK3. The remaining phosphorylated peptides were specifically phosphorylated by LeMPK1, LeMPK2, or LeMPK3 (see also Table II). To determine the specificity of the rMAPKs, the phosphorylated peptides were compared to the nonphosphorylated peptides with Two Sample Logo (TSL) software (Crooks et al., 2004). The peptides consist of 11 residues of which the central residue represents the putative phosphorylation site, which is either a Ser, Thr, or Tyr residue. The TSL plots show for the 11 positions whether a residue is more represented in the phosphorylated peptides as compared to the nonphosphorylated peptides (Fig. 5B; t test, P < 0.05). As expected for MAPKs, which are Ser/Thr-specific kinases (Nakagami et al., 2005), the rMAPKs prefer Ser instead of Tyr phosphorylation (position 6). In addition, rLeMPK1 and rLeMPK2 prefer to phosphorylate sequences containing Pro residues, whereas rLeMPK3 and rAtMPK6 have a preference for sequences containing the positively charged amino acid residues Arg and Lys, in addition to Pro residues. Sequences containing negatively charged residues, such as Asp and Glu, are hardly phosphorylated by any rMAPK (Fig. 5B), which might be caused by static hindrance of the negatively charged phosphate group at the phosphorylation site. These results demonstrate that phosphorylation of the peptides on the PepChips by the various rMAPKs is significantly influenced by the sequence of the peptides and that each of the rLeMPKs phosphorylates a different subset of peptides because clear differences are observed between the TSL plots.
Table II.
Phosphorylation motifs present in the peptides phosphorylated on the PepChips by rLeMPK1, rLeMPK2, rLeMPK3, or rAtMPK6
Phosphorylated peptides were divided into subgroups based on common characteristics and phosphorylation motifs were predicted with TEIRESIAS software (Rigoutsos and Floratos, 1998). Motifs present in at least 40% of the sequences of a subgroup are presented.
| Phosphorylation Motif | rLeMPK1 | rLeMPK2 | rLeMPK3 | rAtMPK6 |
|---|---|---|---|---|
| P x S Pa | 21b | 21 | 14 | 16 |
| S P x [KR]c | 19 | 19 | 19 | 19 |
| [KR] R x S | 10 | –d | – | – |
| [KR] x x x x S | – | – | 21 | 18 |
| S x x [KR] | – | – | 20 | – |
P, Pro; S, Ser; x, any residue. The phosphorylation site (S) is indicated in bold.
The number of phosphorylated peptides containing this motif in the subgroup of sequences.
[KR] refers to the presence of either a Lys (K) or an Arg (R) at this position of the phosphorylation motif.
This motif is not present in the peptides phosphorylated by this kinase.
The sequences of the phosphorylated peptides were loaded into TEIRESIAS software (Rigoutsos and Floratos, 1998) and preferred phosphorylation motifs consisting of three or two residues were predicted for each rMAPK (Table II). These motifs are too short to identify relevant putative in vivo substrates from databases; however, they allow discrimination between the phosphorylation specificities of the individual rMAPKs. To verify whether the results from the PepChip analysis match reported biological substrates, phosphorylation motifs predicted for rAtMPK6 were compared to the phosphorylation sites of its known in vivo substrates, ACS6 and ACS2. The predicted Pro-x-Ser-Pro (PxSP) phosphorylation motif matches for the position of two of the three phosphorylated Ser residues (Ser-483 and Ser-488) described for ACS6 (Liu and Zhang, 2004). However, the third phosphorylated Ser residue (Ser-480) is only followed by a Pro. Such Ser residues were frequently phosphorylated on the PepChip, but motifs matching these Ser-Pro sites did not exceed the threshold set to predict motifs. These data reveal phosphorylation motifs for rLeMPK1, rLeMPK2, and rLeMPK3 that only partially overlap, indicating that LeMPKs share common substrates but also have different substrate specificities (Table II).
VIGS of LeMPK1, LeMPK2, or LeMPK3 Results in Decreased Activity of the Encoded MAPK
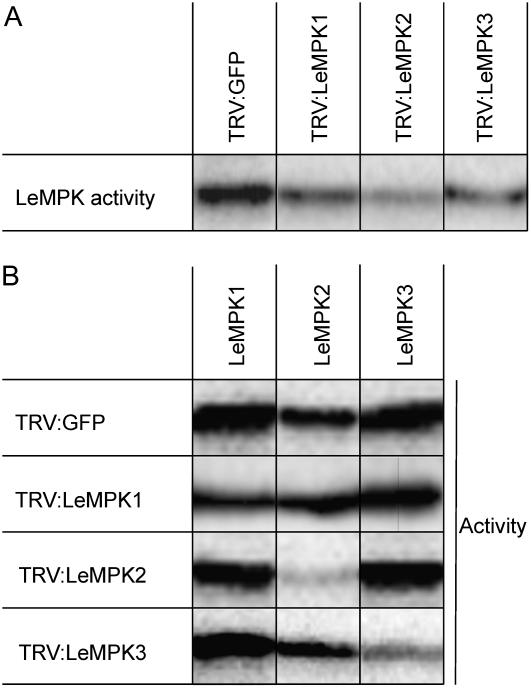
LeMPK1, LeMPK2, and LeMPK3 are activated upon specific Cf-4-mediated recognition of C. fulvum avirulence factor Avr4. To elucidate the role of the individual LeMPKs in HR and resistance of tomato to C. fulvum, VIGS of LeMPK1, LeMPK2, or LeMPK3 was performed. Therefore, ‘Money Maker’-Cf-4 tomato seedlings were inoculated with recombinant tobacco rattle virus (TRV)-silencing constructs (Liu et al., 2002a, 2002b), each containing part of the unique 3′-untranslated region (UTR) of the LeMPK1, LeMPK2, or LeMPK3 genes. LeMPK1, LeMPK2, and LeMPK3 have very low activity in untreated leaf disks (data not shown). However, they can be rapidly activated by wounding (Higgins et al., 2006). To test MAPK activity in silenced plants, we induced MAPK activity by punching leaf disks, which results in a wound stimulus, and floated the disks on water to prevent desiccation (Menke et al., 2004; see “Materials and Methods”). Subsequently, in-gel kinase assays were performed. Overall, LeMPK activity was decreased in TRV:LeMPK1-, TRV:LeMPK2-, and TRV:LeMPK3-inoculated plants when compared to control plants that had been inoculated with TRV containing the ORF of GFP (TRV:GFP; Fig. 6A). To confirm decreased activity of only the targeted LeMPK, immunocomplex assays for LeMPK1, LeMPK2, and LeMPK3 were performed on the TRV:LeMPK- and TRV:GFP-inoculated plants. In the TRV:LeMPK1-inoculated plants, LeMPK1 activity was decreased when compared to the LeMPK1 activity in the TRV:GFP-inoculated plants, whereas the LeMPK2 and LeMPK3 activity did not change when compared to TRV:GFP-inoculated plants (Fig. 6B). Also, a clear specific decrease in LeMPK2 and LeMPK3 activity was observed in TRV:LeMPK2- and TRV:LeMPK3-inoculated plants, respectively. From these observations, we conclude that inoculation of tomato with the different TRV:LeMPK constructs results in specific suppression of the respective MAPK activities.
