Abstract
The biosynthesis of histidine (His) in microorganisms, long studied through the isolation and characterization of auxotrophic mutants, has emerged as a paradigm for the regulation of metabolism and gene expression. Much less is known about His biosynthesis in flowering plants. One limiting factor has been the absence of large collections of informative auxotrophs. We describe here the results of a systematic screen for His auxotrophs of Arabidopsis (Arabidopsis thaliana). Ten insertion mutants disrupted in four different biosynthetic genes (HISN2, HISN3, HISN4, HISN6A) were identified through a combination of forward and reverse genetics and were shown to exhibit an embryo-defective phenotype that could be rescued by watering heterozygous plants with His. Male transmission of the mutant allele was in several cases reduced. Knockouts of two redundant genes (HISN1B and HISN5A) had no visible phenotype. Another mutant blocked in the final step of His biosynthesis (hisn8) and a double mutant altered in the redundant first step of the pathway (hisn1a hisn1b) exhibited a combination of gametophytic and embryonic lethality in heterozygotes. Homozygous mutant seedlings and callus tissue produced from rescued seeds appeared normal when grown in the presence of His but typically senesced after continued growth in the absence of His. These knockout mutants document the importance of His biosynthesis for plant growth and development, provide valuable insights into amino acid transport and source-sink relationships during seed development, and represent a significant addition to the limited collection of well-characterized auxotrophs in flowering plants.
Auxotrophic mutants have long played a pivotal role in the genetic dissection of biosynthetic pathways in microorganisms. Fungal and bacterial mutants unable to produce an essential nutrient have been used to address a wide range of important questions in biology and have become a cornerstone of modern genetics. Repeated attempts over the past several decades to generate an equivalent collection of plant auxotrophs have met with only limited success. Two principal hypotheses have been advanced to explain the scarcity of plant auxotrophs: functional redundancy and early lethality. The genetic dissection of His biosynthesis in Arabidopsis (Arabidopsis thaliana) described here provides compelling evidence in support of both hypotheses, insights into His production and transport in plants, and a fresh perspective on future efforts to establish a comprehensive collection of auxotrophic mutants for research in plant biology.
His is an essential amino acid that becomes incorporated into proteins and is required for cell growth and reproduction. The biosynthesis of His, first defined genetically in Escherichia coli and yeast (Saccharomyces cerevisiae), is highly conserved among microorganisms (Alifano et al., 1996). Information on His biosynthesis in plants has emerged over the past 15 years (Stepansky and Leustek, 2006). Many of the genes were first identified through functional complementation of yeast or bacterial auxotrophs. The plant cDNAs used in these experiments were obtained from a variety of sources, including Arabidopsis (Tada et al., 1994; Fujimori and Ohta, 1998a, 1998b; Ohta et al., 2000), Thlaspi (Persans et al., 1999), Nicotiana (El Malki et al., 1998), cabbage (Brassica capitata; Nagai et al., 1991), and Alyssum (Ingle et al., 2005). The presence of signal sequences on these proteins indicates that His biosynthesis takes place inside the chloroplast.
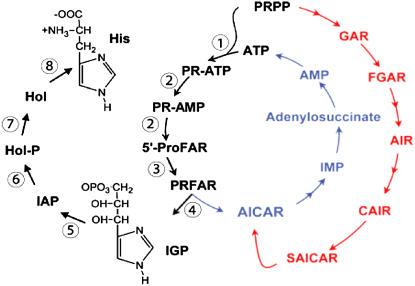
The pathway for His production in plants is summarized in Figure 1. The Arabidopsis enzyme (HISN1) responsible for the first step exhibits feedback inhibition by His. Transgenic plants that constitutively express a variant form of this enzyme accumulate higher levels of free His than wild-type plants and exhibit increased resistance to toxic levels of nickel (Wycisk et al., 2004; Ingle et al., 2005). Plant species that hyperaccumulate nickel also contain increased levels of free His (Krämer et al., 1996) and altered activities of this enzyme (Ingle et al., 2005). His may in addition chelate other metal ions, including zinc (Salt et al., 1999) and copper (Liao et al., 2000). His therefore plays an important role in metal ion homeostasis in plants (Stepansky and Leustek, 2006).
Figure 1.
His biosynthesis (black) and related ATP synthesis (blue, red) and recycling (blue) pathways in Arabidopsis. Numbers refer to specific HISN gene products listed in Table I. His intermediates shown: PRPP, Phosphoribosyl pyrophosphate; PR-ATP, phosphoribosyl ATP; PR-AMP, phosphoribosyl AMP; 5′-ProFAR, pro-phosphoribosyl formimino-5-aminoimidazole-4-carboxamide ribonucleotide; PRFAR, phosphoribulosyl formimino-5-aminoimidazole-4-carboxamide ribonucleotide; IGP, imidazoleglycerol phosphate; IAP, imidazoleacetol phosphate; Hol-P, l-histidinol phosphate; Hol, histidinol; His, l-His.
Several plant mutants disrupted in His biosynthesis have been described to date. One is a haploid cell line of Nicotiana plumbaginifolia with a missense mutation affecting the aminotransferase (HISN6) that catalyzes a late step in the pathway (Negrutiu et al., 1985; El Malki and Jacobs, 2001). A second example is an embryo-defective (emb) mutant of Arabidopsis (emb2196) identified through a forward genetic screen of T-DNA insertion mutants (Tzafrir et al., 2004). This mutant is altered in the same step of the pathway as the Nicotiana auxotroph. A weak mutant allele of this locus (hpa1) exhibits a short-root phenotype in Arabidopsis that can be rescued by supplemental His (Mo et al., 2006). A mutant disrupted in one of the duplicated HISN1 genes responsible for the first step in the pathway also exhibits a root phenotype (Wang et al., 2005). Another mutant (apg10) with a seedling phenotype is defective in the HISN3 enzyme that catalyzes an early step in the pathway (Noutoshi et al., 2005). Knockout alleles of this gene are embryo lethals (DiFraia and Leustek, 2004; Noutoshi et al., 2005). All of these isolated studies are consistent with an essential role for His throughout the life cycle.
We describe in this report a systematic genetic approach to His biosynthesis in plants. We summarize information on Arabidopsis genes encoding seven known enzymes involved in His biosynthesis, describe the phenotypes of 13 insertion mutants disrupted in nine different HISN genes, demonstrate the ability to maintain homozygous mutant plants and callus tissue in the presence of supplemental His, and discuss the relevance of these observations to our understanding of amino acid biosynthesis and transport during plant growth and development. The extensive set of His auxotrophs described here represents a valuable addition to the expanding collection of Arabidopsis mutants with defined alterations in amino acid metabolism. A unique feature of this work is the systematic characterization of knockout alleles for multiple steps in a single pathway. Further analysis of these mutants should provide valuable insights into the regulation of metabolism in flowering plants.
RESULTS
HISN Genes of Arabidopsis
The pathway for His biosynthesis (Fig. 1) involves a total of 11 reactions catalyzed by eight different enzymes. Genes encoding seven of these enzymes have been characterized in plants (for review, see Stepansky and Leustek, 2006). The corresponding Arabidopsis genes have been designated HISN for His biosynthesis and numbered to reflect eight sequential steps in the pathway (Fig. 1; Table I). The HISN7 gene encoding histidinol phosphate phosphatase has not been identified in plants. A candidate gene of Arabidopsis (At3g14890) that exhibits weak sequence similarity to the yeast HIS2 gene and appears to encode a chloroplast-localized protein remains to be functionally characterized (Stepansky and Leustek, 2006). The HISN1, HISN5, and HISN6 genes are duplicated in the Arabidopsis genome. The remaining HISN genes are present in single copies. The HISN1 and HISN5 paralogs exhibit about 80% sequence identity at the protein level. Most of the differences are focused at the N terminus, which includes predicted chloroplast localization signals. Expression data are available for both members of these gene pairs. In contrast, the HISN6 paralogs are so similar at both the protein and genomic levels that expression data cannot distinguish between them. Two single nucleotide polymorphisms differentiate the gene coding sequences. Four additional single nucleotide polymorphisms are found in the 3′ untranslated region and minor differences are present in the 5′ untranslated region. All of the ESTs identified to date that can be uniquely associated with one of the two genes are products of HISN6A. Genomic regions surrounding the two paralogs do not share a high degree of sequence identity. The recent duplication event responsible for generating these HISN6 paralogs therefore appears to have been restricted to a single gene.
Table I.
