Abstract
High-throughput gene expression analysis of genes expressed during salt stress was performed using a novel multiplexed quantitative nuclease protection assay that involves customized DNA microarrays printed within the individual wells of 96-well plates. The levels of expression of the transcripts from 16 different genes were quantified within crude homogenates prepared from Arabidopsis (Arabidopsis thaliana) plants also grown in a 96-well plate format. Examples are provided of the high degree of reproducibility of quantitative dose-response data and of the sensitivity of detection of changes in gene expression within limiting amounts of tissue. The lack of requirement for RNA purification renders the assay particularly suited for high-throughput gene expression analysis and for the discovery of novel chemical compounds that specifically modulate the expression of endogenous target genes.
High-throughput screens using mammalian cells are widely employed within the biomedical community for the discovery of novel pharmacological compounds. In such screens, very many samples of identical cells are treated in vitro with different potential agonists or antagonists, and a single readout is typically employed to record the cellular response of interest. Chemicals are identified that induce the desired response. Design strategies are then applied to these chemicals to identify structural variants that are most effective and selective with respect to the process of interest, and these subsequently become drug candidates. This type of approach is greatly facilitated if the cellular response can be quantitatively measured as a function of the concentration of each test compound to provide a precise EC50 value (the concentration of each compound leading to a half-maximal effect) because EC50 values permit quantitative comparisons of the relative efficacy and selectivity of different compounds.
Biochemical enzyme and receptor assays were the first to provide the high sample throughput required for screening, the sensitivity to differentiate weak effects above that of background, and the quantitative reproducibility required to precisely determine EC50 values from dose-response data (Sittampalam et al., 1997). More recently, attention has turned to gene expression assays because they offer the potential to describe molecular phenotypes, defined as the combination of genes whose expression gives rise to a specific cellular state (Hughes et al., 2000). In many studies, it has been shown that there are unique and consistent clusters of genes whose coordinated regulation of expression is associated with various disease, drug, tissue, or cellular phenotypes (e.g. see Strachan et al., 1997; Golub et al., 1999; Hughes et al., 2000; Schmid et al., 2005; Lamb et al., 2006). These studies have, for the most part, employed microarrays in which DNA sequences specific to individual transcripts are immobilized at defined locations in an array format on a solid surface, either through mechanical spotting or some implementation of in situ synthesis (Fodor et al., 1991; Schena et al., 1995; Singh-Gasson et al., 1999; Deyholos and Galbraith, 2001).
Interest exists in providing high-throughput platforms and readout technologies that can selectively examine changes in the transcript levels of specific genes in response to multiple treatments. Although conventional microarrays, in principle, have the necessary throughput, they are limited with respect to cost, reproducibility, and dynamic range and therefore are not well suited for the task of producing dose-response data for multiple chemicals from which accurate and reliable EC50 values can be calculated. An alternative to microarrays has recently been described (Martel et al., 2002a, 2002b). Based on a quantitative nuclease protection assay (qNPA), this platform has been used with mammalian cells for high-throughput screens to identify small molecules that specifically perturb the expression of defined target genes (Martel et al., 2002a, 2002b). Efficiencies of cost, as compared to microarrays, are provided by using a standard 96-well plate both for growth of the cultured cells and as the assay format, with up to 16 genes being simultaneously measured within each well of the corresponding ArrayPlate (Martel et al., 2002a, 2002b). The strengths of qNPA are that (1) it is highly reproducible, permitting acquisition of precise dose-response data and calculation of EC50 values; (2) it is highly sensitive and has an exceptionally large dynamic range, which provides the ability to simultaneously identify small changes due to weakly active compounds, as well as large changes due to strongly active compounds; and (3) it provides high sample throughput, allowing rapid screening of large libraries of chemical compounds. This platform is therefore ideally suited for comprehensive screens for small-molecule effectors that regulate expression of specific genes of interest. Such molecules represent drug candidates in biomedical research and in the agricultural sector represent a category of molecules of increasing recent interest both at the basic and applied levels (Raikhel and Pirrung, 2005).
High-throughput in vivo screens for small-molecule effectors have been widely and historically employed within the agricultural research and development community. Such screens typically used whole plants and have been most notably employed for the identification of herbicides. However, even though regulation of gene expression is an important level of control of cellular and whole-organism phenotype and function, development of in vivo high-throughput screens based on molecular readouts of plant gene expression has not been reported. In part, this has been due to a lack of platforms for the analysis of gene expression that combine reproducibility with accuracy, high sample throughput, easy automation, sufficient signal-to-noise sensitivity, and cost effectiveness. For most gene expression analysis platforms, including both those based on microarrays and on PCR (Czechowski et al., 2004), or on sequencing-based strategies (for review, see Meyers et al., 2004), costs are prohibitive for processing the very high numbers of samples needed for in-depth sampling of chemical libraries for agrochemical discovery. Furthermore, to pursue gene-based discovery and development programs, it is necessary to have an assay capable of providing accurate dose-response data that is repeatable over time and from which precise EC50 values can be calculated. This allows comparison of the efficacy and specificity of analogs during the subsequent profiling and optimization of leads required for the development of potential commercial products. There is increasing interest emerging within the agricultural sector in discovering novel chemicals having precise in vivo effects upon gene expression. In part, this is due to the recognition that small-molecule interactions modulate many responses of organisms to their environments through mechanisms involving changes in gene expression and that identification of small molecules that induce the expression of specific genes, such as those involved in defense responses, for example, could be a useful, selective, and effective adjuvant to crop production under adverse conditions.