Figure 6.
VIGS of LeMPK1, LeMPK2, or LeMPK3 decreases the activity of the encoded MAPK. Discs were taken from leaflets of TRV-GFP- and TRV-LeMPK-inoculated plants 3 weeks after inoculation and they were floated on water for 15 min, after which protein extracts were made. A, In-gel kinase assay was performed for each individual leaf disc and representative overall LeMPK activities are shown. Bands represent a combination of LeMPK1, LeMPK2, and LeMPK3 activities because the slightly smaller LeMPK3 protein was not separated from LeMPK1 and LeMPK2 on these gels. Equal amounts of protein were loaded in each lane (data not shown). B, LeMPK1, LeMPK2, and LeMPK3 proteins were individually immunoprecipitated from the protein extracts shown in A and subjected to a kinase assay. Representative results from three independent experiments are shown. Note that only the activity of the targeted LeMPK is decreased when compared to the activity levels from TRV:GFP-inoculated plants.
LeMPKs Have Different and Overlapping Roles in Cf-4-Mediated HR and Resistance to C. fulvum
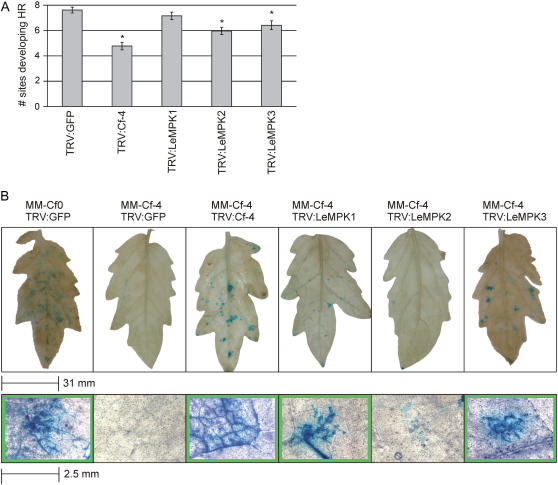
Three weeks after inoculation of ‘Money Maker’-Cf-4 tomato with the various TRV VIGS constructs described above, eight leaflets of compound leaves at similar positions on the plant were injected with Avr4 protein at 10 sites per leaflet. Sites that developed necrosis, reflecting Cf-4/Avr4-induced HR, were counted (see “Materials and Methods” for details). The maximal response to Avr4 of 7.6 necrotic spots per leaflet (±0.2 sem) was obtained in TRV:GFP-inoculated ‘Money Maker’-Cf-4 plants, whereas inoculation with TRV:Cf-4 resulted in a significant decrease of the response to Avr4 (Fig. 7A). Interestingly, inoculation with TRV:LeMPK2 or TRV:LeMPK3 also caused a significant decrease in the responsiveness of the plant, whereas inoculation with TRV:LeMPK1 did not affect the Cf-4/Avr4-induced HR in this experimental setup (P ≤ 0.05; Fig. 7A).
Figure 7.
The role of LeMPK1, LeMPK2, and LeMPK3 in Cf-4-mediated HR and resistance to C. fulvum. Tomato plants were inoculated with TRV:GFP, TRV:Cf-4, TRV:LeMPK1, TRV:LeMPK2, or TRV:LeMPK3 and injected with Avr4 or challenged with C. fulvum-expressing Avr4. A, After 3 weeks, a total of 160 leaflets of the TRV-inoculated ‘Money Maker’-Cf-4 plants were injected with Avr4 protein at 10 sites. The average number of sites per leaf that developed a specific HR, visible as necrosis, is shown (see “Materials and Methods” for details). The asterisks indicate a significantly decreased response as compared to TRV:GFP-inoculated plants (P ≤ 0.05). B, ‘Money Maker’-Cf-0 plants that are fully susceptible to C. fulvum and ‘Money Maker’-Cf-4 plants that are fully resistant were inoculated with the indicated recombinant TRV VIGS constructs. After 3 weeks, the plants were inoculated with a strain of C. fulvum-expressing Avr4 and GUS and leaflets were stained for GUS activity after 14 d. Leaflets representative of five independent experiments are shown at the top and magnification of GUS-stained areas is shown at the bottom; green margins indicate compromised resistance.
To determine whether, in addition to their requirement for a full HR, LeMPKs are also required for Cf-4-mediated resistance, ‘Money Maker’-Cf-4 tomato plants that had been inoculated with the various TRV constructs were challenged with a strain of C. fulvum-expressing Avr4. As a control, fully susceptible ‘Money Maker’-Cf-0 plants lacking functional R genes to C. fulvum were inoculated with TRV:GFP and challenged with the fungus. The C. fulvum strain also expresses the pGPD:GUS transgene, thereby allowing detection of the mycelium of the fungus in the leaves. Two weeks after inoculation with C. fulvum, leaflets from fully developed compound leaves were treated with X-gluc, resulting in blue staining of leaf sections that are successfully colonized. As shown in Figure 7B (top), leaflets of susceptible TRV:GFP-inoculated ‘Money Maker’-Cf-0 tomato plants were colonized by C. fulvum, as reflected by the blue staining in the leaflet. Magnification of the blue-stained areas clearly revealed the presence of fungal mycelium growth in the leaf (bottom). ‘Money Maker’-Cf-4 tomato plants inoculated with TRV:GFP did not show any colonization by C. fulvum, indicating that these TRV-inoculated plants are still fully resistant to the fungus. Inoculation of the ‘Money Maker’-Cf-4 plants with TRV:Cf-4 compromised resistance to C. fulvum as mycelium of the fungus is observed in the leaflets. Interestingly, loss of full resistance to C. fulvum was also observed in the TRV:LeMPK1- and TRV:LeMPK3-inoculated ‘Money Maker’-Cf-4 plants because the intercellular spaces in certain patches of the leaflets were successfully colonized (Fig. 7B). Surprisingly, although VIGS of LeMPK2 significantly affected Cf-4/Avr4-induced HR (Fig. 7A), we did not find compromised resistance to C. fulvum in TRV:LeMPK2-inoculated ‘Money Maker’-Cf-4 plants (Fig. 7B).
