Abstract
In all plant species studied to date, sucrose synthase occurs as multiple isoforms. The specific functions of the different isoforms are for the most part not clear. Six isoforms of sucrose synthase have been identified in the model legume Lotus japonicus, the same number as in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). The genes encoding these isoforms are differentially expressed in all plant organs examined, although one, LjSUS4, is only expressed in flowers. LjSUS1 is the most highly expressed in all plant organs tested, except root nodules, where LjSUS3 accounts for more than 60% of the total SUS transcripts. One gene, LjSUS2, produces two transcripts due to alternative splicing, a feature not observed in other species to date. We have isolated plants carrying ethyl methanesulfonate-induced mutations in several SUS genes by targeting-induced local lesions in genomes reverse genetics and examined the effect of null alleles of two genes, LjSUS1 and LjSUS3, on nodule function. No differences were observed between the mutants and wild-type plants under glasshouse conditions, but there was evidence for a nitrogen-starvation phenotype in the sus3-1 mutant and severe impairment of growth in the sus1-1/sus3-1 double mutant under specific environmental conditions. Nodules of sus3-1 mutant plants retained a capacity for nitrogen fixation under all conditions. Thus, nitrogen fixation can occur in L. japonicus nodules even in the absence of LjSUS3 (the major nodule-induced isoform of SUS), so LjSUS1 must also contribute to the maintenance of nitrogen assimilation.
Nitrogen fixation in bacteroids and conversion of ammonia to amino acids in the infected cells of legume nodules require energy (ATP), carbon skeletons, and reductants that are produced from Suc synthesized in the leaves and imported into the nodules (Vance and Heichel, 1991; Gordon, 1995; Udvardi and Day, 1997). Two types of enzymes could in theory be responsible for the initial cleavage of imported Suc in nodule cells: Suc synthase (EC 2.4.1.13) and invertase (EC 3.2.1.26). Suc synthase cleaves Suc into Fru and UDP-Glc and is reversible in vivo (Geigenberger and Stitt, 1993). In contrast, cleavage by invertases leads to the formation of Fru and Glc and is irreversible (Avigad, 1982). In general, Suc synthase is believed to be the main route of Suc catabolism in organs in which oxygen tension is low (Guglielminetti et al., 1995; Rolletschek et al., 2002) because conversion of Suc to hexose phosphates via Suc synthase requires less ATP than conversion via invertase. Suc synthase activity is also important for the provision of energy for phloem loading (Martin et al., 1993) and in the development of sink strength (Edwards and ap Rees, 1986), two other processes important in nodulation.
Suc synthase is encoded by a small multigene family in all species analyzed to date, including pea (Pisum sativum; Barratt et al., 2001), Arabidopsis (Arabidopsis thaliana; Baud et al., 2004), potato (Solanum tuberosum; Zrenner et al., 1995), and maize (Zea mays; Duncan et al., 2006). Analysis of mutant and transgenic plants with reduced Suc synthase activity has revealed that specific isoforms are essential for normal metabolism in different organs, including maize kernels, pea embryos, and cotton (Gossypium hirsutum) seeds (Chourey et al., 1998; Craig et al., 1999; Ruan et al., 2003), maize roots (Subbaiah and Sachs, 2001), and potato tubers (Zrenner et al., 1995). However, systematic analysis of Arabidopsis mutants lacking individual isoforms of Suc synthase failed to discover specific roles in particular organs for any of the six isoforms (Bieniawska et al., 2007). This suggests either a high level of redundancy within the Suc synthase gene family in this species or that isoforms of invertase can play the same role as Suc synthase in this species (Bieniawska et al., 2007).
Vacuolar and cell wall acid invertases have essential roles in cell expansion (Sergeeva et al., 2006) and pollen development (Koonjul et al., 2005; Oliver et al., 2005) in several species, but there is general agreement that acid invertase is unlikely to be responsible for the metabolism of Suc, which provides ATP and reductant in nodule cells (Gordon et al., 1999). Supporting this, acid invertase transcripts have not been identified from nodule EST libraries of Lotus japonicus and only very low acid invertase activities have been detected in this organ (Flemetakis et al., 2006). There is no consensus, however, about the relative importance of alkaline/neutral invertase and Suc synthase in nodule metabolism. Flemetakis and coworkers (2006) showed that there was an alkaline/neutral invertase gene, LjINV1, with enhanced expression in nodules of L. japonicus, and proposed that it played a role in Suc partitioning and metabolism. This agrees with earlier findings in developing soybean (Glycine max) nodules, where alkaline/neutral invertase seems to be the predominant Suc-cleaving enzyme (Morell and Copeland, 1984). In pea, a specific isoform of Suc synthase, SUS1, is essential for nitrogen fixation in nodules (Craig et al., 1999). Plants carrying mutant alleles at the rug4 locus that specifically lack SUS1 activity could form nodules containing bacteroids (Gordon et al., 1999), but δ15N analysis showed that nitrogen fixation was low or absent and nodules senesced early (Craig et al., 1999). There was Suc synthase activity remaining in the rug4 mutants (Craig et al., 1999), which could have been due to the other two Suc synthase isoforms found later to be present in pea (Barratt et al., 2001). These different results about the relative contributions of Suc synthase and invertase in carbon metabolism in the nodule suggest that there may be species-specific differences in Suc metabolism in legume nodules.
