Abstract
Carbonic anhydrase (CA) catalyzes the interconversion of CO2 and bicarbonate, the forms of inorganic carbon used by the primary carboxylating enzymes of C3 and C4 plants, respectively. Multiple forms of CA are found in both photosynthetic subtypes; however, the number of isoforms and the location and function of each have not been elucidated for any single plant species. Genomic Southern analyses showed that the C4 dicotyledon Flaveria bidentis ‘Kuntze’ contains a small gene family encoding β-CA and cDNAs encoding three distinct β-CAs, named CA1, CA2, and CA3, were isolated. Quantitative reverse transcription-polymerase chain reactions showed that each member of this β-CA family has a specific expression pattern in F. bidentis leaves, roots, and flowers. CA3 transcripts were at least 50 times more abundant than CA2 or CA1 transcripts in leaves. CA2 transcripts were detected in all organs examined and were the most abundant CA transcripts in roots. CA1 mRNA levels were similar to those of CA2 in leaves, but were considerably lower in roots and flowers. In vitro import assays showed CA1 was imported into isolated pea (Pisum sativum) chloroplasts, whereas CA2 and CA3 were not. These results support the following roles for F. bidentis CAs: CA3 is responsible for catalyzing the first step in the C4 pathway in the mesophyll cell cytosol; CA2 provides bicarbonate for anapleurotic reactions involving nonphotosynthetic forms of phosphoenolpyruvate carboxylase in the cytosol of cells in both photosynthetic and nongreen tissues; and CA1 carries out nonphotosynthetic functions demonstrated by C3 chloroplastic β-CAs, including lipid biosynthesis and antioxidant activity.
Carbonic anhydrase (CA; EC 4.2.1.1) catalyzes the reversible hydration of CO2 and appears to be a ubiquitous enzyme, having been found in species of archaebacteria, eubacteria, protists, plants, and animals. Three independent classes of CA, α-CA, β-CA, and γ-CA (Hewett-Emmett and Tashian, 1996; Moroney et al., 2001), have been well characterized. α-CAs are found in plants, animals, protists, and eubacteria, and vertebrate α-CAs have been studied intensely. Plants and protists, as well as archaebacteria and eubacteria, contain β- and γ-CAs (Moroney et al., 2001; Parisi et al., 2004). Although no conservation of amino acid sequence or tertiary and quaternary structures is seen between these different classes, all are Zn2+ metalloenzymes and demonstrate a similar mechanism in the interconversion of CO2 and bicarbonate (Liljas and Laurberg, 2000; Tripp et al., 2001). Recently, three additional types of CA have been identified. A subclass of β-CAs, the ɛ-class of CAs, is found in several marine cyanobacteria and chemolithoautotrophic bacteria (So et al., 2004; Sawaya et al., 2006). A Zn2+ metalloenzyme showing distant homology to α-CAs was characterized from the diatom Thalassiosira weissflogii (Lane and Morel, 2000) and designated the prototype of the δ-CA class (Tripp et al., 2001). Diatoms also contain ζ-CAs in which cadmium replaces zinc in the active site of the enzymes (Lane et al., 2005).
Molecular, biochemical, and genetic studies indicate that most, if not all, C3 and C4 plants contain multiple forms of β-CA, that at least one β-CA is associated with photosynthetic tissues, and that both cytosolic and chloroplastic forms of the enzyme exist (for review, see Burnell, 2000; Coleman, 2000; Moroney et al., 2001). In plants utilizing the C3 photosynthetic pathway, most CA activity is localized to the chloroplast stroma of the mesophyll cells (Poincelot, 1972; Jacobsen et al., 1975; Tsuzuki et al., 1985) and, although the exact role of the enzyme is not clear, it has been suggested that CA may be responsible for maintaining adequate concentrations of CO2 around Rubisco by facilitating the diffusion of CO2 across the chloroplast envelope or by rapidly dehydrating bicarbonate to CO2 (Reed and Graham, 1981; Cowan, 1986; Price et al., 1994). More recently, roles in plant defense (Slaymaker et al., 2002) and lipid synthesis (Hoang and Chapman, 2002) have been demonstrated for chloroplastic β-CAs of C3 plants.
In contrast, the role of CA in the C4 photosynthetic pathway is clear (Hatch and Burnell, 1990). In most terrestrial C4 plants, two cell types are involved in CO2 assimilation, mesophyll and bundle-sheath cells (bscs), and the majority of CA activity in these plants has been localized to the cytosol of the mesophyll cells (Gutierrez et al., 1974; Ku and Edwards, 1975; Burnell and Hatch, 1988). Here, the enzyme catalyzes the first reaction in the C4 photosynthetic pathway, which is the hydration of atmospheric CO2 to bicarbonate, the substrate for the primary carboxylating enzyme of C4 plants phosphoenolpyruvate carboxylase (PEPC; Hatch and Burnell, 1990). C4 acids produced through PEPC activity are subsequently decarboxylated in the bscs, resulting in elevated concentrations of CO2 around Rubisco, which is located in the bundle-sheath chloroplasts. Thus, the C4 photosynthetic pathway functions as a CO2-concentrating mechanism (CCM), allowing Rubisco to function at close to CO2 saturation (von Caemmerer and Furbank, 2003).
Some nongreen tissues, including roots (Demir et al., 1997) and root nodules (Atkins, 1974; Coba de la Peña et al., 1997; Atkins et al., 2001; Flemetakis et al., 2003), etiolated and white sections of variegated leaves (Reed, 1979), cotton (Gossypium hirsutum) embryos (Hoang and Chapman, 2002), and dark-grown seedlings (Hoang et al., 1999), also demonstrate CA activity. Molecular studies have shown that β-CAs account for at least some of this (Coba de la Peña et al., 1997; Hoang et al., 1999; Kavroulakis et al., 2000; Hoang and Chapman, 2002; Flemetakis et al., 2003). The suggested roles of CA in these nonphotosynthetic tissues are varied and comprise lipid biosynthesis, gas exchange, pH balance, and provision of carbon skeletons for amino acid biosynthesis and Kreb's cycle intermediates.
Our understanding of the functional specificity or redundancy of the multiple forms of CA found in plants is limited because the total number of CA genes and the location and function of each isoform they encode have not been fully elucidated for any single plant species. Indeed, Moroney et al. (2001) highlighted these gaps in our knowledge of this ubiquitous enzyme as key unresolved questions.
We have been studying the β-CAs in selected species within the genus Flaveria, which has allowed us to (1) examine the importance of the enzyme in the C4 CCM using a transgenic approach (von Caemmerer et al., 1997, 2004; Ludwig et al., 1998) and (2) investigate the molecular evolution of the C4 photosynthetic pathway (Ludwig and Burnell, 1995) because the genus Flaveria contains individual species demonstrating either C3, C4, or C3-C4 intermediate photosynthesis (Edwards and Ku, 1987). Here, we describe cDNAs encoding three distinct β-CAs from F. bidentis ‘Kuntze’ and show that a small multigene family encodes the enzyme in this C4 species. We also report the differential expression patterns of the three CA genes in F. bidentis leaves, roots, and flowers, as well as identify the subcellular locations of the three CA isoforms and discuss their likely physiological functions.
RESULTS
Isolation and Characterization of cDNAs Encoding F. bidentis β-CA
During the screening of a λgt11 cDNA library of F. bidentis leaf tissue with a radiolabeled fragment encoding F. bidentis CA1 (Cavallaro et al., 1994), plaques exhibiting hybridization signals that differed in intensity were detected (data not shown). Sequence determination of the inserts from plaques demonstrating moderate to low intensities showed they encoded two isoforms of CA that differed significantly from CA1, which were named CA2 and CA3. Examination of the sequences and comparisons with other plant β-CA sequences in the databases suggested that a full-length CA3 clone, with a predicted open reading frame (ORF) of 774 bp, had been isolated; however, the longest cDNA fragment obtained encoding CA2 was truncated at the 5′ end (data not shown). The sequence of this fragment was used to design an oligonucleotide primer to amplify the 5′ region of CA2 from an adaptor-ligated F. bidentis leaf cDNA library using 5′ RACE. Fragments encoding the probable translation initiation codon and an upstream, in-frame stop codon were obtained and resulted in an ORF of 837 bp coding for the full-length CA2 isoform.
