Abstract
A novel phenyltriazole acetic acid compound (DAS734) produced bleaching of new growth on a variety of dicotyledonous weeds and was a potent inhibitor of Arabidopsis (Arabidopsis thaliana) seedling growth. The phytotoxic effects of DAS734 on Arabidopsis were completely alleviated by addition of adenine to the growth media. A screen of ethylmethanesulfonate-mutagenized Arabidopsis seedlings recovered seven lines with resistance levels to DAS734 ranging from 5- to 125-fold. Genetic tests determined that all the resistance mutations were dominant and allelic. One mutation was mapped to an interval on chromosome 4 containing At4g34740, which encodes an isoform of glutamine phosphoribosylamidotransferase (AtGPRAT2), the first enzyme of the purine biosynthetic pathway. Sequencing of At4g34740 from the resistant lines showed that all seven contained mutations producing changes in the encoded polypeptide sequence. Two lines with the highest level of resistance (125-fold) contained the mutation R264K. The wild-type and mutant AtGPRAT2 enzymes were cloned and functionally overexpressed in Escherichia coli. Assays of the recombinant enzyme showed that DAS734 was a potent, slow-binding inhibitor of the wild-type enzyme (I50 approximately 0.2 μm), whereas the mutant enzyme R264K was not significantly inhibited by 200 μm DAS734. Another GPRAT isoform in Arabidopsis, AtGPRAT3, was also inhibited by DAS734. This combination of chemical, genetic, and biochemical evidence indicates that the phytotoxicity of DAS734 arises from direct inhibition of GPRAT and establishes its utility as a new and specific chemical genetic probe of plant purine biosynthesis. The effects of this novel GPRAT inhibitor are compared to the phenotypes of known AtGPRAT genetic mutants.
The developing field of chemical genetics relies on the ability of certain small molecules to mimic the effect of genetic mutations by blocking, or otherwise modulating, specific cellular processes (Stockwell, 2000; Blackwell and Zhao, 2003; Mayer, 2003). The use of small molecules to perturb plant biological processes can have several convenient advantages over genetic methods. Compounds can be applied and removed at specific times and locations to rapidly produce their effects, and they have the potential to be used on a variety of different species, including those that are not genetically tractable. Recent chemical genetic studies using plant systems have identified several new chemical effectors of a variety of plant cellular functions (Asami et al., 2003; Zhao et al., 2003; Armstrong et al., 2004; Surpin et al., 2005; Zheng et al., 2006). Compounds that intervene in primary metabolism can be of special interest for chemical genetic studies modulating plant biosynthetic pathways and their downstream events. Many herbicides have potent and specific action within these pathways and so are instructive chemical probes of plant biosynthetic processes. For example, commercial herbicide targets include enzymes in branched chain and aromatic amino acid, fatty acid, cellulose, and plastoquinone biosynthesis (Wakabayashi and Boger, 2002). Much has been learned about the individual target enzymes, the pathways, and their role in plant metabolism via detailed understanding of the effects of these herbicides. This suite of chemical probes has been expanded by many experimental herbicidal compounds that (for a variety of reasons) have not achieved commercialization but can still serve as informative inhibitors of additional pathways. Examples are inhibitors of His biosynthesis (Cox et al., 1997; Dancer et al., 1999) and the plastidial nonmevalonate isoprenoid pathway (Mueller et al., 2000; Lange et al., 2001).
Currently there are few small molecules that are known to act directly and specifically on purine biosynthesis, particularly in the early steps of the pathway. This important pathway in plant primary metabolism (Fig. 1A) produces purine precursors for DNA and RNA synthesis, for the energy transfer and building block nucleotides ATP and GTP, and as components of important coenzymes such NAD, S-adenosyl-Met, and FMN (Moffatt and Ashihara, 2002; Boldt and Zrenner, 2003; Zrenner et al., 2006). Plants also use purine skeletons for synthesis of plant hormone cytokinins, purine alkaloids, and in several species, particularly within the legumes, for nitrogen utilization via ureide metabolism (Smith and Atkins, 2002). Thus, small molecule inhibitors of purine synthesis may be useful for modulating a wide variety of biologically important plant processes, as well as being potentially useful leads for herbicide design. The Gln antagonists azaserine, acivicin, and 6-diazo-5-oxo-l-norleucine have often been used as purine biosynthesis inhibitors, but these compounds interfere with many amidotransferases and so lack specificity (Lyons et al., 1990). The natural product phytotoxins hydantocidin and ribofuranosyl triazolonone have been shown to exert their toxic effect by bioconversion via phosphorylation into inhibitors of adenylosuccinate synthetase (AdSS), the enzyme catalyzing the penultimate step in AMP biosynthesis (Heim et al., 1995; Cseke et al., 1996; Siehl et al., 1996; Schmitzer et al., 2000). Another natural product, hadacidin, inhibits plant AdSS (Hatch, 1967). Purine biosynthesis is also a well-known target for many antineoplastic and antiviral agents designed for potential pharmaceutical use (Christopherson et al., 2002).
Figure 1.
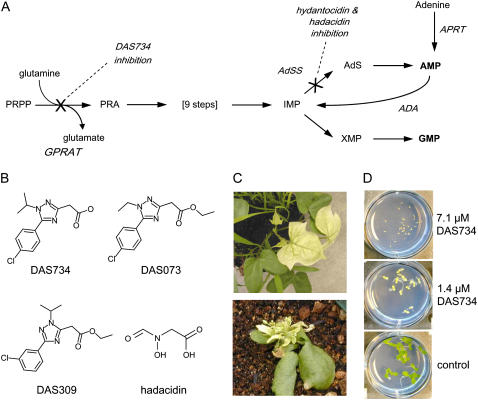
Purine biosynthesis pathway, structures of compounds used in this study, and effect of DAS734 on plants. A, The purine biosynthesis pathway. Abbreviations not defined in the text are as follows: PRA, phosphoribosylamine; AdS, adenylosuccinate; APRT, adenine phosphoribosyltransferase; ADA, AMP deaminase. The X symbols denote the sites of inhibition of DAS734 (GPRAT) and hadacidin (AdSS). B, Chemical structures of compounds used in this study. C, Effect of 1 kg/ha DAS734 on morningglory (top) and Arabidopsis (bottom) 10 d after greenhouse application. Note the bleaching of newly emerged leaves. D, Effect of DAS734 on Arabidopsis seedlings in plate tests.
We found that treatment of plants with a simple but novel phenyltriazole acetic acid compound results in a phytotoxic phenotype of interest in several dicotyledonous plant species by producing distinctive bleaching of developing leaves. The compound is especially potent on Arabidopsis (Arabidopsis thaliana). A better understanding of the observed biological effects (phenotypes) of chemical genetic probes can be gained by knowledge of their specific target site(s). However, determination of the precise biochemical targets of effective compounds is often a difficult and rate-limiting step in the exploitation of new compounds for chemical genetic studies (Burdine and Kodadek, 2004; Tochtrop and King, 2004). We have used a combination of genetic, chemical, and biochemical techniques to elucidate the molecular target of the phenyltriazole acetate compound as Gln phosphoribosylpyrophosphate amidotransferase (GPRAT), the first committed enzyme of de novo purine biosynthesis. The effects of the compound can be compared to the phenotypes of known AtGPRAT genetic mutants (Hung et al., 2004; van der Graaff et al., 2004). We have functionally expressed Arabidopsis GPRAT (AtGPRAT) in Escherichia coli at high levels and have shown that DAS734 is a slow, tight-binding inhibitor of the enzyme. Thus, we have identified a novel specific chemical probe of the first step in plant purine biosynthesis and characterized its mode of interaction with its cognate target.
RESULTS
Phytotoxic Activity of DAS734
The phenyltriazole acetic acid [5-(4-chlorophenyl)-1-isopropyl-1H-[1,2,4]triazol-3-yl]-acetic acid (DAS734; Fig. 1B) exerted modest herbicidal activity on the seedlings of several dicotyledonous weed species in greenhouse postemergent applications. It was most active on morningglory (Ipomoea hederacea), field pansy (Viola arvensis), redroot pigweed (Amaranthus retroflexus), and buckwheat (Polygonum convolvulus). The phytotoxic symptoms were characterized by extensive bleaching of new growth such that newly emerging leaves turned white (Fig. 1C). Although greenhouse activity against commercially relevant weeds was modest, we found that DAS734 was very phytotoxic to Arabidopsis seedlings grown on agarose media containing the compound (Fig. 1D). The concentration of DAS734 required to inhibit root growth by 50% (RI50) was 200 nm. The ethyl ester of DAS734 was even more potent with an RI50 of 30 nm. It is likely that the neutral ethyl ester may have improved uptake relative to the free acid form. The high level of activity in Arabidopsis seedling assays approaches that of some potent commercial herbicides, such as acetolactate synthase inhibitors.
