Abstract
Inositol 1,3,4,5,6-pentakisphosphate 2-kinase, an enzyme encoded by the gene IPK1, catalyzes the terminal step in the phytic acid biosynthetic pathway. We report here the isolation and characterization of IPK1 cDNA and genomic clones from maize (Zea mays). DNA Southern-blot analysis revealed that ZmIPK1 in the maize genome constitutes a small gene family with two members. Two nearly identical ZmIPK1 paralogs, designated as ZmIPK1A and ZmIPK1B, were identified. The transcripts of ZmIPK1A were detected in various maize tissues, including leaves, silks, immature ears, seeds at 12 d after pollination, midstage endosperm, and maturing embryos. However, the transcripts of ZmIPK1B were exclusively detected in roots. A variety of alternative splicing products of ZmIPK1A were discovered in maize leaves and seeds. These products are derived from alternative acceptor sites, alternative donor sites, and retained introns in the transcripts. Consequently, up to 50% of the ZmIPK1A transcripts in maize seeds and leaves have an interrupted open reading frame. In contrast, only one type of splicing product of ZmIPK1B was detected in roots. When expressed in Escherichia coli and subsequently purified, the ZmIPK1 enzyme catalyzes the conversion of myo-inositol 1,3,4,5,6-pentakisphosphate to phytic acid. In addition, it is also capable of catalyzing the phosphorylation of myo-inositol 1,4,6-trisphosphate, myo-inositol 1,4,5,6-tetrakisphosphate, and myo-inositol 3,4,5,6-tetrakisphosphate. Nuclear magnetic resonance spectroscopy analysis indicates that the phosphorylation product of myo-inositol 1,4,6-trisphosphate is inositol 1,2,4,6-tetrakisphosphate. Kinetic studies showed that the Km for ZmIPK1 using myo-inositol 1,3,4,5,6-pentakisphosphate as a substrate is 119 μm with a Vmax at 625 nmol/min/mg. These data describing the tissue-specific accumulation and alternative splicing of the transcripts from two nearly identical ZmIPK1 paralogs suggest that maize has a highly sophisticated regulatory mechanism controlling phytic acid biosynthesis.
Inositol 1,2,3,4,5,6-hexakisphosphate [Ins(1,2,3,4,5,6)P6], commonly known as phytic acid, is an abundant natural product and is found in all eukaryotes (Sasakawa et al., 1995). Mineral salts of phytic acid, called phytate or phytin, have been detected in a wide range of plant tissues including seeds, roots, tubers, leaves, and pollen (Raboy, 2003). The biological functions of phytate in mammals and yeast (Saccharomyces cerevisiae) include involvement in dynamin I-mediated endocytosis (Hoy et al., 2002), DNA repair (Hanakahi and West, 2002), RNA export (Miller et al., 2004), and RNA editing (Macbeth et al., 2005).
In higher plants, it has been demonstrated that phytic acid can mediate abscisic acid-induced guard cell closure by inactivating plasma membrane inward K+ conductance (Lemtiri-Chlieh et al., 2000). During this process, phytic acid appears to act as an endomembrane calcium-releasing signal (Lemtiri-Chlieh et al., 2003). However, the biological significance of phytic acid accumulation in plant seeds remains unclear. Phytate typically represents about 75% of total seed phosphorus (Raboy, 1997). Most, if not all, of this phosphate storage product is dispensable since mutations that cause significant reduction in seed phytate do not affect seed development in barley (Hordeum vulgare; Larson et al., 1998; Rasmussen and Hatzack 1998; Dorsch et al., 2003), maize (Zea mays; Raboy and Gerbasi 1996; Raboy et al., 2000; Shi et al., 2003, 2005), soybean (Glycine max; Wilcox et al., 2000; Hitz et al., 2002), wheat (Triticum aestivum; Guttieri et al., 2004), rice (Oryza sativa; Larson et al., 2000), or Arabidopsis (Arabidopsis thaliana; Stevenson-Paulik et al., 2005). Almost 50% of the phosphorus fertilizer applied worldwide ends up as phytate in crop seeds (Lott et al., 2000). This massive accumulation of phytate in seeds poses a serious challenge to industrial agriculture. Monogastric animals such as pigs, poultry, and fish can't digest phytate. When crop seeds are used to feed these livestock, phosphorus in the form of phytate is released as pollutant waste into the environment. Phytic acid also has an antinutritional effect on feed quality because it chelates minerals and significantly reduces their bioavailability to animals. An additional consequence of the production and consumption of phosphorus fertilizers and phosphorus feeding additives is the depletion of natural reserves of inorganic phosphorus in the environment. These challenges point to a need for development and commercialization of crops with reduced levels of phytate in seeds. To accomplish this goal, gene targets in the phytic acid biosynthesis and regulatory pathways need to be identified for modification.
Two parallel phytic acid biosynthetic pathways, one lipid dependent and the other lipid independent, have been proposed for higher plants (Brearley and Hanke, 2000; Stevenson-Paulik et al., 2002; Raboy, 2003). Radioisotope labeling experiments with duckweed (Spirodela polyrhiza), an aquatic plant, suggest that a lipid-independent pathway may be operational in plants. The intermediates of this phytic acid biosynthetic route were reported to be Ins(3)P1, Ins(3,4)P2, Ins(3,4,6)P3, Ins(3,4,5,6)P4, and Ins(1,3,4,5,6)P5 (Brearley and Hanke, 1996a, 1996b). In a separate study, Ins(3,4,5,6)P4 1-kinase activity was detected in mesophyll protoplasts of Commelina communis using a cell permeabilization method (Brearley and Hanke, 2000). Taken together, these observations support the existence of a functional lipid-independent pathway in higher plants.
Ins(1,3,4,5,6)P5 is the common intermediate in both lipid-dependent and lipid-independent phytic acid biosynthetic pathways. A kinase activity that is able to phosphorylate this substrate at the 2 position on the inositol ring was first detected in mung bean (Vigna radiata; Biswas et al., 1978; Stephens et al., 1991). Purification of a protein corresponding to this kinase activity from immature soybean seeds revealed that the enzyme had a molecular mass of 52,000 D as determined by SDS-PAGE and a pH optimum at 6.8 (Phillippy et al., 1994). More recently, progress on characterization of this kinase at the molecular level has followed the discovery of the Ins(1,3,4,5,6)P5 2-kinase gene (IPK1) in budding yeast (York et al., 1999; Ives et al., 2000). Orthologs of IPK1 were subsequently isolated from Schizosaccharomyces pombe (Ives et al., 2000), human (Verbsky et al., 2002), Drosophila (Seeds et al., 2004), and Arabidopsis (Stevenson-Paulik et al., 2005; Sweetman et al., 2006). T-DNA insertional disruption of the Arabidopsis AtIPK1 gene was shown to result in accumulation of Ins(1,3,4,5,6)P5 in seeds, and Arabidopsis lines containing insertional disruptions in both AtIPK1 and AtIPK2β gave rise to phytate-free seeds (Stevenson-Paulik et al., 2005). These findings suggest that Ins(1,3,4,5,6)P5 2-kinase is a promising target for the manipulation of phytate in crops.
In a previous publication, Stevenson-Paulik et al. (2005) identified two assembled genomic sequences, AZM_26714 and AZM_81106, as maize IPK1 orthologs. We report here the isolation and characterization of maize IPK1 (ZmIPK1) cDNA and genomic clones from a commercial inbred line (5XH751). Different ZmIPK1 splicing variants have been identified and isolated from seed and leaf tissues. We have discovered that two distinct maize IPK1 paralogs, ZmIPK1A and ZmIPK1B, are expressed in a tissue-specific manner. In addition, the detailed biochemical properties of ZmIPK1 kinase encoded by maize ZmIPK1 and its role in phytate biosynthesis will also be discussed.
RESULTS
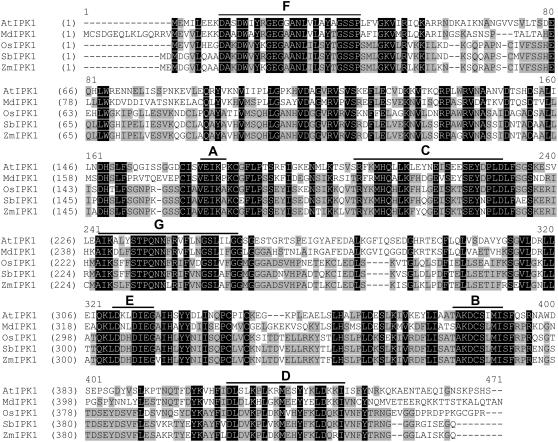
To identify the maize IPK1 ortholog, the amino acid sequence of the human IPK1 (Verbsky et al., 2002) was used as a query to BLAST the maize EST sequence database. A 1.7 kb maize contiguous sequence (contig) with 23% amino acid sequence identity to human IPK1 was identified (ZMtuc02-12-23.4536), and appeared to be a plausible IPK1 ortholog in maize. Cloning of the maize IPK1 ortholog (designated as ZmIPK1) from inbred line 5XH751 was accomplished by two rounds of reverse transcription (RT)-PCR. First, cDNA was amplified from maize seed at 12 d after pollination (DAP) using primers designed from the sequence of ZmTuc02-12-23.4536. This clone is 1.6 kb in length with a predicted open reading frame of 1.3 kb. The nucleotide sequence of this clone is 98% identical to the contig ZMtuc02-12-23.4536. Based on the sequences of 1.6 kb ZmIPK1 cDNA clone, additional PCR primers were designed, and 5′- and 3′-RACE reactions were performed to obtain the 5′ and 3′ untranslated region (UTR) of the ZmIPK1 transcripts. After sequencing the 5′- and 3′-RACE products, the sequences representing the longest 5′ and 3′ UTR fragments were selected to design primers for the amplification of full-length ZmIPK1 cDNA sequences. The resulting 2,012 bp cDNA clone (GenBank accession: DQ431470) contains a large predicted open reading frame of 440 amino acids with a predicted molecular mass of 48,850 D, overall charge of +7.5, and a pI of 7.8. The ZmIPK1 protein has high amino acid sequence identity to the IPK1 orthologs from the two closely related monocots. Rice IPK1 protein (OsIPK1:NP_001054147) is 82% identical to ZmIPK1 at the amino acid level. Sorghum IPK1 (SbIPK1) protein, deduced from a 1.4 kb EST contig (PUT-157a-Sorgum_bicolor-18953) and a 3.8 kb Sorghum genomic survey sequence contig (SbGSStuc11-12-04.5224.1) shares 91% identity to ZmIPK1 protein. In contrast, the degree of amino acid sequence identity between ZmIPK1 and three different Arabidopsis orthologs are 48% (At5g42810), 46% (At1g22100), and 44% (At1g59312), respectively. In addition, a 1.8 kb apple (Malus domestica) EST contig (PUT-157a-Malus_x_domestica-76137, Plant GDB) was identified as an apple IPK1 ortholog. The deduced amino acid sequence of this EST (MdIPK1) shares 52% identity to ZmIPK1. Figure 1 shows the alignment of the predicted amino acid sequences of IPK1 gene products from maize, Arabidopsis, apple, Sorghum, and rice. The conserved motifs known as A, B, C, D, and E boxes, identified in the IPK1 proteins from human (Verbsky et al., 2002) and Arabidopsis (Sweetman et al., 2006), are present in the monocot species examined. In addition, domains we designate as F and G boxes were conserved in such diverse higher plants as Arabidopsis and apple (dicots) and rice, Sorghum and maize (monocots). The F-box domain contains the conserved amino acid residues GEG(G/A)ANL and the G-box domain contains the sequence PQNN(F/L)R(V/I)F. These conserved residues have also been identified in a truncated IPK1 protein from Phaseolus vulgaris (GenBank accession: CAM33431). The functional significance of these conserved residues in the various IPK1 proteins has not been determined.