The 5′-RACE technique and the leaf adaptor-ligated cDNA library were also used to verify the 5′ ends of the cDNA sequences encoding F. bidentis CA1 and CA3. Sequence determination of the CA1 5′-RACE products indicated several sequencing errors in the 5′ region of the previously reported cDNA encoding this CA isoform (Cavallaro et al., 1994; GenBank entry amended). The longest 5′-RACE products generated for CA1 encoded 24 bp upstream of the putative start codon; however, no in-frame termination codon was found in this region. Nevertheless, based on comparisons with other plant β-CA cDNA sequences, we believe we have determined the complete sequence of the F. bidentis CA1 ORF, which is 990 bp in size. Sequence determination of the 5′-RACE products obtained for CA3 confirmed the 5′ sequence of the λgt11 cDNA clone and, in addition, showed that an in-frame stop codon was located 24 bp upstream of the putative start codon.
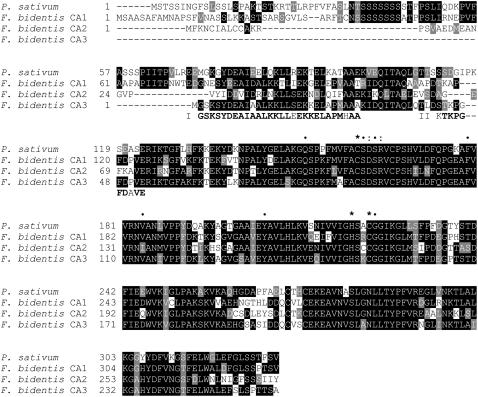
A comparison of the deduced amino acid sequences of the F. bidentis CA cDNAs with that of pea (Pisum sativum; Fig. 1) showed that these proteins share a high level of sequence conservation, especially over the C-terminal regions from Glu-123 of the pea sequence. F. bidentis CA1 is 61% identical to the pea sequence, whereas CA2 and CA3 share slightly less identity at 53% and 55%, respectively. Among the deduced F. bidentis proteins, CA1 and CA3 are 60% identical, whereas CA2 shows 51% and 59% identity with CA1 and CA3, respectively. Figure 1 also illustrates that the F. bidentis CA isoforms are predicted to contain all the residues found, by site-directed mutagenesis (Provart et al., 1993; Bracey et al., 1994) and structural (Kimber and Pai, 2000) studies, to be important for β-CA function. The three residues, His-220, Cys-160, and Cys-223 (numbering based on the pea CA sequence), required for Zn2+ binding in the active site of the pea enzyme are in corresponding positions in the F. bidentis proteins (Fig. 1). Moreover, the residues forming the channel into the catalytic site of pea CA and those making up the hydrophobic surface of the catalytic pocket (Kimber and Pai, 2000) are also in equivalent positions in the F. bidentis CA isoforms (Fig. 1).
Figure 1.
Alignment of the deduced amino acid sequences of pea CA and F. bidentis CA1, CA2, and CA3, and CA3 N-terminal amino acid sequences. Identical residues are boxed in black, whereas conservative changes are shown in gray. Dashes represent gaps inserted to maximize the alignment. Fragments obtained from N-terminal amino acid sequencing, I and II, are shown aligned in bold, except at mismatched residues. *, Residues implicated in Zn2+ binding; •, highly conserved active site residues of higher plant β-CAs; :, D/R pair found thus far in all β-CAs (Provart et al., 1993; Bracey et al., 1994; Kimber and Pai, 2000). GenBank accession number: pea, M63627.
The F. bidentis β-CA Multigene Family
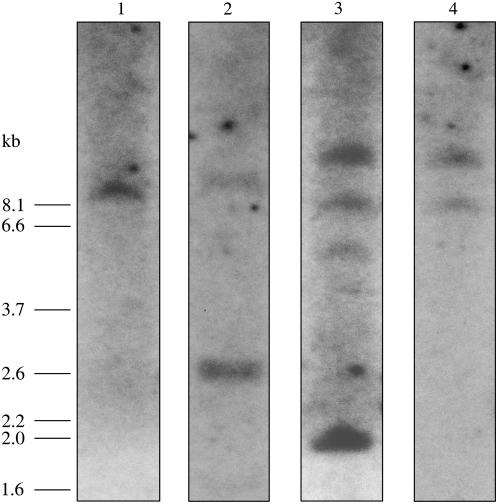
Relatively simple labeling patterns were obtained when F. bidentis genomic DNA digested with XbaI was hybridized with 32P-labeled F. bidentis CA ORF probes and a 118-bp single-exon probe from CA3 (Fig. 2), none of which contained an XbaI site. A single fragment of approximately 9.4 kb was detected with the CA1 ORF probe, whereas the CA2 ORF probe also hybridized to a 9.4-kb fragment as well as a 2.6-kb band. Two genomic fragments of about 12.0 and 8.1 kb labeled with both the CA3 ORF and single-exon probes. Additional fragments of approximately 5.6 and 2.0 kb also labeled with the CA3 ORF probe. No fragments smaller than 1.6 kb were detected in any of the blots. Uncomplicated hybridization patterns were also found using these probes with EcoRI and BamHI-digested genomic DNA (data not shown). The labeling patterns suggest single genes probably encode CA1 and CA2 in F. bidentis, whereas there may be two genes encoding CA3. The results also indicate that it is unlikely an additional CA isoform closely related to CA1, CA2, or CA3 is encoded by the F. bidentis genome. This is also supported by the lack of additional CA sequences detected in multiple screens of λgt11 and adaptor-ligated cDNA libraries, as well as numerous reverse transcription (RT)-PCRs.
Figure 2.
Southern-blot analysis of F. bidentis genomic DNA. Genomic DNA was digested with XbaI, fragments were separated by electrophoresis, transferred to membrane, and then hybridized with radiolabeled CA1 ORF (lane 1), CA2 ORF (lane 2), CA3 ORF (lane 3), or the 118-bp CA3 exon probe (lane 4). Molecular size markers are shown at the left (in kb).
Expression of F. bidentis CA1, CA2, and CA3 in Leaves, Roots, and Flowers
Quantitative (q)RT-PCR assays were developed to measure the expression of the three CA genes in F. bidentis leaves, roots, and flowers. However, before a cDNA preparation was used for qRT-PCR analyses, it was tested in standard PCRs with the CA gene-specific primer pairs to ensure that only fragments of the predicted size were amplified and that it contained no genomic DNA. Only cDNA populations meeting both these criteria were used in the qRT-PCRs.
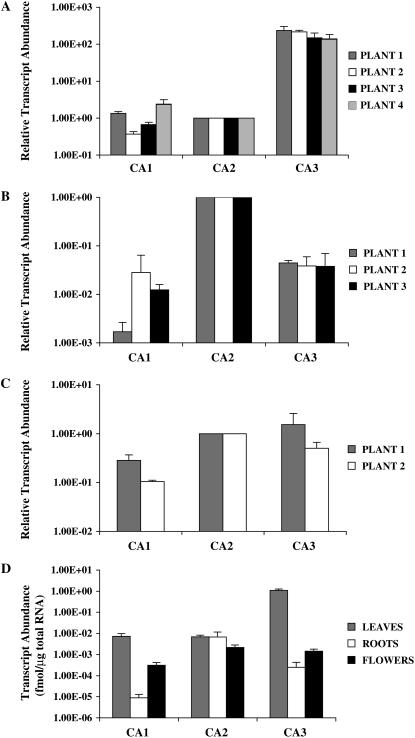
Figure 3, A to C, shows the relative abundance of CA1, CA2, and CA3 transcripts in leaves, roots, and flowers of individual F. bidentis plants. In these figures, CA2 mRNA levels were set at 1 to facilitate comparison of CA transcript abundance in the same organ of different plants. CA3 gene transcripts were the most abundant CA transcripts in the leaf tissues of all plants examined, being at least 50 times more plentiful than either CA1 or CA2 mRNAs (Fig. 3A). Relative to CA2 mRNA abundance, only slight differences in the levels of CA1 and CA3 transcripts were detected in the leaves of the four individual F. bidentis plants.
Figure 3.
CA transcript abundance in F. bidentis organs. Relative transcript levels of the CA1, CA2, and CA3 genes in leaves (A), roots (B), and flowers (C) of individual plants. Values for CA1 and CA3 transcripts were normalized to those of the CA2 gene. Absolute CA transcript levels (D) in F. bidentis organs based on the total RNA used in the qRT-PCRs. A to C, sd of replicates done with the same cDNA preparation from an individual plant. D, se of the combined data from the individual plants shown in A to C.