In this article, we demonstrate the application of qNPA ArrayPlate technology for high-throughput in vivo screening for novel chemical entities that have defined effects on gene expression in higher plants. We demonstrate that the technology is accurate, sensitive, and highly specific and has a large dynamic range. Finally, we show that it can be applied in a cost-effective manner for high-throughput small-molecule discovery and can be used for optimization of leads identified from screening chemical libraries.
RESULTS
Description of the Technology
ArrayPlate qNPA kits (HTG) were used for all components of the assay, from sample lysis to measurement of transcript levels. An overview of the experimental pipeline for the technology is given in Supplemental Figure S1. Individual plants were grown under axenic conditions in 96-well microplates and were treated with the test chemicals. Alternatively, explants, such as leaf punches, may be removed from soil-grown plants and transferred to microplates either before or after chemical treatment. Following addition of lysis buffer included with the qNPA kit, the tissues were mechanically homogenized. Aliquots were transferred into a second plate for RNase protection treatment. Following completion of this step, aliquots from each well were transferred into corresponding wells in the ArrayPlate for quantification of transcript levels, simultaneously measuring the expression of 16 genes per well. A detailed description of the chemical and biochemical manipulations is provided in “Materials and Methods.”
Conceptually central to the technology is the release of total RNA from the plant samples by homogenization in lysis buffer followed by generation of a stoichiometric number of nuclease protection probes from the lysate (Supplemental Fig. S2). Critical to high-throughput considerations, this conversion of unstable RNA to stable DNA does not require that the RNA be purified or extracted from the lysate. Briefly, protection oligonucleotides were added to the lysate and allowed to form RNA/DNA heteroduplexes. Nonhybridized or poorly hybridized mismatched protection oligonucleotides were hydrolyzed using S1 nuclease, as was the overhang S1 control sequence of each probe hybridized to RNA. This leaves the protected target RNA/probe DNA heteroduplexes which, because the protection oligonucleotides were provided in excess of the RNA, were produced stoichiometrically. RNA was then eliminated from the heteroduplexes by alkali treatment, leaving single-strand probe DNA molecules, whose amounts are proportional to the amounts of the corresponding input RNA transcripts. The neutralized digest was then added to the ArrayPlate.
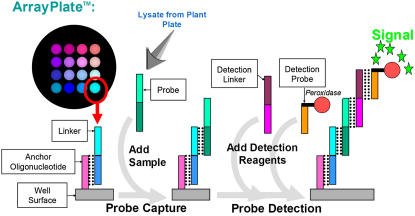
Figure 1 illustrates an array from one well, which was reiterated within all the wells of the 96-well plate. The kit used in this study provides multiplexed measurement of the amounts of up to 16 separate protection fragments based on chemiluminescence. Each well was printed with an identical universal array comprising 16 different 25-mer oligonucleotides, which provide locations addressable by hybridization of subsequent sets of custom oligonucleotides (Fig. 1). These arrays were used in a progressive sandwich assay format: This starts with programming the arrays using sets of 16 specific DNA 50-mers designed such that one-half of each oligonucleotide was complementary to one of the 16 universal anchor sequences printed within the wells and such that the second half of the oligonucleotide was complementary to one-half of the sequence of one of the specific protection probes generated by nuclease protection. Programmed arrays were in this way customized to capture 16 different nuclease protection probes. The next step in the assay employed third and fourth layers of hybridization to detect the amounts of the specific nuclease protection probes immobilized at the programmed locations of the universal array. This involved the addition of a mixture of 16 detection linkers, each comprising a different gene-specific 50-mer, the first half of which was complementary to the remaining half of the individual protection probes, and the second half of which was a universal sequence complementary to a final oligonucleotide sequence attached to the detection probe used to measure the abundance of every gene in the array. The detection probe was a 25-mer oligonucleotide covalently linked to horseradish peroxidase (HRP) and formed the final component of a five-part hybrid complex. Chemiluminescent detection, coupled to high-sensitivity imaging, was used to quantify the amounts of probe bound at the individual array element locations. Previous experiments have consistently demonstrated that addition of RNase to lysate destroys the signal, whereas treatment with DNase has no impact. This has been observed for extracts prepared from mammalian cells, fixed tissue, and organisms (B. Seligmann, unpublished data).
Figure 1.
Means whereby processed nuclease protection probes are captured and detected at specific array elements in the ArrayPlate. The S1-processed lysates (Supplemental Fig. S1) are added to the corresponding wells of a plate previously programmed by addition of linkers to capture each specific nuclease protection probe. These nuclease protection probes hybridize to their complementary array-bound programming linker. Detection linkers are added next, each hybridizing to its respective nuclease protection probe. The media are then aspirated and replaced with media containing detection probes. Detection probes hybridize to each array-bound detection linker. The plate is then washed and substrate for HRP is added, producing chemiluminescence at each array element location, which is then quantitatively imaged.