Figure 1.
Comparison of IPK1 amino acid sequences from Arabidopsis, apple, rice, Sorghum, and maize. Full-length amino acid sequences of IPK1 were derived from publicly available genomic sequences of Arabidopsis (AtIPK1: AT5g42810) and rice (OsIPK1: NP_001054147), or cDNA sequences of apple (MdIPK1: PUT-157a-Malus_x_domestica-76137 from PlantGDB) and maize (ZmIPK1: DQ431470). Sorghum IPK1 (SbIPK1) protein sequence was derived from an EST contig (PUT-157a-Sorgum_bicolor-18953) and a genomic survey sequence contig (SbGSStuc11-12-04.5224.1). Identical amino acid residues are marked with black background and the conserved residues with gray background. The conserved boxes A, B, C, and D, reported in Arabidopsis and human IPK1 proteins (Verbsky et al., 2002; Sweetman et al., 2006), and the conserved E and F domains, are labeled above the relevant sequences.
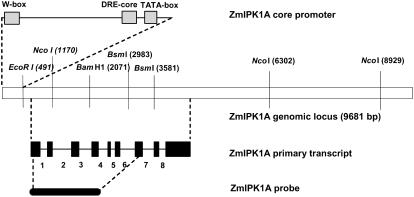
To address the genomic organization of ZmIPK1 in maize inbred 5XH751, we constructed a maize bacterial artificial chromosome (BAC) library from 5XH751 and isolated ZmIPK1 BAC clones as described in the “Materials and Methods” section. Sequence analysis of one BAC genomic clone indicated that it encoded a ZmIPK1 cDNA isolated from leaves and seeds. We have therefore designated this genomic clone as ZmIPK1A (GenBank accession: EF447274). Alignments of the leaf- and seed-derived cDNA sequences with ZmIPK1A defined nine predicted exons and eight predicted introns in the ZmIPK1 genomic sequence (Fig. 2). Comparison of ZmIPK1A DNA sequence to publicly available sequences in the Maize Assembled Genomic Island (MAGI) database revealed several accessions with remarkably high sequence identity to ZmIPK1A: MAGI4_31335, MAGI4_105158, MAGI4_110829, MAGI4_9262, and MAGI4_87135. A conserved TATA box sequence, TATATT, is located at position 362 bp of ZmIPK1A, suggesting that the transcription initiation site for the gene may actually be upstream of the 5′-most end of the cDNAs isolated from leaves and seeds. A conserved W-box cis-element, TTGACC, is located at position 20 to 25 bp of this sequence. It is well documented that a family of plant-specific zinc finger transcription factors, the WRKY transcription factors, is able to bind to this cis-element and regulate gene expression (Ülker and Somssich, 2004). In addition, a dehydration responsive element core sequence (Jia et al., 2006), CCGAC at position 323 to 326 bp, was also identified in the predicted promoter region of clone ZmIPK1A. Further experiments are needed to define the transcriptional start site of ZmIPK1 and to explore if these cis-elements in ZmIPK1A are of any biological significance.
Figure 2.
Genomic organization of ZmIPK1A from 5XH751. Restriction map of a 9.7 kb genomic ZmIPK1A (GenBank accession: EF447274) from maize inbred 5XH751 that contains the core promoter and transcribed regions. Gray boxes represent the putative TATA box, dehydration responsive element core sequence, and W box. Black boxes represent exons. Introns are numbered 1 through 8. The ZmIPK1A probe used for Southern analysis and BAC library screening is illustrated below the primary transcript.
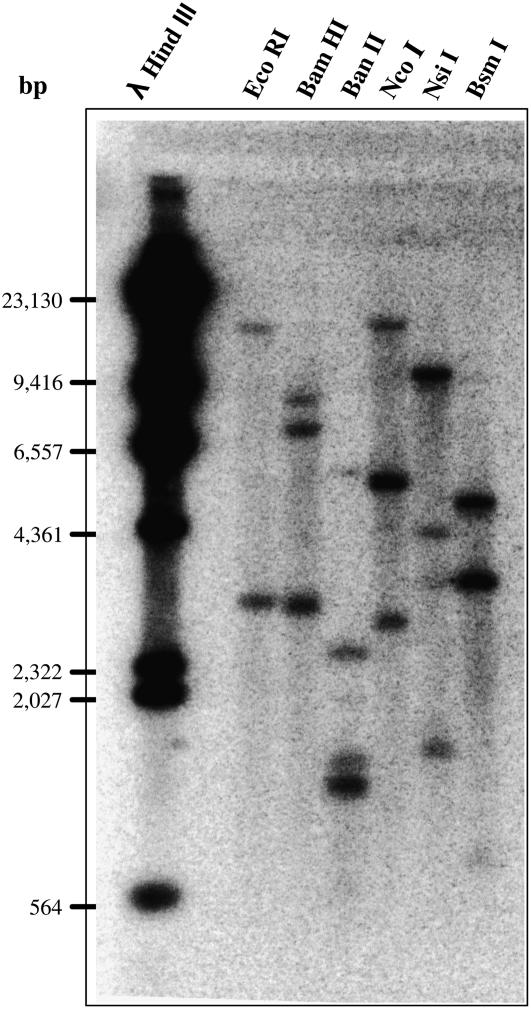
Genomic Southern blotting was performed using a 32P-labeled ZmIPK1 cDNA as a hybridization probe (Fig. 3). The genomic ZmIPK1A sequence contains only one EcoRI site (Fig. 2) in the predicted promoter region of the gene. If the ZmIPK1 gene is present in a single copy in the genome, hybridization with a 32P-labeled cDNA probe (as shown in Fig. 2) should detect only one band larger than 9 kb. As shown in Figure 3, two restriction digestion fragments were able to hybridize with ZmIPK1 cDNA probe when the genomic DNA was digested with EcoRI, including a band smaller than 9 kb in size. Therefore, we hypothesize that at least two IPK1 genes are present in the maize genome. The hybridization patterns of genomic DNA restricted with the enzymes BamHI and NcoI also support this assumption. If the gene is present in a single copy, the ZmIPK1A probe is predicted to hybridize with two genomic ZmIPK1A fragments (based on the restriction map, shown in Fig. 2). However, three maize 5XH751 genomic fragments generated from BamHI and NcoI digestions hybridized with the 32P-labeled ZmIPK1A cDNA probe (Fig. 3), indicating that an additional IPK1 gene is present in maize 5XH751 genome.
Figure 3.
Southern-blot analysis of ZmIPK1A. Genomic DNA was isolated from inbred line 5XH751 and digested with EcoRI, BamHI, BanII, NcoI, NsiI, and BsmI. The digested maize genomic DNA and λ phage DNA were separated on 0.8% agarose and transferred to Nylon membrane. The blot was probed with 32P-dCTP labeled ZmIPK1A and λ phage DNA. Black bars indicate the size and positions of the DNA Mr markers.
