Abstract
Boron (B) phytotoxicity affects cereal-growing regions worldwide. Although B-tolerant barley (Hordeum vulgare) germplasm is available, molecules responsible for this tolerance mechanism have not been defined. We describe and use a new comparative proteomic technique, iTRAQ peptide tagging (iTRAQ), to compare the abundances of proteins from B-tolerant and -intolerant barley plants from a ‘Clipper’ × ‘Sahara’ doubled-haploid population selected on the basis of a presence or absence of two B-tolerance quantitative trait loci. iTRAQ was used to identify three enzymes involved in siderophore production (Iron Deficiency Sensitive2 [IDS2], IDS3, and a methylthio-ribose kinase) as being elevated in abundance in the B-tolerant plants. Following from this result, we report a potential link between iron, B, and the siderophore hydroxymugineic acid. We believe that this study highlights the potency of the iTRAQ approach to better understand mechanisms of abiotic stress tolerance in cereals, particularly when applied in conjunction with bulked segregant analysis.
Boron (B) is an essential plant micronutrient but is toxic at high levels. Elevated soil B is a common feature of soils derived from marine sediments, a feature of the geological history of many cereal-growing regions in Australia. B phytotoxicity also affects soils in North Africa and western Asia. In barley (Hordeum vulgare), yield penalties of up to 17% have been attributed directly to B phytotoxicity (Cartwright et al., 1984). A number of barley accessions, originally isolated from northern Africa, display striking B tolerance (Nable, 1988), and incorporation of these B-tolerance traits into elite barley varieties continues to be part of current breeding programs.
A genetic study examining B toxicity tolerance in barley identified four quantitative trait loci (QTL) contributing to B tolerance (Jeffries et al., 1999). The two strongest QTL were on chromosomes 4H and 6H, and both contribute to a net reduction in B uptake (Jeffries et al., 1999). Thus, although multiple mechanisms may be involved in the reduction of B accumulation in planta, the actual molecular entities that are encoded by the 4H and 6H QTL are unknown. The mechanism(s) appears to be constitutive, with tolerant plants accumulating less B compared to intolerant varieties, regardless of the external B concentration (Nable, 1988; Hayes and Reid, 2004). Following from these previous studies, any mechanism leading to a reduced B uptake in both the roots and the leaves would necessarily have to function at the site of B uptake, namely at the plasma membrane (PM) of cells at the root epidermis.
The involvement of an anion transporter responsible for B efflux has recently been predicted (Hayes and Reid, 2004), although the identity of this protein is unknown. PM-located proteins with both active and passive B-transporting roles have been identified in Arabidopsis (Arabidopsis thaliana; Bor1 [Takano et al., 2002] and NIP5;1 [Takano et al., 2006]), but no B transporter has been demonstrated to be involved in a B-tolerance mechanism. B also has a capacity to form complexes with a variety of hydroxylated molecules (Power and Woods, 1997), most notably in the cell wall, where B is involved in complexing molecules of rhamnogalactouranan II via ester linkages to apiosyl residues (O'Neill et al., 2004). Given the conditions for B-rhamnogalactouranan II complexation exist in the apoplast, other B complexes may also be formed in this region and cannot currently be excluded from an involvement in a B-tolerance mechanism.
Regardless of how B tolerance occurs in planta, it is likely that differences in proteins, either in relative levels or amino acid sequence, will play a key role. Proteins involved in the regulation of membrane-bound transporters, as well as those involved in the synthesis of many low Mr, hydroxylated metabolites, reside in the cytoplasm. With this information in mind, we decided to compare the soluble, cytoplasmic proteins isolated from the roots of B-tolerant and B-intolerant plants using a quantitative mass spectrometry (MS) approach.
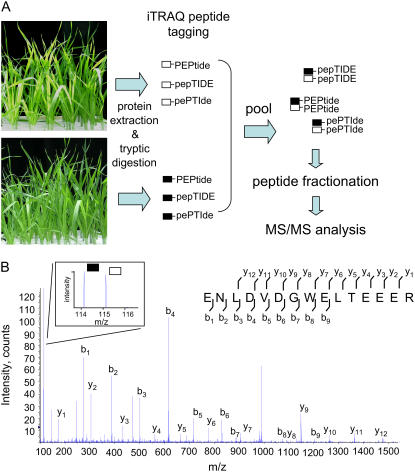
Following the first descriptions of multidimensional peptide chromatography and MS/MS identification (MudPIT) by Yates and colleagues (Washburn et al., 2001), this approach has been rapidly adopted in the plant field as an approach to identify hundreds of proteins from a single sample (Zieske, 2006). A limitation of the current techniques has been the difficulty of performing quantitative comparisons between multiple samples from separate experiments. Recently, a series of isobaric peptide tags (iTRAQ, Applied Biosystems) has allowed the comparative, quantitative analysis of up to four samples in a single MudPIT-style experiment. The chemically identical but isotopically distinct iTRAQ tags are attached to peptides through amine groups prior to pooling samples and analyzing by MS/MS (Fig. 1A). Due to the identical chemistry of the distinct tags, identical peptides (with MS differentiable tags) will coelute into the mass spectrometer during peptide fractionation. During MS/MS peptide fragmentation, ions derived from the distinct tags are detected, with the relative intensities of the different tags corresponding to the relative abundances of the peptides in the different samples (Fig. 1B).
Figure 1.
iTRAQ peptide tagging. A, Flow diagram outlining major steps of workflow leading to the comparative analysis of peptide abundances between samples using iTRAQ peptide tagging. In this example, proteins are extracted from two pools of plants and independently digested. Each pool of peptides is labeled with different iTRAQ tags; in this example, black (m/z 114) and white (m/z 115) are used. After tagging, the samples were pooled. Due to the identical chemistry of the tags, identical peptides from different samples cofractionate and are eluted into and analyzed by the MS simultaneously. B, Representative MS/MS spectra of iTRAQ-tagged peptide. MS/MS spectra of iTRAQ-tagged peptide matching to TC14799, NADH-dependent oxidoreductase. y and b ions are labeled. Inset, An expanded view of reporter ion region. This peptide is present in relatively similar abundances in both samples, as evidenced by the similar peak areas of reporter ions m/z 114 (black) and m/z 115 (white). [See online article for color version of this figure.]
In this study, we describe application of iTRAQ technology to the analysis of soluble proteins isolated from hydroponically grown barley plants. Initially, we tested the system by comparing two pools of proteins isolated from the leaves of replicate barley ‘Golden Promise’ plants. This allowed us to establish that the methodology was robust and sufficiently sensitive to detect small changes (>2.5-fold) in protein abundances between samples. We then used iTRAQ to look for differences in protein abundances between two pools of genetically similar barley plants that are defined by the presence or absence of both the 4H and 6H B-tolerance loci.
Few previous studies have examined the differences in proteins found in different tissues in barley plants. We found little overlap in the proteins identified from the soluble pools of proteins from the roots and leaves of barley plants, ignoring the varietal differences between the two analyses. Unsurprisingly, the soluble protein complement of both tissues was dominated by enzymes involved in metabolic processes in both tissues. Four proteins showed an increase in abundance in the B-tolerant plants. Three of these proteins are involved in production of phytosiderophores, and all four proteins have previously been demonstrated to be increased in expression in response to iron (Fe) deficiency (Negishi et al., 2002). Two of these proteins catalyze the final two steps in the synthesis of the phytosiderophore hydroxymugineic acid (HMA), while the third is involved in the Yang (Met) cycle, which provides S-adenosyl Met (SAM) for HMA production (Mori and Nishizawa, 1987). A possible role for this compound in B tolerance is discussed.
RESULTS
Identification of Proteins from the Leaves of ‘Golden Promise’ Barley Plants
Initially, we examined the variation of the iTRAQ system by comparing the abundances of proteins isolated from the leaves of replicate ‘Golden Promise’ plants. These samples were independently isolated, digested, and labeled with iTRAQ tags mass-to-charge ratio (m/z) 114 and m/z 115. The pools of differentially labeled peptides were combined, fractionated, and analyzed by electrospray ionization (ESI)-MS/MS. A total of 641 peptides were identified after searching the MS/MS spectra against a six-frame translation of the barley gene indices The Institute for Genomic Research (TIGR) database (V9.0). The complete data set is presented in Supplemental Table S1.
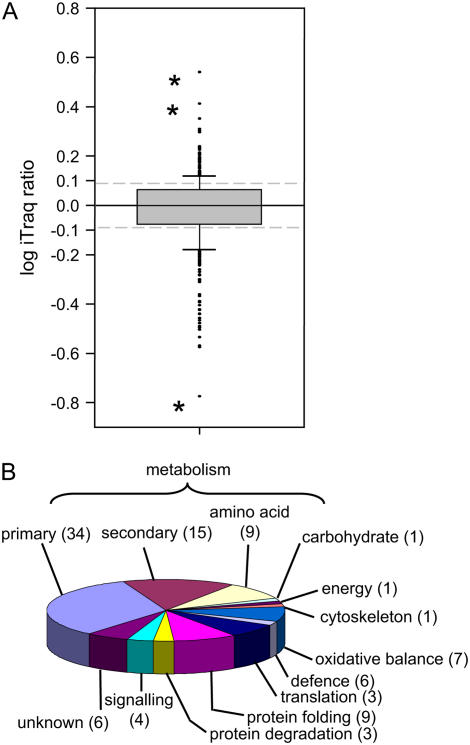
This experiment resulted in the identification of 138 unique proteins, which are listed in Table I. Functional classification of these proteins demonstrated that they were dominated by proteins involved in metabolism (primary and secondary, 58%; Fig. 2B). Eight proteins directly involved in photosynthetic reactions, large and small subunits of Rubisco, two Rubisco activase isoforms, two (23 and 33 kD) oxygen-evolving PSII proteins, and two plastocyanins (including a blue copper-binding protein) were identified, highlighting photosynthesis as one of the dominant metabolic processes occurring in green tissue. Proteins involved in translation (five elongation factors and five ribosomal proteins) and protein folding (including seven heat shock proteins) also made up 16% of the identifications, indicating that protein synthesis was also a major process occurring in this tissue. A small percentage of proteins (6%) were classified as unknown due to a lack of a predicted function.
Table I.
List of proteins identified from leaves isolated from ‘Golden Promise’ seedlings
Proteins are ordered according to functional classification. A pie chart based on percentages represented by each functional group is presented in Figure 2B. Bold entries indicate protein also identified in root tissue.
