Abstract
Cadmium (Cd) tolerance seems to be a constitutive species-level trait in Arabidopsis halleri sp. halleri. Therefore, an interspecific cross was made between A. halleri and its closest nontolerant interfertile relative, Arabidopsis lyrata sp. petraea, and a first-generation backcross population (BC1) was used to map quantitative trait loci (QTL) for Cd tolerance. Three QTL were identified, which explained 43%, 24%, and 16% of the phenotypic variation in the mapping population. Heavy metal transporting ATPases4 (HMA4), encoding a predicted heavy metal ATPase, colocalized with the peak of the major QTL Cdtol-1 and was consequently further studied. HMA4 transcripts levels were higher in the roots and the shoots of A. halleri than in A. lyrata sp. petraea. Furthermore, HMA4 was also more highly expressed in all BC1 genotypes harboring the HMA4 A. halleri allele at the QTL Cdtol-1, independently of the presence of an A. halleri allele at the two other QTL. Overexpression of AhHMA4 in yeast (Saccharomyces cerevisiae) supported a role of HMA4 in zinc (Zn) and Cd transport by reducing the Cd and Zn contents of the yeast cells. In epidermal tobacco (Nicotiana tabacum) cells, AhHMA4:green fluorescent protein was clearly localized in the plasma membrane. Taken together, all available data point to the elevated expression of HMA4 P1B-type ATPase as an efficient mechanism for improving Cd/Zn tolerance in plants under conditions of Cd/Zn excess by maintaining low cellular Cd2+ and Zn2+ concentrations in the cytoplasm.
Pollutants such as heavy metals can occur at very high concentrations in the soil as a consequence of either ancient natural processes or recent human activities such as mining, industrial, and agricultural practices. Although heavy metals are very toxic at high concentrations, some metal-tolerant plant species have developed the ability to grow and reproduce on soils highly polluted by heavy metals (Macnair, 1987). Among those metal-tolerant plants, some possess a remarkable physiological trait called hyperaccumulation that enables them to tolerate and accumulate high heavy metal concentrations in their shoots (about 100 times those occurring in nonaccumulator plants growing in the same substrates). Metal hyperaccumulators are consequently a particularly valuable resource to study the genetic basis of metal homeostasis.
Arabidopsis halleri is an emerging model species for the molecular elucidation of metal tolerance and hyperaccumulation (Becher et al., 2004; Weber et al., 2004; Craciun et al., 2006; Filatov et al., 2006). This species belongs to the family Brassicaceae and shares about 94% DNA sequence identity within coding regions with the non-metal-tolerant plant species Arabidopsis (Arabidopsis thaliana; Becher et al., 2004). A. halleri constitutively tolerates zinc (Zn; Pauwels et al., 2006) and so far all the identified accessions are able to tolerate cadmium (Cd) and to hyperaccumulate Zn (Macnair et al., 1999). Hyperaccumulation of Cd was observed in at least one A. halleri population (Auby, France; Bert et al., 2002, 2003).
Metal tolerance and hyperaccumulation are thought to have evolved through adaptations of metal homeostasis processes including metal uptake, chelation, trafficking, and storage (Pence et al., 2000; Pollard et al., 2002; Krämer, 2005; Clemens, 2006). To identify the genes governing these traits, numerous studies have been performed. For instance, segregation analyses of crosses between nontolerant and tolerant individuals indicated that a single gene or a small number of genes govern metal tolerance (Schat and Ten Bookum, 1992; Macnair et al., 1999). More recently, molecular and functional studies were performed on various metal homeostasis genes to determine their role in metal tolerance and/or hyperaccumulation. Also, microarray and cDNA-amplified fragment length polymorphism analyses were applied to hyperaccumulator species to identify genes differentially regulated under various metal conditions in nontolerant nonhyperaccumulator versus tolerant hyperaccumulator species (Craciun et al., 2006; van de Mortel et al., 2006). Very recently, quantitative trait loci (QTL) mapping was used to identify the chromosomal regions involved in Zn and Cd hyperaccumulation in the hyperaccumulator species Thlaspi caerulescens. Two QTL were identified for Zn and Cd accumulation in roots, while three QTL and one QTL were reported for Zn and Cd accumulation, respectively, in shoots (Deniau et al., 2006). But, in contrast to Thlaspi there is a possibility of identifying individual genes in A. halleri because of its close relationship to Arabidopsis.
In this work, QTL mapping was performed to identify the genes underlying Cd tolerance in A. halleri. Because no Cd-sensitive accession has been identified so far in A. halleri, a first-generation backcross progeny of A. halleri sp. halleri (pollen donor) and its nontolerant relative Arabidopsis lyrata sp. petraea (pollen recipient) was produced (Bert et al., 2003) and used for the QTL analysis of Cd tolerance. The A. halleri genotype used in this study originated from a metallicolous population, previously shown to be tolerant to high Zn and Cd concentrations and able to hyperaccumulate those metals. The gene heavy metal transporting ATPases4 (HMA4) was found to colocalize with the major QTL and was consequently focused on in this study.
The P1B-type ATPases, also known as the HMAs, play an important role in transporting transition metal ions against their electrochemical gradient using the energy provided by ATP hydrolysis. They have been identified in all living organisms from archaea to humans, including yeast (Saccharomyces cerevisiae) and plants. The HMAs cluster into two classes: those transporting monovalent cations (copper [Cu]/silver [Ag] group) and those transporting divalent cations (Zn/cobalt [Co]/Cd/lead [Pb] group; Axelsen and Palmgren, 2001). Although all classes of P1B-type ATPases have been detected in prokaryotes (Rensing et al., 1999), in nonplant eukaryotes only Cu/Ag P1B-type ATPases have been identified to date. Arabidopsis and Oryza sativa contain eight HMA genes (Baxter et al., 2003), suggesting that these enzymes play important roles in the transport of metals in plants. In Arabidopsis, analysis of the complete genome sequence predicts the distribution of these eight HMAs in two classes: HMA1-4 for the transport of Zn/Co/Cd/Pb and HMA5-8 for Cu/Ag.
HMA4 was the first plant P1B-type ATPase of the divalent transport group to be cloned and characterized in Arabidopsis (Mills et al., 2003). A role for HMA4 in Zn homeostasis and Cd detoxification, and in the translocation of these metals from the root to the shoot, has been demonstrated in Arabidopsis (Mills et al., 2003, 2005; Hussain et al., 2004; Verret et al., 2004, 2005).
In this study, HMA4 was cloned and functionally characterized in A. halleri.
RESULTS
QTL Mapping of Cd Tolerance
The QTL for Cd tolerance were identified on the A. halleri × A. lyrata sp. petraea genetic linkage map (henceforth called the Ah × Alp map), which was constructed on 196 individuals of the A. halleri × A. lyrata sp. petraea backcross population (BC1) progeny (henceforth called BC1 progeny; Willems et al., 2007). On this linkage map, a total of 85 markers was distributed on eight linkage groups (LGs), corresponding to the haploid chromosome number of both parental genotypes. Significant segregation at a locus-by-locus significance level of 0.05 was found for 40% of the markers. All markers showed an excess of the homospecific A. lyrata sp. petraea 1/A. lyrata sp. petraea 2 genotypes compared to the heterospecific A. halleri/A. lyrata sp. petraea 2 genotypes. Because we applied stringent goodness-of-fit thresholds to minimize the effects of segregation distortion and observed a high degree of macrosynteny with the Arabidopsis genome and the recently published linkage map of A. lyrata sp. petraea (Kuittinen et al., 2004), the Ah × Alp map is assumed to be robust.
A highly significant genotype effect of Cd tolerance (P < 0.0001) was revealed through an ANOVA analysis, performed on the 100% effective concentration (EC100), i.e. the lowest concentration at which no new root growth was observed, values obtained on 79 BC1 individuals. A high broad-sense heritability of 0.89 was obtained for this trait in the mapping population.
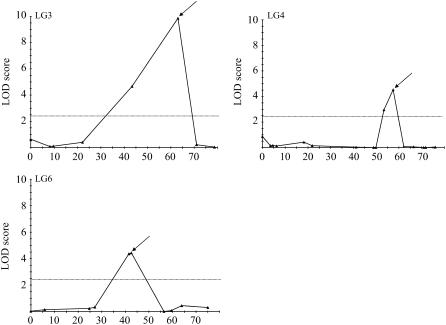
We performed the QTL analysis of Cd tolerance on the EC100max values, corresponding to the maximum tolerance level of the BC1 genotypes characterized for Cd tolerance. Three QTL for Cd tolerance, showing a log of the odds (LOD) score exceeding the significance threshold of 2.4 (α = 0.05), were detected on the LGs LG3, LG4, and LG6 of the Ah × Alp linkage map (Fig. 1) through Multiple QTL Mapping (MQM; MapQTL 4.0; Van Ooijen et al., 2002). Their significance was verified through a nonparametric Kruskal-Wallis test (P < 0.0001, P < 0.0001, P < 0.005). The QTL Cdtol-1, Cdtol-2, and Cdtol-3 explained 42.9%, 23.7%, and 15.9% of the phenotypic variance, respectively (Table I). Together they accounted for 82.5% of the phenotypic variance, or for 92.7% of the genetic variance observed for Cd tolerance in the BC1 progeny. At all three QTL, the A. halleri allele had a positive effect on Cd tolerance. The increase in Cd tolerance due to the presence of the A. halleri allele, however, was greater for the QTL Cdtol-1 than for the other QTL (Table I). BC1 individuals harboring the A. halleri allele at QTL Cdtol-1 showed a nearly 2-fold increase in Cd tolerance when compared to BC1 genotypes carrying the A. lyrata sp. petraea allele, independently of the presence of the A. halleri allele at the two other QTL (Fig. 2). One significant epistatic interaction was revealed between the QTL Cdtol-1 and Cdtol-3 at a low significance threshold (P = 0.022; Table II). However, using log-transformed Cd tolerance data, the significance of the epistatic interaction between the QTL Cdtol-1 and Cdtol-3 was lost (P = 0.294), indicating most probably a statistical artifact (Table II). The QTL for Cd tolerance are consequently inferred to be mainly additive.