DISCUSSION
To reveal differences in LeMPK1, LeMPK2, and LeMPK3 peptide substrate specificity, PepChip analysis was performed. We have incubated two different PepChips, each carrying a triplicate peptide set, with rLeMPK1 in two independent experiments and found that the peptides selected as phosphorylated peptides were identical when both experiments were compared (data not shown). Therefore, the other rMAPKs were incubated on only one PepChip. PepChips that we employed carry peptides containing phosphorylation motifs for human kinases and therefore we focused on the overall phosphorylation patterns rather than on the phosphorylation of individual peptides. Analysis with TEIRESIAS motif prediction software revealed motifs in the sequences of the phosphorylated peptides (Table II). To enable comparison of rMAPK-specific phosphorylation motifs obtained from PepChip analysis, different amounts of the rMAPK proteins with similar MBP kinase activities were applied to the slides. This implies that the relevance of a certain phosphorylation motif identified for a LeMPK could be different in vivo because LeMPK protein concentrations and specific activities differ in the plant tissue. Each rMAPK phosphorylated approximately 80 to 100 peptides consisting of 11 amino acid residues, which allowed identification of motifs of two or three residues. Table II shows motifs for the individual rLeMPKs present in 40% or more of the sequences in a subgroup, which could match with motifs present in in vivo substrates. In contrast to Figure 5B, where residues with a higher frequency in phosphorylated, as compared to nonphosphorylated, peptides are presented, these motifs consist of residues that have high frequency in the phosphorylated peptides. Unfortunately, motifs consisting of two or three residues are not discriminative in database searches. However, manual annotation revealed that the PxSP motif, which has been previously described by Schwartz and Gygi (2005), matches for Ser-483 and Ser-488 of ACS6, which are phosphorylated by AtMPK6 (Liu and Zhang, 2004). The third phosphorylated Ser of ACS6 (Ser-480) was only followed by a Pro. Peptides with a central Ser residue followed by a Pro are frequently phosphorylated on the slides and only a subset of the sequences matches the PxSP motif. The remaining Ser-Pro sequences were not part of a pattern exceeding the threshold set for motif prediction. Furthermore, many potential AtMPK6 substrates have been described by Feilner et al. (2005). However, the exact phosphorylation sites are not known for these substrates, rendering verification of the motifs not possible. In addition to rAtMPK6, rLeMPK1, rLeMPK2, and rLeMPK3 also phosphorylate the PxSP motif (Table II), and, in tomato, this motif matches the orthologs LeACS6 and LeACS2, suggesting that these enzymes are substrates of LeMPK1, LeMPK2, and LeMPK3. The identification of motifs phosphorylated by only one of the tested rLeMPKs (Table II) implies that, in addition to overlapping specificities, these LeMPKs also have different substrate specificities in vivo. Alternatively, the LeMPKs could target different phosphorylation sites of the same protein, which implies a different regulatory function for each of the LeMPKs.
Because our PepChip analysis points to different, and also overlapping, regulatory functions for the LeMPKs in vivo, the role of these MAPKs in the initiation of Cf-4/Avr4-induced HR and disease resistance was studied. The LeMPKs were individually targeted in ‘Money Maker’-Cf-4 tomato plants by VIGS, which resulted in decreased LeMPK activity compared to the control TRV:GFP-inoculated plants. To avoid off-target silencing, the sequences to target the individual LeMPKs were designed on the highly unique 3′-UTR regions and these sequences have less than 21-bp homology to any other tomato gene present in the NCBI or SGN databases (data not shown). Furthermore, standardized immunoprecipitations, with equal amounts of LeMPK antibodies, protein A agarose beads, and input protein for each sample, did not reveal decreased activity of the homologous, nontargeted LeMPKs (Fig. 6B). Therefore, it is unlikely that expression of other genes is affected in the different TRV:LeMPK-inoculated plants.
VIGS in tomato is patchy and usually only results in partial knockdown of gene expression. Therefore, we only observed a slight decrease in the Avr4-induced HR even upon silencing of the Cf-4 R gene itself (Fig. 7A). However, the decrease is significant and also correlates with a clear loss of full resistance to an Avr4-expressing strain of C. fulvum (Fig. 7B). In many cases, silencing of a gene encoding a protein that functions downstream of Cf-4 in the HR signaling cascade even has a smaller effect on the responsiveness of the plant to Avr4, which is probably due to redundancy (Gabriëls et al., 2006). Still, significant suppression of the Avr4-induced HR was found in ‘Money Maker’-Cf-4 plants in which either LeMPK2 or LeMPK3 activity was decreased, whereas decreased LeMPK1 activity did not affect the Avr4-induced HR (Fig. 7A). The latter could be caused by the relatively slight decrease in LeMPK1 activity (Fig. 6B). The TRV:LeMPK-inoculated ‘Money Maker’-Cf-4 plants were also challenged with an Avr4-expressing strain of C. fulvum. Surprisingly, in this assay the LeMPK1-silenced plants showed a phenotype, as localized patches of blue-stained intercellular mycelium were visible upon treatment of the inoculated leaves with X-gluc (Fig. 7B). Although the LeMPK1 activity is only slightly decreased and the HR is not affected (Fig. 7A), it does cause suppressed resistance, indicating that the degree of LeMPK1 silencing is sufficient to observe a phenotype. The lower LeMPK2 activity did not affect resistance, whereas for silencing of LeMPK3, in addition to its effect on the HR, suppressed resistance was found (Fig. 7B).