Validation of the qNPA Platform for Measurement of Alterations in Plant Gene Expression
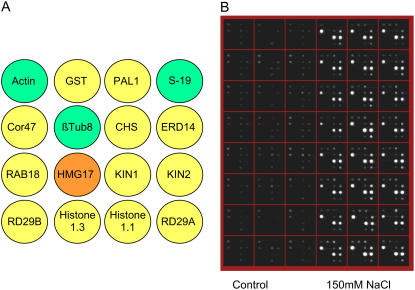
For the qNPA platform to be useful in the high-throughput analysis of plant gene expression, it must be capable of detecting expected changes in gene expression in response to specific treatments, using plants grown within its 96-well plate format or from plant tissues with sufficient reproducibility. To provide this validation, we selected a number of genes known to be up-regulated by NaCl stress and corresponding control genes that are unaffected for design of nuclease protection fragments. Sequence design was done based on identification of specific exonic regions within these genes having similar GC content. We employed software for this purpose commercially available from HTG, but any primer design software can achieve the same results. The genes, nuclease protection probe sequences, and their functions are listed in Supplemental Tables S1 and S3. The universal arrays were programmed to measure three housekeeping genes (actin, tubulin, and S-19), one negative control gene (human β-thromboglobulin), which should never give a signal, and eight genes: GST, COR47, KIN1, KIN2, RD29A, RD29B, ERD14, and PAL1 (Supplemental Tables S1 and S3), which were all known to be induced by osmotic stress, including NaCl treatment (Jenkins et al., 2001; Nylander et al., 2001; Takahashi et al., 2004; Kim et al., 2005). The remaining genes in the array were for other experimental purposes not described in this article. Arabidopsis seedlings, grown for 10 to 14 d within the wells of 96-well plates, were treated for 4 h with 150 mm NaCl and homogenized. The homogenates were analyzed for transcript abundance using the qNPA assay. The primary image of a typical plate, in this case representing 24 biological replicates for the control and treated samples, is provided in Figure 2. Alterations in gene expression monitored by the platform were highly reproducible, as determined by quantification of the data extracted from the image files (Fig. 3).
Figure 2.
Layout of the microarrays within the ArrayPlate wells after programming to detect 16 specific transcripts. A, Green circles contain control housekeeping genes used to normalize the data, the orange circle contains a human gene used as a negative control (which should never provide a signal), and yellow circles represent test genes. B, Image of chemiluminescence emission captured at the positions of the array elements. Analysis was done of transcripts in homogenates from plants following treatment with 150 mm NaCl or the corresponding water control.
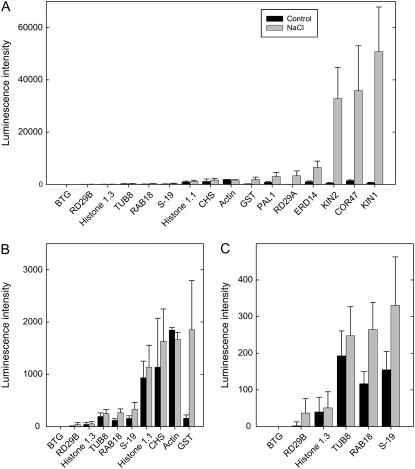
Figure 3.
Graphic analysis of data extracted from the image shown in Figure 2. The intensity values are means (n = 24; one plant per well), normalized to the levels of the transcripts of both actin and S-19 as housekeeping genes (the averaged normalized value is 1,000), with sds depicted by the error bars. A, Data for all transcripts. B, Figure rescaled to accommodate transcripts at medium levels. C, Figure rescaled to accommodate transcripts at low levels.
In terms of differential gene expression, the expected induction of transcript levels was observed for GST, COR47, KIN1, KIN2, RD29A, RD29B, ERD14, and PAL1. These results were concordant with results obtained using Affymetrix-based transcriptional profiling (Nover et al., 2005) and using other array platforms (Nylander et al., 2001; Takahashi et al., 2004; Kim et al., 2005). In comparison with the Affymetrix dataset, with the exception of KIN2, which was not represented on the GeneChip, those genes whose expression was found to be induced using the qNPA assay were all induced at least 2-fold in the Affymetrix dataset. Correspondingly, none of the remaining genes represented by our probe set, which were not highly responsive to NaCl treatment, were induced 2-fold or more in the Affymetrix dataset. Thus, using Affymetrix data as the basis for comparison, neither false-positive nor false-negative hits were detected in our assay. In comparison to data obtained using other microarray platforms, similar levels of induction were observed for COR47, KIN1, and KIN2 (Seki et al., 2002; Ma et al., 2006). We conclude that the qNPA platform provides results that are concordant with other microarray expression platforms.
Assay Sensitivity
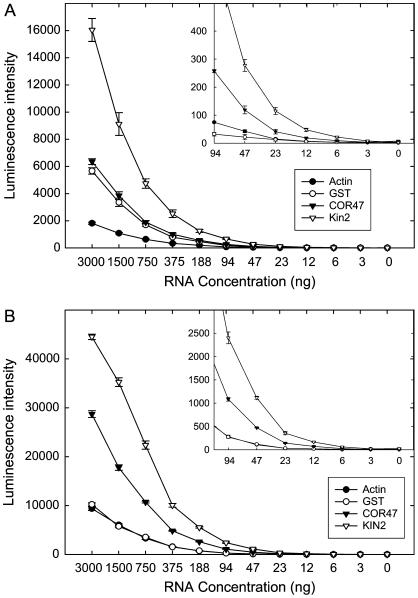
The next series of experiments were designed to evaluate the sensitivity of the qNPA assay. Arabidopsis plants hydroponically grown in bulk were treated with NaCl and separate pools of treated and untreated plants were homogenized in liquid N2 by grinding. Most of the homogenate was employed for purification of total RNA using conventional methods, but a small part (130 mg) was transferred into 340 μL of qNPA lysis buffer. Serial dilutions of the lysate and of conventionally purified total RNA were assayed using the same plate, with four replicate wells per sample (technical replications). The amount of RNA in the lysate was estimated based on the amount of total RNA purified from the tissue. The results are presented for the extracted RNA in Figure 4A and the lysate in Figure 4B. A linear relationship was observed for all serial dilutions, including those corresponding to extremely low RNA concentrations (Fig. 4, A and B, insets). The qNPA lysate preserved about 5-fold more RNA than conventional extraction methods. Because the reagents and concentrations of reagents were identical in all wells, the relative amounts of luminescence therefore corresponded to differences in relative amounts of RNA between the lysate and the total RNA extract. The sd of the measurements for the technical replicates (one sample split four ways for replicate measurement) illustrate that 23 ng total RNA were sufficient for accurate quantification of a signal from all genes in Figure 4 (relative ses of 11.0%–19.9%). For KIN2, a gene whose transcription is highly stimulated by salt stress, 12 ng total RNA was sufficient for reliable measurements (a technical replication CV [coefficient of variation] of 25% in this case). Thus, we conclude the platform can be used to accurately measure expression from very small amounts of tissue.