The phytotoxic effects of many inhibitors of primary metabolism can often be reversed by addition of the end product(s) of the affected pathway (Subramanian et al., 1999). A variety of metabolites were added individually or in pools to the Arabidopsis seedling growth medium in the presence of DAS734. Of these, only the addition of 185 μm adenine was found to completely alleviate all of the phytotoxic effects of DAS734, including bleaching and root inhibition (Fig. 2A), whereas amino acids, pyrimidines, and various other products of primary metabolism had no effect. Alleviation of phytotoxicity was found to a lesser extent with addition of adenosine and adenine nucleotides, but other purines, such as hypoxanthine, guanine, and inosine, had no effect (data not shown). This pattern of symptom alleviation restricted to adenine and its derivatives is similar to that seen with hydantocidin and hadacidin (Heim et al., 1995; Siehl et al., 1996). However, DAS734 had no effect in enzyme assays of AdSS, the target of hydantocidin and hadacidin, or the subsequent enzyme in purine biosynthesis, adenylosuccinate lyase (data not shown). This suggested that DAS734 may act at some novel site involved in purine biosynthesis. As there are 10 additional enzymes in de novo purine biosynthesis, many of which are difficult to directly assay, we elected to take a genetic approach to dissect the precise site of action. Because DAS734 had no inhibitory activity on E. coli, cyanobacteria, green algae, or yeast but was a potent inhibitor of Arabidopsis growth, Arabidopsis was chosen as the genetic model organism of choice for these studies.
Figure 2.
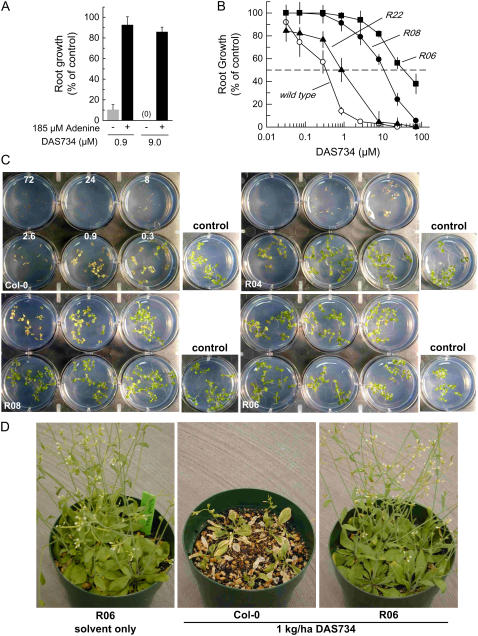
Effects of DAS734 on wild-type (Col-0) and DAS734-resistant Arabidopsis lines. A, Reversal by adenine of root growth inhibition by DAS734. Arabidopsis seedlings were grown in the presence of DAS734 and root lengths measured after 9 d. Typical root length (±sd) of untreated control seedlings was 17 (±1.2) mm. Adenine (185 μm) was added to the growth media in the samples labeled “+.” B, Dose response of three DAS734-resistant mutant lines to DAS734. The dashed line represents 50% inhibition of root growth. Data points are the average (±sd) of five individual root measurements. C, Effect of increasing concentrations of DAS734 on wild-type and resistant line R04, R08, and R06 seedlings grown in agarose media. Each plate contains a series of 3-fold dilutions of DAS734 starting at 72 μm, as shown in the top left plate containing wild-type (Col-0) seedlings. An untreated control well for each line is shown to the right of each plate. Based on root growth measurements, the resistance levels to DAS734 for R04, R08, and R06 are 6-fold, 56-fold, and 145-fold, respectively. The difference in resistance of the mutant lines to the phytotoxic effects of DAS734 on the cotyledon and shoots is also apparent. D, Effect of DAS734 applied as a 1 kg/ha postemergent spray to greenhouse-grown Arabidopsis plants. Plants were sprayed at the five- to six-leaf rosette stage and photographed 10 d later.
Screen for Mutants Resistant to DAS734
To identify mutants resistant to DAS734, ethylmethanesulfonate-mutagenized M2 Arabidopsis seeds were germinated and grown in medium containing sublethal concentrations of the herbicide. Several pilot screens of approximately 42,000 seedlings were performed at DAS734 concentrations between 0.18 and 1.1 μm, and a concentration of 0.54 μm DAS734 was selected to perform a large-scale screen of 480,000 seedlings. At this concentration, seedlings formed purple cotyledons, did not produce true leaves, and had significantly inhibited root growth. Resistant mutants were identified as seedlings with green, fully expanded cotyledons, emerging true leaves, and normal root length. Sixteen mutants were recovered and confirmed as showing some level of resistance after rescreening M3 seed on media containing 0.54 μm DAS734. When assessed on a lower concentration of DAS734 (0.054 μm), nine mutants produced green cotyledons but had some inhibition of growth of roots and true leaf formation. Seven mutants (R01, R03, R04, R06, R07, R08, and R22) showed complete resistance with growth equivalent to control seedlings. These mutants were used in further analyses.
To quantify the levels of resistance, the seven mutants were grown on media containing increasing concentrations of DAS734, and RI50 values were determined for each mutant (Fig. 2, B and C). The mutants could be readily classified into three distinct groups showing strong (>110-fold), moderate (approximately 60-fold), and weak (approximately 5-fold) resistance (Table I) relative to wild-type Columbia (Col)-0 seedlings. All seven mutants were phenotypically normal, healthy, and fertile under our standard growth conditions on agarose media and in the greenhouse. The adult Arabidopsis mutant plants also showed significant resistance to DAS734 in greenhouse postemergent spray applications of the herbicide. The application rate of the compound giving a 50% reduction in growth relative to an untreated control was 80 g/ha for wild-type plants, whereas treatments of up to 2,000 g/ha had little or no effect on the strongly resistant R06 mutant line (Fig. 2D). This corresponds to >25-fold resistance for the adult plants.
Table I.
RI50 values and mutations in AtGPRAT2 nucleotide and polypeptide sequences for seven Arabidopsis mutant lines resistant to DAS734
The asterisks separate three groups with high (***, >110-fold), moderate (**, approximately 58-fold), and low (*, approximately 5-fold) levels of resistance.
| Line | RI50a | Fold Resistance | Changes in AtGPRAT2 Sequence
|
|
|---|---|---|---|---|
| Nucleotideb | Amino Acid | |||
| μm | ||||
| Wild type | 0.2 | – | – | – |
| R03*** | 20 | 111 | G791A | R264K |
| R06*** | 35 | 145 | G791A | R264K |
| R01** | 14 | 58 | C1426T, A388G | P476S, T130A |
| R07** | 14 | 58 | C1426T, A416Gc | P476S |
| R08** | 10 | 56 | C1426T, C1246Tc | P476S |
| R04* | 1.0 | 6 | C793T, A1482T | P265S, Y494F |
| R22* | 0.9 | 5 | G1110A | G371S |
Determined from dose responses of seedlings grown in agarose media.
Numbered from the initiator TAG codon.
Silent mutation.
Resistance to Other Herbicidal Chemistries
To ensure that the observed herbicide resistance was specific to DAS734, the seven mutant lines were tested for resistance to three other herbicidal chemistries (structures shown in Fig. 1B). DAS073 is a close structural analog of DAS734 and shares the same phytotoxic phenotype that can be alleviated by addition of adenine (data not shown). All of the DAS734-resistant mutants tested were cross-resistant to DAS073, consistent with a shared mode of action. DAS309 is an isomer of DAS734 differing only in the attachment position of the isopropyl group on the triazole ring. However, it produces a different phytotoxic phenotype (no bleaching) that is not alleviated by adenine. All of the DAS734-resistant mutants were sensitive to DAS309, suggesting that the resistance mechanism is specific to the mode of action rather than via recognition of similar chemical structures. This provided some reassurance that resistance could be due to mutations at the target site and not from serendipitous chemical recognition by mechanisms involving metabolism or pumping to another cell compartment. Hadacidin is structurally unrelated to DAS734 but is known to inhibit purine biosynthesis via inhibition of AdSS. Its phytotoxic symptoms can be alleviated by adenine (Heim et al., 1995). All of the mutants were sensitive to hadacidin, suggesting that DAS734 has a site of action affecting purine biosynthesis that is different from that of the hadacidin target AdSS. The chemical specificity of the resistance of the mutants to different herbicidal chemistries encouraged us to further genetically characterize the mutants.
