Abstract
Alternative splicing (AS) can add significantly to genome complexity. Plants are thought to exhibit less AS than animals. An algorithm, based on expressed sequence tag (EST) pairs gapped alignment, was developed that takes advantage of the relatively small intron and exon size in plants and directly compares pairs of ESTs to search for AS. EST pairs gapped alignment was first evaluated in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and tomato (Solanum lycopersicum) for which annotated genome sequence is available and was shown to accurately predict splicing events. The method was then applied to 11 plant species that include 17 cultivars for which enough ESTs are available. The results show a large, 3.7-fold difference in AS rates between plant species with Arabidopsis and rice in the lower range and lettuce (Lactuca sativa) and sorghum (Sorghum bicolor) in the upper range. Hence, compared to higher animals, plants show a much greater degree of variety in their AS rates and in some plant species the rates of animal and plant AS are comparable although the distribution of AS types may differ. In eudicots but not monocots, a correlation between genome size and AS rates was detected, implying that in eudicots the mechanisms that lead to larger genomes are a driving force for the evolution of AS.
Alternative RNA processing pathways result in the combining of different splice junctions that are present in pre-mRNA transcripts. In this way, a genetic unit can have a variety of mRNA and protein products, thus expanding the potential informational content of eukaryotic genomes. Recent evidence indicates a high incidence (up to 60%) of alternative splicing (AS) is present in the human genome, predominantly in the form of exon skip while a minor form is of the type called intron retention (5%–16%; Kan et al., 2002; Modrek and Lee, 2002; Carninci et al., 2005; Nagasaki et al., 2005). Although rare, intron retention can play an important biological role. It has been linked to tumor growth and is part of developmental regulation of proinsulin expression (Mansilla et al., 2005).
Plants are thought to exhibit less AS and, unexpectedly, analysis in Arabidopsis (Arabidopsis thaliana) showed that intron retention is the most common type of AS, comprising 45% of the AS types (Iida et al., 2004; Ner-Gaon et al., 2004; Nagasaki et al., 2005; Wang and Brendel, 2006). The plants that were used, Arabidopsis and rice (Oryza sativa), showed comparable results. Furthermore, most AS clusters consist of two isoforms (Campbell et al., 2006). Indeed, plant intron sequence and control of splicing differ from their vertebrate and yeast (Saccharomyces cerevisiae) counterparts (Brown et al., 2002). Plant retained introns were shown to be present in RNA derived from polyribosomes, demonstrating that these intron retention events are not the by-product of incomplete splicing but are found in a potentially translatable context in the cytoplasm (Ner-Gaon et al., 2004). Using a different method, Iida et al. (2004) aligned RIKEN Arabidopsis full-length cDNA/EST sequences to the Arabidopsis genome and observed AS for 11.6% of the transcription units of which 44% were retained introns (Iida et al., 2004). In a more recent study that combined data from a large collection of EST/cDNA, 21.8% of the transcripts showed AS events and approximately 56% of these events were of the intron retention type (Wang and Brendel, 2006). A more comprehensive study revealed a higher percent of total AS (30%; Campbell et al., 2006). Taken together these studies confirmed that a lower percentage of genes are alternatively spliced compared to humans and that intron retention is the most prevalent form of AS in Arabidopsis and rice. A possible consequence of this phenomenon is that in plants a preponderance of transcript isoforms will presumably have negative consequences for protein structure and function, as retained introns tend to include premature stop codons (Campbell et al., 2006).
Comparison of AS between different species is of interest. At the genome level it can teach us about the evolution of AS, the conservation of mechanisms that control AS, and its biological consequences for a species. At the gene level, conservation of AS can teach us about gene function, the evolutionary history of particular genes, and of their gene families (Modrek and Lee, 2003; Sorek and Ast, 2003; Lareau et al., 2004; Kan et al., 2005). For example, plants have large Ser/Arg-rich protein families that are active in splicesosome assembly, which impact on splicing patterns (Lorkovic and Barta, 2002; Isshiki et al., 2006). Cross-species comparison of transcripts within the family revealed evolutionary preservation of intron elements that are correlated with conserved splice forms that serve a biological function (Iida and Go, 2006; Kalyna et al., 2006).
To carry out cross-species comparison on a global scale, rates of AS have been defined as the number of AS events that occur in a set number of loci or genes and by restricting the analysis to a constant number of genes, EST, or both. In this way, species with vastly different total EST coverage can be compared. It is of interest that using 650 cDNA in a comparison of AS rates from Arabidopsis, Caenorhabditis elegans, Drosophila melanogaster, and a few mammals, only small differences in species-specific rates were detected with the exception of Arabidopsis, which showed a much lower AS rate (Brett et al., 2002). The conclusion drawn was that AS expands the coding capacity of genomes irrespective of the perceived organism complexity. However, a similar comparison of four animal species using more EST and a different method of data normalization showed a certain degree of variation between AS rates in different species (Brett et al., 2002; Kim et al., 2004). These contradicting results may be attributed to the fact that the analyses depended on EST coverage that varies between the organisms tested. When EST coverage was normalized in a different manner the percentage of genes and exons undergoing AS was found to be higher in vertebrates compared with invertebrates. Furthermore, in metazoan evolution, intron retention remained the rarest type of AS, whereas exon skipping is more prevalent and exhibits a slight increase, from invertebrates to vertebrates (Kim et al., 2007). Thus, within related groups such as mammals (human and mouse; Kim et al., 2007) and angiosperms (Arabidopsis and rice) less than 30% difference in rates were noted (Nagasaki et al., 2005).