HISN genes of Arabidopsis
| Arabidopsis Gene | Yeast Ortholog | Bacterial Ortholog | Arabidopsis Locus | Enzyme Activity | Functional Analysis | Allele 1 | Allele 2 | Allele 3 |
|---|---|---|---|---|---|---|---|---|
| HISN1A | HIS1 | HisG | At1g58080 | Phosphoribosyltransferase | Ohta et al. (2000) | SK-152420 | SK-3720 | |
| HISN1B | HIS1 | HisG | At1g09795 | Phosphoribosyltransferase | Ohta et al. (2000) | RATM13-4733-1G | SK-65159 | |
| HISN2 | HIS4 | HisIE | At1g31860 | Cyclohydrolase and pyrophosphohydrolase | Fujimori and Ohta (1998b) | SK-47603 | SK-143868 | SK-55503 |
| HISN3 | HIS6 | HisA | At2g36230 | BBMII isomerase | Fujimori et al. (1998) | SK-41416 | SK-97704 | Ds15-1767-1c |
| HISN4 | HIS7 | HisHF | At4g26900 | Amidotransferase/cyclase | Fujimori and Ohta (1998a) | SK-41907 | SK-2283 | SL-556-C04 |
| HISN5A | HIS3 | HisB | At3g22425 | Dehydratase | Tada et al. (1994) | SK-9063 | ||
| HISN5B | HIS3 | HisB | At4g14910 | Dehydratase | Not characterized | None confirmed | ||
| HISN6A | HIS5 | HisC | At5g10330 | Aminotransferase | El Malki et al. (1998)a | emb2196b | SL-750-F01 | hpa1d |
| HISN6B | HIS5 | HisC | At1g71920 | Aminotransferase | No ESTs confirmed | None identified | ||
| HISN7 | HIS2 | HisB | Unknown | Phosphatase | Unknown | Unknown | ||
| HISN8 | HIS4 | HisD | At5g63890 | Dehydrogenase | Nagai et al. (1991) | SL-1230-F01 |
Characterized from Nicotiana tabacum.
Refer to www.seedgenes.org; Tzafrir et al. (2004).
Characterized by Noutoshi et al. (2005).
Characterized by Mo et al. (2006).
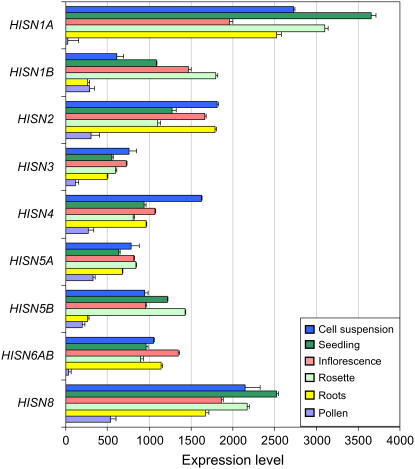
The nonredundant HISN genes of Arabidopsis (HISN2, HISN3, HISN4, and HISN8) must be constitutively expressed because His is required throughout growth and development. More detailed information on spatial and temporal patterns of HISN expression generated from multiple microarray experiments was obtained using Genevestigator (Zimmermann et al., 2004). Results are summarized in Figure 2 and Supplemental Figure S1. Several interesting features emerge from this analysis: (1) high levels of HISN1A and HISN8 transcripts are present throughout the plant; (2) HISN1A transcripts are far more abundant than HISN1B transcripts, especially in the root; (3) HISN5B is expressed at moderate levels in different parts of the plant; and (4) only trace amounts of HISN transcripts have been detected in pollen. These observations are relevant to phenotypic analyses of specific insertion mutants.
Figure 2.
Summary of expression data for HISN genes of Arabidopsis. Results of microarray experiments were obtained from https://www.genevestigator.ethz.ch. HISN6A and HISN6B are combined because they are indistinguishable.
Identification of Informative T-DNA Insertion Mutants
The analysis of His mutants described here represents a collaboration between two laboratories. The Meinke lab is focused on saturating for EMB genes with a knockout phenotype in the seed (www.seedgenes.org). After one mutant identified through a forward genetic screen of T-DNA insertion lines (McElver et al., 2001) was found to be disrupted in a gene involved with His biosynthesis (Tzafrir et al., 2004), additional examples of mutations affecting other steps in the pathway were sought from the Salk insertion database (Alonso et al., 2003). The Leustek lab is focused on amino acid metabolism in plants (Lee et al., 2005; Hudson et al., 2006). The His pathway in Arabidopsis, which includes multiple steps and relatively few redundant genes, was a logical candidate for using reverse genetics to study the physiological and developmental consequences of interfering with amino acid biosynthesis (Stepansky and Leustek, 2006). Eventually these overlapping efforts were coordinated to focus on 13 mutants disrupted in nine different HISN genes of Arabidopsis. Allele designations for these mutants are presented in Table I. Most of the mutants were grown in both laboratories and results were compared for plants maintained under somewhat different conditions. The only confirmed homozygotes identified for putative null alleles grown in the absence of supplemental His (hisn1a-1, hisn1b-1, and hisn5a-1) were for redundant genes.
Disruption of His Biosynthesis Results in Seed Abortion
The most common phenotype of insertion mutants disrupted in His biosynthesis is seed abortion at the preglobular stage of embryo development. Siliques from plants heterozygous for mutant alleles of hisn2, hisn3, hisn4, and hisn6a contain approximately 15% to 25% aborted seeds following self-pollination (Table II). A typical silique is shown in Figure 3A. Aborted seeds are reduced in size and become desiccated and turn brown before wild-type seeds in the same silique. Similar phenotypes were observed in plants maintained at different locations. Typical phenotypes of arrested embryos are shown in Figure 4. Developmental arrest of the mutant embryo occurs without the cell enlargement or aberrant suspensor growth characteristic of some embryo-defective mutants. The mutant embryo and endosperm appear to terminate development over a short period of time. A single mutant line (hisn5a-1) with a confirmed insertion located within the coding region of HISN5A produced viable homozygotes with no visible phenotype. This indicates that expression of the redundant HISN5B gene is sufficient for normal growth and development. The atypical ovule phenotype characteristic of hisn8 knockouts (Fig. 3B) is described in more detail in a later section.
Table II.
Analysis of confirmed mutant alleles
| Mutant Allele | Confirmed Insertion Sitea | Siliques Screened | Seeds Screened | Aborted Seedsb | Top Halfc | Average Seed Size | Terminal Phenotype |
|---|---|---|---|---|---|---|---|
| % | μm | ||||||
| hisn2-1 | Exon 5 (5) | 25 | 898 | 16.0*** | 71.5*** | 320 | Preglobular |
| 50 | 1,997 | 20.8*** | 62.9*** | Preglobular | |||
| hisn2-2 | Exon 5 (5) | 50 | 1,672 | 16.0*** | 69.3*** | 220 | Preglobular |
| 60 | 2,346 | 19.9*** | 65.2*** | Preglobular | |||
| hisn2-3 | Promoter | 50 | 2,117 | 23.9 | 55.9** | 340 | Preglobulard |
| hisn3-1 | Exon 1 (7) | 50 | 2,168 | 21.7*** | 72.9*** | 270 | Preglobular |
| 60 | 2,201 | 22.0** | 66.8*** | Preglobular | |||
| hisn3-2 | Exon 5 (7) | 50 | 2,092 | 20.7*** | 72.3*** | 280 | Preglobular |
| hisn4-1 | Exon 1 (19) | 50 | 2,059 | 21.8** | 64.7*** | 380 | Preglobular |
| 45 | 1,830 | 20.0*** | 65.0*** | Preglobular | |||
| hisn4-2 | Intron 14 (18) | 50 | 2,114 | 23.4 | 65.8*** | 330 | Preglobular |
| 50 | 1,926 | 20.9*** | 64.5*** | Preglobular | |||
| hisn4-3 | Intron 17 (18) | 50 | 2,161 | 23.5 | 58.7*** | 330 | Preglobular |
| hisn6a-1 | Exon 2 (8) | 50 | 1,787 | 28.7*** | 53.2 | 440 | Preglobulare |
| 25 | 1,053 | 25.0 | 54.4 | 340 | Preglobular | ||
| hisn6a-2 | Exon 7 (8) | 25 | 1,228 | 25.4 | 59.2*** | 370 | Preglobular |
| 10 | 396 | 23.4 | 55.6 | Preglobular | |||
| hisn8 | Exon 10 (11) | 110 | 4,756 | [24.5]f | N.D. | <100 | Aborted ovule |
Based on alignment of flanking sequence; total number of exons or introns noted in parentheses.
Present in heterozygous siliques; 25% expected; significant deviation: **, P < 0.01; ***, P < 0.001.
Aborted (homozygous mutant) seeds in top half of heterozygous siliques; 50% expected. Significant deviation indicates probable effect of mutation on pollen tube growth. N.D., Not determined.
Occasional seeds with cotyledon defects of unknown origin also found in this line and included in totals.
Occasional seeds with delayed phenotype attributed to heterozygotes included in totals for this population.
Percentage of aborted ovules and early aborted seeds combined.
