Abstract
The effector protein NIP1 from the barley (Hordeum vulgare) pathogen Rhynchosporium secalis specifically induces the synthesis of defense-related proteins in cultivars of barley expressing the complementary resistance gene, Rrs1. In addition, it stimulates the activity of the barley plasma membrane H+-ATPase in a genotype-unspecific manner and it induces necrotic lesions in leaf tissues of barley and other cereal plant species. NIP1 variants type I and II, which display quantitative differences in their activities as elicitor and H+-ATPase stimulator, and the inactive mutant variants type III* and type IV*, were produced in Escherichia coli. Binding studies using 125I-NIP1 type I revealed a single class of binding sites with identical binding characteristics in microsomes from near-isogenic resistant (Rrs1) and susceptible (rrs1) barley. Binding was specific, reversible, and saturable, and saturation ligand-binding experiments yielded a Kd of 5.6 nm. A binding site was also found in rye (Secale cereale) and the nonhost species wheat (Triticum aestivum), oat (Avena sativa), and maize (Zea mays), but not in Arabidopsis (Arabidopsis thaliana). For NIP1 types I and II, equilibrium competition-binding experiments revealed a correlation between the difference in their affinities to the binding site and the differences in their elicitor activity and H+-ATPase stimulation, indicating a single target molecule to mediate both activities. In contrast, the inactive proteins type III* and type IV* are both characterized by high affinities similar to type I, suggesting that binding of NIP1 to this target is not sufficient for its activities.
All classes of microorganisms—fungi, oomycetes, bacteria, viruses, and nematodes—contain members that have evolved to exploit living plants as a nutrient source. Despite the vast differences in their lifestyles, these plant pathogens share a common dilemma: They need to physically interact with the host plant, thereby generating wounds and releasing components from the plant surface, which allow the plant to recognize the invasion. Furthermore, exposed microbial surface structures and essential components, such as the major protein of bacterial flagellae, flagellin (Gomez-Gomez and Boller, 2002), or the outermembrane lipooligosaccharides of gram-negative bacteria (Gerber and Dubery, 2004; Gross et al., 2005), can serve as vehicles to launch the plant defense response. These molecules possess crucial basic functions for the microbe without being specifically intended for the interaction and have been termed microbe- or pathogen-associated molecular patterns (MAMPs/PAMPs; Nürnberger and Lipka, 2005). Their recognition by plant pattern recognition receptors results in MAMP/PAMP-triggered immunity (Jones and Dangl, 2006). To use a plant as a host, microbes therefore must evolve strategies to avoid this type of recognition or to suppress its consequences. Furthermore, to optimize their nutritional situation, microbes usually produce and secrete effector molecules or even transfer them into the host cytoplasm to manipulate host physiology in their favor, thereby causing effector-triggered susceptibility (Jones and Dangl, 2006). However, this again means leaving cover and running the risk of being picked up by the plant surveillance system through the products of plant resistance (R) genes. Hence, those effectors that are meant to serve the microbe as virulence factors may be turned into specific recognition factors. They now function as avirulence (Avr) determinants for the host (Luderer and Joosten, 2001; van't Slot and Knogge, 2002), mediating effector-triggered immunity (Jones and Dangl, 2006). Genetically, this latter scenario has been described as a gene-for-gene interaction in which plant R genes meet complementary pathogen Avr genes to cause disease resistance (Flor, 1971). Detailed analyses of R-Avr gene interactions in several pathosystems have suggested that, in the majority of cases, this recognition is not based on a simple receptor-ligand model (Baker et al., 1997). Instead, pathogen effectors interact with specific plant virulence targets, thereby inducing conformational changes, which are detected by the R proteins (guard hypothesis; van der Hoorn et al., 2002; Jones and Dangl, 2006). As a consequence, a signaling process is initiated, which leads to plant defense (Baker et al., 1997; Dangl and Jones, 2001).
Many pathogen Avr factors have been detected through their activity as elicitors of plant resistance, which in most cases is manifested as a hypersensitive response. However, the portion of these factors, for which virulence function was later identified, is increasing, particularly in bacterial pathosystems (Luderer and Joosten, 2001; van't Slot and Knogge, 2002). The plant-fungus pathosystem, where this has been best studied at the molecular level, comprises tomato (Solanum lycopersicum) and the leaf mold pathogen Cladosporium fulvum (Joosten and de Wit, 1999; Rivas and Thomas, 2005). Tomato Cf genes conferring resistance to races of C. fulvum encode transmembrane proteins with extensive extracellular Leu-rich repeat domains and very short cytoplasmic tails, suggesting extracellular interaction with the corresponding Avr determinants (Jones and Jones, 1997; Takken and Joosten, 2000). However, Avr proteins AVR2, AVR4, and AVR9 do not bind to their R protein counterparts. AVR2 binds and inhibits the extracellular tomato Cys protease Rcr3, and the enzyme-AVR2 complex is proposed to activate Cf-2-dependent resistance (Rooney et al., 2005). AVR4 was found to bind very efficiently to chitin, thus protecting fungal cell walls from degradation by plant chitinases (Westerink et al., 2002; van den Burg et al., 2003, 2004), but it remains unclear how the Cf-4 protein is activated. The intrinsic function of AVR9, the first Avr protein identified in a fungal pathogen, is not known. However, the peptide shows structural homology to the potato (Solanum tuberosum) carboxy peptidase inhibitor (Vervoort et al., 1997). Extensive studies have failed to demonstrate direct interaction with the matching Cf-9 protein (Luderer et al., 2001). Furthermore, plasma membranes from both resistant (Cf-9) and susceptible (Cf-0) plants, as well as from other solanaceous species, displayed high-affinity AVR9-binding sites with similar properties (Kooman-Gersmann et al., 1996). As with AVR2 and AVR4, the corresponding R gene, Cf-9, therefore does not encode the primary AVR9-binding site. Instead, the proposed models for the AVR9-induced resistance response assume the formation of a heterotrimeric signal perception complex involving a nonspecific primary receptor, the ligand AVR9, and a specific signal transducer to initiate the signaling process in resistant (Cf-9) plants that leads to defense activation (Kooman-Gersmann et al., 1998; Joosten and de Wit, 1999; Luderer et al., 2001).
Tomato Cf gene products are transmembrane proteins. In contrast, the majority of plant R genes characterized to date encode cytoplasmic proteins, suggesting they guard intracellular targets (Takken and Joosten, 2000). Gram-negative phytopathogenic bacteria possess a type III secretion system that mediates the direct delivery of bacterial effector proteins into the host cytoplasm (Galán and Collmer, 1999; Bonas and Lahaye, 2002). For fungal phytopathogens, no such mechanism has been identified to date. Nevertheless, evidence for intracellular recognition of Avr proteins by plant R proteins does exist in fungi and oomycetes. The Avr gene, AVR-Pita, from the rice (Oryza sativa) blast fungus, Magnaporthe grisea (Orbach et al., 2000), encodes a predicted secreted metalloprotease that causes a hypersensitive response in the presence of the intracellular rice R gene product, Pi-ta (Bryan et al., 2000; Jia et al., 2000). Direct interaction between AVR-Pita and Pi-ta was observed using the yeast (Saccharomyces cerevisiae) two-hybrid system and far-western analysis (Jia et al., 2000). In addition, proteolytic cleavage of the R protein may be crucial for Avr function because mutations in the putative protease active site render the protein inactive as a defense elicitor. The products of the Avr genes AvrL567, AvrM, and AvrP4 from the flax (Linum usitatissimum) rust Melampsora lini, when lacking their predicted signal sequence, are recognized inside plant cells when they are coexpressed in planta with the respective R genes (Dodds et al., 2004; Catanzariti et al., 2006). Likewise, expression of the Avr3a gene from Phytophthora infestans in its host plant potato carrying the R gene R3a, as well as coexpression of both genes in Nicotiana benthamiana, resulted in a resistance response after removal of the signal peptide-encoding sequence, suggesting cytoplasmic interaction (Armstrong et al., 2005). Recently, a host-targeting signal with an RxLx(E, D, or Q) core motif was identified in the leader sequence of the Avr3a protein, as well as of other secretory oomycete proteins (Bhattacharjee et al., 2006). This motif was originally defined to enable translocation of effector proteins from the Malaria parasite Plasmodium falciparum across mammalian membranes (Hiller et al., 2004; Marti et al., 2004). Evidence for the existence of a mechanism in the host plasma membrane for the translocation of secreted fungal effector proteins also comes from work on proteins expressed in haustoria of two related rust fungi, Uromyces fabae and Uromyces striatus (Kemen et al., 2005). Using polyclonal antisera, the functionally unknown protein RTP1p was also found inside the cells of the respective hosts, broad bean (Vicia faba) and alfalfa (Medicago sativa). For several of the effector proteins, it was shown that the pathogen is not required for host cell entrance, suggesting a host plant-encoded transport mechanism (Catanzariti et al., 2006). In this context, it may be noteworthy that sensitivity of wheat (Triticum aestivum) toward the intracellularly acting protein toxin Ptr ToxA from Pyrenophora tritici-repentis appears to be mediated through protein import, not through an intracellular target (Manning and Ciuffetti, 2005).