Figure 4.
Analysis of the sensitivity of the qNPA assay platform as indicated by a dilution series. A, Total RNA purified from 10-d-old Arabidopsis plants grown in soil and treated for 24 h with 150 mm NaCl. The inset is the same figure, but rescaled to show the response at lower RNA concentrations. B, Lysate extracted from 10-d-old Arabidopsis plants grown in soil and treated for 24 h with 150 mm NaCl. Each data point is the mean of quadruplicate samples and the error bars indicate ses. Data are not normalized. Only the results of the transcripts from a control gene (actin) and four NaCl-stimulated genes (KIN1, KIN2, COR47, and GST) are shown. RNA concentrations indicated on the abscissa of B are estimated based on the data from A.
Because the RNA quantifications measured by the assay were a consequence of nuclease protection probe hybridization, this means that the absolute levels of each transcript can be compared to each other and quantified through comparison to the signals produced by synthetic target gene sequences of known concentration. It should also be noted that qNPA measurements were performed without the requirement for amplification of the input RNA. Although amplification can provide similar levels of sensitivity of RNA detection using Ribo-SPIA (Dafforn et al., 2004), it can also introduce additional variability as well as significant costs per sample.
Generation of Dose-Response Curves Relative to Treatment with Salt and Abscisic Acid
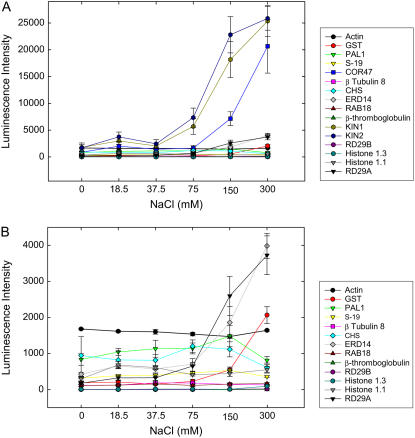
The next set of experiments were designed to generate dose-response curves to further characterize the effects of salt and abscisic acid (ABA) treatment and to illustrate the generation of EC50 values. For these experiments, we employed the same panel of salt-responsive genes in the assay (Fig. 5). The data indicated that saturation in response to NaCl treatment (300 mm) was not reached for any of the genes, with KIN2 exhibiting the closest approach to saturation. Because some of the genes represented in the panel were known to be regulated by ABA (e.g. RD29A, Kin1, Kin2; Nakashima et al., 2006), dose-response curves were also generated for treatment with this phytohormone (Supplemental Fig. S3). In contrast to salt treatment, treatment with different concentrations of ABA resulted in obvious response saturation, with EC50 values for all genes being <25 μm. Interestingly, the NaCl and ABA dose-response curves show contrasting patterns; whereas the response to NaCl treatment initiates at around 75 mm NaCl, continues increasing to the highest level tested, and differentiates three groups of genes based on approximate EC50 values (KIN1, KIN2 induction beginning at 37.5 mm, ERD14, RD29A, COR47 induction beginning at 75 mm, and GST induction beginning at 150 mm), the induction response to ABA peaks at 50 μm, decreases at higher treatment levels, and does not differentiate between the regulated genes. Further, the onset of ABA suppression occurs at different concentrations for different genes. These results suggest that different mechanisms underlie the complex behavior of these three groups of genes and this behavior appears independent of overall transcript abundance. Finally, these experiments serve to confirm the high dynamic range of the assay platform because genes expressed at high and low levels were readily measured in the same experiment using the same array, showing similar relative ses (the global relative square error values for all measurements had a mean of 19% with a sd of 19%).
Figure 5.
Dose-response of Arabidopsis transcripts to NaCl treatment. Arabidopsis plants, grown from seed for 10 d in 96-well plates, were treated with the indicated amounts of NaCl for 4 h and then harvested. A, Data are from quadruplicate biological samples and ses are shown by error bars. Data are normalized to the average of the transcript levels of actin and S-19. Note that two genes (KIN1 and KIN2) exhibit an EC50 of approximately 112 mm, whereas a third, COR47, exhibits an EC50 >175 mm. B, Scale of the ordinate is expanded to illustrate the sensitivity of the assay without loss of reproducibility.