Figure 3.
Aborted seed phenotypes of His auxotrophs. A, Immature silique from hisn2-1 heterozygote with mutant seeds (white, brown) arrested at a preglobular stage of embryo development. Brown seeds began to desiccate before the white seeds. B, Immature siliques from hisn8 heterozygotes showing characteristic ova phenotype (top silique, white spots in bottom valve) and partially rescued white seeds after His supplementation (bottom silique). Scale bar = 1 mm.
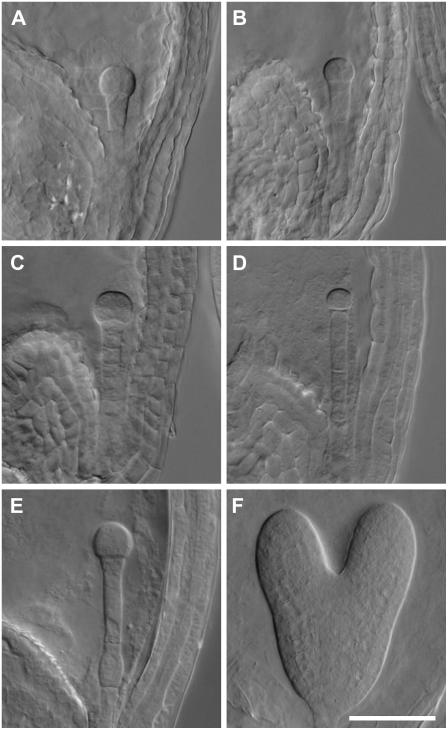
Figure 4.
Arrested embryo phenotypes of His auxotrophs. A to E, Nomarski images of cleared mutant seeds removed from immature siliques heterozygous for hisn3-1 (A and B), hisn4-1 (C), hisn2-1 (D), and hisn6a-1 (E). F, Wild-type embryo at the heart stage of development, when all seeds were examined. Scale bar = 50 μm.
Most of the mutant alleles examined appear to be nulls based on confirmed T-DNA locations within the gene (Table II). Allelism was in each case demonstrated by screening siliques produced from crosses between heterozygotes. Two mutants (hisn2-1 and hisn2-2) were later found to contain insertions at precisely the same location. Although we present these as independent alleles, they may represent sibling lines that resulted from accidental seed contamination before we received the stocks from the Arabidopsis Biological Resource Center (ABRC). The strong phenotype observed in these two lines was at first surprising in light of the 3′ location of the insertion, but it remains consistent with that observed in hisn3 and hisn4 knockouts. Furthermore, several conserved residues are located at the carboxyl terminus of cyclohydrolase/pyrophosphohydrolases.
Two mutants contained a number of seeds arrested at later stages of development. In one case (hisn2-3), the origin and significance of these atypical seeds remains unknown. In a second case (hisn6a), we suspect that most of these seeds are heterozygous for the mutation. The phenotype is subtle and requires that immature siliques be screened at a stage when differences between heterozygous and wild-type seeds are most apparent. Immature siliques produced by crossing wild-type pollen onto flowers of hisn6a-1 and hisn6a-2 heterozygotes, when screened at the appropriate stage of development, contained equal numbers of normal and delayed seeds, as expected if the delayed seeds were heterozygous for the mutation. Results of these crosses are presented in Table III. The original segregation data for selfed heterozygotes shown in Table II were collected over an extended period. The first 50 hisn6a-1 siliques were screened before this distinction was recognized. This may explain the somewhat elevated percentage of aborted seeds found initially (28.7%; n = 1,787) in contrast to subsequent screens of both hisn6a-1 (25.0%; n = 1,053) and hisn6a-2 (25.4%; n = 1,228). What remains to be explained is why delayed development of heterozygous seeds is not an obvious feature of all His mutants.
Table III.
Delayed development of heterozygous hisn6a seedsa
| Female Parent | Siliques Examined | Normal Seeds | Delayed Seeds | Intermediate Seeds |
|---|---|---|---|---|
| hisn6a-1 | 3 | 46 | 39 | 10 |
| hisn6a-2 | 3 | 47 | 42 | 7 |
| Total | 6 | 93 | 81 | 17 |
Immature siliques from crosses of wild type (♂) to hisn6a heterozygous (♀) plants were screened for seed phenotypes. Approximately half of these seeds (presumed heterozygotes) were delayed in development.
Male Transmission of Mutant Alleles Is Often Reduced
One distinctive feature of most hisn2, hisn3, and hisn4 alleles is the reduced percentage of aborted seeds found in selfed heterozygotes (Table II). Because this characteristic often results from reduced transmission of mutant pollen tubes, which can affect the likelihood that mutant ovules at the base of the silique develop into mutant seeds, we examined the distribution of normal and aborted seeds along the length of heterozygous siliques using a method developed for the analysis of other embryo-defective mutants (Meinke, 1982). The results shown in Table II indicate that for all hisn2, hisn3, and hisn4 alleles examined, mutant seeds are underrepresented at the base of the silique, and as a result, more than half of the total aborted seeds are located in the top half of the silique. Further evidence for reduced male transmission was sought by performing outcrosses with a representative mutant allele (hisn3-1). Pollen from heterozygous plants was crossed onto wild-type flowers, and mature seeds from the resulting F1 siliques were planted in the order of their location within the silique. The resulting plants were then genotyped by screening for the presence of aborted seeds following self-pollination. When these genotypes were superimposed on maps of the original F1 siliques, a nonrandom distribution of heterozygous and wild-type seeds was revealed, consistent with reduced transmission of mutant pollen tubes. Results for siliques produced from 13 independent crosses are presented in Table IV. Interfering with the initial steps of His biosynthesis in Arabidopsis therefore results not only in embryo lethality but also in reduced transmission of male gametes. Whether hisn6a knockouts disrupted later in the pathway exhibit a similar defect remains an open question. Results for the two mutant alleles shown in Table II are inconclusive on this point, in part because some heterozygous seeds with a delayed phenotype may have been present. Expression of HISN6B may also limit the severity of the pollen defect.
Table IV.
Distribution of heterozygous seeds in siliques produced after crossing hisn3-1 heterozygotes (♂) to wild type (♀)
| Silique Analyzed | Total Seeds | Resulting Plants Screened | Heterozygous Plantsa | Top Halfb |
|---|---|---|---|---|
| % | ||||
| 1 | 46 | 42 | 45.2 | 73.7** |
| 2 | 44 | 44 | 52.3 | 52.2 |
| 3 | 46 | 43 | 53.5 | 56.5 |
| 4 | 45 | 42 | 40.7 | 76.5** |
| 5 | 45 | 34 | 44.1 | 73.3** |
| 6 | 38 | 37 | 32.4* | 75.0** |
| 7 | 49 | 35 | 42.8 | 66.7* |
| 8 | 42 | 41 | 31.7* | 76.9*** |
| 9 | 34 | 29 | 48.3 | 71.4* |
| 10 | 43 | 33 | 48.5 | 56.2 |
| 11 | 43 | 38 | 47.4 | 44.4 |
| 12 | 39 | 32 | 50.0 | 62.5 |
| 13 | 45 | 37 | 45.9 | 64.7 |
| Total | 559 | 487 | 44.8* | 64.2*** |
Expected 50% if male transmission of the mutant allele was not reduced during the cross. Significant deviation: *, P < 0.05.
Heterozygous seeds present in top half of each silique as determined by phenotyping the resulting plants; 50% expected if mutation has no effect on male transmission. A significant deviation (*, P < 0.05; **, P < 0.01; ***, P < 0.001) indicates a negative effect of the hisn3-1 mutation on pollen tube growth.
Blocking the Final Step in His Biosynthesis Results in Ovule Abortion
A more severe and variable phenotype was observed in hisn8 heterozygotes disrupted in the final step of His biosynthesis. These plants typically produced many aborted ovules (Fig. 3B, top silique) and a few small seeds arrested early in development. A similar ovule abortion (ova) phenotype has been described for knockouts of aminoacyl-tRNA synthetases localized to mitochondria (Berg et al., 2005). The two phenotypic classes combined often accounted for about 25% of total ovules (Table II), much less than the 50% expected for female gametophytic mutants. Some heterozygous plants gave inconsistent results in subsequent experiments, perhaps reflecting changes in developmental and environmental factors affecting the availability of His. Gametophytic transmission of the mutant allele was examined by making reciprocal crosses to wild-type plants (Table V). Female transmission was frequently reduced, consistent with the presence of aborted ovules, whereas male transmission was surprisingly variable. Individual flowers of heterozygous plants therefore differ widely in their ability to produce mutant pollen grains capable of participating in fertilization. Despite the atypical phenotype of this mutant allele, the response of aborted ovules to His was later confirmed through rescue experiments. An example of partial rescue of mutant ovules is shown in Figure 3B (bottom silique).