Interaction between the imperfect fungus Rhynchosporium secalis, the causal agent of barley (Hordeum vulgare) scald, and its host plant also complies with the gene-for-gene scheme (Knogge and Marie, 1997). The Avr gene, AvrRrs1, from R. secalis is one of the still limited number of fungal Avr genes characterized to date. After removal of a 22-amino acid signal peptide, the mature gene product, termed NIP1, is a 60-amino acid elicitor of defense reactions in barley cultivars carrying the R gene, Rrs1 (Hahn et al., 1993; Rohe et al., 1995; Steiner-Lange et al., 2003). However, unlike in many other plant-pathogen interactions, a hypersensitive response is not observed in resistant barley (Lehnackers and Knogge, 1990). In addition to its role as an Avr determinant, NIP1 belongs to a small group of secreted toxic proteins, which cause scald-like lesions in a genotype-unspecific manner when they are injected into leaves of barley (Wevelsiep et al., 1991) and other cereal plant species, such as wheat, rye (Secale cereale), and oat (Avena sativa), but not of Arabidopsis (Arabidopsis thaliana; W. Knogge, unpublished data). Furthermore, at subnecrotic concentrations, NIP1 indirectly stimulates the plasma membrane-localized H+-ATPase regardless of the plant R genotype (Wevelsiep et al., 1993; Fiegen and Knogge, 2002). Therefore, a virulence role of NIP1 has been proposed and targeted disruption of the NIP1 gene indeed slightly reduced fungal virulence on susceptible plants compared to the NIP1-expressing wild-type strain (Knogge and Marie, 1997).
R. secalis strains virulent on Rrs1 barley plants either lack the NIP1 gene or carry point mutations in the coding sequence that result in single amino acid alterations, which strongly reduce or abolish the biological activity of the gene product (Rohe et al., 1995). Comparison of the deduced amino acid sequences encoded by NIP1 alleles from more than 200 fungal isolates so far led to the classification of 18 NIP1 variants (Rohe et al., 1995; Schürch et al., 2004), two of which may not be expressed properly due to mutations in the start codon and in the acceptor splice site, respectively. Of the remaining 16 variants, four have been studied in more detail. NIP1 types I and II, which were found in >50% of all NIP1-carrying fungal isolates (Schürch et al., 2004), are elicitor active on Rrs1 barley, with type II showing somewhat lower activity than type I. In contrast, types III and IV are drastically less active or inactive (Rohe et al., 1995; Fiegen and Knogge, 2002). Interestingly, the Rrs1-specific elicitor activity of these four NIP1 variants appears to correlate with their nonspecific toxic lesion-inducing activity and H+-ATPase stimulation in barley (Fiegen and Knogge, 2002). These observations strongly suggest that elicitor receptor (triggering resistance) and toxin receptor (conditioning disease) are identical.
Here we describe the presence of a single class of high-affinity NIP1-binding sites in microsomal membranes of resistant and susceptible barley and other cereal plant species. Comparison of the affinity of different NIP1 variants to the receptor with their capacities to elicit defense reactions and to stimulate H+-ATPase suggests that both activities are mediated through a single receptor, but that receptor binding is not sufficient for the response.
RESULTS
Expression and Purification of NIP1 Variants
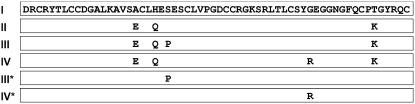
Expression and purification of NIP1 type I has been previously reported (Gierlich et al., 1999). cDNA encoding the type II protein was obtained by reverse transcription-PCR using RNA from R. secalis, isolate AU1 (Rohe et al., 1995), as a template. Types II, III, and IV have three amino acid alterations in common that distinguish them from type I (Fig. 1). In addition, elicitor-inactive types III and IV are specified by the single amino acid alterations Ser-23 → Pro and Gly-45 → Arg, respectively (Rohe et al., 1995). These unique amino acids were introduced into the type I sequence by site-directed mutagenesis to yield proteins termed type III* and type IV*. All constructs were cloned into the pQE-30 expression vector (Qiagen) with a sequence encoding a factor Xa cleavage site 5′ of the sequence coding for the mature NIP1, which allowed the removal of the His tag. All NIP1 variants were expressed in Escherichia coli purified from inclusion bodies and subsequently refolded (Gierlich et al., 1999). Purity and masses of the proteins were confirmed by matrix-assisted laser-desorption ionization (MALDI)-time of flight (TOF) yielding 6,433.66 D for type I, 6,508.0 D for type II, 6,442.60 D for type III*, and 6,531.84 D for type IV*.
Figure 1.
Primary sequence of the mature NIP1 type I and amino acid positions deviating from this sequence in types II to IV, III*, and IV*. Types I to IV are natural variants, whereas types III* and IV* were generated by targeted mutagenesis of the type I sequence.
Elicitor and H+-ATPase Stimulating Activities of NIP1 Variants
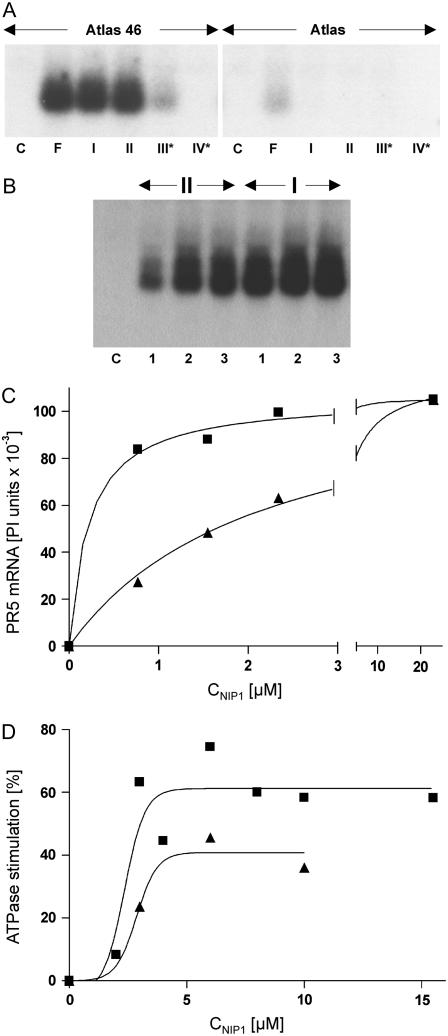
Infection of Rrs1 barley leaves (‘Atlas 46’) by avirulent isolates of R. secalis induces rapid and strong accumulation of mRNA encoding pathogenesis-related protein 5 (PR5). This response was also triggered upon application of purified or recombinant NIP1 type I to the leaf surface of Rrs1 barley, but not after application to rrs1 barley (‘Atlas’; Hahn et al., 1993; Gierlich et al., 1999). Purified NIP1 type II induced the same Rrs1-specific PR5 mRNA accumulation, albeit with lower efficiency, whereas purified protein types III and IV were drastically less active or inactive as elicitors and H+-ATPase stimulators (Rohe et al., 1995; Fiegen and Knogge, 2002). Induction of PR5 mRNA accumulation was again used to characterize the elicitor activity of the different recombinant NIP1 variants. To detect any minor activity, a saturating concentration of 10 μm of the recombinant proteins was used. Under these conditions, type III* retained only very weak activity on ‘Atlas 46’ (Rrs1), whereas type IV* did not show any elicitor activity. In addition, both variants were inactive on ‘Atlas’ (rrs1; Fig. 2A). Therefore, the two independent mutations specifying type III and type IV, respectively, suffice to drastically reduce or even abolish elicitor activity. NIP1 types I and II exhibited similar Rrs1-specific elicitor activity. However, using lower protein concentrations revealed the difference in their elicitor activities on the Rrs1 cultivar (Fig. 2B). Based on phosphoimager-assisted quantification of northern hybridization results and nonlinear regression analysis (Fig. 2C), EC50 values of 0.22 and 2.14 μm, respectively, were calculated for types I and II with similar maximal activities (Table I).
Figure 2.
NIP1-elicited PR5 mRNA accumulation and H+-ATPase stimulation. A, RNA gel blot using RNA from primary leaves of barley ‘Atlas 46’ (Rrs1) and ‘Atlas’ (rrs1) 24 h posttreatment with NIP1. Leaves were either inoculated with spores of the NIP1 type I-expressing fungal isolate UK7 (F) or treated with 10 μL of a NIP1 solution containing 25 μm NIP1 type I, II, III*, IV*, respectively, in 0.05% Tween 20. As a control (C), leaves were treated with 10 μL of 0.05% Tween 20. B, RNA gel blot using RNA from primary leaves of barley ‘Atlas 46’ 24 h posttreatment with 10 μL of NIP1 type I (I) or type II (II) solutions containing increasing protein concentrations (1, 0.77 μm; 2, 1.55 μm; 3, 2.34 μm). Nonlinear regression analysis (C) of a representative RNA gel blot after phosphoimager-assisted quantification and of H+-ATPase stimulation (D; mean values from Wevelsiep et al., 1991, 1993). Type I, ▪; type II, ▴.