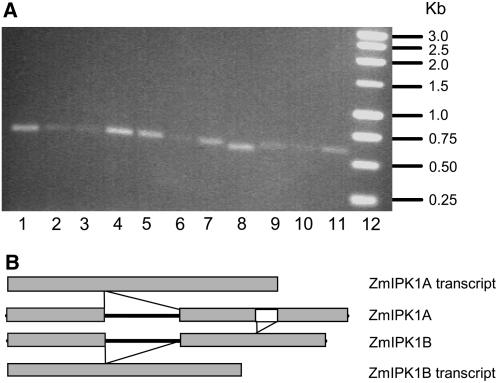
The expression patterns of ZmIPK1 in different tissues were analyzed by RT-PCR. To discriminate transcript-derived PCR products from those amplified from genomic DNA, the PCR primers were designed to flank intron 8 and cover a portion of 3′ UTR of ZmIPK1 as described in the “Materials and Methods” section. As shown in Figure 4A, ZmIPK1 transcripts were detected in leaves, silks, immature ears, seed at 5 and 10 DAP, endosperm at 15 and 25 DAP, and embryos at 20 and 31 DAP. Under the PCR conditions used in this experiment, the relative signal strength of ZmIPK1 transcripts appeared to be higher in embryo at 20 DAP, endosperm at 20 and 25 DAP, silk at flowering, seed at 10 DAP, and root from 3-week-old seedlings than in the other tissues examined. This observation need to be further validated by a more quantitative method such as quantitative RT-PCR. Interestingly, the RT-PCR products amplified from root tissues were slightly smaller in size than those derived from the other tissues. Sequence analysis of the cDNA fragments amplified from roots revealed that they contained a 31-bp deletion in addition to seven nucleotide polymorphisms when compared to transcripts from other tissues. To determine whether these transcripts were transcribed from different copies of the gene, or resulted from alternative splicing, genomic fragments of the corresponding region, 735 bp in size, were amplified by PCR, cloned, and sequenced. Sequence analysis revealed that there are two types of ZmIPK1 genomic sequences. One genomic fragment is identical to the ZmIPK1A genomic sequence obtained from isolated BAC clones (Fig. 2), and the other, designated as ZmIPK1B (GenBank accession: EF527875), contains a 31 bp deletion and a 7 bp polymorphism when compared to ZmIPK1A. As diagrammed in Figure 4B, the ZmIPK1 transcripts in the roots were derived from ZmIPK1B, while the ZmIPK1 transcripts from all the other tested tissues appear to be derived from ZmIPK1A. To further explore the expression of ZmIPK1B, 10 full-length cDNA clones of ZmIPK1B (GenBank accession: EF527876) were subsequently isolated from root tissue of maize inbred 5XH751 by 5′ and 3′ RACE. The full-length transcripts from ZmIPK1A and ZmIPK1B share greater than 99% identity. This high level of identity was also present in the 5′- and 3′-UTR regions. Therefore, we believe that the ZmIPK1A and ZmIPK1B are nearly identical paralogs (NIPs). BLAST searches also identified EST sequences of ZmIPK1A and IPK1B from maize inbred line B73 in the maize sequence public databases. Hence the occurrence of NIPs ZmIPK1A and B in maize is not limited to inbred 5XH751. Furthermore, orthologs of ZmIPK1A genomic and EST sequences, but not orthologs of ZmIPK1B, were identified in Sorghum and rice, indicating that this genome attribute may not be preserved in all monocots.
Figure 4.
Expression of ZmIPK1. A, RT-PCR amplification of ZmIPK1 transcripts from different maize tissues. RTs were initiated with mRNA from embryo at 20 (lane 1) and 31 DAP (lane 2), endosperm at 15 (lane 3), 20 (lane 4), and 25 (lane 5) DAP, seed at 5 (lane 6) and 10 (lane 7) DAP, root (lane 8), leaf (lane 9), immature ear (lane 10), and silk (lane 11). DNA Mr markers were loaded in lane 12. Transcripts of ZmIPK1 were detected in all the tested tissues, but the RT-PCR products from roots were slightly smaller than those of other tissues. B, Diagrammatic representation of the genomic regions of ZmIPK1A and ZmIPK1B corresponding to RT-PCR amplified cDNA fragments shown in section A. ZmIPK1B and its transcripts contained a deletion of 31 bp compared to ZmIPK1A.
By the use of 5′ and 3′ RACE, we have isolated 28 full-length cDNA clones from different maize tissues including leaves, seeds, and roots. Alignment of these cDNA sequences to genomic sequences from clone ZmIPK1A has revealed the presence of multiple splicing variants of ZmIPK1 transcripts (Fig. 5). At intron 1, alternative splicing may be caused by two alternative acceptor sites that are 3 or 13 bp distal from the original site. Consequently, 3 or 13 bp may be inserted into the 5′ UTR of the gene. These insertions have no effect on the open reading frame of ZmIPK1. Among the eight full-length cDNA clones derived from leaves, six contained this 3-bp insertion. In contrast, examination of cDNA clones from seeds revealed that only three out of 10 contained this insertion. Interestingly, all 10 full-length transcripts from roots contain a 13-bp insertion. Studying another position in the gene, the sixth intron may be spliced in four different ways as shown in Figure 5. When GAG↑GTG is used as donor site with CAG↑ATA as the acceptor site, the splicing product has an uninterrupted reading frame. Among 18 sequenced full-length cDNA clones from leaves and seeds, we observed that 10 were processed in this way. Alternatively, the acceptor site CAG↑GTT may be coupled with the donor site GAG↑GTG. As the result of this pairing, a 34 bp insertion would be generated in the transcripts, leading to a reading frame shift. Six of the 18 cDNA clones we examined contained this insertion. Additionally, intron 6 could be processed using the donor site AGG↑GTG and acceptor site CAG↑GTT, or the donor site CAG↑GGT and acceptor site TCA↑GAT. All of the alternatively spliced products at intron 6 are predicted to produce truncated proteins. At intron 7, alternative splicing is predicted to create a truncated open reading frame that would delete the carboxyl terminal of the kinase. Since this region contains amino acids that are highly conserved among the IPK1 proteins from different eukaryotes (Verbsky et al., 2002), such truncated translational products are unlikely to be functional. This type of transcript is assumed to be rare since only one of the 18 cDNA clones we examined contained this deletion. Taken together, our data on splicing variants of introns 6 and 7 indicates that nine of the 18 full-length ZmIPK1A cDNA clones from leaves and seeds contain an interrupted reading frame. However, no alternative splicing variants in introns 6 and 7 were detected among the 10 full-length cDNA clones from roots. Therefore, we hypothesize that alternative splicing of introns 6 and 7 is found predominantly in nonroot tissues.
Figure 5.
Alternative splicing of ZmIPK1 transcripts. Exon/intron junction site sequences of genomic ZmIPK1 at introns 1, 6, and 7 were aligned with the corresponding cDNA sequences from seed, leaf, and root. ZmIPK1 T1 to T9 are segments of cDNA sequences from leaves, seeds, and roots. All 10 transcripts from maize roots were spliced like T3 at intron 1, T6 at intron 6, and T8 at intron 7. The 18 transcripts from leaf and seed included various combinations of T1, T2, T4, T5, T6, T7, T8, and T9.
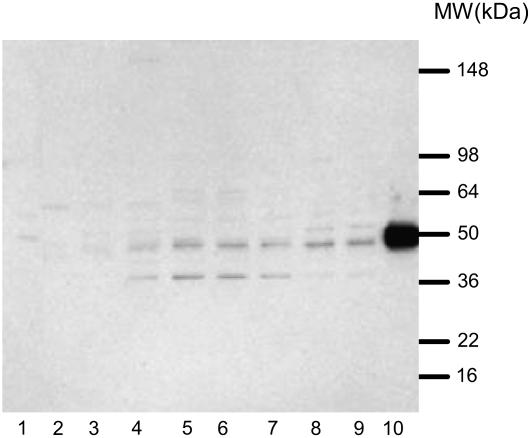
To understand if alternative splicing has contributed to ZmIPK1 protein complexity in different tissues, we have further analyzed the expression of ZmIPK1 with western blots. Polyclonal antibodies against ZmIPK1A protein were produced in rabbits. This antiserum was used to detect the ZmIPK1 protein in different tissues of maize as shown in Figure 6. Appreciable ZmIPK1 protein was detected in maize seed, endosperm, and embryos as a significant band with molecular mass of approximately 50 kD. This band may represent the unmodified ZmIPK1 protein in these tissues since it has the same size as the Escherichia coli expressed ZmIPK1. A protein band of slightly larger size was also detected in the embryo samples. This band presumably represents posttranslationally modified ZmIPK1. Since this band is clearly visible in embryo tissue where massive phytic acid accumulation is found, we speculate that this modification may have functional significance. In addition, a minor band with Mr of approximately 40 kD was detected in the seed and endosperm samples. This band is not likely to be the product of protease degradation since protease inhibitors were included in the protein extraction buffer. The alternative splicing variant of intron 7 described in Figure 5 is predicted to result in a truncated protein of similar size to the band we observed in the western blot. Therefore, we hypothesize that alternative splicing has contributed to the complexity of ZmIPK1 protein expression.
Figure 6.
Detection of ZmIPK1 protein in different maize tissues via western blotting. Proteins were extracted from maize leaf (lane 1), immature ear (lane 2), root (lane 3), seed at 12 DAP (lane 4), endosperm at 15 (lane 5), 20 (lane 6), and 25 (lane 7) DAP, and embryo at 24 (lane 8) and 31 (lane 9) DAP. After SDS-PAGE on a 8% to 16% gradient Tris-Gly polyacrylamide gel, the proteins were transferred onto a nitrocellulose membrane, incubated with polyclonal antibodies against maize ZmIPK1 protein, and detected with goat anti-rabbit antibody conjugated to horseradish peroxidase. ZmIPK1 protein (10 ng) purified from E. coli lysate was used as the reference (lane 10).
Our molecular analysis, as reported above, clearly demonstrated that ZmIPK1 is expressed in maize in a wide range of tissues as both transcript and protein. We then set out to address the biochemical functions of this gene product in maize tissues. To facilitate the purification process, ZmIPK1 protein was expressed in E.coli as a glutathione S-transferase (GST) fusion protein. Initially, accumulation of soluble ZmIPK1 protein was very low under various growing temperatures and induction conditions (data not shown), and the majority of the fusion protein was detected in inclusion bodies. Therefore, an alternative strategy using chaperone protein coexpression to enhance ZmIPK1 protein solubility was exploited. Several chaperone proteins, including DnaK, DnaJ, grpE, groES, groEL, and Trigger Factor (TF) in various combinations were evaluated. Only the chaperone protein TF significantly affected the solubility of GST-ZmIPK1 in E.coli BL21(DE3) cells, in conjunction with induction at a lower growth temperature (15°C–16°C) and lower isopropyl β-d-1-thiogalactopyranoside concentration (20 μm). Therefore, we used this approach to increase the amount of soluble GST-ZmIPK1 protein in cellular extract to approximately 20 mg/L as determined by Bradford assays.