| TIGR Accession | No. of Peptides Defining Group | NCBInr | Annotation | Organism | E Value |
|---|---|---|---|---|---|
| Primary metabolism | |||||
| TC130714 | 8 | gi|125580 | Phosphoribulokinase | Wheat | 0 |
| TC131346 | 7 | gi|62732953 | Fru-bisphosphate aldolase class I | Rice | 0 |
| TC131467 | 3 | gi|34911932 | NADP-specific isocitrate dehydrogenase | Rice | 0 |
| TC131518 | 5 | gi|29367547 | Adenosine kinase-like protein | Rice | 0 |
| TC131556 | 4 | gi|1143500 | ADP-Glc pyrophosphorylase small subunit | Barley | 0 |
| TC132023 | 4 | gi|21741 | Fru-bisphosphatase | Wheat | 0 |
| TC132350 | 4 | gi|2105137 | ADP-Glc pyrophosphorylase large subunit | Barley | 0 |
| TC138581 | 6 | gi|33113259 | Enolase | Rice | 0 |
| TC138582 | 9 | gi|763035 | Glyceraldehyde-3-P dehydrogenase | Zea mays | 0 |
| TC138635 | 13 | gi|108705993 | Glyceraldehyde-3-P dehydrogenase B | Rice | 0 |
| TC138641 | 15 | gi|31087909 | Rubisco large subunit | Barley | 0 |
| TC138666 | 15 | gi|167097 | Ribulose 1,5-bisphosphate carboxylase activase isoform 2 | Barley | 0 |
| TC146737 | 6 | gi|167095 | Ribulose 1,5-bisphosphate carboxylase activase | Barley | 0 |
| TC139062 | 11 | gi|50934283 | Glycolate oxidase | Rice | 0 |
| TC139210 | 4 | gi|1212996 | UDP-Glc pyrophosphorylase | Barley | 0 |
| TC139211 | 5 | gi|50904581 | Hydroxypyruvate reductase | Rice | 0 |
| TC139220 | 3 | gi|77548686 | Pyruvate kinase | Rice | 0 |
| TC139256 | 12 | gi|77554291 | Rubisco subunit binding-protein α-subunit | Rice | 0 |
| TC146378 | 20 | gi|3293043 | Phosphoglycerate kinase | Wheat | 0 |
| TC146528 | 3 | gi|56785335 | Phosphoglycerate mutase | Rice | 0 |
| TC146663 | 12 | gi|28190676 | Transketolase | Rice | 0 |
| TC146784 | 6 | gi|18076790 | Phosphoglucomutase | Wheat | 0 |
| TC146896 | 9 | gi|14265 | Sedoheptulose-1,7-bisphosphatase | Wheat | 0 |
| TC131363 | 1 | gi|18978 | Glyceraldehyde 3-P dehydrogenase | Barley | 0 |
| TC131622 | 3 | gi|76363515 | Fru-1,6-bisphosphatase | Saccharum sp. | 1E-178 |
| TC131870 | 1 | gi|50899346 | Enoyl-acyl carrier protein reductase | Rice | 2E-178 |
| TC131364 | 11 | gi|729003 | Carbonic anhydrase | Barley | 3E-176 |
| TC139061 | 8 | gi|50934283 | Glycolate oxidase | Rice | 1E-172 |
| TC139042 | 5 | gi|21844 | 33-kD oxygen-evolving protein of PSII | Wheat | 5E-170 |
| TC146536 | 4 | gi|50910187 | Glyceraldehyde-3-P dehydrogenase | Rice | 4E-169 |
| TC147698 | 4 | gi|54291349 | PSII stability/assembly factor HCF136 | Rice | 9E-168 |
| TC146289 | 5 | gi|34915204 | Glyoxysomal malate dehydrogenase | Rice | 2E-152 |
| TC138805 | 8 | gi|609262 | Triosephosphate isomerase | Secale cereale | 7E-146 |
| TC147935 | 3 | gi|51535181 | Fructokinase | Rice | 3E-143 |
| TC146529 | 4 | gi|50932771 | Malate dehydrogenase | Rice | 9E-139 |
| TC131409 | 3 | gi|2507469 | Triosephosphate isomerase | Barley | 1E-120 |
| TC131384 | 3 | gi|15240250 | Ribulose-P 3-epimerase | Arabidopsis | 8E-119 |
| TC131383 | 4 | gi|21837 | 23-kD oxygen evolving protein of PSII | Wheat | 6E-118 |
| TC140560 | 4 | gi|50934597 | Rib-5-P isomerase | Rice | 7E-115 |
| TC132198 | 3 | gi|51090360 | Fru-bisphosphate aldolase | Rice | 2E-108 |
| TC138580 | 7 | gi|11990897 | Rubisco small subunit | Wheat | 6E-98 |
| TC132200 | 1 | gi|51090360 | Fru-bisphosphate aldolase | Rice | 1E-77 |
| TC134951 | 1 | gi|56784876 | Phosphoribulokinase/uridine kinase-like | Rice | 4E-76 |
| TC134990 | 1 | gi|18978 | Glyceraldehyde 3-P dehydrogenase | Barley | 1E-56 |
| TC147083 | 1 | gi|37651973 | Blue copper-binding protein | Barley | 1E-54 |
| TC146310 | 1 | gi|431920 | Plastocyanin | Barley | 9E-51 |
| TC139192 | 1 | gi|431920 | Plastocyanin | Barley | 3E-50 |
| Secondary metabolism | |||||
| TC130720 | 3 | gi|68655441 | AdoMet synthase 2 | Barley | 0 |
| TC130859 | 4 | gi|34915052 | Ferredoxin-nitrite reductase | Rice | 0 |
| TC131827 | 3 | gi|50082771 | Hydroxymethylbutenyl 4-diphosphate synthase | Z. mays | 0 |
| TC139066 | 4 | gi|417745 | Adenosylhomocysteinase | Wheat | 0 |
| TC139106 | 4 | gi|52353541 | Ketol-acid reductoisomerase | Rice | 0 |
| TC139229 | 3 | gi|1705612 | Catalase isozyme 1 | Barley | 0 |
| TC146718 | 4 | gi|2493543 | Catalase 1 | Wheat | 0 |
| TC139584 | 3 | gi|52077207 | Monodehydroascorbate reductase | Rice | 0 |
| TC139836 | 4 | gi|34894800 | Dihydrolipoamide dehydrogenase | Rice | 0 |
| TC146253 | 7 | gi|50947367 | Putative aminotransferase | Rice | 0 |
| TC131070 | 3 | gi|34911282 | Guanine nucleotide-binding protein β-subunit-like protein | Rice | 3E-170 |
| TC138583 | 3 | gi|32352138 | Thiamine biosynthetic enzyme | Rice | 7E-167 |
| TC132431 | 1 | gi|19849543 | Porphobilinogen deaminase | Wheat | 8E-166 |
| TC139562 | 1 | gi|55233175 | β-Cyano-Ala synthase | Rice | 3E-159 |
| TC133238 | 3 | gi|7619802 | Putative glyoxalase I | Wheat | 6E-158 |
| TC131549 | 2 | gi|3688398 | Ascorbate peroxidase | Barley | 1E-141 |
| TC139685 | 3 | gi|50909553 | γ Hydroxybutyrate dehydrogenase | Rice | 2E-141 |
| TC132418 | 6 | gi|52076371 | Oxidoreductase like | Rice | 1E-138 |
| TC132249 | 7 | gi|50945155 | Oxidoreductase, Zn binding | Rice | 3E-132 |
| TC146831 | 8 | gi|15808779 | Ascorbate peroxidase | Barley | 8E-128 |
| TC146265 | 4 | gi|28059599 | Thylakoid lumenal 17.4-kD protein | Arabidopsis | 4E-52 |
| Amino acid metabolism | |||||
| TC130708 | 7 | gi|2565305 | Gly decarboxylase P subunit | Triticeae | 0 |
| TC131380 | 6 | gi|68655495 | METS1 enzyme | Barley | 0 |
| TC131397 | 9 | gi|50918513 | Gly hydroxymethyltransferase | Rice | 0 |
| TC131957 | 5 | gi|1707878| | Aminomethyltransferase | Solanum tuberosum | 0 |
| TC139279 | 10 | gi|50510140 | Ferredoxin-dependent Glu synthase | Rice | 0 |
| TC139283 | 4 | gi|71362640 | Plastid Gln synthetase isoform GS2c | Wheat | 0 |
| TC139989 | 4 | gi|37703720 | Aminotransferase AGD2 | Rice | 0 |
| TC140047 | 3 | gi|633095 | Plastidic Asp aminotransferase | Panicum miliaceum | 0 |
| TC146244 | 11 | gi|50510015 | Ala aminotransferase | Rice | 0 |
| TC147191 | 5 | gi|50915564 | Leu aminopeptidase | Rice | 0 |
| TC147233 | 7 | gi|1170029 | Glu-1-semialdehyde 2,1-aminomutase | Barley | 0 |
| TC146634 | 4 | gi|585032 | Cys synthase | Wheat | 2E-165 |
| TC132821 | 5 | gi|57899533 | Putative plastidic Cys synthase 1 | Rice | 7E-126 |
| Carbohydrate metabolism | |||||
| TC132929 | 1 | gi|18025340 | α-l-Arabinofuranosidase/β-d-xylosidase isoenzyme ARA-I | Barley | 0 |
| TC130915 | 1 | gi|3037080 | Glucan endo-1,3-β-glucosidase isoenzyme I | Barley | 2E-160 |
| Energy | |||||
| TC130729 | 4 | gi|525291 | ATP synthase β-subunit | Wheat | 0 |
| TC148629 | 1 | gi|11583 | ATPase, β-subunit | Barley | 5E-60 |
| Cytoskeleton | |||||
| TC131417 | 3 | gi|108864035 | Actin-7 | Rice | 0 |
| TC146790 | 3 | gi|1709779 | Profilin-1 | Barley | 2E-61 |
| Oxidative balance | |||||
| TC131399 | 1 | gi|20302473 | Ferredoxin-NADP(H) oxidoreductase | Wheat | 0 |
| TC131398 | 3 | gi|20302471 | Ferredoxin-NADP(H) oxidoreductase | Wheat | 1E-167 |
| TC130797 | 6 | gi|34901636 | Thioredoxin-like protein CDSP32 | Rice | 8E-111 |
| TC130826 | 4 | gi|3328221 | Thioredoxin peroxidase | S. cereale | 2E-109 |
| TC131780 | 1 | gi|6179600 | GPX12Hv, glutathione peroxidase-like protein | Barley | 2E-97 |
| TC146933 | 4 | gi|4138592 | Thioredoxin M | Wheat | 6E-93 |
| TC132207 | 3 | gi|55833012 | Peroxiredoxin Q | Wheat | 4E-90 |
| TC133526 | 1 | gi|51535721 | Thioredoxin peroxidase 1 | Rice | 1E-79 |
| TC131676 | 4 | gi|1572627 | Copper/Zn superoxide dismutase | Wheat | 2E-78 |
| TC146752 | 3 | gi|108708142 | Superoxide dismutase 1 | Rice | 4E-74 |
| Defense | |||||
| TC138584 | 4 | gi|50909007 | Putative elongation factor 2 | Rice | 0 |
| TC146252 | 7 | gi|2119927 | Translation elongation factor EF-G | Glycine max | 0 |
| TC146566 | 3 | gi|949878 | Elongation factor 1-α | Barley | 0 |
| TC146710 | 3 | gi|50906401 | Elongation factor 1-γ | Rice | 1E-178 |
| TC133131 | 1 | gi|3550485 | cp33Hv | Barley | 1E-149 |
| TC140393 | 5 | gi|56682582 | Thaumatin-like protein TLP5 | Barley | 3E-130 |
| TC147110 | 3 | gi|77556660 | Elongation factor TS family protein | Rice | 1E-126 |
| TC139502 | 5 | gi|3550483 | cp31BHv | Barley | 4E-126 |
| TC148742 | 4 | gi|77556660 | Elongation factor TS family protein | Rice | 6E-48 |
| Translation | |||||
| TC131505 | 5 | gi|14017610 | Ribosomal protein S3 | Wheat | 6E-132 |
| TC147671 | 1 | gi|50905143 | Putative 50S ribosomal protein L3 | Rice | 4E-111 |
| TC131976 | 3 | gi|50917085 | Ribosomal protein | Rice | 3E-98 |
| TC131434 | 4 | gi|968902 | Ribosomal protein S8 | Rice | 1E-84 |
| Protein folding | Rice | ||||
| TC131557 | 9 | gi|77554415 | 70-kD heat shock-related protein | Rice | 0 |
| TC131558 | 9 | gi|92870233 | Heat shock protein Hsp70 | Medicago truncatula | 0 |
| TC138914 | 5 | gi|77552703 | Heat shock cognate 70-kD protein | Rice | 0 |
| TC138915 | 4 | gi|108707463 | Heat shock cognate 70-kD protein | Rice | 0 |
| TC139132 | 8 | gi|34897924 | Chaperonin 60 β | Rice | 0 |
| TC139483 | 3 | gi|556673 | Heat shock protein | S. cereale | 0 |
| TC139572 | 3 | gi|110289207 | Chaperonin CPN60-1 | Rice | 0 |
| TC132470 | 1 | gi|50945195 | Peptidyl-prolyl cis-trans isomerase | Rice | 0 |
| TC146697 | 1 | gi|34897236 | 60S ribosomal protein L1 | Rice | 3E-174 |
| TC146605 | 3 | gi|13925734 | Cyclophilin A-2 | Wheat | 1E-88 |
| TC138916 | 1 | gi|59799993 | Heat shock protein 70 | Z. mays | 7E-85 |
| TC135924 | 1 | gi|50948109 | Immunophilin/FKBP-type peptidyl-prolyl cis-trans isomerase | Rice | 1E-51 |
| Protein degradation | |||||
| TC131728 | 6 | gi|399213 | ATP-dependent protease ATP-binding subunit | Lycopersicon esculentum | 0 |
| TC141753 | 1 | gi|22331173 | ATPREP1/ATZNMP; metalloendopeptidase | Arabidopsis | 1E-139 |
| TC146981 | 1 | gi|11967891 | 20S proteasome α-subunit | Z. mays | 7E-130 |
| TC139286 | 3 | gi|1323748 | Thiol protease | Wheat | 1E-86 |
| Signaling | |||||
| TC146758 | 8 | gi|108862567 | RNA-binding protein | Rice | 0 |
| TC147198 | 12 | gi|50910077 | Translational elongation factor Tu | Rice | 0 |
| TC133717 | 11 | gi|50935225 | Putative mRNA-binding protein precursor | Rice | 1E-139 |
| TC132022 | 1 | gi|34913270 | 29-kD ribonucleoprotein A | Rice | 5E-88 |
| TC138855 | 1 | |dbj|BAA02436.1| | Elongation factor 1 β | Wheat | 7E-70 |
| Unknown | |||||
| TC139280 | 7 | gi|2072727 | Fd-GOGAT protein | Rice | 0 |
| TC139914 | 1 | gi|15235282 | Amino acid-binding/oxidoreductase | Arabidopsis | 0 |
| TC146506 | 3 | gi|50925621 | OSJNBa0084K20.14 | Rice | 3E-114 |
| TC141769 | 1 | gi|54290425 | Unknown protein | Rice | 2E-101 |
| TC131703 | 3 | gi|18394414 | Unknown protein | Arabidopsis | 1E-79 |
| TC150479 | 1 | gi|50947401 | Unknown protein | Rice | 2E-72 |
| TC148625 | 5 | gi|50928389 | OSJNBa0086O06.22 | Rice | 3E-60 |
| TC139271 | 1 | gi|19087 | Unnamed protein product | Barley | 9E-43 |
Figure 2.
Relative abundance and identity of proteins identified from the leaves of barley ‘Golden Promise’ plants. A, Box plot showing distribution of log10-transformed iTRAQ peptide ratios derived from proteins isolated from the leaves of two identical pools of barley (‘Golden Promise’) plants. All 480 peptides with iTRAQ ratios are represented in this segment. Box defines the 25th and 75th percentiles of the population. Error bars define 10th and 90th percentiles of the population. Asterisks indicate extreme outlying values discussed in text. B, Pie chart showing the functional classification of the 138 unique proteins identified in this analysis. Numbers in brackets indicate the percentage of proteins within this category. [See online article for color version of this figure.]
The distribution of numbers of peptides defining a family is shown in Supplemental Figure S1 (white bars). Over 75% of the protein matches included at least three peptides with two proteins, phosphoglycerate kinase, and the large subunit of Rubisco, defined by a large number of peptides (20 and 15, respectively; Table I).
Defining the Variation within the Experimental System
The second aspect of this analysis was the comparison of relative peptide abundances between the two pools of proteins isolated from the replicate plants. iTRAQ ratios were collected for 480 of the 641 peptides (74%). The distribution of these iTRAQ ratios is shown in Figure 2A, with peak area data shown in Supplemental Figure S2. Over 50% of the peptides displayed a variation between pools of less than 0.25-fold from the median (black box), and 80% of the ratios deviated less than 0.5-fold from the median (Fig. 2A). Of the 480 peptides, 19 (3.96%) had ratios that were more than 2.5-fold different between the replicate samples.
Three extreme outliers were apparent in this data set (Fig. 2A, asterisks). The two peptides with the largest ratios of 0.53 and 0.41 (indicating an increase in peptide abundance of 3.38 and 2.57, respectively, in one sample) were derived from the small and large subunits of Rubisco, respectively. Both subunits had multiple peptides with average iTRAQ ratios, excluding the outlying values of 0.09 (small subunit, n = 5) and 0 (large subunit, n = 14). The peptide with the lowest ratio of −0.77 was derived from Met synthase 1 (METS1); three other peptides from the same protein had an average iTRAQ ratio of −0.21. There were 16 other peptides with ratios between −0.4 and −0.6; in each of these cases, the average ratio of the other peptides that matched to the same proteins deviated by less than 0.2 units from zero (i.e. less than 1.5-fold difference). Based on this information, we decided that peptides with ratios of at least 0.4 units either side of zero (i.e. 2.5-fold difference between samples) would be selected for further examination in the subsequent analysis.