We have chosen the model legume L. japonicus to dissect genetically the transit of carbon through nodules and to develop an understanding of the metabolic factors that influence the rate of nitrogen fixation in nodules. In this article, we report the identification of six genes encoding Suc synthase in L. japonicus and their patterns of expression in the plant. We used our recently developed TILLING (for targeting-induced local lesions in genomes) platform (Perry et al., 2003) to obtain mutants by reverse genetics. For the genes most highly expressed in the root nodule, we have characterized two mutant lines in detail.
RESULTS
There Are Six Isoforms of Suc Synthase in L. japonicus
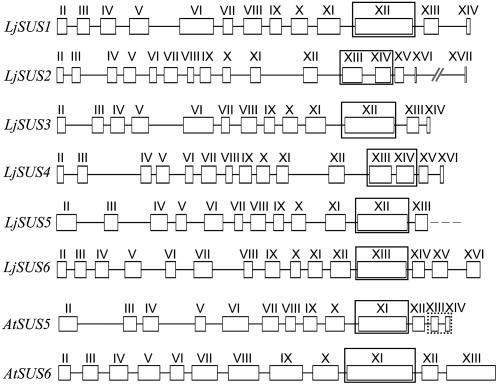
There are at least six genes encoding different isoforms of Suc synthase in L. japonicus (Fig. 1), which have been found by exhaustive analysis of the published genome or sequences that are available at the Kazusa DNA Research Institute. The first three genes were identified via assembly of EST sequences followed by screening of the public databases or transformation-competent artificial chromosome (TAC)/bacterial artificial chromosome libraries for corresponding genomic sequences as described in “Materials and Methods.” LjSUS4 was identified by analyzing TAC clones for sequence similarities to LjSUS1 and LjSUS3. LjSUS5 and LjSUS6 were found by comparison with known Arabidopsis sequences for Suc synthases 5 and 6 (Baud et al., 2004). Only a partial sequence was found for LjSUS5. The encoded isoform is 71% identical to AtSUS5 on the amino acid level. Comparison with the gene structure of AtSUS5 suggested that only two small exons were missing from the LjSUS5 genomic sequence. Comparison of the intron-exon structure, cDNA, and predicted amino acid sequences revealed that the Suc synthase family in L. japonicus consists of three distinct pairs of proteins that are closely related to each other: isoforms 1 and 3, isoforms 2 and 4, and isoforms 5 and 6 (Fig. 1). At the cDNA level, the sequences of the pairs of genes are closer to each other than to the other isoforms, with 85% identity between LjSUS1 and LjSUS3 and LjSUS2 and LjSUS4, and 73% identity between LjSUS5 and LjSUS6. Overall, at the level of predicted amino acid sequences, the identity between all the isoforms is greater than 54%, and it is greatest between LjSUS1 and LjSUS3, with 89%, whereas LjSUS2 and LjSUS4 are 85% identical, and LjSUS5 and LjSUS6 are 72% identical. The sequence encoding the glycosyl-transferase domain in LjSUS2 and LjSUS4 is split by an intron into two exons (exons XIII and XIV), whereas it is encoded by one large exon in the other four Suc synthase genes (exon XII for LjSUS1, LjSUS3, and LjSUS5, and exon XIII for LjSUS6; Fig. 1).
Figure 1.
Comparison of L. japonicus and Arabidopsis Suc synthase gene structures from start to stop codons. Exons are represented by numbered boxes and introns by connecting lines according to Baud et al. (2004). The possible unknown exons for the partial sequence of LjSUS5 are framed by a dashed box in the AtSUS5 structure. The glycosyl transferase domain in all structures is framed. Accession numbers (and nucleotide positions of the beginning and end of coding sequence) for LjSUS1 to LjSUS6 are: AP004481 (3500–7563), AP009335 (83700–77870), AP009336 (27092–31370), AP007582 (50566–56206), AP009338 (4831–0, partial), and AP009337 (78148–83083), respectively.