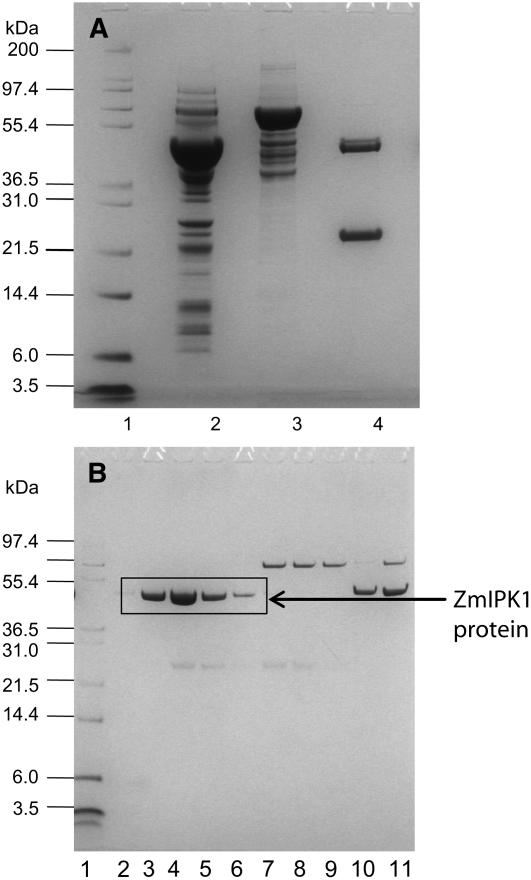
To purify the ZmIPK1 protein, a glutathione affinity column was used to capture intact GST-ZmIPK1 (Fig. 7A). The purified GST-ZmIPK1 fusion was subsequently cleaved with thrombin and further purified by a sequential affinity column and ion-exchange column chromatography (Fig. 7B). ZmIPK1 protein was eluted at 0.1 m NaCl on a high resolution Mono Q column with an estimated purity of 95%. The identity of the purified protein was confirmed via trypsin digestion followed by matrix-assisted laser deposition/ionization (MALDI) time-of-flight mass spectrometry analysis and comparison of the MALDI fingerprint to the amino acid sequences deduced from ZmIPK1 (data not shown).
Figure 7.
Purification of ZmIPK1 protein from E. coli lysate. A, ZmIPK1 protein was purified via glutathione affinity chromatography followed by GST cleavage with Thrombin. Lane 1, protein mass standards; lane 2, total soluble lysate of the recombinant host prior to purification; lane 3, proteins eluted from Glutathione-Sepharose; lane 4, thrombin cleaved samples. B, Purification of Thrombin digested GST-ZmIPK1 fusion protein with Mono Q ion-exchange chromatography. Lane 1, protein mass standards; lanes 2 to 11 are fractions eluted with 0.05 to 0.3 m NaCl.
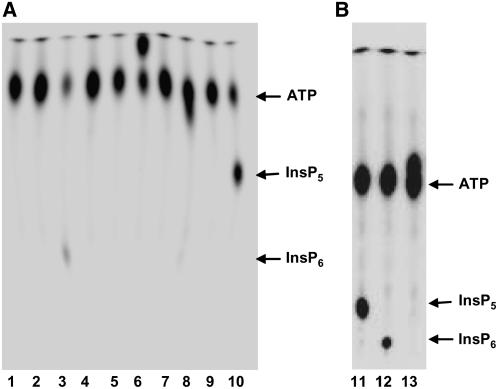
The purified ZmIPK1 protein was used in vitro to catalyze the phosphorylation of a number of inositol phosphate substrates. Ins(1,3,4,5,6)P5, Ins(1,4,6)P3, and Ins(1,4,5,6)P4 were phosphorylated by ZmIPK1 protein as demonstrated by thin-layer chromatography (TLC) analysis of radiolabeled inositol phosphates (Fig. 8). The phosphorylation product of Ins(1,3,4,6)P4 showed a migration similar to that observed for ATP under conditions when the TLC plate was developed with 1 n HCl (Fig. 8, lane 8). This phosphorylation product separated from ATP when 0.5 n HCl was used to develop the TLC plate (data not shown). However, separation of the phosphorylation product of Ins(1,4,6)P3 from ATP was accomplished by the use of 1 n HCl to develop the TLC plates (Fig. 8, lane 6). At a lower HCL concentration (0.75 n), the phosphorylation product comigrated with ATP (Fig. 8B, lane 13).
Figure 8.
Analysis of ZmIPK1 activities via TLC. The activity of ZmIPK1 protein was analyzed in 50 mm Tris-HCl (pH 7.5) containing 6 mm MgCl2, 10 mm LiCl, 1 mm DTT, 50 μm of ATP, and 2.5 μCi of γ-32P ATP in a 20 μL reaction. Ins(1,3,4,5,6)P5 (lane 3 and lane 12), Ins(1,5)P2 (lane 4), Ins(1,5,6)P3 (lane 5), Ins(1,4,6)P3 (lane 6 and lane 13), Ins(2,3,5)P3 (lane 7), Ins(1,3,4,6)P4 (lane 8) and Ins(1,3,5,6)P4 (lane 9), and Ins(1,4,5,6)P4 (lane 10 and lane 11) were tested as substrates of ZmIPK1. Controls were loaded in lane 1 (no enzyme/inositol phosphate added) and lane 2 [Ins(1,3,4,5,6)P5 was added, no enzyme]. The PEI-cellulose plate was developed in 1 n HCl (A) or 0.75 N HCl (B). The identity of the phosphorylation product in lanes 6 and 13 was further determined by NMR.
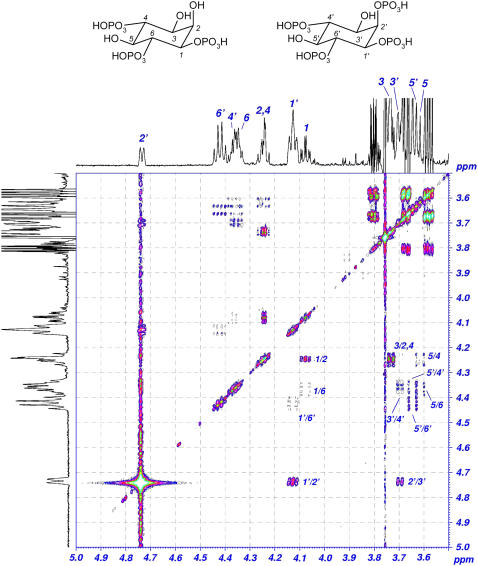
The identity of the phosphorylation product of Ins(1,4,6)P3 was subsequently determined by H+ NMR. After 24 h of incubation with ZmIPK1 protein purified from E. coli lysate, the in vitro kinase reaction was approximately 45% complete as judged from peak integrals. The reaction product was determined to be Ins(1,2,4,6)P4. Peak assignments for both the substrate and product were made using the COSY spectrum (Fig. 9). Proton 2′ of the product is obscured by a strong water peak at 4.7 ppm in the two-dimensional COSY spectrum. However, cross peaks to 1′ and 3′ can be readily observed and the peak corresponding to 2′ is clearly visible in the one-dimensional RECUR-TOCSY spectrum shown as projections. A strong downfield shift for this proton from 4.24 to 4.72 ppm is observed along with a large one-bond 1H-31P coupling, indicating a phosphorylation event at the 2′ position in the product to form Ins(1,2,4,6)P4. Mass spectrometry analysis of the reaction product indicated that a Mr consistent with a tetraphosphate moiety was observed (data not shown). Proton chemical shifts and peak multiplicities for all other protons in the product were similar to those in the starting material and indicated that no additional phosphorylation, dephosphorylation, or epimerization had occurred.
Figure 9.
Six hundred megahertz COSY NMR spectrum of Ins(1,4,6)P3 solution in D2O 21 h after the addition of ZmIPK1 protein. Peaks 1 to 6 correspond to Ins(1,4,6)P3, while peaks 1′ to 6′ correspond to the product Ins(1,2,4,6)P4. Multiplets at 3.81, 3.68, and 3.59 ppm are due to glycerol present as an impurity in the protein. The large cross peak at 4.74 ppm in the COSY spectrum is due to the residual water peak. This peak was eliminated using a RECUR-TOCSY pulse sequence in the proton spectra shown as one-dimensional projections.
To determine the substrate specificity of the ZmIPK1, a comprehensive survey using a wide range of commercially available IP4 and IP5 substrate species was performed. For this study, the degree of relative phosphorylation, normalized to the product of Ins(1,3,4,5,6)P5 (designated as 100%), was used to assess substrate specificity. Of the four different IP5 species tested, the highest activity was found with Ins(1,3,4,5,6)P5 as the substrate (Table I). Low, but detectable, activity was found when Ins(1,2,3,5,6)P5 was used. Of the 11 IP4 species tested, Ins(1,4,5,6)P4 was most efficiently phosphorylated by ZmIPK1 protein. Appreciable activity was detected when Ins(3,4,5,6)P4 was used as substrate in the reaction. Four IP3s were tested in this study, and we observed that Ins(1,4,6)P3 was the only IP3 species that was phosphorylated. Therefore, we conclude that the ZmIPK1 kinase is able to phosphorylate IP3, IP4, and IP5 at various efficiencies. For efficient phosphorylation, availability of an open substrate at the 2 position on the inositol ring is required. Phosphorylation reactions at the other positions of the inositol ring may significantly affect the phosphorylation at the 2 position.
Table I.
