Abstract
Germination of cereals is accompanied by extensive change in the redox state of seed proteins. Proteins present in oxidized form in dry seeds are converted to the reduced state following imbibition. Thioredoxin (Trx) appears to play a role in this transition in cereals. It is not known, however, whether Trx-linked redox changes are restricted to cereals or whether they take place more broadly in germinating seeds. To gain information on this point, we have investigated a model legume, Medicago truncatula. Two complementary gel-based proteomic approaches were followed to identify Trx targets in seeds: Proteins were (1) labeled with a thiol-specific probe, monobromobimane (mBBr), following in vitro reduction by an NADP/Trx system, or (2) isolated on a mutant Trx affinity column. Altogether, 111 Trx-linked proteins were identified with few differences between axes and cotyledons. Fifty nine were new, 34 found previously in cereal or peanut seeds, and 18 in other plants or photosynthetic organisms. In parallel, the redox state of proteins assessed in germinating seeds using mBBr revealed that a substantial number of proteins that are oxidized or partly reduced in dry seeds became more reduced upon germination. The patterns were similar for proteins reduced in vivo during germination or in vitro by Trx. In contrast, glutathione and glutaredoxin were less effective as reductants in vitro. Overall, more than half of the potential targets identified with the mBBr labeling procedure were reduced during germination. The results provide evidence that Trx functions in the germination of seeds of dicotyledons as well as monocotyledons.
A growing body of evidence indicates that the germination of seeds is accompanied by extensive change in the redox state of proteins in cereals. Proteins of both the starchy endosperm and embryo that are present mainly in the oxidized (S-S) form in the dry seed are converted to the reduced or sulfhydryl (SH) state following imbibition (Kobrehel et al., 1992; Lozano et al., 1996; Gobin et al., 1997; Yano et al., 2001a; De Gara et al., 2003; Marx et al., 2003; Rhazi et al., 2003). The regulatory disulfide protein, thioredoxin (Trx), appears to play a central role in the redox conversion. When reduced enzymatically in the presence of NADPH, Trx acts as a signal early in germination to facilitate the mobilization of reserves by (1) reducing storage proteins, enhancing their solubility and susceptibility to proteolysis; (2) reducing and inactivating disulfide proteins that inhibit specific amylases and proteases, thereby facilitating the breakdown of stored starch and proteins; and (3) reductively activating individual enzymes functional in germination (Kobrehel et al., 1991, 1992; Jiao et al., 1993; Besse et al., 1996; Lozano et al., 1996; Yano et al., 2001b; Wong et al., 2004a).
Experiments with transgenic grain have confirmed and extended these conclusions. Trx overexpressed in barley (Hordeum vulgare) endosperm accelerated germination, the accompanying release of starch-hydrolyzing enzymes and the reduction of storage and other proteins (Wong et al., 2002). Perhaps not surprisingly, transgenic wheat (Triticum aestivum) in which Trx expression was suppressed showed the opposite effect, i.e. germination was inhibited, thereby leading to protection against preharvest sprouting (Guo et al., 2007). Finally, recent proteomic studies have revealed that a large number of different types of disulfide seed proteins are linked to Trx; approximately 100 authentic or potential Trx targets have been identified in wheat and barley (Maeda et al., 2003; Marx et al., 2003; Wong et al., 2003, 2004b; Balmer et al., 2006).
In contrast to the comprehensive evidence obtained for cereals during the past 15 years, there is scant information on other types of plants. The question, therefore, arises as to whether protein redox changes are restricted to cereals, or whether they take place more broadly in germinating seeds. To obtain information on this point, we have conducted a study with the dicot, Medicago truncatula, a model legume that differs from cereals in seed architecture as well as germination properties. Using complementary proteomic approaches based on fluorescent gel and mutant affinity column procedures, we have identified 111 potential or previously established Trx targets in embryo axes and cotyledons of M. truncatula. We have further shown that a quarter of these targets are reduced in germinating seeds. These findings, summarized below, suggest that the Trx-linked reduction of disulfide proteins associated with seed germination is a general property of plants.
RESULTS AND DISCUSSION
Identification of Potential Trx Targets in Germinating M. truncatula Seeds
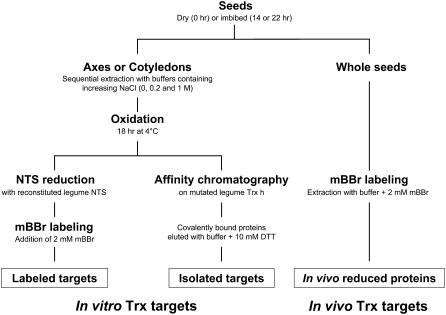
Two approaches were followed to identify potential Trx targets in germinating seeds of M. truncatula (Fig. 1). Proteins of seed extracts were either reduced in vitro with a reconstituted NADP/Trx system (NTS) and then labeled with monobromobimane (mBBr), a fluorescent probe that reacts with thiol groups (Yano et al., 2001b), or, alternatively, proteins were subjected to a mutated Trx affinity column (Motohashi et al., 2001) with bound modified pea (Pisum sativum) Trx h3 (Montrichard et al., 2003). The mBBr-labeled proteins or proteins isolated from the column were identified by liquid chromatography/tandem mass spectrometry (LC/MS/MS) after separation in two-dimensional (2D) gels. The extensive resources currently available for the model legume M. truncatula greatly facilitated the identification of proteins. The data shown were obtained in two separate experiments.
Figure 1.
Procedures for the identification of potential Trx targets and the determination of the redox state of proteins in germinating seeds of M. truncatula. The targets were identified by proteomic procedures.
Preliminary experiments were performed with the mBBr procedure in which labeled proteins were analyzed in one-dimensional (1D) gels. Dry seeds (0 h imbibition) or seeds imbibed for 14 h (before radicle protrusion) or 22 h (after radicle protrusion) were dissected into embryo axes and cotyledons. Because storage proteins were possible targets, preparations were extracted sequentially with 0, 0.2, and 1.0 m NaCl solutions to separate albumin (mainly corresponding to metabolic proteins), vicilin, and legumin enriched fractions, respectively (Krochko and Bewley, 1988). The fractions were incubated aerobically overnight at 4°C to allow the oxidation of proteins that were found to be reduced in germinating seeds (see below). Finally, proteins were reduced with the NTS and labeled with mBBr. Control samples without NTS treatment were prepared in parallel. Proteins of both samples were resolved by SDS-PAGE. The analyses revealed that proteins were not labeled without NTS treatment, indicating that they had been fully oxidized during the overnight incubation (Supplemental Fig. S1). The pattern of proteins labeled with the NTS treatment revealed that Trx-reducible proteins were most abundant in the 0 m NaCl fraction (metabolic proteins). Relatively few proteins were labeled in the 0.2 m NaCl fraction (results not shown). They mainly corresponded to storage proteins such as albumin and legumins, also found in the 0 m NaCl fractions (see below). Essentially none were seen in the 1.0 m NaCl fraction (data not shown). Because of the abundance of Trx-interacting proteins, we further focused on the 0 m NaCl fraction.
Besides NTS, two other redox systems able to modify the redox state of the proteins exist in the cell. They comprise glutathione (GSH) or glutaredoxin (Grx). GSH is the main thiol compound present in aerobic eukaryotes. In addition to its major function in scavenging reactive oxygen species (ROS), particularly peroxides, it has the ability to form mixed disulfides with certain proteins. This reversible process, known as glutathionylation, functions in either regulation of target protein activity or protection of the thiol groups from oxidation (Ghezzi, 2005; Michelet et al., 2006). In contrast, Grx, a small protein related to Trx, has, as Trx, a disulfide reductase activity in the presence of GSH. To see whether the reduction of M. truncatula proteins observed in the presence of Trx was specific, we performed experiments in which the NTS was replaced by GSH or GSH plus Grx. Reduction by GSH or GSH plus Grx were much less effective (data not shown). Thus, the targets seemed to be specific for Trx as noted earlier with cereal seeds.
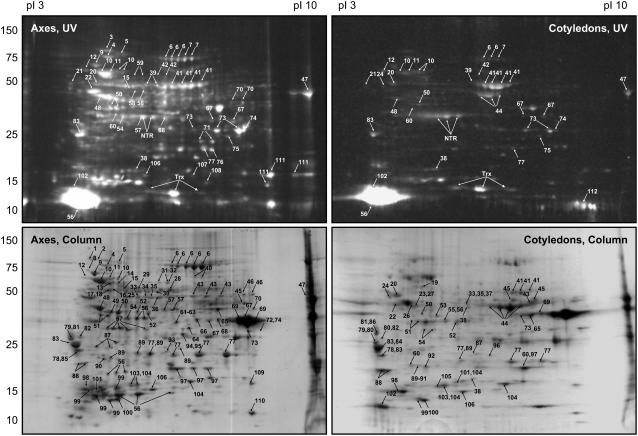
Patterns in 2D gels of proteins labeled with NTS in 0 m NaCl fraction from axes and cotyledons of 14 h-imbibed seeds are shown Figure 2 (top sections). Supplemental Figure S2 demonstrates that, as above, while proteins were labeled after NTS treatment, they were either unlabeled or much less labeled in the corresponding control experiment performed without NTS treatment. Figure 2 also shows that more major fluorescent spots were visible in axis than in cotyledon preparations (154 versus 69 based on a PD-Quest analysis of this representative experiment). In addition, the intensity of fluorescence of the axis spots was generally higher than their cotyledon counterparts. Nevertheless, almost all spots detected in the cotyledon gel matched equivalent spots in the axis gel when compared with PD-Quest software, suggesting that Trx targets in the two parts of the seed are quite similar. The most striking difference was the highly abundant protein that seemed to be unique to cotyledons and that corresponded to the large subunit of Rubisco (spots 44). Surprisingly, relatively few differences were observed in the pattern of labeled proteins during the time course of imbibition in each part of the seed (data not shown). Based on the mBBr results, we elected to analyze only a single time of imbibition (14 h) for Trx targets using the affinity column procedure (Fig. 1). Again, potential targets were more numerous in preparations from axis than cotyledons, 176 versus 65 spots in the representative experiment shown in Figure 2 (bottom sections). Further, although there were minor differences, the Trx-linked proteins isolated from the axes seemed to include those of the cotyledons. Patterns of potential targets obtained with the two strategies were not the same, although they shared many similarities.
Figure 2.
2D gel patterns of potential Trx targets of M. truncatula. Proteins were extracted from embryo axes or cotyledons from seeds imbibed for 14 h. Potential targets were labeled with mBBr (top sections) or isolated by affinity chromatography (bottom sections) then resolved in 2D gels as shown in Figure 1. Fluorescent proteins were detected under UV light at 365 nm and isolated proteins by Coomassie Blue staining. The potential targets identified are numbered and their names and properties appear in Table I. MtNTRA (NTR) and PsTrxh3 (Trx), components of the mBBr labeling procedure, are also indicated.
All potential Trx targets obtained with both the mBBr and affinity column procedures were punched out of the 2D gels of the axes of 14 h-imbibed seeds for identification by LC/MS/MS (Table I). Although most of the proteins identified with cotyledons from 14 h-imbibed seeds could be matched by mobility to those of the axes, the Trx targets in both tissues were analyzed to verify their identities. The matched proteins in the axes and cotyledons were found to be identical for the 14 h-imbibed seeds as well as for the 0 h-imbibed seeds (data not shown). Collectively, these results showed that, in almost all cases, the potential Trx targets were essentially the same in both parts of the seed irrespective of time of imbibition. It should be noted that tissue specificity assignment in Table I is somewhat imprecise. For example, a protein found only in the axis may truly be unique to this tissue if no equivalent spot is present in the cotyledon gel and no similar protein is identified in any other cotyledon spot. However, an equivalent spot may be detected in the cotyledon gel and the protein may be present, but was not identified. Alternatively, in the mBBr procedure, the protein has been identified but mixed with other proteins. In this case, its presence would not be indicated in Table I because this methodology cannot identify the specific target. With respect to the few spots that appeared to be present only in gels of axes or cotyledons or to be specific to a time of imbibition, most were forms differing in pI of proteins identified previously at other positions in the gels. Thus, in the end, very few Trx targets appeared to be unique to axes or cotyledons. This property is mentioned in their description (see below).
Table I.
Identification of potential Trx targets in germinating M. truncatula seeds
The labeled and isolated targets identified in axis (A) or cotyledon (C) extracts showed an E-value <−3. Available tentative consensus (TC), EST, and accession numbers corresponding to M. truncatula proteins are mentioned. Proteins for which only partial sequences are known in M. truncatula, the accession numbers of the nearest matching orthologs, are indicated. Key properties are also listed: theoretical MW/pI, coverage (%), peptide number, conserved/total Cys, and potential localization. Predotar and Psort were used to determine potential localization of complete proteins known in M. truncatula or others species. C, Cytosol; M, mitochondria; ER, endoplasmic reticulum; CP, chloroplasts; N, nucleus. Proteins identified with only one peptide are indicated by §. The presence of the protein in the 0.2 m NaCl fraction is denoted with *. The three proteins that have no Cys residues (spots 63, 101, 108) were not further considered in the study.
| Spot No.
|
Protein
|
E-Value
|
Protein Labeled with mBBr
|
Protein Isolated on Mutant Trx
|
M. truncatula TC, EST, or Accession No.
|
Nearest Ortholog Accession No.
|
Theoretical MW/pI
|
Coverage (%)
|
Peptide No.