Genetic Characterization of the DAS734-Resistant Mutants
In four of the seven DAS734-resistant mutants tested (R01, R06, R07, and R08), M3 progeny segregated for resistance. The ratio of resistant to sensitive progeny was approximately 3:1, suggesting that these lines contained a dominant mutation and the M2 plants were heterozygous for the mutation. The progeny from the remaining three mutants were 100% resistant to DAS734, indicating that the mutations in these lines were homozygous for the mutation. Further test crosses with wild-type plants showed that these DAS734-resistant mutants also contained dominant mutations. To determine if the mutations in the DAS734-resistant mutants were allelic, M3 lines identified as homozygous for the resistance mutation were crossed with each other. In every case, all of the F2 seed was resistant to DAS734. This indicates that the DAS734-resistant phenotypes in all seven lines were caused by mutations in the same or closely linked genes. Because all the mutants appeared to have a mutation at the same locus, only one mutant line was used to map the resistance gene.
Genetic Mapping and Identification of the Resistance Mutations
To identify the gene responsible for the mutant phenotype, the mutation in the strongly resistant line R03 was used for genetic mapping experiments. The wild-type allele was mapped to the lower arm of chromosome 4 between 1.26 × 107 bp and 1.58 × 107 bp using single nucleotide polymorphism markers (Supplemental Fig. S1). Genes in this region were inspected to identify if any were involved in purine biosynthesis or metabolism. One gene (At4g34740) was found with annotations matching these criteria and encodes GPRAT, the first committed enzyme in the purine biosynthetic pathway. There are three isoforms of GPRAT in the Arabidopsis genome, and At4g34740 encodes AtGPRAT2. To determine if mutant line R03 contained a mutation in At4g34740, the gene was PCR amplified from both the R03 mutant and wild-type genomic DNA and the PCR products were sequenced. A single transition mutation of G to A was detected at nucleotide 791 relative to the start codon. This mutation causes Lys-264 to be changed to Arg in the encoded polypeptide (Fig. 3). Because the mutations in the other DAS734-resistant mutants appeared to be allelic with the R03 mutation, the At4g34740 gene was PCR amplified and sequenced from the other mutant lines. All seven DAS734-resistant lines were found to contain mutations affecting the coding sequence of this gene (Fig. 3; Table I); thus, these mutations are likely to be responsible for conferring resistance to DAS734.
Figure 3.
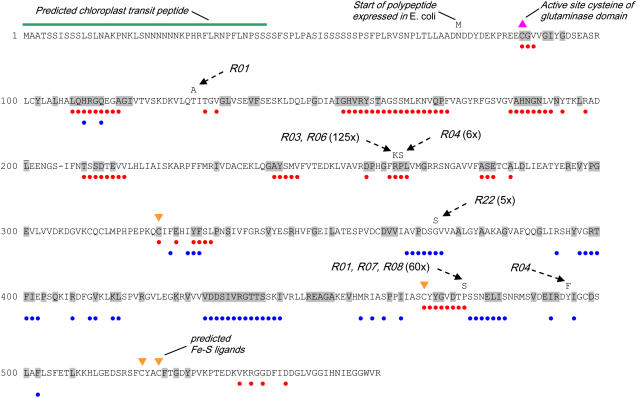
Annotated polypeptide sequence of AtGPRAT2 showing sites of mutations conferring resistance to DAS734. For heterologous expression in E. coli, the AtGPRAT2 polypeptide was truncated by 75 residues, resulting in Asn-76 being converted to a starting Met residue. The conserved Cys that becomes the N-terminal active site residue after autoprocessing is marked. Mutations that were found by sequencing of the AtGPRAT2 genes from the DAS734-resistant mutant lines are shown above the sequence. The resistance level in seedling assays for the lines carrying the mutation is indicated in parentheses. Residues that are identical with the B. subtilis and E. coli GPRAT sequences are shaded gray. Residues within 10 Å of the active site thiol residue in the B. subtilis GPRAT crystal structure, PDB ID 1AO0 (Chen et al., 1997), and within 10 Å of the 6-diazo-5-oxonor-Leu-derivatized active site Cys in the E. coli GPRAT structure, PDB ID 1ECC1 (Krahn et al., 1997), are denoted with red dots below the sequence. These residues are therefore at or close to the glutaminase site of GPRAT. Residues within 10 Å of the bridging oxygen of the Rib pyrophosphate in GMP in the B. subtilis 1AO0 structure and in the carbocyclic PRPP analog in the E. coli 1ECC1 structure are denoted with blue dots below the sequence. These residues are therefore at or close to the PRPP catalytic site. The four Cys residues that are equivalent to the ligands for the 4Fe-S cofactor in B. subtilis GPRAT are marked.
R06 (145-fold resistance to DAS734) contained the same mutation in AtGPRAT2 as R03, an Arg-to-Lys substitution at codon 264. R01, R07, and R08 (approximately 50-fold resistance to DAS734) all contain a Pro-to-Ser mutation at residue 476. These mutants do not appear to be siblings as R01 and R07 each contained another different mutation in AtGPRAT2. R01 contained a Thr-to-Ala mutation at residue 130, and R07 contained a silent A-to-G mutation at nucleotide 416 (relative to the start codon). The T130A mutation in R01 probably does not contribute to resistance as the two other phenotypically indistinguishable mutants that contain only the P476S mutation (R07 and R08) do not contain this mutation. The two mutants displaying the lowest level of resistance to DAS734, R04 and R22, contained unique mutations. R04 contained two mutations, a Pro-to-Ser mutation at amino acid 265 and a Tyr-to-Phe mutation at amino acid 494, whereas R22 contained a single Gly-to-Ser mutation at amino acid 371 (Fig. 3; Table I). The P265S mutation in R04 is perhaps a more likely candidate to confer resistance than Y494F as it is adjacent to the R264K mutation that gives high levels of resistance in R03 and R06.
Heterologous Expression of AtGPRAT2 in E. coli
GPRATs are highly regulated enzymes and they exert significant control over de novo purine biosynthesis via feedback inhibition by purine nucleotides (Chen et al., 1997). At this point in our investigations, it was unclear if DAS734 directly inhibited GPRAT and mutations in the enzyme conferred resistance, or if the mutations altered regulation of the enzyme, allowing the mutant plant to overproduce purines and therefore alleviate the effects of the herbicide, as addition of exogenous adenine does. We therefore cloned and overexpressed AtGPRAT2 in E. coli to ascertain the direct effects of DAS734 on the enzyme.
All eukaryotic and many microbial GPRATs possess a short N-terminal propeptide that is autocatalytically cleaved to yield a conserved N-terminal Cys. The γ-sulfhydryl group of this Cys residue acts as the nucleophile in the Glu transaminase portion of the two-step reaction catalyzed by the enzyme; thus, GPRATs are members of the self-processing Ntn (N-terminal nucleophile)-hydrolase class of enzymes (Smith, 2004). In addition, AtGPRAT2 is predicted to contain a chloroplast transit peptide (Hung et al., 2004; Fig. 3). For expression in E. coli, we truncated the AtGPRAT2 gene to eliminate the chloroplast-targeting sequence, leaving 11 residues before the putative catalytic Cys-87 (Fig. 3). This protein was expressed in E. coli and a novel polypeptide of approximately 54 kD was detected by SDS-PAGE analysis of cell pellet extracts (Fig. 4A). The extracts were assayed for GPRAT activity by measuring the phosphoribosylpyrophosphate (PRPP)-dependent production of Glu from Gln. Extracts from E. coli containing the plasmid encoding the AtGPRAT2 gene contained high levels of GPRAT activity, whereas extracts produced from E. coli containing a control plasmid had negligible activity (<5% of the activity in extracts from cells expressing AtGPRAT2). The Km for Gln of the recombinant enzyme was 1.34 mm (Supplemental Fig. S2). This is considerably lower than that determined for soybean (Glycine max) GPRAT purified from root nodules (18 mm), but similar to that of recombinant GPRATs from microbial sources (Reynolds et al., 1984).
Figure 4.