Table I.
Binding constants of NIP1 variants and comparison with activities of NIP1 types I and II
| NIP1 Type | Binding
|
H+-ATPase Stimulation
|
Elicitor Activity
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Kd, Ki | Ratioa | EC50 | Emaxb | IRAc | IREd | EC50 | Emaxb | IRAc | IREd | |
| nm | μm | % | μm | PI Units | ||||||
| I | 5.6e | 2.35 | 61.2 | 1.82 | 0.22 | 1.06 × 105 | 8.94 | |||
| II | 734f | 130 | 2.86 | 40.9 | 72.0 | 2.14 | 1.15 × 105 | 14.7 | ||
| III | Approximately 730f,g | – | – | |||||||
| IV | Approximately 730f,g | – | – | |||||||
| III* | 13.7f | – | – | |||||||
| IV* | 6.8f | – | – | |||||||
Ki type II/Kd type I.
Maximal response: stimulation of the H+-ATPase (% of unstimulated activity), PR5 mRNA accumulation (phosphoimager [PI] units).
Intrinsic relative activity.
Intrinsic relative efficacy:intrinsic efficacy (ɛ) of type II expressed relative to that of type I.
Kd.
Ki.
Expected.
Types III and IV were also unable to stimulate the barley plasma membrane H+-ATPase, suggesting that the effect on the membrane enzyme is mediated by the same receptor as elicitor activity (Fiegen and Knogge, 2002). When previous data on the stimulatory activity of types I and II (Wevelsiep et al., 1993; Fiegen and Knogge, 2002) were reevaluated, nonlinear regression analysis favored a sigmoidal rather than a hyperbolic concentration response curve, with type II displaying only approximately 65% of the maximal stimulation obtained with type I. In contrast to elicitor activity, similar EC50 values of 2.35 and 2.86 μm were calculated for H+-ATPase stimulation by types I and II, respectively (Fig. 2D; Table I).
Iodination of NIP1
To study the binding of NIP1 to plant plasma membranes, protein was radioactively labeled with iodine-125 (125I). The lactoperoxidase-Glc oxidase method was used, which directly labels the protein by substituting 125I ortho to the hydroxyl group of Tyr phenolic rings (McFarthing, 1992). NIP1 contains three Tyr residues and, hence, six putative iodination sites that may give rise to mixtures of protein labeled at single or multiple sites. Therefore, the method was developed and optimized using nonradioactive 127I. After iodination, several protein peaks were separated by reverse-phase (RP)-HPLC (data not shown). Analysis by MALDI-TOF mass spectroscopy revealed that noniodinated NIP1 (6,443 D) eluted first, followed by a monoiodinated protein form (6,563 D) in the second major fraction. In contrast, the third major fraction and the following minor fractions contained mixtures of mono- and disubstituted and di- and trisubstituted NIP1.
Monoiodinated NIP1 was tested for its biological activity in barley leaves. The position of 127I is not known. However, iodination did not significantly affect the capacity of NIP1 to induce lesion formation in rrs1 barley and PR5 mRNA accumulation in Rrs1 barley (data not shown). In addition, structure and activity of the iodinated elicitor were stable in solution for at least 3 d at room temperature. Using the optimized protocol for labeling and purification, 125I was commercially incorporated into NIP1 type I. This 125I-NIP1 coeluted during RP-HPLC with the monoiodinated protein in fraction 2.
Binding of 125I-NIP1 to Barley Microsomes
Binding of 125I-NIP1 to microsomal membranes from primary leaves of barley ‘Atlas 46’ (Rrs1) was investigated at room temperature. A vacuum filtration technique was used to separate free from bound ligand to decrease the loss of bound radioactivity caused by rapid dissociation of the receptor-ligand complex. Highest binding activity was found between pH 6 and pH 7.5, whereas the optimum temperature was around 21°C. At 37°C, rapid and irreversible precipitation of the microsomes occurred independent of the presence of NIP1. Specific binding of 125I-NIP1 to barley microsomes represented about 60% of total binding at an initial ligand concentration of 250 pM. Less than 5% of the initially applied ligand became bound to microsomal membranes, reducing the possibility that ligand depletion interfered with the characterization of the NIP1-binding site. The amount of specifically bound radioactive ligand increased linearly with increasing amounts of membrane protein ranging from 30 to 200 μg of protein in the standard assay (data not shown).
To localize the specific binding site, binding assays were performed using 125 pM 125I-NIP1 and plasma membrane vesicles prepared by phase partitioning from leaves of barley ‘Atlas 46’ (Kjellbom and Larsson, 1984; Wevelsiep et al., 1993) as well as microsomes obtained from epidermis tissue mechanically peeled off ‘Atlas 46’ primary leaves. Per milligram of membrane protein, the plasma membrane vesicles bound a 2-fold higher amount of radiolabeled ligand compared to microsomal membranes. Similarly, epidermis-derived microsomes showed more than 4-fold higher binding compared to whole-leaf microsomes (data not shown). These results suggest that the binding site may be localized on the plasma membrane, in particular of epidermis cells. This would agree with the previous observation that right-side-out orientation is required for ATPase stimulation by NIP1 (Wevelsiep et al., 1993). Nevertheless, all further experiments were performed with microsomal membranes from whole leaves.
Characterization of the NIP1-Binding Site in Barley
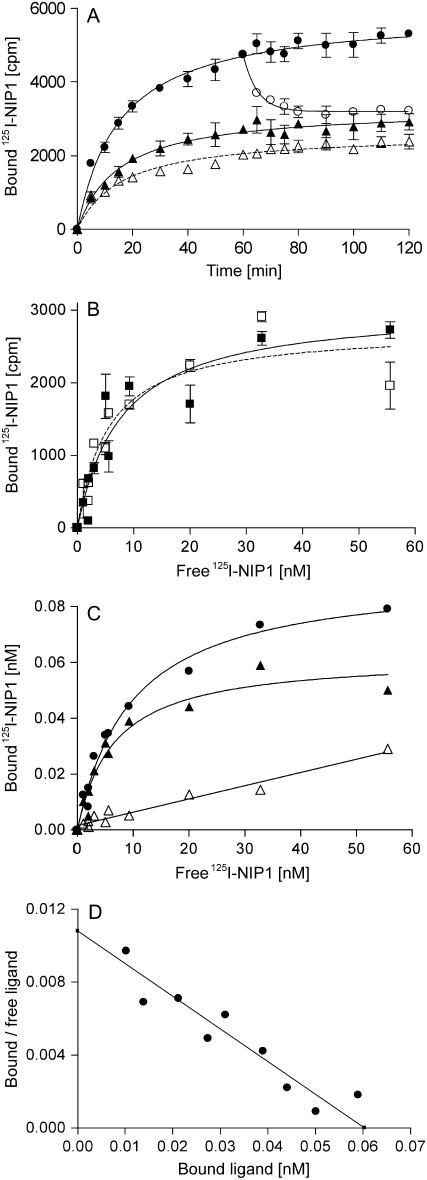
Analysis of elicitor-binding kinetics demonstrated that, before equilibrium is reached, at 125 pM 125I-NIP1 association is faster than dissociation. Half-maximal binding was achieved within 15 min after addition of the ligand and equilibrium between association and dissociation was reached after about 60 min (Fig. 3A). Nonspecific binding only slightly increased during this time. Addition of a 1,000-fold molar excess of unlabeled NIP1 at t = 60 min initiated the dissociation of bound radioactive NIP1 with a dissociation rate constant (Koff) of 3.3 × 10−3/s and a half-life of approximately 3.5 h, demonstrating that binding of 125I-NIP1 to barley microsomes is a reversible process. However, about 25% of the bound ligand was not displaced after 60 min, indicating a stable receptor-ligand complex or incomplete reversibility of binding.
Figure 3.
Binding of the 125I-NIP1 ligand to barley microsomal membranes is saturable and of high affinity. A, Time courses of specific ligand binding to barley membranes and displacement by unlabeled NIP1. Assays were initiated by the addition of 125 pM 125I-NIP1 to barley microsomes (50 μg membrane protein). Total binding (•), nonspecific binding (▵), and binding after displacement of bound 125I-NIP1 by unlabeled NIP1 initiated 60 min after addition of the radioligand (○) were assayed at the times indicated. To determine specific binding (▴), nonspecific binding was subtracted from total binding. Nonspecific binding and displacement of bound 125I-NIP1 were determined in the presence of 125 nm unlabeled NIP1. Data points represent the averages of three independent experiments with comparable results. B, Specific binding of 125I-NIP1 to microsomal membranes from barley ‘Atlas 46’ (▪) and ‘Atlas’ (□), using increasing amounts of radioligand. C, Specific binding of 125I-NIP1 to barley microsomal membranes, using increasing amounts of radioligand. Symbols as in A. D, Scatchard plot displaying the specific binding data derived from C. Kd (5.6 nm) and number of binding sites (255 fmol × mg−1 microsomal protein) were determined by nonlinear regression.