Selection of Lines Containing 4H/6H B-Tolerance Loci from the ‘Clipper’ × ‘Sahara’ Doubled-Haploid Population
In the B-tolerant barley ‘Sahara,’ four distinct QTL have been described that are involved in contributing to B tolerance (Jeffries et al., 1999). Two of these QTL, located on chromosomes 4H and 6H, are both linked to a reduction in B uptake. Reduced B uptake appears to be a constitutive trait, with ‘Sahara’ plants also accumulating less B at low concentrations of B (Nable et al., 1990).
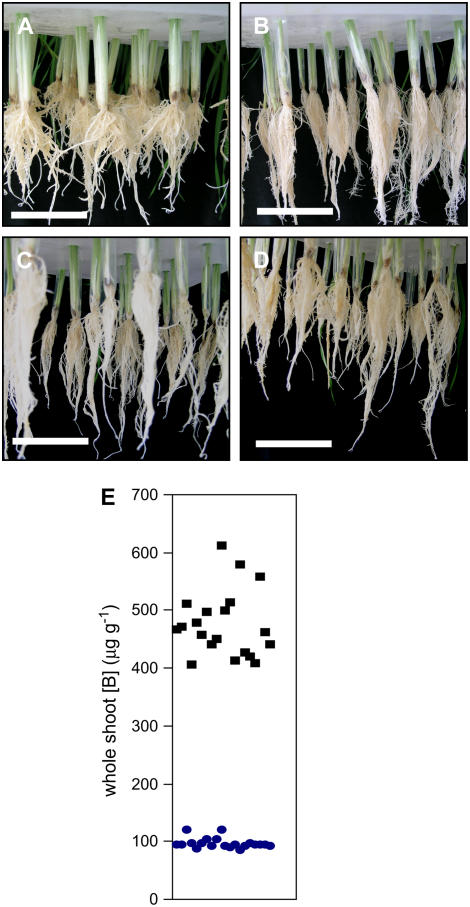
The QTL mapping was performed using a doubled-haploid (DH) population, created from parental ‘Sahara’ and the B-intolerant ‘Clipper.’ These parental lines are distantly related and display distinct growth habits (Roessner et al., 2006). Specifically looking at root morphology, which is the site of B uptake, the ‘Sahara’ plants have distinctly shorter and thicker roots than the elongated, thin roots of the ‘Clipper’ plants (compare Fig. 3, A and B).
Figure 3.
Photographs of roots from 2-week-old hydroponically grown plants. A, Barley ‘Sahara.’ B, Barley ‘Clipper.’ C, Representative plants from B-tolerant bulks. D, Representative plants from B-intolerant bulks. E, B accumulation in whole shoots for individual DH lines used in this study. Square symbols, intolerant lines; circles, tolerant lines. Each data point represents the whole shoot B concentration (μg g−1) for an individual DH line used in the bulked segregant analysis, measured as described in Jeffries et al. (1999) and reproduced with permission from Jeffries (2000). Bar = 10 cm. [See online article for color version of this figure.]
A comparative analysis specifically examining B tolerance would be compounded by the large varietal variation between ‘Clipper’ and ‘Sahara.’ To circumvent this, we have adopted a bulked segregant approach (Michelmore et al., 1991), exploiting the availability of a population of 150 DH lines created from crossing the B-tolerant landrace ‘Sahara 3771’ and the intolerant, improved ‘Clipper’ (Karakousis et al., 2003).
In this study, we selected two pools of plants from the DH population, each composed of 20 lines. The lines in each pool were chosen on the basis of a presence or absence of both the 4H and 6H tolerance loci. Coincident with this genotypic segregation, these lines also segregated on the basis of leaf B levels after growth in elevated levels of B (Fig. 3E; Jefferies, 2000). In terms of root morphology, these two pools of plants were very similar, with a phenotype intermediate between both the parents (compare Fig. 3, C and D).
iTRAQ Comparison of Proteins Isolated from the Roots of B-Tolerant and B-Intolerant Plants
Entire root systems from the B-tolerant and B-intolerant pools of DH plants, grown at a nontoxic concentration of B (50 μm), were harvested and homogenized. Due to the aforementioned constitutive nature of the B exclusion trait, plants were grown in a nontoxic concentration of B to minimize identification of B toxicity-responsive proteins in the intolerant plants. After centrifugation of the homogenate, soluble proteins were collected in the supernatant. Both pools of proteins were digested with trypsin, and the resultant peptides were tagged with iTRAQ tags m/z 114 (B intolerant) and m/z 115 (B tolerant). A total of 1,225 peptides were identified during the comparison of the two pools of tolerant and intolerant plants. Reporter ion peak areas were collected for 1,038 of the 1,225 peptides (84%) and are presented as a box plot in Figure 4A, while relative peak area values are shown in Supplemental Figure S3. The complete set of peptides and their assignments is included in Supplemental Table S2.
Figure 4.
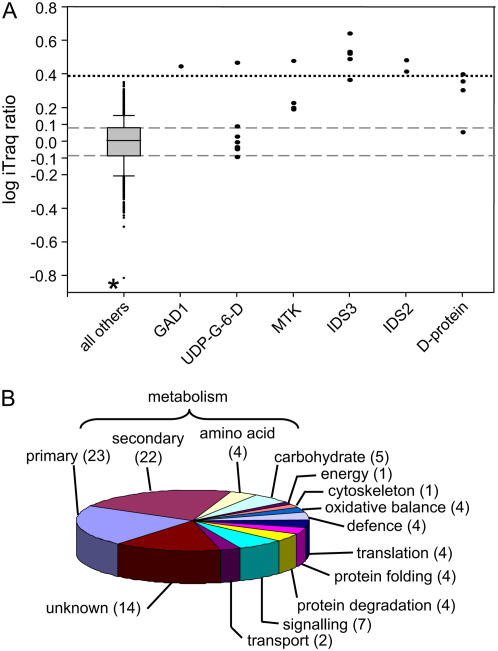
Relative abundance and identity of proteins identified in B-tolerant and B-intolerant plants. A, Box plots showing distribution of log-transformed iTRAQ peptide ratios derived from proteins isolated from the roots of B-tolerant and B-intolerant bulked segregants. All 1,038 peptides with iTRAQ ratios are represented in this segment. Positive values (in segment B) indicate an increase in peptide/protein abundance in the B-tolerant plants. Box defines the 25th and 75th percentiles of the population. Error bars define 10th and 90th percentiles of the population. Asterisk indicates extreme outlying values discussed in text. Dashed lines indicate values less than 0.25-fold different between samples. Dotted line indicates 2.5-fold significant difference between samples. GAD1, TC131033; UDP-G-6-D, UDP-Glc-6-dehydrogenase (TC138934); MTK, TC140103; IDS3, TC142112; IDS2 (TC137786); D-protein (TC134738). B, Pie chart showing the functional classification of the 341 proteins identified in this analysis. Numbers in brackets indicate the percentage of proteins that are present in each category. [See online article for color version of this figure.]
A total of 341 proteins were identified in this experiment (Table II). The proteins were classified according to predicted function, and these classifications are displayed in Figure 4B. Over one-half (54%) of the proteins were involved in metabolic functions, including 19 proteins involved in complex carbohydrate metabolism. Ten distinct proteasome subunits were also identified. From this data set, 50 distinct proteins (representing 15% of the identifications) were identified as having no known function.
Table II.
List of proteins identified from roots isolated from B-tolerant and B-intolerant plants
Proteins are ordered according to functional classification. A pie chart based on percentages represented by each functional group is presented in Figure 4B. Bold entries indicate protein also found in leaf tissue.
| TIGR Accession | No. of Peptides Defining Group | NCBInr | Annotation | Organism | E Value |
|---|---|---|---|---|---|
| Primary metabolism | |||||
| TC147887 | 5 | gi|38141533 | Fructan 1-exohydrolase precursor | Barley | 0 |
| TC147636 | 6 | gi|50900276 | Glc-6-P isomerase | Rice | 0 |
| TC138581 | 14 | gi|90110845 | Enolase (2-phosphoglycerate dehydratase) | Rice | 0 |
| TC147465 | 3 | gi|2182267 | Lipoxygenase | Barley | 0 |
| TC147361 | 4 | gi|34905462 | ATP citrate lyase | Rice | 0 |
| TC147336 | 3 | gi|55847605 | Suc:Suc 1-fructosyltransferase | Wheat | 0 |
| TC147154 | 3 | gi|57900129 | Transaldolase | Rice | 0 |
| TC131363 | 7 | gi|18978 | Glyceraldehyde 3-P dehydrogenase | Barley | 0 |
| TC146895 | 14 | gi|3341490 | Phosphoenolpyruvate carboxylase | Wheat | 0 |
| TC146849 | 6 | gi|34897872 | Phosphogluconate dehydrogenase | Rice | 0 |
| TC146784 | 8 | gi|18076790 | Phosphoglucomutase | Wheat | 0 |
| TC146767 | 3 | gi|50947901 | Diphosphonucleotide phosphatase | Rice | 0 |
| TC146663 | 4 | gi|28190676 | Transketolase | Rice | 0 |
| TC146528 | 17 | gi|56785335 | Phosphoglycerate mutase | Rice | 0 |
| TC146369 | 10 | gi|129916 | Phosphoglycerate kinase | Wheat | 0 |
| TC146300 | 4 | gi|46358940 | Vacuolar invertase1 | Triticum monococcum | 0 |
| TC139173 | 14 | gi|2429087 | Lipoxygenase 2 | Barley | 0 |
| TC139815 | 6 | gi|50912091 | Diphosphate-Fru-6-P 1-phosphotransferase | Rice | 0 |
| TC139326 | 3 | gi|52076758 | Pyrophosphate-dependent phosphofructokinase | Rice | 0 |
| TC139308 | 7 | gi|37694731 | NADP malic enzyme | Rice | 0 |
| TC139301 | 7 | gi|51091407 | Aldehyde dehydrogenase | Rice | 0 |
| TC139220 | 8 | gi|77548686 | Pyruvate kinase | Rice | 0 |
| TC139210 | 6 | gi|1212996 | UDP-Glc pyrophosphorylase | Barley | 0 |
| TC139089 | 5 | gi|94717590 | GDP-Man 3,5-epimerase 2 | Rice | 0 |
| TC138932 | 7 | gi|77554940 | UDP-Glc 6-dehydrogenase | Rice | 0 |
| TC133156 | 5 | gi|4158230 | Amylogenin | Wheat | 0 |
| TC132208 | 5 | gi|50251801 | Phosphoenolpyruvate carboxylase | Rice | 0 |
| TC131693 | 3 | gi|91694275 | Glc-6-P isomerase | Barley | 0 |
| TC131518 | 8 | gi|29367547 | Adenosine kinase-like protein | Rice | 0 |
| TC131467 | 10 | gi|34911932 | NADP-specific isocitrate dehydrogenase | Rice | 0 |
| TC131388 | 12 | gi|88687741 | Suc synthase | Lolium perenne | 0 |
| TC149390 | 1 | gi|49388286 | Acetyl-CoA synthetase | Rice | 0 |
| TC148243 | 1 | gi|34897074 | Inosine monophosphate dehydrogenase | Rice | 0 |
| TC147295 | 1 | gi|50920285 | Rubisco subunit binding-protein α-subunit precursor | Rice | 0 |
| TC146860 | 1 | gi|21263612 | Formate dehydrogenase, mitochondrial precursor | Barley | 0 |
| TC139625 | 2 | gi|77549262 | Pyruvate kinase | Rice | 0 |
| TC139350 | 2 | gi|8918502 | Glc-6-P dehydrogenase | Rice | 0 |
| TC139062 | 1 | gi|50934283 | Glycolate oxidase | Rice | 0 |
| TC134291 | 1 | gi|52077150 | Fru/tagatose bisphosphate aldolase | Rice | 0 |
| TC132727 | 1 | gi|6685803 | Adenylosuccinate synthetase | Wheat | 0 |
| TC132006 | 2 | gi|50912579 | Pyruvate dehydrogenase E1 α-subunit | Rice | 0 |
| TC131953 | 1 | gi|18844860 | dTDP-Glc 4,6-dehydratase | Rice | 0 |
| TC131782 | 1 | gi|34906844 | Phospho-2-dehydro-3-deoxyheptonate aldolase 1 | Rice | 0 |
| TC131346 | 1 | gi|62732953 | Fru-bisphosphate aldolase class I | Rice | 0 |
| TC130807 | 7 | gi|50948183 | Sorbitol dehydrogenase | Rice | 2E-177 |
| TC131237 | 1 | gi|62130764 | Hexokinase 6 | Rice | 4E-175 |
| TC147925 | 1 | gi|27817922 | Transaldolase ToTAL2 | Rice | 2E-172 |
| TC146609 | 6 | gi|18202485 | Malate dehydrogenase | Z. mays | 8E-171 |
| TC131070 | 8 | gi|1346109 | Guanine nucleotide-binding protein β-subunit | Rice | 3E-170 |
| TC139687 | 8 | gi|34907582 | Fructokinase I | Rice | 7E-164 |
| TC147014 | 11 | gi|50940457 | Fructokinase | Rice | 2E-163 |
| TC147086 | 6 | gi|28950668 | Legumin-like protein | Z. mays | 3E-163 |
| TC147455 | 3 | gi|81686712 | Glu dehydrogenase 2 | Rice | 1E-160 |
| TC146537 | 1 | gi|34911788 | Malate dehydrogenase | Rice | 1E-152 |
| TC138805 | 5 | gi|609262 | Triosephosphate isomerase | S. cereale | 7E-146 |
| TC132870 | 3 | gi|108706464 | Hydrolase, carbon-nitrogen family protein | Rice | 1E-142 |
| TC147359 | 3 | gi|34905462 | ATP citrate lyase | Rice | 4E-141 |
| TC146529 | 3 | gi|50932771 | Malate dehydrogenase | Rice | 9E-139 |
| TC146498 | 3 | gi|729003 | Carbonic anhydrase | Barley | 3E-136 |
| TC147237 | 1 | gi|17385742 | d-Isomer specific 2-hydroxyacid dehydrogenase | Rice | 2E-125 |
| TC131409 | 8 | gi|2507469 | Triosephosphate isomerase | Barley | 1E-120 |
| TC139245 | 4 | gi|28192421 | Dehydroascorbate reductase | Wheat | 2E-112 |