Figure 1.
QTL for Cd tolerance in the Ah × Alp BC1 progeny. LOD score profiles are given along the LGs LG3, LG4, and LG6, on which the QTL for Cd tolerance were identified. Map positions are plotted along the abscissa. LOD scores are plotted along the y axis. Dashed lines correspond to the LOD score threshold (2.4) for QTL detection at an error level α = 0.05. QTL are indicated by arrows above the LOD score profile.
Table I.
QTL for Cd tolerance
| QTLa | LG Markerb | LOD Scorec | R2d | ae |
|---|---|---|---|---|
| Cdtol-1 | LG3-HMA4 | 9.87 | 42.9 | −57.8085 |
| Cdtol-2 | LG4-ICE12 | 4.53 | 23.7 | −45.7050 |
| Cdtol-3 | LG6-AthCDPK9 | 4.43 | 15.9 | −37.0000 |
QTL are named by the trait and ordered from 1 to 3.
LG on which QTL were mapped and marker at QTL position.
Maximum LOD score at the QTL position.
Percentage of explained variance of the QTL.
Additive effects of the QTL, corresponding to the differences between the Cd tolerance means (micromolars of Cd) of the two genotypic groups in the BC1 progeny (negative values imply that the A. halleri allele increases Cd tolerance compared to the Alp allele).
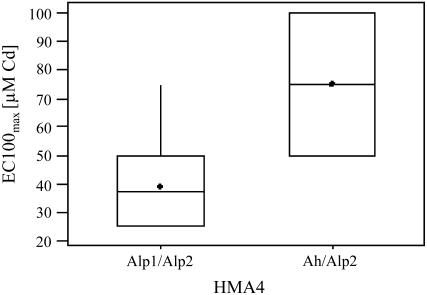
Figure 2.
Box plot for Cd tolerance at the marker HMA4, located at the maximum LOD score position of the QTL Cdtol-1. Box plots for Cd tolerance are given for two groups of BC1 individuals that can be distinguished based on their genotype at the marker HMA4, i.e. those carrying both alleles from the nontolerant parental individuals A. lyrata sp. petraea 1 and A. lyrata sp. petraea 2 at the three QTL or markers HMA4, ICE12, and AthCDPK9 and those carrying one allele from the tolerant parental individual A. halleri and one from the nontolerant parental individual A. lyrata sp. petraea 2 at the marker HMA4, while carrying both alleles from the nontolerant parental individuals A. lyrata sp. petraea 1 and A. lyrata sp. petraea 2 at the markers ICE12 and AthCDPK9 (P = 0.001). The boxes represent the interquartile range, with arithmetic means indicated by black diamonds and medians indicated by horizontal lines. Whiskers connect the nearest observations within 1.5 times the interquartile ranges of the lower and upper quartiles.
Table II.
Epistatic interactions between QTL for Cd tolerance
| Source | dfa | On Raw EC100max Data
|
On Log-Transformed EC100max Data
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| SSb | MSc | Fd | Pe | SSb | MSc | Fd | Pe | ||
| Cdtol-1 | 1 | 46,465 | 46,465 | 75.87 | 0 | 1.42009 | 1.42009 | 70.37 | 0 |
| Cdtol-2 | 1 | 17,030 | 17,030 | 27.81 | 0 | 0.55831 | 0.55831 | 27.67 | 0 |
| Cdtol-3 | 1 | 12,967 | 12,967 | 21.17 | 0 | 0.30622 | 0.30622 | 15.18 | 0 |
| Cdtol-1 × Cdtol-2 | 1 | 709 | 709 | 1.16 | 0.286 | 0.01897 | 0.01897 | 0.94 | 0.336 |
| Cdtol-1 × Cdtol-3 | 1 | 3,379 | 3,379 | 5.52 | 0.022 | 0.02255 | 0.02255 | 1.12 | 0.294 |
| Cdtol-2 × Cdtol-3 | 1 | 503 | 503 | 0.82 | 0.368 | 0.00009 | 0.00009 | 0 | 0.947 |
| Error | 69 | 42,260 | 612 | 1.39236 | 0.020179 | ||||
| Total | 75 | 144,178 | 4.59379 | ||||||
Degrees of freedom.
Sum of squares.
Mean squares.
Values of F statistic.
Significance levels.
Marker Densification in the Major QTL Region
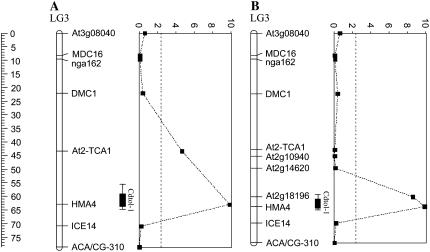
The HMA4 locus was located at the maximum LOD score position of the major QTL for Cd tolerance, Cdtol-1, explaining up to 48.2% of the genetic variance observed for this trait in the Ah × Alp BC1 progeny (Fig. 3A). To narrow down the confidence interval associated with this QTL and, assuming a high degree of synteny between Arabidopsis and A. halleri, the genes At2g10940, At2g14620, and At2g18196 were selected based on their position in the Arabidopsis genome. As expected, these markers mapped to LG3 of the Ah × Alp map in the same order as in Arabidopsis. The mapping of these markers increased the accuracy of the QTL Cdtol-1 and reinforced its colocalization with the HMA4 locus (Fig. 3B).
Figure 3.
Densification of the major QTL region Cdtol-1. A, LG LG3, on which initially eight markers were mapped, is represented. The position in centiMorgans of the markers on LG3 is given by the ruler. The LOD score profile along the LG is represented at the right of the LG. Map positions are plotted along the vertical axis and LOD scores are plotted along the horizontal axis. Dashed lines correspond to the LOD score threshold (2.4) for QTL detection at an error level α = 0.05. The position of a QTL is shown as the interval over which the LOD score is within one or two log units of its maximum value, i.e. at the most likely position of the QTL. Bars indicate the one-LOD (10-fold) support interval and whiskers (lines extending beyond bars) indicate the two-LOD (100-fold) support intervals. The length of the one- and two-LOD support intervals correspond to approximately 5 and 9 cM, respectively. B, Three additional markers At2g10940, At2g14620, and At2g18196 were mapped on LG3 to increase the accuracy of the major QTL position. A reduction in length of the LOD support intervals associated with the QTL Cdtol-1 was obtained: one- and two-LOD support intervals correspond to approximately 4 and 6 cM, respectively.
AhHMA4 Cloning and Sequence Analysis
Because of the importance of HMA4 in the Cd tolerance of A. halleri, we decided to investigate the possible specificities of AhHMA4. To isolate a full-length cDNA for AhHMA4, a reverse transcription (RT)-PCR approach was taken. The 3,486-bp AhHMA4 cDNA was directly cloned into pYES2-GFP plasmid. According to its nucleotide sequence, AhHMA4 belongs to the P1B-type ATPases involved in metal transport.
The deduced 1,161 amino acid sequence of AhHMA4 was aligned with the Arabidopsis sequence of HMA4 (AtHMA4) and with the T. caerulescens sequence of HMA4 (TcHMA4; Fig. 4), with which it shares 80% and 70% identity, respectively. Whereas the sequence of the N-terminal part (AhHMA4-N, amino acid position 1–720, between the start codon and the end of the last transmembrane span) was highly similar in the three HMA4 sequences (95% identity with AtHMA4 and 89% with TcHMA4), the sequence of the C-terminal part (AhHMA4-C, amino acid position 720–1,161, between the end of the last transmembrane span and the stop codon) was more divergent (59% identity with AtHMA4 and 46% with TcHMA4). No specific features were observed for AhHMA4 compared to At- or TcHMA4 homologs.
Figure 4.
Sequence similarity of AhHMA4 (Auby, France) to other HMA4 amino acid sequences. Deduced amino acid sequences for AhHMA4 (accession no. DQ221101), AtHMA4 (NM127468), and TcHMA4 (AJ567384) are shown aligned using the ClustalW method (Thompson et al., 1994). A number of motifs common to P1B-type ATPases are indicated, including the highly conserved CPx motif (boxed), the ATPase phosphorylation site (shaded in dark gray), the transmembrane spans (consensus: continued lines; probable: dotted lines, according to ARAMEMNON predictions softwares). Also, Cys residues in the C terminus are highlighted. The start of the C-terminal fragment used in the yeast tolerance assay is indicated with a bracket. Asterisks (*) indicate the identical residues; “:” indicates that one of the following strong groups is fully conserved: STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, and FYW; “.” indicates that one of the following weaker groups is fully conserved: CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, and HFY.