It is of interest to expand the comparisons of AS rates to additional plant species, however, discovery of AS has relied on aligning EST contigs or cloned cDNA to annotated genomic sequence (Mironov et al., 1999; Croft et al., 2000; Brett et al., 2002). These methods are restricted to the major model species, since they require sequenced genomes and a large sample of reliable EST and mRNA. In an effort to look at trends in the rate of AS for the many plant species that have significantly large EST databases (dbESTs) but do not have sequenced genomes, we conducted an EST-driven AS search comparison. We have previously applied this method to Arabidopsis and showed that a preferred form of AS in Arabidopsis was intron retention (Ner-Gaon et al., 2004). An improved algorithm is applied here and was validated by comparison to Arabidopsis, rice, and tomato genomic databases. In this method, a direct rigorous BLAST-like alignment tool (BLAT) algorithm (Kent, 2002) was first employed to match ESTs. The paired ESTs were then searched for insertion/deletions (indels). The estimation of mean intron and exon size demonstrated that this method is readily applicable to plant genomes but less so to human genomes. The method was applied to 11 plant species that includes 17 cultivars for which enough ESTs are available. The results show up to 3.7-fold differences in AS rates between plant species. This indicates that in some plant species the rates of animal and plant AS are comparable although the types of AS and their distribution may differ. In addition, a correlation was evident between AS rates and genome size of eudicots.
RESULTS
Types of AS Detected by EST Pairs Gapped Alignment
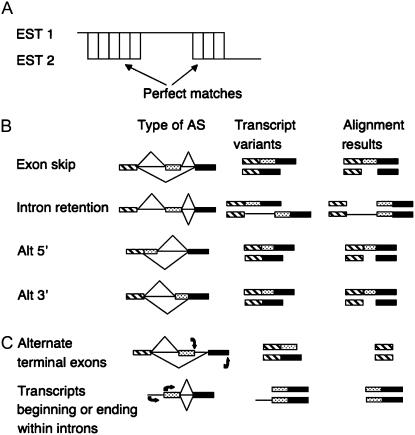
EST pairs gapped alignment (EPGA) is an algorithm meant to search for RNA transcripts that are the result of AS. EPGA looks for ESTs sharing two matching regions that flank a discontinuity in the alignment arising from an indel, as shown in Figure 1A. In theory, EPGA can accurately detect exon skips, intron retention, and 5′ and/or 3′ alternative splice sites (Fig. 1B). In practice, to avoid indels that are a result of errors in EST sequencing, the minimum indel size was set empirically to 5 bp. It has been estimated that alternative 5′ and 3′ of 3 to 4 bp size make up less than 15% of the total alternative 5′ and 3′ type of AS (Campbell et al., 2006). As alternative 5′ and 3′ type is less than 28% of all the AS types; it is calculated that about 4% of the total AS will not be detected using the 5 bp cutoff criteria. Furthermore, as the method requires overlap at both EST ends of at least 25 bp it cannot detect certain types of AS, for example, alternate terminal exons or transcripts beginning or ending within introns (Fig. 1C). These AS types have been estimated to comprise about 30% of the total AS (Ner-Gaon et al., 2004).
Figure 1.
Schematic diagrams of AS types detected by the EPGA algorithm. A, EPGA showing perfectly matched regions of at least 25 bp that flank a 5 bp or longer indel. B, Types of AS that match the EPGA requirements as shown in A include: exon skipping, alternative 5′ splice donor sites, alternative 3′ splice acceptor sites, and intron retention. AS patterns are represented by (top and bottom) diagonal lines. The AS results in two transcript variants that are shown together with the predicted alignment results. Boxes represent exon sequences and lines represent intron sequences. C, Other types of AS including alternate terminal exons or transcripts beginning or ending within introns that do not match the EPGA alignment requirements. The description is as in B where arrows at the bottom represent start and termination of transcription.
Quality-Control Criteria for Selecting EST and dbESTs
The accuracy by which EPGA detects exon skips, intron retention, and 5′ and/or 3′ alternative splice sites will be a function of the quality and uniformity of the dbESTs. Starting with a survey that included all available eudicot and monocot dbEST (Supplemental Table S1) standard quality-control parameters for accepting EST, i.e. elimination or tailoring of EST with high error, vector sequence, or poly A, were applied (see “Materials and Methods”). To control possible DNA contamination, in the absence of available genomes, we hypothesized that libraries that contained a high proportion of multiple indels may be contaminated with genomic sequence. Therefore, libraries containing more than 1% EST pairs with multiple indels were not used. Using these criteria, 57 out of 711 of the libraries were disqualified (Supplemental Table S1). Furthermore, for the same reason, in all our analyses using the EPGA algorithm we reject all EST pairs that contain multiple indels.
To achieve uniformity between dbEST, the average length of ESTs in a database was required to be within 2 sds of the average EST size of all the libraries (dbEST average size = 513). This is important as average EST size will dictate the number of exons/introns queried. Another basis for uniformity is requiring that dbEST represent cDNA made from diverse tissue types. This was examined in two ways. First, ESTs from each ecotype/cultivar were subjected to clustering as the number of clusters reflects the dbEST diversity. Low cluster number can represent enrichment for particular tissue types while high cluster number can represent genomic contamination. Thus, ecotype/cultivar with clusters that are more than 2 sds from the average cluster number were not used (average dbEST cluster number = 2,929 using 20,000 ESTs). Second, only libraries containing a robust mixture of EST from diverse plant elements were selected. dbEST in which more than 20% of the libraries were from flowers or fruits were not used to avoid tissue-specific bias in AS (Supplemental Table S1).
Feasibility of Applying EPGA to dbEST
EPGA analysis can be meaningful for comparative analysis of different species as long as the indel size, which represents a full or partial exon or intron, is shorter than the typical EST size. To establish this, a variety of species for which full or partial genomic sequence is available were examined for constitutive median intron and exon size by alignment of EST to genomic data (see “Materials and Methods”). As shown in Table I for selected eudicot and monocot plants, the intron and exon median size was found to have a range of 100 to 200 bp. The values obtained here for Arabidopsis and rice exon and intron median size are similar to sizes found in recent analysis of whole genomes (Collins and Penny, 2006). Significantly, as shown in Table I, the proportion of ESTs in which constitutive introns could be detected varied from 45% to 63% and was not correlated with the intron size of the species. This indicates that within these intron sizes the length of EST is sufficient for making plant genomes amenable to EPGA analysis even for the extreme case of complete intron retention. In contrast, in humans, the much larger median size of introns, 1,422 bp (Collins and Penny, 2006), obviates application of such search algorithms to dbESTs for intron retention, although exon skipping should be detectable.
Table I.