|
Conserved/Total Cys
|
Potential Localization
|
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | A | C | ||||||||||
| 1 | UK1 | −15 | • | TC101829 | – | 15 | 3 | ||||||
| 2 | UK2 | −7 | • | TC97992 | – | 6.5 | 2 | ||||||
| 3 | HSP90 | −60 | • | TC94880 | 95.5/5.1 | 12 | 8 | 12/14 | C | ||||
| 4 | CDC48 | −147 | • | • | TC94474 | 89.7/5.1 | 32 | 17 | 13/15 | C | |||
| −117 | • | • | TC97683 | P54774 | 89.7/4.9 | 4.1 | 14 | 14/14 | C | ||||
| 5 | ADP-ribosyltransferase or PM38 | −92 | • | • | BQ124797 | ADD51626 | 91.6/5.2 | 15 | 11 | 18/20 | C | ||
| 6 | EF2 | −181 | • | • | • | TC93936 | 94.1/5.8 | 39 | 21 | 15/16 | C | ||
| −14 | • | TC94039 | Xp_465992 | 94.0/5.7 | 3.7 | 3 | 15/18 | C | |||||
| −72 | • | • | TC94015 | 94.1/5.8 | 7.2 | 10 | 15/17 | C | |||||
| 7 | Lipoxygenase | −270 | • | • | TC94816 | P09439 | 97.1/6.2 | 49 | 28 | 4/5 | C | ||
| 8 | HSP90 (putative)§ | −5 | • | TC96263 | NP_187434 | 90.9/5.0 | 1.7 | 1 | 3/4 | M | |||
| 9 | HSP83 | −232 | • | • | TC106622 | 80.1/4.9 | 40 | 28 | 5/5 | C | |||
| 10 | HSP70 (cytosol) | −187 | • | • | • | TC100383 | 71.0/5.1 | 42 | 22 | 7/7 | C | ||
| −163 | • | • | • | TC100389 | 70.9/5.0 | 10 | 20 | 7/7 | C | ||||
| −240 | • | • | • | TC108198 | 71.5/5.2 | 50 | 27 | 7/7 | C | ||||
| −49 | • | • | BQ139103 | AAS57912 | 71.0/4.9 | 5.3 | 6 | 7/7 | C | ||||
| 11 | HSP70 (endoplasmic reticulum) | −63 | • | • | • | TC100382 | 73.5/5.0 | 12 | 9 | 4/5 | ER | ||
| −7 | • | TC100386 | CAB72128 | 73.4/4.8 | 2.4 | 2 | 3/3 | ER | |||||
| 12 | HSP70 (chloroplasts) | −280 | • | • | • | TC108910 | CAA49147 | 75.5/5.0 | 31 | 30 | 2/2 | CP | |
| 13 | HSP70 (mitochondria) | −75 | • | TC106412 | 70.9/5.3 | 15 | 10 | 3/4 | CP | ||||
| −29 | • | TC106356 | 72.3/5.8 | 6.9 | 4 | 3/3 | M | ||||||
| 14 | Leukotriene-A4 hydrolase like | −49 | • | TC95368 | 68.6/5.0 | 12 | 7 | 3/7 | C | ||||
| 15 | CPN60-2 | −8 | • | • | TC100626 | CAA50217 | 61.0/5.4 | 3.7 | 2 | 6/6 | M | ||
| −9 | • | • | TC100627 | CAA50217 | 61.0/5.4 | 3.1 | 2 | 6/6 | M | ||||
| 16 | Arg-methyltransferase | −10 | • | TC110222 | NP_850528 | 59.7/4.8 | 20 | 2 | 3/7 | C | |||
| −9 | • | TC95822 | NP_850528 | 59.7/4.8 | 4.3 | 2 | 3/7 | C | |||||
| 17 | TCP CPN60 chaperonin subunit ɛ§ | −3 | • | TC106794 | 52.9/5.3 | 3.8 | 1 | 7/9 | C | ||||
| 18 | TCP CPN60 chaperonin subunit τ | −32 | • | TC96153 | Q75HJ3 | 58.7/5.0 | 20 | 5 | 7/9 | C | |||
| 19 | ATPase subunit α (vacuole) | −9 | • | TC106363 | Q9SM09 | 68.6/5.1 | 4 | 2 | 5/5 | C | |||
| 20 | PDI | −311 | • | • | • | TC106707 | 57.0/4.9 | 54 | 34 | 4/4 | ER | ||
| 21 | Calreticulin | −63 | • | • | TC106673 | 48.4/4.4 | 15 | 8 | 3/3 | ER | |||
| 22 | Tubulin β | −133 | • | • | TC106434 | 50.5/4.7 | 38 | 14 | 8/10 | C | |||
| 23 | Tubulin α | −33 | • | TC100409 | 49.5/4.8 | 13 | 5 | 11/11 | C | ||||
| 24 | CPN60 subunit α | −345 | • | • | TC100730 | 61.9/4.8 | 61 | 46 | 1/1 | CP | |||
| 25 | CPN60 subunit β | −34 | • | TC100508 | 62.7/5.3 | 8 | 5 | 6/6 | CP | ||||
| 26 | ATP synthase subunit β (mitochondria) | −180 | •* | • | TC93945 | 59.8/5.6 | 37 | 19 | 2/2 | M | |||
| −5 | • | CX526674 | O04275 | 60.1/6.5 | 6.3 | 1 | 1/3 | M | |||||
| 27 | UDP-Glc pyrophosphorylase | −136 | • | TC94328 | 51.4/5.0 | 26 | 15 | 2/2 | C | ||||
| 28 | Pyruvate decarboxylase | −35 | • | TC100918 | 63.1/4.4 | 11 | 5 | 14/15 | C | ||||
| −116 | • | TC100561 | 65.0/5.6 | 29 | 13 | 14/17 | C | ||||||
| 29 | Phosphoglycerate dehydrogenase | −5 | • | NM_101636 | 66.4/5.6 | 1.3 | 2 | 3/6 | CP | ||||
| −15 | • | NM_119583 | 63.3/6.0 | 4.8 | 3 | 3/5 | CP | ||||||
| 30 | Ketol acid reductoisomerase§ | −3 | • | TC94305 | 62.9/6.2 | 2.2 | 1 | 6/7 | CP | ||||
| 31 | Succinate dehydrogenase | −15 | • | TC100922 | NM_126074 | 69.6/5.8 | 3.3 | 2 | 8/11 | M | |||
| 32 | Stress-induced protein (Sti1) | −40 | • | TC102206 | Q9STH1 | 63.7/5.8 | 13 | 6 | 6/6 | C | |||
| 33 | Selenium-binding protein | −90 | • | • | TC107269 | 54.2/5.5 | 28 | 11 | 7/7 | C | |||
| 34 | UDP-Glc dehydrogenase | −5 | • | TC106325 | 52.9/6.7 | 4.2 | 2 | 10/11 | C | ||||
| 35 | Succinic semialdehyde dehydrogenase (mitochondria) | −108 | • | • | TC101970 | 52.6/5.4 | 27 | 12 | 5/7 | CP | |||
| 36 | 4-Coumarate-CoA ligase | −21 | • | TC95226 | 55.8/5.5 | 7 | 4 | 4/6 | C | ||||
| 37 | Leu aminopeptidase | −100 | • | TC94615 | Q8GZD8 | 60.2/7.7 | 25 | 12 | 5/10 | CP | |||
| 38 | Vicilin (47 kD) | −180 | • | • | • | TC100301 | 53.1/5.1 | 29 | 21 | 1/2 | ER | ||
| −121 | • | • | • | TC100302 | 53.7/5.5 | 28 | 14 | 1/1 | ER | ||||
| 39 | PM51 | −6 | • | • | TC100258 | 55.1/5.7 | 3.4 | 2 | 1/1 | ER | |||
| 40 | SBP65 | −6 | • | TC102224 | Q41060 | 59.5/5.7 | 4.6 | 2 | 0/2 | C | |||
| 41 | Suc-binding protein P54 | −106 | • | • | • | TC94339 | 55.8/5.9 | 21 | 13 | 6/7 | ER | ||
| 42 | Malic enzyme (cytosol) | −136 | • | • | TC106706 | 65.3/5.9 | 33 | 16 | 5/6 | C | |||
| 43 | Aldehyde dehydrogenase | −97 | • | • | TC108200 | Q84V96 | 59.0/8.2 | 19 | 11 | 6/7 | M | ||
| −39 | • | • | TC106752 | 60.2/7.9 | 3.9 | 5 | 7/8 | M | |||||
| 44 | Rubisco large subunit | −46 | • | AJ847602 | O03042 | 52.9/5.7 | 18 | 5 | 9/9 | CP | |||
| 45 | RNA-binding protein | −65 | • | • | TC94137 | 39.4/6.7 | 17 | 7 | 1/1 | N | |||
| −25 | • | • | TC94494 | 40.5/6.1 | 6.3 | 4 | 1/1 | N | |||||
| 46 | Catalase | −15 | • | TC100988 | CAA42736 | 57.3/6.7 | 6.2 | 3 | 3/5 | C | |||
| 47 | EF1 subunit α | −145 | • | • | • | TC106485 | 49.1/9.0 | 8.2 | 16 | 6/6 | C | ||
| 48 | 26S proteasome regulatory subunit 6A | −89 | • | • | • | TC94498 | 47.4/4.7 | 24 | 10 | 4/4 | C | ||
| 49 | PDI like§ | −4 | • | TC106884 | 40.1/5.1 | 5.5 | 1 | 6/6 | ER | ||||
| 50 | Actin | −66 | • | • | • | • | TC106785 | 41.7/5.2 | 14 | 7 | 4/4 | C | |
| −94 | • | • | • | TC107326 | 41.7/5.3 | 23 | 11 | 4/4 | C | ||||
| −5 | • | TC109603 | XP_469569 | 41.8/5.1 | 5.7 | 2 | 4/4 | C | |||||
| −47 | • | • | • | TC106786 | XP_469569 | 41.8/5.1 | 2.7 | 7 | 4/4 | C | |||
| 51 | 4-Methyl-5(b-hydroxyethyl)- thiazole monophosphate biosynthesis protein | −185 | • | • | TC107759 | 46.8/6.7 | 38 | 20 | 3/3 | CP | |||
| 52 | SNF4b | −50 | • | • | AAO61675 | 41.9/5.3 | 31 | 7 | 5/6 | C | |||
| 53 | EF-tu | −133 | • | TC100566 | 53.1/6.1 | 26 | 14 | 1/2 | CP | ||||
| 54 | Gln synthetase (cytosol) | −61 | • | • | • | TC106729 | 39.1/5.2 | 24 | 7 | 3/3 | C | ||
| −56 | • | • | TC106808 | P04771 | 39.2/5.2 | 26 | 7 | 3/3 | C | ||||
| 55 | Gln synthetase (chloroplasts) | −33 | • | TC106913 | 47.0/6.2 | 16 | 5 | 3/6 | CP | ||||
| 56 | Legumin A | −96 | • | • | • | TC100259 | Q41702 | 56.5/6.5 | 7.9 | 12 | 6/7 | ER | |
| −137 | • | • | • | • | TC100252 | 60.4/6.2 | 23 | 16 | 5/7 | ER | |||
| 57 | Legumin J | −45 | • | •* | • | TC100250 | P05692 | 56.9/5.5 | 15 | 6 | 5/6 | ER | |
| −70 | • | •* | • | TC100253 | P05692 | 56.9/5.5 | 21 | 8 | 5/6 | ER | |||
| 58 | Enolase | −140 | • | TC100309 | 47.8/5.5 | 30 | 14 | 3/4 | C | ||||
| 59 | Phosphoglycerate mutase | −107 | • | TC100321 | 60.6/5.4 | 25 | 13 | 3/3 | C | ||||
| 60 | Allergen 28 K | −60 | • | • | • | TC101005 | 50.7/5.4 | 18 | 7 | 2/3 | C | ||
| 61 | Aldose-1-epimerase | −66 | • | TC95178 | 35.7/5.6 | 20 | 8 | 2/3 | C | ||||
| 62 | Cinnamoyl-CoA reductase | −4 | • | TC101159 | 36.0/5.7 | 5.3 | 2 | 7/8 | C | ||||
| 63 | 26S proteasome subunit 7 | −9 | • | TC109249 | O24412 | 34.7/6.0 | 15 | 2 | 0/1 | C | |||
| 64 | Chalcone reductase | −63 | • | TC100398 | 34.8/5.9 | 33 | 8 | 2/3 | C | ||||
| 65 | Glycinin subunit G7 | −31 | • | • | AJ498697 | Q6DR94 | 60.5/6.6 | 29 | 5 | 1/10 | ER | ||
| 66 | Aldo/keto auxin- induced reductase | −16 | • | TC94944 | 37.9/5.9 | 7.9 | 3 | 4/6 | C | ||||
| 67 | GAPDH (cytosol) | −349 | • | • | • | TC106518 | 36.6/6.5 | 53 | 34 | 2/2 | C | ||
| −134 | • | • | • | TC106519 | 37.0/6.9 | 26 | 14 | 2/2 | C | ||||
| 68 | Malate dehydrogenase | −78 | • | • | TC100430 | 35.6/6.1 | 25 | 9 | 5/5 | C | |||
| −78 | • | • | TC100429 | 35.6/6.1 | 27 | 10 | 5/6 | C | |||||
| 69 | Dehydrin | −160 | • | • | • | TC100921 | 31.2/6.4 | 41 | 14 | 0/1 | C | ||
| 70 | GTP-binding protein | −81 | • | • | TC95094 | Q9M7P3 | 44.4/6.3 | 40 | 9 | 5/7 | C | ||
| −44 | • | • | TC103575 | Q9M7P3 | 44.4/6.3 | 15 | 6 | 5/7 | C | ||||
| 71 | Ran | −113 | • | TC93955 | 25.2/6.3 | 51 | 12 | 6/6 | C | ||||
| 72 | 5,10-Methylene tetrahydrofolate dehydrogenase§ | −6 | • | TC107067 | 31.5/7.0 | 3.7 | 1 | 3/3 | C | ||||
| 73 | PM34 | −187 | • | • | • | • | TC107621 | 31.8/6.9 | 58 | 20 | 2/5 | C | |
| −57 | • | • | BQ122292 | Q9LLQ6 | 31.7/6.5 | 59 | 7 | 1/5 | C | ||||
| 74 | G-protein subunit β | −155 | • | • | • | TC100539 | 35.6/7.0 | 41 | 17 | 7/7 | C | ||
| 75 | ESP | −207 | • | • | TC103893 | 27.4/6.7 | 72 | 22 | 3/5 | C | |||
| 76 | GST (mitochondria) | −57 | • | TC95046 | 24.8/6.1 | 30 | 7 | 2/4 | M | ||||
| 77 | 1-Cys Prx | −196 | • | • | • | • | TC108877 | 24.3/6.0 | 57 | 22 | 2/3 | C | |
| 78 | EF1 subunit β | −129 | • | • | TC94335 | 24.2/4.4 | 39 | 15 | 1/1 | C | |||
| 79 | EF1 subunit δ | −86 | • | • | TC106814 | 25.3/4.2 | 43 | 11 | 1/1 | C | |||
| −48 | • | • | TC106815 | 25.3/4.2 | 4.4 | 7 | 1/1 | C | |||||
| 80 | DNA polymerase d auxiliary protein | −20 | • | AJ497831 | O82134 | 29.4/4.4 | 22 | 3 | 5/5 | N | |||
| 81 | PM24 | −43 | • | • | AW559621 | Q9SEL0 | 26.8/4.9 | 48 | 6 | 1/2 | N | ||
| 82 | Ran-binding protein | −33 | • | • | TC94495 | 24.5/4.5 | 16 | 5 | 4/4 | N | |||
| 83 | PM25 | −134 | • | • | • | • | ABB16353 | 26.4/4.5 | 69 | 15 | 1/1 | N | |
| 84 | 14-3-3 protein§ | −6 | • | TC106436 | 29.3/4.5 | 7.8 | 1 | 2/2 | C | ||||
| 85 | UK3 | −16 | • | TC108880 | 27.3/4.5 | 13 | 3 | 1/1 | M | ||||
| 86 | LEA protein (group 5) | −57 | • | TC96862 | P09444 | 26.9/4.8 | 40 | 7 | 0/1 | C | |||
| 87 | Wali 7 | −32 | • | TC100852 | 26.9/5.1 | 17 | 5 | 4/4 | C | ||||
| −45 | • | TC100620 | 28.0/5.6 | 17 | 6 | 4/6 | C | ||||||
| 88 | 2-Cys Prx | −122 | • | • | TC94151 | 29.1/5.8 | 34 | 14 | 2/3 | CP | |||
| −82 | • | • | TC93962 | 29.0/6.0 | 11 | 10 | 2/4 | CP | |||||
| 89 | Ferritin | −84 | • | • | TC96963 | Q1SCB6 | 20.3/5.9 | 30 | 10 | 3/4 | CP | ||
| 90 | UK4 | −20 | • | • | TC109465 | – | 22 | 3 | |||||
| 91 | Isopropylmalate dehydratase | −9 | • | TC101142 | 26.4/5.6 | 6.8 | 2 | 3/4 | CP | ||||
| 92 | SOUL-heme-binding protein | −76 | • | TC107473 | 25.3/5.4 | 20 | 9 | 2/3 | ER | ||||
| 93 | Triose phosphate isomerase | −43 | • | TC93925 | Q6GW08 | 27.2/5.6 | 20 | 6 | 3/4 | C | |||
| 94 | Sorbitol-6-P dehydrogenase§ | −4 | • | TC101758 | AAL86677 | 34.9/6.7 | 4.5 | 1 | 6/6 | C | |||
| 95 | Ni-binding urease accessory protein§ | −3 | • | TC94960 | 29.9/5.9 | 6.9 | 1 | 3/3 | C | ||||
| 96 | Short-chain alcohol dehydrogenase | −11 | • | TC106262 | Q9SQF9 | 28.1/5.1 | 13 | 2 | 4/5 | C | |||
| 97 | Gly-rich protein | −90 | • | • | TC98399 | Q75QN8 | 21.5/5.5 | 59 | 10 | 6/9 | C | ||
| 98 | Prx IIE | −7 | • | • | TC95578 | 23.2/9.6 | 8.9 | 2 | 2/2 | CP | |||
| 99 | Gly-rich RNA-binding protein | −32 | • | TC96436 | P49311 | 14.8/4.7 | 33 | 4 | 1/1 | C | |||
| −14 | • | TC100232 | P49311 | 16.3/4.9 | 21 | 3 | 1/1 | C | |||||
| −34 | • | • | TC93939 | Q41518 | 15.7/5.0 | 2.3 | 5 | 1/1 | C | ||||
| 100 | PM28 | −18 | • | • | CA989687 | Q9LE44 | 9.5/5.5 | 14 | 3 | 0/1 | C | ||
| 101 | Small HSP (18.2 kD) | −16 | • | TC100459 | 18.0/5.0 | 23 | 3 | 0/1 | C | ||||
| −16 | • | TC108710 | 17.1/4.9 | 17 | 3 | 0/1 | C | ||||||
| 102 | 2S albumin | −23 | • | • | • | TC100254 | 16.8/6.2 | 6.5 | 3 | 8/8 | ER | ||