To obtain further information on the nature of the binding site and to test whether NIP1-binding correlates with the presence of the R gene, Rrs1, saturation-binding experiments were performed. Microsomal membranes from the resistant and the susceptible barley ‘Atlas 46’ (Rrs1) and ‘Atlas’ (rrs1), respectively, were incubated with increasing amounts of 125I-NIP1. No significant difference between the binding curves was observed regardless of the origin of the membranes and a saturation state was achieved at a ligand concentration of approximately 60 nm (Fig. 3, B and C). Using the combined binding data, the dissociation constant Kd was determined by nonlinear regression to be 5.6 nm (se = 1.0 nm). A linear display of the data in a Scatchard plot (Fig. 3D) suggests a single class of NIP1-binding sites, whose concentration was calculated to be 255 fmol (se = 15 fmol) per milligram of microsomal membrane protein.
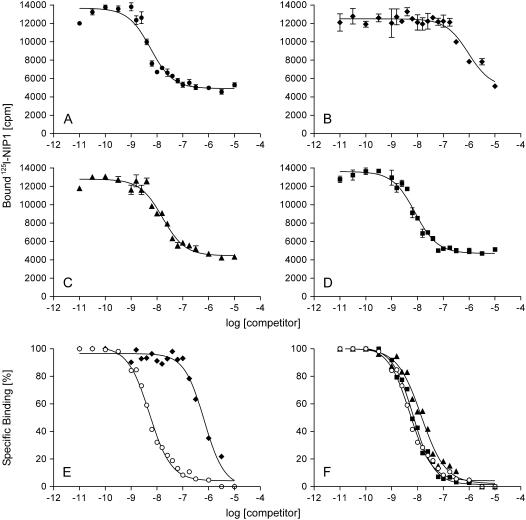
Equilibrium competition-binding experiments were performed and Ki values calculated to compare the affinity of the binding site to different competitors. Competition of both nonradioactive 127I-NIP1 (data not shown) and noniodinated NIP1 (Fig. 4A) with 125 pM 125I-NIP1 yielded very similar Ki values. This indicated that iodination does not affect the binding properties of NIP1, thus allowing the use of noniodinated NIP1 as a competitor in binding assays. In contrast, 2-mercaptoethanol-reduced HPLC-purified NIP1 did not compete for binding at a concentration of 1 μm (data not shown). This is in agreement with the observation that reduction of NIP1 abolishes its elicitor activity. The Hill slope of the competition curve was determined to be −1.03, again indicating a single class of binding sites.
Figure 4.
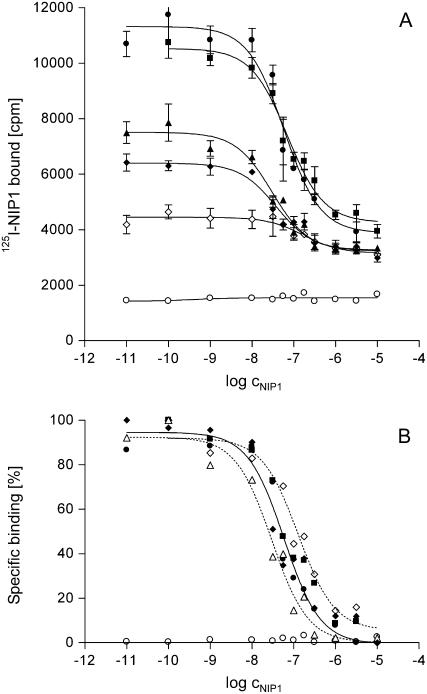
Competition for 125I-NIP1 binding to barley microsomal membranes with different variants of unlabeled NIP1 as competitors. A, Type I. B, Type II. C, Type III*. D, Type IV*. All samples were taken from the same experiment. All data points represent the average of three independent measurements. E and F show normalized competition curves for type I (○) and type II (♦, compare with A and B) and for type I (○), type III* (▴), and type IV* (▪; compare with A, C, and D), respectively. For transparency reasons, the error bars are not shown in E and F.
NIP1 Types as Competitors in Binding Experiments
Equilibrium competition-binding experiments were also performed using the naturally occurring NIP1 type II and the two mutant proteins types III* and IV* (Fig. 4, B–D). The elicitor-active protein type II has a Ki of 734 nm, a value 130-fold higher than the Kd of type I (Fig. 4E; Table I). In contrast, with Ki values of 13.7 and 6.6 nm, respectively, both inactive mutant protein types III* and IV* have affinities for the NIP1-binding site resembling that of type I (Fig. 4F). Very similar results were obtained with microsomes from the susceptible barley ‘Atlas’ and from barley plants heterozygous for the Rrs1 locus (data not shown). Types III and IV were not used in competition experiments. However, from their structural similarity to type II, their Ki values can be expected to be close to the Ki of type II (approximately 730 nm).
Comparison of the Relative Efficacies of the NIP1 Variants
The affinity of type I to the binding site by far exceeds that of type II, whereas their EC50 values for elicitor activity differ only by a factor of 10 with similar maximal PR5 mRNA accumulation (Emax). In the other functional assay of NIP1, stimulation of the barley plasma membrane H+-ATPase, similar EC50 values were obtained for types I and II. However, in contrast to elicitor activity, the two variants differ in their maximal activities. Therefore, the structural differences between types I and II appear to affect affinity and efficacy in an opposing way. Calculation of the intrinsic activities and estimation of the intrinsic efficacies relative to each other using EC50, Emax, and equilibrium dissociation constants (Kd, Ki) provides a means to quantitatively compare and assess the contribution of the efficacy factor to activity (Table I; Ehlert et al., 1999). The results reveal that both as H+-ATPase stimulator and even more distinct as elicitor type I shows a higher intrinsic activity (IRA) relative to type II. When the greatly differing equilibrium dissociation constants are taken into account, however, the relationship is reversed, with type II displaying a substantially higher intrinsic efficacy (IRE) relative to type I in both responses.
Binding of NIP1 to Microsomes from Other Cereal Plant Species
NIP1 is a genotype-unspecific toxin inducing scald-like lesions in other cereal plant species including nonhosts, such as wheat (Wevelsiep et al., 1993). Therefore, microsomal membranes were also isolated from wheat, rye, oat, and maize (Zea mays), as well as from Arabidopsis. Equilibrium competition-binding experiments revealed the presence of a NIP1-binding site in microsomes from all tested monocotyledonous species with Ki values in a narrow window of ligand concentration ranging from 50% (rye) to 290% (maize) of that of barley (Fig. 5). The concentration of binding sites per milligram of microsomal protein varied in the different species (wheat, 220 fmol; rye, 150 fmol; oat, 110 fmol; maize, 40 fmol). In contrast, under the conditions applied, NIP1 did not show any affinity for microsomes from the dicotyledonous species Arabidopsis (Fig. 5).
Figure 5.
A, NIP1-binding site in different cereals. Equilibrium competition-binding experiments were carried out using microsomal membranes from barley (•), wheat (▪), rye (▵), oat (♦), maize (⋄), and Arabidopsis (○). B, Normalized competition data with the dotted lines showing the competition curves for rye with the lowest and maize with the highest IC50 value, whereas the solid line represents barley. For transparency reasons, error bars are not shown.
DISCUSSION
Fungal Effector Protein NIP1 Interacts with a Single Class of High-Affinity Binding Sites
The small secreted effector protein NIP1 from the barley pathogen R. secalis has dual functions. It is a potential virulence factor as reflected by its necrosis induction in leaves of barley and of other cereals and its stimulation of the plasma membrane-localized H+-ATPase, both regardless of the plant R genotype (Wevelsiep et al., 1991, 1993). Additional evidence for this role was provided by a NIP1 gene replacement mutant that exhibited a slightly weaker virulence phenotype producing reduced and delayed lesions on barley leaves compared to the wild-type strain (Knogge and Marie, 1997). In addition, however, NIP1 is an Avr factor triggering defense reactions specifically in barley plants carrying R gene Rrs1, but not in rrs1 plants (Hahn et al., 1993; Rohe et al., 1995; Steiner-Lange et al., 2003). These opposing functions prompted the question as to how the protein is perceived by the plant (Knogge, 1996). Different activities may result from NIP1 binding to two separate receptors on the host membrane. Alternatively, a single receptor may mediate both functions of the protein. In this case, the Rrs1 gene may not encode the NIP1 receptor, but rather an additional factor located downstream of binding in the signaling pathway. The data presented in this article demonstrate the presence of a single class of high-affinity NIP1-binding sites for which all four NIP1 variants compete. In conjunction with the observation that amino acid alterations that affect the H+-ATPase-stimulating and necrosis-inducing activity of NIP1 also influence its elicitor activity (Rohe et al., 1995; Fiegen and Knogge, 2002), this finding suggests that a single membrane-localized target molecule is involved in both triggering resistance and mediating virulence.