Median size of exons and introns in humans and select plant species
The ESTs were aligned to GenBank genomic data using BLAT (Kent, 2002). The lengths of the exons and introns were extracted from the alignment results (see “Materials and Methods”). The data source and methods are described in the “Materials and Methods.” The data for humans is as described (Collins and Penny, 2006).
| Arabidopsis | Tomato | Oryza-‘indica’ | Oryza-‘japonica’ | Maize | Human | |
|---|---|---|---|---|---|---|
| Exons median (mean) | 99 (116) | 105 (123) | 98 (115) | 99 (123) | 111 (150) | 124 |
| No. of exons | 23,156 | 3,090 | 21,585 | 36,145 | 706 | 283,216 |
| Introns median (mean) | 100 (177) | 201 (545) | 133 (358) | 139 (396) | 92 (688) | 1,422 |
| No. of introns | 40,827 | 4,833 | 38,224 | 60,773 | 2,338 | 252,375 |
| No. of ESTs | 94,278 | 13,075 | 46,833 | 109,619 | 16,696 | – |
| ESTs with constitutive introns | 51,534 | 8,224 | 29,488 | 61,853 | 7,518 | – |
| % ESTs with constitutive introns | 55 | 63 | 63 | 56 | 45 | – |
Calibration of EPGA
The EPGA algorithm was calibrated by comparing the results from direct EST comparisons to the results of EST genomic sequence alignments for the species Arabidopsis, rice (‘japonica’ group), and tomato. EST from Arabidopsis Columbia ecotype (National Center for Biotechnology Information [NCBI] ESTs databases 23/12/2006) yielded 111,142 ESTs after quality-control processing (see “Materials and Methods”; Supplemental Table S1). EPGA was then applied to the ESTs and detected 6,161 alternatively derived EST pairs. Additionally, all of the Arabidopsis ESTs were clustered in 25,616 clusters of which 11,466 contained more than one EST. Out of these, 467 clusters contained one or more alternatively derived EST pairs (Table II).
Table II.
Summary of alignment to the genome for EPGA pairs from Arabidopsis, rice, and tomato
| Arabidopsis Strain Columbia | Rice japonica ‘Nipponbare’ | Tomato Strain ta496 | |
|---|---|---|---|
| No. of EST analyzed | 111,142 | 111,502 | 89,186 |
| Percent identity to genome | 95 | 95 | 95 |
| Clusters from all ESTs | 25,616 | 29,915 | 20,689 |
| Clusters with >1 EST | 11,466 | 14,682 | 10,614 |
| No. of ESTs in the AS pairs | 4,110 | 6,750 | 8,538 |
| AS pairs | 6,161 | 7,720 | 11,064 |
| AS clusters | 467 | 813 | 1,087 |
| Genome-aligned pairs (%) | 417 (89) | 708 (87) | 95 (86)a |
| Genome-aligned pairs with consensus intron borders (%) | 353 (76) | 570 (70) | 87 (81) |
| Intron retention (%) | 240 (68) | 347 (61) | 53 (61) |
| Retained introns median size | 93 bp | 104 bp | 139 bp |
| Exon skip (%) | 7 (2) | 28 (5) | 3 (3) |
| Exon skip median size | 51 bp | 90 bp | 158 bp |
| Alt 5′ or 3′ (%) | 93 (26) | 160 (28) | 22 (25) |
| Alt 5′ or 3′ median size | 19 bp | 33 bp | 26 bp |
| Other (%)b | 13 (4) | 35 (6) | 9 (10) |
| AS ratec | 4.1 | 5.5 | 10.2 |
The lower number of genome aligned pairs in tomato is due to the limited availability of genome sequence (about 2% of the genome).
Other includes transcript pairs with more than one type of AS in the same pair.
The AS rate is computed by the percent of AS clusters out of clusters >1 EST.
The veracity of EST processing by EPGA was examined by comparison of each EST pair to the genome, at a minimum identity of 95% using the BLAT program. Pairs aligning to the same genome location indicate that EPGA identified transcripts originating from the same loci. Of the 467 splice sites, 76% (353 pairs) were found to be authentic AS candidates by the following criteria: Each EST in the pair aligned to the same genome location, with consensus intron signals (Table II). Another 64/467 (14%) pairs align to the same genome location but contained nonconsensus intron border (not GT-AG, GC-AG, AT-AC). It should be noted that nonconsensus intron borders are present in The Institute for Genomic Research (http://www.tigr.org/tdb/e2k1/ath1/Arabidopsis_nonconsensus_splice_sites.shtml) and in other databases (Larkin and Park, 1999; Zhu et al., 2003). Significantly, genes with noncanonical intron borders were shown to undergo AS and are not necessarily the result of sequencing errors (Li et al., 2006). Thus, by including nonconsensus intron borders, 89% of the EPGA pairs could be confirmed as bona fide AS. The rest, 11%, aligned to more than one genome location, suggesting EST pairs that originate from multigene families. This can potentially cause false positives when searching for AS using ESTs alone. For example, in Arabidopsis 65% of the genes belong to multigene families (Arabidopsis Genome Initiative, 2000). However, due to the stringent requirements for exact pairing in the EPGA algorithm, interference from EST pairs arising from genes of a multigene family was at most 11%.
A similar process was carried out using EST from the rice (‘japonica’ group) database (Nipponbare strain). Of 892,016 ESTs available a random sample of 111,502 were chosen, comparable in size to that in Arabidopsis, and used for genome verification. In this case, the accuracy of EPGA is 70% without accepting noncanonical intron borders or 87% if noncanonical intron borders are accepted (Table II). For each pair the AS type was determined, as illustrated in Figure 1B, and the results are summarized in Table II. This process was also carried out for tomato for which only partial genomic sequence is available. The distribution of AS types was found to be similar to Arabidopsis and rice.
The proportion of intron retention reported by EPGA is high (above 60% of the total AS types) mainly due to the lack of retrieval of splicing events at the transcript termini. As shown previously (Ner-Gaon et al., 2004), transcript termini comprise 30% to 46% of the total AS. When this is taken into consideration the corrected proportion of intron retention type is closer to 45%. These results are similar to recent publications that indicate intron retention as the major type of AS in Arabidopsis (Iida et al., 2004; Ner-Gaon et al., 2004; Nagasaki et al., 2005) but differ in the estimation of exon skip that was reported to be 8.8% (Nagasaki et al., 2005) but here is only 2%.