| 103 | EIF5 A-2 | −95 | • | • | TC94209 | 17.2/5.1 | 70 | 11 | 4/4 | C | |||
| −51 | • | TC94697 | 17.5/5.2 | 14 | 7 | 4/4 | C | ||||||
| 104 | GSH peroxidase | −72 | • | • | TC94412 | 25.8/9.8 | 23 | 10 | 3/4 | M | |||
| 105 | Prx IIF§ | −5 | • | TC107761 | 21.4/9.4 | 4.9 | 1 | 2/2 | M | ||||
| 106 | Prx II | −81 | • | • | • | TC106854 | 17.5/5.5 | 40 | 9 | 2/2 | C | ||
| 107 | Ripening-related protein | −50 | • | TC106511 | 18.2/6.0 | 24 | 6 | 2/4 | C | ||||
| 108 | Nucleoside diphosphate kinase | −47 | • | TC106697 | 16.4/6.2 | 23 | 6 | 0/1 | C | ||||
| 109 | UK 5 | −62 | • | TC106727 | 22.6/9.7 | 27 | 8 | 1/1 | C | ||||
| 110 | 40S ribosomal protein S21 | −14 | • | TC98256 | 9.02/8.0 | 14 | 3 | 1/2 | C | ||||
| 111 | FKBP | −159 | • | TC106311 | 18.2/8.3 | 49 | 16 | 4/4 | C | ||||
| 112 | Nonspecific LTP | −9 | • | TC94138 | 11.3/9.0 | 14 | 2 | 9/10 | ER | ||||
| 113 | Conglutin | −123 | •* | AJ498597 | CAA46552 | 48.9/9.0 | 46 | 13 | 9/12 | ER | |||
| 114 | Sali 3.2 | −41 | •* | TC106693 | 30.5/6.0 | 20 | 6 | 6/6 | ER | ||||
Finally, some mBBr-labeled proteins observed in gels developed with the 0.2 m NaCl fraction were analyzed. Again, they were found, in the main, to correspond to proteins previously identified in the 0 m NaCl fraction. Two potential targets unique to the 0.2 m NaCl fraction were nevertheless uncovered. They are indicated at the end of the Table I (spots 113 and 114).
Approximately 800 spots were analyzed by LC/MS/MS. It is noted that some spots containing proteins present as several forms differing in pI gave the same identification while others gave no identification. Only proteins identified with an expectation score or E-value <−3 and, with the exception of the nine proteins designated with a § in Table I, with at least two different peptides (Supplemental Table S1). We elected to retain those nine proteins for potential value for future studies as pointed out by Veenstra et al. (2004).
mBBr and Trx Affinity Column Approaches Are Complementary
The two approaches together yielded a total of 114 different potential Trx targets in M. truncatula seeds. This number is higher than the totals reported for similar screens in other plant systems, e.g. approximately 70 potential targets were identified in wheat with the same two approaches (Wong et al., 2004b). The names of the proteins identified in M. truncatula are given in Table I. Key properties, theoretical MW/pI, percent coverage, number of peptides, conserved Cys/total Cys residues and potential localization, as well as accession number are also indicated. Many proteins are represented by more than one isoform, i.e. isoforms encoded by different genes. When the M. truncatula primary sequence was unknown or only partially known, the accession number of the complete sequence of the nearest ortholog and its properties appear in Table I. Each protein listed contains one or more Cys residues, except for small heat shock protein (HSP) 18.2 kD (spot 101), subunit 7 of 26S proteasome (spot 63), and nucleoside diphosphate kinase (spot 108). Small HSP and subunit 7 of 26S proteasome were isolated by the affinity column procedure. Since they are part of protein complexes comprising at least one previously reported potential Trx target containing conserved Cys residues, they may have been trapped due to this association. Thus, small HSP could have been associated with HSP70 (Jinn et al., 1995) and subunit 7 with the complementary subunit 6A of the proteasome. The only explanation in the case of nucleoside diphosphate kinase, that was in contrast uncovered in the mBBr procedure, is that the fluorescent spot contained more than one protein and the one identified is possibly not the target. These three proteins having no Cys residues are listed in Table I but were not further considered in the study. The remaining proteins harbored one or more Cys residues and, in almost all cases, at least one of these was conserved (Table I).
Among the 111 remaining Trx-linked proteins found in M. truncatula, 45 were recovered with mBBr labeling in 0 and 0.2 m NaCl fractions (41%), 96 with the affinity column (86%), and 30 by both procedures (27%). The set of proteins identified by the two approaches is basically similar to that reported for wheat seeds (Wong et al., 2004b) and isolated amyloplasts (Balmer et al., 2006). These results thus confirm the previous conclusion that the two approaches are complementary, each having advantages and drawbacks. The labeling procedure has the advantage of ensuring that reduction of the targets actually took place. Further, as seen below, the proteins that are reduced by Trx in vitro can be compared directly to those reduced in vivo, thereby allowing the validation of potential targets. The labeling approach, however, has drawbacks. It highlights only the more abundant targets and, in some cases, spots contain more than one protein, hampering the identification of the actual target. The affinity method also has strengths and weaknesses. Trx targets of low abundance can often be identified, and the presence of more than one protein per spot is generally not a problem. However, some proteins that are not actual targets may be captured as part of a Trx-linked complex. Because of a general lack of specificity of Trx isoforms under in vitro conditions as noted earlier (Motohashi et al., 2001; Balmer et al., 2003, 2004b; Maeda et al., 2004; Yamazaki et al., 2004), both methods lead accordingly to the isolation of targets from all cell compartments (Table I).
Of the Trx targets identified in germinating seeds of M. truncatula, 59 were new, 34 were common to cereal or peanut (Arachis hypogaea) seeds, and 18 were reported for other plant organs or organisms (Table II). Thus, almost half of the targets were previously known, thereby confirming the reliability of the experimental procedures. On the other hand, half were not earlier associated with Trx. The large number of new targets is likely, at least in part, due to the nature of the material analyzed, i.e. a legume seed whose redox biology has not been previously explored. An in silico analysis of gene expression indicated that 25 of the 111 candidate targets listed in Table I may be specifically synthesized or highly accumulated in seeds, including several storage proteins and late embryogenesis abundant (LEA) proteins (M. truncatula gene indices [MtGI]: http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=medicago; release 8 from January 26, 2005). These proteins are identified by # in Table II.
Table II.
Potential and true Trx targets identified in M. truncatula germinating seeds have diverse functions
The proteins are divided in two groups: the previously unrecognized and the previously recognized targets, already reported for other photosynthetic organisms. Targets already reported in screens of cereal or peanut seeds are indicated with *. Targets that are reduced in germinating seeds of M. truncatula are indicated in bold, and those whose gene expression is associated with seeds are indicated with #.
| Previously Unrecognized Targets | Previously Reported Targets |
|---|---|
| Metabolism | Metabolism |
| Carbon metabolism | Carbon metabolism |
| Malic enzyme (cytosol) | Enolase* |
| Amino acid synthesis | GAPDH (cytosol)* |
| Isopropylmalate dehydratase | Malate dehydrogenase (cytosol)* |
| Vitamin synthesis | Phosphoglycerate mutase* |
| 5,10-Methylene tetrahydrofolate dehydrogenase | Triose phosphate isomerase* |
| 4-Methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis protein | PM34*# |
| Cell wall synthesis | Succinate dehydrogenase |
| UDP-Glc dehydrogenase | Rubisco large subunit |
| Cinnamoyl-CoA reductase | Amino acid synthesis |
| 4-Coumarate-CoA ligase | Ketol acid reductoisomerase* |
| DNA synthesis | Phosphoglycerate dehydrogenase |
| DNA polymerase d auxiliary protein | Gln synthetase (chloroplasts) |
| Protein biogenesis and degradation | Gln synthetase (cytosol) |
| Translation | ATP-linked energy |
| EF1α | ATP synthase subunit β (mitochondria)* |
| EF1δ | ATPase subunit α (vacuole) |
| EIF5 A-2 | Cell wall synthesis |
| 40S ribosomal protein S21 | UDP-Glc pyrophosphorylase* |
| Folding | Cell structure |
| HSP90 | Actin* |
| HSP83 | Tubulin α* |
| HSP90 (putative) | Tubulin β* |
| PDI like | Protein biogenesis and degradation |
| TCP CPN60 chaperonin ɛ | Translation |
| TCP CPN60 chaperonin τ | EF1β* |
| FKBP (cytosol) | EF2* |
| Targeting | EF-tu (chloroplast) |
| Ran-binding protein | Folding |
| Degradation | PDI* |
| Ni-binding urease accessory protein | HSP70 (cytosol) |
| Storage proteins | HSP70 (endoplasmic reticulum)* |
| Allergen 28 K# | HSP70 (mitochondria) |
| Glycinin subunit G7# | HSP70 (chloroplasts) |
| Legumin J# | CPN60-2 (mitochondria) |
| Vicilin 47 kD (whole protein and fragments)# | CPN60α (chloroplasts) |
| Binding proteins | CPN60β (chloroplasts)* |
| Calreticulin | Stress-induced protein Sti1* |
| Selenium-binding protein | Targeting |
| Ferritin# | Ran |
| SOUL-heme-binding protein | Degradation |
| P54# | Leu aminopeptidase* |
| SBP65# | Proteasome subunit 6A* |
| Response to stress | CDC48* |
| Oxidative stress | Storage proteins |
| GST (mitochondria)# | 2S Albumin*# |
| Desiccation and osmotic stress | Conglutin*# |
| Dehydrin# | Legumin A (whole protein and fragments)*# |
| LEA protein (group 5)# | Response to stress |
| PM24# | Oxidative stress |
| PM25# | Catalase* |
| PM51# | 1-Cys Prx*# |
| Sorbitol-6-P dehydrogenase | 2-Cys Prx* |
| Hypoxia | Prx II (cytosol) |
| Pyruvate decarboxylase | Prx IIF (mitochondria) |
| Succinic semialdehyde dehydrogenase | Prx IIE (chloroplasts) |
| Aluminum stress | GSH peroxidase* |
| Sali 3.2# | Aldo/keto auxin-induced reductase* |
| Wali 7 | Hypoxia |
| Biotic stress | Aldehyde dehydrogenase* |
| Chalcone reductase | Alcohol dehydrogenase |
| Lipoxygenase# | Biotic stress |
| Leukotriene-A4 hydrolase like | Nonspecific LTP*# |
| Signal transduction | Signal transduction |
| Arg-methyltransferase | Gly-rich RNA-binding protein* |
| ADP-ribosyltransferase# | G-protein subunit β* |
| GTP-binding protein | Unknown processes |
| RNA-binding protein | Aldose-1-epimerase |
| 14-3-3 protein | ESP*# |
| SNF4b# | |
| Unknown processes | |
| PM28# | |
| Gly-rich protein | |
| Ripening-related protein | |
| UK1 | |
| UK2 | |
| UK3 | |
| UK4 | |
| UK5 |
Redox State of Potential Trx Targets during Germination
Experiments performed with cereals support the view that proteins present mainly in the S-S form in the dry seed are converted to the −SH state following imbibition. This is particularly obvious for major storage proteins. However, while storage proteins have been shown to undergo reduction, data for most other proteins is lacking. To help fill this gap and, at the same time, determine whether seeds of dicotyledons undergo redox change similar to cereals, we examined the redox state of individual proteins during the course of germination using the mBBr/fluorescent gel approach (Fig. 1). In addition to extending the germination redox changes to dicots, as seen below, the data provide additional evidence for the function of Trx in vivo as the other disulfide reductants (GSH and Grx) showed only marginal activity in reducing proteins in vitro as mentioned above.