| TC146774 | 4 | gi|23504745 | Glutathione transferase F5 | Wheat | 3E-106 |
| TC132874 | 4 | gi|50941197 | Monodehydroascorbate reductase | Rice | 2E-101 |
| TC139680 | 4 | gi|9711921 | Adenine phosphoribosyltransferase | Barley | 2E-96 |
| TC133105 | 1 | gi|38426301 | 6-Phosphogluconate dehydrogenase | Rice | 2E-86 |
| TC139131 | 3 | gi|9652119 | Nucleoside diphosphate kinase | L. perenne | 6E-79 |
| TC150725 | 1 | gi|55773816 | Ribokinase like | Rice | 7E-76 |
| TC148941 | 1 | gi|50917631 | Deoxyuridine triphosphatase | Rice | 3E-72 |
| TC146554 | 11 | gi|18496065 | Fru 1-,6-biphosphate aldolase | Wheat | 5E-68 |
| TC144012 | 3 | gi|5052007 | Apoplastic invertase | Rice | 4E-59 |
| BG309197 | 1 | gi|50904517 | UMP synthase 1 | Rice | 4E-55 |
| TC147083 | 3 | gi|37651973 | Blue copper-binding protein | Barley | 1E-54 |
| TC133349 | 1 | gi|37651973 | Blue copper-binding protein | Barley | 5E-40 |
| TC147221 | 1 | gi|6007803 | d-Ribulose-5-P 3-epimerase | Rice | 7E-40 |
| CA019619 | 1 | gi|52077150 | Fru/tagatose bisphosphate aldolase | Rice | 1E-36 |
| TC142991 | 1 | gi|37730876 | Legumin-like protein | Z. mays | 1E-26 |
| TC144813 | 1 | gi|50919645 | AMP-binding protein | Rice | 1E-13 |
| Secondary metabolism | |||||
| TC147448 | 5 | gi|4566505 | β-d-Glucan exohydrolase isoenzyme ExoI | Barley | 0 |
| TC147311 | 3 | gi|50401177 | Met S-methyltransferase | Rice | 0 |
| TC147191 | 10 | gi|50915564 | Leu aminopeptidase | Rice | 0 |
| TC147167 | 4 | gi|12407304 | IDI2 | Barley | 0 |
| TC146955 | 9 | gi|2506825 | Lipoxygenase 1 | Barley | 0 |
| TC146875 | 11 | gi|50941891 | Aconitate hydratase | Rice | 0 |
| TC146792 | 9 | gi|18904 | Aspartic proteinase | Barley | 0 |
| TC146761 | 3 | gi|17887465 | Phosphoethanolamine methyltransferase | Wheat | 0 |
| TC142112 | 5 | gi|9711238 | IDS3 | Barley | 0 |
| TC141775 | 4 | gi|34912652 | Acetyl transferase | Rice | 0 |
| TC141288 | 3 | gi|50919455 | Leukotriene A-4 hydrolase | Rice | 0 |
| TC140255 | 4 | gi|50906357 | Aminopeptidase M | Rice | 0 |
| TC140156 | 4 | gi|34906304 | Taxadien-5-α-ol O-acetyltransferase | Rice | 0 |
| TC140103 | 4 | gi|46883147 | MTK | Rice | 0 |
| TC139584 | 6 | gi|52077207 | Monodehydroascorbate reductase | Rice | 0 |
| TC139517 | 3 | gi|73913047 | Δ-1-Pyrroline-5-carboxylate dehydrogenase | Barley | 0 |
| TC139434 | 5 | gi|322833 | Glu-ammonia ligase | Barley | 0 |
| TC139408 | 4 | gi|92429669 | Aconitate hydratase 1 | Sorghum bicolor | 0 |
| TC139106 | 3 | gi|52353541 | Ketol-acid reductoisomerase | Rice | 0 |
| TC138810 | 17 | gi|50910709 | Phe ammonia lyase | Rice | 0 |
| TC137786 | 5 | gi|285634 | IDS2 | Barley | 0 |
| TC131815 | 3 | gi|50948547 | Aminopeptidase N | Rice | 0 |
| TC131783 | 6 | gi|34334010 | Cytosolic glutathione reductase | T. monococcum | 0 |
| TC131701 | 9 | gi|50919385 | Methylenetetrahydrofolate reductase | Rice | 0 |
| TC131524 | 7 | gi|15236375 | Gly hydroxymethyltransferase | Arabidopsis | 0 |
| TC131046 | 5 | gi|68655435 | AdoMet synthase 1 | Barley | 0 |
| TC131477 | 5 | gi|50659026 | UDP-d-glucuronate decarboxylase | Barley | 0 |
| TC131451 | 3 | gi|50915842 | Alcohol dehydrogenase class III | Rice | 0 |
| TC147775 | 1 | gi|63021727 | 12-Oxo-phytodienoic acid reductase | Z. mays | 0 |
| TC147691 | 1 | gi|54290767 | Dehydroquinate dehydratase | Rice | 0 |
| TC147596 | 2 | gi|60686892 | Δ 1-Pyrroline-5-carboxylate synthetase | Wheat | 0 |
| TC147044 | 2 | gi|32400295 | Hydroxyanthranilate hydroxycinnamoyltransferase 3 | Avena sativa | 0 |
| TC146990 | 1 | gi|77551313 | Metallopeptidase family M24-containing protein | Rice | 0 |
| TC141625 | 1 | gi|45510867 | N-myristoyl transferase | Wheat | 0 |
| TC139989 | 1 | gi|37703720 | Aminotransferase AGD2 | Rice | 0 |
| TC139595 | 1 | gi|15238398 | Oxysterol binding (Arabidopsis) | Arabidopsis | 0 |
| TC139279 | 1 | gi|50510140 | Ferredoxin-dependent Glu synthase | Rice | 0 |
| TC132714 | 1 | gi|77554110 | Aspartyl aminopeptidase | Rice | 0 |
| TC132209 | 1 | gi|77556036 | Metalloenzyme superfamily | Rice | 0 |
| TC132132 | 1 | gi|108707229 | Chorismate synthase 2, chloroplast precursor | Rice | 0 |
| TC131599 | 1 | gi|50915896 | 3-Ketoacyl-CoA thiolase; acetyl-CoA acyltransferase | Rice | 0 |
| TC130859 | 1 | gi|34915052 | Ferredoxin-nitrite reductase | Rice | 0 |
| TC139402 | 5 | gi|50899020 | Acetyl-CoA C-acyltransferase | Rice | 1E-179 |
| TC132326 | 1 | gi|50913253 | 1-Aminocyclopropane-1-carboxylate deaminase | Rice | 3E-179 |
| TC142387 | 1 | gi|29466964 | Secretory acid phosphatase precursor | Rice | 1E-178 |
| TC132270 | 1 | gi|108711425 | Ferredoxin-NADP reductase, root isozyme | Rice | 3E-173 |
| TC132684 | 1 | gi|34912654 | Acetyl transferase | Rice | 2E-171 |
| TC141237 | 5 | gi|27531337 | O-methyltransferase | Barley | 8E-168 |
| TC131211 | 5 | gi|50941905 | Glyoxalase I | Rice | 7E-156 |
| TC131671 | 1 | gi|50932765 | Lipase | Rice | 6E-155 |
| TC140063 | 3 | gi|45735967 | 41-kD chloroplast nucleoid DNA-binding protein | Rice | 2E-153 |
| TC131287 | 1 | gi|52077048 | Molybdenum cofactor sulfurase protein like | Rice | 1E-147 |
| TC141301 | 1 | gi|50912077 | NADPH-thioredoxin reductase | Rice | 3E-142 |
| TC139685 | 5 | gi|50909553 | γ Hydroxybutyrate dehydrogenase | Rice | 2E-141 |
| TC139567 | 3 | gi|57900400 | S-formylglutathione hydrolase | Rice | 1E-137 |
| TC146961 | 3 | gi|62734422 | O-methyltransferase | Rice | 2E-132 |
| TC133095 | 6 | gi|62734422 | O-methyltransferase | Rice | 1E-129 |
| TC130741 | 2 | gi|50915968 | Fibrillarin | Rice | 2E-128 |
| TC146831 | 8 | gi|15808779 | Ascorbate peroxidase | Barley | 8E-128 |
| TC146925 | 1 | gi|108706322 | 1,2-Dihydroxy-3-keto-5-methylthiopentene dioxygenase | Rice | 2E-110 |
| TC140546 | 4 | gi|34897892 | Methylthioadenosine | Rice | 2E-109 |
| TC139390 | 3 | gi|34909214 | ADP-ribosylation factor | Rice | 3E-101 |
| TC146548 | 1 | gi|50947279 | Caffeoyl-CoA O-methyltransferase 1 | Rice | 1E-100 |
| TC147322 | 4 | gi|53749369 | 1,4-Benzoquinone reductase | Rice | 2E-99 |
| TC130725 | 2 | gi|32401384 | Cyclophilin | Wheat | 2E-92 |
| TC151803 | 2 | gi|21212950 | Glutathione-S-transferase, I subunit | Barley | 7E-89 |
| TC132414 | 1 | gi|50913035 | S-adenosyl-methionine methyltransferase | Rice | 1E-88 |
| TC150875 | 3 | gi|50916004 | O-diphenol-O-methyl transferase | Rice | 3E-85 |
| TC147986 | 3 | gi|22022398 | Glutathione-S-transferase Cla47 | Wheat | 1E-65 |
| TC144930 | 1 | gi|22202676 | Dioxygenase extradiol | Rice | 6E-65 |
| TC146383 | 5 | gi|51536102 | Formate-tetrahydrofolate ligase | Rice | 9E-58 |
| TC131587 | 1 | gi|54111525 | Immunophilin | Z. mays | 2E-52 |
| TC147423 | 1 | gi|50940931 | Blue copper-binding protein | Rice | 6E-39 |
| TC143987 | 1 | gi|50916927 | Oxidoreductase | Rice | 2E-33 |
| TC137024 | 3 | gi|21593610 | Globulin-like protein | Arabidopsis | 1E-21 |
| BQ471723 | 1 | gi|63021725 | 12-Oxo-phytodienoic acid reductase | Z. mays | 3E-18 |
| Amino acid metabolism | |||||
| TC139066 | 10 | gi|417745 | Adenosylhomocysteinase | Wheat | 0 |
| TC131380 | 22 | gi|68655495 | METS1 enzyme | Barley | 0 |
| TC130910 | 5 | gi|89511843 | Asp aminotransferase | Barley | 0 |
| TC130906 | 6 | gi|57900353 | Asp aminotransferase | Rice | 0 |
| TC130774 | 3 | gi|50540685 | GAD | Rice | 0 |
| TC147620 | 1 | gi|56784224 | Asp aminotransferase | Rice | 0 |
| TC140390 | 1 | gi|50937181 | β-Ala synthases | Rice | 0 |
| TC140047 | 1 | gi|633095 | Plastidic Asp aminotransferase | P. miliaceum | 0 |
| TC146634 | 7 | gi|585032 | Cys synthase | Wheat | 2E-165 |
| TC147456 | 1 | gi|81686712 | Glu dehydrogenase 2 | Rice | 1E-160 |
| TC132821 | 1 | gi|57899533 | Plastidic Cys synthase 1 | Rice | 7E-126 |
| TC146732 | 1 | gi|469148 | Ala aminotransferase | Barley | 4E-81 |
| TC134795 | 1 | gi|108708268 | Branched-chain amino acid aminotransferase | Rice | 4E-48 |
| Carbohydrate metabolism | |||||
| TC133521 | 3 | gi|108864437 | Glycosyl hydrolases family 38 protein, expressed | Rice | 0 |
| TC133163 | 3 | gi|37535638 | α-Galactosidase | Rice | 0 |
| TC133155 | 9 | gi|50899994 | β-Glucan-binding protein | Rice | 0 |
| TC132929 | 5 | gi|18025340 | α-l-Arabinofuranosidase/β-d-xylosidase | Barley | 0 |
| TC132139 | 3 | gi|13398414 | Arabinoxylan arabinofuranohydrolase | Barley | 0 |
| TC131885 | 3 | gi|37535646 | α-Galactosidase preproprotein | Rice | 0 |
| TC150244 | 1 | gi|50510227 | 4-α-Glucanotransferase | Rice | 0 |
| TC133712 | 1 | gi|50510292 | α-Glucosidase II | Rice | 0 |
| TC149802 | 5 | gi|1352328 | Endo-1,3-β-glucosidase | Barley | 9E-174 |
| TC131099 | 1 | gi|295806 | (1-3,1-4)-β-d-Glucanase | Barley | 1E-168 |
| TC135072 | 1 | gi|73622088 | Xylanase inhibitor protein 1 | Wheat | 2E-164 |
| TC130915 | 4 | gi|3037080 | Glucan endo-1,3-β-glucosidase isoenzyme I | Barley | 2E-160 |
| TC130923 | 1 | gi|18865 | Glucan endo-1,3-β-glucosidase | Barley | 1E-155 |
| TC143154 | 1 | gi|50934913 | GlcNAc-P mutase | Rice | 4E-149 |
| TC140649 | 1 | gi|55168332 | β-N-acetylhexosaminidase | Rice | 2E-105 |
| TC147598 | 4 | gi|20160766 | Xylanase inhibitor | Rice | 3E-50 |
| BG300456 | 1 | gi|50938049 | β-1,3-Glucanase | Rice | 4E-48 |
| BF627009 | 1 | gi|55168332 | β-N-acetylhexosaminidase | Rice | 3E-38 |
| Energy | |||||
| TC139468 | 3 | gi|2493132 | ATP synthase B-subunit isoform 2 | Barley | 0 |
| TC139247 | 8 | gi|11527563 | Vacuolar proton-ATPase | Barley | 0 |
| TC130729 | 4 | gi|525291 | ATP synthase β-subunit | Wheat | 0 |
| TC132069 | 1 | gi|50932993 | Vacuolar ATP synthase subunit C | Rice | 2E-175 |
| Cytoskeleton | |||||
| TC146478 | 5 | gi|4165488 | α-Tubulin 3 | Barley | 0 |
| TC132044 | 4 | gi|77548264 | Clathrin heavy chain | Rice | 0 |
| TC131561 | 5 | gi|1743277 | β-Tubulin 1 | Barley | 0 |
| TC133559 | 1 | gi|6094430 | Tubulin α-2 chain | Eleusine indica | 6E-94 |
| TC146790 | 3 | gi|1229169 | Profilin | Barley | 2E-61 |
| Oxidative balance | |||||
| TC140370 | 3 | gi|2759999 | Peroxidase | Barley | 0 |
| TC148287 | 6 | gi|50940483 | Oxidase like | Rice | 5E-174 |
| TC131790 | 1 | gi|57635161 | Peroxidase 8 | T. monococcum | 8E-171 |
| TC139150 | 8 | gi|57635151 | Peroxidase 3 | T. monococcum | 3E-149 |
| TC147991 | 5 | gi|108707054 | NADH-dependent oxidoreductase 1 | Rice | 1E-148 |
| TC148196 | 4 | gi|57635165 | Peroxidase 10 | T. monococcum | 3E-135 |
| TC139337 | 1 | gi|37530466 | Peroxidase | Rice | 1E-129 |
| TC139146 | 1 | gi|55700995 | TPA: class III peroxidase 64 precursor | Rice | 1E-104 |