HMA4 Expression in the BC1 Parental Genotypes and Individuals
The expression of HMA4 was studied by RT-PCR in the root and shoot of the BC1 parental genotypes A. halleri, A. lyrata sp. petraea, and the F1 individual with or without exposure to CdSO4 (0 or 10 μm for 72 h, Fig. 5A; 100 μm for 72 h or 7 d for A. halleri, data not shown). The analysis was performed with primers designed in a perfectly conserved coding region between AhHMA4 and AlpHMA4 cDNAs.
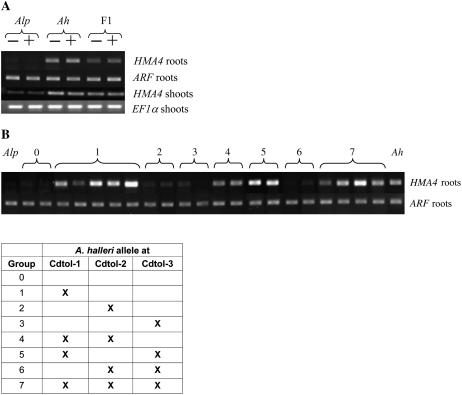
Figure 5.
RT-PCR analysis of HMA4 on A. halleri, A. lyrata sp. petraea, F1, and BC1 progeny. A, RT-PCR was used to compare transcript levels for HMA4 or EF1α (as a control) in roots and shoots of A. halleri, A. lyrata sp. petraea, and the F1 individual. Plants were grown on hydroponic media containing 0 (−) or 10 μm CdSO4 (+) for 72 h. The PCR conditions were the same for HMA4 in roots and EF1α in roots and in shoots (24 cycles), but 29 cycles were used to detect HMA4 expression in shoots. B, RT-PCR was used to compare transcript levels of HMA4 or ARF (as a control) in roots of A. halleri, A. lyrata sp. petraea, and 21 BC1 individuals belonging to eight different classes characterized by the presence or the absence of an A. halleri allele at one, two, or three QTL for Cd tolerance. Plants were grown on hydroponic media. Amplification was performed for 24 cycles for HMA4 and 20 cycles for ARF. The table represents the different groups of the BC1 individuals based on the presence or the absence of an A. halleri allele at one, two, or three QTL for Cd tolerance. The presence of the A. halleri allele at the QTL Cdtol-1, Cdtol-2, and/or Cdtol-3 is indicated by X.
HMA4 was highly expressed in the root and shoot of A. halleri and, to a lesser extent, in the root and shoot of the F1 individual. Almost no signal was detected in the root and shoot of the Cd-sensitive parental genotype A. lyrata sp. petraea at the same number of amplification cycles. Overall, HMA4 was more highly expressed in the roots compared to the shoots. Four additional PCR cycles were needed to observe a band intensity in the shoots similar to the one obtained in roots. Transcript levels of HMA4 were not affected by the Cd concentration (10 μm or 100 μm CdSO4) or the duration (72 h or 7 d) of the treatment.
HMA4 expression was further studied in the roots of 21 BC1 genotypes distributed among eight different classes characterized by the presence or absence of the A halleri allele at one or more of the QTL for Cd tolerance (Fig. 5B). This analysis revealed that HMA4 was highly expressed independently of the presence of the A. halleri allele at the QTL Cdtol-2 and Cdtol-3. Indeed, high expression levels of HMA4 were obtained in the BC1 individuals harboring the A. halleri allele at the QTL Cdtol-1, while carrying two A. lyrata sp. petraea alleles at the QTL Cdtol-2 and Cdtol-3 (class 1). On the contrary, low expression levels, comparable to those observed in A. lyrata sp. petraea roots, were observed for the BC1 individuals belonging to classes 2 and 3, carrying the A. halleri allele at the QTL Cdtol-2 and Cdtol-3, respectively, and the A. lyrata sp. petraea alleles at the other QTL (Fig. 5B).
Role of AhHMA4 in Cd and Zn Tolerance and Transport in Yeast
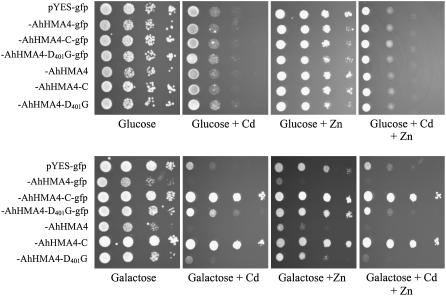
To investigate the function of AhHMA4, the full-length and AhHMA4-C sequences were cloned in pYES2-GFP under control of the GAL1 promoter and expressed in the yeast strains BY4741 (wild type) and the Zn-sensitive zrc1/cot1 double mutant. HMA4 inserts were also cloned without translational fusion with GFP. Growth tests were performed using two Cd and Zn concentrations depending on the yeast strain: 75 μm Cd and/or 10 mm Zn for BY4741 and 50 μm Cd and/or 10 mm Zn for zrc1/cot1. The expression of AhHMA4 increased the sensitivity of BY4741 to Cd and Zn, whereas the expression of AhHMA4-C enhanced the tolerance of this strain to Cd and Zn (Fig. 6). Similar phenotypes were observed for zrc1/cot1 (data not shown). Interestingly, on Gal medium, yeast cells expressing AhHMA4 grew more slowly than the other constructs, even without Cd. Translational fusions of the AhHMA4 and AhHMA4-C constructs with GFP did not modify the phenotype of transformed yeast (Fig. 6). Replacement of D401 by a G in the phosphorylation site of AhHMA4 (AhHMA4D401G) led to loss of the metal-sensitive phenotype, suggesting that this phenotype was due to transport activity (Fig. 6).
Figure 6.
Growth of yeast strain BY4741 on selective media. Yeast cells were transformed with the empty pYES2-GFP vector, with the full-length AhHMA4 sequence, with the C-terminal sequence, or with the AhHMA4-D401G sequence fused or not with GFP at the C-terminal end. Yeast cells were grown in SD induction medium to an OD600 of 1.0, serially diluted to an OD600 of 0.1 to 0.0001 (from left to right in each section), and then 10 μL drops spotted on plates containing Glc or Gal and 0 or 75 μm of CdSO4 and 0 or 10 mm ZnSO4. Plates were incubated for 5 d at 30°C. Drop tests were repeated at least three times with similar results.
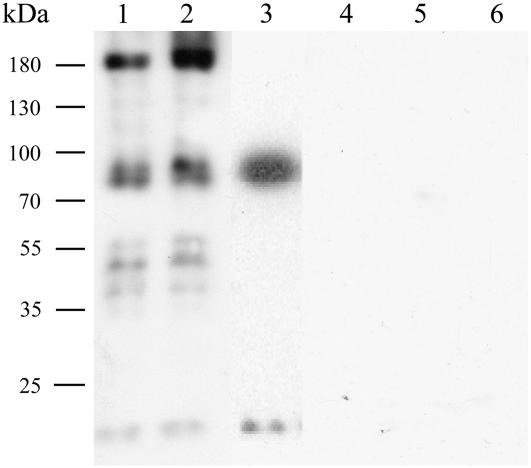
To confirm the expression of the recombinant proteins, immunoblots blots were performed on cytoplasmic and membrane fractions of transformed yeast (grown on Gal) using anti-GFP antibodies. In the membrane fraction (lanes 1 and 2) and in the soluble fraction (lane 3) of yeast extracts, proteins were detected of the correct predicted size (151.8 kD for AhHMA4:GFP and 73.9 kD for AhHMA4-C:GFP; Fig. 7). The size of the lower bands in the membrane extracts of AhHMA4-expressing cells most probably corresponds to degradation products.
Figure 7.
Western-blot analysis of GFP fusion proteins. Yeast containing pYES2-GFP vector (1 and 5) or expressing either the AhHMA4:GFP (2 and 6), the AhHMA4-D401G:GFP (3 and 7), or the AhHMA4-C:GFP (4 and 8) fusion protein were grown on medium with 2% Gal (1–4) or with 2% Glc (5–8). Proteins were extracted (membrane proteins: 2, 3, 6, and 7; soluble proteins: 1, 4, 5, and 8) and separated on 8% SDS-PAGE gels and further analyzed using anti-GFP antibodies. Numbers beside the gel indicate molecular mass in kilodaltons.
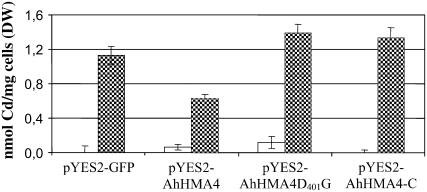
Measurements of intracellular Cd indicated reduced accumulation of Cd in yeast overexpressing AhHMA4 compared to yeast cells transformed with empty plasmid (Fig. 8). No difference in Cd content was observed between cells expressing AhHMA4-C or AhHMA4D401G compared to cells transformed with the empty vector.
Figure 8.
Cd content in BY4741 expressing AhHMA4, AhHMA4D401G, or AhHMA4-C. Yeast BY4741 cells transformed with the pYES2-GFP plasmid and with the pYES2-GFP containing AhHMA4, AhHMA4G401D, or AhHMA4-C were grown in liquid SD induction media with (hatched bars) or without 20 μm CdSO4 for 48 h (white bars). Metal contents were analyzed by atomic absorption spectroscopy. Results are an average (±se) from three independent experiments.