A very different pattern of CA gene expression was found in F. bidentis roots (Fig. 3B), with the CA2 gene being transcribed at a higher steady-state level than either of the other CA genes in these organs. CA2 mRNA was at least 20 times more abundant than CA1 or CA3 gene transcripts in the root tissue of the three plants examined. The levels of CA3 mRNA were consistent among the plants examined, whereas transcript levels of the CA1 gene tended to be more variable and just at the limit of detection of our assays.
F. bidentis CA2 and CA3 genes demonstrated similar levels of expression in flowers, whereas CA1 transcript abundance was consistently lower (Fig. 3C). These data, however, may not accurately reflect CA transcript abundance in the organs because F. bidentis has composite flowers and it is likely that some of the individual flowers used from both plants were immature and contained associated photosynthetic tissue.
When CA gene transcript abundance in each organ was calculated based on the amount of total RNA used in the qRT-PCRs and the data from the individual plants were combined, the results showed CA3 gene expression was highest in F. bidentis leaves, being at least 1,000 times greater in these organs than in roots or flowers (Fig. 3D). In contrast, CA2 transcripts were essentially equally abundant in all organs examined (Fig. 3D). The levels of CA1 mRNA were highest in F. bidentis leaves, albeit about 100-fold lower than the levels of CA3 transcripts in these organs (Fig. 3D). As mentioned above, CA1 mRNA was detected at very low levels in F. bidentis roots and, although higher levels of CA1 transcripts were found in flowers (Fig. 3D), these values are probably inflated due to the presence of photosynthetic tissue.
Several criteria were used to monitor and confirm the robustness of the qRT-PCR results. Reaction efficiencies of the CA gene-specific primer pairs, as determined from the slopes of qRT-PCR standard curves, ranged between 1.6 and 1.9 and deviated less than 15% for a given primer pair over all experiments. All standard curves had error values of less than 0.3 and regression coefficients of −0.99 or −1.00 (data not shown). Melting-curve analyses and gel electrophoresis were routinely carried out on the qRT-PCR products and indicated only specific products of the predicted sizes were amplified with a given CA primer pair (data not shown).
Subcellular Localization of F. bidentis β-CA Isoforms
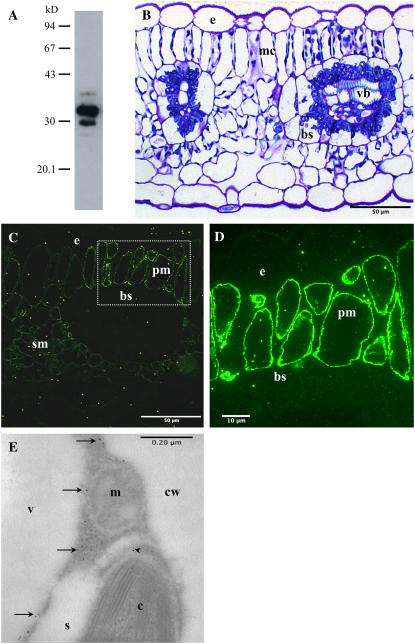
An antiserum generated against F. bidentis CA3 was affinity purified for use in immunocytochemical studies. The resulting CA-specific antiserum detected four polypeptides of approximately 35, 32, 30, and 28 kD in F. bidentis leaf extracts on denaturing polyacrylamide gels (Fig. 4A), with the 32- and 30-kD species being intensely labeled. This labeling pattern is the same as that detected in a previous study using an anti-tobacco (Nicotiana tabacum) CA antiserum (Ludwig et al., 1998). When the affinity-purified antiserum was used to label F. bidentis leaf sections for fluorescence microscopy, intense fluorescence was detected along the periphery of the mesophyll cells (Fig. 4, C and D). This labeling coincides with the location of the mesophyll cell cytoplasm, which is pushed up against the internal surface of the cell wall due to a large central vacuole (compare Fig. 4, B–D). No fluorescence was detected in the epidermal cells or bscs or the vascular tissues of F. bidentis leaves (Fig. 4, C and D). To resolve the subcellular structures or regions that labeled with the affinity-purified anti-CA antiserum, F. bidentis leaf sections were prepared for immunogold electron microscopy. Immunolabeling in these sections was concentrated in the cytoplasm of the mesophyll cells (Fig. 4E). Labeling above background level was also detected in some sections of the chloroplast stroma. No labeling was seen in other mesophyll cell structures (Fig. 4E) or in other cell types of F. bidentis leaves (data not shown).
Figure 4.
Immunolabeling of F. bidentis CA isoforms. A, F. bidentis leaf proteins were separated by 12% SDS-PAGE, transferred to nitrocellulose, and labeled with an affinity-purified anti-F. bidentis CA3 antiserum. Immunoreactive proteins of approximately 35, 32, 30, and 28 kD were detected. Molecular mass markers shown in kD. B, A transverse section through a leaf of F. bidentis stained with methylene blue illustrates the characteristic Kranz anatomy of this C4 dicot. Vascular bundles (vb) are surrounded by bundle-sheath (bs) cells, which are surrounded by mesophyll cells (mc). The densely stained structures at the periphery of the mc and in a centripetal location in the bs are chloroplasts. e, Epidermal cell. C, An F. bidentis leaf in transverse section labeled with the affinity-purified anti-F. bidentis CA3 antiserum. Immunofluorescence was detected in the palisade (pm) and spongy (sm) mesophyll cells. Only nonspecific background fluorescence was seen in epidermal (e) and bundle-sheath (bs) cells. D, An immunolabeled adjacent section of the palisade mesophyll (pm) cells boxed in C at higher magnification. Bright fluorescence was seen at the periphery of the cells, corresponding to the location of the cytoplasm. Little or no fluorescence was detected in the epidermal (e) or bundle-sheath (bs) cells. E, Transverse section through part of a F. bidentis mesophyll cell labeled with the affinity-purified anti-F. bidentis CA3 antiserum followed by a secondary antibody coupled to colloidal gold particles. Gold particles (arrows) labeled the cytosol of the cells, and the chloroplast stroma (arrowhead). Mitochondria (m), starch grains (s), vacuole (v), and cell wall (cw) were not labeled.
As shown in Figure 1, the N terminus of F. bidentis CA1, like that of pea CA, has characteristics of a chloroplast transit peptide, being enriched in hydroxylated amino acids and having relatively few acidic residues (von Heijne et al., 1989). N-terminal sequencing showed the mature form of pea CA begins at Gln-106 (Roeske and Ogren, 1990) and a high degree of amino acid sequence conservation is seen in the corresponding regions of all the deduced F. bidentis CA sequences (Fig. 1); however, TargetP (Emanuelsson et al., 2000) and ChloroP (Emanuelsson et al., 1999) predicted only F. bidentis CA1 to localize to the chloroplast, whereas CA2 and CA3 were calculated to be cytosolic enzymes (data not shown).
High-quality N-terminal amino acid sequence supported the hypothesis that F. bidentis CA3 is a cytosolic enzyme. Twenty-eight amino acids were determined from a 32-kD polypeptide that labeled with an anti-tobacco CA antiserum (Ludwig et al., 1998) and the residues aligned with the predicted N-terminal sequence of CA3, differing at only two residues (Fig. 1). Ten amino acids were resolved from the N terminus of a 30-kD immunoreactive CA polypeptide (Ludwig et al., 1998) and this sequence also shared high identity with the predicted amino acid sequence of CA3; however, the sequence aligned at residues 43 to 52 of the CA3 sequence and showed differences at two positions (Fig. 1). That both the 32- and 30-kD polypeptides are forms of CA3 is consistent with the immunoblot labeling pattern of the anti-F. bidentis CA3 antiserum described above. No obvious precursor-product relationship was observed for the 32- and 30-kD polypeptides when leaf protein extracts were incubated for extended periods at 37°C (data not shown). It is possible that, during the isolation procedure, the 32-kD polypeptide was specifically degraded at its N terminus, resulting in the formation of the 30-kD species. No N-terminal amino acid sequence was obtained for the 35- and 28-kD CA polypeptides that also labeled with the anti-tobacco CA antiserum (Ludwig et al., 1998), presumably due to blocked N termini and/or insufficient protein.