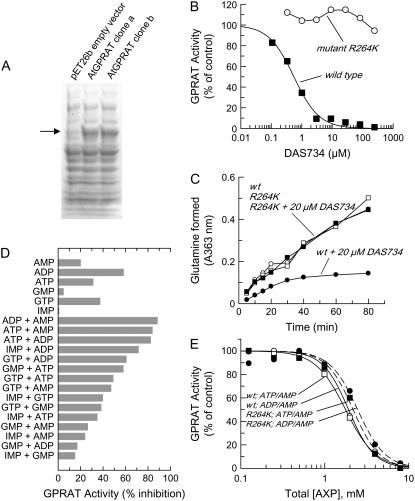
Expression of AtGPRAT2 in E. coli, effect of DAS734 on recombinant wild-type and mutant R264K AtGPRAT2 enzyme activity, and effect of purine nucleotides on wild-type and mutant enzymes. A, SDS-PAGE analysis of extracts from E. coli cells expressing AtGPRAT2 induced by the addition of 75 μm isopropyl-d-thiogalactopyranoside. A control extract with the empty pET26b vector and extracts from cells with expression plasmids containing from two separate AtGPRAT2 clones are shown The arrow denotes the presence of a new band at approximately 54 kD in the AtGPRAT2-expressing extracts. B, Inhibition of recombinant wild-type AtGPRAT2 and mutant enzyme R264K by DAS734. Extracts of E. coli expressing AtGPRAT2 were assayed by monitoring the PRPP-dependent production of Glu in the presence of increasing concentrations of DAS734. The level of activity is expressed relative to an untreated control reaction. Enzyme extracts were preincubated with DAS734 for 10 min prior to addition of substrates. The I50 for DAS734 inhibition in this experiment was 0.5 μm. Subsequent experiments with a number of enzyme preparations gave I50 values of approximately 0.2 μm. C, Time course of inhibition of wild-type and mutant enzymes. Substrates and 20 μm DAS734 were simultaneously added to AtGPRAT2 solutions and aliquots of the reaction mixture were analyzed for PRPP-dependent Gln production over time. Shown are wild-type AtGPRAT2 with no DAS734 (white circles), wild type plus 20 μm DAS734 (black circles), AtGPRAT2 R264K with no DAS734 (white boxes), and R264K plus 20 μm DAS734 (black boxes). D, Wild-type AtGPRAT2 activity was assayed in the presence of various nucleotides, alone or in pairs. All nucleotides were assayed at a concentration of 5 mm; thus, the total nucleotide concentration in the paired combinations was 10 mm. E, Dose response of wild-type AtGPRAT2 (solid lines) and AtGPRAT2 R264K (dashed lines) to 1:1 mixtures of AMP/ADP and AMP/ATP. Wild-type AtGPRAT2 with AMP/ADP (white circles), wild-type AtGPRAT2 with AMP/ATP (white boxes), AtGPRAT2 R264K with AMP/ADP (black circles), and AtGPRAT2 R264K with AMP/ATP (black boxes) are shown.
Inhibition of AtGPRAT2 by DAS734
The enzyme activity of AtGPRAT2 was potently inhibited by the addition of DAS734 to the assays (Fig. 4B). Initial Michaelis-Menten kinetic characterization showed that DAS734 behaved like a noncompetitive inhibitor with respect to Gln (Supplemental Fig. S2). However, this type of kinetic behavior can be manifested by slow, tight-binding inhibitors. To evaluate whether DAS734 exhibited slow, tight-binding characteristics, the enzyme reaction progress was monitored over time after simultaneous addition of substrates and inhibitor. The time course was clearly curved, indicating that inhibition of the enzyme increased with time (Fig. 4C). I50 curves generated by preincubating the inhibitor with enzyme over increasing time intervals also showed increasing potency (data not shown). Maximal inhibition could be achieved by a 10-min preincubation of DAS734 with AtGPRAT2 without any other substrate additions being required, yielding an average I50 value under our assay conditions of 0.2 μm. This suggests that DAS734 binds to GPRAT without requirement for Gln or PRPP binding. Enzyme that was fully inhibited by DAS734 was passed over a desalting column to remove unbound ligand. Forty-seven percent of the enzyme activity could be recovered relative to an untreated control, indicating that the dissociation rate of DAS734 from the enzyme is relatively slow but reversible and accounts for the slow, tight-binding behavior.
Having established that DAS734 was a potent inhibitor of AtGPRAT2, we cloned the mutant enzyme AtGPRAT2 R264K from the resistant line R03, one of the two lines showing the highest levels of DAS734 resistance. The mutant enzyme was successfully overexpressed at high levels in E. coli. AtGPRAT2 R264K was exceptionally resistant to DAS734, showing >500-fold increase in I50 over that of the wild type (Fig. 4B). Also, DAS734 had no effect on the reaction time course catalyzed by the mutant enzyme (Fig. 4C), in marked contrast to the wild-type form. Although the mutant enzyme was uninhibited by DAS734 at 100 μm in biochemical assays of GPRAT activity, root growth of the Arabidopsis lines harboring this mutated GPRAT (R03 and R06) was somewhat inhibited at DAS734 concentrations above 20 μm (Fig. 2B). This could be due to additional nonspecific effects on root growth at these high levels of compound.
Inhibition of Other AtGPRAT Isoforms by DAS734
There are three expressed GPRAT homologs in the Arabidopsis genome that are differentially expressed in various plant tissues (Ito et al., 1994; Boldt and Zrenner, 2003; Hung et al., 2004). AtGPRAT1 shares 93% amino acid identity with AtGPRAT2 over the length of the mature polypeptide, whereas AtGPRAT3 has 72% identity. Genes for AtGPRATs 1 and 3 were PCR amplified from an Arabidopsis seedling cDNA library and expression was assessed in E. coli as was done for AtGPRAT2. High levels of GPRAT activity were detected in extracts of E. coli expressing the AtGPRAT3 gene. This activity was inhibited by DAS734 with an I50 of 0.2 μm, demonstrating that this isoform was as sensitive to DAS734 as AtGPRAT2. In contrast, we were unable to detect recombinant GPRAT activity from the AtGPRAT1 gene. To verify this result, a hexahistidine tag was added to the C terminus of both AtGPRAT1 and AtGPRAT2 to facilitate purification of the proteins. High levels of GPRAT activity could be obtained from AtGPRAT2-6xHis after purification of the protein via nickel column chromatography, whereas no significant GPRAT activity or visible protein could be recovered from AtGPRAT1-6xHis expressions.
Site of DAS734-Resistance Mutations in the GPRAT Structure and Effect on Allosteric Inhibition by Nucleotides
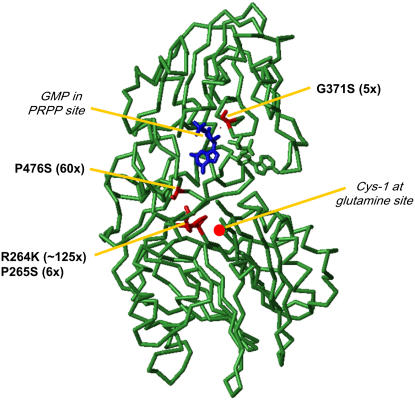
The Bacillus subtilis and E. coli GPRAT protein sequences share 47.4% and 33.3% amino acid identity, respectively, with AtGPRAT2 (Supplemental Fig. S3). Using the available crystal structures of the microbial enzymes (Chen et al., 1997; Krahn et al., 1997), we determined the AtGPRAT2 residues that are equivalent to those in the vicinity of the glutaminase and PRPP-binding sites. These are marked in Figure 3. The relative sites of the mutations conferring resistance to DAS734 can also be positioned in the structure of the B. subtilis enzyme (Fig. 5). Three mutations (R264K, P265S, and P476S) occur in residues that lie within 10 Å of the glutaminase site of the enzyme, whereas the weakly resistant mutation (5-fold, G371S) is positioned on the opposite domain in the periphery of the PRPP site (approximately 8 Å from the Rib substrate).
Figure 5.
Position of mutations conferring resistance to DAS734 mapped onto the structure of B. subtilis GPRAT (PDB ID 1AO0) from Chen et al. (1997). The residues in B. subtilis GPRAT that are equivalent to the mutation sites conferring resistance to DAS734 in AtGPRAT2 in the alignment in Supplemental Figure S3 are highlighted in red. The location of the Gln site can be identified by the N-terminal active site Cys residue (red dot). A GMP molecule (in blue) occupies the PRPP catalytic site and an ADP molecule occupies an adjacent allosteric site.