Localization of AhHMA4 in Yeast and in Planta
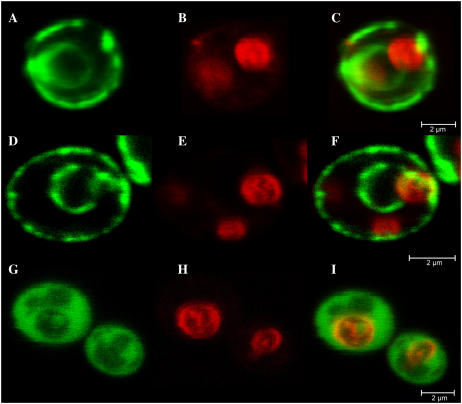
Because the expression of AhHMA4-GFP and AhHMA4-C-GFP fusion proteins in yeast cells gave a similar phenotype to the GFP unfused proteins, we used GFP to investigate the subcellular localization of HMA4. At the standard image acquisition settings used for GFP visualization, autofluorescence from cells transformed with the untagged AhHMA4 was absent (data not shown), so all detectable fluorescence in the transformants was GFP specific. When exponentially growing cells were analyzed, green fluorescence resulting from the expression of AhHMA4:GFP and AhHMA4 D401G:GFP showed a ring-like pattern around the cells and the nucleus (Fig. 9). AhHMA4-C:GFP was only visible in the cytosol. To verify the absence of the fused proteins in the tonoplast, this membrane was stained for a short term with the lipophilic dye FM4-64, which selectively stains yeast vacuolar membrane (Vida and Emr, 1995). Superposition of the GFP and FM4-64 images demonstrated the absence of detectable GFP fused protein in the tonoplast (Fig. 9).
Figure 9.
Localization of AhHMA4:GFP, AhHMA4D401G:GFP, and AhHMA4-C:GFP fusion proteins in yeast, viewed by confocal laser scanning microscopy. Cells were visualized after 20 h of induction. Three images from the same cell are shown. A, GFP fluorescence from a AhHMA4:GFP-expressing strain. B, FM4-64 staining the vacuolar membrane of the same cell. C, A pseudocolored merged image with GFP in green and FM4-64 in red. D, E, and F and G, H, and I are as for A, B, and C, but expressing the AhHMA4D401G:GFP and AhHMA4-C:GFP proteins, respectively. Bars = 2 μm.
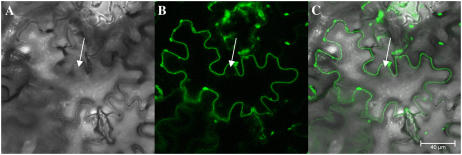
The AhHMA4:GFP fusion protein was transiently overexpressed in tobacco (Nicotiana tabacum) epidermal cells, which were then analyzed by confocal laser scanning microscopy. The superposition of the bright-field image and the corresponding fluorescence image showed an unambiguous localization of GFP to the plasma membrane (Fig. 10).
Figure 10.
Plasmalemma localization of the AhHMA4:GFP fusion protein in tobacco plants, viewed by confocal laser scanning microscopy. Transient expression of AhHMA4:GFP in tobacco leaf epidermal cells. A, Bright-field image of the tobacco leaf epidermis. The nucleus is indicated with an arrow. B, GFP fluorescence in the same cell concentrated to the plasmalemma, which followed the cell contour, outside the nucleus. C, Pseudocolored merged image with GFP in green. Bar = 40 μm.
DISCUSSION
QTL Analysis of Cd Tolerance in A. halleri
Three QTL for Cd tolerance were detected in the Ah × Alp BC1 progeny through MQM. These QTL explained the major part of the phenotypic (82.5%) and genetic variance (92.7%) observed for this trait in the mapping population. However, some caution should be adopted in the interpretation of this result. Due to the relatively small progeny size (<100 individuals), as well as the segregation distortion observed at the QTL Cdtol-3 (markers ICE2 and AthCDPK9), QTL of minor effect probably remained undetected and the effects of the detected QTL might be overestimated (Beavis et al., 1994; Bradshaw et al., 1998).
Genetic Architecture of Cd Tolerance in A. halleri
The genetic architecture of Cd tolerance was previously addressed by Bert et al. (2003) through the analysis of the segregation of Cd tolerance in 66 of the 79 BC1 individuals used in this QTL experiment. The segregation ratios were shown to be significantly different from the 1:1 ratio expected for a character controlled by a single major gene, while the hypothesis of Cd tolerance controlled by two or three major genes with additive effect could not be rejected (Bert et al., 2003). The QTL analysis performed in this study provided evidence for the involvement of one major QTL explaining almost half (48.2%) of the genetic variance observed for Cd tolerance in the BC1 progeny, and two QTL of less major effect on Cd tolerance. As we did not detect any significant epistatic interaction between the QTL, the three QTL presumably have a largely additive effect. At all three QTL, the trait-enhancing allele was found to originate from the Cd-tolerant parental genotype A. halleri. The A. halleri individual used in this study originated from an industrial polluted site characterized by elevated concentrations of Zn, Cd, and Pb (Van Rossum et al., 2004). The high selective pressures represented by the occurrence of toxic ambient Cd concentrations most probably led to the selection of genes increasing Cd tolerance, which should give rise to the positive effect on Cd tolerance of the A. halleri alleles (Orr, 1998). Whereas this explanation is plausible for metallicolous A. halleri individuals, it remains to be verified for A. halleri individuals growing on unpolluted sites, which are not subject to a high selective pressure for Cd tolerance.
The A. halleri origin of the trait-enhancing allele at all three QTL might indicate the constitutive presence of Cd tolerance in A. halleri, as observed for Zn tolerance (Bert et al., 2002; Pauwels et al., 2006). However, this should be confirmed through an extensive survey of Cd tolerance in metallicolous and nonmetallicolous A. halleri populations. Interestingly, the genetic architecture of Zn tolerance, studied through a QTL analysis on the same mapping population, revealed three QTL for Zn tolerance, among which one showed colocalization with the major QTL Cdtol-1 for Cd tolerance. The involvement of this QTL in both metal tolerances might indicate that Cd and Zn tolerance initially evolved in A. halleri through the fixation of the QTL conferring tolerance to both metals (Willems et al., 2007).
Cloning and Analysis of AhHMA4 Expression
The HMA4 locus was located at the peak of the major QTL region for Cd tolerance in the Ah × Alp BC1 progeny. In silico analysis of the full-length coding sequence of AhHMA4 did not reveal common features between the sequences of AhHMA4 and TcHMA4 that could differentiate these metal-tolerant species from Arabidopsis. Rather, the HMA4 sequences of A. halleri and Arabidopsis were most similar to each other, reflecting the phylogenetic relatedness of these taxa. Indeed, although the Cys- and His-rich C terminus of HMA4 may play a role in metal chelation or regulation of activity, the positions of these potential metal-binding residues were not more highly conserved between the Cd/Zn hyperaccumulator species A. halleri and T. caerulescens than between A. halleri and Arabidopsis. This might indicate that rather than their position, the presence of these potential metal-binding residues is important.
HMA4 was more highly expressed in the roots and shoots of A. halleri than in A. lyrata sp. petraea or, to a lesser extent, the F1 individual. Moreover, in the BC1 progeny the presence of the AhHMA4 allele was always accompanied by a higher HMA4 expression even in the absence of the A. halleri allele at the QTL Cdtol-2 and Cdtol-3. No significant modification of HMA4 expression level was observed in A. halleri or in the F1 individual upon exposure to increased Cd concentrations or upon prolonged exposure to Cd.
In A. halleri, a higher expression of HMA4 was recently reported and suggested to be related to the presence of several copies of this gene in the A. halleri genome (Talke et al., 2006). In this work, the amplification of HMA4 for its mapping on the Ah × Alp genetic linkage map revealed a single fragment in A. halleri different in size from the one amplified in A. lyrata sp. petraea. Consequently, only one locus was mapped on the Ah × Alp linkage map, which was located on its expected position based on the synteny of the Ah × Alp map with the Arabidopsis genome. If all copies were located in a small region, this result would not be not in contradiction with the hypothesis of the presence of several copies of HMA4 in the A. halleri genome. Such genomic amplification could be explained by a duplication event at the same locus in A. halleri.
Role of HMA4 in Metal Transport: Functional Expression in Yeast and Localization in Yeast and Plant Cells
To address the critical question of the physiological function of HMA4, both in vivo and in vitro metal transport studies were performed and showed that P1B-ATPases drive the export of ions from the cell cytoplasm (Eren and Arguello, 2004). Another approach, which was undertaken to characterize HMA4, consisted of the functional expression of HMA4 in yeast. Heterologous expression of AtHMA4 in yeast resulted in an increased tolerance to Cd (Mills et al., 2003), but increased (or did not alter) also the sensitivity to Zn, Co, Cu, or nickel (Verret et al., 2005). AtHMA4:GFP fusion protein was observed essentially in endocytic vesicles and to a lesser extent at the plasma membrane (Verret et al., 2005). Enhanced Cd tolerance was also observed in yeast cells transformed with TcHMA4 (Papoyan and Kochian, 2004). On the contrary, Bernard et al. (2004) expressed TcHMA4 as well as AtHMA4 in two different wild-type yeast strains and observed a higher sensitivity to high Zn and Cd concentrations.