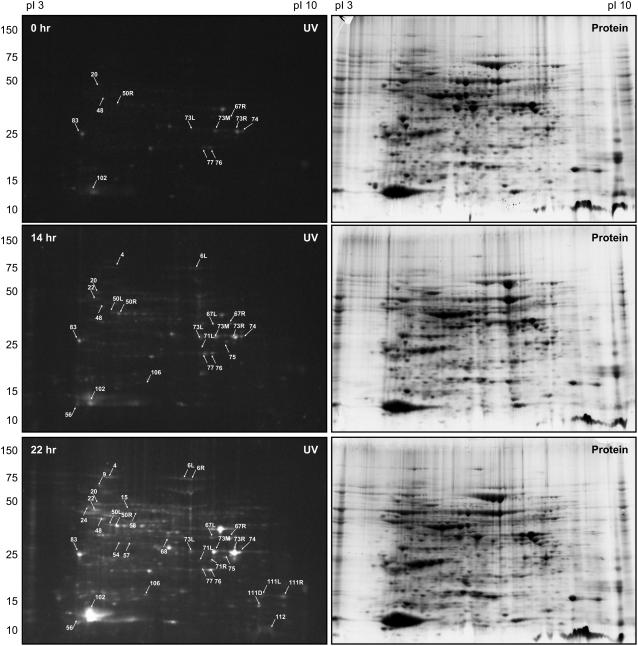
As most of the targets were found to be common to axes and cotyledons, we used whole seeds to examine the redox state of proteins during germination. The use of whole seeds also limited the necessary experimental manipulations, thereby minimizing artifactual redox changes. Initially, seeds imbibed for 0, 14, and 22 h were rapidly extracted in the presence of mBBr (Fig. 1), allowing the labeling of proteins that had been reduced in vivo prior to separation on 2D gels.
Two separate experiments leading to similar results were performed. While relatively few proteins were reduced in dry (0 h) seeds, their number increased progressively with the time of imbibition. An analysis with PD-Quest of the results obtained in the experiment presented Figure 3 led to the detection of 26, 44, and 118 fluorescent spots in the 0, 14, and 22 h gels, respectively. For many spots, an increase in fluorescence intensity with imbibition time was also visible. As discussed below, this fluorescence change (number and intensity of spots) was not due to an increase in protein amount as the pattern of all proteins stained with Coomassie Blue was quite similar at all stages examined. These results are consistent with previous findings with seeds of another dicot, Arabidopsis (Arabidopsis thaliana), that showed almost no change in the nature or abundance of proteins during germination (Gallardo et al., 2001; Rajjou et al., 2004).
Figure 3.
Analysis of the redox state of the proteins in whole seeds during germination. Proteins were extracted from dry (0 h), 14, or 22 h-imbibed seeds in the presence of mBBr and resolved in 2D gels. Fluorescent proteins were detected under UV light at 365 nm (left sections) and total proteins were stained with Coomassie Blue (right sections).
As can be seen by comparing Figures 2 and 3, many proteins found to be reduced in 22 h imbibed seeds were previously shown to be reduced by Trx in the mBBr-labeling experiments. Among them, 26 of 43 potential targets identified in the 0 m NaCl fraction were readily recognized as undergoing reduction during germination. These proteins are identified by arrows in Figure 3 and in bold print in Table II. It is clear, nevertheless, that additional Trx targets remain to be identified, perhaps with a higher resolution approach.
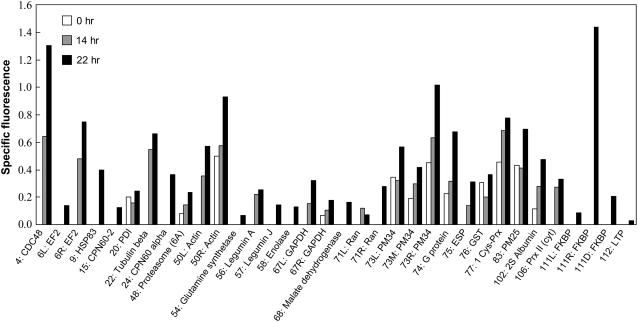
We examined in detail the redox state of the 26 Trx targets that were recognized by assessing the ratio of fluorescence intensity to protein content (i.e. the specific fluorescence) at the three times of imbibition (Fig. 4). It should be noted that some of these targets were present as two to three forms differing in pI or mass. Except in the case of a fragment of legumin A, that appeared at 14 h imbibition and increased in quantity between 14 and 22 h, the relative protein amount of the majority of these targets changed less than 2-fold during germination, indicating that protein synthesis contributed minimally to the appearance of SH groups during this period (Supplemental Fig. S3). In addition, Figure 4 shows that the specific fluorescence of these targets was greater at 22 h than at 0 h imbibition. This result confirms that these proteins underwent an increase in reduction in vivo during germination and indicates that they are true Trx targets. Of the 26 proteins shown in Figure 4, six were already known to be linked to Trx: 2S albumin (Shin et al., 1993), enolase as well as cytosolic glyceraldehyde 3-P dehydrogenase (GAPDH), malate dehydrogenase (Wong et al., 2003), Gln synthetase (Florencio et al., 1993), and peroxiredoxin (Prx) II (Choi et al., 1999), while 20 represent proteins that were not previously validated as authentic Trx targets (Table II, targets appearing in bold in the right column). Among them, 15 were, nevertheless, present in other plant screens whereas five are newly identified here: HSP83, a cytosolic peptidyl-prolyl cis-trans isomerase of FK506-binding protein (FKBP) type, legumin J, mitochondrial glutathione-S-transferase (GST), and postmaturation protein 25 (PM25) (Table II, targets appearing in bold in the left column).
Figure 4.
Specific fluorescence of Trx targets during germination. The ratio of fluorescence to normalized density/spot was calculated for the targets recognized in Figure 3 at three germination times (0, 14, and 22 h).
The Trx targets of M. truncatula seeds fall into two distinct categories. The first category includes the majority of proteins that appear to be largely oxidized in dry seeds and undergo reduction during imbibition. The second category is comprised of proteins that are partially reduced in dry seeds. For some, a transient decrease in the level of reduction is visible at 14 h imbibition. This is notably the case for protein disulfide isomerase (PDI) and GST. This change could be due to oxidative conditions that predominate early in germination (Bailly, 2004), resulting in the oxidation of certain SH groups. In the case of actin and GAPDH, both present in two different spots, one form belongs to the first category and the other form to the second category. Finally, 17 of the proteins identified as potential Trx targets in the 0 m NaCl fraction by the mBBr procedure and having conserved Cys were not detected in the gels possibly because they were oxidized at the three times analyzed or because reduction was transient. Alternatively, Trx may target them in a physiological context other than germination.
The data indicate that, in M. truncatula, many metabolic proteins present in the oxidized or partly reduced state in dry seeds became reduced or more reduced as germination progressed. Furthermore, the increase in both number and intensity of spots that was higher between 14 and 22 h than between 0 and 14 h, suggests that reduction is more active or efficient late in germination. It is perhaps not surprising that the unfolding of the proteins promoted by reducing disulfide bridges follows the alleviation of oxidative conditions that predominate early in germination (Bailly, 2004). Earlier reduction would possibly lead to irreversible oxidation of SH groups beyond the S-S state, i.e. to the sulfinic and sulfonic acid derivatives and potentially to denaturation and degradation of the protein.
Proteins Identified as Trx Targets Have a Spectrum of Functions
The Trx targets identified in this study function in major processes of the seed, including metabolism, cell structure, protein biogenesis and degradation, storage proteins, binding proteins, response to stress, signal transduction, as well as unknown processes (Table II). These processes are discussed individually below.
Metabolism
Carbon Metabolism
Several enzymes functional in carbon metabolism were identified as Trx targets in M. truncatula seeds, among them a cytosolic isoform of malic enzyme (Table II). A precursor of this enzyme was recently reported to be a target in isolated pea and potato (Solanum tuberosum) mitochondria (Balmer et al., 2004b). As in the starchy endosperm of wheat, we identified enolase, two cytosolic dehydrogenases, GAPDH (two isoforms) and malate dehydrogenase (two isoforms), phosphoglycerate mutase, triose phosphate isomerase and, as in peanut seeds (Yano et al., 2001b), PM34 (two isoforms). The PM34 gene and its ortholog are specifically expressed in seeds of M. truncatula (EST data) and barley (Alexander et al., 1994). Further, the recombinant barley protein was found to have Glc dehydrogenase activity and was considered to function specifically in seeds (Alexander et al., 1994). Partly reduced in dry seeds, PM34 (three spots) is progressively further reduced during germination (Fig. 4). In contrast, enolase and malate dehydrogenase were reduced only toward the end of germination while GAPDH existed under two different forms, one reduced and the other not, in dry seeds. GAPDH, enolase, and malate dehydrogenase as well as triose phosphate isomerase were previously identified as Trx targets in wheat seed by showing an enhancement of their in vitro activity upon Trx reduction (Wong et al., 2003). The effect of reduction on the activity of PM34 remains to be addressed.
Succinate dehydrogenase was recovered in M. truncatula as in prior Trx target screens of mitochondria (Balmer et al., 2004b). Interestingly, this enzyme, together with enolase, phosphoglycerate mutase, and triose phosphate isomerase, was found exclusively in axis preparations (Table I). The data suggest that glycolysis and the citric acid cycle may be more active in axes than in cotyledons during germination. In contrast, the large subunit of Rubisco (spots 44, Fig. 2), was observed only associated with cotyledons, perhaps in preparation for transition from storage organs to photosynthetic first leaves of the young seedling. The exclusive presence of Rubisco in cotyledons represents the most striking difference in the Trx target patterns between axes and cotyledons (Fig. 2). Other investigators have also identified either one or both Rubisco subunits as Trx targets, i.e. in spinach (Spinacia oleracea) chloroplasts (Motohashi et al., 2001; Balmer et al., 2003), a green alga Chlamydomonas reinhardtii (Lemaire et al., 2004), and cyanobacteria (Lindahl and Florencio, 2003). While Rubisco activase is a well known Trx-regulated enzyme that modulates the activity of Rubisco (Zhang and Portis, 1999), the effect of reduction on Rubisco itself remains an unanswered question.
Amino Acid Synthesis
Five potential or confirmed Trx targets were found to function in amino acid synthesis in M. truncatula seeds: isopropylmalate dehydratase (Leu), ketol acid reductoisomerase (Val and Ile), two isoforms of phosphoglycerate dehydrogenase (Ser), and one chloroplastic and two cytosolic isoforms of Gln synthetase (Table II). Isopropylmalate dehydratase represents a new Trx target. Ketol acid reductoisomerase was previously reported as a potential target in cereal seeds (Wong et al., 2004b), phosphoglycerate dehydrogenase in spinach chloroplasts (Balmer et al., 2003), and Gln synthetases in C. reinhardtii and Arabidopsis (Motohashi et al., 2001; Lemaire et al., 2004; Marchand et al., 2004; Yamazaki et al., 2004). The activity of both chloroplastic and cytosolic isoforms of Gln synthetase have long been known to be activated by Trx (Tischner and Schmidt, 1982; Florencio et al., 1993). The cytosolic isoform was found to undergo reduction in germinating M. truncatula seeds in this study. Phosphoglycerate dehydrogenase and ketol acid reductoisomerase were recovered only in axes, while isopropylmalate dehydratase and chloroplast Gln synthetase were only in cotyledons. These differences may, once again reflect the relative abundance of the enzymes and the potential of the cotyledons to undergo early greening with the accompanying onset of photorespiration (accumulation of Gln synthetase).
ATP-Linked Energy
In our screens, we identified two isoforms of the β-subunit of ATP synthase, the enzyme functional in the synthesis of ATP in mitochondria, as well as the α-subunit of vacuolar ATPase, the enzyme that converts ATP to an electrochemical proton gradient across the tonoplast membrane. Both proteins were previously reported to be Trx targets: mitochondrial ATP synthase for cereals and vacuolar ATPase for C. reinhardtii (Lemaire et al., 2004; Wong et al., 2004b). As ATP synthase of chloroplasts is also regulated by Trx (Buchanan, 1980; He et al., 2000), it appears that both the synthesis and hydrolysis of ATP are tightly regulated by reduction throughout the cell.
Vitamin Synthesis
Two new potential Trx targets were identified in the synthesis of folic acid (5,10-methylene tetrahydrafolate dehydrogenase) and thiamine biosynthesis [4-methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis protein]. This is the first evidence for a link between Trx and folic acid synthesis. Previously, other enzymes of thiamine synthesis were reported as Trx targets in chloroplasts (Balmer et al., 2003) and C. reinhardtii cells (Lemaire et al., 2004).
Cell Wall Synthesis
Four enzymes associated with cell wall synthesis were identified as Trx targets in M. truncatula seeds. UDP-Glc pyrophosphorylase and UDP-Glc dehydrogenase produce nucleotide sugars for polysaccharide synthesis, while cinnamoyl-CoA reductase and 4-coumarate-CoA ligase function in the synthesis of phenylpropanoids, intermediates that yield a variety of products, including flavonoids and lignin. Among these, only UDP-Glc pyrophosphorylase was previously reported, with wheat seeds (Wong et al., 2004b). The other three are new, making this, to our knowledge, the first report of a link between Trx and the phenylpropanoid pathway. Interestingly, the enzymes of this pathway were recovered only in axis preparations.
DNA Synthesis
A DNA polymerase auxiliary subunit was identified as a new potential target in our screens, evidence of a link between DNA polymerization and Trx.
Cell Structure
Actin (four isoforms) and tubulin (α- and β-subunits), both previously linked to Trx in cereals (Wong et al., 2004b), were identified in these screens. In addition, actin and β-tubulin were found here to be reduced during germination. Actin has been reported to be regulated by glutathionylation, hampering its polymerization (Wang et al., 2001). The removal of protein-bound glutathione is likely catalyzed in vivo by Grx as this redox protein appears to be more effective than Trx in this capacity (Ghezzi, 2005). The evidence suggests that deglutathionylation during germination favors the polymerization of actin, leading to cell enlargement.