Table V.
Gametophytic transmission of the hisn8 mutant allele in reciprocal crosses to wild type
| Heterozygous Parent | Silique Examined | Progeny Plants Analyzeda | Heterozygotes to Wild Types |
|---|---|---|---|
| Male | 1 | 38* | 1.1 |
| 2 | 44 | 0.4 | |
| 3 | 36 | 0.9 | |
| 4 | 23 | 0.2 | |
| 5 | 31 | 0.1 | |
| 6 | 28* | 0.1 | |
| Total | 6 | 200 | 0.5 (0.43)b |
| Female | 1 | 34 | 0.8 |
| 2 | 32 | 0.9 | |
| 3 | 38 | 0.6 | |
| 4 | 32 | 0.9 | |
| 5 | 38 | 0.7 | |
| 6 | 41 | 0.5 | |
| 7 | 33 | 0.5 | |
| 8 | 43 | 0.6 | |
| 9 | 38 | 0.7 | |
| Total | 9 | 329 | 0.7 (0.15)b |
Asterisks denote plants genotyped with PCR. Remaining plants were genotyped by plating on selection media.
Averages with sds in parentheses. Normal transmission should result in equal numbers of heterozygous and wild-type plants. Note the variable rates of male transmission.
Mutant Seeds Are Rescued by Watering Heterozygous Plants with His
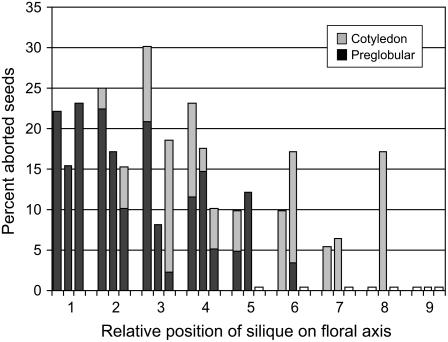
The His requirement of mutant seeds altered in six different steps of the pathway was confirmed by feeding heterozygous plants supplemental His. The best results were obtained when His was supplied to the roots by daily watering. Foliar application of His often resulted in leaf damage at higher concentrations and was less effective overall. Two different strategies were used to identify the desired heterozygotes targeted for rescue. One involved PCR genotyping of individual plants before flowering. The other required an initial screen of selfed siliques from plants grown in the absence of His. Removal of excess stems and branches helped to direct the His into rescued siliques. His feeding began once heterozygotes were identified. Solutions containing 1 mm His were often sufficient for complete rescue. The success of individual experiments was first evaluated by looking for the absence of aborted seeds in siliques that formed after His was added. Results of a successful experiment are presented in Figure 5. Three classes of siliques were typically found in progression: those with a characteristic mutant seed phenotype, followed by an intermediate group with some mutant seeds at a later stage of development, and finally those that were completely rescued and contained no aborted seeds. Siliques that were fully rescued had not reached the preglobular stage of development when His supplementation began. Although success rates varied depending on growth conditions, plant vigor, and His concentration and method of application, complete rescue was observed for at least one mutant allele of hisn2, hisn3, hisn4, hisn6a, and hisn8.
Figure 5.
His rescue of hisn4-1 mutant seeds. Nine consecutive siliques from a single floral axis on each of three heterozygous plants were screened for aborted seeds after daily watering with 1 mm His. At the onset of each experiment, silique number 1 contained normal seeds at a green cotyledon stage, whereas silique number 9 was developing within a flower. Screening of the first silique began 10 d after the start of His supplementation. Subsequent siliques were screened after they had reached an equivalent stage of development. Note the progressive decline in the frequency of preglobular aborted seeds, the appearance of seeds with a later (cotyledon) phenotype, and the presence of siliques with no aborted seeds (white bars at the baseline).
Rescued Mutant Seeds Produce Plants and Callus in the Presence of His
Siliques from rescued heterozygotes should contain 20% to 25% homozygous mutant seeds that exhibit continued growth in the presence of His and rapid senescence in the absence of His. This expected result was confirmed in part by germinating on a basal medium all of the mature seeds from individual, rescued siliques. The percentage of arrested seedlings observed was then compared with that obtained when seeds from equivalent siliques were germinated on a medium with His. Results of these experiments (Table VI) were consistent with complete rescue of mutant seeds and rapid depletion of stored reserves of His following germination on a basal medium. Rescued mutant seedlings exhibited a range of phenotypes in the absence of His (Fig. 6). Some seeds failed to germinate (Fig. 6A), others arrested shortly after germination and remained white (Fig. 6, B and C), and some had green cotyledons that later turned white (Fig. 6, E–H). These differences likely reflect variations in the amount of His made available to the developing seed during the initial feeding experiments. Arrested seedlings for all hisn2, hisn3, and hisn4 mutant alleles examined failed to produce leaves. A different result was obtained with hisn6a-1, where mutant seedlings on a basal medium formed a small rosette with leaves that remained green for more than a month (Fig. 6, I–L). This difference is attributed to the presence of a functional HISN6B gene that provides at least some of the required enzyme. This observation provides the first definitive evidence of HISN6B function in Arabidopsis.
Table VI.
Germination response of mature seeds from rescued heterozygotes
| Mutant Allele | Medium | Siliques Tested | Total Seeds Plated | Seeds Not Germinated | Arrested Seedlings |
|---|---|---|---|---|---|
| % | |||||
| hisn2-1 | Basal | 3 | 105 | 3.8 | 18.1 |
| His | 5 | 190 | 3.7 | 2.1 | |
| hisn3-1 | Basal | 3 | 149 | 0.0 | 23.4 |
| His | 4 | 194 | 7.7 | 0.0 | |
| hisn4-1 | Basal | 3 | 124 | 0.8 | 16.1 |
| His | 6 | 246 | 3.6 | 0.0 | |
| hisn6a-1 | Basal | 3 | 122 | 0.0 | 28.7a |
| His | 5 | 168 | 0.5 | 0.0 | |
| hisn8 | Basal | 6 | 270 | 1.1 | 20.0 |
| His | 5 | 198 | 1.0 | 0.0 | |
These mutant seedlings were delayed but not arrested in growth and development.
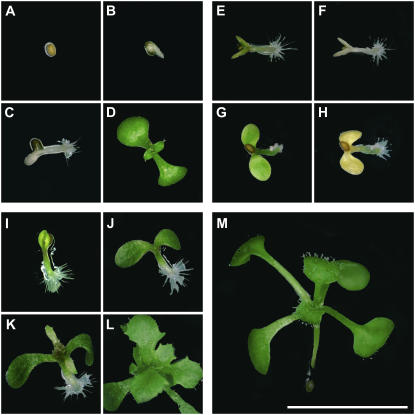
Figure 6.
Responses of rescued mutant seeds germinated on media without His. Rescued mutant seeds and wild-type controls were dry harvested from heterozygous plants watered daily with 1 mm His, surface sterilized, and plated on agar media containing 3% Glc and observed for 4 weeks. A to D, Examples of hisn2-1 rescued mutant seeds that failed to germinate (A) or arrested early in germination and remained white (B and C) after 7 d, in contrast to wild-type controls on the same plate (D). E to H, Examples of hisn3-1 (E and F) and hisn2-1 (G and H) rescued mutant seeds that germinated to produce a small seedling that was green after 7 d (E and G) but then stopped growing and turned white or yellow at 14 d (F and H). I to L, Example of a hisn6a-1 rescued mutant seed that germinated to produce a small green seedling that continued to grow slowly after 7 (I), 14 (J), 21 (K), and 28 d (L) without His, reflecting a low level of redundant HISN6B activity. M, Wild-type seedling after 14 d in culture. Scale bar = 5 mm.
A second approach to the identification of rescued homozygotes involved germinating rescued seeds in the presence of His and then testing cotyledon explants from these seedlings for their ability to produce callus that required His for continued growth. Results of these experiments are shown in Figure 7 and Table VII. When parental plants were not watered with His, all of the resulting seedlings produced callus that grew in the absence of His because homozygotes were eliminated as aborted seeds. In contrast, 10% to 20% of the seedlings derived from plants watered with His produced callus that required His for continued growth. The one exception (hisn6a-1) can be attributed to residual HISN6B function. A final approach was to take all of the rescued seedlings germinated in the presence of His, transfer these seedlings either to soil or to a medium lacking His, and then look for gradual senescence of the homozygotes. Results of these experiments are summarized in Figure 8 and Table VIII. With the exception of hisn6a-1 homozygotes, which were reduced in size but continued to grow after transplantation, all of the mutants examined produced seedlings that senesced over several weeks. Mature plants were not completely rescued by His supplementation and often exhibited reduced apical dominance, sensitivity to wilting, and reduced fertility, consistent with limited efficiency of His translocation throughout the plant.