In this work, heterologous expression of AhHMA4 in yeast resulted in an increased sensitivity to Cd and Zn. A mutation in the conserved phosphorylated Asp of AhHMA4 restored the capacity of growth of the strains in presence of high Zn and Cd concentrations, indicating that the higher sensitivity to Zn and Cd was related to a transport activity (ATP hydrolysis coupled to transport activity). The sensitivity of yeast upon HMA4 overexpression might be due to the transport of metals not only to the outside of the cells, but also from the cytoplasm to metal-sensitive organelles due to the lack of a specific targeting sequence for yeast. The first transport process would result in the reduced metal content while the second would be responsible for the sensitive phenotype. Our hypothesis is supported by the localization of GFP-tagged AhHMA4 at the plasma membrane as well as at the endoplasmic reticulum (ER) around the nucleus in the yeast. In agreement with this, immunoblot analysis of AhHMA4 indicated a good translation and accumulation of the protein. Strong evidence for the high sensitivity of the ER to Cd accumulation was previously published by Clemens et al. (2002), who showed that the zhf deletion mutant of Schizosaccharomyces pombe, impaired in an ER-localized cation diffusion facilitator transporter capable of transporting Cd into this organelle, was more tolerant to Cd than the wild type. This suggests that Cd ions may exert their toxic effects on cellular metabolism in the ER.
In plant cells, however, the AhHMA4-GFP fusion was only detected in the plasma membrane, consistent with a role in metal detoxification by export from the cell. This localization was previously reported for AtHMA4 (Verret et al., 2005).
Conclusion: Role of HMA4 in Metal Tolerance
In Arabidopsis, HMA4 is mainly expressed in the vascular tissues of roots, leaves, and stems (Hussain et al., 2004; Verret et al., 2004). Analysis of hma4 null mutant lines revealed an increased sensitivity to Zn and Cd (Verret et al., 2004; Mills et al., 2005), as well as a decreased Zn and Cd accumulation in shoot tissues when compared to the wild type under high Zn concentrations (Hussain et al., 2004; Verret et al., 2004). On the contrary, AtHMA4-overexpressing plants had an increased tolerance to high Zn and Cd, as well as higher Cd and Zn levels in the leaves compared to the wild type. Based on these results, a role of HMA4 was proposed in the root-to-shoot translocation of Cd and Zn, in the redistribution of metals in the whole plant, and in the detoxification of the cytosol through the exclusion of metals from sensitive cells by acting as an efflux pump (Hussain et al., 2004; Verret et al., 2004; Mills et al., 2005). Up to now, experimental data suggest that HMA4 transports free metal ions, probably delivered to HMAs in a coordinated form, but by means that remain currently unknown (Eren and Arguello, 2004; Verret et al., 2005).
In the Cd/Zn hyperaccumulators A. halleri and T. caerulescens (Bernard et al., 2004), HMA4 is more highly expressed in both roots and shoots compared with Cd/Zn-sensitive close relatives. The elevated expression of HMA4 in two different Zn/Cd hyperaccumulator species that evolved independently strongly supports the idea that HMA4 plays an important role in tolerance to both metals. In A. halleri at least, this hypothesis is reinforced by the QTL analysis of Cd (this study) and Zn tolerance (Willems et al., 2007) performed on the Ah × Alp BC1 progeny, indicating the colocalization of the QTL Cdtol-1 and Zntol-1, both of which are located at the marker HMA4 on the Ah × Alp linkage map. As observed in AtHMA4-overexpressing lines (Verret et al., 2004), the increased expression of HMA4 in A. halleri or T. caerulescens might be related to the increased Cd and Zn tolerance and hyperaccumulation phenotype of these species. However, we cannot exclude the possibility that the activity of HMA4 proteins also displays different characteristics or differential regulation in Arabidopsis and the hyperaccumulator species.
In this work, we have shown that in addition to the major QTL Cdtol-1 at least two other QTL are involved in Cd tolerance in A. halleri. Among the candidate genes responsible for metal tolerance in A. halleri, MTP1 was reported to be capable of transporting Zn into the vacuole and two of the three MPT1 copies present in A. halleri were shown to cosegregate with Zn tolerance (Dräger et al., 2004; Willems et al., 2007). However, none of these copies colocalized with the QTL for Cd tolerance. Consequently, HMA4 is currently the only gene for which there is genetic evidence for a role in both Zn and Cd tolerance.
Taken together, all available data point to the elevated expression of HMA4 P1B-type ATPase as an efficient mechanism for improving Cd/Zn tolerance in plants under conditions of Cd/Zn excess by maintaining low cellular Cd2+ and Zn2+ concentrations in the cytoplasm.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Treatments
A cross was performed between an individual of the Zn/Cd-tolerant Arabidopsis halleri sp. halleri species (pollen donor), collected on a site highly contaminated with Zn, Cd, and Pb (Auby, France [Van Rossum et al., 2004]), and an individual of the nontolerant Arabidopsis lyrata sp. petraea (Alp) species (pollen recipient; Unhost, Central Bohemia, Czech Republic [Macnair et al., 1999]). To obtain a first-generation backcross (BC1) progeny, one randomly chosen F1 individual was crossed with a second A. lyrata sp. petraea individual (pollen recipient; Unhost, Central Bohemia, Czech Republic; Bert et al., 2003).
Cuttings from mother plants were grown on sand for 6 weeks in a glasshouse (light and humidity conditions changed according to the season, the temperature being kept above 5°C at night in winter). Hydroponic plant culture was performed in a modified Murashige and Skoog solution consisting of K2SO4 (0.88 mm), KH2PO4 (0.25 mm), NaCl (10 μm), Ca(NO3)2 (2 mm), MgSO4 (1 mm), FeEDDHA (20 μm), H3BO3 (10 μm), ZnSO4 (1 μm), MnSO4 (0.6 μm), CuSO4 (0.1 μm), and (NH4)6Mo7O24 (0.01 μm), adjusted to pH 5.8 in a climate-controlled growth chamber (temperature cycle of 20°C/17°C and a light [100 μmol m−2 s−1] cycle of 16 h light/8 h dark). The hydroponic solutions used were continuously aerated and changed every week. After 6 weeks in nutrient solution, 10 μm CdSO4 was added. Roots and shoots were collected after 72 h or 7 d of treatment and immediately frozen in liquid nitrogen until use.
Phenotypic Evaluation of Cd Tolerance
Cd tolerance was assessed on 79 individuals of the A. halleri × A. lyrata sp. petraea BC1 progeny. Two to three clonal replicates of each BC1 individual were obtained through vegetative propagation. After growth for 6 weeks on sand moistened with deionized water, rooted cuttings were transferred to 4 L vessels containing a nutrient solution to which Cd was added as CdCl2. To measure Cd tolerance, plants were sequentially transferred to increasing concentrations of Cd. The range of Cd concentrations tested was 10, 25, 50, 75, 100, 150, and 250 μm (Bert et al., 2003). At the end of each week, roots of each plant were gently dried with tissue paper and the whole plant was weighed. In addition, root length was measured. Cd tolerance was determined as the lowest concentration at which no new root growth was observed (the EC100; Bert et al., 2003).
QTL Mapping of Cd Tolerance
Statistical Analysis
The genotype effect of Cd tolerance in the BC1 progeny was estimated by a one-way ANOVA using the generalized linear model procedure in SAS (SAS, 1999). The broad-sense heritability of this trait was calculated using the mean square values (MS) of ANOVA analysis (h2 = MSgenot/[MSgenot + MSerror]). Type III sums of squares were used because the data set was unbalanced due to the unequal number of replicates of each BC1 individual. We performed an Anderson-Darling test (Minitab) on the EC100max values (the maximum value of the EC100 values of the clones) of the BC1 genotypes to test for deviation from a normal distribution. We tested for significant pairwise interactions between the QTL using the generalized linear model procedure in Minitab. The markers at the QTL position were considered as fixed factors in this analysis. The box plot for Cd tolerance at the marker HMA4 was obtained using Minitab. We performed a Kruskal-Wallis test, based on Wilcoxon rank scores of the data, to test for significant differences of Cd tolerance between both groups (Minitab).
QTL Analysis
The set of markers (65 Arabidopsis [Arabidopsis thaliana]-anchored markers and 18 amplified fragment length polymorphism markers) and the genetic map of the Ah × Alp BC1 progeny (henceforth called the Ah × Alp map) obtained with the program Joinmap 3.0 (Van Ooijen and Voorips, 2001; Willems et al., 2007) were used for the QTL analysis of Cd tolerance, performed with the MapQTL 4.0 program (Van Ooijen et al., 2002). QTL of Cd tolerance were identified using EC100max values of the BC1 genotypes. The significance threshold of the LOD score for QTL detection was obtained by performing a permutation test on the quantitative data and set at a LOD score of 2.4, corresponding to an error level α = 0.05. A first QTL analysis was performed using interval mapping (IM). In IM, LOD scores were calculated every centiMorgan (cM) within the intervals along the LGs. The markers for which the LOD score exceeded the significance threshold were included in an automatic cofactor selection analysis and the selected markers were then used as cofactors in MQM analysis. In MQM mapping, a one-dimensional search over the genome is performed as in IM, while simultaneously fitting the selected cofactors that take over the role of the other QTL. In the QTL analysis of Cd tolerance, MQM mapping was performed twice while adjusting the set of cofactors to obtain the best possible set of QTL, i.e. showing maximal LOD scores. One- and two-LOD support intervals were obtained using Mapchart 2.1 (Voorrips, 2002). The percentage of variance explained by each QTL (R2) was calculated in IM. Additive genetic effects correspond to the differences between the mean Cd tolerance levels of the BC1 individuals characterized by homospecific (i.e. Alp1/Alp2) and heterospecific (i.e. Ah/Alp2) allelic combinations at each QTL.