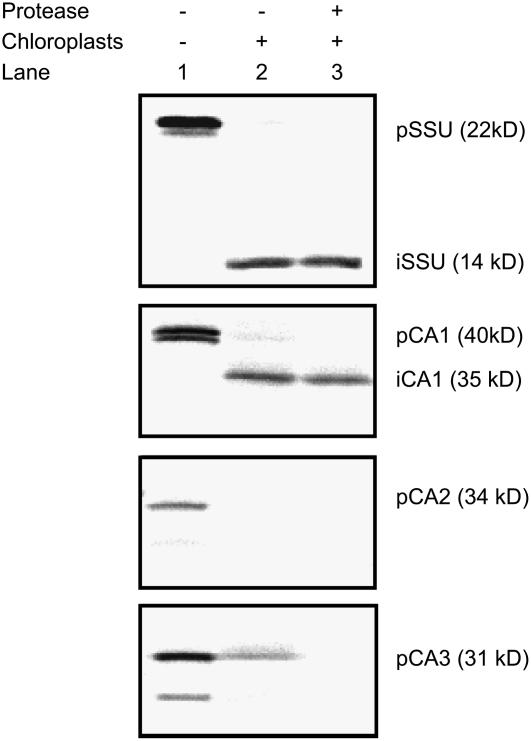
To resolve which CA isoforms were responsible for the immunolabeling detected in F. bidentis mesophyll cell chloroplasts and cytosol, we carried out in vitro import assays with isolated pea chloroplasts. In vitro transcription and translation of the F. bidentis CA1 and CA3 ORFs generated two precursor proteins (Fig. 5), the sizes of which corresponded to initiation of translation at an internal Met as well as the N-terminal Met (compare Figs. 1 and 5). A similar result was obtained for the control protein, the small subunit (SSU) of pea Rubisco, whereas a single precursor protein was made from the CA2 ORF (Fig. 5). As shown in Figure 5, F. bidentis CA1, like pea Rubisco SSU, was imported into pea chloroplasts and processed to a lower molecular mass form, which was protected from externally added protease. The hypothesis that F. bidentis CA2 and CA3 are cytosolic proteins was also supported by the assays because these proteins were not imported into isolated pea chloroplasts and were susceptible to externally added thermolysin (Fig. 5). The CA precursor proteins consistently appeared larger on the import gels than expected from their deduced amino acid sequences, presumably because of the conformations in which they migrated through the gels and/or posttranslational modifications made in the rabbit reticulocyte lysate used.
Figure 5.
Chloroplast import assays of in vitro transcribed and translated F. bidentis CA1, CA2, and CA3. Lane 1, Precursor (p) proteins generated for CA1, CA2, CA3, and the pea Rubisco SSU, which served as an import control. A single precursor was obtained for CA2, whereas two polypeptides were generated for the other constructs due to initiation of translation at internal Met residues as well as at the N termini. Lane 2, Chloroplast fractions following incubation of the precursor proteins under conditions favoring import. Imported (i) polypeptides were detected for CA1 and the SSU. Lane 3, As for lane 2, except protease was externally added to the chloroplasts prior to their collection.
DISCUSSION
cDNAs encoding full-length ORFs of three distinct β-CA isoforms have been isolated from the C4 dicot F. bidentis. The deduced amino acid sequences of these enzymes indicate that they all share significant identity with the β-CA sequences of other dicotyledonous plants and they contain a His and two Cys residues in comparable positions to the pea CA (His-220, Cys-160, and Cys-223), which were shown to be important in binding the catalytic zinc ion by both mutational studies (Provart et al., 1993; Bracey et al., 1994) and crystallography (Kimber and Pai, 2000). The F. bidentis enzymes also contain the Asp-162 and Arg-164 pair, which is conserved in all β-CAs so far examined (Burnell, 2000; Kimber and Pai, 2000) and, with one exception, the conserved key active site residues identified by Kimber and Pai (2000) as being present in all known higher plant β-CAs. The F. bidentis CA2 sequence has an Ile residue at position 184, whereas the other two F. bidentis CAs and the pea enzyme have Val at this position. A survey of other dicot β-CA sequences indicates that either Ile or Val is found at this amino acid position. The crystal structure of pea CA (Kimber and Pai, 2000) indicates Val-184 forms part of the hydrophobic surface of the binding pocket in the enzyme's active site, an environment that would be maintained with the substitution of an Ile residue at this position.
The relatively simple labeling pattern of F. bidentis genomic Southern blots indicates that a small multigene family encodes β-CA in this species. Further evidence that F. bidentis β-CA is encoded by a small gene family comes from immunoblots of whole-leaf extracts probed with anti-spinach (Spinacia oleracea) CA (Ludwig and Burnell, 1995), anti-tobacco CA (Ludwig et al., 1998), and affinity-purified anti-F. bidentis CA antisera. These antisera detected two to four immunoreactive polypeptides in F. bidentis leaf extracts, with differences in the labeling patterns likely due to differing titers and affinities of the individual anti-CA antibodies composing them. As N- terminal sequencing results indicated the 32- and 30-kD polypeptides detected with the anti-tobacco CA antiserum were both products of the CA3 gene, only two other polypeptides in F. bidentis leaf extracts appear to share significant cross-reactivity with the anti-CA antisera. Although we cannot yet say with absolute certainty that the CA1 and CA2 genes encode the other two immunoreactive polypeptides, taken together the above data do suggest that it is unlikely there are additional CA genes in the F. bidentis genome with high homology to CA1, CA2, and/or CA3.
Biochemical studies have shown that most of the CA activity in C4 plants is contained in the mesophyll cell cytosol where it catalyzes the conversion of atmospheric CO2 to HCO3−, which is used by PEPC, the primary carboxylase in the C4 photosynthetic pathway (Gutierrez et al., 1974; Ku and Edwards, 1975; Burnell and Hatch, 1988). Our immunocytochemical results also demonstrated a higher level of CA labeling in the cytosol than in other compartments of F. bidentis mesophyll cells. Import assays using isolated pea chloroplasts and in vitro transcribed and translated F. bidentis CA ORFs unequivocally showed that CA2 and CA3 do not localize to the chloroplast, confirming the cytosolic location suggested for CA3 by N-terminal amino acid sequencing and predicted for both isoforms by subcellular protein localization programs (Emanuelsson et al., 1999, 2000).
We recently proposed that F. bidentis CA3 was likely to be the isoform responsible for the hydration of CO2 in the mesophyll cell cytosol of this C4 plant (von Caemmerer et al., 2004). Using the CA3 ORF in an antisense construct to transform wild-type F. bidentis plants, we obtained transformants with severely impaired photosynthetic rates and a requirement for elevated CO2 concentrations for growth. However, because the ORFs of CA2 and CA3 share 60% nucleic acid sequence identity, we could not discount the possibility that a decrease in CA2 activity was responsible for, or contributed to, the antisense phenotype. Indeed, there was no strict correlation between the lack of CA activity and the abundance of individual CA isoforms in the transformants (von Caemmerer et al., 2004), and some of the transformants showed a reduction in CA2 transcripts as well as CA3 mRNA (S. Tanz and M. Ludwig, unpublished data). The qRT-PCR results presented here, however, make a strong case for the reduced activity of CA3 being responsible for the high CO2-requiring phenotype of the CA antisense plants. Steady-state levels of CA3 gene transcripts were highest in F. bidentis leaves and more than 50 times greater than the levels of CA1 and CA2 mRNAs in all the individual plants examined. Although we cannot say that CA transcript levels absolutely reflect the levels of CA protein, the affinity-purified anti-F. bidentis CA antiserum labeled the 32- and 30-kD CA3 polypeptides most intensely in F. bidentis leaf protein extracts.
The results of our Southern-blot analyses are suggestive of the presence of two CA3 genes in the F. bidentis genome. In the closely related C4 plant Flaveria trinervia, the ppcA gene family, which encodes the PEPC isoform important in the C4 pathway, consists of two genes and results from cDNA analyses indicate both these genes are expressed (Hermans and Westhoff, 1990; Poetsch et al., 1991). It has been suggested that the increased expression of C4 ppcA PEPC, which is clearly evident in F. trinervia (Hermans and Westhoff, 1990), is a simple way to overcome the decreased catalytic activity exhibited by this enzyme relative to nonphotosynthetic PEPCs (Bläsing et al., 2002). We have no evidence for two distinct CA3 transcripts in F. bidentis and thus we cannot rule out the possibility that some of the labeled fragments on our genomic blots may be derived from a pseudogene. However, if indeed the C4 photosynthetic pathway in Flaveria has evolved to contain two active CA3 genes, which may encode identical transcripts, a likely outcome of this would also be the mitigation of some of the inefficiency of the C4 primary carboxylase (PEPC) by ensuring that adequate concentrations of bicarbonate are available, especially when C4 cycle activity is high. CA3 antisense F. bidentis plants demonstrated the importance of ample bicarbonate levels for proper C4 pathway function (von Caemmerer et al., 2004). The CO2 hydration rates of these plants were compromised, which limited the availability of cytosolic bicarbonate and resulted in reduced CO2 assimilation rates relative to wild-type plants at the same intercellular CO2 partial pressure (von Caemmerer et al., 2004).