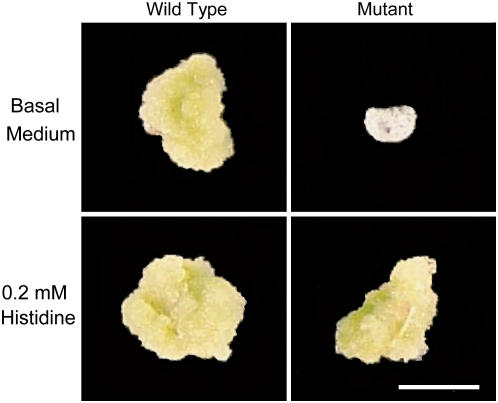
Figure 7.
Callus response of cotyledon segments from hisn2-1 rescued seedlings. Cotyledon explants were removed from mutant and wild-type seedlings grown on 0.2 mm His and cultured on a callus induction medium with either 0.2 mm His or no supplement. Cultures were photographed after 24 d. Scale bar = 5 mm.
Table VII.
Identification of rescued mutant plants using a callus induction assay with cotyledon explantsa
| Mutant Allele | Progeny Plants Forming Callus on His | Plants with Auxotrophic Callus Response |
|---|---|---|
| % | ||
| hisn2-1 | 50 (50) | 16.0 (0.0) |
| hisn3-1 | 50 (50) | 18.0 (0.0) |
| hisn4-1 | 31 (60) | 19.4 (0.0) |
| hisn6a-1 | 58 (42) | 0.0 (0.0) |
| hisn8 | 34 (36) | 11.8 (0.0) |
Seeds from rescued heterozygotes were germinated on a medium containing His. Explants were then tested for callus production on media with and without His. Numbers in parentheses represent controls from parental plants not supplemented with His.
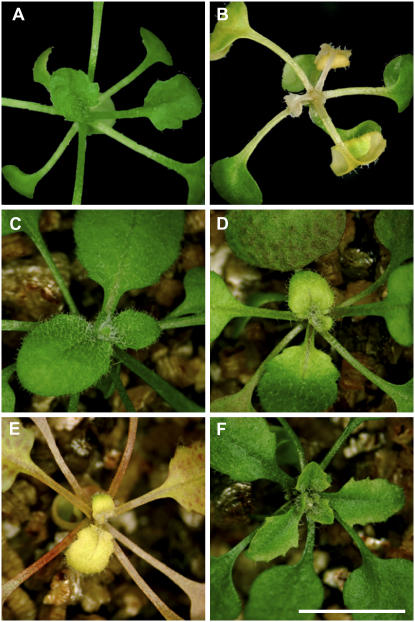
Figure 8.
Senescence of mutant seedlings transferred from His media. Wild-type (A) and hisn4-1 (B) seedlings 3 weeks after transfer to basal media without His. Wild-type (C), hisn4-1 (D), hisn2-1 (E), and hisn6a-1 (F) seedlings 3 weeks after transfer to soil without His.
Table VIII.
Senescence of mutant seedlings transplanted from His mediaa
| Mutant Allele | No. Tested | Senescent |
|---|---|---|
| % | ||
| Seedlings transplanted to soil | ||
| hisn2-1 | 30 | 16.7 |
| hisn3-1 | 48 | 31.3 |
| hisn4-1 | 42 | 19.1 |
| hisn6a-1 | 39 | 20.5b |
| Seedlings transplanted to basal medium | ||
| hisn4-1 | 41 | 26.8 |
| hisn6a-1 | 31 | 0.0c |
| hisn8 | 77 | 14.3 |
Mature seeds from rescued heterozygotes were germinated on a medium with 1 mm His. The resulting seedlings were transplanted after 2 to 3 weeks in culture and the number of senescent plants determined after 2 to 3 weeks without His.
Mutant plants were smaller than normal but remained viable much longer than with other rescued knockouts.
Mutant plants could not be identified by phenotype until several weeks later.
Double Knockouts of the Redundant First Step in His Biosynthesis
Analysis of the redundant first step in His biosynthesis catalyzed by ATP phosphoribosyl transferase required the construction of plants heterozygous for knockout alleles of both HISN1A (At1g58080) and HISN1B (At1g09795). The mutant alleles examined contained insertions near the beginning (hisn1b-1) or middle (hisn1a-1) of the coding region. Double knockouts for other redundant HISN genes could not be constructed because confirmed insertions remain to be identified for HISN5B and HISN6B. Twenty progeny plants from a selfed double heterozygote (AaBb) segregating for both hisn1a-1 (a) and hisn1b-1 (b) were PCR genotyped and screened for defects in seed and ovule development. The following plants were identified: AABb (5), AAbb (2), AaBB (5), and AaBb (8). Despite the small sample size, these results are consistent with two important conclusions supported by further studies: reduced transmission of double knockout gametes (ab) and embryo lethality of double homozygotes (aabb). No viable double homozygotes were identified in subsequent experiments.
Siliques from AABb plants did not contain an increased number of aborted ovules or defective seeds. Homozygous mutant plants (AAbb) identified by PCR appeared normal. Expression of HISN1B is therefore not required for completion of the life cycle. In contrast, siliques from AaBB plants often contained a low frequency (3.5%; n = 2,000) of aborted seeds with variable phenotypes. The frequency of aborted ovules in these siliques (7.9%) was also marginally elevated. Homozygous mutant plants (aaBB) did not survive to maturity but were identified by PCR as minute plantlets with short roots and rudimentary leaves on a basal germination medium. This phenotype was rescued in the presence of 2 mm His. Sibling plants with normal phenotypes were either heterozygous (AaBB) or wild type (AABB). Disruption of HISN1A may therefore occasionally result in ovule or seed abortion but is most often associated with seedling lethality.
Siliques of double heterozygotes also exhibited an increased frequency of aborted seeds (3.2%) and ovules (12.4%; n = 2,560). Although these results may reflect in part the reduced viability of single knockout gametes (a) and seeds (aa) noted above, we suspected that failure to identify more aborted seeds of the expected phenotype and the complete absence of double homozygous mutant plants might reflect an additional problem with male transmission of the double knockout (ab) gametes. This hypothesis was tested by analyzing progeny from crosses between AaBb (♂) and AABB (♀) plants. Seeds from nine different crosses were plated on hygromycin to select for the mutant (b) allele. The observed ratio of sensitive to resistant plants (1.85; n = 296) was close to that expected (2.0) with no male transmission of the double mutant allele. This assumes that HISN1A and HISN1B are too far apart on chromosome 1 to exhibit significant linkage. Progeny plants from six crosses involving the same male parent were then PCR genotyped. Among the 83 hygromycin-resistant plants (Bb) genotyped for the mutant (a) allele, 80 were AA and only three were Aa, a significant deviation from the 1:1 ratio of AaBb to AABb plants expected with normal transmission of male (ab) gametes. Simultaneous disruption of HISN1A and HISN1B therefore results in a dramatic reduction in male gamete transmission, slight reduction in female transmission, and early lethality of the double knockout embryos that are occasionally produced.
Double heterozygous plants were then treated with 2 mm His to evaluate the ability of His to improve transmission of double mutant gametes and rescue double mutant seeds. The frequency of ovule and seed abortion in these plants was reduced but not eliminated. Some of the mature seeds produced were then germinated on His and tested for the ability to form callus in the presence and absence of His. Seven of the 65 progeny plants tested (10.8%) showed a strict His growth requirement. Six of these plants were then PCR genotyped and all were found to be aaBb. The absence of aabb seedlings among the 65 progeny plants grown in the presence of His suggests that His supplementation of parental double heterozygotes was unable to fully rescue ab pollen and aabb seeds. Alternatively, the sample size may have been too small to detect rare double homozygotes. Because these experiments were performed under somewhat different growth conditions than those described for the single gene knockouts, we cannot say for certain whether the apparent failure of His to rescue double mutants reflects an additional requirement of cells blocked at the initial step in the pathway or minor differences in experimental protocols. The auxotrophic response of aaBb cotyledon explants in these experiments nevertheless demonstrates that a single functional (B) allele is insufficient to sustain callus growth in the absence of His. In contrast, aaBB plants identified by their short root phenotype produced callus in both the presence and absence of His. These observations are consistent with differential expression levels of HISN1A and HISN1B.