Employing the qNPA Platform in Screens for Discovery of Chemical Compounds That Affect Expression of Stress-Inducible Genes
The next experiments implemented a small-scale screen of a library of potentially bioactive molecules produced via combinatorial chemistry (details of the library and its availability are provided in “Materials and Methods”). Plants grown in 96-well plates were treated in vivo with different chemical compounds, employing a pooling strategy in which 10 different compounds were tested per well. Within the plates, the first 11 columns were employed for testing of library compounds and the last column was employed for positive and negative controls. The first four wells within this column represented untreated plants and the last four wells represented plants treated with 150 mm NaCl. A single well (position E2) was also treated under double-blind conditions, with 150 mm NaCl, to determine whether a positive effector could be detected within a background of 10 different chemical compounds. The data in each well were normalized to the level of expression of two control genes, in this case Elongation Factor1α (EF-1α) and actin, whose expression was not expected to change with treatment. This allowed comparison of expression of genes from well to well even if the amounts of total RNA vary across wells.
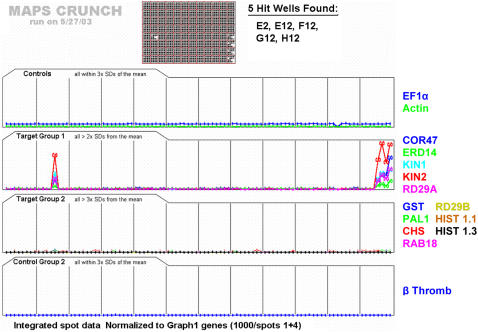
We grouped the target genes into four categories (Fig. 6): control group 1 genes (housekeeping controls, in this case EF-1α and actin), target group 1 genes (the NaCl stress marker genes COR47, ERD14, KIN1, KIN2, and RD29A), target specificity group 2 genes (other genes of interest not as strongly associated with NaCl stress: GST, PAL1, CHS, RAB18, RD29B, histone 1.1, and histone 1.3), and control group 2 genes (a negative control, in this case a single human gene, β-thromboglobulin, whose protection fragment used does not appear in the Arabidopsis genome). Target group 1 genes represent the gene signature required for identification of a positive compound during screening. Specifically, criteria were set such that a hit compound was scored as present if the expression levels of all the genes in target group 1 were more than 2 times the sd from the global mean for these genes across the entire plate (e.g. signal intensity ≥3 sd of background). In addition, compounds of interest were defined as those not affecting the expression of target group 2 genes. As indicated, the platform was able to identify the single spiked well and those corresponding to the positive controls, for a total of five wells. No hits were found out of the total of 880 novel compounds (10 per well) that were tested in this plate. Single-target whole-cell (much less whole organism) screens are typically plagued by high false-positive rates. Therefore, the data show that this technology is suitable for implementing more extensive screens.
Figure 6.
Example of implementation of a screen of Arabidopsis plants involving 880 different chemical compounds. One Arabidopsis plant was grown within each well. After 10 d, each plant was treated for 4 h with a mixture of 10 different chemical compounds. In this example, well E2 was spiked with 150 mm NaCl. The last four wells are positive controls treated with 150 mm NaCl. The transcript level data are normalized to the average of the values for EF-1α and actin transcripts. The well spiked with NaCl was correctly identified and no false positives were detected. One quadrant of the figure shows the housekeeping control genes, one quadrant shows the negative control gene, and the other two quadrants show the rest of the genes. The software allowed us to separate the genes as desired.
Measurement of Gene Expression within Specific Arabidopsis Organs
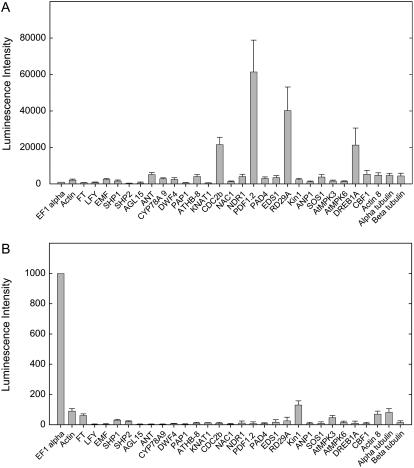
To further explore the sensitivity of the assay platform, we examined its applicability for the analysis of gene expression within specific plant organs. Samples comprising 20 to 30 dry seeds, or one green silique, weighing 4 to 5 mg, were placed within each well of the 96-well plate and were homogenized in the standard manner. An ArrayPlate was programmed to contain two different arrays, each of 16 genes, 48 wells/array, representing controls (EF-1α) and genes implicated in various developmental and stress-responsive pathways. Details of the oligonucleotide sequences representing the genes are found in Supplemental Table S2 and of the gene functions in Supplemental Table S4. EF-1α, Actin2, Actin8, α-tubulin, and β-tubulin were included as generally expressed controls. In these particular experiments, the data was normalized to EF-1α and Actin2. Gene expression from a few seeds and a single silique was readily and accurately measured (Fig. 7, A and B). The lack of overall correlation between the transcript levels detected in the seeds and siliques is consistent with the observation that they represent very different tissue types and that the mass of the seeds is small relative to that of the siliques.
Figure 7.
Analysis of transcript levels in seeds and siliques. A, Twenty to 30 dry seeds were placed in each well and homogenized in lysis buffer. The experiment was repeated eight times and means and sds (error bars) normalized to EF-1α levels are presented. B, One silique was placed in each well prior to homogenization. The experiment was repeated eight times and means and sds (error bars) normalized to EF-1α levels are presented.