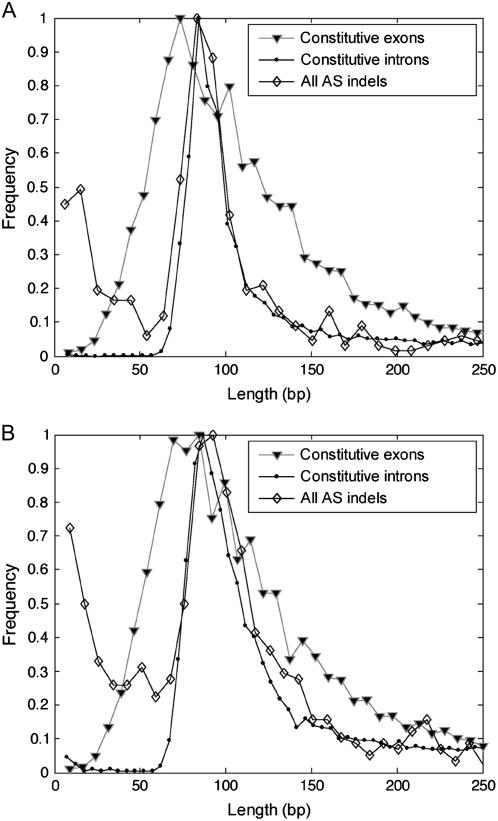
The distribution of all indel sizes detected by EPGA was analyzed and a graphical presentation of all indel sizes retrieved from Arabidopsis and rice is illustrated together with constitutive introns and exons (Fig. 2). The distribution for indel sizes between 60 and 120 bp follows the distribution of the constitutive intron size in both species more closely than that of the exon sizes. Another peak in the indel size at below 40 bp in size is the result of 5′ and 3′ alternative splice sites. Taken together, these results indicate that EPGA can faithfully report genome-wide splicing events within the median distribution of exons and introns for the model eudicot and monocot species.
Figure 2.
Distribution of indels detected by EPGA and the distribution of constitutive introns and exons in the genomes of Arabidopsis and rice. A, Distribution of constitutive exons, constitutive introns, and indel sizes in Arabidopsis. The ESTs were aligned to the genome and constitutive exon and constitutive intron sizes were calculated from the alignment results. Constitutive introns were extracted from the genomes in the places were the ESTs had gaps in the alignment to the genome and contained the canonical borders (GT-AG, GC-AG, AT-AC). Constitutive exons were alignments to the genome situated between two constitutive introns. Indel sizes were calculated from EPGA results. B, Distribution as in A but for rice (‘japonica’ group).
Comparative AS Rates and EST Database Size
To be able to compare AS rate in other plant species for which sequenced genomes are not available, we define the AS rate as the percentage of clusters that contain an AS event out of the total number of clusters retrieved with more than one EST. Thus, in Arabidopsis and rice the AS rate, using more than 110,000 ESTs, was 4.1 and 5.5, respectively (Table II). In tomato, the AS rate was 10.2, when using about 90,000 ESTs. Clusters do not necessarily indicate gene units, hence, the values of AS rates as defined do not specify the rate per gene (i.e. 10%–20% in Arabidopsis and rice; Wang and Brendel, 2006) but is useful to compare the relative AS activity between different plant species. Obviously, the number of AS detected will increase with the number of EST in a database, as with increasing number there is a greater chance of matching an indel-containing EST to an existing cluster (Kan et al., 2001; Brett et al., 2002). Thus, different species can be compared only when EPGA is carried out with an identical EST database sampling size. Since the number of EST in most plant species is less than that available in the model plant species we first examined if lower sampling size would maintain the relative differences in AS rates for Arabidopsis, rice, and tomato. Hence, the AS rate was computed by normalizing to a fixed EST sampling number of 20,000 ESTs. The computations were repeated six times by random selection from the specific plant dbEST. The results in Tables III and IV show that the relative AS rate ratio between Arabidopsis and rice (‘japonica’) was similar whether the sampling size was 20,000 or 110,000 ESTs (approximately 1.4). The AS rate ratio between Arabidopsis and tomato is approximately 2.7 in both sampling sizes.
Table III.
AS and EST clusters in eudicot dbEST
| Species | Common Name | Cultivar | Genome Sizea | AS Rateb | dbESTsc |
|---|---|---|---|---|---|
| Mbp | std | ||||
| Arabidopsis thaliana | Thale cress | ‘columbia’ | 120 | 1.9 (0.14) | 111,142 |
| ‘wassilewskija’ | 120 | 2.7 (0.23) | 49,667 | ||
| Lotus japonicus | ‘gifu b-129’ | 470 | 3.5 (0.31) | 48,654 | |
| Medicago truncatula | Barrel medic | ‘a17’ | 500 | 4.6 (0.33) | 66,752 |
| Malus domestica | Apple | ‘Royal gala’ | 750 | 4.6 (0.25) | 117,790 |
| Solanum lycopersicum | Tomato | ‘micro-tom’ | 950 | 5.7 (0.38) | 92,074 |
| ‘ta496’ | 950 | 5.6 (0.19) | 89,186 | ||
| Glycine max | Soybean | ‘williams’ | 1,200 | 6.0 (0.44) | 91,691 |
| ‘williams82’ | 1,200 | 5.2 (0.26) | 80,218 | ||
| Lactuca sativa | Lettuce | ‘salinas’ | 2,597 | 6.9 (0.3) | 80,144 |
Data for 1C value of genome size were obtained from Plant DNA C-values Database release 5.0, December, 2004 (http://www.kew.org/cvalues/CvalServlet?querytype=2) and, where possible, from direct sequencing results from NCBI Entrez Genome Project database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=genomeprj).
Selections of 20,000 EST were done in six random passes and were matched against each other. The alignment results were parsed for AS pairs and clusters and the AS rate was computed as described in the “Materials and Methods.” The complete list appears in Supplemental Table S3. The average AS rate and sd are shown here.
dbEST indicates the number of ESTs greater than 100 bp size, after removal of poly A and vector contamination, from a specific cultivar. The cultivar used was in all cases the cultivar that provided at least 40,000 ESTs.
Table IV.