| TC151783 | 1 | gi|55701007 | TPA: class III peroxidase 70 precursor | Rice | 6E-78 |
| TC146841 | 4 | gi|34911078 | Peroxiredoxin | Rice | 6E-77 |
| TC146754 | 4 | gi|6018682 | Superoxide dismutase-4AP | Z. mays | 1E-73 |
| TC146479 | 5 | gi|32186040 | Thioredoxin h isoform 1; HvTrxh1 | Barley | 8E-62 |
| TC146902 | 1 | gi|32401362 | Glutaredoxin | Wheat | 2E-45 |
| Defense | |||||
| TC139711 | 3 | gi|50938485 | Insulin-degrading enzyme | Rice | 0 |
| TC139653 | 3 | gi|50916138 | Oligopeptidase A like | Rice | 0 |
| TC132290 | 4 | gi|50945443 | Puromycin-sensitive aminopeptidase | Rice | 0 |
| TC131055 | 4 | gi|6682829 | Cys protease | Z. mays | 0 |
| TC147009 | 3 | gi|18146827 | Chitinase 2 | Wheat | 3E-153 |
| TC148826 | 1 | gi|34913680 | DNA-damage-repair/toleration protein DRT102 | Rice | 3E-123 |
| TC147216 | 1 | gi|90959771 | Multidomain cystatin | Rice | 2E-114 |
| TC150881 | 3 | gi|50915254 | Subtilisin-like proteinase | Rice | 4E-98 |
| TC139537 | 1 | gi|3550467 | cp31AHv protein | Barley | 2E-95 |
| TC139845 | 3 | gi|62733218 | Chitinase III C10701 | Rice | 1E-94 |
| TC131676 | 1 | gi|1572627 | Copper/Zn superoxide dismutase | Wheat | 2E-78 |
| TC143082 | 1 | gi|34910862 | Pathogenesis-related protein | Rice | 2E-72 |
| TC140501 | 1 | gi|62861391 | Cold acclimation-induced protein 2-1 | Wheat | 1E-57 |
| TC147802 | 1 | gi|1617121 | Subtilisin-chymotrypsin inhibitor 2 | Barley | 5E-32 |
| Protein translation | |||||
| TC146747 | 12 | gi|37534770 | Endoplasmic reticulum membrane fusion protein | Rice | 0 |
| TC140222 | 3 | gi|50948039 | Glycyl-tRNA synthetase | Rice | 0 |
| TC146725 | 1 | gi|77556802 | 60S ribosomal protein l2 | Rice | 3E-143 |
| TC138849 | 5 | gi|50940807 | 60S acidic ribosomal protein P0 | Rice | 1E-134 |
| TC130710 | 3 | gi|50252099 | Ribosomal protein S4 | Rice | 2E-127 |
| TC138786 | 1 | gi|108862547 | 40S ribosomal protein S3a, expressed | Rice | 1E-119 |
| TC130754 | 1 | gi|50939279 | 40S ribosomal protein | Rice | 2E-119 |
| TC131114 | 1 | gi|57471706 | Ribosomal protein L13a | Wheat | 2E-105 |
| TC146724 | 1 | gi|77551804 | 40S ribosomal protein S9 | Rice | 7E-96 |
| TC139071 | 1 | gi|34893994 | 40S ribosomal protein S5 | Rice | 4E-95 |
| TC139174 | 3 | gi|50911805 | 60S ribosomal protein L12 | Rice | 2E-81 |
| TC146756 | 1 | gi|50934241 | Ribosomal protein S12 | Rice | 3E-65 |
| CV061576 | 1 | gi|56783875 | Acidic ribosomal protein P3a | Rice | 1E-23 |
| Protein folding | |||||
| TC147982 | 3 | gi|34895466 | 66-kD stress protein | Rice | 0 |
| TC147147 | 6 | gi|4056568 | Protein disulfide isomerase-like protein | Z. mays | 0 |
| TC147130 | 5 | gi|476003 | Heat shock protein 70 | Barley | 0 |
| TC146888 | 10 | gi|34906196 | Heat shock protein | Rice | 0 |
| TC146674 | 7 | gi|1709617 | Protein disulfide-isomerase precursor | Barley | 0 |
| TC138926 | 10 | gi|50919489 | Heat shock protein cognate 70 | Rice | 0 |
| TC131381 | 9 | gi|32765549 | Heat shock protein 90 | Barley | 0 |
| TC131558 | 3 | gi|92870233 | Heat shock protein Hsp70 | M. truncatula | 0 |
| TC139542 | 2 | gi|50919217 | TCP-1/cpn60 chaperonin family protein | Rice | 0 |
| TC139525 | 1 | gi|3023751 | 70-kD peptidyl-prolyl isomerase | Wheat | 0 |
| TC139412 | 1 | gi|50913271 | dnaK-type molecular chaperone precursor | Rice | 0 |
| TC146605 | 4 | gi|13925734| | Cyclophilin A-2 | Wheat | 1E-88 |
| Protein degradation | |||||
| TC148201 | 3 | gi|50942477 | Proteasome 26S non-ATPase subunit 1 | Rice | 0 |
| TC131750 | 12 | gi|401237 | Ubiquitin-activating enzyme E1 2 | Wheat | 0 |
| TC131582 | 3 | gi|40643250 | Cathepsin B | Barley | 0 |
| TC147593 | 1 | gi|53792862 | Proteasome activator subunit 4 like | Rice | 0 |
| TC134348 | 1 | gi|50905317 | 26S proteasome regulatory subunit S2 | Rice | 0 |
| TC132495 | 3 | gi|50905317 | 26S proteasome regulatory subunit S2 | Rice | 8E-166 |
| TC146450 | 3 | gi|52548240 | 20S proteasome β 5 subunit | Wheat | 3E-143 |
| TC146981 | 5 | gi|11967891 | 20S proteasome α-subunit | Z. mays | 7E-130 |
| TC131955 | 1 | gi|66271075 | β1 Proteasome-7D | Aegilops tauschii | 6E-125 |
| TC132063 | 4 | gi|1709758 | Proteasome α-subunit type 1 | Rice | 6E-124 |
| TC139363 | 2 | gi50931867 | Proteasome α-subunit type 3 | Rice | 6E-124 |
| TC132038 | 4 | gi|17380182 | Proteasome β-subunit type 1 | Rice | 2E-118 |
| TC148954 | 1 | gi|50918591 | Sec63 domain-containing protein | Rice | 8E-71 |
| TC130753 | 6 | gi|167073 | Ubiquitin | Barley | 3E-63 |
| Signaling | |||||
| TC146926 | 10 | gi|2499708 | Phospholipase D α1 | Z. mays | 0 |
| TC138584 | 14 | gi|50909007 | Elongation factor 2 | Rice | 0 |
| TC132548 | 6 | gi|50909927 | Glycosylphosphatidylinositol-anchored protein | Rice | 0 |
| TC131653 | 3 | gi|62997485 | Protein phosphatase 2A regulatory a-subunit | Z. mays | 0 |
| TC130804 | 5 | gi|53792733 | Eukaryotic initiation factor 4A | Rice | 0 |
| TC149773 | 1 | gi|3023693 | Elongation factor 1-α | Aureobasidium pullulans | 0 |
| TC147364 | 1 | gi|50919526 | Phospholipase | Rice | 0 |
| TC146917 | 1 | gi|50909061 | RNA-binding protein Rp120 | Rice | 0 |
| TC140422 | 1 | gi|52353695 | N-ethylmaleimide sensitive fusion protein | Rice | 0 |
| TC139879 | 1 | gi|50920113 | Translational elongation factor Tu | Rice | 0 |
| TC139323 | 1 | gi|1737492 | Poly(A)-binding protein | Wheat | 0 |
| TC146710 | 4 | gi|50906401 | Elongation factor 1-γ | Rice | 1E-178 |
| TC140179 | 2 | gi|33146739 | GPI-anchored protein like | Rice | 2E-158 |
| TC139070 | 5 | gi|22607 | 14-3-3 Protein homolog | Barley | 3E-142 |
| TC139287 | 1 | gi|51535961 | Protein phosphatase 2C | Rice | 2E-140 |
| TC139604 | 3 | gi|50920031 | Late embryogenesis abundant protein | Rice | 2E-135 |
| TC140571 | 5 | gi|20804751 | Cytosolic factor-like protein | Rice | 4E-122 |
| TC146854 | 3 | gi|52346236 | Acid phosphatase | Barley | 4E-121 |
| TC131959 | 3 | gi|108711028 | Stem-specific protein TSJT1 | Rice | 1E-108 |
| TC139972 | 1 | gi|108707683 | Pathogenesis-related protein 1 | Rice | 5E-78 |
| TC138855 | 3 | gi|232033 | Elongation factor 1-β | Wheat | 7E-70 |
| TC146685 | 1 | gi|728594 | Gly-rich protein, RNA-binding protein | Barley | 5E-42 |
| TC147175 | 6 | gi|54778542 | Horcolin | Barley | 3E-31 |
| Transport | |||||
| TC152581 | 3 | gi|29123368 | High-affinity phosphate transporter | Barley | 0 |
| TC147625 | 3 | gi|50934997 | Coatomer protein γ 2-subunit | Rice | 0 |
| TC146833 | 3 | gi|53982658 | GDP dissociation inhibitor | Rice | 0 |
| TC139045 | 4 | gi50400847 | H+-ATPase | Wheat | 0 |
| TC131757 | 2 | gi|62900380 | Importin α-1b-subunit | Rice | 0 |
| TC146836 | 1 | gi|6691629 | HvPIP1;3 | Barley | 2E-151 |
| TC148527 | 1 | gi|23954314 | Transportin | Rice | 4E-123 |
| BJ464951 | 1 | gi|7339699 | Importin-α reexporter | Rice | 3E-90 |
| Unknown | |||||
| TC134738 | 6 | gi|18146791 | D protein | Barley | 0 |
| TC130945 | 6 | gi|4158232 | Reversibly glycosylated polypeptide | Wheat | 0 |
| TC148380 | 1 | gi|52077208 | Unknown protein | Rice | 0 |
| TC147606 | 1 | gi|19071 | Protein zx | Barley | 0 |
| TC146779 | 1 | gi|50921575 | OSJNBa0027H09.17 | Rice | 0 |
| TC146718 | 1 | gi|1408512 | |dbj|BAA13068.1| | Rice | 0 |
| TC139280 | 1 | gi|2072727 | Fd-GOGAT protein | Rice | 0 |
| TC138693 | 1 | gi|50926280 | OSJNBa0014K14.18 | Rice | 0 |
| TC131623 | 1 | gi|50931573 | |ref|XP_475314.1| | Rice | 0 |
| TC131033 | 1 | gi|31296711 | GAD1 | Barley | 0 |
| TC139573 | 1 | gi|37535140 | |ref|NP_921872.1| | Rice | 6E-178 |
| TC130742 | 4 | gi|108707930 | Expressed protein | Rice | 8E-177 |
| TC131324 | 3 | gi|50923165 | OSJNBa0008A08.11 | Rice | 1E-174 |
| TC133361 | 1 | gi|57899406 | Helicase-B-associated transcript 1 | Rice | 4E-161 |
| TC131647 | 1 | gi|46805936 | |dbj|BAD17230.1| | Rice | 2E-160 |
| TC140535 | 2 | gi|50929321 | OSJNBa0011F23.4 | Rice | 3E-159 |
| TC147335 | 1 | gi|51963828 | P0575F10.14 | Rice | 1E-158 |
| TC132188 | 5 | gi|50923829 | OSJNBa0044M19.9 | Rice | 1E-154 |
| TC139943 | 1 | gi|50915982 | KH domain-containing protein NOVA like | Rice | 6E-148 |
| TC147431 | 1 | gi|54291831 | Unknown protein | Rice | 4E-140 |
| TC131062 | 1 | gi|50948271 | |ref|XP_483663.1| | Rice | 3E-118 |
| TC132224 | 4 | gi|50915640 | SPATULA like | Rice | 4E-111 |
| TC147192 | 1 | gi|50923623 | OSJNBa0069D17.2 | Rice | 2E-107 |
| TC140155 | 1 | gi|50511386 | Unknown protein | Rice | 4E-106 |
| AV835541 | 1 | gi|50938581 | Karyopherin-β 3 variant | Rice | 2E-98 |
| TC147995 | 1 | gi|77552020 | Patatin-like protein | Rice | 3E-96 |
| TC131723 | 1 | gi|3702665 | |emb|CAA07474.1| | Rice | 5E-87 |
| TC135855 | 2 | gi|50932957 | Unknown protein | Rice | 3E-82 |
| TC142088 | 1 | gi|37537066 | Unknown protein | Rice | 3E-81 |
| TC136407 | 1 | gi|6815075 | MAWD-binding protein | Rice | 8E-80 |
| TC147301 | 1 | gi|55700995 | |tpe|CAH69306.1| | Rice | 1E-73 |
| TC132108 | 1 | gi|50939495 | |ref|XP_479275.1| | Rice | 2E-73 |
| TC153068 | 1 | gi|27261082 | Unknown protein | Rice | 3E-70 |
| TC139514 | 1 | gi|32492140 | |emb|CAE03373.1| | Rice | 5E-68 |
| TC147815 | 1 | gi|34908928 | Latex-abundant protein | Rice | 4E-66 |
| TC136015 | 1 | gi|50911579 | GAMM1 protein | Rice | 4E-66 |
| TC131126 | 1 | gi|21322752 | |dbj|BAB78536.2| | Rice | 7E-62 |
| TC141421 | 1 | gi|34910236 | Unknown protein | Rice | 6E-54 |
| TC131726 | 1 | gi|34899866 | Unknown protein | Rice | 1E-48 |
| TC141742 | 1 | gi|51091938 | PrMC3 | Rice | 3E-46 |
| BQ768779 | 1 | gi|52353425 | Unknown protein | Rice | 6E-44 |
| TC136111 | 1 | gi|50926656 | OSJNBa0074L08.23 | Rice | 2E-41 |
| BQ763407 | 1 | gi|50252172 | Senescence-associated protein like | Rice | 4E-36 |
| AL511164 | 1 | gi|90399278 | H0306F03.6 | Rice | 2E-33 |
| TC147252 | 1 | gi|50915240 | |ref|XP_468084.1| | Rice | 1E-28 |
| TC134176 | 1 | gi|83647364 | FAD/FMN-containing dehydrogenase | Hahella chejuensis | 1E-26 |
| TC131063 | 1 | gi|50904847 | Gly-rich protein 2 | Rice | 1E-26 |
| TC145521 | 1 | gi|50929757 | OSJNBa0088H09.2 | Rice | 9E-26 |
| TC146830 | 1 | gi|50919281 | |ref|XP_470037.1| | Rice | 2E-25 |
The distribution of number of peptides that define a family is shown in Supplemental Figure S1 (black bars). Less than one-half (44%) of the protein families in this experiment were defined by a single peptide that was identified using two distinct search algorithms. At the other extreme, METS1 was defined by 22 peptides, and two proteins, phosphoglycerate mutase and Phe ammonia lyase, were identified on the basis of 17 matching peptides.