Studies on other GPRATs from microbes and vertebrates show that the enzyme is allosterically inhibited by end products of the purine biosynthesis pathway (Chen et al., 1997). We wished to ascertain if the highly resistant mutant AtGPRAT2 R264K was altered in its ability to respond to feedback regulators. This could affect its biological function and possibly contribute to the resistance of intact mutant plants to DAS734 treatment. The precise combination and concentration of purine nucleotides that exert maximal effect on GPRATs appears to vary according to the source organism. Little information is available for plant enzymes, although soybean GPRAT purified from root nodules was found to be inhibited by AMP, IMP, and GMP (Reynolds et al., 1984). We examined the effect of purine nucleotides on wild-type AtGPRAT2 and found that maximal inhibition of the enzyme was produced by combinations of adenine nucleotides (Fig. 4D), whereas minimal inhibition was seen with IMP or GMP under our assay conditions. The mutant enzyme AtGPRAT2 R264K had the same response to adenine nucleotides as the wild-type enzyme, indicating that the R264K mutation conferring DAS734 resistance did not affect allosteric inhibition by purine nucleotides (Fig. 4E).
Treatment with DAS734 Phenocopies atd2
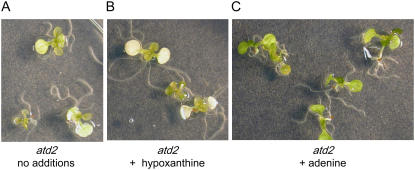
The Arabidopsis genetic mutant atd2 (amidotransferase-deficient2) has a T-DNA insertion within the AtGPRAT2 gene and was identified by its variegated bleached seedling phenotype and strong growth retardation. The wild-type phenotype could be restored by addition of 5 mm IMP (van der Graaff et al., 2004). We assessed if low concentrations of adenine could also restore normal growth to this mutant. We found that bleaching of atd2 was completely alleviated and normal growth restored by addition of 185 μm adenine to the medium (Fig. 6). In contrast, addition of 185 μm hypoxanthine had no effect. Thus, the phenotype of atd2 showing bleaching and reduced growth (van der Graaff et al., 2004) is similar to that found by treatment of wild-type seedlings with DAS734.
Figure 6.
Reversal of bleaching of atd2 mutant seedlings by adenine. Eight-day-old atd2 seedlings on media with no supplementation (A), 184 μm hypoxanthine (B), or 185 μm adenine (C) are shown. The atd2 bleached mutant phenotype is reversed to a normal green color by addition of adenine, whereas addition of hypoxanthine has no effect.
DISCUSSION
Reversal of Phytotoxic Effects of DAS734 by Adenine
Through a combination of chemical and genetic techniques, we have determined the precise biochemical site of action of DAS734 as AtGPRAT, the first committed enzyme in de novo purine biosynthesis. This identification was facilitated by the observation that the purine adenine completely alleviated the phytotoxic symptoms from germination to formation of the first true leaves. The linear purine biosynthetic pathway splits after the formation of IMP to produce either AMP or GMP via different enzymes (Fig. 1A). Thus, it might be expected that alleviation of inhibition of purine biosynthesis prior to IMP formation may require addition of guanine or xanthine in addition to adenine. Indeed, this logic was used to initially define the site of action of hydantocidin as AdSS (Heim et al., 1995). With our discovery of a potent and specific inhibitor of GPRAT, we observed that addition of a relatively low concentration (185 μm) of exogenous adenine is sufficient to restore normal seedling growth. This is likely due to the presence of efficient purine recycling mechanisms in plants such that adenine is readily taken up and converted to AMP by adenine phosphoribosyltransferase (Lee and Moffatt, 1994; Ashihara et al., 2000; Katahira and Ashihara, 2006). AMP can then be converted to IMP by AMP deaminase to feed the GMP biosynthesis branch of the pathway (Fig. 1A). Unlike vertebrates and microbes, plants do not seem to contain high levels of hypoxanthine/guanine phosphoribosyltransferase activity to efficiently recover exogenous hypoxanthine or guanine to form IMP or GMP (Ashihara et al., 2001; Katahira and Ashihara, 2006). Thus, adenine (rather than adenosine, inosine, hypoxanthine, or guanine) is the most efficient exogenous reagent to reverse the toxic effects of DAS734.
GPRAT Redundancy in Arabidopsis
There are three functional GPRAT genes in the Arabidopsis genome (Ito et al., 1994; Boldt and Zrenner, 2003; Hung et al., 2004; van der Graaff et al., 2004). This is in contrast to most of the other genes encoding enzymes of purine biosynthesis that are present in single copies (Boldt and Zrenner, 2003). However, all of the DAS734-resistance mutations that we identified occur in only one GPRAT isoform, AtGPRAT2. Thus, restoration of normal growth can be achieved by resistance mutations in this single GPRAT isoform, even though we showed that AtGPRAT3 was also susceptible to DAS734 inhibition. (We were unable to express AtGPRAT1 in E. coli to enable inhibitor testing.) This is consistent with the observation that AtGPRAT2 is the major GPRAT expressed in roots, leaves, and flowers (Hung et al., 2004). AtGPRAT3 is expressed at much lower levels in these tissues, whereas AtGPRAT1 is expressed at low levels in roots and flowers only (Ito et al., 1994; Hung et al., 2004). Mutants lacking AtGPRAT2 (atd2, chloroplast import apparatus1 [cia1]) are severely stunted with bleached leaves but are viable and fertile under low light conditions. The bleached seedling phenotype of cia1 and atd2 can be alleviated by the addition of 5 mm AMP or IMP to the medium (Hung et al., 2004; van der Graaff et al., 2004). We found that 185 μm adenine was sufficient to abolish bleaching of atd2, similar to the result found with DAS734 inhibition. The viability of atd2 and cia1 indicates that Arabidopsis can survive but does not thrive via the function of AtGPRATs 1 and/or 3. The AtGPRAT1 knockout mutant is indistinguishable from wild type. Double mutants lacking both AtGPRAT1 and AtGPRAT2 have the same phenotype as those lacking AtGPRAT2, again suggesting that AtGPRAT2 is the primary GPRAT (Hung et al., 2004). However, treatment of Arabidopsis seedlings with >5 μm DAS734 is lethal. This may be explained by our observation that AtGPRAT3 is also strongly inhibited by DAS734 and hence the compound can shut down additional GPRAT activity in the plant. Although the genetic and chemical data are consistent with AtGPRAT2 being the predominant isoform supplying nucleotides for growth and development of Arabidopsis, AtGPRAT3 may be required for more specialized but important secondary functions. Thus, treatment by DAS734 may be equivalent to double or triple knockout mutants lacking AtGPRATs 2 and 3 or all GPRAT activity.
Bleaching Effect from GPRAT Inhibition
The phenotype produced by DAS734 is distinctive in comparison with many other herbicidal inhibitors of primary metabolism. New leaves are bleached white, suggesting damage by photooxidative effects or a deleterious effect on chloroplast development or function. Treatment with the compound phenocopies mutants lacking AtGPRAT2 that were identified by their variegated bleached appearance (Hung et al., 2004; van der Graaff et al., 2004). Transgenic tobacco (Nicotiana tabacum) plants expressing a GPRAT silencing construct also exhibit white leaves (van der Graaff et al., 2004). Hung et al. (2004) have shown that chloroplasts isolated from a mutant defective in AtGPRAT2 are less efficient at importing proteins in in vitro assays. As ATP and GTP are required for transport of proteins into plastids (Soll and Schleiff, 2004), this defect may be because of lower availability of purine nucleotides in the AtGPRAT2 mutant chloroplasts. However, poor import efficiency could also arise as a secondary effect of defective chloroplast development. Lack of chloroplast development and characteristic bleaching of developing leaves is also produced by the phytotoxin tagetitoxin, an inhibitor of RNA polymerase (Lukens and Durbin, 1985; Mathews and Durbin, 1990). RNA synthesis could be reduced by a block in purine nucleotide availability, and so this may offer another mechanism whereby loss of GPRAT function leads to the observed physiological symptoms. van der Graaff et al. (2004) hypothesized that the bleached appearance of AtGPRAT2 mutants could be due to photooxidative damage, but it is unclear why loss of GPRAT function should lead to this phenomenon. Treatment of plants with hydantocidin, an herbicidal inhibitor that blocks the penultimate step in AMP biosynthesis after the bifurcation of the purine biosynthesis pathway at IMP, does not produce the same bleaching effect as DAS734 on Arabidopsis or other plant species (Heim et al., 1995). Bleaching symptoms from loss of GPRAT function (induced either genetically or chemically) may therefore be a consequence of a specific depletion of guanine nucleotides, or a combined deficiency of both adenine and guanine nucleotide pools. Addition of exogenous adenine can supplement both AMP and GMP pools (via adenine phosphoribosyltransferase and AMP deaminase) to recover the normal phenotype.