Relative substrate specificity of ZmIPK1 kinase
The activity of ZmIPK1 kinase catalyzing Ins(1,3,4,5,6)P5 phosphorylation was used as the reference (100%) to calculate the relative activity. The data are averages from two repeated assays. ND refers to not detected.
| Reaction | Substrates | Relative Activity |
|---|---|---|
| % | ||
| 1 | Ins(1,2,3,4,6)P5 | ND |
| 2 | Ins(1,3,4,5,6)P5 | 100 |
| 3 | Ins(1,2,3,5,6)P5 | 15.3 |
| 4 | Ins(2,3,4,5,6)P5 | ND |
| 5 | Ins(2,3,5,6)P4 | ND |
| 6 | Ins(1,2,4,5)P4 | ND |
| 7 | Ins(3,4,5,6)P4 | 32.6 |
| 8 | Ins(1,2,3,5)P4 | 2.1 |
| 9 | Ins(1,2,3,4)P4 | 0.5 |
| 10 | Ins(1,2,3,6)P4 | 0.9 |
| 11 | Ins(1,2,5,6)P4 | 0.6 |
| 12 | Ins(1,3,4,5)P4 | 0.8 |
| 13 | Ins(1,4,5,6)P4 | 76.8 |
| 14 | Ins(1,3,5,6)P4 | 1.3 |
| 15 | Ins(1,3,4,6)P4 | 14.9 |
| 16 | Ins(1,4,6)P3 | 93.9 |
| 17 | Ins(1,4,5)P3 | ND |
| 18 | Ins(2,3,5)P3 | ND |
| 19 | Ins(1,5,6)P3 | ND |
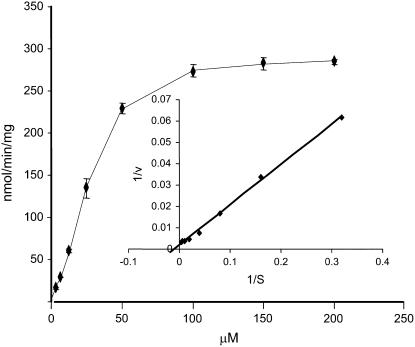
The Michaelis-Menten kinetics of ZmIPK1 protein was investigated according to experimental conditions described by Sweetman et al. (2006). To determine the Km for Ins(1,3,4,5,6)P5, 400 μm of ATP was used in the reactions. Results indicate that the consumption of ATP was less than 10% under these conditions. Figure 10 shows a plot of reaction velocity versus Ins(1,3,4,5,6)P5 concentration. We observed characteristic Michaelis-Menten kinetics. The double reciprocal plot of 1/v against 1/S gave a typical Lineweaver-Burk linear relationship with R2 of 0.997. The apparent Km for Ins(1,3,4,5,6)P5 and Vmax were derived from the plot as 119 μm and 625 nmol/min/mg, respectively. Both Km and Vmax of ZmIPK1 are significantly higher than that reported for the enzyme from Arabidopsis (Stevenson-Paulik et al., 2005; Sweetman et al., 2006). This difference may reflect the catalytic characteristics of the IPK1 kinase from the two divergent plant species.
Figure 10.
Kinetic studies of ZmIPK1. ZmIPK1 protein was purified from E. coli lysate and the GST tag was removed as described under “Materials and Methods.” Phosphorylation of Ins(1,3,4,5,6)P5 was analyzed in a total volume of 20 μL reactions containing 50 mm Tris-HCl (pH 7.5), 10 mm LiCl, 6 mm MgCl2, 400 μm ATP, and 3 μCi 32P-γ-ATP. The concentrations of Ins(1,3,4,5,6)P5 varied from 3.1 to 200 μm. After incubating at 30°C for 15 min, 0.5 μL of the reaction mix was spotted on PEI cellulose plates and developed with 1 n HCl. The error bar represents the deviation from two independent kinase assays and the insert is the Lineweaver-Burk plot of the average values with 1/v as (nmol/min/mg)−1 and 1/S as μm−1 of Ins(1,3,4,5,6)P5.
DISCUSSION
Ins(1,3,4,5,6)P5 2-kinase in maize is encoded by a small gene family with two duplicated members, ZmIPK1A and ZmIPK1B. This differs from the situation in Arabidopsis that contains multiple diverse genes (AT5g42810, AT1g22100, At1g59312, and AT5g59900) that show appreciable similarities to human IPK1 (Stevenson-Paulik et al., 2005; Sweetman et al., 2006). Transcript derived from AT5g42810, which has the highest sequence identity to ZmIPK1A, was detected in Arabidopsis seeds by DNA microarray analysis with peak expression detected at 7 d after flowering (Girke et al., 2000). RT-PCR amplification has detected AT5g42810 transcript in Arabidopsis leaf, cauline leaf, and siliques, while At5g59900 transcript was detected primarily in flower and stem (Sweetman et al., 2006). In contrast, in maize inbred line 5XH751, ZmIPK1A encodes the transcripts in leaves, silks, immature ears, seeds at 12 DAP, midstage endosperm, and maturing embryos, while ZmIPK1B specifically encodes transcripts in the roots. Thus, the two NIPs of ZmIPK1 encompass the diverse tissue-specific expression patterns in maize, while Arabidopsis appears to express a set of multiple diverse genes across tissue types. Since transcripts derived from ZmIPK1A and ZmIPK1B were also identified in inbred line B73 in the public EST database (Plant GDB), we assume that the duplicated ZmIPK genes exist in other domesticated maize lines as well. Interestingly, only the ZmIPK1A ortholog was identified in the publicly available genomic and EST sequences of Sorghum, a closely related monocot species. Based on the sequence information available so far, we hypothesize that ZmIPK1A is the ancestral copy of ZmIPK1 and the duplication presumably happened after the divergence of Sorghum from maize. This is in agreement with the assumption that Sorghum (n = 10) and maize (n = 10) evolved from a common ancestor with a haploid genome of five chromosomes (n = 5) by a genome splitting and a duplication process, respectively (Swigonova et al., 2004). Tetraploidization of maize after the divergence from Sorghum created a large collection of duplicated genes. It has been reported that at least one-third of maize genes are tandemly duplicated (Messing et al., 2004), 1% of maize genes have at least one NIP, and 80% of maize NIP families are differentially expressed in at least one tissue (Emrich et al., 2007). ZmIPK1 can provide a system to explore the biological significance of widespread gene duplications in maize. In Arabidopsis, AtIPK1-1 is required for both phytate accumulation in the seed and phosphorus sensing in the root (Stevenson-Paulik et al., 2005). In maize, due to the specificity of ZmIPK1B for roots, we assume that deletion of ZmIPK1A will affect phytate biosynthesis in the seed without disturbing the phosphorus sensing in the root. One can conceive that the duplication leading to ZmIPK1B increased the plasticity of the maize genome in the course of evolution.
Alternative splicing is a widespread phenomenon in higher eukaryotes. It has been estimated that over two-thirds of human genes and over 40% of Drosophila genes contain one or more alternative exons (Johnson et al., 2003; Stolc et al., 2004). A recent genome-wide analysis showed that about 20% of the genes in Arabidopsis and rice are alternatively spliced (Wang and Brendel, 2006). It was also reported that alternatively spliced genes in higher plants often encode proteins involved in signaling, regulation, and stress responses (Kazan, 2003). Although the phenomenon of alternative splicing is well documented in maize seed (Sun et al., 1997), there is no comprehensive genomic survey of the extent of alternative splicing in the maize transcriptome. We have found that, among 18 full-length ZmIPK1A cDNA clones from leaves and seeds, 50% of ZmIPK1A transcripts have interrupted reading frames as the consequence of alternative splicing of introns 6 and 7. So far, we do not know if these alternatively spliced transcripts are involved in biological functions other than catalysis of the phosphorylation of inositol phosphates. Interestingly, in contrast to multiple splicing variants of the transcripts of ZmIPK1A in leaves and seeds, only one type of splicing product was detected for ZmIPK1B in maize roots. Since alternative splicing has been implicated in accelerating the evolution of protein sequences (Xing and Lee, 2005), the two ZmIPK1 paralogs are presumably on different evolutionary routes. In the paradigm of functional specialization for duplicated genes described by Lynch and Katju (2004), one might imagine that ZmIPK1A is becoming subfunctionalized for both a phosphorus sensing and phosphate storage role, while ZmIPK1B is becoming subfunctionalized to perform the specific biological role of phosphorus sensing in roots (Stevenson-Paulik et al., 2005). This subfunctionization also implies that the biological function of IPK1 in root is of vital importance to higher plants in the course of evolution. Alternative splicing of intron 1 has no affect on the reading frame, but the presence of additional 13 bases in 5′ UTR may affect the efficiency of translation or the stability of the transcript by altering the structure in this region. Further investigations are needed to illustrate the biological significance of the alternative splicing we have observed.
Although both ZmIPK1 and AtIPK1 are able to phosphorylate Ins(1,3,4,5,6)P5 effectively, differences in catalytic kinetics and substrate specificity were revealed from our studies. First, the Km of ZmIPK1 for Ins(1,3,4,5,6,)P5 is 119 μm. This is significantly higher than the Km of AtIPK1 for the same substrate reported previously. Sweetman et al. (2006) reported that the Km for AtIPK1 is 22 μm for Ins(1,3,4,5,6)P5 while Stevenson-Paulik et al. (2005) reported that the Km for the same substrate is 7.6 μm. The Km for ZmIPK1 was derived from a series of kinetic studies with various ATP and enzyme concentrations at different temperature. Therefore, we believe our data reflect the kinetic characteristics of the ZmIPK1 kinase in maize. Several factors may have directly contributed to the difference of Km between ZmIPK1 and AtIPK1 proteins. First, the sequence similarity of IPK1 between maize and Arabidopsis is much lower than the similarity between IPK1 from maize and Sorghum. The difference may reflect the fact that ZmIPK1 kinase evolved to adapt the requirement of inositol phosphate flux in maize. Second, ATP concentration in the kinetic study may significantly affect the Km of Ins(1,3,4,5,6)P5 (Sweetman et al., 2006). To compare the Km of Ins(1,3,4,5,6)P5 IPK1 from maize and Arabidopsis, we have used the enzyme assay conditions of ATP at 400 μm and reaction temperature of 30°C described by Sweetman et al. (2006). In contrast, Stevenson-Paulik et al. (2005) performed their AtIPK1 kinetic studies at the significantly different ATP concentration of 2 mm and reaction temperature of 37°C. Another factor that may affect the Km is that in the previous studies with Arabidopsis IPK1 (Stevenson-Paulik et al., 2005; Sweetman et al., 2006), tagged proteins were directly used for the kinetic studies. In our experiments, the GST tag of ZmIPK1 was cleaved prior to enzyme assay. It is not clear at this time how the N-terminal GST or 6His/FLAG may affect the kinetic properties of the enzyme, though we have observed that the substrate specificity of ZmIPK1 was not affected by the GST tag. Apart from the difference in Km, ZmIPK1 exhibited strong phosphorylation activity when Ins(1,4,6)P3 was provided as a substrate. The product of this phosphorylation was further identified as Ins(1,2,4,6)P4 by NMR. In addition, ZmIPK1 kinase also showed appreciable activity toward Ins(3,4,5,6)P4, leading us to conclude that ZmIPK1 may act as an inositol polyphosphate 2-kinase with Ins(1,3,4,5,6)P5 as the most preferred substrate. IPK1 from Arabidopsis did not show phosphorylation activity on Ins(1,4,6)P3 and Ins(3,4,5,6)P4 (Sweetman et al., 2006). These differences could be caused by the divergence of the amino acid sequences from the two plant species. It is also likely to be caused by the differences in TLC conditions, such as types of plate and nature of developing reagent, of the enzyme assays. Under our experimental conditions, we have observed that the migration of different inositol phosphates on polyethyleneimine (PEI)-cellulose plates was highly sensitive to the concentration of HCl of the TLC developing solution. Under conditions of certain HCl concentrations, the phosphorylation products may comigrate with ATP and give false negative results. Although the phosphorylation of Ins(1,4,6)P3 was confirmed with NMR and mass spectrometry studies, the physiological significance of the phosphorylation of Ins(1,4,6)P3 is unclear since this intermediate is not in the proposed phytic acid biosynthesis pathway of plants (Raboy, 2003). Ins(3,4,5,6)P4 is an intermediate in the lipid-independent phytic acid biosynthesis pathway (Brearley and Hanke, 1996a, 1996b). Phosphorylation of Ins(3,4,5,6)P4 by ZmIPK1 kinase suggests that an even more complicated network of conversions among the inositol phosphates exists. This network may connect the lipid-dependent and lipid-independent phytic acid biosynthesis pathways and maintain homeostasis of inositol phosphates.