Eleven of the 1,038 peptides (1.05%) had iTRAQ ratios of 0.4 (indicating 2.5-fold increase in abundance in the tolerant plants) or greater. Seven of the peptides with increased abundance in the tolerant plants could be assigned to just three proteins: a methylthio-Rib kinase (MTK), Iron Deficiency Sensitive2 (IDS2), and IDS3 (Fig. 4A). These proteins are all involved in the formation of HMA, a phytosiderophore secreted by barley plants to increase the uptake of Fe (Negishi et al., 2002). An eighth peptide was assigned to a D protein. The transcript encoding this protein has previously been identified as increasing in abundance in response to Fe deficiency (Negishi et al., 2002). The ninth peptide was assigned to a UDP-Glc-6-dehydrogenase, a protein with six other peptides also matching the amino acid sequence. The ratios of the other six peptides were close to 0 (Fig. 4A). The 10th peptide with elevated abundance in the B-tolerant plants was assigned to a Glu decarboxylase, GAD1 (Glu decarboxylase isozyme1), a protein responsible for the synthesis of γ-aminobutyrate. One peptide with a negative iTRAQ ratio of 0.153 (Fig. 4A, asterisk) was derived from an O-methyltransferase (TC146961). This protein was identified on the basis of three peptides in total, one lacking a valid iTRAQ ratio and the other having an iTRAQ ratio of 0.94. Aside from this peptide, there were no other peptides displaying a significant decrease in the B-tolerant plants.
Plant Fe Status and B Uptake
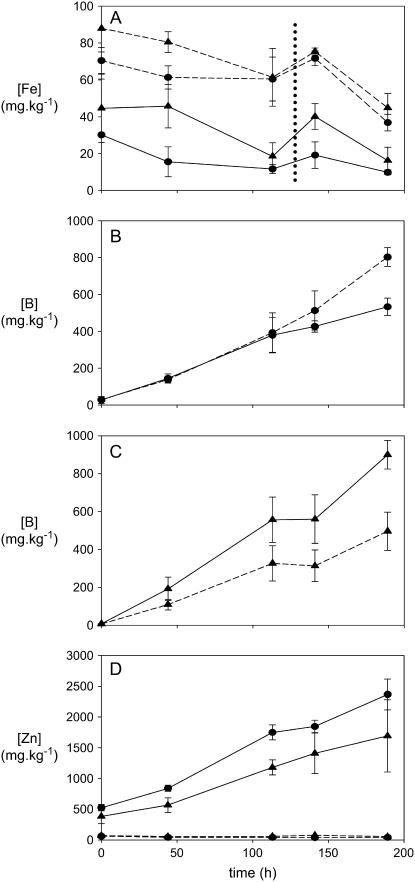
The identification of elevated levels of proteins involved in Fe acquisition in the B-tolerant plants led us to examine if there was a relationship between Fe and B in planta. ‘Clipper’ and ‘Sahara’ plants were grown in Fe-deficient conditions, and we examined how B and zinc (Zn) accumulation in the oldest leaf and Fe accumulation in the youngest leaf was affected, using inductively coupled plasma-optical emission spectrometry.
‘Sahara’ plants accumulated slightly more Fe than ‘Clipper’ plants in both Fe-replete and Fe-depleted conditions (Fig. 5A). In ‘Clipper,’ Fe deficiency resulted in accumulation of similar amounts of B initially, although after 110 h, less B accumulated in the Fe-deficient plants (Fig. 5B). The opposite effect was observed in ‘Sahara’ plants. Compared to Fe-replete plants, Fe-deficient ‘Sahara’ plants accumulated significantly more B, with the difference apparent after 48 h (Fig. 5C). Fe deficiency also had a significant effect on the rate of Zn accumulating in the oldest leaves in both cultivars compared to the Fe-replete plants (Fig. 5D).
Figure 5.
Inductively coupled plasma analysis of elemental abundance in barley leaves from B-tolerant (‘Sahara’) and B-intolerant (‘Clipper’) plants. A, Fe concentrations in the youngest leaves. B, Accumulation of B in the oldest leaf of ‘Clipper.’ C, Accumulation of B in the oldest leaf of ‘Sahara’ plants. D, Zn accumulation in oldest leaves. Circles, ‘Clipper’ plants; triangles, ‘Sahara.’ Dashed lines, Fe-replete plants; solid lines, Fe-deficient plants. Dotted vertical line in A indicates emergence of fourth (new) leaf. Error bars indicate sd (n = 3).
Siderophore Analysis
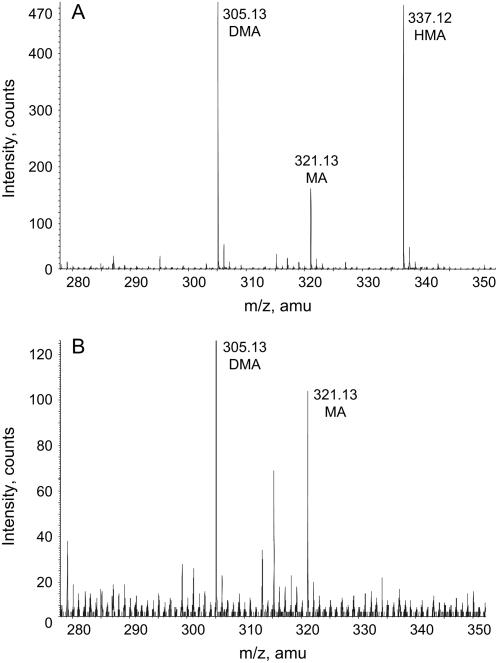
Siderophores were collected from the root secretions of ‘Clipper’ and ‘Sahara’ plants grown in low Fe conditions, which are known to result in elevated siderophore production (Negishi et al., 2002). Both cultivars produced deoxymugineic acid and mugineic acid (MA; Mori and Nishizawa, 1987; Fig. 6). In addition to these two compounds, ‘Sahara’ plants produced HMA (Fig. 6). Currently, we cannot determine if the HMA species produced in the ‘Sahara’ plants is the 3-hydroxy (HMA) or the 3-epi-hydroxy (eHMA) isomer.
Figure 6.
MS analysis of siderophores collected from the root secretions of ‘Sahara’ and ‘Clipper’ plants. ‘Sahara’ (A) and ‘Clipper’ (B) plants were grown in Fe-deficient conditions. Secretions were collected after 10 d of growth in Fe-deficient conditions. All siderophores were identified as singly charged species in the positive ion mode. Samples were purified by fractionation using cation-exchange resin, dried, and resuspended in methanol/1% formic acid (v/v) and infused directly into the MS. We cannot discount the possibility of other coextracted compounds potentially suppressing the siderophore signals in either sample. As such, relative signal intensities between extracts were not considered.
DISCUSSION
iTRAQ and Two-Dimensional Liquid Chromatography MS/MS Analysis of Soluble Proteins Isolated from Barley
In this study, we established a comparative proteomic approach that allowed us to compare the abundances of 479 proteins from the roots and leaves of barley plants. A total of 138 of these proteins was identified from leaf tissue and 341 were identified from root tissues.
The analysis of proteins isolated from leaves of replicate ‘Golden Promise’ plants demonstrated that the iTRAQ approach is sufficiently sensitive to detect differences of 2.5-fold or greater between the samples under comparison. With this information in hand, protein abundances were then compared between two pools of barley plants differing in their B tolerance. Peptides with the greatest relative abundance in the B-tolerant plants, coincident with elevated abundances of greater than 2.5-fold in the tolerant plants, were all derived from proteins that had previously been demonstrated to be involved in an Fe deficiency response, with two of these proteins, IDS2 and IDS3, specifically involved in the formation of the phytosiderophore HMA.
The iTRAQ approach used in this study represents a robust and accurate method of comparing protein abundances between proteins isolated from plants of differing genotypes or variable treatments. This method compares favorably with other proteomic approaches, notably two-dimensional (2D)-PAGE, particularly in relation to the quantitative aspect of the iTRAQ analysis. In terms of the functions of the proteins identified in this study, strong similarities exist in data sets from other cereals, namely wheat (Triticum aestivum) and rice (Oryza sativa; Koller et al., 2002; Donnelly et al., 2005; Nozu et al., 2006). Enzymes involved in biosynthetic metabolic pathways dominated the identifications, while 10 distinct components of the proteasome were also identified in the root protein complement.
In monocotyledonous crop species, proteomic studies have focused on rice for a range of reasons, one of which is the availability of a complete genome sequence (Goff et al., 2002; Rossignol et al., 2006). The ever-increasing size of EST and genomic sequence databases for other cereal species is rapidly creating a situation where proteomic analyses in other graminaceous crop plants are feasible. In this study, we searched MS/MS spectra exclusively against the EST assemblies in the TIGR barley gene index (V9.0). Of the 479 barley proteins matched against this database, only 86 (18%) matched barley proteins in the National Center for Biotechnology Information (NCBI) NCBInr protein database. Sixty percent of the 479 proteins matched to rice orthologs in the NCBInr protein database, highlighting the usefulness of EST-derived databases for proteomic studies (Tables I and II) in cereals that lack a completely sequenced genome.
Notably, there were fewer large differences (>2.5-fold) between peptides identified in the bulked segregant analysis (1.05%) compared to the analysis of peptides from the leaves of ‘Golden Promise’ plants (3.96%). This was despite a fractionally larger spread across the majority of iTRAQ ratios in the bulked segregant analysis, as evidenced by comparison of the width of boxes and error bars in Figures 2A and 4A. It appears that using a bulked segregant approach, in combination with a proteomic comparison, may represent a fruitful avenue to investigate the nature of novel QTL in barley as well as other cereals.
Previous studies comparing the protein profiles of root and leaf tissues have identified variable overlaps in the percentages of shared proteins. In a recent 2D-PAGE analysis of rice tissues, Nozu et al. (2006) identified similar numbers of protein identifications to that described in this study but found a 22% overlap in the identity of proteins found in root and leaf tissue. An earlier study, while identifying more proteins using a combination of 2D-PAGE and MudPIT approaches, found a 5% match between root and leaf proteins (Koller et al., 2002). The 2% overlap in protein expression described in this study is in approximate agreement with the latter study.
Contrasting with this relatively low overlap at the level of protein abundance, metabolite profiles of different tissues have a much higher degree of similarity (Roessner et al., 2006). Disregarding the small differences in metabolite abundances between varieties, comparison of the metabolite profiles of both ‘Clipper’ and ‘Sahara’ root and leaf tissues shows a high degree of overlap. Of the 68 metabolites identified and measured in roots of each cultivar, 63 were also present in leaves, representing an over 90% overlap in metabolite production between tissues (Roessner et al., 2006). This comparison reinforces the notion of tissue-specific expression of enzymes responsible for production of the same metabolite.
It is of note that a recently described tissue-specific barley transcript database reports over 12,000 expressed genes in both root and leaf tissues (Druka et al., 2006). It would be interesting to correlate the overlap in transcript and protein levels between these two tissues, particularly given the potential of this type of analysis to provide insights into posttranslational regulation of protein expression.
Proteins Involved in Siderophore Production Pathway Elevated in B-Tolerant Plants
Along with the identification of elevated levels of IDS2 and IDS3, we also identified a number of enzymes mediating upstream steps in the Yang cycle (Negishi et al., 2002). This pathway produces nicotianamine, the precursor of HMA. Levels of MTK were also elevated in the tolerant plants. Other proteins involved in the Yang cycle, namely METS1, IDI2, adenosyl phoshoribosyl transferase, and SAM sythetase did not notably differ in abundance between tolerant and intolerant plants. METS is notable because of the large number of identified peptides matching to this protein (Table II), indicating that this protein may be relatively abundant in the roots of barley plants. This is despite the product of this enzyme, Met, occurring at relatively low steady-state levels in the roots of barley plants (Ma et al., 1995).