Regardless of the number of CA3 genes that are expressed in F. bidentis, taken together, the above data indicate that CA3 is the enzyme catalyzing the first step of the C4 photosynthetic pathway in the mesophyll cytosol and the activity of this isoform is essential for the proper functioning of the pathway. It is likely that high levels of CA3 are confined to the cytosol of C4 leaf mesophyll cells because we have shown, using a transgenic approach, that, although there is some endogenous CA activity in the bundle sheath of wild-type F. bidentis plants, increased levels of CA in the bundle-sheath cytosol leads to disruption of the C4 CCM (Ludwig et al., 1998).
Because CA2 is a cytosolic CA isoform that does not appear to be directly involved in C4 photosynthesis and because our qRT-PCR assays readily detected CA2 gene transcripts in all F. bidentis organs examined, it is very likely that this CA is the housekeeping form of the enzyme that is potentially found in all cell types. Nonphotosynthetic, cytosolic forms of PEPC require a supply of bicarbonate for anaplerotic processes, such as replenishment of tricarboxylic acid cycle intermediates, carbon skeletons for amino acid biosynthesis, seed maturation, and pH balance (Raven and Newman, 1994; Chollet et al., 1996), and CA2 is the likely supplier of this substrate in F. bidentis plants. In support of this role, CA2 gene transcripts were most prevalent in root tissues, being at least 20 times more abundant than CA1 or CA3 mRNAs in the three individual plants examined. However, no immunoreactive polypeptides were detected when F. bidentis root extracts were probed with the anti-F. bidentis CA3 antiserum. This is not surprising given the relative CA transcript abundance in this organ compared to that found in leaves and is in agreement with earlier studies that reported CA enzyme activity (Atkins, 1974; Reed and Graham, 1981) and polypeptides (Gálvez et al., 2000) are typically not detected in root extracts.
Steady-state CA1 mRNA levels were similar to those of CA2 transcripts in F. bidentis leaves, whereas in flowers they were the lowest of the three CA genes and, in root tissues, were just at the limit of detection of our qRT-PCR assay, which probably accounts for the variability in CA1 mRNA values seen in this organ among the individuals examined. The results of in vitro import assays demonstrated that CA1 was transcribed and translated as a precursor protein that was processed upon import into isolated pea chloroplasts and was then resistant to externally added protease. A chloroplast location for this isoform is consistent with the low-level CA labeling we detected in sections of F. bidentis mesophyll cell chloroplasts, as well as with predictions of intracellular location based on N-terminal sequence similarities with the chloroplast transit peptide of pea CA and subcellular protein localization prediction programs (Emanuelsson et al., 1999, 2000).
A comparative study examining the proteome of the chloroplast stroma from mesophyll cells and bscs of maize (Zea mays) failed to detect CA polypeptides (Majeran et al., 2005). Thus, as suggested by our qRT-PCR results for F. bidentis CA1, it appears that CA is not an abundant protein in the chloroplasts of C4 plants. This is in stark contrast to the situation in C3 species, where CA may make up 2% of the total soluble leaf protein (Okabe et al., 1984) and the majority of CA activity is localized to the chloroplast stroma of mesophyll cells (Poincelot, 1972; Jacobsen et al., 1975; Tsuzuki et al., 1985). Several roles have been suggested for the chloroplastic β-CAs of C3 plants, including supplying adequate levels of CO2 for Rubisco (Reed and Graham, 1981; Cowan, 1986; Price et al., 1994), lipid biosynthesis (Hoang and Chapman, 2002), and antioxidant activity with involvement in the hypersensitive defense response (Slaymaker et al., 2002). It is likely that CA1 is performing the two nonphotosynthetic roles in the chloroplasts of F. bidentis because little CA activity would be required for maintaining adequate supplies of CO2 around Rubisco in bundle-sheath chloroplasts because of the C4 CCM, and no Rubisco-mediated CO2 fixation occurs in C4 mesophyll cells. The detection of CA1 transcripts in F. bidentis roots and flowers, albeit at low levels, is also consistent with involvement of CA1 in lipid biosynthesis and a defense mechanism because these processes would also occur in nongreen plastids.
MATERIALS AND METHODS
Plant Growth
Flaveria bidentis ‘Kuntze’ plants were grown from cuttings or seed throughout the year in soil in a naturally lit glasshouse with day and night temperatures of 28°C and 20°C, respectively. A slow-release fertilizer was used and replenished every 6 months. Tissues used in the analyses included the first four sets of fully expanded leaves, flowers, and young lateral roots. Upon harvesting, tissues, except those for microscopic methods (see below), were immediately frozen in liquid N2 and then stored at −80°C until use.
Pea (Pisum sativum ‘Green Feast’) seeds were soaked in deionized water for 2 h and then planted in vermiculite (grade 3; The Perlite & Vermiculite Company). Plants were maintained in controlled-environment chambers (Thermoline Scientific Equipment) at 24°C, with 65% relative humidity on a 16-h light/8-h dark cycle with a light intensity of 700 μmol m−2 s−1. Plants were watered daily and leaves from 10-d-old plants were harvested and used immediately for in vitro import assays.
Isolation of cDNAs Encoding F. bidentis CA Isoforms
Fragments encoding a partial CA2 cDNA and what appeared to be the full-length CA3 cDNA were initially isolated from a F. bidentis leaf cDNA library constructed in λgt11 using a radiolabeled fragment encoding F. bidentis CA1 as described previously (Cavallaro et al., 1994).
To isolate the 5′ end of the F. bidentis CA2 cDNA and to confirm the 5′ sequences of CA1 and CA3 cDNAs from this species, an adaptor-ligated cDNA library was constructed. mRNA was isolated from F. bidentis leaves as described previously (Ludwig and Burnell, 1995). Synthesis of cDNA and the addition of adaptors were carried out using the Marathon cDNA amplification kit (CLONTECH). The 5′ ends of the cDNAs were amplified using the 5′-RACE method described by the manufacturer (CLONTECH) in combination with one of the following CA gene-specific primers: CA1, 5′-GGAGCTTTGGTGTCGGGTGCTGCGG; CA2, 5′-TTTAAACTCACCGGCGTCTGACACCTCC; CA3, 5′-GGTCAAATCCGGGTTTGGTACTGTCAAG. AmpliTaq Gold buffer II (Applied Biosystems) and 1 unit AmpliTaq Gold DNA polymerase were used in each reaction.
The veracity of all the CA cDNA sequences was confirmed by determining the sequence of several RT-PCR products derived from the mRNA of several individual F. bidentis plants.
Genomic Southern Blots
F. bidentis genomic DNA was extracted from 2.5 g of leaf tissue as described by Marshall et al. (1996) and 20-μg aliquots were digested with restriction enzymes following standard protocols (Sambrook et al., 1989). Genomic DNA fragments were separated on 0.8% (w/v) agarose gels and blotted to Hybond N+ (GE Healthcare Life Sciences) as described by Sambrook et al. (1989). Prehybridization and hybridization were carried out in 5× SSC containing 5× Denhardt's reagent, 0.5% (w/v) SDS, and 100 μg/mL fragmented and denatured salmon sperm DNA at 65°C (Sambrook et al., 1989). Blots were washed twice in 2× SSC containing 0.1% (w/v) SDS at room temperature, twice in the same buffer at 65°C for 15 min, and twice in 1× SSC containing 0.1% (w/v) SDS at 65°C for 15 min.