Binding of various elicitors, in particular of MAMPs/PAMPs, to plasma membrane-localized receptors of several plants has been studied. For instance, a 13-amino acid elicitor peptide (Pep-13), a surface-exposed fragment of a Ca2+-dependent cell wall transglutaminase from Phytophthora sojae (Brunner et al., 2002), binds to its receptor on parsley (Petroselinum crispum) plasma membranes with a Kd of 2.4 nm (Nürnberger et al., 1994). For elicitins, another class of secreted Phytophthora proteins triggering defense reactions in tobacco (Nicotiana tabacum), Kd values between 2 and 13.5 nm have been reported (Wendehenne et al., 1995; Bourque et al., 1998). For one of the most active bacterial elicitors, flg22, a 22-amino acid flagellin-derived peptide, a Kd value of 1 to 2 nm was estimated for binding to tomato membranes (Meindl et al., 2000). Recently, it was demonstrated that flg22 in Arabidopsis is bound by FLS2, a transmembrane receptor-like kinase with an extracellular Leu-rich repeat domain (Gomez-Gomez and Boller, 2000; Chinchilla et al., 2006). Of saccharide elicitors, a hepta-β-glucoside from cell walls of P. sojae displayed a low Kd (0.75 nm) for binding to soybean (Glycine max) membranes (Cheong and Hahn, 1991). A β-glucan-binding protein was identified (Umemoto et al., 1997; Mithöfer et al., 2000) that contains a 1,3-β-glucanase domain releasing oligoglucoside fragments that constitute ligands for the high-affinity binding domain (Fliegmann et al., 2004). Furthermore, chitin perception has been studied in several plants. A Kd of 23 nm was described for the binding of a chitin fragment with a degree of polymerization (DP) of 5 to tomato membranes (Baureithel et al., 1994), whereas for membranes from soybean cell cultures and root tissues, Kd values between 35 and 75 nm were reported for two chitin ligands (DP = 5 and DP = 8; Day et al., 2001) and a Kd of 29 nm for rice membranes (Shibuya et al., 1996). In rice cells, a transmembrane chitin oligosaccharide elicitor-binding protein was isolated that carries two extracellular LysM motifs (Kaku et al., 2006). These latter motifs also occur in Ser-Thr receptor kinases involved in specific recognition of nod factors (Radutoiu et al., 2003; Knogge and Scheel, 2006). Very low Kd values of 0.07 and 0.05 nm, respectively, were described for the Avr gene products AVR9 and AVR4 from the tomato pathogen C. fulvum (Kooman-Gersmann et al., 1996). Whereas the plant-binding site for AVR9 remains to be characterized, AVR4 was shown to bind to fungus-derived nonproteinaceous components (Westerink et al., 2002) and with lower affinity to chitin fragments (DP = 6; Kd = 6.3 μm; van den Burg et al., 2004). However, in both cases, it remains unclear how plant defense is activated. The Kd of 5.6 nm that was determined for NIP1 type I is within the range of most elicitor-receptor systems analyzed to date. In contrast, the Ki of NIP1 type II was calculated to be 734 nm, a value substantially higher than most published data. The type II protein therefore displays much lower affinity to the binding site than NIP1 type I.
Half-maximal binding of type I and type II is reflected by their Kd and Ki values. The concentrations required for half-maximal response induced by type II exceed its Ki by a factor of only 3 to 4. In contrast, substantially higher concentrations of type I relative to its Kd are needed for half-maximal elicitor activity (approximately 40×) and H+-ATPase stimulation (approximately 400×). Similar apparent discrepancies between binding and response have also been described for other receptor-ligand interactions. For the fungal phytotoxin fusicoccin, a concentration 100-fold higher than the Kd of 1 nm was necessary to stimulate the plant plasma membrane H+-ATPase (Basel et al., 1994). An even larger difference (approximately 4,000-fold) between Kd value and the minimal elicitor concentration required for inducing a hypersensitive response in tomato leaves was described for the AVR9 elicitor from C. fulvum (Kooman-Gersmann et al., 1996). However, the quantitative comparison of affinities and activities needs to be interpreted with caution. First, different types of assays were used. In binding studies, the direct molecular interaction between NIP1 and its microsomal-binding site is quantified. In contrast, EC50 values result from indirect functional assays in which ligand-receptor interaction, at least in defense activation, initiates a multifactor signaling process that eventually leads to the downstream response. Second, binding conditions, such as ionic strength and pH that were optimized for in vitro assays, may differ from those in the plant apoplast. Third, the observed differences may be due to inherent properties of the different NIP1 types (see below). Fourth, binding and response may not show a one-to-one relationship. This was shown for flagellin binding and flagellin-induced extracellular alkalinization, where the full response occurred already when only a fraction of the receptor was occupied (Meindl et al., 2000). In addition, although the ligand is applied to the leaf surface in a known concentration, its effective concentration at the receptor sites inside the leaf cannot be determined, but can be expected to be lower. Nevertheless, binding of NIP1 types I and II to the microsomal-binding site correlates with their efficacies as elicitor, at least at low concentrations, and as H+-ATPase stimulators. This provides good evidence that the binding site is relevant for biological activity.
The NIP1-Binding Site Is Present in Barley and Other Cereal Species
The apparent concentration of NIP1-binding sites in barley microsomal preparations of 255 fmol × mg−1 of microsomal protein is in the same order of magnitude as described for other elicitor-binding sites in plants. For the Pep-13 elicitor from P. sojae, 88 fmol × mg−1 were found in parsley cells (Nürnberger et al., 1994), for different elicitins in tobacco, 234 to 403 fmol × mg−1 (Bourque et al., 1998), for the hepta-β-glucoside from P. sojae in soybean cells, 1.2 pmol × mg−1 (Cheong and Hahn, 1991; Cosio et al., 1998), for chitin fragments in tomato, 2.45 pmol × mg−1 (Baureithel et al., 1994), and for soybean cells and roots, 7.6 to 12.2 pmol × mg−1(Day et al., 2001). In comparison, the number of binding sites for the AVR9 elicitor from C. fulvum on tomato membranes is 800 fmol × mg−1 (Kooman-Gersmann et al., 1996).
The presence of a NIP1-binding site in microsomes from barley, regardless of the R genotype, as well as from rye, oat, wheat, and maize, the latter three being nonhosts of R. secalis, but not from Arabidopsis, correlates with the toxic effect of NIP1 on leaves of these plants (Wevelsiep et al., 1991, 1993). In addition, their similar binding characteristics indicate that the binding sites in those crops may be related. This suggests that the binding site represents the virulence target of NIP1, which appears to be common in cereals. Similar results were obtained with the AVR9 elicitor from C. fulvum, for which binding sites are also present in plasma membranes from resistant (Cf-9) and susceptible (Cf-0) tomato genotypes, as well as from other solanaceous species (Kooman-Gersmann et al., 1996).
Binding of NIP1 to Barley Membranes Is Not Sufficient to Trigger the Responses
Two single amino acid alterations, Ser-23 → Pro in NIP1 type III and Gly-45 → Arg in NIP1 type IV are sufficient to render NIP1 nontoxic and inactive as an H+-ATPase stimulator and elicitor (Rohe et al., 1995; Fiegen and Knogge, 2002). However, the inactive mutant protein types III* and IV* still compete efficiently for the NIP1-binding site, displaying Ki values that are close to the Kd of the active NIP1 type I. In contrast, NIP1 type II, differing in three amino acid positions from type I, is a much weaker competitor of type I in competitive-binding experiments. However, despite its relatively low affinity compared to type I, type II is capable of efficiently inducing the synthesis of PR5 mRNA when applied to Rrs1 barley leaves (Rohe et al., 1995; Fiegen and Knogge, 2002). In addition, isolates of R. secalis carrying the type II-encoding NIP1 allele are avirulent on Rrs1 plants. Furthermore, type II has H+-ATPase-stimulating activity. In contrast, type III, type III*, type IV, and type IV* are all inactive as elicitors and fail to stimulate H+-ATPase activity (Fiegen and Knogge, 2002). Furthermore, Ki values similar to that of type II can be expected for the native NIP1 variants type III and type IV, which also contain the three type II-specific amino acids. These results suggest that the affinity of NIP1 variants to the binding site is not proportional to its capacity to trigger the responses. The sigmoidal concentration-response curves for H+-ATPase stimulation indicate that the as-yet-unknown mechanism of H+-ATPase stimulation differs from the induction of PR protein biosynthesis, which yielded hyperbolic concentration-response curves. In particular, some kind of cooperativity appears to be involved in translating NIP1 binding into the stimulatory response.