A recent gas chromatography-MS-based analysis compared the abundances of metabolites isolated from the roots and leaves of ‘Clipper’ and ‘Sahara’ (Roessner et al., 2006). The precursor metabolites for siderophore production that were measured in this study, Met and Asp, as well as γ-aminobutyrate, were found to be present in comparable amounts in both the roots and leaves of each genotype. Despite these steady-state similarities, however, it is quite possible that the fluxes of metabolites passing through these pathways may differ significantly between cultivars. It may also be interesting to examine the levels of SAM and nicotiamine in the two cultivars, as these metabolites are the immediate precursors for siderophore production.
The chromosomal locations of ids2 and ids3 genes have been identified; ids2 maps to the long arm of chromosome 7H, while ids3 maps to the long arm of chromosome 4H (Nakanishi et al., 2000). Both appear to be single copy genes (Nakanishi et al., 2000). The chromosomal locations of genes encoding MTK and the D protein are currently unknown. Nonetheless, it is more probable that the genetic difference underpinning the elevated abundance of these proteins is a regulatory factor that coordinately controls the abundances of these proteins, perhaps functioning as a transcription factor. It is noteworthy that the transcripts of the significantly elevated proteins in this study have all previously been identified as being Fe inducible in barley roots (Negishi et al., 2002 and refs. therein). The genes encoding these proteins all share the same Fe Deficiency Response Element1 (IDE1)-like upstream elements (Kobayashi et al., 2003, 2005). Any candidate transcription factor may share some similarities to the recently identified IDE1-recognizing, Fe-regulated transcription factor IRO2 (Ogo et al., 2006). Significantly, however, IDE1-type sequences are also present upstream of proteins that were not elevated in the B-tolerant plants, specifically SAM sythetase and IDI2.
Plant Fe Status and B Uptake
To begin to differentiate if the increased abundance of siderophore-producing enzymes in the B-tolerant plants was merely associated with the B tolerance loci rather than being responsible for the tolerance trait per se, we examined the effects of Fe availability on B uptake. Fe-deficient ‘Sahara’ plants accumulated more B than the Fe-replete plants, highlighting a potential breakdown of the B-tolerance mechanism in this situation. The effect was observed immediately upon removal of Fe from the growing medium. In contrast, Fe deficiency had no effect on the rate of B accumulation in ‘Clipper’ plants initially, although over time (>110 h), the rate of B accumulation decreased. This situation is reminiscent of studies showing similar increases in B accumulation during Zn deficiency in barley (Graham et al., 1987).
The increased rate of leaf Zn accumulation supported the notion that Fe deficiency resulted in an increase in siderophore production, supporting the recent demonstration of the involvement of MA-related compounds in the uptake of Zn (von Wiren et al., 1996; Suzuki et al., 2006). Notably, high B alone had no effect on leaf Zn levels (Fig. 5). Although graminaceous monocots primarily employ a siderophore-dependent Fe acquisition strategy (Mori, 1999), it is also possible that Fe deficiency results in increased uptake of Zn through nonspecific Fe(II) transporters, possibly similar to OsIRT1 (Bughio et al., 2002), which may be up-regulated in Fe-deficient conditions.
Modeling an Interaction between HMA and B
The proteomics-based identification of elevated levels of IDS2 and IDS3 in the B-tolerant plants led us to consider any possible interactions between B, siderophores (particularly HMA or eHMA), and Fe. As an initial step, we used molecular modeling based on the available crystallographic data from a Cu(II) complex of MA (Nomoto et al., 1981) to determine the feasibility of any interaction between B, Fe, and HMA or eHMA. This analysis suggested that Fe(III) HMA, which has the carboxylate group and the hydroxyl group on the same side of the four-membered ring (a cis arrangement), is more likely to be able to bind a B center than the Fe(III) eHMA.
The recent identification of B complexation with vibrioferrin, a bacterial siderophore (Amin et al., 2007), provides support for the notion of B-siderophore interactions occurring in extracellular environments. A key difference between the B-vibrioferrin interaction and the B-HMA interaction proposed here, however, is the dependence upon complexed Fe(III) in the HMA model. In conditions where Fe is available, an Fe(III) HMA-B interaction may result in a decreased B influx, analogous to the mechanism of aluminum-malate chelation described in wheat (Delhaize et al., 1993). Localized, high concentrations of Fe(III) HMA adjacent to the site of B movement across the PM may be sufficient to partially decrease the rate of B accumulation in planta. Further work will be needed to test this proteomics-driven hypothesis, and we intend to explore this model in the future.
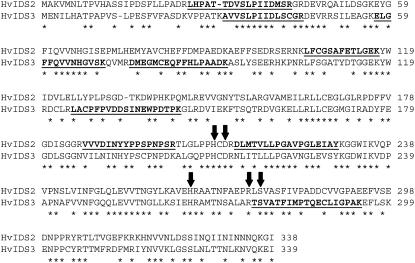
The enzyme responsible for the production of HMA, via the hydroxylation of MA at C3, has not yet been described. Although the hydroxylation reactions catalyzed by IDS2 (producing eHMA) and IDS3 are similar and each protein contains the requisite residues for Fe2+ and 2-oxoglutarate binding (Fig. 7), each protein catalyzes addition of hydroxyl groups to distinct carbon residues. Despite this catalytic selectivity, the two proteins are 55% identical at the amino acid level (Fig. 7). It is highly likely that IDS2 and the protein responsible for the production of HMA, tentatively named IDS2b, share an even greater level of amino acid identity. It is therefore feasible that the peptides identified as matching to IDS2 may indeed be derived from regions of identity within the uncharacterized IDS2b protein.
Figure 7.
Sequence alignment of barley IDS2 (gi 285634) and IDS3 (gi 9711238) proteins, with peptides identified by MS/MS underlined. Identical residues are indicated by asterisks. Arrows indicate conserved residues required for Fe(II) and 2-oxoglutarate binding.
CONCLUSION
We are currently working toward verifying any interaction between B, Fe, and HMA. We are also in the process of defining which tolerance locus (4H or 6H) may be responsible for this trait, although we believe it is more likely that HMA production may be linked to the weaker 6H tolerance locus. This postulate is based on the proposal of Hayes and Reid (2004) that the major tolerance locus may encode a protein responsible for B efflux. To this end, we intend to compare the PM protein profiles from the bulked segregants analyzed in this study using the iTRAQ approach we described.
In conclusion, we have described a robust and reliable new comparative proteomic methodology. This approach has wide-ranging applications, particularly in the field of cereal functional genomics. Protein abundance data collected using this method will be able to be interpreted in conjunction with the increasingly large metabolomic and transcriptomic data sets continuing to appear in the literature.
MATERIALS AND METHODS
All chemicals were purchased from Sigma-Aldrich unless otherwise specified.
Plant Growth
Barley (Hordeum vulgare) seeds (‘Golden Promise,’ ‘Clipper,’ and ‘Sahara’ and selected lines from the ‘Clipper’ × ‘Sahara’ DH population, selected as described in “Results”; Jeffries et al., 1999) were surface sterilized with 70% (v/v) ethanol and 0.5% (v/v) sodium hypochlorite prior to imbibing for 16 h in distilled water with aeration. Seeds were then transferred to moist filter paper and grown until coleoptiles were approximately 30 mm in length. Seedlings were then suspended over 15 L of hydroponic growth solution (36 plants per container) composed of 5 mm NH4NO3, 5 mm KNO3, 2 mm Ca(NO3)2, 2 mm MgSO4, 0.1 mm KH2PO4, 0.5 mm Na2SiO3, 50 μm NaFe(III) EDTA, 5 μm MnCl2, 10 μm ZnSO4, 0.5 μm CuSO4, 0.1 μm Na2MoO3, and 50 μm H3BO3. The solution was gently aerated and changed after 1 week initially and every 3 d subsequently. Plants were grown in a growth chamber using a 13°C, 10-h dark period and an 18°C, 14-h light period (180 μmol m−2 s−1 photon intensity).
For B tissue accumulation experiments, plants were grown as described, except for the Fe-deficient plants, which were grown in solutions lacking NaFe(III) EDTA. After seedling establishment for 1 week, all ‘Clipper’ plants were transferred to solutions containing 1 mm H3BO3, while ‘Sahara’ plants were transferred to solutions containing 5 mm H3BO3. Oldest and youngest leaves were harvested at indicated time points, dried, and elemental composition was determined using inductively coupled plasma optical emission spectrometry as described in Roessner et al. (2006). Earlier studies had determined that the rate of B accumulation in the oldest leaf in ‘Clipper’ plants grown in 1 mm B was similar to that of ‘Sahara’ plants grown in 5 mm B (Fig. 2 from Roessner et al., 2006). Using this information, the effect of Fe nutritional status on the rate of B accumulation in each cultivar was followed over similar time frames by growing each cultivar in different levels of B: 1 mm for ‘Clipper’ and 5 mm for ‘Sahara’ (Fig. 5). In the experiments described here, the rate of B accumulation in the ‘Clipper’ plants grown at 1 mm B was approximately double that of ‘Sahara’ plants grown in 5 mm B in Fe-replete conditions. Initial experiments also indicated that elevated levels of B (1 mm for ‘Clipper,’ and 5 mm for ‘Sahara’) had no effect on Fe accumulation in the youngest leaves of either cultivar (data not shown).
Protein Isolation
After 2 weeks of growth, roots and leaves were harvested 3 h after the beginning of the light period. Tissues were weighed and suspended in 2 volumes of chilled homogenization buffer containing 50 mm phosphate buffer, pH 7.5, 20 mm KCl, 0.5 m Suc, 10 mm dithiothreitol, 0.2 mm phenylmethylsulfonyl fluoride, 10 mm EDTA, and 10 mm EGTA. Tissues were homogenized with a curved, hand-held blade, filtered through a 50-μm nylon mesh, and centrifuged at 6,000g for 10 min. The supernatant from this step was centrifuged at 100,000g for 1 h. The final supernatant was concentrated by precipitation with two volumes of −20°C equilibrated 10% (w/v) TCA in acetone for 16 h at −20°C. The resulting pellet was washed twice with −20°C equilibrated 90% (v/v) acetone before resuspension in 0.5 m triethylammonium bicarbonate, pH 8.5, containing 0.1% SDS. The protein concentration was determined at this stage using a 2D Quant kit (GE Healthcare).
Protein Digestion and iTRAQ Labeling
Protein (100 μg) was reduced by addition of 5 mm tris-(2-carboxyethyl) phosphine and incubation at 60°C for 1 h. Cys residues were then blocked by incubation with 90 mm methyl methanethiosulfonate (MMTS) for 10 min at room temperature. CaCl2 (1 mm) was then added, and proteins were digested with modified trypsin (4 μg, sequencing grade, porcine, Promega) for 16 h at room temperature in a final volume of 40 μL. iTRAQ tags (Applied Biosystems) were resuspended in ethanol, and digestion was stopped by addition of iTRAQ tag/ethanol solution to a final concentration of 70% (v/v) ethanol. iTRAQ labeling was allowed to proceed for 1 h at room temperature. Labeled peptide mixtures were then pooled for chromatography.
Peptide Chromatography
iTRAQ-labeled peptide mixtures were dried under a stream of N2 and resuspended in 100 μL of 25 mm ammonium formate, pH 3.5, containing 5% acetonitrile (buffer A). Peptides were fractionated as described in Wagner et al. (2003) with some modifications. Samples were injected onto a strong-cation exchange (SCX) column (polysulfoethyl A 200 × 4.6 mm, 5 μm, 300  ) using an Agilent HPLC with an automatic fraction collector. Peptides were eluted with a gradient from 100% buffer A to 100% 0.5 m ammonium formate, pH 3.5, containing 25% acetonitrile over 30 min with a flow rate of 0.7 mL min−1. Generally, 60 fractions (350 μL each) were collected. These fractions were dried under vacuum and stored at −20°C.
) using an Agilent HPLC with an automatic fraction collector. Peptides were eluted with a gradient from 100% buffer A to 100% 0.5 m ammonium formate, pH 3.5, containing 25% acetonitrile over 30 min with a flow rate of 0.7 mL min−1. Generally, 60 fractions (350 μL each) were collected. These fractions were dried under vacuum and stored at −20°C.
Individual SCX fractions were resuspended in 0.1% formic acid (60 μL) and loaded onto a 300-μm × 5-mm C18 precolumn. After washing the precolumn with 0.1% formic acid, peptides were eluted from the precolumn onto an in-line C18 column (75 μm i.d. × 15 cm, 3 μm/100 Å Vydac) and fractionated using a gradient of 0% to 70% (v/v) acetonitrile in 0.1% formic acid over 60 min, using a flow rate of 0.25 μL min−1. This column eluted directly into a QSTAR XL hybrid quadropole-time of flight instrument (Applied Biosystems/MDS Sciex) using a nanospray source.
MS
The mass spectrometer was operated in the positive ion mode, ion source voltage of 1,750 V, using 10-μm uncoated SilicaTips (New Objectives). Data were collected using AnalystQS software in a data-dependent acquisition mode for the three most intense ions fulfilling the following criteria: m/z between 450 and 2,000; ion intensity >40 counts; and charge state between 2+ and 4+. After MS/MS analysis, these ions were dynamically excluded for 20 s, using a mass tolerance of 250 ppm. MS scans were accumulated for 0.5 s, and MS/MS scans were accumulated for 2 s. A mass and charge state-dependent rolling collision energy was used and was 20% to 30% greater than was used for an iTRAQ-unlabeled peptide. The MS was calibrated daily with [Glu]-fibrinopeptide B.