AS and EST clusters in monocot dbEST
Notes as in Table III.
| Strain | Common Name | Cultivar | Genome Sizea | AS Rateb | dbESTsc |
|---|---|---|---|---|---|
| Mbp | std | ||||
| Oryza sativa (‘japonica’ group) | Rice | ‘nipponbare’ | 390 | 3.0 (0.41) | 893,777 |
| Sorghum bicolor | Sorghum | ‘btx623’ | 760 | 7.2 (0.27) | 128,043 |
| Zea mays | Corn | ‘b73’ | 2,400 | 5.6 (0.43) | 645,237 |
| ‘f2’ | 2,400 | 5.5 (0.21) | 58,150 | ||
| ‘oh43’ | 2,400 | 4.5 (0.32) | 54,168 | ||
| Hordeum vulgare | Barley | ‘barke’ | 5,000 | 3.1 (0.31) | 96,462 |
| ‘haruna nijo’ | 5,000 | 2.6 (0.15) | 86,597 |
Estimation of AS Rates in Plant Species
The EPGA algorithm was applied to other species to ascertain their relative AS rates. Species with recent polyploid ancestry (e.g. potato [Solanum tuberosum] and wheat [Triticum aestivum]) were avoided to prevent potential complications arising from comparisons of transcripts of genes arising from homologous chromosomes. To maintain sufficient sampling size, only ecotypes/cultivars that include at least 40,000 high quality ESTs were used. In addition, the different ecotypes/cultivars were processed separately to avoid polymorphisms that may originate from evolution-derived divergence rather than from AS and to enable cross-cultivar comparison.
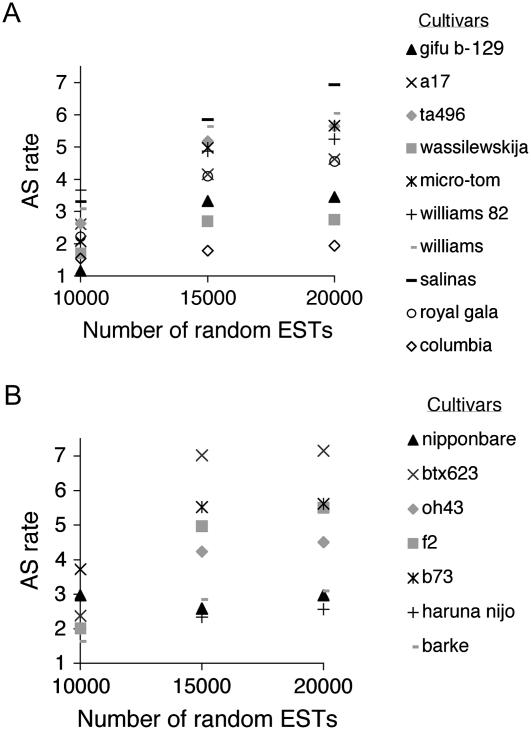
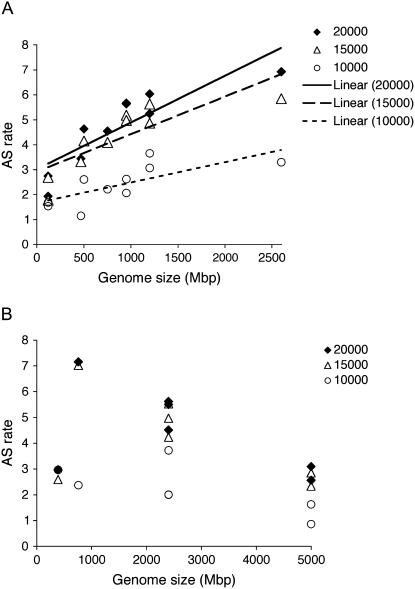
We first examined the variation in AS rate as a function of recurrent sampling size. As shown in Figure 3, A and B, for all monocot and eudicot species, the ratio of AS rates between species remains relatively constant. This indicates that the rate of AS discovery is similar within these sampling sizes and a sampling size of 20,000 was adopted. Table III summarizes the results for eudicots (groups Rosids and Asterids) that were applied to all species with sufficient EST. Due to their more recent evolutionary origin one may expect that different cultivars of the same species would exhibit similar AS rates. Indeed, the AS rates of the different strains were very similar, differing by 2% in tomato (‘micro-tom’ compared to ‘ta496’), 13% in soybean (Glycine max; ‘Williams’ and ‘williams82’), and 30% in Arabidopsis (‘columbia’ versus ‘wassilewskija’). The relatively similar AS rates detected between cultivars lends credence to the significance of AS rates reported by the EPGA algorithm. In comparison, the differences in AS rates between species can be higher and reaches 3.6-fold (e.g. compare lettuce [Lactuca sativa] to Arabidopsis strain Columbia). Interestingly, when AS rates are graphed relative to the genome size, a linear correlation is obtained (R2 = 0.75, P value = 0.001; Fig. 4A). Analysis of the lower sampling sizes of 15,000 and 10,000 ESTs showed a similar linear correlation with reduced R values of 0.67 and 0.53, respectively. Taken together, the results indicate that the majority of variance in AS rate can be attributed to the increase in eudicot genome size.
Figure 3.
AS rate as a function of EST sampling size in eudicot and monocot species. A, Variation in the AS rate as a function of EST sampling size in eudicot species. The AS rate was calculated using EPGA for randomly selected sampling sizes as indicated (see “Materials and Methods”). The AS rate shown is the average of six trials. The following species and cultivars are shown: Lotus japonicus (‘gifu b-129’), Medicago truncatula (‘a17’), Solanum lycopersicum (‘ta496’), Arabidopsis (‘wassilewskija’), tomato (Solanum lycopersicum; ‘Micro-tom’), soybean (‘williams 82’), soybean (‘williams’), lettuce (‘salinas’), Malus x domestica (‘royal gala’), and Arabidopsis (‘columbia’). The sds are smaller than the size of the symbols and are not shown. B, Distribution as in A but for monocot species. The following species and cultivars are shown: rice (‘Nipponbare’), sorghum (‘Btx623’), maize (‘Oh43’), maize (‘F2’), maize (‘b73’), barley (‘Haruna nijo’), and barley (‘Barke’). The sds are smaller than the size of the symbols and are not shown.