The role in disease resistance of various orthologs of the LeMPKs studied here appears to match with our results. VIGS of NtSIPK and NtWIPK, the tobacco orthologs of LeMPK1 and LeMPK3, respectively, in Nicotiana benthamiana compromised resistance to the bacterial pathogen Pseudomonas cichorii and Tobacco mosaic virus (Jin et al., 2003; Sharma et al., 2003). Furthermore, enhanced susceptibility to Peronospora parasitica was found upon silencing of the LeMPK1 ortholog AtMPK6 in Arabidopsis (Menke et al., 2004). Silencing of LeMPK1 and LeMPK2 or silencing of LeMPK3 only was reported to result in a loss of full Mi-1-mediated aphid resistance (Li et al., 2006), and inoculation of tomato with TRV:NtWIPK compromised resistance to P. syringae pv tomato (Ekengren et al., 2003). Finally, constitutive overexpression of StMEK1, thereby activating the LeMPK1 ortholog StMPK1, enhanced resistance to Phytophthora infestans and Alternaria solani (Yamamizo et al., 2006). Interestingly, TRV:NtSIPK inoculation, which should cause simultaneous silencing of LeMPK1 and LeMPK2 in tomato, did not affect the resistance response to P. syringae pv tomato (Ekengren et al., 2003).
Solanaceous species like tomato and tobacco possess two homologous MAPKs in group A, LeMPK1/2 and NtSIPK/NTF4, and it is not clear whether these homologs are fully redundant or have different specificities. Here, we show that LeMPK1 and the 95.4% identical LeMPK2 protein have overlapping but also different peptide phosphorylation specificities in vitro and that both MAPKs are clearly involved in the defense response. The VIGS data indicate that LeMPK1 and LeMPK2 may have different functions with regard to HR and full resistance in tomato. However, a definite result would require complete knockouts of LeMPK1 and/or LeMPK2, which are not available. VIGS of LeMPK3 affects execution of the HR and in this case also full resistance is lost (Fig. 7), suggesting that LeMPK3 has a role in both the initiation of the HR and other defense responses. This hypothesis is supported by the broader phosphorylation specificity of LeMPK3 (Table II).
In transgenic tobacco cell suspensions expressing Cf-9, MAPKs are activated within 5 min after elicitation with the Avr9 avirulence factor of C. fulvum (Romeis et al., 1999; De Jong et al., 2002). Such cell suspensions are also temperature sensitive and it was found that this sensitivity resides at the level of elicitor perception because the amount of Avr9 binding sites was significantly decreased at 33°C (De Jong et al., 2002). The cell suspensions required at least 45 min to regain their ability to perceive Avr9 when transferred from 33°C to 15°C, indicating that de novo protein synthesis is required for this recovery. The Cf-4/Avr4 seedlings also need to recover when shifted from 33°C to 20°C, in this case resulting in a lag phase of 90 to 120 min before MAPK activation is observed (Fig. 1). Furthermore, significantly higher basal MAPK activity was observed in Cf-4/Avr4 seedlings at 33°C as compared to the parents (t = 0 min; Fig. 1B), although immunocomplex assays did not reveal increased activity for LeMPK1, LeMPK2, or LeMPK3 at this time point. Possibly, at this stage during which HR is suppressed, other MAPKs that act as negative regulators of the HR are active. Putative candidates are LeMPK4 and/or LeMPK7 from group B (Fig. 2), which are orthologs of the negative regulator of resistance AtMPK4 (Ichimura et al., 2006). Due to their similar size (Table I), these MAPKs are indistinguishable from LeMPK1, LeMPK2, and LeMPK3 on the gel kinase assays shown in Figure 1. We did not further separate LeMPK3 from the other LeMPKs as this allowed to quantify the total MAPK activity present in one band. Correspondingly, LeMPK4 and/or LeMPK7 or other LeMPKs might be activated simultaneously with LeMPK1, LeMPK2, and LeMPK3 from 120 min onward because the immunoprecipitation data do not provide absolute qualitative data as this depends on the titer and affinity of the antibodies. Recently, for example, activation of the negative regulator AtMPK4 simultaneously with AtMPK3 and AtMPK6 has been reported upon elicitation of Arabidopsis with the bacterial elicitor flagellin (Mészáros et al., 2006).
Cotyledons of Cf-4/Avr4 seedlings develop localized necrotic lesions that become macroscopically visible at about 12 h after the temperature shift and eventually spread over the complete surface of the cotyledons. Interestingly, Cf-4/Avr4 seedlings that are incubated at 20°C for 240 min and subsequently shifted back to 33°C survive and do not develop necrosis (data not shown). In addition, Cf-4/Avr4 seedlings incubated at 20°C for 24 h develop localized necrotic lesions, but, when shifted back to 33°C, these lesions do not further expand and the remaining tissue survives (data not shown). Alvarez et al. (1998) observed the initiation of systemic micro-HRs at certain confined locations in the tissue, leaving no visible trace, upon inoculation of Arabidopsis with avirulent P. syringae. Furthermore, it was found that reactive oxygen intermediates that are generated at defined sites by the plant upon perception of an avirulent pathogen are able to suppress the spread of cell death (Torres et al., 2005). Our observations indicate that similar phenomena take place in the Cf-4/Avr4 seedlings upon temperature shift. The reversibility of the system and the more or less constant total LeMPK activity level after 180 min suggest that, at least during the early stages after the temperature shift, a controlled HR takes place in the Cf-4/Avr4 seedlings. This control mechanism prevents superfluous cell death in Cf-4/Avr4 seedlings and illustrates that the response of the seedlings is a proper reflection of the response of a resistant host plant to invasion by an avirulent pathogen.
MATERIALS AND METHODS
Plants
To generate tomato (Solanum lycopersicum) offspring expressing both the Hcr9-4D (= Cf-4) gene and its cognate avirulence gene Avr4 from Cladosporium fulvum, transgenic ‘Money Maker’-Cf-0 plants expressing Avr4 (‘Money Maker’-Cf-0:Avr4) were crossed to transgenic ‘Money Maker’-Cf-0:Hcr9-4D (‘Money Maker’-Cf-4) plants, as described earlier (Thomas et al., 1997; Cai et al., 2001;). In addition, the ‘Money Maker’-Cf-4 and ‘Money Maker’-Cf-0:Avr4 parental lines were selfed. The resulting Cf-4/Avr4 and parental seeds were isolated from the fruits and germination was stimulated by treatment with 25% (v/v) Lodik (containing 4% [v/v] sodium hypochlorite) for 20 min. After germination under normal daylight conditions at room temperature (RT) for approximately 7 d, seedlings were incubated at 33°C under a 16-h-light/8-h-dark regime (Elbanton) for at least another 7 d. For the activation of Cf-4/Avr4-induced defense signaling, seedlings were shifted to 20°C, and, at several time points after this temperature shift, cotyledons were harvested, immediately frozen in liquid nitrogen, and stored at −80°C. Parental lines were subjected to the same treatment. ‘Money Maker’-Cf-4 and ‘Money Maker’-Cf-0 plants used for VIGS assays were grown under standard greenhouse conditions.