Structural differences between types I and II appear to affect both affinity and activity, albeit in an opposing way. Type I is characterized by a substantially higher affinity to the binding sites than type II. This is, however, only partly reflected at the activity level, where the EC50 value of type I for elicitor activity exceeds that of type II only by a relatively low factor, whereas the EC50 values for H+-ATPase are very similar. In addition, maximal elicitor activities of the two NIP1 variants are almost identical, whereas the H+-ATPase stimulation of type II is lower than that of type I. This suggests that the amino acids leading to stronger binding negatively affect the intrinsic efficacies of the protein, which are substantially higher for type II as compared to type I.
The lack of correlation between binding and response of the different NIP1 variants cannot easily be explained. The three-dimensional structure of NIP1 has been elucidated by 1H- and 14N-NMR spectroscopy (van't Slot et al., 2003). The protein consists of two domains containing two and three antiparallel β-strands, respectively, which are linked by two disulfide bridges each. The fifth disulfide bridge connects the two protein domains, rendering the structure rather rigid, except for two flexible loops. The Gly-45 → Arg mutation occurs near the base of one of these loops, whereas the Ser-23 → Pro mutation is located in a solvent-exposed β-turn on the same side of the molecule. Both mutations are flanking a groove between the two protein domains, which may represent the effector site. Two of the type II-specific amino acids are located in the N-terminal domain of NIP1, the third in the C-terminal domain. At least two of these amino acids appear to be solvent exposed and may be involved in binding to the primary target site.
A possible model for the signal transduction mechanism assumes bipartite functions of NIP1 and its interaction with a plasma membrane-localized proteinaceous binding site. NIP1 interacts through a region containing one or more of the type II-specific amino acids with a specific domain of the binding protein. This causes a conformational change of the target protein, allowing its interaction with the effector site of NIP1 that contains the type III- and type IV-specific amino acids. As a consequence, transduction of the signal across the membrane is triggered (Knogge, 1996). This model is analogous to the address message concept proposed for the activation of the flagellin receptor complex in tomato; in a two-step mechanism involving conformational changes of both receptor and ligand, binding of the flagellin N terminus (address) to the receptor represents the first step that is followed by activation of the response with the C terminus (message) as the second step (Meindl et al., 2000).
An alternative model would be similar to those proposed for the AVR9/Cf-9 interaction (Joosten and de Wit, 1999). Signal transduction may require an additional protein to be involved in the NIP1/NIP1 target protein complex. This protein may be prevented from interacting with the target-ligand complex by the Ser-23 → Pro and Gly-45 → Arg mutations of NIP1, which, however, do not affect the affinity for the primary binding site. In contrast, type II-specific amino acids negatively affect the primary binding event, but augment the capacity to interact with the putative third component of the target protein complex.
In both models, primary binding of NIP1 is not the crucial event distinguishing plant resistance and susceptibility and the Rrs1 gene is therefore unlikely to encode the NIP1 target (Knogge, 1996). Because mutations in NIP1 affect all activities of the protein, it remains unclear where the signaling pathways leading to defense gene activation and to H+-ATPase stimulation and necrosis induction branch off and, hence, where the Rrs1 gene product is positioned in the signaling chain. It is, however, tempting to speculate that, in analogy to other systems, the Rrs1 protein may guard the NIP1 target, which mediates H+-ATPase stimulation (van der Biezen and Jones, 1998).
MATERIALS AND METHODS
Plant Material and Growth Conditions
The near-isogenic barley (Hordeum vulgare) cultivars ‘Atlas 46’ (Rrs1) and ‘Atlas’ (rrs1), as well as fungal isolates were grown as previously described (Lehnackers and Knogge, 1990).
Isolation of Microsomal Fractions and Plasma Membrane Vesicles
Microsomal fractions and plasma membrane vesicles were isolated from 10-d-old barley leaves (Widell et al., 1982; Wevelsiep et al., 1993). All isolation steps were performed at 4°C. Briefly, barley leaves were shortly infiltrated under vacuum with ice-cold 50 mm HEPES buffer, pH 7.5 (0.5 m Suc, 5 mm ascorbic acid, 1 mm dithiothreitol, 0.6% [w/v] water-insoluble polyvinylpyrrolidone phosphate, 1 mm phenylmethylsulfonyl fluoride). Subsequently, the material was ground in a blender. Microsomal membranes were obtained by filtration through four layers of Miracloth (Calbiochem) and two differential centrifugation steps at 10,000g and 100,000g, respectively. Plasma membranes were purified from the microsomal membranes by aqueous two-phase partitioning (Larsson et al., 1988; Palmgren et al., 1990). After isolation, both the microsomal membrane fractions and the plasma membrane-enriched fractions were dissolved in 5 mm potassium phosphate buffer, pH 7.8 (0.33 m Suc, 3 mm KCl) and subsequently homogenized in a glass potter (Braun). Membranes were frozen in liquid nitrogen and stored at −80°C.
Preparation of NIP1
NIP1 was expressed in Escherichia coli as a His-tagged fusion protein (Gierlich et al., 1999). Briefly, fusion protein was obtained from solubilized inclusion bodies by affinity chromatography using a nickel-nitrilotriacetic acid agarose affinity chromatography column (Qiagen). Cystine bond shuffling was facilitated using a previously described folding procedure (Beiboer et al., 1996) that involves a Cys-cystine redox couple. Correctly folded peptide was separated from misfolded peptides by RP-HPLC. A factor Xa cleavage site between the mature NIP1 protein and the His tag allowed for the preparation of a peptide with identical biochemical properties as the fungus-derived protein. The amount of purified NIP1 was determined by OD280 measurement using a molar extinction coefficient of 5,040 L × mol−1 × cm−1, as calculated based on the primary sequence (Gill and von Hippel, 1989), as well as by using Bradford's reagent (Bio-Rad).
Cloning, Expression, and Purification of NIP1 Variants
The cDNA encoding NIP1 type II was obtained by reverse transcription-PCR on total RNA from Rhynchosporium secalis, isolate AU1, using primer 1 and oligo(dT). The amplified fragment was subjected to a second PCR reaction using primers 1 and 2 (see “Primer Table”). Primer 1 is a modification of earlier described primers (Gierlich et al., 1999). The amino acids specific for variants III and IV were introduced into the type I sequence by site-directed mutagenesis to yield the variants type III* and type IV*. 3′ primers 5 and 6 containing the mutations (underlined) were used along with 5′ primer 3 in a first PCR step (see “Primer Table”). The products of this PCR were used as megaprimers in combination with 3′ primer 4 in a second PCR step. Restriction sites (underlined) for cloning were introduced through primers 1 and 4 (SmaI) and primers 2 and 3 (BamHI). The final amplification products were purified from agarose gels, digested with BamHI and SmaI, and subsequently cloned into the pQE30 expression vector (Qiagen). Expression of the three variants in E. coli strain DHB4 (Boyd et al., 1987) and their purification was performed as described above. MALDI-TOF analysis was used to verify the molecular masses of the peptides. Elicitor activity was analyzed as previously described (Hahn et al., 1993), routinely using a concentration of 20 ng × μL−1 (approximately 3 μm).
Primer Table
Primer 1, 5′-GCGGCCCGGGTTAACATTGGCGAAATCCCGTCG-3′ (SmaI); primer 2, 5′-GCGCGGATCCATCGAAGGTAGAGATCGATGCAGATACACCCTTGTTGC-3′ (BamHI); primer 3 (pQE-30 5′), 5′-GCGCGGATCCATCGAAGGTAGAGATCGATGCAGATACACCCTTTGTTGC-3′ (BamHI); primer 4 (pQE-30 3′), 5′-GCGGCCCGGGTTAACATTGGCGGTATCCCGTCG-3′ (SmaI); primer 5 (III*, Ser → Pro), 5′-GGATTCTGGCTCATGTAGGCATG-3′; primer 6 (IV*, Gly → Arg), 5′-GCCACCTTCACGAATGAGC-3′.
Iodination of NIP1
NIP1 type I was modified with the nonradioactive isotope 127I by using the lactoperoxidase-Glc oxidase system (Sigma; McFarthing, 1992). One hundred microliters of a 1% d-Glc solution were added to 300 μL phosphate-buffered saline, pH 7.0, containing 2 μg NIP1, 10 μL of 100 μm NaI, 1 μg Glc oxidase, and 1 μg lactoperoxidase. After 10 min at room temperature, the reaction was stopped by adding 20 μL of a mixture of 100 mg/mL metabisulfate and 10 mg/mL sodium azide. Reaction products were separated on an analytical RP-HPLC column (Vydac protein C4, 0.46 × 25 cm; Macherey-Nagel) that was eluted with an acetonitrile gradient using a Hewlett-Packard chromatograph and subjected to MALDI-TOF mass spectrometric analysis. In addition, elicitor activity of the protein fraction was analyzed as described previously (Hahn et al., 1993). 125I-NIP1 was obtained from ANAWA Laboratories with specific activity of 2,130 Ci/mmol and used in binding assays.