Protein Biogenesis and Degradation
Translation
Several elongation factors (EFs) were identified as potential Trx targets in M. truncatula seeds, including EF1 (α-, β-, and δ-subunits) and three isoforms of EF2 (Table II). Both proteins were also reported to be potential Trx targets in wheat, although only the β-subunit of EF1 was detected in that study (Wong et al., 2004b). Chloroplast EF-tu was also recovered in the M. truncatula screens, as was earlier the case with spinach chloroplasts and C. reinhardtii cells (Balmer et al., 2003; Lemaire et al., 2004). EIF5 A-2 (two isoforms) as well as a protein belonging to the 40S ribosomal particle (S21), previously unreported, represent two new potential Trx targets. EF2 (two spots) was reduced during germination (Fig. 4). The results suggest that the level of protein synthesis needed to sustain postgerminative growth (Bewley, 1997) requires the reduction of EFs. It is noted that while protein synthesis in mammalian systems has been found to be inhibited at the elongation step in response to oxidative stress (Ayala et al., 1996), a corresponding effect in plants is yet to be demonstrated.
Folding
Protein folding was found to be a rich source of Trx-linked proteins. Sixteen potential targets functional in this process were identified in this study, some being also reported in earlier studies on cereals or other types of plants (Table II). In addition to PDI, a well known foldase with two Trx domains, we found Trx to target several chaperones: HSP70 from cytosol (four isoforms), endoplasmic reticulum (two isoforms), mitochondria (two isoforms), and chloroplasts (one isoform); chaperonin 60 (CPN60) from mitochondria (CPN60-2; two isoforms) and chloroplasts (CPN60α and CPN60β); as well as a stress-induced protein (Sti1), a protein interacting with HSP70 and 90 (Johnson et al., 1998). At least one isoform of each of these proteins was previously found to be linked to Trx in other systems (Buchanan and Balmer, 2005). An interaction of yeast (Saccharomyces cerevisiae) HSP70 with a Trx-like protein from Arabidopsis was demonstrated by complementation (Vignols et al., 2003). The interaction is disrupted under oxidative stress by a redox-dependent regulation event involving Trx.
In this study, PDI was found to be reduced in both dry and germinating seeds, whereas CPN60-2 and CPN60α were initially oxidized and underwent reduction late in germination. Finally, new potential targets were also identified in this category: HSP90 and related proteins (HSP83 and putative HSP90), a PDI-like protein, two T-complex protein (TCP) CPN60 chaperonins (ɛ and τ), as well as the FKBP previously mentioned. The HSP83 and FKBP (three spots) were found to be reduced late in germination.
Two different types of peptidyl-prolyl cis-trans isomerases have been identified in plants using Trx screens. A cyclophilin (inhibited by cyclosporin A) was initially found in the screen of spinach chloroplasts (Motohashi et al., 2001). Biochemical evidence later confirmed that the enzyme is redox regulated by Trx (Motohashi et al., 2003). A cyclophilin was also found in screens of cereal seeds and Arabidopsis leaves (Wong et al., 2003; Maeda et al., 2004; Marchand et al., 2004; Wong et al., 2004b). In Arabidopsis, a chloroplast isomerase of FKBP type was recovered (Marchand et al., 2004). In contrast, the FKBP we identified is probably cytosolic. This isoform harbors four conserved Cys residues and is reduced during germination. The results support the view that, in addition to FKBP, cyclophilins of both chloroplasts and the cytosol are regulated by Trx in plants.
Targeting
We identified two Trx-linked proteins functional in targeting to the nucleus in M. truncatula: a Ras-related nuclear protein, Ran, and a Ran-binding protein (Table II). First proposed as a Trx target in C. reinhardtii (Lemaire et al., 2004), Ran is a small GTP-binding protein involved not only in nucleocytoplasmic transport, but also in nuclear envelope assembly and mitotic spindle formation (Quimby and Dasso, 2003). Interestingly, in yeast and mammals, the transport is dedicated mainly to Trx-linked transcription factors that are active in the oxidative stress response (Masutani and Yodoi, 2002). The Ran-binding protein we identified was possibly captured by its association with Ran. Another possibility is that the Ran-binding protein itself is a Trx target as it contains four conserved Cys. Significantly, Ran (two spots) was also found to be reduced during germination (Fig. 4). Overall, the data suggest a strong link between Trx and nuclear targeting.
Degradation
In addition to synthesis, protein degradation was targeted by Trx in this study. This is not surprising since Trx has long been known to enhance proteolysis in cereals by its ability to activate proteases reductively and increase the solubility and proteolytyic susceptibility of storage proteins on the one hand, and inactivate protease inhibitors on the other (Buchanan and Balmer, 2005). In this M. truncatula study, we identified the subunit of urease that binds nickel (Freyermuth et al., 2000) as a potential new Trx target. Based on an inhibitor study with Arabidopsis seeds, urease has been reported to function in germination, possibly in coordination with arginase in the utilization of nitrogen reserves (Zonia et al., 1995). We also identified Leu aminopeptidase as well as the regulatory subunit 6A of the 26S proteasome and two isoforms of cell division cycle 48 (CDC48) as potential Trx targets, each previously reported in wheat seeds (Wong et al., 2003, 2004b). Highly conserved among organisms, CDC48 has been proposed to function in the cell cycle, membrane fusion, and proteasome proteolysis, including endoplasmic reticulum associated degradation. A human ortholog, valosin-containing protein, an ATPase reportedly inhibited under oxidative stress conditions, is induced by degenerative disorders. Inhibition is due to the oxidation of a Cys residue that is conserved only in the enzyme from multicellular organisms (Noguchi et al., 2005). It seems possible, therefore, that the CDC48 is also redox regulated in plants. It is noteworthy that subunit 6A of the 26S proteasome and CDC48 of M. truncatula were reduced during germination (Fig. 4).
Storage Proteins
All major types of seed storage proteins seem to be linked to Trx. Trx was shown initially to reduce the major storage proteins of wheat starchy endosperm in vitro, the soluble albumins and globulins and insoluble gliadins and glutenins (Kobrehel et al., 1992). These proteins were also found to undergo reduction during germination. Similar results were obtained with barley (Marx et al., 2003) and other types of seeds, notably an albumin of castor and an albumin, a conglutin, and a glycinin of peanut (Shin et al., 1993; Yano et al., 2001a). It is thus not surprising that water-soluble storage proteins were found to be Trx targets in M. truncatula: 2S albumin, conglutin, and several legumins (Table II); albumin and legumins A and J being reduced during germination (Fig. 4).
However, surprisingly, legumins were identified as Trx targets in the 0 and 0.2 m NaCl fractions but not in 1 m counterpart that contained the bulk of these proteins. This finding suggests that legumins undergo a change (e.g. partial proteolysis) that renders them amenable to digestion following reduction by Trx, thereby increasing their solubility and resulting in their recovery in the 0 and 0.2 m NaCl fractions as already proposed (Wong et al., 2004b). The reduction probably facilitates their subsequent digestion. This, therefore, contrasts with previous findings made with the major storage proteins gliadins and glutenins found in cereals that were oxidized in the dry grain and insoluble in aqueous solution, but showed increased solubility and protease susceptibility following reduction by Trx (Jiao et al., 1992, 1993; Kobrehel et al., 1992; Marx et al., 2003; Wong et al., 2004a).
It is noted that fragments of legumins were present in axis of 14 h imbibed seeds but not in cotyledons, suggesting that legumin degradation occurs later in this tissue, following radicle protrusion. The presence of these fragments together with other small proteins in axis (but not in cotyledons) after 14 h imbibition accounts for another striking difference in the patterns shown in Figure 2.
Concerning the 2S proteins found in dicots, they are related to the α-amylase and trypsin inhibitors that are widely distributed in cereals, and earlier shown to be deactivated and rendered protease sensitive following reduction by Trx (Jiao et al., 1992, 1993).
Binding Proteins
Six binding proteins were identified as previously undescribed Trx targets: calreticulin, a selenium-binding protein, ferritin, SOUL-heme-binding protein, P54, and a seed biotinylated protein, SBP65 (Table II). Three out of the six proteins bind metals: calreticulin, the selenium-binding protein, and ferritin. A calcium-binding protein, calreticulin, may act in calcium signaling and protein folding (Persson et al., 2001), and the selenium-binding protein has been reported to act in selenium tolerance or symbiosis (Flemetakis et al., 2002). A link between selenium detoxification and Trx has previously been highlighted in barley seeds overexpressing a Trx h isoform (Kim et al., 2003).
In addition to ferritin that binds iron and is associated with seeds, we identified SOUL, a heme-binding protein reported in mammalian cells where it is thought to function in heme transfer or in heme binding to prevent damage by ROS (Taketani et al., 1998). In M. truncatula, the protein was found only in cotyledons, again perhaps in preparation for the developmental shift to photosynthesis. Two other binding proteins were identified: P54 and a vicilin-related seed protein that possibly functions in Suc binding (Overvoorde et al., 1997). In pea, P54 undergoes further maturation into P16, a possible participant in chromatin condensation induced by dehydration (Castillo et al., 2000), whereas in M. truncatula, the protein seems not to be further processed and presumably only acts in Suc binding. Finally, we identified SBP65, a LEA protein that can bind biotin (Duval et al., 1994).
Response to Stress
Trx appears to be linked to an impressive number of proteins active in different stress responses in M. truncatula seeds. Those proteins are discussed below under the appropriate type of stress.
Oxidative Stress
In addition to photosynthesis, abiotic and biotic stresses are major contributors to ROS production in plants, resulting in oxidative damage to proteins, lipids, and DNA. Certain physiological processes can also generate ROS. Indeed, loss of water during maturation of seed, and, as already mentioned, uptake of water during germination are associated with oxidative conditions that produce ROS (Bailly, 2004). Trx is well known to play both protective and signaling roles in reactions elicited by ROS in yeast, mammals, and plants (Fernando et al., 1992; Chae et al., 1994; Muller, 1995; Hirota et al., 1997; Lee et al., 1999; Dietz et al., 2002).
In addition to catalase, we identified the expected four major types of redox proteins functional in hydrogen peroxide detoxification as Trx targets: 1-Cys Prx, a well known protein accumulating specifically in seeds; chloroplast 2-Cys-Prx (two isoforms); type II Prxs (isoforms from cytosol, mitochondria, and chloroplasts); and GSH peroxidase. As Trx can serve as an electron (hydrogen) donor for Prxs, its role as a substrate could account for recovery of certain of these proteins as targets. However, the recovery of 1-Cys-Prx and cytosolic Prx II may be due to the previously mentioned lack of specificity of Trx isoforms in vitro as the preferred substrate is either unknown (1-Cys Prx) or Grx (cytosolic Prx II) as shown by Brehelin et al. (2004). These two Prxs are, nonetheless, reduced during germination (Fig. 4). Another possibility is that Trx may exert a regulatory redox effect on their activity or assembly. A nonspecific interaction may also take place between GSH peroxidase and Trx. Alternatively this enzyme can use Trx as substrate (Herbette et al., 2002). With respect to catalase, the link to Trx would appear to be regulatory based on experiments with the C. reinhardtii protein that was shown to be deactivated on reduction (Lemaire et al., 2004).
GSTs are a large enzyme family that catalyze the transfer of glutathionyl groups to a wide variety of substrates, including other enzymes. Glutathionylation plays a major role in cell xenobiotic detoxification as well as in the protection of SH groups of proteins that otherwise would be irreversibly denatured by oxidation. The mitochondrial isoform of GST identified here as a new Trx target was found to be reduced in dry and germinating seeds as mentioned above (Fig. 4). This isoform of GST, possibly accumulated in seeds, may participate in protection of proteins against oxidative conditions that predominate early after imbibition. A cytosolic isoform of GST was earlier proposed as a Trx target in leaves of Arabidopsis (Marchand et al., 2004). Finally, we found in axis preparations an aldo/keto reductase that is induced by auxin in cereal endosperm. This enzyme belongs to a class that functions in detoxification of toxic carbonyls, such as aldehydes, and products resulting from lipid peroxidation.
Desiccation and Osmotic Stress
Several proteins implicated in response to desiccation were among the new potential Trx targets identified in M. truncatula seeds. All are LEA proteins whose messengers are highly or specifically expressed in seeds according to an in silico analysis of the gene expression in MtGI. For others described for soybean (Glycine max), e.g. PM proteins, synthesis is correlated with seed maturation. In addition to the previously mentioned SBP65, we found dehydrin, a LEA protein of group 5, PM24, PM25, and PM51. It is noted that three of these proteins have no conserved Cys: SBP65, dehydrin, and the LEA protein of group 5 (Table I). Because they were only recovered in the affinity procedure, it seems likely that they were retained due to an association with actual Trx targets. Another possibility is that they interact electrostatically with Trx. Such a structural interaction is known to occur, for example, between Trx and a number of proteins such as T7 DNA polymerase (Mark and Richardson, 1976), although in many cases, the proteins bound can be released with a buffer salt wash prior to elution of the targets with dithiothreitol (DTT; Balmer et al., 2004a), a step carried out in these experiments.
The function of the proteins identified in this aspect of our study, for the most part, is still under investigation. It has been proposed that the LEA proteins function in gaining tolerance to oxidative conditions resulting from water loss. Dehydrin was shown to be expressed in leaves subjected to drought or cold stress. PM25, which was found to be reduced in dry seeds and undergo further reduction during germination (Fig. 4), was recently linked to desiccation tolerance in seedlings of M. truncatula (Boudet et al., 2006). As PM25 resides in the nucleus, it has been proposed to protect DNA from oxidative damage. Finally, sorbitol-6-P dehydrogenase, an enzyme found here as a new potential Trx target, participates in osmotolerance (Gao et al., 2001; Tang et al., 2005).
Hypoxia
Four enzymes implicated in hypoxia were recovered in M. truncatula screens: aldehyde dehydrogenase, alcohol dehyrogenase (two isoforms), pyruvate decarboxylase (two isoforms), and succinic semialdehyde dehydrogenase (Table II). Two of these, aldehyde and alcohol dehydrogenases, were previously reported as potential targets in pea mitochondria (Balmer et al., 2004b) and cereal seeds (Wong et al., 2004b); the remaining two enzymes, pyruvate decarboxylase and succinic semialdehyde dehydrogenase, were identified as two new potential targets in this work. The effect of reduction on their activity remains to be investigated.
Aluminum Stress
Two proteins whose expression is induced by aluminum, Sali 3.2 and Wali 7 (two isoforms), were identified as new potential targets in M. truncatula seeds. While their specific function is not known, it has been shown that the expression of one of them, Sali 3.2, is associated with seeds. This constitutes the first evidence of a link between Trx and aluminum stress.