MATERIALS AND METHODS
Plant Materials
Maize (Zea mays; Dow AgroSciences commercial inbred 5XH751, hereafter referred to 5XH751) plants were grown in 5-gallon pots using 95% Metro-Mix from Sun Gro and 5% mineral soil. Greenhouse temperatures were set at 27°C daytime and 22°C night with relative humidity at 30% to 50%. Lighting was provided by High Pressure Sodium/Metal Halide mix and the day length was set for 16 h. 5XH751 maize kernels were harvested at 5, 10, 15, 20, 25, and 31 DAP. Endosperm and intact embryos were dissected from developing kernels at 15, 20, 25, and 31 DAP. Maize leaf and roots were sampled from 3-week-old seedlings. Immature ear and silk tissues were sampled at flowering. All the samples were frozen in liquid nitrogen and stored at −80°C until use.
Reagents
d-myo-inositol 1,2,3,4,6-pentakisphosphate, d-myo-inositol 1,3,4,5,6-pentakisphosphate, d-myo-inositol 1,2,3,5,6-pentakisphosphate, d-myo-inositol 2,3,4,5,6-pentakisphosphate, d-myo-inositol 2,3,5,6-tetrakisphosphate, d-myo-inositol 1,2,4,5-tetrakisphosphate, d-myo-inositol 3,4,5,6-tetrakisphosphate, d-myo-inositol 1,2,3,5-tetrakisphosphate, d-myo-inositol 1,2,3,4-tetrakisphosphate, d-myo-inositol 1,2,3,6-tetrakisphosphate, d-myo-inositol 1,2,5,6-tetrakisphosphate, d-myo-inositol 1,3,4,5-tetrakisphosphate, d-myo-inositol 1,4,5,6-tetrakisphosphate, d-myo-inositol 1,3,5,6-tetrakisphosphate, d-myo-inositol 1,3,4,6-tetrakisphosphate, d-myo-inositol 1,4,6-trisphosphate, d-myo-inositol 1,4,5-trisphosphate, d-myo-inositol 2,3,5-trisphosphate, and d-myo-inositol 1,5,6-trisphosphate were purchased from A.G. Scientific. Deuterated reagents were obtained from Cambridge Isotope Labs. All other chemicals were obtained from Sigma-Aldrich.
Isolation of cDNA Clones Encoding ZmIPK1 from Maize Seeds
Total RNA was isolated from maize (5XH751) seeds at 12 DAP using TRIzol (Invitrogen) and mRNA was subsequently purified with a MACS kit (Miltenyi Biotec) according to the manufacturer's recommendations. RT-PCR amplifications were performed to obtain cDNA clones encoding ZmIPK1. The RT reaction was carried out on mRNA isolated from maize seeds at 12 DAP and directed by a gene-specific primer derived from the sequence of ZMtuc02-12-03.4536, an EST contig in a public database (MaizeGDB), using the Superscriptase II system (Invitrogen) as suggested by the manufacturer. Subsequent amplification of the ZmIPK1 cDNA was accomplished by using primers derived from ZMtuc02-12-03.4536. The amplified cDNA (1.6 kb in length) was cloned into vector pCR2.1 (Invitrogen) using the TA Cloning kit from Invitrogen, and sequenced with the CEQ Dye Terminator Cycle Sequencing kit from Beckman Coulter.
Identification of the 5′- and 3′-Flanking Sequences of ZmIPK1
To ascertain the DNA sequence of the 5′ and 3′ UTRs of cDNAs encoding ZmIPK1 in maize seeds, leaves, and roots, RACE experiments were performed using the GeneRacer kit from Invitrogen. For 5′ RACE, the RNA oligo supplied by the manufacturer was ligated to mRNA isolated from maize (5XH751) seeds at 12 DAP, or leaves and roots from 3-week-old seedlings, and treated with calf intestinal phosphatase and tobacco (Nicotiana tabacum) acid pyrophosphatase. The 5′ end of ZmIPK1 transcripts were subsequently amplified with the anchor sequence of the RNA oligo and a gene-specific primer. The PCR products were then cloned into plasmid vector pCR2.1 (Invitrogen) using the TA Cloning kit (Invitrogen) and sequenced. To obtain 3′-UTR sequence, RT was directed with an oligo(dT) primer including anchor sequence at 3′ end (GeneRacer kit, Invitrogen). The 3′ ends of ZmIPK1 transcripts were then amplified with gene-specific primers derived from the 1.6 kb ZmIPK1 cDNA clone and from the 3′ anchor sequence flanking the oligo(dT) primer. The resulting PCR products were cloned into pCR2.1 and sequenced.
Isolation of Full-Length ZmIPK1 cDNA Clones from Leaf, Seed, and Root
The 5′- and 3′-end sequences of ZmIPK1 cDNAs generated from the 5′- and 3′-RACE experiments were used to design PCR primers. Full-length ZmIPK1 cDNA was amplified from mRNA isolated from seeds at 12 DAP, leaves, or roots from 3-week-old seedlings and subjected to RT directed by oligodT primers. The resulting PCR products were then cloned into pCR2.1 (Invitrogen) and a total of 28 full-length ZmIPK1 cDNA clones (10 from seeds, 10 from roots, and eight from leaves) were sequenced.
Isolation and Characterization of ZmIPK1 BAC Clones
A BAC library of genomic DNA from maize (5XH751) was prepared according to methods described by Tao et al. (2002). Ligation of megabase genomic DNA into BAC vector pECBAC1 was carried out as follows: genomic DNA eluted from the agarose plug was dialyzed two times against one liter of ice-cold 0.5× Tris-EDTA on ice, for 1 h. The concentration of collected DNA was estimated on a 1% agarose gel. Ligation reactions were performed at a vector:DNA Mr ratio of 1:4 with T4 DNA ligase enzyme according to standard procedures as described in Tao et al. (2002). Transformation of ligation mixtures into competent Escherichia coli cells (DHB10B, Invitrogen) was performed according to Tao et al. (2002) via electroporation using a Cell Porator system with a Voltage Booster and 0.15-cm gap Cell Porator cuvettes (Labrepco). Individual bacterial clone colonies were picked using a Q-Bot robot (Genetix) and arrayed into 300 384-well plates. Titer testing of this BAC library derived from inbred variety 5XH751 indicated that it contained approximately 115,000 clones with an average genomic fragment insertion size of 130 kb.
The arrayed 5XH751 BAC library was spotted onto 22 cm2 nylon membranes in 4 × 4 grids using a Q-Bot robot (Genetix). Filters were grown on Luria-Bertani (LB) agarose at 37°C overnight, denatured, fixed, and dried as per Sambrook and Sambrook (2001) except that an additional lysis step was added prior to hybridization with a 32P-labeled probe consisting of a 916-bp ZmIPK1 fragment generated by PCR using ZmIPK1 specific primers (5′-AGTCCCTTTCCCCGGGCTGTGGTAC-3′ and 5′-TTAAGTTGTTCTGAGGAGTTGAGAAAAGGGA-3′). The filters were washed twice with 1× SSC, 0.1% SDS and twice with 0.2× SSC, 0.1% SDS at 65°C. Visualization of positive clones was carried out via phosphorimaging for a 16-h exposure with storage-phosphor screens followed by Storm phosphorimager (Molecular Dynamics) analysis running Incogen High Density Filter Reader software. Positive clone cultures were retrieved from the library plate array and grown overnight at 37°C in LB media. BAC DNA was extracted from isolated clones using a Qiagen Large Construct kit as per the manufacturer's instructions.
The isolated ZmIPK1 BAC clones are approximately 180 kb in length, corresponding to the genomic region of maize chromosomes containing ZmIPK1. A 9.7 kb fragment of genomic ZmIPK1 sequence was obtained either through direct sequencing of BAC DNA or via shotgun subcloning of the BAC followed by plasmid sequencing (Lark Technologies).
Expression of ZmIPK1 in E. coli
A fragment of the putative ZmIPK1 cDNA sequence corresponding to the predicted open reading frame of 1.32 kb was cloned into the pGEX-2T plasmid expression vector (Amersham Pharmacia Biotech) resulting in a GST-ZmIPK1 fusion protein plasmid construct. The plasmid was used to cotransform E.coli BL21(DE3) cells harboring pTf16 plasmids from the Chaperone Plasmid set (TakaRa Bio Inc.) as per the manufacturer's recommendation. Approximately 20 mL of seed culture was transferred to 1 L of fresh LB medium containing 0.2% l-Ara. Once the suspension culture reached an OD600 of 1.0, the inducer isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 20 μm. The induced culture was then incubated at 15°C to 16°C with constant shaking at 225 RPM overnight.