Probes encoding an exon from F. bidentis CA3 (CA3 cDNA nucleotides 280–397) that showed 87% and 75% identity to the corresponding regions of F. bidentis CA1 and CA2, respectively, and the full-length ORFs of F. bidentis CA1, CA2, and CA3 were generated by PCR and labeled with [α-32P]dATP and random primers (DECAprime II; Ambion).
qRT-PCR
A standard template for qRT-PCR was constructed by inserting fragments encoding the ORFs of F. bidentis CA1, CA2, and CA3 in tandem into pBluescript II KS− (Stratagene) using established methods (Sambrook et al., 1989). The fragments were generated using RT-PCR and the following primer pairs: CA1 forward, 5′-TAGAAAGCAATTCTGCAGACATCGAAC and CA1 reverse, 5′-GGTCATACAGAGGTAGGAGACG; CA2 forward, 5′-GTGGTCGAAATGCTGCAGAATTGTATC and CA2 reverse, 5′-GGTTGGGTGATGAATGAAAGTTC; CA3 forward, 5′-ACTATCTTTTGCAGAGCTCAGACATGG and CA3 reverse, 5′-ACTCGATTATGCCCGGGTAGTAGGTG. To facilitate the cloning steps, PstI sites were introduced into the CA1 and CA2 forward primers, whereas SacI and SmaI sites were introduced into the CA3 forward and reverse primers, respectively. Total RNA was extracted from F. bidentis leaf tissue (Perfect RNA mini kit; Eppendorf) and cDNA synthesis was carried out at 42°C for 60 min in 20-μL reactions of 50 mm Tris-HCl, pH 8.3, containing 1 to 5 μg RNA, 1 mm dNTPs, 2.5 μm oligo(dT)15 primer, 15 mm dithiothreitol (DTT), 20 units RNase inhibitor (RNasin; Promega), 20 units Moloney murine leukemia virus reverse transcriptase (RNase H−; Promega), 75 mm KCl, and 3 mm MgCl2. Aliquots (2 μL) of the RT reaction were used in the PCRs along with the MasterTaq kit (Eppendorf). Amplified fragments were purified using the Wizard PCR Preps DNA purification system (Promega) before restriction enzyme digestion and insertion into pBluescript II KS−.
cDNA templates for qRT-PCR were synthesized from F. bidentis leaf, root, and flower total RNA as described above, except that, prior to the RT reaction, the RNA was digested with DNase (RQ1 RNase-free DNAse; Promega), according to the manufacturer's instructions, to remove genomic DNA. The following gene-specific primers were used to amplify regions in the 3′ halves of F. bidentis CA cDNAs, yielding fragments that ranged in size from 232 to 275 bp and could be distinguished easily from one another by agarose gel electrophoresis. CA1 forward, 5′-GTCCCTCCCTTTGACAAGACC and CA1 reverse, 5′-GTTCACAGCTTCCTTTTCACATAG; CA2 forward, 5′-ATTGCAGCGATTTGGAGTACTCG and CA2 reverse, 5′-GGTTGGGTGATGAATGAAAGTTC; CA3 forward, 5′-GAGCACCGACTTCATTGAGGAC and CA3 reverse, 5′-GGTGAAAGGCTGAATTCAAGCG.
Prior to qPCR, the integrity of all cDNA templates was tested in standard PCRs using the gene-specific primers described above. Control reactions included aliquots of the RNA preparations before and after DNase digestion and a no-template control. PCR conditions were 1 cycle of 95°C for 10 min; 35 cycles of 94°C for 15 s; 56°C for 45 s; 72°C for 1.5 min; and one cycle of 72°C for 7 min.
For q assays, the standard template was linearized with SmaI and CA1, CA2, and CA3 standard curves were generated using either four or five 10-fold dilutions of the template with the appropriate primer pair. Amplification of the standard template dilutions and cDNA templates from F. bidentis leaves, roots, and flowers was done using SYBR Green (QuantiTect SYBR Green PCR kit; Qiagen) with an activation cycle of 95°C for 15 min followed by 35 cycles of 95°C for 15 s, 64°C for 30 s, 72°C for 30 s. The specificity and fidelity of the PCRs were monitored by melting curve analyses and agarose gel electrophoresis of the products.
cDNA Sequence Determination and Analysis
Plasmid DNA was prepared by standard methods (Sambrook et al., 1989). Sequences were determined using dye terminator cycle sequencing (Applied Biosystems) at the Macquarie University DNA Analysis Facility, Macquarie University, or the Western Australian Genome Resource Centre, Royal Perth Hospital. Sequences were analyzed with the MacVector and AssemblyLIGN programs designed for MacIntosh (Accelrys). Multiple sequences alignments were done using ClustalW (Thompson et al., 1994) and subcellular protein localization was predicted using TargetP (Emanuelsson et al., 2000) and ChloroP (Emanuelsson et al., 1999).
Isolation of CA-Enriched F. bidentis Leaf Fractions and N-Terminal Amino Acid Sequencing
All procedures were carried out at 4°C unless otherwise noted. Approximately 100 g of F. bidentis leaf tissue were ground with a mortar and pestle in 50 mm HEPES-KOH, pH 7.5, containing 10 mm MgSO4, 5 mm DTT, 1 mm EDTA, 2.5% (w/v) polyvinylpolypyrollidone, 1 mm phenylmethylsulfonyl fluoride, and 0.1% (v/v) Triton X-100. Following filtration through Miracloth (CalBiochem) and centrifugation at 20,000g for 30 min, polyethylene glycol (Mr 8,000) was added to the supernatant to a final concentration of 7% (w/v). Precipitated protein was collected by centrifugation (as above), resuspended in column buffer (25 mm HEPES-KOH, pH 7.5, 5 mm DTT, 10 mm MgSO4, 1 mm EDTA) containing 1% (v/v) Triton X-100, incubated overnight, and then clarified by centrifugation (as above). The supernatant was loaded on a DEAE-cellulose 52 column and washed with 200-mL column buffer. Protein was eluted from the column with a linear gradient of 0 to 400 mm NaCl and fractions were assayed for CA activity as described previously (Burnell and Hatch, 1988). Proteins in fractions with peak activity were concentrated by acetone precipitation, resolved in duplicate by SDS-PAGE on 12% (w/v) gels (Laemmli, 1970) at room temperature, and transferred to polyvinylidene difluoride membrane using semidry transfer as described previously (Ludwig et al., 1998). CA isoforms were detected on one of the blots with an anti-tobacco (Nicotiana tabacum) CA antiserum (Ludwig et al., 1998) at room temperature, whereas the other blot was stained with 0.01% (w/v) Coomassie Blue R250 in 50% (v/v) methanol. Regions of the Coomassie-stained blot corresponding to the positions of the CA isoforms on the immunoblot were excised and submitted for N-terminal amino acid sequencing by the Biomolecular Resource Facility at the John Curtain School of Medical Research, Australian National University.
In Vitro Import Assays
Intact chloroplasts were isolated from 50 g of 10-d-old pea leaves according to Bruce et al. (1994) and purified by a 50% continuous Percoll gradient using standard procedures (Waegemann and Soll, 1996). Chlorophyll concentration was determined according to Arnon (1949) and calculated using the following equation: chlorophyll concentration (μg mL−1) = [(A645 × 202) + (A663 × 80.2)] × 10.5.
Individual plasmids containing inserts encoding the ORFs of F. bidentis CA1, CA2, CA3, or the SSU of pea Rubisco were used to synthesize precursor proteins in the presence of [35S]-Met in a coupled transcription/translation rabbit reticulate lysate system (TNT; Promega) according to the manufacturer's instructions. Precursor proteins were stored at −80°C until use in import assays.
Import assays contained chloroplasts equivalent to 25 μg chlorophyll, 10 μL of radiolabeled precursor protein, import buffer (50 mm HEPES-KOH, pH 8.0, 330 mm sorbitol), 6.25 mm MgCl2, 6.25 mm ATP, 6.25 mm Met, 15.6 mm potassium acetate, and 15.6 mm sodium bicarbonate. After incubation for 25 min at 25°C in the light, reactions were transferred to 4°C and divided into two equal aliquots. EDTA was added to one aliquot to a final concentration of 10 mm, whereas the other aliquot was treated at 4°C for 30 min with thermolysin (120 mg/mL) supplemented with 0.1 mm CaCl2, after which EDTA was added to a final concentration of 10 mm. Chloroplasts were collected by centrifugation at 830g for 2 min and lysed by boiling for 5 min in SDS-PAGE sample buffer (62.5 mm Tris-HCl, pH 6.8, 2% [w/v] SDS, 10% [v/v] glycerol, 0.25% [w/v] bromphenol blue, 10% [v/v] β-mercaptoethanol). Chloroplast proteins were separated on denaturing 12% (w/v) polyacrylamide gels (Laemmli, 1970). Labeled proteins were detected by exposing dried gels to a BAS TR2040 phosphoimaging plate (Fuji) for 24 h followed by imaging with a BAS2500 phosphoimager (Fuji). Images were analyzed using ImageGauge, version 3.0, software and adjusted using Adobe Photoshop CS software.