DISCUSSION
This article represents the description of the application to plants of a microplate-based high-throughput array-based gene expression system (the qNPA ArrayPlate platform and associated technology). Although this platform has been used successfully with animal cells and tissues (Martel et al., 2002a, 2002b), its use in plants presented several new challenges. One is that plants comprise a complex organism rather than a homogeneous cell line. Another is that the presence of the tough cellulosic cell wall impedes homogenization and cell lysis and thereby might reduce the recovery of RNA. Further, the presence of plant secondary compounds and multiple ribonuclease isoforms might adversely affect the methods of homogenization and the effectiveness of the lysis buffer optimized for animal systems.
Our results indicate these concerns are not warranted. The extraction methods used in this study, coupled to devising an automated method for mechanical bead-based breakage of the tissues, made it possible to release RNA from the plant cells in amounts sufficient for measurement of the levels of specific transcript without requiring RNA purification. The results further demonstrate the high sensitivity of the qNPA assay in that transcript levels can be accurately measured within samples containing as little as 12 ng of total RNA. In comparison, for plant applications, Affymetrix Genechips routinely require 5 μg total RNA for labeling and assay (Schmid et al., 2005), about 400-fold more.
A feature of the qNPA platform is its technical accuracy, which is a result of the combination of nuclease protection, the use of an optimized lysis buffer to minimize the effects of endogenous RNases, and the sandwich read-out assay coupled to HRP-based luminescence. Platform reproducibility is evident in the low relative ses that are seen across the dynamic range of the assay using pooled samples (Fig. 4). A higher level of variation, corresponding to biological (i.e. plant-to-plant) variation, is observed when individual plants or tissues are employed in each well, as would be expected. However, the whole-plant assay remains very reproducible and can be used for high-throughput screening (Fig. 6) and dose-response mechanistic and efficacy studies (Fig. 5; Supplemental Fig. S3). Our results also indicate that very small amounts of tissue can be employed for the analyses and that tissue water content does not appear to affect the applicability of the platform because gene expression analyses were accurately conducted using small amounts of dry seeds.
An important additional point relates to the method of analysis employed by the qNPA platform. The stoichiometric conversion of unstable RNA to stable DNA through hybridization and S1 nuclease protection, coupled with the use of the same generic detection probe for measurement of every transcript gene, means that quantitative comparisons can be made of the level of RNA transcripts across different genes once a normalization curve is established. This is not true for any other array-based assay platform, where between-probe hybridization differences prevent absolute quantification of RNA levels and direct cross comparison of target amounts. Although quantitative comparisons across genes can be done using nonmicroarray platforms, such as PCR (Czechowski et al., 2004), serial analysis of gene expression (Velculescu et al., 1995), or massively parallel signature sequencing (Brenner et al., 2000), these platforms are not suited for the high-throughput applications described here.
Many applications in plant research appear immediately suited for study using this assay platform. A rapidly increasing amount of information is becoming available concerning specific genes involved in a number of different pathways related to plant growth and development and responses to the environment. Considerable interest exists in the use of transgenic technologies for specific manipulation of these pathways. However, there are many processes that would appear to be best controlled by application of inducer chemicals, rather than via transgenes, particularly those involving timing decisions that are made in the field, being based on actual or predicted weather conditions, length of growing season as a function of geographic location, incidence of biotic stress, or other factors. Because the qNPA platform employs plants treated in vivo, with all native regulatory elements and response pathways in place, it is ideal for discovery of compounds that are specific and either direct or indirect regulators of the gene activities of interest. Further, because the platform measures transcript abundances in high throughput, it has the potential to discover compounds that affect specific mRNA stability and not simply those that promote increased or decreased transcript biosynthesis. Finally, because measurements are multiplexed within the wells, genes can be included whose transcript levels report general toxicity to cells or tissues and this provides the advantage that compounds that cause toxic responses can be eliminated during the first phase of the screening.
Although our results indicate the qNPA platform offers the agricultural industry an efficient and cost-effective method for discovery of specific chemical agents having desired and highly specific effects on plant gene expression, the most important part of screening involves deciding which of the initial hits to pursue. In general, functional testing of all lead compounds is cost and time prohibitive and is also limited by the availabilities of the different compounds. To reduce the number of leads, one strategy can be to select only the most potent compounds for further functional characterization. However, this runs the risk of discarding weaker, albeit more specific, chemical leads. The platform described here allows improved decision making by providing more accurate information at the level of the primary screen through the use of clusters of genes as signatures, rather than a single gene, in that such clusters are robust to false positives, and by the fact that it has a very large dynamic range. The data also demonstrate how the platform can be efficiently employed in secondary profiling assays (such as dose-response and temporal assays) to further characterize each potential lead before making the selection of those to advance to full functional assays. Furthermore, it can be readily envisaged how the qNPA platform can be employed for establishing general metabolism and toxicity profiling assays, both to further explore specificity, and to delineate the effects of lead compounds on a wide variety of regulatory pathways other than that used in the initial screening assay. This approach should maximize the probabilities that lead chemicals will successfully negotiate the regulatory pathways required for release of novel chemical effectors.
A final note is that the platform can be readily adapted to provide validation of transgenic species in the form of a high-throughput, high-sensitivity screen for transgenic transcripts (or combinations of transcripts) at desired levels, in the proper organ, and at the proper time.
MATERIALS AND METHODS
Plant Culture and Treatments
Arabidopsis (Arabidopsis thaliana; ecotype Col-0) seeds were purchased from Lehle Seeds. The seeds were placed on sterile filter paper, saturated with growth medium (.5× Murashige and Skoog salts supplemented with 0.5% Suc), then vernalized for 48 h by placing them at 4°C. They were transferred to a Revco model I22LTPA growth chamber at 22°C and a 12-h light/12-h dark cycle at 1.2 μE m−2 s−1 intensity of light.