MS/MS Spectra Interrogation
Peak lists from individual data files were created using the MASCOT.dll script in AnalystQS 1.1 and interrogated using MASCOT (Matrix Science, Perkins et al., 1999) and X!Tandem (Robertson and Beavis, 2004). In both cases, the spectra were searched against a six-frame translation of the Barley V9.0 Gene Index (TIGR, released September 15, 2004). MASCOT parameters were as follows: MS peptide tolerance ±0.25 D, MS/MS tolerance ±0.15 D, trypsin cleavage, allowing one missed cleavage, fixed modifications of iTRAQ tags on N terminus, and Lys residues and MMTS and variable modification of iTRAQ on Tyr residues. The X!Tandem parameters were fragment mass error 0.2 D, parent mass tolerance ±100 ppm, minimum peak 15, maximum peak of 500, complete modification of N terminus with iTRAQ and MMTS on Cys, and potential modifications were iTRAQ on Lys and Tyr, with a maximum of one missed cleavage.
We used an in-house Linux script that collated the MASCOT, X!Tandem, and i-Tracker outputs (see below) from all data files representing an entire series of SCX fractions into a single file. Peptides were only reported if they had a MASCOT score greater than 37 (P < 0.05). Peptides that lacked a C-terminal Lys or Arg residue were rejected. The X!Tandem output was formatted into a single column, showing whether the MASCOT-identified peptide was identified by X!Tandem. When multiple spectra from the SCX dataset were matched to the same peptide, only the match with the highest MASCOT score was reported.
Based on preliminary work comparing MS/MS search algorithm outputs against manually checked MS/MS spectra (n = 400), we found that automatic acceptance of protein matches based on one peptide can result in a high proportion of false positives. As such, we adopted a conservative approach to protein identification in this study. Accessions were accepted using a two-tiered criteria. If three or more peptides (identified solely by MASCOT, score >37) matched to a single accession, the identification was automatically accepted. Using a three-peptide-matches criteria, reverse database searching gave a false positive rate of <1%. Accessions containing one or two peptides were then accepted if at least one peptide was also matched using X!Tandem (peptide accepted by two independently developed algorithms). Only peptides matched by the two algorithms were reported.
Peptide Grouping
Accessions were manually grouped into protein families, or groups of accessions that shared peptides. All peptides within a protein family are unique to that family. This feature of multiple accessions matching to groups of peptides is exacerbated in cereal genomes, where polyploidy results in large families of genes with very similar sequences.
To generate protein identifications from the accession matches, the nucleotide sequence of each matched accession was searched against the NCBInr protein database (BLASTX), using the default parameters. BLAST searches were performed between April and August, 2006.
iTRAQ Ratio Determinations
The tagged peptides were then pooled and fractionated using a SCX column, followed by C18 reversed-phase HPLC, directly interfaced via an ESI source to the MS. Due to the identical chemical nature of the iTRAQ tags, identical peptides from the differentially labeled samples cofractionate and enter the MS and are analyzed simultaneously. During MS/MS analysis of peptides, the reporter ions, derived from the isotopically distinct iTRAQ tags, are released from the differentially tagged peptides. The relative abundance of the reporter ions is representative of the relative abundance of the peptides in the starting mixtures.
iTRAQ tags attach to secondary amine groups, such that in the case of trypsin-derived peptides, tags are attached to both N- and C-terminal ends of each peptide. This increases the amount of energy required to fragment the peptide to generate amino acid information but also improves the quality of the MS/MS spectra, such that more y- and b-ions are observed (Fig. 1B).
iTRAQ ratios were generated using the open source software i-Tracker (Shadforth et al., 2005). We used a reporter ion peak intensity threshold of 10, rejecting ratios with an associated error of greater than 4. We also rejected ratios derived from reporter ions which, after summing the relative peak areas for the tags employed in that experiment, contributed to less than 80% to the total area of all four reporter ion peaks (m/z 114–m/z 117). Ratios were then normalized around 1 by multiplication of all ratios by the average value of the ratios. Normalized ratios were finally log transformed to generate a normally distributed set of data.
Siderophore Purification and Analysis
Siderophores were collected as described by Takagi et al. (1984). Plants were removed from the hydroponic growing solution, and roots were placed in gently aerated distilled water. Root secretions were collected over 3 h. This solution was concentrated 10-fold using rotary evaporation. The concentrated root secretions were fractionated on an Amberlite IR120 cation exchange resin, washed with water, and siderophores were then eluted from the resin with 1 m NH4OH. The eluate was dried under vacuum and resuspended in 50% methanol and 0.1% formic acid. ESI-MS analysis was performed in the positive ion mode, with samples directly infused into the QSTAR XL hybrid quadropole-time of flight instrument. The MS/MS fragmentation profile of the siderophores shared features with the collision-induced dissociation profiles of MA described by Kenny and Nomato (1994; data not shown).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Number of peptides that match to distinct proteins.
Supplemental Figure S2. Plot of relative peak areas of reporter ions 115 and 114 from 480 peptides with iTRAQ ratios from ‘Golden Promise’ leaves.
Supplemental Figure S3. Plot of relative peak areas of reporter ions 115 and 114 from the 1,038 peptides with iTRAQ ratios from B-tolerant and B-intolerant barley roots.
Supplemental Table S1. Complete list of peptides identified from barley ‘Golden Promise’ leaf tissue.
Supplemental Table S2. Complete list of peptides identified from barley root tissue (B-tolerant and B-intolerant ‘Clipper’ × ‘Sahara’ DH lines).
Supplementary Material
Acknowledgments
We thank Ed Newbigin, Brendan Abrahams, Tim Sutton, Andreas Schreiber, Mark Tester, Ute Baumann, and Margie Pallotta for helpful suggestions and discussions.
This work was supported by the Australian Research Council, the Grain Research and Development Corporation, and the Victorian and South Australian State Governments.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: John Patterson (johnhp@unimelb.edu.au).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Amin SA, Kupper FC, Green DH, Harris WR, Carrano CJ (2007) Boron binding by a siderophore isolated from a marine bacteria associated with the toxic dinoflagellate Gymnodium catenatum. J Am Chem Soc 129 478–479 [DOI] [PubMed] [Google Scholar]
- Bughio N, Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S (2002) Cloning an iron-regulated metal transporter from rice. J Exp Bot 53 1677–1682 [DOI] [PubMed] [Google Scholar]
- Cartwright B, Zarcinas BA, Mayfield AH (1984) Toxic concentrations of boron in a red-brown earth at Gladstone, South Australia. Aust J Soil Res 22 261–272 [Google Scholar]
- Delhaize E, Ryan PR, Randall PJ (1993) Aluminum tolerance in wheat (Triticum aestivum L.). II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol 103 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly BE, Madden RD, Ayoubi P, Porter DR, Dillwith JW (2005) The wheat (Triticum aestivum L.) leaf proteome. Proteomics 5 1624–1633 [DOI] [PubMed] [Google Scholar]
- Druka A, Muehlbauer G, Druke I, Caldo R, Baumann U, Rostoks N, Schreiber A, Wise R, Close T, Kleinhofs A, et al (2006) An atlas of gene expression from seed to seed through barley development. Funct Integr Genomics 6 202–211 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan T, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296 92–100 [DOI] [PubMed] [Google Scholar]
- Graham RD, Welch RM, Grunes DL, Cary EE, Norvell WA (1987) Effect of zinc deficiency on the accumulation of boron and other mineral nutrients in barley. Soil Sci Soc Am J 51 652–657 [Google Scholar]
- Hayes JE, Reid RJ (2004) Boron tolerance in barley is mediated by efflux of boron from the roots. Plant Physiol 136 3376–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries SP (2000) Marker assisted backcrossing for gene introgression in barley (Hordeum vulgare L.). PhD thesis. Department of Plant Science, University of Adelaide, Australia
- Jeffries SP, Barr AR, Karakousis A, Kretschmer JM, Manning S, Chalmers KJ, Nelson JC, Islam AKMR, Langridge P (1999) Mapping of chromosome regions conferring boron toxicity tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 98 1293–1303 [Google Scholar]
- Karakousis A, Barr AR, Kretschmer JM, Manning S, Jeffries SP, Chalmers KJ, Islam AKM, Langridge P (2003) Mapping and QTL analysis of the barley population Clipper x Sahara. Aust J Agric Res 54 1137–1140 [Google Scholar]
- Kenny PTM, Nomato K (1994) Tandem mass spectrometric investigation of phytosiderophores mugineic acid, deoxymugineic acid and nicotianamine. Analyst 119 891–895 [Google Scholar]
- Kobayashi T, Nakayama Y, Itai RN, Nakanishi H, Yoshihara T, Mori S, Nishizawa NK (2003) Identification of novel cis-acting elements, IDE1 and IDE2, of the barley IDS2 gene promoter conferring iron-deficiency-inducible, root-specific expression in heterogeneous tobacco plants. Plant J 36 780–793 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Suzuki M, Inoue H, Itai R, Takahashi M, Nakanashi H, Mori S, Nishizawa NK (2005) Expression of iron acquisition-related genes in iron-deficient rice is co-ordinately induced by partially conserved iron-deficiency-responsive elements. J Exp Bot 56 1305–1316 [DOI] [PubMed] [Google Scholar]
- Koller A, Washburn MP, Langer BM, Andon NL, Deciu C, Haynes PA, Hays L, Schiletz D, Ulaszek R, Wei J, et al (2002) Proteomic survey of metabolic pathways in rice. Proc Natl Acad Sci USA 99 11969–11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Shinada T, Matsuda C, Nomoto K (1995) Biosynthesis of phytosiderophores, mugineic acids, associated with methionine cycling. J Biol Chem 270 16549–16554 [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88 9828–9832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S (1999) Iron acquisition by plants. Curr Opin Plant Biol 2 250–253 [DOI] [PubMed] [Google Scholar]
- Mori S, Nishizawa N (1987) Methionine as a dominant precursor of phytosiderophores in Graminaceae plants. Plant Cell Physiol 28 1081–1092 [Google Scholar]
- Nable RO (1988) Resistance to boron toxicity amongst several barley and wheat cultivars: a preliminary examination of the resistance mechanism. Plant Soil 112 45–52 [Google Scholar]
- Nable RO, Cartwright B, Lance RC (1990) Genotypic differences in boron accumulation in barley: relative susceptibilities to boron deficiency and toxicity. In N El Bassam, M Dambroth, B Loughman, eds, Genetic Aspects of Plant Mineral Nutrition. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 243–251
- Nakanishi H, Yamaguchi H, Sasakuma T, Nishizawa NK, Mori S (2000) Two dioxygenase genes, Ids3 and Ids2 from Hordeum vulgare are involved in the biosynthesis of mugineic acid family siderophores. Plant Mol Biol 44 199–207 [DOI] [PubMed] [Google Scholar]
- Negishi T, Nakanishi H, Yazaki J, Kishimoto N, Fujii F, Shimbo K, Yamamoto K, Sakata K, Sasaki T, Kikuchi S, et al (2002) cDNA microarray analysis of gene expression during Fe-deficiency stress in barley suggests that polar transport of vesicles is implicated is phytosiderophore secretion in Fe deficient barley roots. Plant J 30 83–94 [DOI] [PubMed] [Google Scholar]
- Nomoto K, Mino Y, Ishida T, Yoshioka H, Ota N, Inoue M, Takagi S, Takemoto T (1981) X-ray crystal structure of the Copper(II) complex of mugineic acid, a naturally occurring metal chelator of graminaceous plants. J Chem Soc Chem Comm 10 338–339 [Google Scholar]
- Nozu Y, Tsugita A, Kamijo K (2006) Proteomic analysis of rice leaf, stem and root tissues during growth course. Proteomics 6 3665–3670 [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Nakanashi H, Inoue H, Kobayashi T, Suzuki M, Takahashi M, Mori S, Nishzawa NK (2006) Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J Exp Bot 57 2876–2878 [DOI] [PubMed] [Google Scholar]
- O'Neill MA, Ishii T, Albersheim P, Darvill AG (2004) Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu Rev Plant Biol 55 109–139 [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, Creasey DM, Cotrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20 3551–3557 [DOI] [PubMed] [Google Scholar]
- Power P, Woods WG (1997) The chemistry of boron and its speciation in plants. Plant Soil 193 1–13 [Google Scholar]
- Robertson C, Beavis RC (2004) Tandem: matching proteins with mass spectra. Bioinformatics 20 1466–1467 [DOI] [PubMed] [Google Scholar]
- Roessner U, Patterson JH, Forbes MG, Fincher GB, Langridge P, Bacic A (2006) An investigation of boron toxicity in barley using metabolomics. Plant Physiol 142 1087–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol M, Peltier J, Mock H, Matros A, Maldonado AM, Jorrin JV (2006) Plant proteome analysis: a 2004-2006 update. Proteomics 6 5529–5548 [DOI] [PubMed] [Google Scholar]
- Shadforth IP, Dunkley TPJ, Lilley KS, Bessant C (2005) i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics 6 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Takahashi M, Tsukamoto T, Watanabe S, Matsuhashi S, Yazaki J, Kishimoto N, Kikucho S, Nakanishi H, Mori S, et al (2006) Biosynthesis and secretion of mugineic acid family phytosiderophores in zinc deficient barley. Plant J 48 85–97 [DOI] [PubMed] [Google Scholar]
- Takagi S, Nomoto K, Takemoto T (1984) Physiological aspect of mugineic acid, a possible siderophore of graminaceous plants. J Plant Nutr 7 469–477 [Google Scholar]
- Takano J, Noguchi K, Yasumori M, Kobayashi M, Gajdos Z, Miwa K, Hayashi H, Yoneyama T, Fujiwara T (2002) Arabidopsis boron transporter for xylem loading. Nature 420 337–340 [DOI] [PubMed] [Google Scholar]
- Takano J, Wada M, Ludewig U, Schaaf G, von Wiren N, Fujiwara T (2006) The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18 1498–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wiren N, Marschner H, Romheld V (1996) Roots of iron-efficient maize also adsorb phytosiderophore-chelated zinc. Plant Physiol 111 1119–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner Y, Sickmann A, Meyer HE, Daum G (2003) Multidimensional nano-HPLC for analysis of protein complexes. J Am Soc Mass Spectrom 14 1003–1011 [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates III Jr (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol 19 242–247 [DOI] [PubMed] [Google Scholar]
- Zieske LR (2006) A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot 57 1501–1508 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.