Purification of Maize ZmIPK1 Protein from E. coli Lysates
Cells from a 6 L culture were harvested by centrifugation, resuspended in the lysis buffer containing 100 mL of phosphate-buffered saline (PBS) plus 5 mm dithiothreitol (DTT) and 2 mL protease inhibitor cocktail (Sigma-Aldrich), lysed on ice via sonication using a Branson Sonifier 450 (Branson Ultrasonic Corporation), and subsequently centrifuged at 20,000g for 20 min at 4°C. The pellet was resuspended in lysis buffer plus Triton X-100 (Sigma-Aldrich) at a final concentration of 4%, and subsequently incubated at room temperature for 1 h. The extracts were then further diluted to 400 mL with PBS plus 5 mm DTT to reduce the Triton X-100 concentration to 1%, filtered through a 0.45 μm filter, loaded onto a 20 mL GSTPrep 16/10 Glutathione-Sepharose column (Amersham), and washed with PBS plus 5 mm DTT. The glutathione transferase-ZmIPK1 fusion protein (GST-ZmIPK1) was eluted using 25 mm reduced glutathione in 50 mm Tris-HCl, pH 8.0, 5 mm DTT, and 0.1% Triton X-100. The pooled fractions containing GST-ZmIPK1 protein were dialyzed against 1 L of PBS plus 1 mm DTT at 4°C overnight. The total protein concentration was determined by using a Bio-Rad protein assay (Bio-Rad) and adjusted to approximately 1 to 2 mg/mL. Thrombin (Amersham), at 50 units or 5 μg per mg of GST-ZmIPK1 fusion protein, was used to cleave the GST domain from the fusion protein. The cleaved ZmIPK1 protein was subsequently purified via a two-step procedure. First, the final cleavage reaction was applied to a 5 mL GSTrap affinity column (Amersham). The unbound fractions containing ZmIPK1 protein were pooled and dialyzed against 20 mm Tris-HCl, pH 7.5, 1 mm DTT at room temperature for 2 h. Then, the sample was applied to an 8 mL Mono Q column (Amersham). A linear salt gradient (0.05–0.3 m NaCl, at a flow rate of 2 mL/min) was used to elute ZmIPK1 protein. Fractions containing ZmIPK1 protein were pooled, concentrated, and stored at −80°C.
ZmIPK1 Kinase Activity Assay
The activity of purified ZmIPK1 protein was analyzed according to Shi et al. (2003) with modifications. The kinase assays were carried out in buffer containing 50 mm Tris-HCl (pH 7.5), 6 mm MgCl2, 10 mm LiCl, 1 mm DTT, 40 μm inositol phosphate substrates, 40 μm ATP, and 2.5 μCi of γ-32P labeled ATP (3,000 Ci/mmol) in a total volume of 20 μL. After 15 min of incubation at 37°C, the reactions were stopped by addition of 5 μL 1 n HCl. A 0.5 μL aliquot of the reaction mixture was spotted on a PEI cellulose TLC plate (Merck). After developing in 1 n HCl and drying at 60°C, the plate was exposed to a Phosphor-Imager screen for 60 min. The intensity of the spots corresponding to the reaction products was quantified using a Personal FX scanner and Quantity One software (Bio-Rad). When Ins(1,3,4,6)P4 was used as substrate in the enzyme assay, 0.5 n HCl was used to develop the TLC plate for the enhanced separation of the phosphorylation product from ATP.
H+ NMR Identification of the Phosphorylation Product
A stock solution was prepared that contained 1 mm Ins(1,4,6)P3, 10 mm LiCl, 6 mm MgCl2, 50 mm d11-tris(hydroxymethyl)-methylamine, 1 mm d10-dl-1,4-DTT, and 1 mm d14-ATP in D2O. A total of 600 μL of this solution was placed in a 5-mm NMR tube and a baseline proton-NMR spectrum was obtained prior to addition of ZmIPK1 protein. The kinase reaction was then initiated by adding 45 μg of purified ZmIPK1 protein. Spectra were obtained as follows: the sample was maintained at 30°C and the NMR was carried out on a Bruker DRX-600 NMR (600 MHz proton frequency) using a 5-mm 1H-13C-15N triple-resonance inverse probe. Proton NMR data was collected using a RECUR-TOCSY pulse sequence to eliminate the large residual water peak at 4.8 ppm while retaining the substrate peaks lying underneath according to the method of Liu et al. (2001). A total of 440 scans were obtained with 32 K data points and a 2 s relaxation delay. Data collection and processing was carried out using the standard Bruker software XWIN-NMR. COSY spectra were obtained with presaturation of the water resonance using the standard BRUKER pulse sequence.
Kinetic Studies
The enzymatic activity of ZmIPK1 protein was analyzed in a 20 μL reaction volume containing 50 mm Tris-HCl (pH 7.5), 10 mm LiCl, 6 mm MgCl2, 1 mm DTT, 400 μm ATP, and 3 μCi 32P-γ-ATP. The concentration of Ins(1,3,4,5,6)P5 varied from 3.1 to 200 μm. The reactions were incubated in a 30°C water bath for 15 min and 0.5 μL of the reaction mix was spotted on PEI cellulose plates. After developing with 1 n HCl, the plates were dried at 60°C for 30 min and exposed to Phosphor-Imager screen for 2 h. The screen was subsequently scanned with a Personal FX scanner (Bio-Rad) and quantified with Quantity One software (Bio-Rad). The intensity of each spot was converted to radioactivity (disintegrations per min) by direct comparison to 32p-γ-ATP standards spotted on the same PEI cellulose plate.
Southern-Blot Analysis
Genomic DNA from inbred 5XH751 was isolated from 3-week-old leaf tissues as described by Moller et al. (1992). Approximately 12 μg genomic DNA was digested overnight with the following restriction enzymes: EcoRI, BamHI, BanII, NcoI, NsiI, and BsmI (New England Biolabs). The digested DNAs and HindIII digested λ DNA markers were separated on a 0.8% agarose gel and transferred to a Nylon membrane. Hybridization was performed at 65°C overnight with a probe consisting of a PCR-amplified 916 bp fragment of ZmIPK1 cDNA (the same probe described for BAC library screening) and HindIII digested λ DNA labeled with 32P-dCTP using the Random Priming DNA labeling system from Invitrogen. The hybridization buffer consisted 250 mm sodium phosphate, 1 mm EDTA, and 7% SDS at pH 7.4. After three washings with 0.2× SSC at 65°C, the blot was exposed to Phosphor Imager screen (Bio-Rad) and the image was captured with a personal FX scanner (Bio-Rad).
Expression of ZmIPK1
RT-PCR was carried out to analyze the expression pattern of ZmIPK1 in different tissues. For the RT reaction, poly(A)+ RNAs were isolated from leaves, silks, seed at 5 and 10 DAP, endosperm at 15 and 25 DAP, embryos at 20 and 31 DAP of maize inbred 5XH751, using the FastTrack mRNA isolation kit from Invitrogen. To isolate mRNA from roots and immature ears, total RNA was first isolated using TriZol (Invitrogen) and poly(A)+ RNA was subsequently isolated with MACS kit (Miltenyi Biotec). After RT directed by oligodT primer, ZmIPK1A transcripts were amplified using the primers (5′-ACCGCAAAGGACTGTAGCCT-3′ and 5′-GGTAGCCTGTGAGGGAAACTTCAGC-3′) under the following PCR conditions: one cycle at 94°C for 2 min, 25 cycles at 94°C for 1 min, one cycle at 62°C for 1 min, one cycle at 72°C for 1 min, and one cycle at 72°C for 7 min.
Production of Polyclonal Antibodies against ZmIPK1 Protein
Five milligrams of ZmIPK1 protein purified from E. coli was delivered to Custom Antibody Services at Invitrogen for polyclonal antibody production in rabbits. Two rabbits received their first injection of 0.5 mg of the ZmIPK1 protein mixed in Complete Freund's Adjuvant. After a 2 week rest period, booster shots were given in alternate weeks with 0.5 mg protein mixed in Incomplete Freund's Adjuvant. After a total of 5 injections, approximately 50 mL of serum was collected from each rabbit, which was tested in both ELISA and western-blotting experiments to confirm specificity.
Western-Blot Analysis
Total proteins were extracted from maize roots, leaves, immature ears, and seed at 10 DAP, endosperm at 15, 20, and 25 DAP, and embryos at 24 and 31 DAP. The extraction was carried out by grinding the tissues into fine powders in liquid nitrogen and mixing 0.5 g of the powder with 2 mL of extraction buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2 mm DTT, 5 mm EDTA, 0.1% Triton X-100, and 1× protease inhibitor cocktail from Sigma-Aldrich). After homogenization with a Polytron (Kinematica AG) three times for 10 s, the mixture was spun in a benchtop microcentrifuge at 14,000 rpm for 15 min at 4°C. The concentration of total soluble proteins in the supernatant was determined using the Bradford Protein Assay (Bio-Rad). Each sample was subsequently diluted with PBS to 1 mg/mL and incubated with 10 μL Laemmli buffer (Bio-Rad) for 10 min at 95°C. SDS-PAGE was conducted in an 8% to 16% gradient Tris-Gly polyacrylamide gel (Invitrogen). The proteins were then electrotransferred onto nitrocellulose membranes for antibody detection. The blot was first blocked for 1 h with 4% nonfat dry milk (Invitrogen) in PBS followed by incubation for 1 h with anti-ZmIPK1 rabbit antiserum diluted 5,000-fold in the blocking solution. After three 5-min washes with 50 mm sodium phosphate buffer (pH 7.5) containing 155 mm NaCl and 0.05% Tween 20, the blot was incubated with goat anti-rabbit antibody conjugated to horseradish peroxidase (Pierce) followed by another three washes in 50 mm sodium phosphate buffer (pH 7.5) containing 155 mm NaCl and 0.05% Tween 20. The detected protein was visualized by incubating the blot with chemiluminescence substrate ECL western Analysis Reagent (Amersham Pharmacia Biotech) and exposing to x-ray film.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ431470, EF447274, EF527875, and EF527876.