Figure 4.
AS rates in eudicot and monocot species as a function of genome size. Randomly selected 20,000, 15,000, and 10,000 ESTs of each cultivar (average and sd of six independent samplings) were used. The AS were normalized to the number of clusters containing more than one EST. Results are an average of the recurrent AS rate measurements plotted as a function of genome size. Cultivar-specific databases were examined as described in the text. A, AS rates in eudicot species. B, AS rates in monocot species.
Similar analysis was carried out in monocot species and their cultivars for which sufficient EST are available (Table IV). Comparison of AS rates between monocot species (Poaceae) show 20% variance in rates between strains of the same species. Furthermore, AS rates in monocots are not correlated with genome size (Fig. 4B).
DISCUSSION
AS Rates and Genome Complexity
AS can increase the complexity of a genome, it is therefore expected that increased AS rates would be correlated with increased organism complexity. Indeed, simple eukaryotes like yeast show reduced introns and AS rate relative to animals (Brett et al., 2002; Kim et al., 2004) while a comparison of AS rates in higher animal species, e.g. mouse and man, show only small differences (Nagasaki et al., 2005). The difference in the level of AS suggests that AS may contribute greatly to the higher level of phenotypic complexity in mammals (Kim et al., 2007). The human and mouse genomes have diverged 100 million years ago, an extent of time that is similar to the divergence within the eudicot groups examined here (Rosids and Asterids) that have diverged over the last 110 million years (Sanderson et al., 2004). However, in these species, and despite comparable temporal evolutionary divergence, AS rates differ significantly. While humans and mice show relatively small differences in AS rate, the rates in plants vary over 3.6-fold. Indeed, the AS rates that are commonly cited for comparison are based on the two model plants that have sequenced genomes. However, as shown here, Arabidopsis and rice actually represent the lower end of AS rates in vascular plants. In a few eudicot and monocot species, e.g. lettuce and sorghum (Sorghum bicolor), the expectation is that the extent of AS will be comparable to that seen in humans, although the distribution of the types and the regulation may differ. Interestingly, based on partial genome sequence, the distribution of AS types in tomato is similar to that detected in rice and Arabidopsis. Thus, higher rates of AS may maintain similar profiles of AS type distribution.
The species surveyed represent not only a large span of vascular plant evolutionary divergence, i.e. monocots and eudicots, but also includes more than a 40-fold range of genome sizes. Remarkably, the AS rate was correlated with eudicot genome size. Larger genome size does not necessarily indicate more genes. For example, soybean that has a genome 10-fold larger than Arabidopsis, is considered to have no more than 25% of additional genes (Young et al., 2005). One possible explanation for the relationship of AS rates to genome size is that the mechanisms that lead to genome size increments contributes to the molecular platform for enhanced AS. In primates, Alu element sequences have been shown to contain weak splice sites and to undergo a process of exonization (Sorek et al., 2002; Singer et al., 2004). The human transposable Alu elements were found to be responsible for more than 15% of the splice variants (Nekrutenko and Li, 2001). Retrotransposon proliferation is a common mechanism for plant genome size expansion (SanMiguel et al., 1996; Vicient et al., 1999; Shirasu et al., 2000; Meyers et al., 2001; Terol et al., 2001; Wessler, 2001) and in maize (Zea mays) up to 78% of the genome is made up of diverse retrotransposon families (SanMiguel et al., 1998). These elements have been directly implicated in causing AS in both maize and tobacco (Nicotiana tabacum; Varagona et al., 1992; Marillonnet and Wessler, 1997; Leprinc et al., 2001). Thus, AS-linked transposon processes may also be responsible for the trend of increased AS in eudicot species.
A major exception to this trend is in monocots, in which the increased size seems to have saturated in many of the species for additional AS increment. With respect to the increase in genome size in cereals, it has been noted that the preponderance of expansion is due to nested retrotransposons, i.e. insertions within each other and not within genes (Shirasu et al., 2000; Kalendar et al., 2004). Hence, beyond a certain size further growth of the genome will be within futile noncoding regions and any transposable element-associated AS rate rise will tend to reach a plateau.
Divergence in AS Rates within Species
Inspection of AS rates between different cultivars of the same species reveals similar AS rates within the majority of eudicot species. As most cultivars are of very recent origin the similarity in rates is to be expected. When comparing AS rate in one species, the two ecotypes of Arabidopsis, Columbia and Wassilewskija, show the highest difference (30%) in their AS rate. Genetic diversity between Arabidopsis ecotypes as shown by amplified fragment length polymorphism analysis points to distinct subgroups or genotypes. The intraecotypic differences reflect natural variation that is fixed in discrete genotypes because of the self-fertilizing nature of Arabidopsis (Breyne et al., 1999). Wassilewskija has previously been shown to contain altered red/far-red responses and respond differently to stress than Columbia (Aukerman et al., 1997; Kalbina and Strid, 2006). This work highlights another difference as described by AS rates. Cereals show about 20% differences in AS rates between cultivars. This may be due to the intense selection pressure affected by ancient human-mediated cultivation and natural selection. Cultivated barley (Hordeum vulgare) exhibits multicentric origin as an arch starting in Morocco and ending in Tibet that would contribute to great diversity among existing cultivars (Molina-Cano et al., 1999). Maize strains are phenotypically and genetically diverse as well. For more than 6,000 years of cultivation, farmers and breeders have exploited this diversity, which is 2- to 5-fold higher than that of other domesticated grass crops (Buckler et al., 2001; Tenaillon et al., 2001). Hence, diverse AS rate is correlated with niche specialization brought upon by extended domestication in different geographical regions.
Despite the lack of a complex developmental style and sophisticated neuronal and adaptive immune systems that typify mammalians, we show here that plants can host a high degree of genome complexity. Plants, as sessile organisms, must adapt their growth and metabolic style to a changing environment. The great diversity in AS rates indicates that it can play a role in plant adaptation. Interestingly, environmental stress has been shown to activate plant retrotransposon mobility and activation of genes, suggesting an adaptive advantage to this type of genome restructuring (Kalendar et al., 2000; Wessler, 2001; Kashkush et al., 2003). This change may also impact on gene diversity afforded by AS and be an additional element of fitness benefit appropriated by a larger genome size. Future work that delineates and compares the dynamic aspects of AS in normal development and during stress will contribute to the understanding of its biological significance.