Other herbicidal compounds that produce similar distinctive bleached-white phenotypes via photooxidative mechanisms are metabolic inhibitors of pathways that are either directly or indirectly involved in producing photoprotectant carotenoids. These include phytoene desaturase inhibitors (e.g. norflurazon; Sandmann and Boger, 1997), 4-hydroxyphenylpyruvate dioxygenase inhibitors (e.g. mesotrione; Mitchell et al., 2001), and inhibitors of the nonmevalonate isoprenoid pathway (e.g. fosmidomycin and clomazone; Mueller et al., 2000; Lange et al., 2001). It will be of interest to determine if these pathways are particularly sensitive to depletion of purines or intermediates of the purine pathway, either directly or indirectly. In addition to having a bleaching effect on emerging leaves, DAS734 is a potent inhibitor of root growth. This is in contrast to the above compounds that act exclusively by photooxidative mechanisms and so have little direct effect on nonphotosynthetic tissues (T. Walsh, P. Schmitzer, and R. Neal, unpublished data). Thus, DAS734 has additional effects beyond bleaching of new growth, as would be expected from interruption of purine biosynthesis. DAS734 will be a useful chemical probe to ascertain the full scope of these metabolic consequences.
Facile Heterologous Overexpression of AtGPRAT
Many previous biochemical studies of GPRATs have been hindered by the extreme oxygen sensitivity of the enzymes (Itakura and Holmes, 1979; Bernlohr and Switzer, 1981). This arises from the lability of the structural 4Fe-4S cluster present in some microbial and all eukaryotic GPRATs. The 4Fe-4S cluster is presumably present in plant GPRATs as the essential ligand residues for the iron-sulfur center are conserved (Fig. 3; Supplemental Fig. S3). We were successful in functionally overexpressing AtGPRAT2 at high levels in E. coli without the chloroplast transit peptide but retaining an 11-residue propeptide preceding the predicted active site Cys residue. Mass spectrometric analysis of purified recombinant AtGPRAT2-His expressed in E. coli indicated that the propeptide was removed to expose the N-terminal catalytic Cys residue. Work with the B. subtilis enzyme has shown that this processing can occur autocatalytically (Brannigan et al., 1995). We took no special precautions to protect AtGPRAT2 from oxygen exposure and found the recombinant plant enzyme to be relatively stable to daily handling. No significant loss in activity was apparent after several weeks of storage at −70°C. The extraordinary stability of recombinant AtGPRAT2 to oxygen relative to other GPRATs from nonphotosynthetic organisms could perhaps be an adaptive consequence of its location in the oxygen-rich environment of the chloroplast.
Sites of the DAS734-Resistance Mutations in GPRAT
Detailed studies of the B. subtilis and E. coli GPRATs have shown that they are highly functionalized enzymes composed of two domains in tight allosteric communication with each other (Chen et al., 1997; Smith, 1998; Bera et al., 2000). One domain contains the glutaminase site where Gln is hydrolyzed to yield ammonia. The ammonia traverses an intramolecular tunnel, formed during the catalytic cycle, to the adjacent domain where it is condensed with PRPP to form the unstable product phosphoribosylamine (Krahn et al., 1997). The PRPP domain is capable of binding two purine nucleotides, one overlapping the active site and another at a peripheral allosteric site (Chen et al., 1997).
Three resistance mutations occur in vicinity of the glutaminase site. R264K confers the highest level of resistance and Arg-264 (or its equivalent) is conserved in almost every GPRAT sequence that we have inspected in a phylogenetic comparison of sequences from 200 species of microbes and higher organisms. Nevertheless, a conservative substitution of this residue to Lys produces a fully functional enzyme with remarkable (approximately 125-fold) resistance to DAS734. The only naturally occurring GPRATs that we found that do not have Arg at this position are those from a small clade of six species of methanotrophic bacteria that also possess a Lys residue at this location. The mutation conferring 50-fold resistance (P476S) occurs on the periphery of the glutaminase site (approximately 10 Å from the active site thiol group) and could therefore be linked to changes in this pocket due to DAS734 binding at or near this site. Because the mutations giving the highest levels of resistance occur around the glutaminase site and we found no effect of the R264K mutation on inhibition by purine nucleotides acting on the PRPP domain, the glutaminase domain is perhaps the most likely site of interaction. However, as there is clear evidence for extensive interdomain and intradomain allosteric communication in GPRAT function (Smith, 1998; Bera et al., 1999), mutations at locations quite distal to the inhibitor binding site such as G371S could play a role in conferring inhibitor resistance in this enzyme. Crystallographic studies of the plant enzyme in complex with DAS734 will shed further light on this.
DAS734 as a Novel Chemical Probe of Plant Purine Biosynthesis
The combination of chemical and genetic approaches used in this study has provided a novel and unique small molecule ligand for probing GPRAT function and de novo purine biosynthesis in plants. Unlike many other nonspecific inhibitors of purine biosynthesis (Lyons et al., 1990), DAS734 appears to be unusually specific for GPRAT from certain dicotyledonous plant species, in particular Arabidopsis. Its utility as a biochemical probe and an herbicidal lead is enhanced by the fact that it does not require bioactivation to a phosphorylated form as do inhibitors of other enzymes in the purine biosynthesis pathway such as hydantocidin (Cseke et al., 1996; Siehl et al., 1996). DAS734 is a good example of one of the potential advantages of chemical genetics because it inhibits at least two of the three functional isoforms of GPRAT in Arabidopsis and so can overcome GPRAT genetic redundancy. Thus, the compound will be of further utility in dissecting how chemical or genetic disruption of GPRAT activity leads to impaired chloroplast development and function and leaf bleaching.
MATERIALS AND METHODS
Materials
Seeds of atd2 were obtained from the Arabidopsis Biological Resource Center. DAS734 was synthesized as described in Supplemental Materials and Methods S1. All other reagents were obtained from Sigma.
Mutant Screening and Root Growth Assays
Ethylmethanesulfonate-mutagenized M2 Col-0 seed (Lehle Seeds) were screened for resistance as described by Walsh et al. (2006). DAS734-resistant seedlings were identified as plants with green, fully expanded cotyledons and developing true leaves. Arabidopsis (Arabidopsis thaliana) seedling assays and metabolite reversal studies were also conducted as described by Walsh et al. (2006). For reversal experiments, adenine was added to the growth media to a final concentration of 185 μm. Root measurements were made by carefully extracting five individual plants from each treatment and measuring the length of the tap root. Data are reported as percentages of the root length of untreated control plants.
Greenhouse Tests
Seeds of test species were planted in Metromix 360 (Brehob) 8 to 12 d prior to spraying with compound and grown in a greenhouse with a 16-h photoperiod at 24°C to 29°C. Compounds were dissolved in acetone:dimethyl sulfoxide (97:3, v/v) and dilutions made with water:isopropanol:crop oil concentrate (78:20:2, v/v/v) containing 0.02% Triton X-155. Compounds were applied using a DeVilbiss compressed air sprayer.
Genetic Analyses
To determine whether resistance mutations were dominant or recessive, M3 plants resistant to DAS734 were crossed with wild-type Col-0 plants. F1 seed were tested for resistance to DAS734 by plating the seed on modified Murashige and Skoog medium containing 0.54 μm DAS734. To determine if the DAS734-resistant lines contained allelic mutations, five M3 plants resistant to DAS734 from each line were crossed with five M3 plants resistant to DAS734 from every other line. To identify crosses between homozygous DAS734-resistant lines, M4 seed resulting from self-fertilization of each parent used in the crosses were harvested and tested for resistance to DAS734. M3 homozygotes produced M4 seed that were 100% resistant to DAS734. F1 progeny from crosses between the identified homozygous parents resistant to DAS734 were grown and allowed to self-fertilize. At least 300 F2 seedlings from each cross were tested for resistance to DAS734. Lines containing independently segregating resistance genes should produce 93% resistant F2 seedlings, whereas allelic lines should produce 100% resistant seedlings. In all cases where the parent was homozygous for the resistance gene, F1 progeny from the test cross were 100% resistant to DAS734. When the herbicide-resistant parent was heterozygous for the resistance gene, approximately 50% of the F1 progeny showed herbicide resistance.