Binding of NIP1 to Membrane Fractions
Membranes were preincubated at room temperature for 15 min in 30 mm HEPES, pH 7.0, 5 mm MgCl2, 1 g/L fatty acid-free bovine serum albumin (binding buffer) at a membrane protein concentration of 0.5 μg/μL. Binding was initiated by adding different concentrations of 125I-NIP1 in a volume of 10 μL. Nonspecific binding was determined in the presence of a 1,000-fold excess of unlabeled NIP1. Glass fiber microfilters (Whatman) were soaked for several hours in 0.5% polyethylenimine and transferred to a sampling manifold (Millipore). Filters were rinsed with 4 mL of ice-cold washing buffer (binding buffer containing 0.5 m KCl). Filtration of the samples was carried out under vacuum and filters were washed with 2 × 4 mL of binding buffer. The filters were subsequently transferred to 6-mL scintillation vials and 5 mL Ultima Gold scintillation cocktail (Ducheva) were added. After overnight incubation, radioactivity was counted in a scintillation counter (LS1800; Beckman Instruments). Analysis of the data was performed using GraphPad Prism for Windows, version 4.02 (GraphPad Software; www.graphpad.com). From IC50 values that represent the competitor concentration competing for 50% of specific binding under the assay conditions, Ki values were calculated that represent the competitor concentration binding to 50% of the binding sites at equilibrium in the absence of the radioligand using the Cheng-Prusoff equation (Cheng and Prusoff, 1973).
Quantitative Comparison of Active NIP1 Variants
To allow a quantitative comparison of the activities of NIP1 types I and II, the IRA and IRE were calculated using EC50 values, maximal activities (Emax), and equilibrium dissociation constants (Kd, Ki) according to Ehlert et al. (1999). The IRA of type I, which is equivalent to the product of its affinity (1/Kd) and IRE relative to type II, was calculated using the following equation: IRA = EC50-II × Emax-I/EC50-I × Emax-II. To estimate the contribution of the IRE to activity, Kd and Ki were included in the equation. The IRE (ɛ) of type II relative to that of type I was estimated using the equation ɛII/ɛI = EC50-I × Emax-II × Ki/EC50-II × Emax-I × Kd.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF683095 to EF683100.
Acknowledgments
We would like to thank Harrold van den Burg, Laboratory of Biochemistry, Department of Biomolecular Sciences, Wageningen University, for performing mass spectrometric analyses. In addition, the excellent technical assistance of Annette Böttcher, University of Adelaide, Waite Campus, is greatly appreciated.
This work was supported by the Deutsche Forschungsgemeinschaft (research grant to W.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Wolfgang Knogge (wknogge@ipb-halle.de).
References
- Armstrong MR, Whisson SC, Pritchard L, Bos JIB, Venter E, Avrova AO, Rehmany AP, Bohme U, Brooks K, Cherevach I, et al (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci USA 102 7766–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP (1997) Signaling in plant-microbe interactions. Science 276 726–733 [DOI] [PubMed] [Google Scholar]
- Basel LE, Zukowski AT, Cleland RE (1994) Modulation of fusicoccin-binding protein activity in mung bean (Vigna radiata L.) hypocotyls by tissue maturation and by fusicoccin. Plant Physiol 104 691–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baureithel K, Felix G, Boller T (1994) Specific, high affinity binding of chitin fragments to tomato cells and membranes: competitive inhibition of binding by derivatives of chitooligosaccharides and a Nod factor of Rhizobium. J Biol Chem 269 17931–17938 [PubMed] [Google Scholar]
- Beiboer SHW, van den Berg B, Dekker N, Cox RC, Verheij HM (1996) Incorporation of an unnatural amino acid in the active site of porcine pancreatic phospholipase A2: substitution of histidine by 1,2,4-triazole-3-alanine yields an enzyme with high activity at acidic pH. Protein Eng 9 345–352 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S, Hiller NL, Liolios K, Win J, Kanneganti TD, Young C, Kamoun S, Haldar K (2006) The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathogens 2 e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonas U, Lahaye T (2002) Plant disease resistance triggered by pathogen-derived molecules: refined models of specific recognition. Curr Opin Microbiol 5 44–50 [DOI] [PubMed] [Google Scholar]
- Bourque S, Ponchet M, Binet MN, Ricci P, Pugin A, Lebrun-Garcia A (1998) Comparison of binding properties and early biological effects of elicitins in tobacco cells. Plant Physiol 118 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D, Manoil C, Beckwith J (1987) Determinants of membrane protein topology. Proc Natl Acad Sci USA 84 8525–8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner F, Wirtz W, Rose JKC, Darvill AG, Govers F, Scheel D, Nürnberger T (2002) A b-glucosidase/xylosidase from the phytopathogenic oomycete, Phytophthora infestans. Phytochemistry 59 689–696 [DOI] [PubMed] [Google Scholar]
- Bryan GT, Wu KS, Farrall L, Jia YL, Hershey HP, McAdams SA, Faulk KN, Donaldson GK, Tarchini R, Valent B (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12 2033–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH (1973) Relationship between inhibition constant (KI) and concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22 3099–3108 [DOI] [PubMed] [Google Scholar]
- Cheong JJ, Hahn MG (1991) A specific, high affinity binding site for the hepta-β-glucoside elicitor exists in soybean membranes. Plant Cell 3 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosio EG, Pöpperl H, Schmidt WE, Ebel J (1998) High-affinity binding of fungal β-glucan fragments to soybean (Glycine max L.) microsomal fractions and protoplasts. Eur J Biochem 175 309–315 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411 826–833 [DOI] [PubMed] [Google Scholar]
- Day RB, Okada M, Ito Y, Tsukada K, Zaghouani H, Shibuya N, Stacey G (2001) Binding site for chitin oligosaccharides in the soybean plasma membrane. Plant Physiol 126 1162–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG (2004) The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16 755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlert FJ, Griffin MT, Sawyer GW, Bailon R (1999) A simple method for estimation of agonist activity at receptor subtypes: comparison of native and cloned M3 muscarinic receptors in guinea pig ileum and transfected cells. J Pharmacol Exp Ther 289 981–992 [PubMed] [Google Scholar]
- Fiegen M, Knogge W (2002) Amino acid alterations in isoforms of the effector protein NIP1 from Rhynchosporium secalis have similar effects on its avirulence- and virulence-associated activities on barley. Physiol Mol Plant Pathol 61 299–302 [Google Scholar]
- Fliegmann J, Mithöfer A, Wanner G, Ebel J (2004) An ancient enzyme domain hidden in the putative β-glucan elicitor receptor of soybean may play an active part in the perception of pathogen-associated molecular patterns during broad host resistance. J Biol Chem 279 1132–1140 [DOI] [PubMed] [Google Scholar]
- Flor HH (1971) Current status of the gene-for-gene concept. Annu Rev Phytopathol 9 275–296 [Google Scholar]
- Galán JE, Collmer A (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284 1322–1328 [DOI] [PubMed] [Google Scholar]
- Gerber IB, Dubery IA (2004) Protein phosphorylation in Nicotiana tabacum cells in response to perception of lipopolysaccharides from Burkholderia cepacia. Phytochemistry 65 2957–2966 [DOI] [PubMed] [Google Scholar]
- Gierlich A, van't Slot KAE, Li VM, Marie C, Hermann H, Knogge W (1999) Heterologous expression of the avirulence gene product, NIP1, from the barley pathogen Rhynchosporium secalis. Protein Expr Purif 17 64–73 [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 182 319–326 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T (2002) Flagellin perception: a paradigm for innate immunity. Trends Plant Sci 7 251–256 [DOI] [PubMed] [Google Scholar]
- Gross A, Kapp D, Nielsen T, Niehaus K (2005) Endocytosis of Xanthomonas campestris pathovar campestris lipopolysaccharides in non-host plant cells of Nicotiana tabacum. New Phytol 165 215–226 [DOI] [PubMed] [Google Scholar]
- Hahn M, Jüngling S, Knogge W (1993) Cultivar-specific elicitation of barley defense reactions by the phytotoxic peptide NIP1 from Rhynchosporium secalis. Mol Plant Microbe Interact 6 745–754 [DOI] [PubMed] [Google Scholar]
- Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, Lopez-Estrano C, Haldar K (2004) A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306 1934–1937 [DOI] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Jones JDG (1997) The role of leucine-rich repeat proteins in plant defences. Adv Bot Res 24 89–167 [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Joosten MHAJ, de Wit PJGM (1999) The tomato-Cladosporium fulvum interaction: a versatile experimental system to study plant-pathogen interactions. Annu Rev Phytopathol 37 335–367 [DOI] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N (2006) Plant cells recognize chitin fragments for defense signaling through a novel plasma membrane receptor. Proc Natl Acad Sci USA 103 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemen E, Kemen AC, Rafiqi M, Hempel U, Mendgen K, Hahn M, Voegele RT (2005) Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol Plant Microbe Interact 18 1130–1139 [DOI] [PubMed] [Google Scholar]