Acknowledgments
The authors would like to thank Jianhua Wang and Megan Sopko for their technical contributions, Kathryn Kirkwood-Rodriguez for helping with greenhouse experiments, Xiaopin Xu and Weiting Ni for assistance with MALDI-time-of-flight analysis, and Ignacio Larrinua for bioinformatic analysis. We appreciate helpful discussions with Dr. Manyuan Long and critical suggestions from our colleagues Tom Greene, Tim Hey, Joe Petolino, Beth Rubin-Wilson, Sam Reddy, and Terry Walsh.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yuejin Sun (ysun@dow.com).
Open Access articles can be viewed online without a subscription.
References
- Biswas S, Maity IB, Chakrabarti S, Biswas BB (1978) Purification and characterization of myo-inositol hexaphosphate-adenosine diphosphate phosphotransferase from Phaseolus aureus. Arch Biochem Biophys 185 557–566 [DOI] [PubMed] [Google Scholar]
- Brearley CA, Hanke DE (1996. a) Metabolic evidence for the order of addition of individual phosphate esters to myo-inositol moiety of inositol hexakisphosphate in the duckweed Spirodela polyrhiza L. Biochem J 314 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brearley CA, Hanke DE (1996. b) Inositol phosphates in the duckweed Spirodela polyrhiza L. Biochem J 314 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brearley CA, Hanke DE (2000) Metabolic relations of inositol 3,4,5,6-tetrakisphosphate revealed by cell permeabilization: identification of inositol 3,4,5,6-tetrakisphosphate 1-kinase and inositol 3,4,5,6 tetrakisphosphate phosphatase activities in mesophyll cells. Plant Physiol 122 1209–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsch JA, Cook A, Young KA, Anderson JM, Bauman AT, Volkmann CJ, Murthy PP, Raboy V (2003) Seed phosphorus and inositol phosphate phenotype of barley low phytic acid genotypes. Phytochemistry 62 691–706 [DOI] [PubMed] [Google Scholar]
- Emrich SJ, Li L, Wen TJ, Yandeau-Nelson MD, Fu Y, Guo L, Chou HH, Aluru S, Ashlock DA, Schnable PS (2007) Nearly identical paralogs: implications for maize (Zea mays L.) genome evolution. Genetics 175 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttieri M, Bowen D, Dorsch J, Raboy V, Souza E (2004) Identification and characterization of a low phytic acid wheat. Crop Sci 44 418–424 [Google Scholar]
- Hanakahi LA, West SC (2002) Specific interaction of IP6 with human Ku70/80, the DNA-binding subunit of DNA-PK. EMBO J 21 2038–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitz WD, Carlson TJ, Kerr PS, Sebastian SA (2002) Biochemical and molecular characterization of a mutation that confers a decreased raffinosaccharide and phytic acid phenotype on soybean seeds. Plant Physiol 128 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy M, Efanov AM, Bertorello AM, Zaitsev SV, Olsen HL, Bokvist K, Leibiger B, Leibiger IB, Zwiller J, Berggren PO, et al (2002) Inositol hexakisphosphate promotes dynamin I-mediated endocytosis. Proc Natl Acad Sci USA 99 6773–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives EB, Nichols J, Wente SR, York JD (2000) Biochemical and functional characterization of inositol 1,3,4,5, 6-pentakisphosphate 2-kinases. J Biol Chem 275 36575–36583 [DOI] [PubMed] [Google Scholar]
- Jia J, Fu J, Zheng J, Zhou X, Huai J, Wang J, Wang M, Zhang Y, Chen X, Zhang J, et al (2006) Annotation and expression profile analysis of 2073 full-length cDNAs from stress-induced maize (Zea mays L.) seedlings. Plant J 48 710–727 [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Sciences (New York) 302 2141–2144 [DOI] [PubMed] [Google Scholar]
- Kazan K (2003) Alternative splicing and proteome diversity in plants: the tip of the iceberg has just emerged. Trends Plant Sci 8 468–471 [DOI] [PubMed] [Google Scholar]
- Larson SR, Rutger JN, Young KA, Raboy V (2000) Isolation and genetic mapping of a non-lethal rice low phytic acid 1 mutation. Crop Sci 40 1397–1405 [Google Scholar]
- Larson SR, Young KA, Cook A, Blake TK, Raboy V (1998) Linkage mapping 2 mutations that reduce phytic acid content of barley grain. Theor Appl Genet 97 141–146 [Google Scholar]
- Lemtiri-Chlieh F, MacRobbie EA, Brearley CA (2000) Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc Natl Acad Sci USA 97 8687–8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F, MacRobbie EA, Webb AA, Manison NF, Brownlee C, Skepper JN, Chen J, Prestwich GD, Brearley CA (2003) Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc Natl Acad Sci USA 100 10091–10095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Tang H, Nicholson JK, Lindon JC (2001) Recovery of underwater resonances by magnetization transferred NMR spectroscopy (RECUR-NMR). J Magn Reson 153 133–137 [DOI] [PubMed] [Google Scholar]
- Lott JNA, Ockenden I, Raboy V, Batten GD (2000) Phytic acid and phosphor use in crop seeds and fruits: a global estimate. Seed Sci Res 10 11–33 [Google Scholar]
- Lynch M, Katju V (2004) The altered evolutionary trajectories of gene duplicates. Trends Genet 20 544–549 [DOI] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL (2005) Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, Yu Y, Wei F, Fuks G, Soderlund CA, Mayer KF, et al (2004) Sequence composition and genome organization of maize. Proc Natl Acad Sci USA 101 14349–14354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AL, Suntharalingam M, Johnson SL, Audhya A, Emr SD, Wente SR (2004) Cytoplasmic inositol hexakisphosphate production is sufficient for mediating the Gle1-mRNA export pathway. J Biol Chem 279 51022–51032 [DOI] [PubMed] [Google Scholar]
- Moller EM, Bahnweg G, Sandermann H, Geiger HH (1992) A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res 20 6115–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillippy BQ, Ullah AHJ, Ehrlich KC (1994) Purification and some properties of inositol 1,3,4,5,6-petakisphosphate 2-kinase from immature soybean seeds. J Biol Chem 269 28393–28399 [PubMed] [Google Scholar]
- Raboy V (1997) Accumulation and storage of phosphate and minerals. In BA Larkins, IK Vasil, eds, Cellular and Molecular Biology of Plant Seed Development. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 441–477
- Raboy V (2003) Myo-inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64 1033–1043 [DOI] [PubMed] [Google Scholar]
- Raboy V, Gerbasi P (1996) Genetics of myo-inositol phosphate synthesis and accumulation. Subcell Biochem 26 257–285 [DOI] [PubMed] [Google Scholar]
- Raboy V, Gerbasi PF, Young KA, Stoneberg SD, Pickett SG, Bauman AT, Murthy PP, Sheridan WF, Ertl DS (2000) Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol 124 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S, Hatzack F (1998) Identification of low-phytate barley (Hordeum vulgare L.) grain mutants by TLC and genetic analysis. Hereditas 129 107–112 [Google Scholar]
- Sasakawa N, Sharif M, Hanley MR (1995) Metabolism and biological activities of inositol pentakisphosphate and inositol hexakisphosphate. Biochem Pharmacol 50 137–146 [DOI] [PubMed] [Google Scholar]
- Seeds AM, Sandquist JC, Spana EP, York JD (2004) A molecular basis for inositol polyphosphate synthesis in Drosophila melanogaster. J Biol Chem 279 47222–47232 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Sambrook DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Shi J, Wang H, Hazebroek J, Ertl DS, Harp T (2005) The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J 42 708–719 [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Wu Y, Hazebroek J, Meeley RB, Ertl DS (2003) The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol 131 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens LR, Hawkins PT, Stanley AF, Moore T, Poyner DR, Morris PJ, Hanley MR, Kay RR, Irvine RF (1991) Myo-inositol pentakisphosphates: structure, biological occurrence and phosphorylation to myo-inositol hexakisphosphate. Biochem J 275 485–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Bastidas RJ, Chiou ST, Frye RA, York JD (2005) Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc Natl Acad Sci USA 102 12612–12617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson-Paulik J, Odom AR, York JD (2002) Molecular and biochemical characterization of two plant inositol polyphosphate 6-/3-/5-kinases. J Biol Chem 277 42711–42718 [DOI] [PubMed] [Google Scholar]
- Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE, et al (2004) A gene expression map for the euchromatic genome of Drosophila melanogaster. Sciences (New York) 306 655–660 [DOI] [PubMed] [Google Scholar]
- Sun Y, Flannigan BA, Madison JT, Setter TL (1997) Alternative splicing of cyclin transcripts in maize endosperm. Gene 195 167–175 [DOI] [PubMed] [Google Scholar]
- Sweetman D, Johnson S, Caddick SE, Hanke DE, Brearley CA (2006) Characterization of an Arabidopsis inositol 1,3,4,5,6-pentakisphosphate 2-kinase (AtIPK1). Biochem J 394 95–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonova Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J (2004) Close split of sorghum and maize genome progenitors. Genome Res 14 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q, Wang A, Zhang HB (2002) One large-insert plant-transformation-competent BIBAC library and three BAC libraries of Japonica rice for genome research in rice and other grasses. Theor Appl Genet 105 1058–1066 [DOI] [PubMed] [Google Scholar]
- Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7 491–498 [DOI] [PubMed] [Google Scholar]
- Verbsky JW, Wilson MP, Kisseleva MV, Majerus PW, Wente SR (2002) The synthesis of inositol hexakisphosphate: characterization of human inositol 1,3,4,5,6-pentakisphosphate 2-kinase. J Biol Chem 277 31857–31862 [DOI] [PubMed] [Google Scholar]
- Wang BB, Brendel V (2006) Genomewide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci USA 103 7175–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox JR, Premachandra PS, Young KA, Raboy V (2000) Isolation of high inorganic phosphorus, low-phytate soybean mutants. Crop Sci 40 1601–1605 [Google Scholar]
- Xing Y, Lee C (2005) Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc Natl Acad Sci USA 102 13526–13531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR (1999) A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285 96–100 [DOI] [PubMed] [Google Scholar]