Mapping of the R03 Mutation
To generate the mapping population, a resistant homozygous M3 line of R03 (from the Col-0 accession) was crossed with a wild-type plant of the Landsberg erecta accession. The resulting F1 plants were allowed to self-fertilize and produce F2 seed. Because the resistance phenotype is caused by a dominant mutation, the wild-type allele from the Landsberg erecta background was mapped. The F2 seed was germinated in modified Murashige and Skoog medium containing 0.54 μm DAS734. Plants sensitive to the herbicide were identified, rescued onto medium without herbicide, and transplanted to soil after 7 d. When plants were at the rosette stage, one leaf was removed for genomic DNA isolation. Mapping was performed using single nucleotide polymorphism markers as described by Walsh et al. (2006).
For sequencing, the At4g34740 (AtGPRAT2) gene was PCR amplified by Pfu Turbo DNA polymerase (Stratagene) using the primers TCAACTTTTCATAATTGGTTTGTGTGTATTT and AATTTCGTAGAACTCGCCACAAGCA. The PCR products were sequenced using Applied Biosystems BigDye Terminator version 3.1 cycle sequencing kit and run on an ABI 3100.
Expression of AtGPRAT in Escherichia coli
The gene encoding AtGPRAT2 was PCR amplified from an Arabidopsis cDNA library using the forward DNA 5′-CATATGGATGATTATGACGAGAAGCCTCGGGAAGAGTGTGGAG-3′ and the reverse primer 3′-TAGGTGTTGTAACTTCCTCCAACCCATGCCATCATTCCTAGG-5′. To facilitate gene cloning, the forward and reverse primers encoded an NdeI and BamHI site, respectively (underlined). In Bacillus subtilis, propeptide autoprocessing to reveal an N-terminal Cys is required to generate an active GPRAT enzyme. The 5′ primer was therefore designed to encode a comparable 11-amino acid propeptide (in bold) and deletion of the chloroplast transit peptide of AtGPRAT2. The amplification product was gel purified and cloned using TOPO TA cloning vector pCR2.1 TOPO. Four individual clones were sequenced using Beckman CEQ 2000 Dye Terminator chemistry. DNA sequencing verified that the encoded protein sequence was identical to the published sequence. One clone, AtGPRAT2.1, was selected for E. coli expression. The AtGPRAT2 gene was released from AtGPRAT2.1 as an NdeI/BamHI fragment and cloned into identical sites of the E. coli expression vector pET26b.
Following verification by DNA restriction analysis, two clones (pET2.1.2 and pET2.1.8) were selected for E. coli expression experiments. DNA from pET2.1.2, pET2.1.8, and pET26b (empty vector control), respectively, was transformed into BL21(DE3) cells. Transformation reactions were plated on L agar containing 50 μg/mL kanamycin and 50 mm Glc and grown overnight at 37°C. For expression of individual clones, a loopful of fresh cells was inoculated into 250 mL of Luria-Bertani broth containing 50 μg/mL kanamycin and 75 μm isopropyl-d-thiogalactopyranoside. Cultures were grown for 24 h at 28°C. Cells were harvested by centrifugation and the resulting cell pellets were frozen in dry ice and stored at −80°C.
GPRAT Enzyme Activity
For enzyme extraction, pelleted E. coli cells expressing AtGPRAT were resuspended in 0.1 m Tris, pH 7.4, containing 1 mg/mL lysozyme (5 mL/cells from 250 mL culture) at room temperature. After approximately 15 min, the suspension was frozen in liquid N2 and then thawed. DNase was added to 0.02 mg/mL final concentration and MgCl2 to 1 mm. After the extract was no longer viscous, dithiothreitol (DTT) was added to 10 mm and the extract was centrifuged. The supernatant was passed over a Bio-Rad 10DG column preequilibrated with 50 mm Tris-Cl, pH 7.8, 5 mm MgCl2, 10 mm DTT and the eluant stored in aliquots at −70°C.
GPRAT activity was determined by monitoring the PRPP-dependent production of Glu from Gln. Aliquots of extracts were assayed in 50 μL of 50 mm Tris-Cl, pH 7.8, 5 mm MgCl2, 10 mm DTT and the reaction initiated by adding 50 μL of 40 mm Gln, 5 mm PRPP. Control assays were performed in parallel by adding 50 μL of 40 mm Gln with no PRPP. After 30 min, reactions were quenched in a 100°C sandbath for 2.5 min and precipitated protein removed by brief centrifugation. The Glu present in the reaction mixture was quantitated using Glu dehydrogenase and 3-acetylpyridine dinucleotide as a cofactor. Fifty microliters of each assay supernatant was placed in the wells of a 96-well plate, then 150 μL of a mixture containing 1.36 mm 3-acetylpyridine dinucleotide and 33 units/mL Glu dehydrogenase in 125 mm potassium phosphate, pH 8.0, was added. After 30 min, the A363 was read in a Molecular Devices SpectroMax plate reader. The absorbance in the control sample was subtracted from that in the PRPP-containing assay to determine the amount of PRPP-dependent glutaminase activity in the sample. Enzyme kinetic and I50 values were determined by fitting the data to the appropriate equations using Grafit software (Erithacus Software).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_179247 (AtGPRAT1), NP_195200 (AtGPRAT2), and NP_195599 (AtGPRAT3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Map-based cloning of the DAS734-resistance mutation in resistant line R03.
Supplemental Figure S2. Enzyme kinetic characterization of E. coli-expressed AtGPRAT2 and inhibition by DAS734.
Supplemental Figure S3. Comparison of the amino acid sequences of Arabidopsis and microbial GPRATs.
Supplemental Materials and Methods S1. Chemical synthesis of phenyltriazole acetate DAS734.
Supplementary Material
Acknowledgments
We are grateful to several Dow AgroSciences scientists for invaluable assistance. Lowell Markley first conceptualized and synthesized the phenyltriazole acetate herbicides, Nick Irvine provided additional chemical synthesis expertise, and Debby Camper provided molecular modeling help. At Exelixis Plant Sciences, we thank Cathy Hironaka, Darcie Otter, Michelle Leal, Karin Conners, and Alan Lammers for technical assistance. We also thank Dr. Eric van der Graaff (University of Cologne, Germany) for kindly providing unpublished information on the atd2 insertional Arabidopsis mutant.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Terence A. Walsh (tawalsh@dow.com).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Armstrong JI, Yuan S, Dale JM, Tanner VN, Theologis A (2004) Identification of inhibitors of auxin transcriptional activation by means of chemical genetics in Arabidopsis. Proc Natl Acad Sci USA 101 14978–14983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T, Nakano T, Nakashita H, Sekimata K, Shimada Y, Yoshida S (2003) The influence of chemical genetics on plant science: shedding light on functions and mechanism of action of brassinosteroids using biosynthesis inhibitors. J Plant Growth Regul 22 336–349 [DOI] [PubMed] [Google Scholar]
- Ashihara H, Stasolla C, Loukanina N, Thorpe TA (2000) Purine and pyrimidine metabolism in cultured white spruce (Picea glauca) cells: metabolic fate of 14C-labeled precursors and activity of key enzymes. Physiol Plant 108 25–33 [Google Scholar]
- Ashihara H, Stasolla C, Loukanina N, Thorpe TA (2001) Purine metabolism during white spruce somatic embryo development: salvage of adenine, adenosine, and inosine. Plant Sci 160 647–657 [DOI] [PubMed] [Google Scholar]
- Bera AK, Chen S, Smith JL, Zalkin H (1999) Interdomain signaling in glutamine phosphoribosylpyrophosphate amidotransferase. J Biol Chem 274 36498–36504 [DOI] [PubMed] [Google Scholar]
- Bera AK, Smith JL, Zalkin H (2000) Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel. Interdomain signaling and intermediate channeling. J Biol Chem 275 7975–7979 [DOI] [PubMed] [Google Scholar]
- Bernlohr DA, Switzer RL (1981) Reaction of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase with oxygen: chemistry and regulation by ligands. Biochemistry 20 5675–5681 [DOI] [PubMed] [Google Scholar]
- Blackwell HE, Zhao Y (2003) Chemical genetic approaches to plant biology. Plant Physiol 133 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt R, Zrenner R (2003) Purine and pyrimidine biosynthesis in higher plants. Physiol Plant 117 297–304 [DOI] [PubMed] [Google Scholar]