Monitoring HR Development in Cf-4/Avr4 Seedlings
Cf-4/Avr4 seedlings and seedlings of the parents were rescued at 33°C as described above. Seedlings were transferred to 20°C and a Web cam, which was placed in the incubator, took photographs from the seedlings every 5 min over a period of 5 d. Images were cropped by Irfanview software, version 3.98 (http://www.irfanview.com), batch conversed to centralize the seedlings in the photograph, and merged to avi-format by VideoMach 2.7.2 software (http://www.gromada.com).
Kinase Assays
Cotyledons of the seedlings were homogenized in immunoprecipitation buffer (10 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm Na3VO4, 1 mm NaF, 10 mm β-glycerophosphate, 1% [w/v] Triton X-100, 0.5% [w/v] Nonidet P-40, 2 mm dithiothreitol [DTT], and one complete protease inhibitor tablet [Roche]) and the homogenate was centrifuged at 16,000g for 20 min at 4°C, after which the supernatant was recovered. For in-gel kinase assays with MBP as an artificial substrate (Shibuya et al., 1992), a volume containing 25 μg of total protein (Bradford protein assay; Bio-Rad) was loaded per lane. MAPK activity was measured by phosphor imaging (Storm; Molecular Dynamics) and quantified with ImageQuant software (Amersham). Data obtained from five individual in-gel kinase assays were subjected to a two-way design ANOVA (Genstat, release 8.1). Furthermore, 25 μg of protein were loaded on SDS-PAGE gels and stained with Coomassie Brilliant Blue to verify even loading.
Immunocomplex kinase assays were performed as described earlier (Holley et al., 2003) with minor modifications. For immunoprecipitation, 200 μg of protein were incubated with LeMPK1, LeMPK2, or LeMPK3 antiserum in a 100:1 dilution and the antibodies were pulled down with 15 μL of protein A beads (3 mg/mL). To determine the activity of recombinant MAPKs (see below), dilution series of these proteins were incubated with 20 μL of kinase reaction buffer (20 mm HEPES, pH 7.5, 15 mm MgCl2, 2 mm EGTA, 1 mm DTT, 0.25 mg/mL MBP, 25 μm ATP, and 1 μL of 10 μCi [γ-32P]ATP) for 30 min at 30°C. Proteins were subsequently separated on 15% SDS-PAGE gels and a phosphor-imaging screen was exposed to the dried gel. MAPK activity was measured by phosphor imaging and quantified with ImageQuant software.
Analysis of Relationships between MAPK Protein Sequences
To identify the sequences of all putative LeMPK homologs, BLAST searches with the DNA sequence of LeMPK1, LeMPK2, and LeMPK3 ORFs were performed on TIGR Tomato Gene Index (LeGI), the NCBI, and the SGN databases. Each newly found homolog was subsequently BLASTed until no new sequences were identified and ORFs from 13 putative additional LeMPK homologs were obtained. The sequences were translated to protein sequences with the Expasy Proteomics server translate tool (http://us.expasy.org/tools/dna.html) and protein sequences encoded by the ORFs were aligned in ClustalX (Supplemental Fig. S2; Thompson et al., 1997) with the sequences encoded by the ORFs of AtMPK1 to AtMPK20 from Arabidopsis (Arabidopsis thaliana), NtSIPK, NtWIPK, NtNTF4, and NtNTF6 from tobacco (Nicotiana tabacum), and the Homo sapiens HsERK1, which also encodes a MAPK (Zhang and Klessig, 2001; Ichimura et al., 2002). Pairwise distances between sequences were calculated with neighbor joining in ClustalX and a cladogram rooted with HsERK1 was made with Treeview software (Page, 1996).
Cloning and Expression of rMAPKs
To express LeMPK1, LeMPK2, and LeMPK3 as soluble His-tagged proteins, primers were designed to PCR amplify the ORFs of the encoding genes. Respective primers were for LeMPK1, forward 5′-GATCGGATCCATGGATGGTTCCGTTCCGC-3′ and reverse 5′-GATCCTCGAGTCACATGCGCTGGTATTCAGG-3′; LeMPK2, forward 5′-GATCGGATCCATGGATGGTTCAGCTCCGC-3′ and reverse 5′-GATCCTCGAGTCACATGTGCTGGTATTCGGG-3′; and for LeMPK3, forward 5′-GATCGGATCCATGGTTGATGCTAATATGGG-3′ and reverse 5′-GATCCTCGAGTTAAGCATATTCAGGATTCAACG-3′; BamHI and XhoI sites are underlined in the forward and reverse primers, respectively. Amplification products were ligated into BamHI/XhoI-digested pET28a+ vector (Novagen). Plasmids were transformed to Escherichia coli BL21 (DE3) cells and the integrity of the constructs was confirmed by sequencing. The pET28a+-AtMPK6 construct has been described previously (Liu and Zhang, 2004; Menke et al., 2004). Bacteria were cultured in Luria-Bertani medium at 37°C and protein expression was induced at OD 0.6 by adding isopropylthio-β-galactoside to a final concentration of 1 mm. Cells were cultured for another 4 h, washed in cold 20 mm Tris, pH 7.5 (in 25% of the original volume), and stored at −80°C as a cell pellet.
Proteins were recovered from the cell pellet by adding 10 mL/g pellet of CellLytic B bacterial cell lysis extraction reagent (Sigma) plus complete protease inhibitor cocktail (EDTA-free; Roche) and subsequent incubation at RT for 20 min. After centrifuging at 25,000g (4°C), the soluble His-tagged proteins present in the supernatant were bound to 1 mL nickel-nitrilotriacetic acid agarose superflow resin that had been pretreated with 4 volumes of MilliQ water and 10 volumes of buffer (20 mm Tris-HCl, pH 7.9, 0.5 m NaCl) containing 5 mm imidazole. The resin was washed with 10 volumes of the buffer containing 20 mm imidazole and the protein was eluted with 4 volumes of the buffer containing 200 mm imidazole. The eluate was dialyzed against kinase storage buffer (25 mm HEPES, pH 7.5, 2 mm DTT, 50 mm KCl, 5% [v/v] glycerol) using Vivaspin 4 columns and stored in aliquots at −80°C. Protein concentrations were determined using the bicinchoninic acid reducing agent kit (Pierce) and kinase activity was determined from dilution series of the MAPK proteins as described above. The intensity of MBP phosphorylation, quantified by phosphor imagings per microgram of the different MAPK proteins (the specific activities), shows a ratio of 8:5:40:4 for rLeMPK1:rLeMPK2:rLeMPK3:rAtMPK6. Nonactive control rLeMPK3* was obtained by storage of the eluted protein in elution buffer instead of kinase storage buffer.