Marker Densification the Major QTL Region
Three additional markers (At2g10940, At2g14620, and At2g18196) selected using their position in the Arabidopsis genome, were mapped to increase the accuracy of the major QTL position. One primer was designed in an exonic region of the gene and the second primer was placed in the following exon (Table III). To obtain labeled PCR products detectable on the automated genotyper Li-Cor 4200 (Li-Cor-ScienceTec), the forward and reverse primer contained a 5′ tail of 19 bp (forward primer) or 20 bp (reverse primer) homologous to the universal consensus M13 primer sequence, followed by the locus-specific sequence (Oetting et al., 1995). The following PCR conditions were applied: PCR reactions were carried out in a total volume of 15 μL containing 20 ng of template DNA, 2 mm MgCl2, 0.2 mg/mL bovine serum albumin, 0.2 mm dNTP, 0.15 μm of each M13 fluorescently labeled primer (either IRD-700 or IRD-800), 20 mm Tris-HCl (pH 8.3), 50 mm KCl, and 0.4 units of AmpliTaq DNA Polymerase (Applied Biosytems). PCR was performed on a Perkin-Elmer Gene-Amp system 9700: 94°C for 5 min, followed by locus-specific amplification: 94°C for 30 s, annealing temperature for 45 s, 72°C for 40 s (except for marker At2g18196: 72°C for 1 min 15 s), for eight cycles, followed by M13 labeling amplification: 94°C for 30 s, 50°C for 20 s, 72°C for 40 s, for 30 cycles, and a final extension 72°C for 7 min. The annealing temperature of the locus-specific cycles corresponded to 52°C, 48°C, and 58°C for the markers At2g10940, At2g14620, and At2g18196, respectively. Length polymorphisms of the markers At2g10940 and At2g18196 were revealed on polyacrylamide gels using a Li-Cor genotyper. For cleaved-amplified polymorphic sequence marker At2g14620, restriction was carried out in a total volume of 20 μL containing 10 μL of PCR product, 0.2 mm spermidine, 1× specific enzyme buffer provided by the supplier, and 1 unit of restriction enzyme AluI. Restriction was carried out by incubation for at least 4 h at the appropriate temperature in a Perkin-Elmer Gene-Amp system 9700. The polymorphism was revealed on agarose gels.
Table III.
List of primers used with their positions (base pair) on At2g10940, At2g14620, At2g18196, AhHMA4, EF1α, and ARF sequences in order of their first mention in “Materials and Methods”
Restriction sites are underlined. Lowercase letters in primers AhHMA-ATPXF and AhHMA-ATPXR indicate the mismatched nucleotides designed for specific mutations.
| Name | Sequences (5′-3′) | Positions |
|---|---|---|
| At2g10940F | AAGCTACCCGTTCCTCC | 351–367 |
| At2g10940R | CGACTTCAACGAGGCCTTTA | 794–813 |
| At2g14620F | CACAAGCTTCCGTTGTTTCA | 188–207 |
| At2g14620R | ATGGACCCTTTGACCAATCA | 751–769 |
| At2g18196F | ATGCCAATGGCTCGTCCACT | 106–125 |
| At2g18196R | CACATCCACTGCAACACATC | 156–175 |
| EF1αF | ATTGTGGTCATTGGCCACGTCGA | 27–50 |
| EF1αR | CTCCTTCTCAATCTCCTTACCAG | 1,191–1,214 |
| AtTcHMA4F | TGACCTGAAAATGAAAGGTGGTTC | 2,474–2,498 |
| AtTcHMA4R | TGCATAACTCCTGCAACAGCT | 3,237–3,258 |
| AhHMA3′RACE | AGCTGTTGCAGGAGTTATGCA | 3,408–3,429 |
| AhHMA5′RACE | CTCGAGGGAACGATGACGG | 145–164 |
| AhHMA4F | CCAAGCTTATGGCGTCACAAAACAAAGAAG | 1–22 |
| AhHMA4R | CGGATCCTCAAGCACTCACATGGTGATG | 3,465–3,483 |
| AhHMA4nostpR | CCGGATCCCCAGCACTCACATGGTGATGGG | 3,463–3,480 |
| AhHMA4C | CCAAGCTTATGTTGAATGGTAGGAAACTCG | 2,160–2,182 |
| AhHMA-ATPXF | TTCGgCCAAACCGGGACTATTACC | 1,197–1,221 |
| AhHMA-ATPXR | GTTTGGcCGAAAGCAGCGATTTTGA | 1,183–1,208 |
| AhHMA4BamF | CCGGATCCATGGCGTCACAAAACAAAGAAG | 1–22 |
| AhHMAKpnR | CCGGTACCAGCACTCACATGGTGATGGG | 3,463–3,480 |
| ARF_F | GGTCTCGATGCAGCTGGTAAGACTAC | 177–202 |
| ARF_R | TGTTAGAGAGCCAGTCAAGTCCCTCA | 608–633 |
| HMA4_290F | TAGTGGCAAAGAGCTGTTGTGA | 2,537–2,558 |
| HMA4_290R | TGCAGTTTCCTTCATCTTTAATCAG | 2,800–2,825 |
Yeast Cultures, Transformation, and Growth Assays
The yeast (Saccharomyces cerevisiae) strains used for the heterologous expression of AhHMA4 were BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, and ura3Δ0) obtained from Euroscarf (http://www.uni-frankfurt.de/fb15/mikro/euroscarf/) and the zrc1/cot1 mutant (Mat a, zrc1∷natMX3, cot1∷kanMX4, his3Δ1, leu2Δ0, met15Δ0, and ura3Δ0) provided by U. Krämer (Golm, Germany). Yeast cells were transformed as described elsewhere (Gietz and Woods, 2002). They were grown at 30°C in yeast potato (Solanum tuberosum) dextrose or in synthetic defined (SD) medium (0.17% yeast nitrogen base [Sigma]) without amino acids, containing 1% (w/v) Glc or 2% (w/v) Gal (induction medium), supplemented with yeast synthetic dropout without uracyl (Sigma), pH 5.3. For metal tolerance assays, yeast was grown on SD medium (with 2% [w/v] Gal) to an OD600 of 1 to perform further dilutions. The drop assays were performed on SD plates containing 50 μm CdSO4 for zrc1/cot1 mutants and 75 μm for BY4741 with or without 10 mm ZnSO4.
Cloning
Total RNA was extracted from A. halleri roots as described in Sambrook and Russell (2001). RT reactions were performed from 2 μg of total RNA using the Superscript III reverse transcriptase (Invitrogen Life Technologies) as recommended by the manufacturer (oligo dT, at 50°C). cDNAs were used immediately in PCR or kept frozen until use. Two microliters of cDNAs were used for PCR amplification. The suitability of the extracted RNAs for RT-PCR amplification was checked by performing RT-PCR control experiments with Elongation Factor 1 α (EF1α; At1g07920) using the primers EF1αF and EF1αR (Table III) in the following conditions: 94°C for 3 min followed by 35 cycles at 94°C for 30 s, 62°C for 30 s, and 72°C for 2 min using an Eppendorf Mastercycler (Eppendorf). Oligonucleotide primers (AtTcHMA4F; AtTcHMA4R: Table III) were designed to conserved regions of HMA cDNAs from Arabidopsis and Thlaspi caerulescens. The 1 kb sequence obtained was used to design the specific primer (AhHMA3′ RACE) to perform 3′ RACE amplification (following Invitrogen Life Technologies procedure). To perform the 5′ RACE amplification, the primer AhHMA5′ RACE was designed using sequences of AhHMA4 cDNAs from Arabidopsis and T. caerulescens. These amplifications were cloned in pTZ57R (MBI Fermentas) and sequenced. Two specific primers AhHMA4F and AhHMA4nostpR introducing HindIII and BamHI sites were designed to clone the full-length AhHMA4 cDNA into the pYES2-GFP plasmid (Blaudez et al., 2003). The PCR conditions were the following: 95°C for 5 min followed by 24 cycles at 95°C for 30 s, 62°C for 30 s, and 68°C for 5 min using the Pfx polymerase (Invitrogen).
The direct cloning into pYES2-GFP plasmid avoided subcloning and reamplification processes leading to mutations in the sequence. A very limited number of cycles (24) was also used to limit mutations possibilities during the amplification. The insert was subsequently sequenced in both directions and checked for GFP expression in BY4741 yeast strains.
Using the AtTcHMA4F and R primers, a 1,282-bp fragment was amplified from Alp cDNA under the same PCR conditions as above (although 30 cycles were necessary due to the lower expression level of HMA4), cloned into pTZ57R, and sequenced.