- Brannigan J, Dodson G, Duggleby HJ, Moody PCE, Smith JL, Tomchick DR, Murzin AG (1995) A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378 416–419 [DOI] [PubMed] [Google Scholar]
- Burdine L, Kodadek T (2004) Target identification in chemical genetics: the (often) missing link. Chem Biol 11 593–597 [DOI] [PubMed] [Google Scholar]
- Chen S, Tomchick DR, Wolle D, Hu P, Smith JL, Switzer RL, Zalkin H (1997) Mechanism of the synergistic end-product regulation of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase by nucleotides. Biochemistry 36 10718–10726 [DOI] [PubMed] [Google Scholar]
- Christopherson RI, Lyons SD, Wilson PK (2002) Inhibitors of de novo nucleotide biosynthesis as drugs. Acc Chem Res 35 961–971 [DOI] [PubMed] [Google Scholar]
- Cox JM, Hawkes TR, Bellini P, Ellis RM, Barrett R, Swanborough JJ, Russell SE, Walker PA, Barnes NJ, Knee AJ, et al (1997) The design and synthesis of inhibitors of imidazoleglycerol phosphate dehydratase as potential herbicides. Pestic Sci 50 297–311 [Google Scholar]
- Cseke C, Gerwick BC, Crouse GD, Murdoch MG, Green SB, Heim DR (1996) 2a-phosphohydantocidin: the in vivo adenylosuccinate synthetase inhibitor responsible for hydantocidin phytotoxicity. Pestic Biochem Physiol 55 210–217 [Google Scholar]
- Dancer J, Lindell S, Ford MJ (1999) Inhibitors of histidine biosynthesis. In BK Singh, ed, Plant Amino Acids. Marcel Dekker, New York, pp 417–444
- Hatch MD (1967) Inhibition of plant adenylosuccinate synthetase by hadacidin and the mode of action of hadacidin and structurally related compounds on plant growth. Phytochemistry 6 115–119 [Google Scholar]
- Heim DR, Cseke C, Gerwick BC, Murdoch MG, Green SB (1995) Hydantocidin: a possible pro-herbicide inhibiting purine biosynthesis at the site of adenylosuccinate synthetase. Pestic Biochem Physiol 53 138–145 [Google Scholar]
- Hung WF, Chen LJ, Boldt R, Sun CW, Li HM (2004) Characterization of Arabidopsis glutamine phosphoribosyl pyrophosphate amidotransferase-deficient mutants. Plant Physiol 135 1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura M, Holmes EW (1979) Human amidophosphoribosyltransferase. An oxygen-sensitive iron-sulfur protein. J Biol Chem 254 333–338 [PubMed] [Google Scholar]
- Ito T, Shiraishi H, Okada K, Shimura Y (1994) Two amidophosphoribosyltransferase genes of Arabidopsis thaliana expressed in different organs. Plant Mol Biol 26 529–533 [DOI] [PubMed] [Google Scholar]
- Katahira R, Ashihara H (2006) Profiles of purine biosynthesis, salvage and degradation in disks of potato (Solanum tuberosum L.) tubers. Planta 225 115–126 [DOI] [PubMed] [Google Scholar]
- Krahn JM, Kim JH, Burns MR, Parry RJ, Zalkin H, Smith JL (1997) Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site. Biochemistry 36 11061–11068 [DOI] [PubMed] [Google Scholar]
- Lange BM, Ketchum RE, Croteau RB (2001) Isoprenoid biosynthesis. Metabolite profiling of peppermint oil gland secretory cells and application to herbicide target analysis. Plant Physiol 127 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Moffatt BA (1994) Adenine salvage activity during callus induction and plant growth. Physiol Plant 90 739–747 [Google Scholar]
- Lukens JH, Durbin RD (1985) Tagetitoxin affects plastid development in seedling leaves of wheat. Planta 165 311–321 [DOI] [PubMed] [Google Scholar]
- Lyons SD, Sant ME, Christopherson RI (1990) Cytotoxic mechanisms of glutamine antagonists in mouse L1210 leukemia. J Biol Chem 265 11377–11381 [PubMed] [Google Scholar]
- Mathews DE, Durbin RD (1990) Tagetitoxin inhibits RNA synthesis directed by RNA polymerases from chloroplasts and Escherichia coli. J Biol Chem 265 493–498 [PubMed] [Google Scholar]
- Mayer TU (2003) Chemical genetics: tailoring tools for cell biology. Trends Cell Biol 13 270–277 [DOI] [PubMed] [Google Scholar]
- Mitchell G, Bartlett DW, Fraser TEM, Hawkes TR, Holt DC, Townson JK, Wichert RA (2001) Mesotrione: a new selective herbicide for use in maize. Pest Manag Sci 57 120–128 [DOI] [PubMed] [Google Scholar]
- Moffatt BA, Ashihara H (2002) Purine and pyrimidine nucleotide synthesis and metabolism. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0018, www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Mueller C, Schwender J, Zeidler J, Lichtenthaler HK (2000) Properties and inhibition of the first two enzymes of the non-mevalonate pathway of isoprenoid biosynthesis. Biochem Soc Trans 28 792–793 [PubMed] [Google Scholar]
- Reynolds PHS, Blevins DG, Randall DD (1984) 5-Phosphoribosylpyrophosphate amidotransferase from soybean root nodules: kinetic and regulatory properties. Arch Biochem Biophys 229 623–631 [DOI] [PubMed] [Google Scholar]
- Sandmann G, Boger P (1997) Phytoene desaturase as a target for bleaching herbicides. In RM Roe, JD Burton, RJ Kuhr, eds, Herbicide Activity: Toxicology, Biochemistry and Molecular Biology. IOS Press, Fairfax, VA, pp 1–10
- Schmitzer PR, Graupner PR, Chapin EL, Fields SC, Gilbert JR, Gray JA, Peacock CL, Gerwick BC (2000) Ribofuranosyl triazolone: a natural product herbicide with activity on adenylosuccinate synthetase following phosphorylation. J Nat Prod 63 777–781 [DOI] [PubMed] [Google Scholar]
- Siehl DL, Subramanian MV, Walters EW, Lee SF, Anderson RJ, Toschi AG (1996) Adenylosuccinate synthetase: site of action of hydantocidin, a microbial phytotoxin. Plant Physiol 110 753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL (1998) Glutamine PRPP amidotransferase: snapshots of an enzyme in action. Curr Opin Struct Biol 8 686–694 [DOI] [PubMed] [Google Scholar]
- Smith JL (2004) Self-processing cysteine-dependent N-terminal nucleophile hydrolases. In A Barrett, N Rawlings, J Woessner, eds, Handbook of Proteolytic Enzymes, Ed 2. Academic Press, New York, pp 2049–2052
- Smith PMC, Atkins CA (2002) Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiol 128 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll J, Schleiff E (2004) Protein import into chloroplasts. Nat Rev Mol Cell Biol 5 198–208 [DOI] [PubMed] [Google Scholar]
- Stockwell BR (2000) Chemical genetics: ligand-based discovery of gene function. Nat Rev Genet 1 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian MV, Walters EW, Lyzwanski O, Siehl DL (1999) Arabidopsis thaliana in culture: a powerful tool to decipher the mode of action/target sites of herbicides with antimetabolite activity. Curr Plant Sci Biotechnol Agric 36 553–556 [Google Scholar]
- Surpin M, Rojas-Pierce M, Carter C, Hicks GR, Vasquez J, Raikhel NV (2005) The power of chemical genomics to study the link between endomembrane system components and the gravitropic response. Proc Natl Acad Sci USA 102 4902–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochtrop GP, King RW (2004) Target identification strategies in chemical genetics. Comb Chem High Throughput Screen 7 677–688 [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Hooykaas P, Lein W, Lerchl J, Kunze G, Sonnewald U, Boldt R (2004) Molecular analysis of “de novo” purine biosynthesis in solanaceous species and in Arabidopsis thaliana. Front Biosci 9 1803–1816 [DOI] [PubMed] [Google Scholar]
- Wakabayashi K, Boger P (2002) Target sites for herbicides: entering the 21st century. Pest Manag Sci 58 1149–1154 [DOI] [PubMed] [Google Scholar]
- Walsh TA, Neal R, Merlo AO, Honma M, Hicks GR, Wolff K, Matsumura W, Davies JP (2006) Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol 142 542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J (2003) SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301 1107–1110 [DOI] [PubMed] [Google Scholar]
- Zheng W, Zhai Q, Sun J, Li CB, Zhang L, Li H, Zhang X, Li S, Xu Y, Jiang H, Wu X, Li C (2006) Bestatin, an inhibitor of aminopeptidases, provides a chemical genetics approach to dissect jasmonate signaling in Arabidopsis. Plant Physiol 141 1400–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57 805–836 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.