DISCUSSION
The Search for Plant Auxotrophic Mutants
Several different strategies have been used over the past 50 years to search for plant auxotrophs: (1) screening mutagenized populations of Arabidopsis seedlings for defective plants that can be rescued by nutritional supplements (Langridge, 1958; Li and Rédei, 1969); (2) using negative selection strategies to identify auxotrophic cell cultures that can be maintained in the presence of the required nutrient (Blonstein, 1986); (3) incorporating positive selection strategies into forward genetic screens of mutant seedlings (Last and Fink, 1988); and (4) focusing on informative plant phenotypes and identifying the altered metabolic pathway by cloning the disrupted gene or rescuing mutant plants with chemical intermediates (Schneider et al., 1989; Wright et al., 1992; Patton et al., 1998). An alternative approach that became possible with sequencing of the Arabidopsis genome is to use reverse genetics to determine the effects of knocking out specific genes known to participate in the biosynthesis of an essential nutrient. This targeted strategy was pursued here in the analysis of His auxotrophs. The results obtained confirm that multiple auxotrophic mutants disrupted in amino acid biosynthesis can indeed be found when heterozygotes segregating for null alleles of nonredundant genes are examined for early defects in reproductive development.
Plant Auxotrophs Exhibit Diverse Phenotypes
A recurring problem with forward genetic screens for plant auxotrophs has been determining what phenotype to expect when a biosynthetic pathway is disrupted. Vitamin auxotrophs of Arabidopsis, for example, exhibit a variety of phenotypes depending on the locus, pathway, allele strength, and level of functional redundancy involved. Participation of the gene product in other cellular processes may also influence the terminal phenotype (Lukowitz et al., 2001). Seedling phenotypes have been described for mutant alleles of TH1, TZ, and PY (Li and Rédei, 1969); PDX1 (Shi et al., 2002; Chen and Xiong, 2005); VTE1 (Porfirova et al., 2002); VTE2 (Sattler et al., 2004); and VTC1 and VTC2 (Conklin et al., 2000). Embryo lethality has been reported for knockout alleles of BIO1 (Schneider et al., 1989), BIO2 (Patton et al., 1998), PDX2 (Rueschhoff and Daub, 2005), and CYT1 (Nickle and Meinke, 1998). A complete disruption of biotin or pyridoxine biosynthesis therefore results in embryo lethality, whereas the elimination of thiamine production results in seedling lethality, revealing different capacities of the maternal plant to provide mutant embryos with the missing nutrient. Gametophytic lethality of vitamin auxotrophs has not been reported in plants. The most probable explanation is that mutant gametophytes obtain sufficient amounts of the required vitamin from surrounding heterozygous tissues.
Expected Phenotypes of Amino Acid Auxotrophs
Amino acid biosynthesis in plants has been more difficult to address from a genetic perspective. Isolated examples of auxotrophs have been reported in maize (Zea mays; Racchi et al., 1978), Arabidopsis (Last and Fink, 1988; Niyogi et al., 1993; Noutoshi et al., 2005), Nicotiana (Negrutiu et al., 1985, 1992), and Physcomitrella (Schween et al., 2005) and in cell cultures from a variety of plants (Blonstein, 1986). A comprehensive collection of amino acid auxotrophs in a model angiosperm, however, remains to be established. Based on the extensive analysis of His auxotrophs presented here, we propose that most amino acid auxotrophs of Arabidopsis will exhibit gametophytic or embryonic lethality in the absence of functional redundancy at the genomic or biochemical levels and delayed embryonic or seedling phenotypes in the presence of partial redundancy. Consistent with this model, mutations affecting two different steps in Pro biosynthesis have recently been found to exhibit an embryo-defective phenotype (T. Leustek and D. Meinke, unpublished data; Szabados et al., 2005). Disruption of another gene (AGD2) encoding a novel aminotransferase required for Lys biosynthesis (Hudson et al., 2006) also results in embryo lethality (Song et al., 2004). In contrast, weak alleles of HISN3 and HISN6A have a seedling phenotype (Noutoshi et al., 2005; Mo et al., 2006). Collections of Arabidopsis auxotrophs are not sufficiently robust at present to enable a definitive comparison of knockout phenotypes for each pathway. In contrast to vitamin biosynthesis, however, null mutations in nonredundant genes required for amino acid biosynthesis do not appear to be rescued by surrounding tissues during seed development. It should therefore be possible to design a comprehensive reverse genetic screen of single and multiple knockouts of candidate genes involved in amino acid biosynthesis, focused on defects in gametogenesis and embryo development, and saturate for amino acid auxotrophs in a model plant.
Mutant Seed Phenotypes and His Transport
The most common phenotype observed in plants heterozygous for knockout alleles of His biosynthetic genes is embryo lethality. This indicates that surrounding maternal tissues are unable to provide sufficient His to support continued embryo development. Although immature embryos do not have a direct vascular connection to the maternal plant, it is generally presumed that embryo growth and storage product accumulation are reliant in part on maternal sources of amino acids and other metabolites (Hirner et al., 1998; Bewley et al., 2000). In support of this model, a variety of transport proteins has been characterized and implicated in the delivery of metabolites to developing seeds (Gillissen et al., 2000; Tegeder et al., 2000). His has been measured in both xylem and phloem sap, suggesting that it is mobile (Frommer et al., 1995; Lohaus and Moellers, 2000). In addition, His and Lys transport proteins from Arabidopsis (LHT1 and CAT1) have been proposed to function in delivery of these amino acids to seeds (Frommer et al., 1995; Chen and Bush, 1997). The hisn2, hisn3, and hisn4 seed phenotypes, however, demonstrate that mutant embryos are unable to obtain sufficient His from maternal tissues, except when heterozygous plants are watered with high concentrations of His. One possible explanation is that the level of His in maternal tissues is so low that it is unavailable for embryo growth. Indeed, the His concentration of Arabidopsis tissues including flowers has been reported to be in the range of 0.1 to 3 nmol mg−1 fresh weight (Wycisk et al., 2004; T. Leustek, unpublished data). Whether the movement of supplemental His from roots to developing embryos takes place via the normal selective transporters or through general amino acid transporters remains to be determined.
Mutant Phenotypes and Expression Patterns of Redundant Genes
Phenotypic data on the consequences of disrupting one member of a duplicated pair of His biosynthetic genes can in some cases provide valuable information on the effects of reducing but not eliminating His levels in different parts of the plant. In other cases, the absence of a mutant phenotype in a single gene knockout indicates that expression of the other gene is sufficient to meet the overall need for His biosynthesis. The normal phenotype described here for HISN1B knockouts is consistent with high levels of HISN1A expression throughout the plant (Fig. 2; Supplemental Fig. S1). The ability of HISN1A expression to compensate for a complete loss of HISN1B function is not unexpected. The normal phenotype of a HISN5A knockout is more surprising given the modest level of HISN5B expression revealed in microarray experiments. We can nevertheless conclude from the absence of a visible hisn5a phenotype that HISN5B expression is sufficient to meet the requirement for His throughout growth and development. In contrast, HISN6B gene function cannot fully compensate for a complete loss of HISN6A gene function. This case is particularly intriguing because the two genes are so similar in sequence that expression data have not previously been able to distinguish between them. The moderately weak phenotype of HISN6A knockouts when compared with other hisn null alleles provides compelling evidence for expression of HISN6B during plant growth and development.
The root phenotype of HISN1A knockouts described here and elsewhere (Wang et al., 2005) is consistent with high expression levels of HISN1A relative to HISN1B in the wild-type radicle and root (Fig. 2). The loss of HISN1A function in homozygotes therefore first becomes critical during root development. Mo et al. (2006) observed a similar phenotype in a weak allele (hpa1) of HISN6A and concluded that His performs an important role in root meristem maintenance. An alternative explanation is that HISN6B expression is unable to meet cellular needs for His biosynthesis in root development but can compensate more fully for a loss of HISN6A function elsewhere. This alternative model could also explain why a weak allele of HISN3 (apg10) exhibits a different phenotype altogether (pale seedling). The future isolation of weak alleles of HISN2 and HISN4 may help to resolve this inconsistency and determine whether His indeed plays a unique role in root development.