Expression Analysis
The primer pair (HMA4_290F and R) used for the expression analysis was designed in a perfectly conserved coding region between AhHM4 and AlpHMA4 cDNAs, which were partially sequenced. The primers were tested on AhHMA4 and AlpHMA4 cDNAs cloned in plasmids. Both fragments (288 bp of AhHMA4 and 291 bp of AlpHMA4) were amplified with equal efficiency. For RT-PCR expression analysis, total RNA was isolated from shoot and root of A. halleri, A. lyrata sp. petraea, the F1 individual, and the BC1 individuals with or without Cd treatment, and cDNA was obtained as described above. The amplifications were done with GoTaq Flexi DNA polymerase (Promega) using 1 μL of cDNA in a 25 μL reaction with an annealing temperature of 62°C. Two housekeeping genes were chosen to compare the relative abundance of the HMA4 mRNAs: the EF gene EF1α and the ADP ribosylation factor gene ARF (At3g62290; Craciun et al., 2006). PCR samples were taken at successive cycles and visualized on agarose gel. The number of cycles at which a minimal signal was observed for all of the genotypes tested was chosen (24 cycles for the roots, 28 cycles for the shoots). To ensure that we were still in the linear part of the PCR amplification curve, we verified that the signal could be increased by performing more cycles. The resulting fragments were cloned into the pTZ57R plasmid (MBI Fermentas) for sequencing.
Heterologous Expression in Yeast
The truncated AhHMA4-C was amplified on the pYES2-AhHMA4-GFP plasmid using AhHMA4 specific primers designed for the C-terminal part (amino acids 720–1,161; between the end of the last transmembrane span and the stop codon) of AhHMA4. To amplify the C-terminal part, AhHMA4C/AhHMA4R or nostpR (lacking the stop codon) were used. The resulting PCR products were cloned into the HindIII-BamHI-digested pYES2-GFP plasmid.
The mutation of the phosphorylation site was performed by oligonucleotide-directed PCR mutagenesis. First, a fragment was amplified by PCR with the primer pair AhHMA4F and the mutated primer AhHMA-ATPXR replacing the D401 by a G in the phosphorylation site. A second fragment was amplified with the primer pair AhHMA-ATPXF introducing the same mutations and AhHMA4R or −nostpR. DNA fragments were gel purified, mixed in a PCR mix without primers, denaturated at 95°C for 3 min, annealed at 60°C for 30 s, elongated at 68°C for 4 min, and subjected to 20 PCR cycles after the addition of AhHMA4F and AhHMA4R or −nostpR primers. The resulting HindIII-BamHI fragment was cloned in pYES2-GFP plasmid. The presence of targeted mutation in all plasmid constructs was verified by DNA sequencing.
Subcellular Localization of AhHMA4 in Plant Cells
Cellular localization in plant cells was performed by transient transformation. Tobacco (Nicotiana tabacum ‘Petit Havana’) seeds were sown on soil in culture chamber and allowed to germinate and grow under long-day conditions (16-h-light/8-h-dark photoperiod) at approximately 20°C. Twenty centimeters in height tobacco plants were used for Agrobacterium-mediated transient expression.
To create the 35S∷AhHMA4-GFP transgene, full-length AhHMA4 cDNA was amplified by PCR with primers AhHMA4BamF and AhHMA4KpnR (Table III) that incorporated BamHI and KpnI sites at the 5′ and 3′ ends of the gene, respectively. The resulting fragment was digested with BamHI and KpnI and cloned into corresponding sites of pBI35S-GFP (Tillemans et al., 2005) to yield pBI35S∷AhHMA4-GFP. This construct as well as a control containing the EGFP sequence was used for transient transformation.
Transient transformation in tobacco leaves was performed essentially as described previously (Tillemans et al., 2005). Briefly, Agrobacterium tumefaciens bearing the appropriate binary vector was grown at 28°C overnight in 10 mL yeast extract broth liquid medium (0.5% Suc, 0.1% yeast extract, 0.5% Bactopeptone, 0.5% Invitrogen beef extract, 2 mm MgSO4, pH 7.4) supplemented with 25 μg/mL gentamycin, 100 μg/mL rifampicin, and 50 μg/mL kanamycin. After centrifugation, the bacteria were washed with infiltration medium (50 mm MES, 2 mm NaH2PO4, 0.5% Glc, and 100 μm acetosyringone) and then resuspended in the same medium at the desired final OD600. Leaves were infiltrated using a 5-mL syringe by applying gentle pressure through the stomata of the abaxial epidermis. The infiltrated areas were cut at 48 to 72 h after inoculation and further processed for confocal imaging.
Yeast Cell Staining
Yeast vacuolar membranes were selectively stained with red fluorescence probe FM4-64 (Molecular Probes, Invitrogen). This dye has been reported to stain yeast vacuolar membranes selectively (Vida and Emr, 1995). Cells (250 μL at OD600 0.5) were incubated with 6 μL of FM4-64 (2 mg/mL) during 30 min at 30°C, washed twice with SD medium, and incubated 2 h before observation by confocal microscopy.
Confocal Imaging
The GFP fluorescence of yeast and plant cells was visualized by confocal laser scanning microscopy. Yeast cells and lower epidermis of agroinfiltrated leaf fragments were analyzed with an inverted Leica TCS SP2 confocal laser scanning microscope (Leica Microsystems), using a 63 × NA 1.3 Plan-Apo water-immersion objective at 1,024 × 1,024 pixel resolution. For single GFP fluorescence analyses, we used the 488 nm excitation line of an argon ion laser and the emission light was dispersed and recorded at 500 to 535 nm. For GFP and FM4-64 colocalization experiments, a second detector was activated for recording emission fluorescence at 650 to 800 nm. Images were processed with Leica software (version 2.5) and Photoshop 7.0 (Adobe Systems) software.
Protein Extraction and Gel-Blot Analysis
Yeast cells were collected by centrifugation at 5,000g for 1 min. Cell lysates were obtained by vortexing cells at full speed for 2 × 1 min with glass beads (420–600 μm; Sigma) in ice-cold lysis buffer containing 100 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 10% (v/v) glycerol, 100 μg/mL phenylmethylsulfonyl fluoride, 1 μg/mL leupeptin, and 10 μg/mL pepstatin A. Lysates were centrifuged at 12,000g for 15 min at 4°C to pellet cell debris and beads and the supernatants were centrifuged subsequently at 125,000g for 60 min at 4°C. Pellets (membrane fraction) were resuspended subsequently in ice-cold lysis buffer containing 10 mg/mL n-dodecyl-d-maltoside and incubated for 1 h on ice with occasional mixing. The extracts were centrifuged at 200,000g for 30 min to remove insoluble material.
Protein extracts were mixed with sample buffer (60 mm Tris-HCl, pH 6.8, 25% [v/v] glycerol, 2% [w/v] SDS, and 0.0125% [v/v] bromphenol blue) with 2-mercaptoethanol and subjected to SDS-PAGE (Laemmli, 1970) and proteins were transferred to polyvinylidene difluoride membranes (Amersham Pharmacia Biotech). Blots were probed with mouse anti-GFP antibodies (Roche; 1:1,000) and rabbit antimouse HRP-conjugated antibodies (Perbio Science; 1:10,000) were used as the secondary reagent. Proteins were revealed by enhanced chemiluminescence using the western pico revelation kit (Perbio Science) according to the manufacturer's instructions.
Determination of Intracellular Cd Content
Cells were grown for 24 h in the SD induction medium, then diluted to OD600 = 0.5 and grown for 48 h in 30 mL of 20 μm CdSO4-containing induction medium. After centrifugation, supernatants were collected, cells were washed once with water, once with 50 mm EDTA, and then once again with water, pelleted, dried for 48 h at 60°C, and mineralized (with hydrochloric acid at 95°C). The Cd content was determined using flame atomic absorption spectroscopy (Perkin Elmer 3100 series atomic absorption spectrometer).
Accession Number and Sequence Analysis
The GenBank accession numbers for the sequence described in this article are as follows: A. halleri sp. halleri: AhHMA4, DQ221101; Arabidopsis: AtHMA4, NM127468; T. caerulescens: TcHMA4, AJ567384; A. lyrata sp. petraea: AlpHMA4, EF176604. Alignment of HMA4 sequences was performed using ClustalW (Thompson et al., 1994) and was manually adjusted.
Acknowledgments
We thank Ute Krämer (Max Planck Institut, Golm, Germany) for the gift of the zrc1/cot1 mutant yeast strain and Damien Blaudez (Université Henri Poincare, Nancy, France) for providing the pYES2-GFP plasmid. We acknowledge J.A.C. Smith (Oxford University, Oxford) for proofreading and discussion and Piétro Salis and Geoffrey Gosset (Université Libre de Bruxelles, Brussels) for practical help.
This work was supported by the European Union (Research Training Network METALHOME, grant no. HPRN–CT–2002–00243), the Belgian Science Policy (Interuniversity Attraction Pole Programme V/13 and VI/33), and the Fonds National de la Recherche Scientifique (grant no. Fonds de la Recherche Fondamentale Collective 2.4565.02) at the Université Libre de Bruxelles. At the Université de Lille (Unité Mixte de Recherche Centre National de la Recherche Scientifique 8016), research was supported by the European Union (Research Training Network, METALHOME, grant no. HPRN–CT–2002–00243), Contrat de Plan Etat/Région Nord-Pas de Calais, the European Fonds Européens de Développement Régional (contract no. 79/1769), the Programme National Action Concentrée Incitative-Fonds National de la Science “Ecosphère Continentale” (contract no. 04 2 9 FNS), and a Marie Curie Intra-European Fellowship (proposal no. 024683 METOLEVOL).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Nathalie Verbruggen (nverbru@ulb.ac.be).