- Kjellbom P, Larsson C (1984) Preparation and polypeptide composition of chlorophyll-free plasma membranes from leaves of light-grown spinach and barley. Physiol Plant 62 501–509 [Google Scholar]
- Knogge W (1996) Fungal infection of plants. Plant Cell 8 1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knogge W, Marie C (1997) Molecular characterization of fungal avirulence. In IR Crute, EB Holub, eds, The Gene-for-Gene Relationship in Plant-Parasite Interactions. CAB International, Wallingford, UK, pp 329–346
- Knogge W, Scheel D (2006) LysM receptors recognize friend and foe. Proc Natl Acad Sci USA 103 10829–10830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooman-Gersmann M, Honée G, Bonnema G, de Wit PJGM (1996) A high-affinity binding site for the AVR9 peptide elicitor of Cladosporium fulvum is present on plasma membranes of tomato and other solanaceous plants. Plant Cell 8 929–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooman-Gersmann M, Vogelsang R, Vossen P, van den Hooven HW, Mahé E, Honée G, de Wit PJGM (1998) Correlation between binding affinity and necrosis-inducing activity of mutant AVR9 peptide elicitors. Plant Physiol 117 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Widell S, Sommarin M (1988) Inside-out plant plasma membrane vesicles of high purity obtained by aqueous two-phase partitioning. FEBS Lett 229 289–292 [Google Scholar]
- Lehnackers H, Knogge W (1990) Cytological studies on the infection of barley cultivars with known resistance genotypes by Rhynchosporium secalis. Can J Bot 68 1953–1961 [Google Scholar]
- Luderer R, Joosten MHAJ (2001) Avirulence proteins of plant pathogens: determinants of victory and defeat. Mol Plant Pathol 2 355–364 [DOI] [PubMed] [Google Scholar]
- Luderer R, Rivas S, Nürnberger T, Mattei B, van den Hooven HW, van der Hoorn RAL, Romeis T, Wehrfritz JM, Blume B, Nennstiel D, et al (2001) No evidence for binding between resistance gene product Cf-9 of tomato and avirulence gene product AVR9 of Cladosporium fulvum. Mol Plant Microbe Interact 14 867–876 [DOI] [PubMed] [Google Scholar]
- Manning VA, Ciuffetti LM (2005) Localization of Ptr ToxA produced by Pyrenophora tritici-repentis reveals protein import into wheat mesophyll cells. Plant Cell 17 3203–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti M, Good RT, Rug M, Knuepfer E, Cowman AF (2004) Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306 1930–1933 [DOI] [PubMed] [Google Scholar]
- McFarthing KG (1992) Selection and synthesis of receptor-specific radioligands. In EC Hulme, ed, Receptor-Ligand Interactions. A Practical Approach. IRL Press, Oxford, pp 1–18
- Meindl T, Boller T, Felix G (2000) The bacterial elicitor flagellin activates its receptor in tomato cells according to the address-message concept. Plant Cell 12 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithöfer A, Fliegmann J, Neuhaus-Url G, Schwarz H, Ebel J (2000) The hepta-β-glucoside elicitor-binding proteins from legumes represent a putative receptor family. Biol Chem 381 705–713 [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Lipka V (2005) Non-host resistance in plants: new insights into an old phenomenon. Mol Plant Pathol 6 335–345 [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Nennstiel D, Jabs T, Sacks WR, Hahlbrock K, Scheel D (1994) High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 78 449–460 [DOI] [PubMed] [Google Scholar]
- Orbach MJ, Farrall L, Sweigard JA, Chumley FG, Valent B (2000) A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG, Askerlund P, Fredrikson K, Widell S, Sommarin M, Larsson C (1990) Sealed inside-out and right-side-out plasma membrane vesicles: optimal conditions for formation and separation. Plant Physiol 92 871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Gronlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592 [DOI] [PubMed] [Google Scholar]
- Rivas S, Thomas CM (2005) Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu Rev Phytopathol 43 395–436 [DOI] [PubMed] [Google Scholar]
- Rohe M, Gierlich A, Hermann H, Hahn M, Schmidt B, Rosahl S, Knogge W (1995) The race-specific elicitor, NIP1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J 14 4168–4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney HCE, van't Klooster JW, van der Hoorn RAL, Joosten MHAJ, Jones JDG, de Witt PJGM (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308 1783–1786 [DOI] [PubMed] [Google Scholar]
- Schürch S, Linde CC, Knogge W, Jackson LF, McDonald BA (2004) Molecular population genetic analysis differentiates two virulence mechanisms of the fungal avirulence gene NIP1. Mol Plant Microbe Interact 17 1114–1125 [DOI] [PubMed] [Google Scholar]
- Shibuya N, Ebisu N, Kamada Y, Kaku H, Cohn J, Ito Y (1996) Localization and binding characteristics of a high-affinity binding site for N-acetylchitooligosaccharide elicitor in the plasma membrane from suspension-cultured rice cells suggest a role as a receptor for the elicitor signal at the cell surface. Plant Cell Physiol 37 894–898 [Google Scholar]
- Steiner-Lange S, Fischer A, Böttcher A, Rouhara I, Liedgens H, Schmelzer E, Knogge W (2003) Differential defense reactions in leaf tissues of barley in response to infection by Rhynchosporium secalis and to treatment with a fungal avirulence gene product. Mol Plant Microbe Interact 16 893–902 [DOI] [PubMed] [Google Scholar]
- Takken FLW, Joosten MHAJ (2000) Plant resistance genes: their structure, function and evolution. Eur J Plant Pathol 106 699–713 [Google Scholar]
- Umemoto N, Kakitani M, Iwamatsu A, Yoshikawa M, Yamaoka N, Ishida I (1997) The structure and function of a soybean β-glucan-elicitor-binding protein. Proc Natl Acad Sci USA 94 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg HA, Spronk CAEM, Boeren S, Kennedy MA, Vissers JPC, Vuister GW, de Wit PJGM, Vervoort J (2004) Binding of the AVR4 elicitor of Cladosporium fulvum to chitotriose units is facilitated by positive allosteric protein-protein interactions—the chitin-binding site of AVR4 represents a novel binding site on the folding scaffold shared between the invertebrate and the plant chitin-binding domain. J Biol Chem 279 16786–16796 [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Westerink N, Francoijs KJ, Roth R, Woestenenk E, Boeren S, de Wit PJGM, Joosten MHAJ, Vervoort J (2003) Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. J Biol Chem 278 27340–27346 [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Jones JDG (1998) The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol 8 R226–R227 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RAL, de Wit PJGM, Joosten MHAJ (2002) Balancing selection favors guarding resistance proteins. Trends Plant Sci 7 67–71 [DOI] [PubMed] [Google Scholar]
- van't Slot KAE, Knogge W (2002) A dual role of microbial pathogen-derived proteins in plant disease and resistance. CRC Crit Rev Plant Sci 21 229–271 [Google Scholar]
- van't Slot KAE, van den Burg HA, Kloks CPAM, Hilbers CW, Knogge W, Papavoine CHM (2003) Solution structure of the plant disease resistance-triggering protein NIP1 from the fungus Rhynchosporium secalis shows a novel β-sheet fold. J Biol Chem 278 45730–45736 [DOI] [PubMed] [Google Scholar]
- Vervoort J, van den Hooven HW, Berg A, Vossen P, Vogelsang R, Joosten MHAJ, de Wit PJGM (1997) The race-specific elicitor AVR9 of the tomato pathogen Cladosporium fulvum: a cystine knot protein. FEBS Lett 404 153–158 [DOI] [PubMed] [Google Scholar]
- Wendehenne D, Binet MN, Blein JP, Ricci P, Pugin A (1995) Evidence for specific, high-affinity binding sites for a proteinaceous elicitor in tobacco plasma membrane. FEBS Lett 374 203–207 [DOI] [PubMed] [Google Scholar]
- Westerink N, Roth R, van den Burg HA, de Wit PJGM, Joosten MHAJ (2002) The AVR4 elicitor protein of Cladosporium fulvum binds to fungal components with high affinity. Mol Plant Microbe Interact 15 1219–1227 [DOI] [PubMed] [Google Scholar]
- Wevelsiep L, Kogel K-H, Knogge W (1991) Purification and characterization of peptides from Rhynchosporium secalis inducing necrosis in barley. Physiol Mol Plant Pathol 39 471–482 [Google Scholar]
- Wevelsiep L, Rüpping E, Knogge W (1993) Stimulation of barley plasmalemma H+-ATPase by phytotoxic peptides from the fungal pathogen Rhynchosporium secalis. Plant Physiol 101 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widell S, Lundborg T, Larsson C (1982) Plasma membranes from oats prepared by partitioning in an aqueous polymer two-phase system. Plant Physiol 70 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]