Gametophytic Defects in His Mutants
The transmission defects observed in knockouts of HISN2, HISN3, HISN4, HISN8, and HISN1A/B suggest that developing gametophytes in heterozygous plants fail to receive sufficient His from surrounding floral tissues to rescue the mutant phenotype. This conclusion is consistent with the gametophytic defects observed in a double mutant (trp1-110 trp4) of Arabidopsis that is more completely deficient in Trp biosynthesis than either single mutant (Niyogi et al., 1993) and with the transmission problems observed in His auxotrophs of Nicotiana regenerated from cell cultures (Negrutiu et al., 1992). However, the gametophytic defects described here seem inconsistent with the trace amounts of HISN transcripts found in wild-type pollen (Fig. 2). The normal transmission rates observed with hisn6a may reflect residual function of the redundant HISN6B gene. An alternative explanation for mutations affecting early steps in the His pathway is that reduced ATP synthesis is partially responsible for the observed defects in gametophytic transmission. This would then require a different explanation for the gametophytic abnormalities observed in hisn8. One possibility is that these gametophytes accumulate histidinol, which is a potent cytotoxin of animal cells (Hansen et al., 1972). Another model is that other His precursors are responsible for partial rescue of gametophytes blocked elsewhere in the pathway but these same precursors are unable to rescue hisn8 mutant cells, which are blocked in the final step of His biosynthesis. The future identification of knockout alleles of HISN5B, HISN6B, and HISN7 should help to distinguish between these models and complete the establishment of a comprehensive collection of His auxotrophs in a flowering plant.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seed stocks for Salk (Alonso et al., 2003) and SAIL (Sessions et al., 2002) insertion lines (Columbia ecotype) were obtained from the ABRC at Ohio State University. The emb2196 (hisn6a-1) mutant allele (Wassilewskija ecotype) described by Tzafrir et al. (2004) was obtained from internal seed stocks but is also available from ABRC. The RATM (Ito et al., 2005) insertion line (Nössen ecotype) was obtained from the RIKEN Bioresource Center in Japan. Plants in the Meinke laboratory were grown in a soil mixture and placed in a growth room (24°C ± 2°C) under fluorescent lights (16-h-light/8-h-dark cycles) as described by Berg et al. (2005). Plants in the Leustek laboratory were grown in a soil mixture (Promix BX, Premier Horticulture), placed in a growth chamber, and watered with Peters (20:20:20 N:P:K) fertilizer (Grace-Sierra) at the dilution recommended for indoor plants. Light intensity was 100 μmol photons m−2 s−1 supplied from cool-white fluorescent bulbs.
Primer Design and PCR Analysis
Gene-specific primers for each mutant line were designed using the SIGnAL iSect Primer Design program at http://signal.salk.edu and were purchased from either IDT (Meinke) or Invitrogen (Leustek). Primers for the left T-DNA border in Salk (SK) and SAIL (SL) lines and the Dissociation (Ds) border in the Riken (RATM) line were used in combination with the appropriate gene-specific primers to detect and confirm insertions. A complete listing of primers used is presented in Supplemental Table S1. Genomic DNA was isolated in the Meinke lab using a modified cetyl trimethyl ammonium bromide protocol (Lukowitz et al., 2000) and in the Leustek lab using the REDExtract-N-AMP Plant PCR kit (Sigma). Two different PCR parameters were used: 94°C for 2 min followed by 30 cycles of 94°C for 30 s, 56°C for 40 s, 72°C for 80 s, and a final elongation step of 72°C for 10 min (Meinke); and 94°C for 3 min followed by 36 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 1 min, and a final cycle at 72°C for 5 min (Leustek). Reactions were performed with a Biometra Uno II (Meinke) or a Perkin Elmer 9700 (Leustek) thermocycler. Amplified products were separated in 1.0% agarose gels, stained with ethidium bromide, gel purified (Qiagen), and sequenced at the Oklahoma State University Recombinant DNA/Protein Resource Facility or the Rutgers' Cook College DNA Sequencing Facility to confirm insert locations.
Genetic and Phenotypic Analyses
Heterozygous plants were identified by screening siliques for the presence of aborted seeds and ovules. Allelism tests were performed by crossing two heterozygotes and screening immature F1 siliques for the presence of aborted seeds. Detailed information on the methods used to characterize mutant seeds is presented in the tutorial section at www.seedgenes.org. Additional details on the ovule abortion (ova) phenotype can be obtained from Berg et al. (2005).
His Rescue Experiments
Several different strategies were tested for His rescue of mutant seeds in heterozygous plants (Meinke lab). The best results were obtained by daily watering of plants (40 mL/pot) with a standard nutrient solution (Berg et al., 2005) supplemented with 1 mm l-His (Product H-8125, Sigma). The solution was refrigerated between applications to limit microbial contamination. The alternative method of spraying leaves and inflorescences with an aqueous solution of 5 mm His often resulted in partial rescue of mutant seeds. Heterozygous plants selected to receive treatments were first identified by screening immature siliques for aborted seeds. Mature stems were then removed from each plant, leaving a single young stem as the sink for His supplementation. Plants received treatment for about 2 weeks. Mature siliques were harvested when possible from plants that appeared to be fully rescued based on the absence of aborted seeds. Seeds from these mature siliques were examined under a dissecting microscope to confirm full rescue before they were surface sterilized and plated on germination media (Murashige and Skoog salts, 3% [w/v] Glc, 0.8% [w/v] agar) in the presence or absence of His. To determine the effects of His depletion on plant growth, rescued mutant and wild-type seedlings were transplanted to soil or to a culture medium lacking His after 2 to 3 weeks of axenic growth in the presence of 1 mm His.
His Rescue and Callus Growth Assays
Plants in the Leustek lab were grown to the rosette stage without His. Heterozygotes identified by PCR were then watered daily with 5 mL of a solution containing 1 to 10 mm l-His (Sigma). Seedlings derived from seeds of rescued heterozygotes were grown on Murashige and Skoog medium, pH 5.8, with 2.0% (w/v) Glc, 0.8% (w/v) agar, and 0.2 mm His. After the cotyledons were fully expanded, one cotyledon was excised, cut laterally into two pieces, and each explant placed on a callus induction medium composed of Murashige and Skoog salts, 2.0% (w/v) Glc, 0.8% agar, 2.3 μm 2,4-dichlorophenoxyacetic acid, 0.25 μm kinetin, and 0.2 mm His, pH 5.8, and filter sterilized as noted. Cultures were incubated in a Percival growth chamber (24°C; 16-h-light/8-h-dark cycles) for a minimum of 10 d before callus growth was scored. Leaves from the initial seedlings were then PCR genotyped as described above. His auxotrophy was confirmed by testing the growth dependence of subcultured callus and by analyzing progeny seedlings derived from rescued homozygotes.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Temporal patterns of HISN gene expression in Arabidopsis based on microarray data obtained from https://www.genevestigator.ethz.ch.
Supplemental Table S1. Primer sequences for genotype analysis.
Supplementary Material
Acknowledgments
We thank Patricia Nugent, Christopher DiFraia, Nirav Patel, Devin Camenares, Sandrine Casanova, and Shipra Mittal for technical assistance and plant maintenance. Allan Dickerman assisted with the analysis of HISN6 paralogs.
This work was supported by the National Science Foundation (Integrative Plant Biology program grant to T.L. and Arabidopsis 2010 program grant to D.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: David Meinke (meinke@okstate.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alifano P, Fani R, Lio P, Lazcano A, Bazzicalupo M, Carlomagno MS, Bruni CB (1996) Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol Rev 60 44–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Berg M, Rogers R, Muralla R, Meinke D (2005) Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J 44 866–878 [DOI] [PubMed] [Google Scholar]
- Bewley JD, Hempel FD, McCormick S, Zambryski P (2000) Reproductive development. In B Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Biologists, Rockville, MD, pp 988–1043
- Blonstein AD (1986) Auxotroph isolation in vitro. In AD Blonstein, PJ King, eds, A Genetic Approach to Plant Biochemistry. Springer-Verlag, New York, pp 259–279
- Chen H, Xiong L (2005) Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J 44 396–408 [DOI] [PubMed] [Google Scholar]
- Chen L, Bush DR (1997) LHT1, a lysine- and histidine-specific amino acid transporter in Arabidopsis. Plant Physiol 115 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Saracco SA, Norris SR, Last RL (2000) Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 154 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFraia C, Leustek T (2004) Functional genomics study in Arabidopsis thaliana of histidine biosynthesis. The Rutgers Scholar 6 1–5 [Google Scholar]
- El Malki F, Frankard V, Jacobs M (1998) Molecular cloning and expression of a cDNA sequence encoding histidinol phosphate aminotransferase from Nicotiana tabacum. Plant Mol Biol 37 1013–1022 [DOI] [PubMed] [Google Scholar]