Biotic Stress
Flavonoids perform diverse functions in plants by acting as attractant, repulsion, or toxic molecules. In legumes, two members of this family, apigenin and luteolin, are secreted as signal molecules by germinating seeds to facilitate interactions with Rhizobium bacteria. Another member, medicarpin, participates in inducible plant defense (Hirsch et al., 2001). Three enzymes of the flavonoid pathway were identified as new potential Trx targets in axis preparations from M. truncatula, the two mentioned above (in cell wall synthesis) as well as chalcone reductase, the enzyme catalyzing the first committed step of flavonoid synthesis. This suggests a link between Trx and symbiosis or pathogen interaction.
Lipoxygenase and leukotriene-A4 hydrolase like, two enzymes functional in the synthesis of jasmonate, a hormone associated with host-pathogen interactions, were also detected as new Trx targets in this study. Another enzyme of the jasmonate pathway, allene oxide cyclase, was previously reported as a potential target in mitochondria (Balmer et al., 2004b). Interestingly, the isoform of lipoxygenase found here is seed specific. Thus, the increase in lipoxygenase activity found for seeds (Wang et al., 1999) may be due not only to the expression of a specific isoform, but also to its activation by reduction. As in earlier wheat and barley screens (Maeda et al., 2004; Wong et al., 2004b), nonspecific lipid transfer protein (LTP) was found to be Trx linked in M. truncatula seeds. Further, the protein potentially accumulated in seeds was reduced during germination (Fig. 4).
Signal Transduction
Several proteins associated with signaling were identified as potential Trx targets in this study, most being new: Arg-methyltransferase (two isoforms), ADP-ribosyltransferase (also described as PM38), GTP-binding protein (two isoforms), two types of RNA-binding protein (either Gly rich [three isoforms] or not [two isoforms]), G-protein (β-subunit), a 14-3-3 protein, and a regulatory subunit of the Suc nonfermenting-related kinase complex (SNF4b). The gene expression of two of them, ADP-ribosyltransferase and SNF4b, is associated with seeds, while Arg-methyltransferase, ADP-ribosyltransferase, and GTP-binding protein as well as certain isoforms of Gly-rich RNA-binding protein accumulated specifically in axes. The reduction of the axis proteins may represent a step in signal transduction that leads to radicle protrusion or to preparation for biotic interactions. The G-protein β-subunit and Gly-rich RNA-binding protein were previously detected in Trx screens from wheat seeds (Wong et al., 2004b). In this study, the G-protein β-subunit was found to be partly reduced in dry seeds and further reduced during germination (Fig. 4).
The function of several of these signaling proteins has been well described. G-protein is involved in signal transduction across the plasma membrane, ADP-ribosyltransferase is implicated in the activation of DNA transcription upon histone ADP-ribosylation, whereas Arg-methyltransferase appears to act via inhibition of histone methylation. The function of highly conserved GTP-binding proteins may be to transduce signals by their ability to bind GTP. RNA-binding proteins both stabilize and prevent the translation of mRNA. Interestingly, in our study, fragments of a Gly-rich RNA-binding protein specific to the axis were isolated by affinity chromatography, indicating degradation during germination. As most of the RNAs present in the dry seed are synthesized during development, one can envisage that this RNA-binding protein is involved in the protection of mRNAs so they cannot be translated. Isoforms of 14-3-3 protein, known to regulate targets by protein-protein interaction, were recently shown to interact with Trx and Prx (Meek et al., 2004) or Grx (Rouhier et al., 2005).
Another signaling protein, SNF4b, is a regulatory subunit of SnRK1, a protein kinase complex that is the ortholog of yeast SNF1. In yeast, the kinase plays an essential role in metabolic adaptation to different carbon sources and to environmental stress (Honigberg and Lee, 1998). It is also involved in developmental processes such as sporulation (Honigberg and Lee, 1998), life span, and ageing (Ashrafi et al., 2000). The plant counterpart regulatory subunit, SNF4b, has been shown to participate in seed and desiccation tolerance (Buitink et al., 2004). The finding of SNF4b among our potential targets suggests that these processes are linked to Trx either through assembly of the sugar signaling complex or regulation of its activity. It is possible that Trx may influence metabolism according to the carbon sources available in seeds via SNF4b. To our knowledge, this is the third instance that a plant protein kinase has been identified as a Trx target, others being associated with phosphoenolpyruvate carboxylase of C4 plants (Saze et al., 2001) and self-incompatibility (Bower et al., 1996). A junction with a chloroplast protein phosphatase functional in starch metabolism has also recently been uncovered (Sokolov et al., 2006).
Unknown Processes
We identified a number of potential Trx targets of unknown function, of which these were new: aldose-1-epimerase, that was previously described as a Trx target in C. reinhardtii (Lemaire et al., 2004), an ortholog to soybean PM28, Gly-rich proteins, ripening-related protein, and an ortholog to the embryo-specific protein (ESP) from barley (Maeda et al., 2003), the latter being reduced in germinating M. truncatula seeds (Fig. 4). While the function of ESPs is not defined, they appear during seed maturation and are hydrolyzed during germination, likely as a result of Trx reduction (Maeda et al., 2004; Yano and Kuroda, 2006). Finally, five unknown proteins (UK1–UK5) were also identified as potential new targets.
Interpretation of Results
Prior to this study little was known of the role of Trx in dicot seeds with respect either to proteins targeted or to a possible role in germination. Two complementary proteomic approaches enabled us to identify 111 potential targets in seeds of the model legume, M. truncatula, and uncover six pathways or functions that were not previously known to be linked to Trx: folic acid synthesis, phenylpropanoid pathway, DNA polymerization, ion or metabolite binding, aluminum, and desiccation tolerance.
We further demonstrated that a number of metabolic proteins (extracted in 0 m NaCl buffer) that are oxidized or partly reduced in dry seeds became reduced or more reduced as germination progressed, Trx appearing to play a central role in this transition. This finding first underlines that reduction promoted by Trx during germination may be a general process that concerns not only storage proteins as earlier found in cereals but also metabolic proteins. It also further allowed us to validate 26 of the potential targets as authentic Trx targets.
In M. truncatula, many metabolic proteins stored in dry seed are oxidized, i.e. present in the S-S inactive and, typically, less soluble state. Yet, the disulfide state can be advantageous to seeds in that, while conferring stability, proteins can be readily retrieved and, in most cases, regain metabolic function, i.e. following reduction back to the SH state. Thus, redox regulation of metabolic proteins could ensure quiescence in the oxidized state occurring in the dry seed and the resumption of metabolic activity upon hydration and reduction during germination. As a component of the NTS, Trx makes possible a simple, efficient redox mechanism to control metabolic activity.
The disulfide state may also protect the metabolic proteins from an oxidation of not only Cys, but also other amino acid residues sensitive to more extreme oxidative modification, e.g. carbonylation. Apropos this point, it is noted that many orthologs of the Trx targets currently identified in M. truncatula can be altered by carbonylation in germinating Arabidopsis seeds and further degraded (Job et al., 2005).
The Trx targets identified in this study have a spectrum of functions, reflecting the myriad events accompanying the germination of legume seeds. Certain changes are common to cereals, e.g. the mobilization of major storage proteins (albumin, conglutin, several globulins, Leu aminopeptidase, CDC48, and proteasome). Others so far seem unique to legumes, e.g. the participation of the Trx target urease accessory subunit in nitrogen mobilization. In concert with nitrogen mobilization, we observed that Trx target enzymes functional in glycolysis and the citric acid cycle are more abundant in axes than cotyledons and are reduced during germination. This finding is consistent with the conclusion that active catabolism provides energy for cell elongation in axes, leading to radicle protrusion and subsequent growth.
Proteins participating in cell elongation that were previously reported to be Trx targets in cereals, i.e. actin and tubulin, were also identified in our screens with M. truncatula seeds. Enzymes of associated processes, DNA polymerase auxiliary protein, UDP-Glc dehydrogenase, cinnamoyl-CoA reductase, and 4-coumarate-CoA ligase, appear to be new potential targets. As in cereals, the resumption of protein synthesis and folding needed to sustain postgerminative growth may be triggered by the reduction of EFs and chaperonins. We found significantly more proteins participating in these processes (EF1, EF2, EF-tu, EIF5-A-2, 40S ribosomal protein S21, FKBP, Sti1, HSP70, HSP90, putative HSP90, HSP83, PDI, PDI like, CPN60, TCP CPN60) than reported for cereals, indicating that protein synthesis and assembly are active in legumes and are under Trx regulation in the major cell compartments.
In another capacity, Trx appears to prepare cells for survival against stress conditions that predominate during and after the initiation of germination. Many proteins reported to link Trx to oxidative and hypoxic conditions in cereals were identified in M. truncatula in this study. Additionally, we found previously unreported potential targets active in response to desiccation, aluminum stress, and biotic stress. The abundance of proteins of this type may be linked to the fact that legumes establish symbioses with bacteria while cereals do not. Thus, aside from LTP that is common to cereals, we identified lipoxygenase, leukotriene-A4 hydrolase, and chalcone reductase as new targets in M. truncatula. The latter enzyme links Trx to the phenylpropanoid pathway, an association not previously recognized. This pathway functions in the synthesis of flavonoids that are necessary not only for normal root development, but are also involved in symbiosis signaling. The apparent restriction of Trx targets of this pathway to axes supports the idea that Trx has a role in root development or establishing symbiosis.
We also identified several previously unreported binding proteins as Trx targets in M. truncatula seeds (P54, SBP65, calreticulin, selenium-binding protein, ferritin, and SOUL-heme-binding protein). They bind ions or metabolites probably necessary for seed development, germination, postgerminative growth, and/or symbiosis. Among them, ferritin and SOUL-heme-binding protein are both seemingly essential for the limitation of ROS damage. It remains to be seen whether reduction of these proteins during germination allows their degradation and the release of their ligands. Similarly, the incidence of the reduction on the stability of the LEA proteins uncovered here has also to be investigated. These proteins, involved in desiccation tolerance, are accumulated during seed maturation and disappear after germination.
Finally, a number of proteins functioning in signal transduction were highlighted in this investigation, e.g. Arg-methyltransferase, ADP-ribosyltransferase, GTP-binding protein, and two kinds of RNA-binding proteins, gene expression of two of them being associated with seeds. Several of these proteins were accumulated only in embryo axes, suggesting a function in the signaling pathway that leads to radicle protrusion, growth, or preparation for biotic interactions. However, as in previous attempts (Buchanan and Balmer, 2005), we failed to identify Trx-linked transcription factors, proteins well known in yeast and animals (Hirota et al., 1997; Paget et al., 1998; Izawa et al., 1999). We did, however, identify Trx targets closely connected to transcription, including Ran, a protein that, in other organisms, participates in the transfer of transcription factors to the nucleus (Masutani and Yodoi, 2002). We also identified a subunit of a protein kinase (SNF4b), the first detection of a protein kinase in a Trx target screen, to our knowledge. It seems possible that the reduction of SNF4b by Trx enables the seed to adjust metabolism in response to available carbon source. The interaction between Trx and this kinase highlights the interconnection that exists between redox regulation and protein phosphorylation in germination, a key step in plant biology.
CONCLUDING REMARKS
This evidence extends our understanding of the role of redox in two ways. First, seed proteins of the model legume, M. truncatula, were shown to undergo changes during germination akin to those of cereals, thereby extending the role of redox to the germination of dicotyledons, a major plant group. Second, Trx was linked to an extensive group of proteins functional in processes not previously known to involve redox. These findings underscore the significance of both redox and Trx to the biology of seeds throughout the plant kingdom.
The work also raises questions for the future. For example, it will be interesting to analyze the germination capacity of mutants deficient in the newly identified signaling proteins such as SNF4. Further, the question of how Trx interacts with these targets to influence associated cell processes becomes an increasingly important question. As discussed previously, this interaction can assume several forms, ranging from classical regulation to protein assembly (Buchanan and Balmer, 2005). Whatever their nature, it seems almost certain that elucidation of these interactions will add fundamentally to our knowledge of plants and potentially to technology and medicine as well.
MATERIALS AND METHODS
Materials
Seeds of Medicago truncatula ‘Jemalong A17’ were harvested from plants grown in controlled conditions in a growth chamber at 19°C with 16 h of light and 8 h of dark (lots 2003 and 2004; Institut National de la Recherche Agronomique Dijon, France). Seeds were imbibed without prior scarification or sterilization and germinated in the dark on filter paper moistened with 3.5 mL water in petri dishes for 0, 14, or 22 h at 20°C. Embryo axes and cotyledons were dissected from the seeds and either used immediately or frozen in liquid nitrogen and stored at −80°C. Tissues were ground with a pestle in a mortar containing extraction buffer (50 mm Tris-HCl, pH 7.5; 1 mm EDTA; 1 mm phenylmethylsulfonyl fluoride; 1 g/10 mL). Proteins were extracted sequentially by increasing NaCl concentration in the extraction buffer from 0 (initial fraction) to 0.2 and finally to 1.0 m to obtain fractions enriched in albumins, vicilins, and legumins, respectively. After centrifugation (30,000g, 1 h, 4°C), protein was determined (Bradford, 1976) with bovine serum albumin as a standard. Biochemicals were obtained from Sigma Chemical.
Methods
Potential Trx Target Labeling or Isolation
Trx target proteins were either labeled with the mBBr procedure following reduction with the NTS (Yano et al., 2001b) or isolated on the Trx mutant affinity column (Motohashi et al., 2001), then separated by 1D electrophoresis (preliminary experiments) or 2D electrophoresis for an identification by LC/MS/MS. To minimize differences in levels of protein reduction observed at different times of imbibition, fresh extracts were incubated in air overnight at 4°C prior to target labeling or isolation.
mBBr Fluorescent Gel Procedure
Proteins extracted from axes of 100 to 500 seeds either dry or imbibed for 0, 14, or 22 h were labeled with mBBr before or after reduction by the reconstituted NTS. For reduction by the NTS, proteins were incubated for 4 h at 37°C in the presence of a reconstituted legume NTS consisting of 0.2 mm NADPH, 20 μg/mL of recombinant NTR from M. truncatula (MtNTRA; Alkhalfioui et al., 2007), and 10 μg/mL Trx h3 from pea (Pisum sativum; Montrichard et al., 2003) in 50 mm Tris-HCl, pH 7.5 (final volume, 1 mL). mBBr in acetonitrile was then added to a final concentration of 2 mm and incubation continued for an additional 15 min at 25°C. In some experiments, the NTS was replaced by 5 mm GSH plus 5 mm Grx or GSH alone (Wong et al., 2004b). Control experiments without NTS, Grx, and GSH were prepared in parallel. After incubation, proteins were separated in 2D gels (one gel per time point). The experiment was repeated for the three time points. According to PD-Quest analysis, fluorescent spot patterns were similar at the three times of imbibition. The potential Trx targets corresponding to fluorescent spots were in identical positions in five or six of the six gels. The same approach was applied to cotyledons. The protein spot patterns were similar for the three times of imbibition. The mBBr-labeled spots of cotyledons were found to match the corresponding set observed in axis gels except for Rubisco, which was present only in the cotyledon gels. Most of the Trx targets (41 out of 45) were identified in two different tissues (in axes and cotyledons), at two different times of imbibition (0 and 14 h), or in two different fractions (0 and 0.2 m NaCl), all in duplicate experiments.