Generation and Affinity Purification of an Anti-F. bidentis CA3 Antiserum
The cDNA encoding the ORF of F. bidentis CA3 was inserted into pTrcHisB (Invitrogen) and this construct was used to generate a His6-F. bidentis CA3 fusion protein in Escherichia coli DH10B cells. Fusion protein, purified from inclusion bodies using the B-PER II reagent (Pierce) and HiTrap chelating HP columns (GE Healthcare Life Sciences) charged with Ni2+ ions, served as the antigen to generate a rabbit anti-F. bidentis CA3 antiserum (Institute of Medical and Veterinary Science). The antiserum was affinity purified using a column (NHS-activated HiTrap HP column; GE Healthcare Life Sciences) to which the purified His6-F. bidentis CA3 fusion protein was bound.
Immunoblotting
Immunodetection of CA in protein extracts of F. bidentis leaf tissue was done as described previously (Ludwig et al., 1998), except a 12% (w/v) polyacrylamide gel and a 1/3,000 dilution of the affinity-purified anti-F. bidentis CA3 antibody were used.
Microscopy
F. bidentis leaf tissue was fixed overnight at 4°C in 4% (w/v) paraformaldehyde and 0.5% (v/v) glutaraldehyde in 0.1 m PIPES, pH 7.2. Tissue was then rinsed in 0.1 m PIPES, pH 7.2, dehydrated through a graded ethanol series, and infiltrated with LR White resin (ProSciTech):ethanol mixtures of 1:3 (v/v), 1:2 (v/v), 1:1 (v/v), 2:1 (v/v), followed by three changes of LR White resin. All dehydration and infiltration steps were done at 4°C. Tissue was embedded in LR White resin by polymerization at 60°C for 24 h.
For anatomical studies, semithin sections (1 μm) of F. bidentis leaf tissue were cut with a Reichert Ultracut microtome (Leica), fixed onto microscope slides by heating for 2 min at 80°C, stained with 1% (w/v) methylene blue in 40% (v/v) glycerol and 1% (w/v) sodium bicarbonate, and then mounted in Biomount (ProSciTech). Sections were examined at the Microscopy Unit, Macquarie University, with a BH2-RFCA Olympus microscope (Olympus Australia) and images were captured using a Sony DFW-X700 digital camera (Sony Australia) and BTV Pro, version 5.41, image capture software (Ben Bird).
For immunocytochemistry, ultrathin sections (50 nm) were cut with a microtome (Leica) and mounted on 0.1% (w/v) poly-l-Lys-coated microscope slides or 0.2% (w/v) piloform-coated nickel grids. All labeling steps were done at 25°C in a humid chamber. For immunofluorescence, sections on slides were blocked in phosphate-buffered saline (PBS) containing 10% (w/v) fetal bovine serum for 30 min, labeled with a 1/50 dilution of the affinity-purified anti-F. bidentis CA3 antiserum in PBS for 1 h, and then incubated in 5 μg mL−1 AlexaFluor 488-conjugated goat anti-rabbit antibody (Invitrogen) in PBS for 1 h in the dark. Sections were mounted in Gel/Mount (ProSciTech) and examined, with images captured, as described above. For immunoelectron microscopy, sections were incubated in 50 mm Gly in PBS for 15 min followed by 30 min in PBS containing 5% (w/v) bovine serum albumin and 5% (w/v) fetal bovine serum (w/v). After rinsing in PBS containing 0.1% (w/v) bovine serum albumin, sections were labeled with the anti-F. bidentis CA3 antiserum, as described for immunofluorescence, followed by a 1/50 dilution of a goat anti-rabbit IgG antibody conjugated to 5-nm gold particles (ProSciTech) in PBS for 1 h. Antigen-antibody complexes were stabilized with 2% (v/v) glutaraldehyde in PBS for 5 min and sections were then stained with 2% aqueous uranyl acetate for 5 min and lead citrate for 2 min. Sections were examined and photographed with a Philips CM10 transmission electron microscope (Microscopy Unit, Macquarie University).
All images were compiled and adjusted using ImageJ, version 1.30 (National Institutes of Health) and Adobe Photoshop, version 5.0 (Adobe Systems).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers U08398, AY167112, and AY167113 for F. bidentis CA1, CA2, and CA3, respectively.
Acknowledgments
We thank Dr. G. Dean Price from the Research School of Biological Sciences, Australian National University, Canberra, Australian Capital Territories, Australia, for the generous gift of the anti-tobacco CA antiserum and Prof. J. Whelan from the School of Biomedical, Biomolecular, and Chemical Sciences, University of Western Australia, Crawley, Western Australia, Australia, for the pea Rubisco SSU clone. We are also very grateful to Nancy Hancock and Debra Birch (Department of Biological Sciences, Macquarie University, Sydney) for excellent technical assistance.
This work was supported by the Australian Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Martha Ludwig (mludwig@cyllene.uwa.edu.au).
Open Access articles can be viewed online without a subscription.
References
- Arnon DI (1949) Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol 24 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins C, Smith P, Mann A, Thumfort P (2001) Localization of carbonic anhydrase in legume nodules. Plant Cell Environ 24 317–326 [Google Scholar]
- Atkins CA (1974) Occurrence and some properties of carbonic anhydrases from legume root nodules. Phytochemistry 13 93–98 [Google Scholar]
- Bläsing OE, Ernst K, Streubel M, Westhoff P, Svensson P (2002) The non-photosynthetic phosphoenolpyruvate carboxylases of the C4 dicot Flaveria trinervia—implications for the evolution of C4 photosynthesis. Planta 215 448–456 [DOI] [PubMed] [Google Scholar]
- Bracey MH, Christiansen J, Tovar P, Cramer SP, Bartlett SG (1994) Spinach carbonic anhydrase: investigation of the zinc-binding ligands by site-directed mutagenesis, elemental analysis, and EXAFS. Biochemistry 33 13126–13131 [DOI] [PubMed] [Google Scholar]
- Bruce BD, Perry S, Froelich J, Keegstra K (1994) In vitro import of proteins into chloroplasts. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp J1–J15
- Burnell JN (2000) Carbonic anhydrases of higher plants: an overview. In WR Chegwidden, ND Cater, YH Edwards, eds, The Carbonic Anhydrases. Birkhauser Verlag, Basel, pp 501–518 [DOI] [PubMed]
- Burnell JN, Hatch MD (1988) Low bundle sheath carbonic anhydrase is apparently essential for effective C4 pathway operation. Plant Physiol 86 1252–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallaro A, Ludwig M, Burnell J (1994) The nucleotide sequence of a complementary DNA encoding Flaveria bidentis carbonic anhydrase. FEBS Lett 350 216–218 [DOI] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O'Leary MH (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 47 273–298 [DOI] [PubMed] [Google Scholar]
- Coba de la Peña T, Frugier F, McKhann HI, Bauer P, Brown S, Kondorosi A, Crespi M (1997) A carbonic anhydrase gene is induced in the nodule primordium and its cell-specific expression is controlled by the presence of Rhizobium during development. Plant J 11 407–420 [DOI] [PubMed] [Google Scholar]
- Coleman JR (2000) Carbonic anhydrase and its role in photosynthesis. In RC Leegood, TD Sharkey, S von Caemmerer, eds, Advances in Photosynthesis, Vol 9. Photosynthesis: Physiology and Metabolism. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 353–367
- Cowan IR (1986) Economics of carbon fixation in higher plants. In TJ Givnish, ed, On the Economy of Plant Form and Function. Cambridge University Press, Cambridge, UK, pp 133–170
- Demir N, Demir Y, Yildirim A (1997) Carbonic anhydrases from leaves and roots of Daucus carota. Phytochemistry 44 1247–1250 [Google Scholar]
- Edwards GE, Ku MSB (1987) Biochemistry of C3-C4 intermediates. In MD Hatch, NK Boardman, eds, The Biochemistry of Plants: A Comprehensive Treatise, Vol 10. Photosynthesis. Academic Press, New York, pp 275–325
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemetakis E, Dimou M, Cotzur D, Aivalakis G, Efrose RC, Kenoutis C, Udvardi M, Katinakis P (2003) A Lotus japonicus β-type carbonic anhydrase gene expression pattern suggests distinct physiological roles during nodule development. Biochim Biophys Acta 1628 186–194 [DOI] [PubMed] [Google Scholar]