MATERIALS AND METHODS
Data Sources
Plant ESTs for eudicots and monocots were obtained from NCBI dbESTs 23/12/2006 (ftp://ftp.ncbi.nih.gov/genbank/). Based on their annotation, plant species were separated by cultivar to create a cultivar-specific database. For the Arabidopsis (Arabidopsis thaliana) genome sequence, the January 22, 2004 version was obtained from The Arabidopsis Information Resource center: ftp.arabidopsis.org/home/tair/Sequences/whole_chromosomes/.
For the rice (Oryza sativa) genome sequence, IRGSP Releases Build 4.0, Pseudomolecules of the Rice Genome (rice sp. japonica ‘Nipponbare’) was used (http://rgp.dna.affrc.go.jp/E/IRGSP/Build4/build4.html). The Build 4.0 Pseudomolecules were constructed based on the data freeze on January 25, 2005. Programs to extract EST details and genomic location are available upon request from H.N.-G.
Quality Control of ESTs and Libraries
As ESTs are the bases for comparison it was important that the quality and the uniformity of the dbEST be rigorously examined according to the following criteria: (1) Poly A from the end of the ESTs was deleted. (2) ESTs were aligned to a vector database that was obtained from NCBI 26/11/03 (ftp://ftp.ncbi.nih.gov/blast/db/FASTA/vector.gz) using BLAT (Kent, 2002) with minimum identity of 95%. The default conditions were used for the other BLAT parameters. Vector contamination from the edge of the ESTs was removed while ESTs containing vector sequence within the sequence were not used. (3) Only ESTs equal to or above 100 bp size and with less than 10% Ns were used. (4) The ESTs were collected into a database by their cultivar classification. To reduce the possibility of genomic contamination, specific libraries that include more than 1% ESTs containing multiple indels, as measured by the EPGA algorithm, were not used. Libraries listed as EST but prepared from the assembled 5′- and 3′-RACE product nucleotide sequences to obtain full-length cDNA were not used (Xiao et al., 2002). (5) To ensure uniformity cultivar databases were used only if their average EST length was within 2 sd of the average EST length computed from all the databases used. (6) The ESTs of each cultivar were clustered by simple transitive clustering. To ensure uniformity the cultivar databases were used only if their average cluster number was within 2 sd of the average cluster number computed from all the databases used.(7) Cultivars that had an unusually high amount (>20%) of ESTs that originated from flowers, fruit, or berries, or that had less than 40,000 high quality ESTs were not used.
A summary of the above analysis is shown in Supplemental Table S1.
Description of the EPGA Algorithm and Calculation of AS Rate
EPGA method was used to identify AS. Pairs of ESTs that share two sequentially matching regions that flank a discontinuity in the alignment were considered to be AS pair. For each dbESTs, BLAT (Kent, 2002) was used to identify all pairs of ESTs that show alignment for at least 75 bp (with a minimum identity of 95% and a tile size of 18). A pair was reported to indicate AS if it had at least 25 bp long alignment, followed by an indel of at least 5 bp, followed by a subsequent match of at least 25 bp. All pairs that were obtained in each species appear in Supplemental Table S2.
The ESTs of each cultivar were clustered by simple transitive clustering. An EST was considered to belong to a cluster if it aligned to one of the ESTs in that cluster with an overlap of at least 75 bp in total, with minimum identity of 95%. The clusters were defined as AS clusters if they contained at least one AS pair.
The percent AS rate was calculated by dividing the number of AS clusters by the total number of clusters with more than one EST. EPGA algorithm was applied to six different random samples, each of 10,000, 15,000, and 20,000 ESTs. The programs for filtering and clustering were all written in PERL, and are available upon request from H.N.-G. The complete analysis of the 20,000 random samples is shown in Supplemental Table S3.
Genome Size
Data for 1C value of genome size were obtained from Plant DNA C-values Database (http://www.kew.org/cvalues/CvalServlet?querytype=2) or from direct sequencing results from NCBI Entrez Genome Project database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=genomeprj).
Median Size of Constitutive Exons and Introns
The list of all the plant genome accession numbers (GI) is available at ftp://ftp.ncbi.nlm.nih.gov/genomes/PLANTS/BLASTDB/. The tomato (Solanum lycopersicum) genome sequence was downloaded from http://www.sgn.cornell.edu/bulk/input.pl?mode=bac. The ESTs obtained after quality control were applied and were aligned to the genomic sequences using BLAT. Only ESTs that match the genome for at least 75 bp, with a minimum identity of 95%, and had less than 5 bp gap in the EST were used. Gaps of at least 5 bp length in the alignment were considered as constitutive introns. The sizes of the aligned regions between the gaps were registered as constitutive exons. The lengths of the exons and introns were extracted from the alignment results by a program written in PERL.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Quality-control processing of ESTs.
Supplemental Table S2. List of all EST pairs from all plant species and cultivars analyzed.
Supplemental Table S3. Cluster and AS pair results obtained from six random samplings of 20,000 ESTs each.