Mutant Affinity Column Procedure
Potential Trx targets in an oxidized state were trapped by affinity column chromatography using mutated Trx h3 C42S from pea. This method relies on the molecular mechanism of disulfide reductase activity of Trx that proceeds through a two-step reaction (Kallis and Holmgren, 1980). In the first step, the more terminal Cys (the catalytic residue) attacks the disulfide bridge of the target protein, reducing one Cys and establishing a disulfide bridge with the second. In the next step, the more C-terminal Cys of Trx attacks the mixed disulfide intermediate, allowing the release of both the reduced target and oxidized Trx. The replacement of the more terminal Cys by an analog such as Ser or Ala allows stabilization of the mixed disulfide intermediate (Qin et al., 1995; Wynn et al., 1995). The potential target protein is then released with DTT.
The mutant Trx was generated by changing the coding sequence of PsTrx h3 inserted into the expression plasmid pASK-IBA3 (Montrichard et al., 2003) using the QuickChange site-directed mutagenesis kit (Stratagene) and the following primers (forward primer: ttcttggtgcggtccaagccgttttattgc, and reverse primer: gcaataaaacggcttggaccgcaccaagaa) according to the manufacturer's instructions. The recombinant mutated Trx was overproduced and then bound to CNBr activated Sepharose (Amersham) using 5 mg protein/g matrix. Two grams of the Trx-Sepharose was prepared and equilibrated with extraction buffer (50 mm Tris-HCl, pH 7.5) containing 5 mm DTT and then washed with extraction buffer just prior to use.
Five milligrams of oxidized protein, extracted from axes or cotyledons of 500 to 1,000 seeds imbibed for 14 h, were added to the washed Trx-Sepharose and the mixture was incubated for 4 h at room temperature. Proteins noncovalently bound to the Trx-Sepharose were removed by extensive washing with extraction buffer supplemented with 200 mm NaCl. Subsequently, potential Trx targets were eluted from the Trx-Sepharose with extraction buffer containing 200 mm NaCl and 10 mm DTT. The experiment was repeated for each part of the seed. The 2D patterns of the duplicates were almost identical and differed only by the quantity of protein eluted from the column. Protein spots were analyzed from two gels developed for axes and one gel developed for cotyledons. Thus, the majority of the proteins were identified twice.
Gel Electrophoresis
Proteins labeled with mBBr or isolated by affinity chromatography were precipitated by the addition of 5 volumes cold acetone and incubation for 18 h at −20°C. Proteins were pelleted by centrifugation (14,000g, 10 min, 4°C), washed twice with acetone, and resolved in either 1D (preliminary experiments) or 2D gels. Separation in 1D gels was performed by SDS-PAGE, according to Laemmli (1970). For 2D separation, proteins were solubilized in rehydration buffer [6 m urea, 2 m thiourea, 4% CHAPS (3-[3-(cholamidopropyl) dimethylammonio]-1-propane-sulfonate), 20 mm DTT, and 0.2% ampholytes]. For 2D electrophoresis, 400 μg protein in 200 μL rehydration buffer was loaded onto an 11 cm, pI 3-10NL immobilized pH gradient strip (Bio-Rad). Isoelectric focusing was performed using the Protean isoelectric focusing cell (Bio-Rad) with the following parameters: 0 to 250 V linear gradient for 15 min; 250 to 8,000 V linear gradient for 3 h, and 8,000 V until 38,000 V h were reached. Next, the strips were equilibrated for 20 min at room temperature in 2 mL equilibration buffer (375 mm Tris-HCl [pH 8.8], 8 m urea, 20% glycerol, 130 mm DTT, and 2% SDS) followed by 30 min in the same buffer in which DTT was replaced with 135 mm iodoacetamide. The strips were placed on the top of Criterion Tris-HCl Precast gels (10%–20%; Bio-Rad) for electrophoresis at 150 V for 80 min. Fluorescent proteins were observed under UV light (365 nm) and digitalized images of the gels were captured with identical integration times using a Gel Doc 1000 with Quantity One program (Bio-Rad). Protein was stained with colloidal Coomassie Blue G-250 overnight at 25°C. The gels were scanned using identical sensitivity parameters on a UMAX PowerLook 100 scanner with Photoshop.
Identification of Potential Trx Targets
Coomassie Blue-stained spots were excised manually from the 2D gels and proteins were identified essentially as previously described (Vensel et al., 2005) and as outlined below. Excised spots were deposited in a 96-well reaction plate that was inserted into a Digest-Pro (Itavis) where destaining reduction, alkylation, and tryptic digestion were carried out in preparation for analysis of the peptides by mass spectrometry. The plate from the Digest-Pro was placed in the autosampler of the nano flow reversed phase (RP)-HPLC system that was interfaced to a QSTAR PULSAR i quadrupole time-of-flight (TOF) mass spectrometer (Applied Biosystems/MDS Sciex). The TOF mass analyzer was calibrated using the 187.0713 and the 1,245.5444 m/z fragment ions from the collision-induced dissociation of the +2 charge state (m/z 785.8) of glufibrogen. Tryptic peptides separated by nano flow gradient RP-HPLC were introduced into the mass spectrometer by electrospray ionization. Data collection was conducted using the information-dependent acquisition (IDA) mode of Analyst QS software. The most abundant ion of double or triple charge in the survey scan of mass range m/z 400 to 1,500 was selected for fragmentation. The quadrupole mass filter (Q1) was adjusted so that only ions selected during the survey scan entered the collision cell of Q2. Collision-induced dissociation was carried out in Q2 using ultrahigh purity nitrogen. Fragment ion analysis by the TOF mass analyzer was set to a range of 70 to 2,000 m/z. The precursor ion was precluded from further MS/MS experiments. An IDA script was used to determine the optimum collision energy for each precursor ion. Following the 3 s MS/MS fragmentation period, the MS survey scan was repeated until another MS/MS period was triggered. Gradient RP-HPLC and QSTAR instrument parameters for operation and data collection were as previously described.
In preparation for data analysis, the spectra from binary data acquisition files (WIFF files) created by Analyst QS software were converted to text files (DTA) using a WIFF-to-DTA converter (Genomic Solutions) or by use of the Mascot Daemon (http://www.matrixscience.com). Visualization and analysis of the data were performed using a local installation of open source software obtained from the Global Proteome Machine (GPM) organization (http://www.thegpm.org/). X! TANDEM (version: 2005.06.01.2) the spectrum modeler that is a part of the GPM software was used to match MS/MS fragmentation data to peptide sequences. The data-containing text file for each gel spot was searched against a flat file containing amino acid sequences of all plant proteins in the transcript assembly of M. trunculata (MtGI), all plant sequences in the National Center for Biotechnology Information redundant database, and the common Repository of Adventitious Proteins known to occur as unavoidable contamination (http://www.thegpm.org/crap/index.html). The translation of the nucleotide sequences into amino acid sequences in all six reading frames was carried out using the Knexus suite of software (Genomic Solutions). Data analysis results were stored in a locally installed version of the GPM database (http://www.thegpm.org/GMPDB/index.html). Only proteins detected with an expectation score or E-value <−3 and were retained for the study. In accord with our experience, approximately 90% of the spots analyzed were identified in this study.
Redox State of Potential Trx Targets
To determine the initial redox state of proteins in dry seeds and the change in redox during the course of imbibition, proteins were extracted in the presence of 2 mm mBBr to label SH groups. The derivatized preparation was separated by 2D electrophoresis as outlined above. Following electrophoresis, gel images were captured as above.
Gel Image Analyses
Digitized images were analyzed using the PD-Quest 7.1 software (Bio-Rad). Images were filtered (mode pepper outlier 5 × 5), parameters for background subtraction and spot detection optimized, and spots matched. Spot density was normalized using the total density in the valid spot method and fluorescence intensity (mBBr) determined.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Oxidation of protein extracts.
Supplemental Figure S2. Negative controls in the mBBr labeling procedure shown in Figure 1.
Supplemental Figure S3. Protein content in spots of the targets recognized in Figure 3 at three times of germination (0, 14, and 22 h).
Supplemental Table S1. Peptide sequences allowing the identification of the Trx-linked proteins in Table I.
Supplementary Material
Acknowledgments
We thank Christine Le Signor (Institut National de la Recherche Agronomique, Dijon, France) for providing the M. truncatula ‘Jemalong’ seeds.
This work was supported by the France-Berkeley fund, the Contrat Etat Region des Pays de la Loire 2000 to 2006, and the California Agricultural Experiment Station. F.A. was supported by a fellowship from the Conseil Général du Maine et Loire.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Françoise Montrichard (francoise.montrichard@univ-angers.fr).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alexander R, Alamillo JM, Salamini F, Bartels D (1994) A novel embryo-specific barley cdna clone encodes a protein with homologies to bacterial glucose and ribitol dehydrogenase. Planta 192 519–525 [DOI] [PubMed] [Google Scholar]
- Alkhalfioui F, Renard M, Montrichard F (2007) Unique properties of NADP-thioredoxin reductase C in legumes. J Exp Bot 58 969–978 [DOI] [PubMed] [Google Scholar]
- Ashrafi K, Lin SS, Manchester JK, Gordon JI (2000) Sip2p and its partner Snf1p kinase affect aging in S. cerevisiae. Genes Dev 14 1872–1885 [PMC free article] [PubMed] [Google Scholar]
- Ayala A, Parrado J, Bougria M, Machado A (1996) Effect of oxidative stress, produced by cumene hydroperoxide, on the various steps of protein synthesis: modifications of elongation factor-2. J Biol Chem 271 23105–23110 [DOI] [PubMed] [Google Scholar]
- Bailly C (2004) Active oxygen species and antioxidants in seed biology. Seed Sci Res 14 93–107 [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schurmann P, Buchanan BB (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci USA 100 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Koller A, Val GD, Schurmann P, Buchanan BB (2004. a) Proteomics uncovers proteins interacting electrostatically with thioredoxin in chloroplasts. Photosynth Res 79 275–280 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schurmann P, Hurkman WJ, Buchanan BB (2006) A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc Natl Acad Sci USA 103 2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Tanaka CK, Hurkman WJ, Gelhaye E, Rouhier N, Jacquot JP, Manieri W, Schurmann P, Droux M, et al (2004. b) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci USA 101 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse I, Wong JH, Kobrehel K, Buchanan BB (1996) Thiocalsin: a thioredoxin-linked, substrate-specific protease dependent on calcium. Proc Natl Acad Sci USA 93 3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD (1997) Seed germination and dormancy. Plant Cell 9 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet J, Buitink J, Hoekstra FA, Rogniaux H, Larre C, Satour P, Leprince O (2006) Comparative analysis of the heat stable proteome of radicles of Medicago truncatula seeds during germination identifies late embryogenesis abundant proteins associated with desiccation tolerance. Plant Physiol 140 1418–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower MS, Matias DD, Fernandes-Carvalho E, Mazzurco M, Gu T, Rothstein SJ, Goring DR (1996) Two members of the thioredoxin-H family interact with the kinase domain of a Brassica S locus receptor kinase. Plant Cell 8 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantity of protein utilizing the principle of protein dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Brehelin C, Laloi C, Setterdahl A, David B, Meyer Y (2004) Cytosolic, mitochondrial thioredoxins and thioredoxin reductases in Arabidopsis thaliana. Photosynth Res 79 295–304 [DOI] [PubMed] [Google Scholar]
- Buchanan BB (1980) Role of light in the regulation of chloroplast enzymes. Annu Rev Plant Physiol 31 341–374 [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56 187–220 [DOI] [PubMed] [Google Scholar]
- Buitink J, Thomas M, Gissot L, Leprince O (2004) Starvation, osmotic stress and desiccation tolerance lead to expression of different genes of the regulatory β and γ subunits of the Snrk1 complex in germinating seeds of Medicago truncatula. Plant Cell Environ 27 55–67
- Castillo J, Rodrigo MI, Marquez JA, Zuniga A, Franco L (2000) A pea nuclear protein that is induced by dehydration belongs to the vicilin superfamily. Eur J Biochem 267 2156–2165 [DOI] [PubMed] [Google Scholar]
- Chae HZ, Chung SJ, Rhee SG (1994) Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem 269 27670–27678 [PubMed] [Google Scholar]
- Choi YO, Cheong NE, Lee KO, Jung BG, Hong CH, Jeong JH, Chi YH, Kim K, Cho MJ, Lee SY (1999) Cloning and expression of a new isotype of the peroxiredoxin gene of chinese cabbage and its comparison to 2cys-peroxiredoxin isolated from the same plant. Biochem Biophys Res Commun 258 768–771 [DOI] [PubMed] [Google Scholar]
- De Gara L, de Pinto MC, Moliterni VMC, D'Egidio MG (2003) Redox regulation and storage processes during maturation in kernels of Triticum durum. J Exp Bot 54 249–258 [DOI] [PubMed] [Google Scholar]
- Dietz K, Horling F, Konig J, Baier M (2002) The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J Exp Bot 372 1321–1329 [PubMed] [Google Scholar]
- Duval M, Job C, Alban C, Douce R, Job D (1994) Developmental patterns of free and protein-bound biotin during maturation and germination of seeds of Pisum sativum: characterization of a novel seed-specific biotinylated protein. Biochem J 299 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando MR, Nanri H, Yoshitake S, Nagata-Kuno K, Minakami S (1992) Thioredoxin regenerates proteins inactivated by oxidative stress in endothelial cells. Eur J Biochem 209 917–922 [DOI] [PubMed] [Google Scholar]
- Flemetakis E, Agalou A, Kavroulakis N, Dimou M, Martsikovskaya A, Slater A, Spaink HP, Roussis A, Katinakis P (2002) Lotus japonicus gene Ljsbp is highly conserved among plants and animals and encodes a homologue to the mammalian selenium-binding proteins. Mol Plant Microbe Interact 15 313–322 [DOI] [PubMed] [Google Scholar]