Immunoblot Analysis
Proteins present in extracts obtained as described for the in-gel kinase assays or recombinant MAPK proteins were separated on 10% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes (Bio-Rad). For detection of pY in rMAPKs, membranes were blocked in 50 mm Tris, pH 7.5, 150 mm NaCl, 0.1% (w/v) Tween 20, and 3% bovine serum albumin and incubated with 1:2,000 diluted monoclonal Phospho-Tyrosine IgG (CST no. 9411) in the same solution o/n. After three washes with 50 mm Tris, pH 7.5, 150 mm NaCl, and 0.1% (w/v) Tween 20, the membranes were incubated with horseradish peroxidase-linked anti-mouse IgG (1:3,000 diluted; CST no. 7076) and developed using the ECL detection kit (Pierce).
PepChip Analysis
PepChips (Pepscan) are spotted with a triplicate set of 976 peptides (excluding controls) that resemble experimentally verified phosphorylation sites for human kinases (PhosphoBase) and their original surrounding residues (sequences available at http://www.pepscan.nl/index5.htm). The peptides mostly consist of 11 amino acids, of which the central position is the putative phosphorylation site. Six peptides are spotted that consist of only 9 or 10 amino acids in which the phosphorylation site is not centralized.
For incubation of PepChips, 11, 3.6, 0.8, and 1.1 μg, respectively (representing equal kinase activities), of purified rLeMPK1, rLeMPK2, rLeMPK3, or rAtMPK6 protein in kinase storage buffer was mixed with 5 μL of 33P-γ-ATP (3,000 Ci/mmol; 50 μCi/PepChip) in a final volume of 30 μL, and added to 30 μL of 2× PepChip Mastermix (40 mm HEPES, pH 7.5, 30 mm MgCl2, 4 mm EGTA, 2 mm DTT, 40% [v/v] glycerol, 0.02 mg/mL bovine serum albumin, 0.02% [v/v] Brij-35, and 0.56 mm ATP). The mix was brought onto a cover slide, after which the PepChip was posed over the sample and turned around. The PepChip was incubated for 4 h at 30°C in a closed box with wet paper to prevent drying of the chip. The cover slides were rinsed off the PepChip with 50 mm Tris, pH 7.5, 150 mm NaCl, 0.02% Tween 20 (TBST) and the PepChips were washed twice with 2 m NaCl, once with 2 m urea, twice with 10% SDS, and three times with MilliQ water in a washing tube (provided with the PepChips) by manual shaking. Phosphorylation intensity of the various spotted peptides was determined by phosphor imaging (50-μm scan resolution) and quantitative values were obtained with ImageQuant software by adding the numerical values of each pixel within a prescribed area (=spot), subtracted by the background value. Data were exported and collected in a Microsoft Excel worksheet and the average phosphorylation intensity per set was calculated, after which the sets were normalized. In addition, data obtained from the different PepChips were normalized based on the average PepChip phosphorylation intensity.
Average phosphorylation intensity from the triplicates was calculated for each peptide, and peptides with an average phosphorylation intensity ≥1.5 times the average PepChip phosphorylation intensity were selected for further phosphorylation pattern analysis (referred to as phosphorylated peptides; approximately 10% of all peptides). Phosphorylated peptides with a sd exceeding 1.5 times the average PepChip phosphorylation intensity were removed from the dataset. The subset of nonphosphorylated peptides represents the peptides that had a phosphorylation intensity of zero (approximately 10% of all peptides). Significant differences between the sequences of the phosphorylated and nonphosphorylated peptides were calculated by TSL software with a t test (P ≤ 0.05) and TSL plots were drawn (Crooks et al., 2004).
Sequences of the phosphorylated peptides were combined in subsets based on common phosphorylation by one or more LeMPKs and TEIRESIAS software (Rigoutsos and Floratos, 1998) calculated phosphorylation motifs in these sequences. Peptides phosphorylated by AtMPK6 were combined in subsets based on the overlapping phosphorylation with LeMPK1. Calculated phosphorylation motifs present in at least 40% of the sequences in the respective subset were included in Table II. Motifs indicated in Table II as not being phosphorylated were absent in the motif prediction for the respective kinase.
VIGS of LeMPKs in Tomato
The TRV-based binary VIGS vectors TRV:RNA1 and TRV:RNA2 (pYL156) have been described before (Liu et al., 2002a, 2002b). For TRV:LeMPK construction, the following primers were used to PCR amplify LeMPK sequences of genomic DNA isolated from ‘Money Maker’-Cf-0 tomato (BamHI [forward] and Acc65I [reverse] sites are underlined): for LeMPK1, forward 5′-CAGGATCCATAATTGCTGACAGATTGTTGCAG-3′ and reverse 5′-CAGGTACCGTACTCGCTCGTTTGCTGTTGGAT-3′; for LeMPK2, forward 5′-CAGGATCCCAGTTCTTCTCTTGCTTACCTAGT-3′ and reverse 5′-CAGGTACCCTCTCCATACATAAGTCAGCTTC-3′; and for LeMPK3, forward 5′-CAGGTACCTGAACCACTTTCTTGGAGTACAG-3′ and reverse 5′-CAGGTACCACACAAGCTAGCCCGAACACCAC-3′. This resulted in fragments of 166, 199, and 205 bp corresponding to the 3′-UTR region of LeMPK1, LeMPK2, and LeMPK3, respectively. Fragments were ligated into BamHI/Acc65I-digested TRV:RNA2 and the resulting TRV:LeMPK constructs were transformed to Agrobacterium tumefaciens strain GV3101. Construction of TRV:GFP, TRV:Cf-4, and TRV:phytoene desaturase was described previously (Liu et al., 2002a; Gabriëls et al., 2007). For VIGS, cultures of A. tumefaciens containing TRV:RNA1 were mixed 1:1 with the various TRV:LeMPK cultures to an OD of 1.0 and infiltrated in cotyledons of 10-d-old tomato seedlings. In each experiment, four plants were infiltrated per TRV:LeMPK construct. Phytoene desaturase-silenced plants, which develop white patches on the leaflets upon successful silencing, were used to visually monitor the development of the silencing process.