Open Access articles can be viewed online without a subscription.
References
- Axelsen KB, Palmgren MG (2001) Inventory of the superfamily of P-type ion pumps in Arabidopsis Plant Physiol 126 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis WD, Smith OS, Grant D, Fincher R (1994) Identification of quantitative trait loci using a small sample of topcrossed and F4 progeny from maize. Crop Sci 34 882–896 [Google Scholar]
- Becher M, Talke IN, Krall L, Kramer U (2004) Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J 37 251–268 [DOI] [PubMed] [Google Scholar]
- Bernard C, Roosens N, Czernic P, Lebrun M, Verbruggen N (2004) A novel CPx-ATPase from the cadmium hyperaccumulator Thlaspi caerulescens. FEBS Lett 569 140–148 [DOI] [PubMed] [Google Scholar]
- Bert V, Bonnin I, Saumitou-Laprade P, de Laguerie P, Petit D (2002) Do Arabidopsis halleri from nonmetallicolous populations accumulate zinc and cadmium more effectively than those from metallicolous populations? New Phytol 155 47–57 [DOI] [PubMed] [Google Scholar]
- Bert V, Meerts P, Saumitou-Laprade P, Salis P, Gruber W, Verbruggen N (2003) Genetic basis of Cd tolerance and hyperaccumulation in Arabidopsis halleri. Plant Soil 249 9–18 [Google Scholar]
- Blaudez D, Kohler A, Martin F, Sanders D, Chalot M (2003) Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell 15 2911–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw HD, Otto KG, Frewen BE, McKay JK, Schemske DW (1998) Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics 149 367–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S (2006) Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88 1707–1719 [DOI] [PubMed] [Google Scholar]
- Clemens S, Bloss T, Vess C, Neumann D, Nies DH, zur Nieden U (2002) A transporter in the endoplasmic reticulum of Schizosaccharomyces pombe cells mediates zinc storage and differentially affects transition metal tolerance. J Biol Chem 277 18215–18221 [DOI] [PubMed] [Google Scholar]
- Craciun AR, Courbot M, Bourgis F, Salis P, Saumitou-Laprade P, Verbruggen N (2006) Comparative cDNA-AFLP analysis of Cd-tolerant and -sensitive genotypes derived from crosses between the Cd hyperaccumulator Arabidopsis halleri and Arabidopsis lyrata ssp. petraea. J Exp Bot 57 2967–2983 [DOI] [PubMed] [Google Scholar]
- Deniau A, Pieper B, Ten Bookum WM, Lindhout P, Aarts MGM, Schat H (2006) QTL analysis of cadmium and zinc accumulation in the heavy metal hyperaccumulator Thlaspi caerulescens. Theor Appl Genet 113 907–920 [DOI] [PubMed] [Google Scholar]
- Dräger DB, Desbrosses-Fonrouge AG, Krach C, Chardonnens AN, Meyer RC, Saumitou-Laprade P, Kramer U (2004) Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. Plant J 39 425–439 [DOI] [PubMed] [Google Scholar]
- Eren E, Arguello JM (2004) Arabidopsis HMA2, a divalent heavy metal-transporting P-IB-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol 136 3712–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov V, Dowdle J, Smirnoff N, Ford-Lloyd B, Newbury HJ, Macnair MM (2006) Comparison of gene expression in segregating families identifies genes and genomic regions involved in a novel adaptation, zinc hyperaccumulation. Mol Ecol 15 3045–3059 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350 87–96 [DOI] [PubMed] [Google Scholar]
- Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, Young J, Camakaris J, Harper JF, Cobbett CS (2004) P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis Plant Cell 16 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer U (2005) Phytoremediation: novel approaches to cleaning up polluted soils. Curr Opin Biotechnol 16 133–141 [DOI] [PubMed] [Google Scholar]
- Kuittinen H, de Haan AA, Vogl C, Oikarinen S, Leppälä J, Koch M, Mitchell-Olds T, Langley CH, Savolainen O (2004) Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature 227 680–681 [DOI] [PubMed] [Google Scholar]
- Macnair MR (1987) Heavy metal tolerance in plants: a model evolutionary system. Trends Ecol Evol 2 354–359 [DOI] [PubMed] [Google Scholar]
- Macnair MR, Bert V, Huitson SB, Saumitou-Laprade P, Petit D (1999) Zinc tolerance and hyperaccumulation are genetically independent characters. Proc Biol Sci 266 2175–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RF, Francini A, da Rocha PSCF, Baccarini PJ, Aylett M, Krijger GC, Williams LE (2005) The plant P-1B-type ATPase AtHMA4 transports Zn and Cd and plays a role in detoxification of transition metals supplied at elevated levels. FEBS Lett 579 783–791 [DOI] [PubMed] [Google Scholar]
- Mills RF, Krijger GC, Baccarini PJ, Hall JL, Williams LE (2003) Functional expression of AtHMA4, a P-1B-type ATPase of the Zn/Co/Cd/Pb subclass. Plant J 35 164–176 [DOI] [PubMed] [Google Scholar]
- Oetting WS, Lee HK, Flanders DJ, Wiesner GL, Sellers TA, King RA (1995) Linkage analysis with multiplexed short tandem repeat polymorphism using infrared fluorescence and M13 tailed primers. Genomics 30 450–458 [DOI] [PubMed] [Google Scholar]
- Orr HA (1998) Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics 149 2099–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoyan A, Kochian LV (2004) Identification of Thlaspi caerulescens genes that may be involved in heavy metal hyperaccumulation and tolerance: characterization of a novel heavy metal transporting ATPase. Plant Physiol 136 3814–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels M, Frérot H, Bonnin I, Saumitou-Laprade P (2006) A broad-scale analysis of population differentiation for Zn tolerance in an emerging model species for tolerance study: Arabidopsis halleri (Brassicaceae). J Evol Biol 19 1838–1850 [DOI] [PubMed] [Google Scholar]
- Pence NS, Larsen PB, Ebbs SD, Letham DL, Lasat MM, Garvin DF, Eide D, Kochian LV (2000) The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc Natl Acad Sci USA 97 4956–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard AJ, Powell KD, Harper FA, Smith JAC (2002) The genetic basis of metal hyperaccumulation in plants. CRC Crit Rev Plant Sci 21 539–566 [Google Scholar]
- Rensing C, Ghosh M, Rosen BP (1999) Families of soft-metal-ion-transporting ATPases. J Bacteriol 181 5891–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- SAS (1999) SAS OnLineDoc V8. Statistical Analysis System Institute, Cary, NC
- Schat H, Ten Bookum WM (1992) Genetic control of copper tolerance in Silene vulgaris. Heredity 70 142–147 [Google Scholar]
- Talke I, Hanikenne M, Krämer U (2006) Zinc dependent global transcriptional control, transcriptional de-regulation and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol 142 148–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillemans V, Dispa L, Remacle C, Collinge M, Motte P (2005) Functional distribution and dynamics of Arabidopsis SR splicing factors in living plant cells. Plant J 41 567–582 [DOI] [PubMed] [Google Scholar]
- van de Mortel J, Villanueva LA, Schat H, Kwekkeboom J, Coughlan S, Moerland PD, Ver Loren van Themaat E, Koornneef M, Aarts MGM (2006) Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol 142 1127–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW, Boer MP, Jansen RC, Maliepaard C (2002) MapQTL 4.0, Software for the Calculation of QTL Positions on Genetic Maps. Plant Research International, Wageningen, The Netherlands
- Van Ooijen JW, Voorips RE (2001) Joinmap 3.0, Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands
- Van Rossum F, Bonnin I, Fenart S, Pauwels M, Petit D, Saumitou-Laprade P (2004) Spatial genetic structure within a metallicolous population of Arabidopsis halleri, a clonal, self-incompatible and heavy-metal-tolerant species. Mol Ecol 13 2959–2967 [DOI] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Leonhardt N, David P, Nussaume L, Vavasseur A, Richaud P (2004) Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett 576 306–312 [DOI] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Preveral S, Forestier C, Vavasseur A, Richaud P (2005) Heavy metal transport by AtHMA4 involves the N-terminal degenerated metal binding domain and the C-terminal His(11) stretch. FEBS Lett 579 1515–1522 [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93 77–78 [DOI] [PubMed] [Google Scholar]
- Weber M, Harada E, Vess C, von Roepenack-Lahaye E, Clemens S (2004) Comparative microarray analysis of Arabidopsis thaliana and Arabidopsis halleri roots identifies nicotianamine synthase, a ZIP transporter and other genes as potential metal hyperaccumulation factors. Plant J 37 269–281 [DOI] [PubMed] [Google Scholar]
- Willems G, Dräger DB, Courbot M, Godé C, Verbruggen N, Saumitou-Laprade P (2007) Quantitative trait loci mapping of zinc tolerance in the metallophyte Arabidopsis halleri ssp. halleri. Genetics doi/10.1534/genetics.106.064485 [DOI] [PMC free article] [PubMed]