- Florencio FJ, Gadal P, Buchanan BB (1993) Thioredoxin-linked activation of the chloroplast and cytosolic forms of Chlamydomonas reinhardtii glutamine synthase. Plant Physiol Biochem 31 649–655 [Google Scholar]
- Freyermuth SK, Bacanamwo M, Polacco JC (2000) The soybean Eu3 gene encodes an Ni-binding protein necessary for urease activity. Plant J 21 53–60 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol 126 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Tao R, Miura K, Dandekar AM, Sugiura A (2001) Transformation of Japanese persimmon (Diospyros Kaki Thunb.) with apple cDNA encoding NADP-dependent sorbitol-6-phosphate dehydrogenase. Plant Sci 160 837–845 [DOI] [PubMed] [Google Scholar]
- Ghezzi P (2005) Regulation of protein function by glutathionylation. Free Radic Res 39 573–580 [DOI] [PubMed] [Google Scholar]
- Gobin P, Ng PKW, Buchanan BB, Kobrehel K (1997) Sulfhydryl-disulfide changes in proteins of developing wheat grain. Plant Physiol Biochem 35 777–783 [Google Scholar]
- Guo H, Yin J, Ren J, Wang Z, Chen H (2007) Changes in proteins within germinating seeds of transgenic wheat with an antisense construct directed against the thioredoxin. Mol Biol 33 18–24 [PubMed] [Google Scholar]
- He X, Miginiac-Maslow M, Sigalat C, Keryer E, Haraux F (2000) Mechanism of activation of the chloroplast ATP synthase: a kinetic study of the thiol modulation of isolated ATPase and membrane-bound ATP synthase from spinach by Eschericia coli thioredoxin. J Biol Chem 275 13250–13258 [DOI] [PubMed] [Google Scholar]
- Herbette S, Lenne C, Leblanc N, Julien JL, Drevet JR, Roeckel-Drevet P (2002) Two Gpx-like proteins from Lycopersicon esculentum and Helianthus annuus are antioxidant enzymes with phospholipid hydroperoxide glutathione peroxidase and thioredoxin peroxidase activities. Eur J Biochem 269 2414–2420 [DOI] [PubMed] [Google Scholar]
- Hirota K, Matsui M, Iwata S, Nishiyama A, Mori K, Yodoi J (1997) Ap-1 transcriptional activity is regulated by a direct association between thioredoxin and ref-1. Proc Natl Acad Sci USA 94 3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch AM, Lum MR, Downie JA (2001) What makes the Rhizobia-legume symbiosis so special? Plant Physiol 127 1484–1492 [PMC free article] [PubMed] [Google Scholar]
- Honigberg SM, Lee RH (1998) Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol Cell Biol 18 4548–4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa S, Maeda K, Sugiyama K, Mano J, Inoue Y, Kimura A (1999) Thioredoxin deficiency causes the constitutive activation of Yap1, an Ap-1-like transcription factor in Saccharomyces cerevisiae. J Biol Chem 274 28459–28465 [DOI] [PubMed] [Google Scholar]
- Jiao J, Yee BC, Wong JH, Kobrehel K, Buchanan BB (1993) Thioredoxin-linked changes in regulatory properties of barley α-amylase/subtilisin inhibitor protein. Plant Physiol Biochem 31 799–804 [Google Scholar]
- Jiao JA, Yee BC, Kobrehel K, Buchanan BB (1992) Effect of thioredoxin-linked reduction on the activity and stability of the kunitz and bowman-birk soybean inhibitor proteins. J Agric Food Chem 40 2333–2336 [Google Scholar]
- Jinn TL, Chen YM, Lin CY (1995) Characterization and physiological function of class I low-molecular-mass, heat-shock protein complex in soybean. Plant Physiol 108 693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job C, Rajjou L, Lovigny Y, Belghazi M, Job D (2005) Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol 138 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BD, Schumacher RJ, Ross ED, Toft DO (1998) Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem 273 3679–3686 [DOI] [PubMed] [Google Scholar]
- Kallis GB, Holmgren A (1980) Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J Biol Chem 255 10261–10265 [PubMed] [Google Scholar]
- Kim YB, Garbisu C, Pickering IJ, Prince RC, George GN, Cho MJ, Wong JH, Buchanan BB (2003) Thioredoxin H overexpressed in barley seeds enhances selenite resistance and uptake during germination and early seedling development. Planta 218 186–191 [DOI] [PubMed] [Google Scholar]
- Kobrehel K, Wong JH, Balogh A, Kiss F, Yee BC, Buchanan BB (1992) Specific reduction of wheat storage proteins by thioredoxin H. Plant Physiol 99 919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobrehel K, Yee BC, Buchanan BB (1991) Role of the NADP/thioredoxin system in the reduction of alpha-amylase and trypsin inhibitor proteins. J Biol Chem 266 16135–16140 [PubMed] [Google Scholar]
- Krochko JE, Bewley JD (1988) Use of electrophoretic techniques in determining the composition of seed storage proteins in alfalfa. Electrophoresis 9 751–763 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lee J, Godon C, Lagniel G, Spector D, Garin J, Labarre J, Toledano MB (1999) Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J Biol Chem 274 16040–16046 [DOI] [PubMed] [Google Scholar]
- Lemaire SD, Guillon B, Le Marechal P, Keryer E, Miginiac-Maslow M, Decottignies P (2004) New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 101 7475–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Florencio FJ (2003) Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc Natl Acad Sci USA 100 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano RM, Wong JH, Yee BC, Peters A, Kobrehel K, Buchanan BB (1996) New evidence for a role for thioredoxin H in germination and seedling development. Planta 200 100–106 [Google Scholar]
- Maeda K, Finnie C, OStergaard O, Svensson B (2003) Identification, cloning and characterization of two thioredoxin H isoforms, Hvtrxh1 and Hvtrxh2, from the barley seed proteome. Eur J Biochem 270 2633–2643 [DOI] [PubMed] [Google Scholar]
- Maeda K, Finnie C, Svensson B (2004) Cy5 maleimide labelling for sensitive detection of free thiols in native protein extracts: identification of seed proteins targeted by barley thioredoxin H isoforms. Biochem J 378 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand C, Le Marechal P, Meyer Y, Miginiac-Maslow M, Issakidis-Bourguet E, Decottignies P (2004) New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics 4 2696–2706 [DOI] [PubMed] [Google Scholar]
- Mark DF, Richardson CC (1976) Escherichia coli thioredoxin: a subunit of bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA 73 780–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C, Wong JH, Buchanan B (2003) Thioredoxin and germinating barley: targets and protein redox changes. Planta 216 454–460 [DOI] [PubMed] [Google Scholar]
- Masutani H, Yodoi J (2002) Thioredoxin: overview. Methods Enzymol 347 279–286 [DOI] [PubMed] [Google Scholar]
- Meek SEM, Lane WS, Piwnica-Worms H (2004) Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J Biol Chem 279 32046–32054 [DOI] [PubMed] [Google Scholar]
- Michelet L, Zaffagnini M, Massot V, Keryer E, Vanacker H, Miginiac-Maslow M, Issakidis-Bourguet E, Lemaire S (2006) Thioredoxins, glutaredoxins, and glutathionylation: new crosstalks to explore. Photosynth Res 89 225–245 [DOI] [PubMed] [Google Scholar]
- Montrichard F, Renard M, Alkhalfioui F, Duval DF, Macherel D (2003) Identification and differential expression of two thioredoxin H isoforms in germinating seeds from pea. Plant Physiol 132 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci USA 98 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Koyama F, Nakanishi Y, Ueoka-Nakanishi H, Hisabori T (2003) Chloroplast cyclophilin is a target protein of thioredoxin: thiol modulation of the peptidyl-prolyl cis-trans isomerase activity. J Biol Chem 278 31848–31852 [DOI] [PubMed] [Google Scholar]
- Muller E (1995) A redox-dependent function of thioredoxin is necessary to sustain a rapid rate of DNA synthesis in yeast. Arch Biochem Biophys 318 356–361 [DOI] [PubMed] [Google Scholar]
- Noguchi M, Takata T, Kimura Y, Manno A, Murakami K, Koike M, Ohizumi H, Hori S, Kakizuka A (2005) ATPase activity of P97/valosin-containing protein is regulated by oxidative modification of the evolutionally conserved cysteine 522 residue in walker a motif. J Biol Chem 280 41332–41341 [DOI] [PubMed] [Google Scholar]
- Overvoorde PJ, Chao WS, Grimes HD (1997) A plasma membrane sucrose-binding protein that mediates sucrose uptake shares structural and sequence similarity with seed storage proteins but remains functionally distinct. J Biol Chem 272 15898–15904 [DOI] [PubMed] [Google Scholar]
- Paget MS, Kang JG, Roe JH, Buttner MJ (1998) Sigmar, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). EMBO J 17 5776–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wyatt SE, Love J, Thompson WF, Robertson D, Boss WF (2001) The Ca(2+) status of the endoplasmic reticulum is altered by induction of calreticulin expression in transgenic plants. Plant Physiol 126 1092–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Clore GM, Kennedy WP, Huth JR, Gronenborn AM (1995) Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NfκB. Structure 3 289–297 [DOI] [PubMed] [Google Scholar]
- Quimby BB, Dasso M (2003) The small GTPase ran: interpreting the signs. Curr Opin Cell Biol 15 338–344 [DOI] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D (2004) The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol 134 1598–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhazi L, Cazalis R, Aussenac T (2003) Sulfhydryl-disulfide changes in storage proteins of developing wheat grain: influence on the SDS-unextractable glutenin polymer formation. J Cereal Sci 38 3–13 [Google Scholar]
- Rouhier N, Villarejo A, Srivastava M, Gelhaye E, Keech O, Droux M, Finkemeier I, Samuelsson G, Dietz KJ, Jacquot JP, et al (2005) Identification of plant glutaredoxin targets. Antioxid Redox Signal 7 919–929 [DOI] [PubMed] [Google Scholar]
- Saze H, Ueno Y, Hisabori T, Hayashi H, Izui K (2001) Thioredoxin-mediated reductive activation of a protein kinase for the regulatory phosphorylation of C4-form phosphoenolpyruvate carboxylase from maize. Plant Cell Physiol 42 1295–1302 [DOI] [PubMed] [Google Scholar]
- Shin S, Wong JH, Kobrehel K, Buchanan BB (1993) Reduction of castor-seed 2S albumin by thioredoxin. Planta 189 557–560 [Google Scholar]
- Sokolov L, Dominguez-Solis J, Allary A, Buchanan B, Luan S (2006) A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc Natl Acad Sci USA 103 9732–9737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketani S, Adachi Y, Kohno H, Ikehara S, Tokunaga R, Ishii T (1998) Molecular characterization of a newly identified heme-binding protein induced during differentiation of urine erythroleukemia cells. J Biol Chem 273 31388–31394 [DOI] [PubMed] [Google Scholar]
- Tang W, Peng X, Newton RJ (2005) Enhanced tolerance to salt stress in transgenic loblolly pine simultaneously expressing two genes encoding mannitol-1-phosphate dehydrogenase and glucitol-6-phosphate dehydrogenase. Plant Physiol Biochem 43 139–146 [DOI] [PubMed] [Google Scholar]
- Tischner R, Schmidt A (1982) A thioredoxin-mediated activation of glutamine synthetase and glutamate synthase in synchronous Chlorella sorokiniana. Plant Physiol 70 113–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra T, Conrads T, Issaq H (2004) What to do with “one-hit wonders”? Electrophoresis 25 1278–1279 [DOI] [PubMed] [Google Scholar]
- Vensel W, Tanaka C, Cai N, Wong J, Buchanan B, Hurkman W (2005) Developmental changes in the metabolic protein profiles of wheat endosperm. Proteomics 5 1594–1611 [DOI] [PubMed] [Google Scholar]
- Vignols F, Mouaheb N, Thomas D, Meyer Y (2003) Redox control of Hsp70-co-chaperone interaction revealed by expression of a thioredoxin-like Arabidopsis protein. J Biol Chem 278 4516–4523 [DOI] [PubMed] [Google Scholar]
- Wang C, Croft KPC, Jarlfors U, Hildebrand DF (1999) Subcellular localization studies indicate that lipoxygenases 1 to 6 are not involved in lipid mobilization during soybean germination. Plant Physiol 120 227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Boja ES, Tan W, Tekle E, Fales HM, English S, Mieyal JJ, Chock PB (2001) Reversible glutathionylation regulates actin polymerization in A431 cells. J Biol Chem 276 47763–47766 [DOI] [PubMed] [Google Scholar]
- Wong JH, Balmer Y, Cai N, Tanaka CK, Vensel WH, Hurkman WJ, Buchanan BB (2003) Unraveling thioredoxin-linked metabolic processes of cereal starchy endosperm using proteomics. FEBS Lett 547 151–156 [DOI] [PubMed] [Google Scholar]
- Wong JH, Cai N, Balmer Y, Tanaka CK, Vensel WH, Hurkman WJ, Buchanan BB (2004. b) Thioredoxin targets of developing wheat seeds identified by complementary proteomic approaches. Phytochemistry 65 1629–1640 [DOI] [PubMed] [Google Scholar]
- Wong JH, Cai N, Tanaka CK, Vensel WH, Hurkman WJ, Buchanan BB (2004. a) Thioredoxin reduction alters the solubility of proteins of wheat starchy endosperm: an early event in cereal germination. Plant Cell Physiol 45 407–415 [DOI] [PubMed] [Google Scholar]
- Wong JH, Kim YB, Ren PH, Cai N, Cho MJ, Hedden P, Lemaux PG, Buchanan BB (2002) Transgenic barley grain overexpressing thioredoxin shows evidence that the starchy endosperm communicates with the embryo and the aleurone. Proc Natl Acad Sci USA 99 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn R, Cocco MJ, Richards FM (1995) Mixed disulfide intermediates during the reduction of disulfides by Escherichia coli thioredoxin. Biochemistry 34 11807–11813 [DOI] [PubMed] [Google Scholar]
- Yamazaki D, Motohashi K, Kasama T, Hara Y, Hisabori T (2004) Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol 45 17–24 [DOI] [PubMed] [Google Scholar]
- Yano H, Kuroda M (2006) Disulfide proteome yields a detailed understanding of redox regulations: a model study of thioredoxin-linked reactions in seed germination. Proteomics 6 294–300 [DOI] [PubMed] [Google Scholar]
- Yano H, Wong JH, Cho MJ, Buchanan BB (2001. a) Redox changes accompanying the degradation of seed storage proteins in germinating rice. Plant Cell Physiol 42 879–883 [DOI] [PubMed] [Google Scholar]
- Yano H, Wong JH, Lee YM, Cho MJ, Buchanan BB (2001. b) A strategy for the identification of proteins targeted by thioredoxin. Proc Natl Acad Sci USA 98 4794–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Portis AR Jr (1999) Mechanism of light regulation of rubisco: a specific role for the larger rubisco activase isoform involving reductive activation by thioredoxin-F. Proc Natl Acad Sci USA 96 9438–9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia LE, Stebbins NE, Polacco JC (1995) Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds. Plant Physiol 107 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.