Assessment of Avr4-Induced HR Development and Statistical Analysis
Three weeks after agroinfiltration of the VIGS constructs, leaflets of comparable compound leaves were injected parallel to the midvein with 150 μg/mL Pichia pastoris-produced His-FLAG-Avr4 protein (Rooney et al., 2005), using a microsyringe (Ito Corporation). Per day, two leaflets of four plants were injected at 10 sites and each plant was injected on four different days (80 sites/plant, resulting in 320 sites/pTRV:LeMPK construct/experiment). The experiment was repeated five times. After 4 to 7 d, the number of sites per leaflet showing HR, visible as necrosis, was scored. An arcsin√x transformation was performed to obtain a normally distributed dataset. Data were analyzed with a split-plot design ANOVA, after which multiple comparisons of all constructs were performed with a Student-Newman-Keuls test in Genstat (version 8.1.0.155; VSN International Ltd.).
C. fulvum Inoculations and GUS Staining
A strain of C. fulvum race 5, expressing Avr4 and containing a pGPD∷GUS transgene, which contains the GUS gene under control of the constitutive glyceraldehyde-3-P dehydrogenase promoter, was subcultured on 2% (w/v) potato dextrose agar to which 1.5% (w/v) technical agar was added. Conidia were obtained from 10-d-old plates, washed three times in water by centrifuging (4,000g) and decanting the supernatant, and diluted to 6 × 105 spores/mL water. Plants were dip inoculated 3 weeks after agroinfiltration with the VIGS constructs described above and were kept in closed cages covered with transparent plastic for 2 d. Two weeks after inoculation, two to four leaflets were harvested and vacuum infiltrated with X-gluc buffer (0.1 m NaPi, pH 7.0, 1% [v/v] Triton X100, 1% [v/v] dimethyl sulfoxide, 10 mm EDTA, and 1 mg/mL X-gluc). Leaves were incubated overnight in the dark at 37°C and destained with 70% ethanol at RT after which photographs were taken using an Axioskop Zeiss microscope equipped with a Coolsnap camera. This experiment was repeated five times.
Confirmation of MAPK Silencing
Discs with a diameter of 1.5 cm were taken close to the midvein from different leaflets at 4 (HR assays) or 6 (C. fulvum-inoculation studies) weeks after TRV inoculation of plants of which the other leaflets were used for HR scoring or GUS analysis, respectively. Punching leaf disks allows for a targeted sampling in regions that probably have the most pronounced silencing, based on the phytoene desaturase results. The leaf discs were floated on water for 15 min to allow MAPK activation by the wound response from punching the disks and to prevent the leaf disks from desiccating (Menke et al., 2004). Leaf disks were individually analyzed for total inducible LeMPK activity by in-gel kinase assays. For extracts that showed decreased LeMPK activity in the in-gel kinase assays, the activity of LeMPK1, LeMPK2, and LeMPK3 was determined by immunocomplex kinase assays. This experiment was repeated three times.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Movie S1. Monitoring systemic HR development in a Cf-4/Avr4 seedling after a temperature shift.
Supplemental Figure S1. Phosphorylation intensities of rLeMPK1 phosphorylated and nonphosphorylated peptides.
Supplemental Figure S2. Alignment of MAPK protein sequences.
Supplementary Material
Acknowledgments
We thank D. Klessig and S. Zhang for the pET28a+-AtMPK6 construct, G. van den Berg for cloning the TRV:LeMPK constructs, H. Smid for horticultural assistance, and M. Hartog of the Department of Molecular Biology (WUR) for the use of the phosphor imager. We thank B. Thomma and P. de Wit for critically reading the manuscript.
This work was supported by the Dutch Organization for Scientific Research (VIDI grant to M.H.A.J.J.) and the National Science Foundation (grant no. 0321453 to J.W.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Matthieu H.A.J. Joosten (matthieu.joosten@wur.nl).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92 773–784 [DOI] [PubMed] [Google Scholar]
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NHT, Zhu S, Qiu J, Micheelsen P, Rocher A, Petersen M, et al (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 24 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Takken FLW, Joosten MHAJ, De Wit PJGM (2001) Specific recognition of AVR4 and AVR9 results in distinct patterns of hypersensitive cell death in tomato, but similar patterns of defense-related gene expression. Mol Plant Pathol 2 77–86 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814 [DOI] [PubMed] [Google Scholar]
- Crews CM, Alessandrini AA, Erikson RL (1991) Mouse Erk-1 gene product is a serine/threonine protein kinase that has the potential to phosphorylate tyrosine. Proc Natl Acad Sci USA 88 8845–8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong CF, Honée G, Joosten MHAJ, De Wit PJGM (2000) Early defense responses induced by AVR9 and mutant analogues in tobacco cell suspensions expressing the Cf-9 resistance gene. Physiol Mol Plant Pathol 56 169–177 [Google Scholar]
- De Jong CF, Takken FLW, Cai X, De Wit PJGM, Joosten MHAJ (2002) Attenuation of Cf-mediated defense responses at elevated temperatures correlates with a decrease in elicitor-binding sites. Mol Plant Microbe Interact 15 1040–1049 [DOI] [PubMed] [Google Scholar]
- Del Pozo O, Pedley KF, Martin GB (2004) MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekengren SK, Liu Y, Schiff M, Dinesh-Kumar SP, Martin GB (2003) Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 36 905–917 [DOI] [PubMed] [Google Scholar]
- Feilner T, Hultschig C, Justin L, Meyer S, Immink RGH, Koenig A, Possling A, Seitz H, Beveridge A, Scheel D, et al (2005) High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol Cell Proteomics 4 1558–1568 [DOI] [PubMed] [Google Scholar]
- Flor HH (1942) Inheritance of pathogenicity in Melampsora lini. Phytopathology 32 653–669 [Google Scholar]
- Gabriëls SHEJ, Takken FLW, Vossen JH, De Jong CF, Turk SC, Wachowski LK, Peters J, Witsenboer HMA, De Wit PJGM, Joosten MHAJ (2006) cDNA-AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Mol Plant Microbe Interact 19 567–576 [DOI] [PubMed] [Google Scholar]
- Gabriëls SHEJ, Vossen JH, Ekengren SK, Van Ooijen G, Abd-El-Haliem AM, Van den Berg GCM, Rainey DY, Martin GB, Takken FLW, De Wit PJGM, et al (2007) An NB-LRR protein required for HR signaling mediated by both extra- and intracellular resistance proteins. Plant J 50 14–28 [DOI] [PubMed] [Google Scholar]