For bulk cultures, seedlings were grown in a shaking incubator at 25°C for 10 d. Plants were removed from the liquid and put in a mortar for homogenization. Liquid nitrogen was added and the plants homogenized with a pestle. Homogenized plants were stored at −80°C until used.
For plate assays, after 5 d on filter paper, seedlings were individually transferred into single wells of a half-size-deep well plate (Corning type 3956 plates; sterile, RNase free) containing 50 μL of the same medium. Plates were sealed with Parafilm, secured with plastic tape on the sides, and replaced in the growth chamber for 5 to 10 more days. Treatments were done by removing the liquid from the wells and replacing it with 50 μL liquid medium containing the various test compounds.
Preparation of Homogenates for the ArrayPlate Assay
96-Well Format
After treatment, liquid was removed and a sterile stainless steel ball (4-mm diameter) was added to each well followed by 200 μL lysis buffer (HTG). Plates were covered with a polypropylene mat (Beckman Coulter capmat [no. 267005]) and the samples immediately homogenized for 30 s, using a GenoGrinder (SPEX) set at 1,400 rpm. Plates were then heated at 95°C for 10 min in a plate heater. After cooling, 25-μL aliquots of the homogenates were removed to sterile 96-well v-bottom plates (Corning), covered with 60 μL of oil (HTG), covered with plastic tape (Rainin), and frozen at approximately 80°C.
Plants Grown in Bulk
Treated and control Arabidopsis plants were collected, placed in liquid N2, and homogenized using a mortar and pestle. RNA was prepared using the Qiagen RNeasy maxi kit. Total RNA was measured in one experiment to be 1.5 mg RNA per 3.5 g of tissue.
The ArrayPlate Assay
qNPA ArrayPlate kits are commercially available (HTG) and contain all the reagents required for processing the assay, from sample lysis to read out of transcript levels. Plates were prepared containing aliquots of homogenate or purified RNA, and 5 μL of protection fragments (Supplemental Tables S1 and S2) were added per well. In the first experiments, protection fragments (Supplemental Table S1) that were 75 bases long, 60 bases being complementary to the RNA target sequence, and 15 nonhybridizing bases acting as a S1 cleavable overhang as a control for S1 activity, were used and, in the later experiments, protection fragments of 65 bases were used, 50 bases being complementary to the RNA target sequence, and 15 nonhybridizing bases acting as a S1 cleavable overhang as a control for S1 activity (Supplemental Table S2). Currently, 65 bases are used for protection fragments. The GC% of oligonucleotides is between 48% to 54% and the melting temperature is between 69°C to 73°C. Plates were resealed with plastic tape (Rainin), heated at 95°C for 10 min, and incubated at 70°C for 6 h. Nonhybridized DNA was removed by addition (20 μL/well) of 50 units S1 nuclease (Promega) dissolved in 1.4 m NaCl, 22.5 mm zinc sulfate, 250 mm sodium acetate, pH 4.5. Incubation was continued at 50°C for 30 min. The reaction was terminated by addition (10 μL/well) of 1.6 m NaOH containing 135 mm EDTA, followed by heating for 15 min at 95°C, and cooling to room temperature. This step also serves to degrade any residual RNA. Ten microliters per well of neutralization solution (1 m HEPES, pH 7.5, 1.6 m HCl, 6× SSC) were added. Aliquots (60 μL) of the aqueous phases were transferred to the wells of the ArrayPlates for quantification of RNA levels. The ArrayPlates were incubated at 50°C overnight, washed with SSCS (1× SSC, 0.1% [w/v] SDS). Detection linkers (5 nm) were added and the plates incubated at 50°C for 1 h. Plates were then washed and a HRP detection probe (10 nm) was added. Following incubation at 37°C for 30 min, plates were washed and HRP chemiluminescent peroxidase substrate (Atto-PS; Lumigen) was added. The luminescence was captured following transfer of the plates into an OMIX imager (HTG). Typical imaging times were 30 s to 10 min, depending on signal intensity, within 30 min of substrate addition. The intensity values were extracted from the resultant TIFF images using OMIX imaging software (HTG). Gene signatures and control (housekeeping) genes were designated and the level of each transcript was normalized to those of the controls measured simultaneously within each well. This was carried out by calculating a normalization factor for each sample equal to 1,000 divided by the intensity measured for the transcripts of the controls for that sample. The transcript intensity values for every gene in the array from that well were then multiplied by this normalization factor. When two or more housekeeping genes were used for normalization controls, the normalization factor was calculated from the corresponding mean of the intensities representing these genes. Control genes included actin, S-19, and EF-1α, and the locus numbers and sequences are provided in Supplemental Tables S1 and S2.
Chemical Compound Library
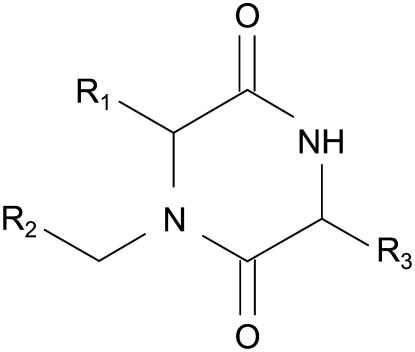
The compound library used for screening is H001 from HTG. The structure of the library is as follows.