- Gálvez S, Hirsch AM, Wycoff KL, Hunt S, Layzell DB, Kondorosi A, Crespi M (2000) Oxygen regulation of a nodule-located carbonic anhydrase in alfalfa. Plant Physiol 124 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M, Huber SC, Ku SB, Kanai R, Edwards GE (1974) Intracellular localization of carbon metabolism in mesophyll cells of C4 plants. In M Avron, ed, Proceedings of the Third International Congress on Photosynthesis. Elsevier Science Publishers, Amsterdam, pp 1219–1230
- Hatch MD, Burnell JN (1990) Carbonic anhydrase activity in leaves and its role in the first step of C4 photosynthesis. Plant Physiol 93 825–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans J, Westhoff P (1990) Analysis of expression and evolutionary relationships of phosphoenolpyruvate carboxylase genes in Flaveria trinervia (C4) and F. pringlei (C3). Mol Gen Genet 224 459–468 [DOI] [PubMed] [Google Scholar]
- Hewett-Emmett D, Tashian RE (1996) Functional diversity, conservation, and convergence in the evolution of the α-, β-, and γ-carbonic anhydrase gene families. Mol Phylogenet Evol 5 50–77 [DOI] [PubMed] [Google Scholar]
- Hoang CV, Chapman KD (2002) Biochemical and molecular inhibition of plastidial carbonic anhydrase reduces the incorporation of acetate into lipids in cotton embryos and tobacco cell suspensions and leaves. Plant Physiol 128 1417–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang CV, Wessler HG, Local A, Turley RB, Benjamin RC, Chapman KD (1999) Identification and expression of cotton (Gossypium hirsutum L) plastidial carbonic anhydrase. Plant Cell Physiol 40 1262–1270 [DOI] [PubMed] [Google Scholar]
- Jacobsen BS, Fong F, Heath RL (1975) Carbonic anhydrase of spinach: studies on its location, inhibition, and physiological function. Plant Physiol 55 468–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavroulakis N, Flemetakis E, Aivalakis G, Katinakis P (2000) Carbon metabolism in developing soybean root nodules: the role of carbonic anhydrase. Mol Plant Microbe Interact 13 14–22 [DOI] [PubMed] [Google Scholar]
- Kimber MS, Pai EF (2000) The active site architecture of Pisum sativum β-carbonic anhydrase is a mirror image of that of α-carbonic anhydrases. EMBO J 19 1407–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku SB, Edwards GE (1975) Photosynthesis in mesophyll protoplasts and bundle sheath cells of various C4 plants. V. Enzymes of respiratory metabolism and energy utilizing enzymes of photosynthetic pathways. Z Pflanzenphysiol 77 16–32 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lane TW, Morel FMM (2000) Regulation of carbonic anhydrase expression by zinc, cobalt, and carbon dioxide in the marine diatom Thalassiosira weissflogii. Plant Physiol 123 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane TW, Saito MA, George GN, Pickering IJ, Prince RC, Morel FMM (2005) A cadmium enzyme from a marine diatom. Nature 435 42. [DOI] [PubMed] [Google Scholar]
- Liljas A, Laurberg M (2000) A wheel invented three times. EMBO Rep 1 16–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M, Burnell JN (1995) Molecular comparison of carbonic anhydrase from Flaveria species demonstrating different photosynthetic pathways. Plant Mol Biol 29 353–365 [DOI] [PubMed] [Google Scholar]
- Ludwig M, von Caemmerer S, Price GD, Badger MR, Furbank RT (1998) Expression of tobacco carbonic anhydrase in the C4 dicot Flaveria bidentis leads to increased leakiness of the bundle sheath and a defective CO2-concentrating mechanism. Plant Physiol 117 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W, Cai Y, Sun Q, van Wijk KJ (2005) Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17 3111–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JS, Stubbs JD, Taylor WC (1996) Two genes encode highly similar chloroplastic NADP-malic enzymes in Flaveria. Plant Physiol 111 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Bartlett SG, Samuelsson G (2001) Carbonic anhydrase in plants and algae. Plant Cell Environ 24 141–153 [Google Scholar]
- Okabe K, Yang SY, Tsuzuki M, Miyachi S (1984) Carbonic anhydrase: its content in spinach leaves and its taxonomic diversity studied with anti-spinach leaf carbonic anhydrase antibody. Plant Sci Lett 33 145–153 [Google Scholar]
- Parisi G, Perales M, Fornasari MS, Colaneri A, González-Schain N, Gómez-Casati D, Zimmermann S, Brennicke A, Araya A, Ferry JG, et al (2004) Gamma carbonic anhydrases in plant mitochondria. Plant Mol Biol 55 193–207 [DOI] [PubMed] [Google Scholar]
- Poetsch W, Hermans J, Westhoff P (1991) Multiple cDNAs of phosphoenolpyruvate carboxylase in the C4 dicot Flaveria trinervia. FEBS Lett 292 133–136 [DOI] [PubMed] [Google Scholar]
- Poincelot RP (1972) Intracellular distribution of carbonic anhydrase in spinach leaves. Biochim Biophys Acta 258 637–642 [DOI] [PubMed] [Google Scholar]
- Price GD, von Caemmerer S, Evans JR, Yu JW, Lloyd J, Oja V, Kell P, Harrison K, Gallagher A, Badger MR (1994) Specific reduction of chloroplast carbonic anhydrase activity by antisense RNA in transgenic tobacco plants has a minor effect on photosynthesis. Planta 193 331–340 [Google Scholar]
- Provart NJ, Majeau N, Coleman JR (1993) Characterization of pea chloroplastic carbonic anhydrase: expression in Escherichia coli and site-directed mutagenesis. Plant Mol Biol 22 937–943 [DOI] [PubMed] [Google Scholar]
- Raven JA, Newman JR (1994) Requirement for carbonic anhydrase activity in processes other than photosynthetic inorganic carbon assimilation. Plant Cell Environ 17 123–130 [Google Scholar]
- Reed ML (1979) Intracellular location of carbonate dehydratase (carbonic anhydrase) in leaf tissue. Plant Physiol 63 216–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed ML, Graham D (1981) Carbonic anhydrase in plants: distribution, properties and possible physiological roles. In L Reinhold, JB Harborne, T Swain, eds, Progress in Biochemistry, Vol 7. Pergamon Press, Oxford, pp 47–94
- Roeske CA, Ogren WL (1990) Nucleotide sequence of pea cDNA encoding chloroplast carbonic anhydrase. Nucleic Acids Res 18 3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sawaya MR, Cannon GC, Heinhorst S, Tanaka S, Williams EB, Yeates TO, Kerfeld CA (2006) The structure of β-carbonic anhydrase from the carboxysome shell reveals a distinct subclass with one active site for the price of two. J Biol Chem 281 7546–7555 [DOI] [PubMed] [Google Scholar]
- Slaymaker DH, Navarre DA, Clark D, del Pozo O, Martin GB, Klessig DF (2002) The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc Natl Acad Sci USA 99 11640–11645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AKC, Espie GS, Williams EB, Shively JM, Heinhorst S, Cannon GC (2004) A novel evolutionary lineage of carbonic anhydrase (ɛ class) is a component of the carboxysome shell. J Bacteriol 186 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp BC, Smith K, Ferry JG (2001) Carbonic anhydrase: new insights for an ancient enzyme. J Biol Chem 276 48615–48618 [DOI] [PubMed] [Google Scholar]
- Tsuzuki M, Miyachi S, Edwards GE (1985) Localization of carbonic anhydrase in mesophyll cells of terrestrial C3 plants in relation to CO2 assimilation. Plant Cell Physiol 26 881–891 [Google Scholar]
- von Caemmerer S, Furbank RT (2003) The C4 pathway: an efficient CO2 pump. Photosynth Res 77 191–207 [DOI] [PubMed] [Google Scholar]
- von Caemmerer S, Ludwig M, Millgate A, Farquhar GD, Price D, Badger M, Furbank RT (1997) Carbon isotope discrimination during C4 photosynthesis: insights from transgenic plants. Aust J Plant Physiol 24 487–494 [Google Scholar]
- von Caemmerer S, Quinn V, Hancock N, Price GD, Furbank RT, Ludwig M (2004) Carbonic anhydrase and C4 photosynthesis: a transgenic analysis. Plant Cell Environ 27 697–703 [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG (1989) Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem 180 535–545 [DOI] [PubMed] [Google Scholar]
- Waegemann K, Soll J (1996) Phosphorylation of the transit sequence of chloroplast precursor proteins. J Biol Chem 271 6545–6554 [DOI] [PubMed] [Google Scholar]