- Hamel LP, Nicole M, Sritubtim S, Morency M, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, et al (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11 192–198 [DOI] [PubMed] [Google Scholar]
- Higgins R, Lockwood T, Holley S, Yalamanchili R, Stratmann JW (2006) Changes in the extracellular pH are neither required nor sufficient for activation of mitogen-activated protein kinases (MAPKs) in response to systemin and fusicoccin in tomato. Planta 225 1535–1546 [DOI] [PubMed] [Google Scholar]
- Holley SR, Yalamanchili RD, Moura DS, Ryan CA, Stratmann JW (2003) Convergence of signaling pathways induced by systemin, oligosaccharide elicitors, and ultraviolet-B radiation at the level of mitogen-activated protein kinases in Lycopersicon peruvianum suspension-cultured cells. Plant Physiol 132 1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Shinozki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang S, Hirt H, Wilson C, et al (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7 301–308 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K (2006) MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J Biol Chem 281 36969–36976 [DOI] [PubMed] [Google Scholar]
- Jin H, Liu Y, Yang K, Kim CY, Baker B, Zhang S (2003) Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J 33 719–731 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Krens SFG, Spaink HP, Snaar-Jagalska E (2006) Functions of the MAPK family in vertebrate-development. FEBS Lett 580 4984–4990 [DOI] [PubMed] [Google Scholar]
- Lee S, Hirt H, Lee Y (2001) Phosphatidic acid activates a wound-activated MAPK in Glycine max. Plant J 26 479–486 [DOI] [PubMed] [Google Scholar]
- Li Q, Xie Q, Smith-Becker J, Navarre DA, Kaloshian I (2006) Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling cascades. Mol Plant Microbe Interact 6 655–664 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP (2002. a) Virus-induced gene silencing in tomato. Plant J 31 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002. b) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30 415–429 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose M, Bonshtien A, Sessa G (2004) LeMPK3 is a mitogen-activated protein kinase with dual specificity induced during tomato defense and wounding responses. J Biol Chem 279 14819–14827 [DOI] [PubMed] [Google Scholar]
- Menke FLH, Van Pelt JA, Pieterse CMJ, Klessig DF (2004) Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell 16 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke FLH, Kang H, Chen Z, Park JM, Kumar D, Klessig DF (2005) Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol Plant Microbe Interact 18 1027–1034 [DOI] [PubMed] [Google Scholar]
- Mészáros T, Helfer A, Hatzimasoura E, Magyar Z, Serazetdinova L, Rios G, Bardóczy V, Teige M, Koncz C, Peck S, et al (2006) The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J 48 485–498 [DOI] [PubMed] [Google Scholar]
- Mishra NS, Tuteja R, Tuteja N (2006) Signaling through MAP kinase networks in plants. Arch Biochem Biophys 452 55–68 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signaling. Trends Plant Sci 10 339–349 [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12 357–358 [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2004) Identification of MAPKs and their possible MAPK kinase activators involved in the Pto-mediated defense response of tomato. J Biol Chem 279 49229–49235 [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2005) Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol 8 541–547 [DOI] [PubMed] [Google Scholar]
- Rigoutsos I, Floratos A (1998) Combinatorial pattern discovery in biological sequences: the TEIRESIAS Algorithm. Bioinformatics 14 55–67 [DOI] [PubMed] [Google Scholar]
- Rivas S, Thomas CM (2005) Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu Rev Phytopathol 43 395–436 [DOI] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Jones JDG (2000) Resistance gene-dependent activation of a calcium-dependent protein kinase in the plant defense response. Plant Cell 12 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Piedras P, Zhang S, Klessig DF, Hirt H, Jones JDG (1999) Rapid Avr9- and Cf-9-dependent activation of MAP kinases in tobacco cell cultures and leaves: convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11 273–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney HCE, Van't Klooster JW, Van der Hoorn RAL, Joosten MHAJ, Jones JDG, De Wit PJGM (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308 1783–1786 [DOI] [PubMed] [Google Scholar]
- Schwartz D, Gygi SP (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 23 1391–1398 [DOI] [PubMed] [Google Scholar]
- Sharma PC, Ito A, Shimizu T, Terauchi R, Kamoun S, Saitoh H (2003) Virus-induced silencing of WIPK and SIPK genes reduces resistance to a bacterial pathogen, but has no effect on the INF1-induced hypersensitive response (HR) in Nicotiana benthamiana. Mol Genet Genomics 269 583–591 [DOI] [PubMed] [Google Scholar]
- Shibuya EK, Boulton TG, Cobb MH, Ruderman JV (1992) Activation of p42 MAP kinase and the release of oocytes from cell cycle arrest. EMBO J 11 3963–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Chen J, Song F, Zheng Z (2006) A novel rice MAPK gene, OsBIMK2, is involved in disease-resistance responses. Plant Biol 8 587–596 [DOI] [PubMed] [Google Scholar]
- Thomas CM, Jones DA, Parniske M, Harrison K, Balint-Kurti PJ, Hatzixanthis K, Jones JDG (1997) Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Van Esse HP, Crous PW, De Wit PJGM (2005) Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol Plant Pathol 6 379–393 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL (2005) Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nature 37 1130–1134 [DOI] [PubMed] [Google Scholar]
- Yamamizo C, Kuchimura K, Kobayashi A, Katou S, Kawakita K, Jones JDG, Doke N, Yoshioka H (2006) Rewiring mitogen-activated protein kinase cascade by positive feedback confers potato blight resistance. Plant Physiol 140 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (2001) MAPK cascades in plant defense signalling. Trends Plant Sci 15 2285–2295 [DOI] [PubMed] [Google Scholar]
- Zhang T, Yang T, Zhang L, Xu S, Xue L, An L (2006) Diverse signals converge at MAPK cascades in plant. Plant Physiol Biochem 44 274–283 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.