Figure .
Each compound is a variant with various substitutions at the R1, R2, and R3 positions. Ten building blocks were employed for the R1 position, 12 for the R2 position, and eight for the R3 position. In this article, a total of 880 different compounds were screened, the library being employed as a pool of compounds with 10 compounds per well of a 96-well microplate. This left one column of the microplate available for controls.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers in Supplemental Tables S1 and S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Pipeline.
Supplemental Figure S2. ArrayPlate nuclease protection.
Supplemental Figure S3. Dose-response of Arabidopsis plants to ABA treatment.
Supplemental Table S1. Array 1 oligonucleotide sequences.
Supplemental Table S2. Array 2 oligonucleotide sequences.
Supplemental Table S3. Gene function array 1.
Supplemental Table S4. Gene function array 2.
Supplementary Material
Acknowledgments
Nuvogen Research has a financial interest in High Throughput Genomics, Inc. (HTG). J. Hinton, I. Botros, R. Martel, and B. Seligmann are employees of HTG. M.K. Deyholos, G. Lambert, and D.W. Galbraith declare no competing interests.
This work was supported by grants from the National Science Foundation Small Business Innovative Research Program (grant no. DMI–0110472 to R.M.K.) and the National Science Foundation Plant Genome Research Program (grant no. DBI 98–13360 to D.W.G.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Richard Martin Kris (rkris@nuvogenresearch.com).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18 630–634 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38 366–379 [DOI] [PubMed] [Google Scholar]
- Dafforn A, Chen P, Deng G, Herrler M, Iglehard D, Koritala S, Lato S, Pillarisetty S, Purohit R, Wang M, et al (2004) Linear mRNA amplification from as little as 5 ng total RNA for global gene expression analysis. Biotechniques 37 854–857 [DOI] [PubMed] [Google Scholar]
- Deyholos MK, Galbraith DW (2001) High-density DNA microarrays for gene expression analysis. Cytometry 43 229–238 [PubMed] [Google Scholar]
- Fodor SPA, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D (1991) Light-directed, spatially addressable parallel chemical synthesis. Science 251 767–773 [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286 531–537 [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai HY, He YDD, et al (2000) Functional discovery via a compendium of expression profiles. Cell 102 109–126 [DOI] [PubMed] [Google Scholar]
- Jenkins GI, Long JC, Wade HK, Shenton MR, Bibikova TN (2001) UV and blue light signalling: pathways regulating chalcone synthase gene expression in Arabidopsis. New Phytol 151 121–131 [DOI] [PubMed] [Google Scholar]
- Kim HS, Yu Y, Snesrud EC, Moy LP, Linford LD, Haas BJ, Nierman WC, Quackenbush J (2005) Transcriptional divergence of the duplicated oxidative stress-responsive genes in the Arabidopsis genome. Plant J 41 212–220 [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al (2006) The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313 1929–1935 [DOI] [PubMed] [Google Scholar]
- Ma SS, Gong QQ, Bohnert HJ (2006) Dissecting salt stress pathways. J Exp Bot 57 1097–1107 [DOI] [PubMed] [Google Scholar]
- Martel RR, Botros IW, Rounseville MP, Hinton JP, Staples RR, Morales DA, Farmer JB, Seligmann BE (2002. a) Multiplexed screening assay for mRNA combining nuclease protection with luminescent array detection. Assay Drug Dev Technol 1 61–71 [DOI] [PubMed] [Google Scholar]
- Martel RR, Rounesville MP, Botros IW, Kris R, Felder S, Seligmann BE (2002. b) Multiplexed molecular profiling transcription assay in ArrayPlates for high-throughput measurement of gene expression. In MP Weiner, Q Lu, eds, Gene Cloning and Expression Technologies. Easton Publishing, Westborough, MA, pp 549–564
- Meyers BC, Galbraith DW, Nelson T, Agrawal V (2004) Methods for transcriptional profiling in plants: be fruitful and replicate. Plant Physiol 135 637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Transcriptional regulation of ABI3-and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol Biol 60 51–68 [DOI] [PubMed] [Google Scholar]
- Nover L, Weigel D, Altmann T (2005) AtGenExpress project: arrays 3611-3622. http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenex.htm (December 29, 2006)
- Nylander M, Svensson J, Palva ET, Welin BV (2001) Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol 45 263–279 [DOI] [PubMed] [Google Scholar]
- Raikhel N, Pirrung M (2005) Adding precision tools to the plant biologist's toolbox with chemical genomics. Plant Physiol 138 563–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270 467–470 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31 279–292 [DOI] [PubMed] [Google Scholar]
- Singh-Gasson S, Green RD, Yue Y, Nelson C, Blattner F, Sussman MR, Cerrina F (1999) Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat Biotechnol 17 974–978 [DOI] [PubMed] [Google Scholar]
- Sittampalam GS, Kahl SD, Janzen WP (1997) High throughput screening: advances in assay technologies. Curr Opin Chem Biol 1 384–391 [DOI] [PubMed] [Google Scholar]
- Strachan T, Abitbol M, Davidson D, Beckmann JS (1997) A new dimension for the human genome project: towards comprehensive expression maps. Nat Genet 16 126–132 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Seki M, Ishida J, Satou M, Sakurai T, Narusaka M, Kamiya A, Nakajima M, Enju A, Akiyama K, et al (2004) Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Mol Biol 56 29–55 [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW (1995) Serial analysis of gene expression. Science 270 484–487 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.