Supplementary Material
This work was supported by the Israel Science Foundation (grant no. 388/02) and the Binational Agriculture Research and Development (grant no. IS–3454–03).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert Fluhr (robert.fluhr@weizmann.ac.il).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA (1997) A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett D, Pospisil H, Valcarcel J, Reich J, Bork P (2002) Alternative splicing and genome complexity. Nat Genet 30 29–30 [DOI] [PubMed] [Google Scholar]
- Breyne P, Rombaut D, Van Gysel A, Van Montagu M, Gerats T (1999) AFLP analysis of genetic diversity within and between Arabidopsis thaliana ecotypes. Mol Gen Genet 261 627–634 [DOI] [PubMed] [Google Scholar]
- Brown JW, Simpson CG, Thow G, Clark GP, Jennings SN, Medina-Escobar N, Haupt S, Chapman SC, Oparka KJ (2002) Splicing signals and factors in plant intron removal. Biochem Soc Trans 30 146–149 [DOI] [PubMed] [Google Scholar]
- Buckler ES, Thornsberry JM, Kresovich S (2001) Molecular diversity, structure and domestication of grasses. Genet Res 77 213–218 [DOI] [PubMed] [Google Scholar]
- Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR (2006) Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics 7 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al (2005) The transcriptional landscape of the mammalian genome. Science 309 1559–1563 [DOI] [PubMed] [Google Scholar]
- Collins L, Penny D (2006) Proceedings of the SMBE Tri-National Young Investigators' Workshop 2005: investigating the intron recognition mechanism in eukaryotes. Mol Biol Evol 23 901–910 [DOI] [PubMed] [Google Scholar]
- Croft L, Schandorff S, Clark F, Burrage K, Arctander P, Mattick JS (2000) ISIS, the intron information system, reveals the high frequency of alternative splicing in the human genome. Nat Genet 24 340–341 [DOI] [PubMed] [Google Scholar]
- Iida K, Go M (2006) Survey of conserved alternative splicing events of mRNAs encoding SR proteins in land plants. Mol Biol Evol 23 1085–1094 [DOI] [PubMed] [Google Scholar]
- Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K (2004) Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res 32 5096–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki M, Tsumoto A, Shimamoto K (2006) The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of pre-mRNA. Plant Cell 18 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbina I, Strid A (2006) Supplementary ultraviolet-B irradiation reveals differences in stress responses between Arabidopsis thaliana ecotypes. Plant Cell Environ 29 754–763 [DOI] [PubMed] [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH (2000) Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA 97 6603–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoy A, Schulman AH (2004) Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166 1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M, Lopato S, Voronin V, Barta A (2006) Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res 34 4395–4405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, Garrett-Engele PW, Johnson JM, Castle JC (2005) Evolutionarily conserved and diverged alternative splicing events show different expression and functional profiles. Nucleic Acids Res 33 5659–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, Rouchka EC, Gish WR, States DJ (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res 11 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, States D, Gish W (2002) Selecting for functional alternative splices in ESTs. Genome Res 12 1837–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33 102–106 [DOI] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G (2007) Different levels of alternative splicing among eukaryotes. Nucleic Acids Res 35 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Klein R, Majewski J, Ott J (2004) Estimating rates of alternative splicing in mammals and invertebrates. Nat Genet 36 915–916; author reply 916–917 [DOI] [PubMed] [Google Scholar]
- Lareau LF, Green RE, Bhatnagar RS, Brenner SE (2004) The evolving roles of alternative splicing. Curr Opin Struct Biol 14 273–282 [DOI] [PubMed] [Google Scholar]
- Larkin PD, Park WD (1999) Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant Mol Biol 40 719–727 [DOI] [PubMed] [Google Scholar]
- Leprinc AS, Grandbastien MA, Christian M (2001) Retrotransposons of the Tnt1B family are mobile in Nicotiana plumbaginifolia and can induce alternative splicing of the host gene upon insertion. Plant Mol Biol 47 533–541 [DOI] [PubMed] [Google Scholar]
- Li J, Li X, Guo L, Lu F, Feng X, He K, Wei L, Chen Z, Qu LJ, Gu H (2006) A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. J Exp Bot 57 1263–1273 [DOI] [PubMed] [Google Scholar]
- Lorkovic ZJ, Barta A (2002) Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res 30 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansilla A, Lopez-Sanchez C, de la Rosa EJ, Garcia-Martinez V, Martinez-Salas E, de Pablo F, Hernandez-Sanchez C (2005) Developmental regulation of a proinsulin messenger RNA generated by intron retention. EMBO Rep 6 1182–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillonnet S, Wessler SR (1997) Retrotransposon insertion into the maize waxy gene results in tissue-specific RNA processing. Plant Cell 9 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Tingey SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11 1660–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AA, Fickett JW, Gelfand MS (1999) Frequent alternative splicing of human genes. Genome Res 9 1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B, Lee C (2002) A genomic view of alternative splicing. Nat Genet 30 13–1911753382 [Google Scholar]
- Modrek B, Lee CJ (2003) Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet 34 177–180 [DOI] [PubMed] [Google Scholar]
- Molina-Cano JL, Moralejo M, Igartua E, Romagosa I (1999) Further evidence supporting Morocco as a centre of origin of barley. Theor Appl Genet 98 913–918 [Google Scholar]
- Nagasaki H, Arita M, Nishizawa T, Suwa M, Gotoh O (2005) Species-specific variation of alternative splicing and transcriptional initiation in six eukaryotes. Gene 364 53–62 [DOI] [PubMed] [Google Scholar]
- Nekrutenko A, Li WH (2001) Transposable elements are found in a large number of human protein-coding genes. Trends Genet 17 619–621 [DOI] [PubMed] [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39 877–885 [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, Thorne JL, Wikstrom N, Bremer K (2004) Molecular evidence on plant divergence times. Am J Bot 91 1656–1665 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nat Genet 20 43–45 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, et al (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274 765–768 [DOI] [PubMed] [Google Scholar]
- Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P (2000) A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res 10 908–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SS, Mannel DN, Hehlgans T, Brosius J, Schmitz J (2004) From “junk” to gene: curriculum vitae of a primate receptor isoform gene. J Mol Biol 341 883–886 [DOI] [PubMed] [Google Scholar]
- Sorek R, Ast G (2003) Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res 13 1631–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Ast G, Graur D (2002) Alu-containing exons are alternatively spliced. Genome Res 12 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS (2001) Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc Natl Acad Sci USA 98 9161–9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terol J, Castillo MC, Bargues M, Perez-Alonso M, de Frutos R (2001) Structural and evolutionary analysis of the copia-like elements in the Arabidopsis thaliana genome. Mol Biol Evol 18 882–892 [DOI] [PubMed] [Google Scholar]
- Varagona MJ, Purugganan M, Wessler SR (1992) Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell 4 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Suoniemi A, Anamthawat-Jonsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH (1999) Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11 1769–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, Brendel V (2006) Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA 103 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler SR (2001) Plant transposable elements: a hard act to follow. Plant Physiol 125 149–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YL, Malik M, Whitelaw CA, Town CD (2002) Cloning and sequencing of cDNAs for hypothetical genes from chromosome 2 of Arabidopsis. Plant Physiol 130 2118–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S (2005) Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol 137 1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Schlueter SD, Brendel V (2003) Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol 132 469–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.