- El Malki F, Jacobs M (2001) Molecular characterization and expression study of a histidine auxotrophic mutant (his1−) of Nicotiana plumbaginifolia. Plant Mol Biol 45 191–199 [DOI] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Unseld M, Ninnemann O (1995) Seed and vascular expression of a high-affinity transporter for cationic amino acids in Arabidopsis. Proc Natl Acad Sci USA 92 12036–12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori K, Ohta D (1998. a) An Arabidopsis cDNA encoding a bifunctional glutamine amidotransferase/cyclase suppresses the histidine auxotrophy of a Saccharomyces cerevisiae his7 mutant. FEBS Lett 428 229–234 [DOI] [PubMed] [Google Scholar]
- Fujimori K, Ohta D (1998. b) Isolation and characterization of a histidine biosynthetic gene in Arabidopsis encoding a polypeptide with two separate domains for phosphoribosyl-ATP pyrophosphohydrolase and phosphoribosyl-AMP cyclohydrolase. Plant Physiol 118 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori K, Tada S, Kanai S, Ohta D (1998) Molecular cloning and characterization of the gene encoding N′-[(5′-phosphoribosyl)-formimino]-5-aminoimidazole-4-carboxamide ribonucleotide (BBM II) isomerase from Arabidopsis thaliana. Mol Gen Genet 259 216–223 [DOI] [PubMed] [Google Scholar]
- Gillissen B, Burkle L, Andre B, Kuhn C, Rentsch D, Brandl B, Frommer WB (2000) A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell 12 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen BS, Vaughan MH, Wang L (1972) Reversible inhibition by histidinol of protein synthesis in human cells at the activation of histidine. J Biol Chem 247 3854–3857 [PubMed] [Google Scholar]
- Hirner B, Fischer WN, Rentsch D, Kwart M, Frommer WB (1998) Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J 14 535–544 [DOI] [PubMed] [Google Scholar]
- Hudson AO, Singh BK, Leustek T, Gilvarg C (2006) An LL-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol 140 292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle RA, Mugford ST, Rees JD, Campbell MM, Smith JA (2005) Constitutively high expression of the histidine biosynthetic pathway contributes to nickel tolerance in hyperaccumulator plants. Plant Cell 17 2089–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Motohashi R, Kuromori T, Noutoshi Y, Seki M, Kamiya A, Mizukado S, Sakurai T, Shinozaki K (2005) A resource of 5,814 Dissociation transposon-tagged and sequence-indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant Cell Physiol 46 1149–1153 [DOI] [PubMed] [Google Scholar]
- Krämer U, Cotter-Howells JD, Charnock J, Baker A, Smith JAS (1996) Free histidine as a metal chelator in plants that accumulate nickel. Nature 379 635–638 [Google Scholar]
- Langridge J (1958) A hypothesis of developmental selection exemplified by lethal and semi-lethal mutants of Arabidopsis. Aust J Biol Sci 11 58–68 [Google Scholar]
- Last RL, Fink GR (1988) Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240 305–310 [DOI] [PubMed] [Google Scholar]
- Lee M, Martin MN, Hudson AO, Lee J, Muhitch MJ, Leustek T (2005) Methionine and threonine synthesis are limited by homoserine availability and not the activity of homoserine kinase in Arabidopsis thaliana. Plant J 41 685–696 [DOI] [PubMed] [Google Scholar]
- Li SL, Rédei GP (1969) Thiamine mutants of the crucifer, Arabidopsis. Biochem Genet 3 163–170 [DOI] [PubMed] [Google Scholar]
- Liao MT, Hedley MJ, Woolley DJ, Brooks RR, Nichols MA (2000) Copper uptake and translocation in chicory (Cichorium intybus L. cv Grasslands Puna) and tomato (Lycopersicon esculentum Mill. Cv Rondy) plants grown in NFT system. II. The role of nicotianamine and histidine in xylem sap copper transport. Plant Soil 223 243–252 [Google Scholar]
- Lohaus G, Moellers C (2000) Phloem transport of amino acids in two Brassica napus L. genotypes and one B. carinata genotype in relation to their seed protein content. Planta 211 833–840 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, Somerville CR (2001) Arabidopsis cyt1 mutants are deficient in mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci USA 98 2262–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW (1982) Embryo-lethal mutants of Arabidopsis thaliana: evidence for gametophytic expression of the mutant genes. Theor Appl Genet 63 543–552 [DOI] [PubMed] [Google Scholar]
- Mo X, Zhu Q, Li X, Li J, Zeng Q, Rong H, Zhang H, Wu P (2006) The hpa1 mutant of Arabidopsis reveals a crucial role of histidine homeostasis in root meristem maintenance. Plant Physiol 141 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai A, Ward E, Beck J, Tada S, Chang JY, Scheidegger A, Ryals J (1991) Structural and functional conservation of histidinol dehydrogenase between plants and microbes. Proc Natl Acad Sci USA 88 4133–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutiu I, De Brouwer D, Dirks R, Jacobs M (1985) Amino acid auxotrophs from protoplast cultures of Nicotiana plumbaginifolia, Viviani. Mol Gen Genet 199 330–337 [Google Scholar]
- Negrutiu I, Hinnisdaels S, Cammaerts D, Cherdshewasart W, Gharti-Chhetri G, Jacobs M (1992) Plant protoplasts as genetic tool: selectable markers for developmental studies. Int J Dev Biol 36 73–84 [PubMed] [Google Scholar]
- Nickle TC, Meinke DW (1998) A cytokinesis-defective mutant of Arabidopsis (cyt1) characterized by embryonic lethality, incomplete cell walls, and excessive callose accumulation. Plant J 15 321–332 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Last RL, Fink GR, Keith B (1993) Suppressors of trp1 fluorescence identify a new Arabidopsis gene, TRP4, encoding the anthranilate synthase β subunit. Plant Cell 5 1011–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutoshi Y, Ito T, Shinozaki K (2005) ALBINO AND PALE GREEN 10 encodes BBMII isomerase involved in histidine biosynthesis in Arabidopsis thaliana. Plant Cell Physiol 46 1165–1172 [DOI] [PubMed] [Google Scholar]
- Ohta D, Fujimori K, Mizutani M, Nakayama Y, Kunpaisal-Hashimoto R, Munzer S, Kozaki A (2000) Molecular cloning and characterization of ATP-phosphoribosyl transferase from Arabidopsis, a key enzyme in the histidine biosynthetic pathway. Plant Physiol 122 907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW (1998) An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol 116 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persans MW, Yan X, Patnoe JM, Krämer U, Salt DE (1999) Molecular dissection of the role of histidine in nickel hyperaccumulation in Thlaspi goesingense (Halacsy). Plant Physiol 121 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porfirova S, Bergmuller E, Tropf S, Lemke R, Dormann P (2002) Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc Natl Acad Sci USA 99 12495–12500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racchi ML, Gavazzi G, Monti D, Manitto P (1978) An analysis of the nutritional requirements of the pro mutant in Zea mays. Plant Sci Lett 13 357–364 [Google Scholar]
- Rueschhoff B, Daub ME (2005) PDX2, a de novo vitamin B6 biosynthetic pathway gene, is essential for seed development in Arabidopsis (abstract no. 286). In 16th International Conference on Arabidopsis Research. University of Wisconsin Conference Services, Madison, WI
- Salt DE, Prince RC, Baker AJM, Raskin I, Pickering IJ (1999) Zinc ligands in the metal hyperaccumulator Thlaspi caerulescens as determined using x-ray absorption spectroscopy. Environ Sci Technol 33 713–717 [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D (2004) Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Dinkins R, Robinson K, Shellhammer J, Meinke DW (1989) An embryo-lethal mutant of Arabidopsis thaliana is a biotin auxotroph. Dev Biol 131 161–167 [DOI] [PubMed] [Google Scholar]
- Schween G, Egener T, Fritzowsky D, Granado J, Guitton MC, Hartmann N, Hohe A, Holtorf H, Lang D, Lucht JM, et al (2005) Large-scale analysis of 73,329 Physcomitrella plants transformed with different gene disruption libraries: production parameters and mutant phenotypes. Plant Biol (Stuttg) 7 228–237 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Xiong L, Stevenson B, Lu T, Zhu JK (2002) The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 14 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JT, Lu H, Greenberg JT (2004) Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, ABERRANT GROWTH AND DEATH2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepansky A, Leustek T (2006) Histidine biosynthesis in plants. Amino Acids 30 127–142 [DOI] [PubMed] [Google Scholar]
- Szabados L, Szekely G, Abraham E, Rigo G, Csiszar J, Koncz C (2005) Proline biosynthesis in Arabidopsis: a model for stress responses (abstract no. 516). In 16th International Conference on Arabidopsis Research. University of Wisconsin Conference Services, Madison, WI
- Tada S, Volrath S, Guyer D, Scheidegger A, Ryals J, Ohta D, Ward E (1994) Isolation and characterization of cDNAs encoding imidazoleglycerolphosphate dehydratase from Arabidopsis thaliana. Plant Physiol 105 579–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M, Offler CE, Frommer WB, Patrick JW (2000) Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiol 122 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lin Y, Guo J (2005) Molecular genetics of Arabidopsis seed development and storage product synthesis (abstract no. 264). In 16th International Conference on Arabidopsis Research. University of Wisconsin Conference Services, Madison, WI
- Wright AD, Moehlenkamp CA, Perrot GH, Neuffer MG, Cone KC (1992) The maize auxotrophic mutant orange pericarp is defective in duplicate genes for tryptophan synthase β. Plant Cell 4 711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wycisk K, Kim EJ, Schroeder JI, Krämer U (2004) Enhancing the first enzymatic step in the histidine biosynthesis pathway increases the free histidine pool and nickel tolerance in Arabidopsis thaliana. FEBS Lett 578 128–134 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.