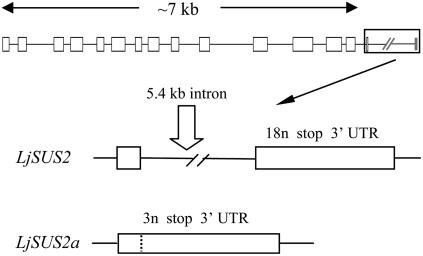
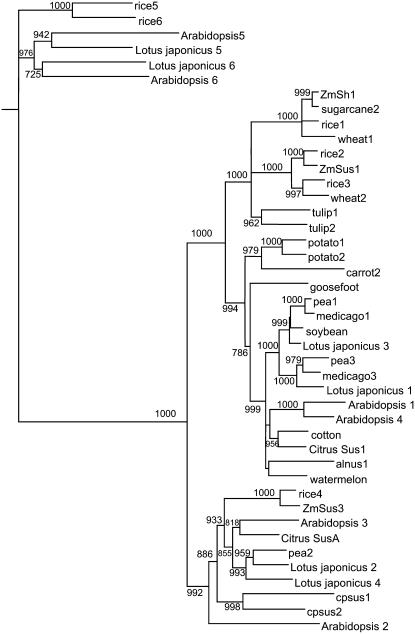
LjSUS2 differed from the genes encoding other family members with regard to a very long intron at its 3′ end. Analysis of EST clones revealed that LjSUS2 can give rise to two different mRNAs, referred to as LjSUS2 and LjSUS2a (Fig. 2). In LjSUS2, the last intron, with a length of 5.4 kb, is spliced. The last exon of LjSUS2 consists of 21 nucleotides, coding for six amino acids and a stop codon, and the 3′-untranslated region (UTR). In LjSUS2a, the last intron is retained in the mRNA. This alternative splicing transforms the intron of LjSUS2 to an exon consisting of six nucleotides, coding for one amino acid and a stop codon, and the 3′ UTR. The two mRNAs are predicted to give rise to two different proteins, LjSUS2 being five amino acids longer than LjSUS2a. The phylogenetic relationship between the L. japonicus Suc synthase isoforms and those of other species was constructed from their deduced amino acid sequences (Fig. 3). It showed clearly that the three pairs of L. japonicus isoforms fit into the three respective classes of isoforms as noted elsewhere (Komatsu et al., 2002).
Figure 2.
Detailed gene structure of LjSUS2. Processed LjSUS2 transcript also occurs in the form of LjSUS2a, possibly due to alternative splicing. Part of the long (5.4 kb) intron in LjSUS2 represents an exon of LjSUS2a. The protein encoded by LjSUS2a is five amino acids shorter than that encoded by LjSUS2. n, Nucleotides; stop, stop codon.
Figure 3.
Comparison of deduced amino acid sequences of plant Suc synthases. A phylogenetic dendrogram was generated and bootstrap analysis with 1,000 replicates performed as described in “Materials and Methods.” The deduced amino acid sequences of plants came from the following sources (with abbreviation in the tree in brackets and the number indicating the isoform of Suc synthase followed by accession numbers): Alnus glutinosa (alnus1, X92378); Arabidopsis (Arabidopsis1–6, At5g20830; At5g49190; At4g02280; At3g43190; At5g37180; At1g73370), Citrullus vulgaris (watermelon, Q9SBL8 Q9SBL8|Q9SBL8); Citrus reticulate (Citrus Sus1, Q9SLY1|Q9SLY1; Citrus SusA, Q9SLY2|Q9SLY2); Chenopodium rubrum (goosefoot, X82504); Craterostigma plantagineum (cpsus1–2, AJ1319999; AJ1320000); Daucus carota (carrot2, Y16091); Glycine max (soybean, AF030231); Gossypium hirsutum (cotton, Q9XGB7); Lotus japonicus (Lotus japonicus1–6, AP004481; AP009335; AP009336; AP007582; AP009338; AP009337); Medicago truncatula (medicago1, TC67957; medicago3, TC67958); Oryza sativa (rice1–3, P30298; P31924; Q43009); Pisum sativum (pea1–3, AJ012080; AJ001071; AJ311496); Saccharum officinalis (sugarcane2, AF263384); Solanum tuberosum (potato1–2, P10691; P49039), Triticum aestivum (wheat1–2, AJ001117; AJ000153); Tulipa gesneriana (tulip1–2, Q41608; Q41607); Zea mays (ZmSus1, L22296; ZmSh1, X02400; ZmSus3, AY124703). Isoforms 5 and 6 form the “New Group” according to Komatsu et al. (2002).
Suc Synthase Genes Are Differentially Expressed in L. japonicus
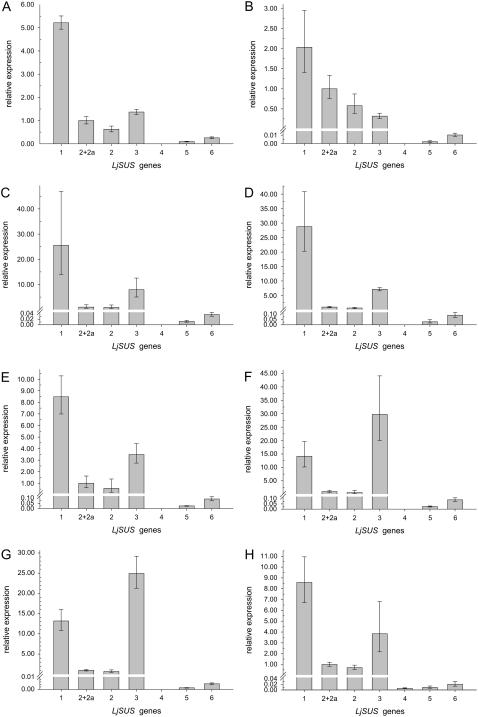
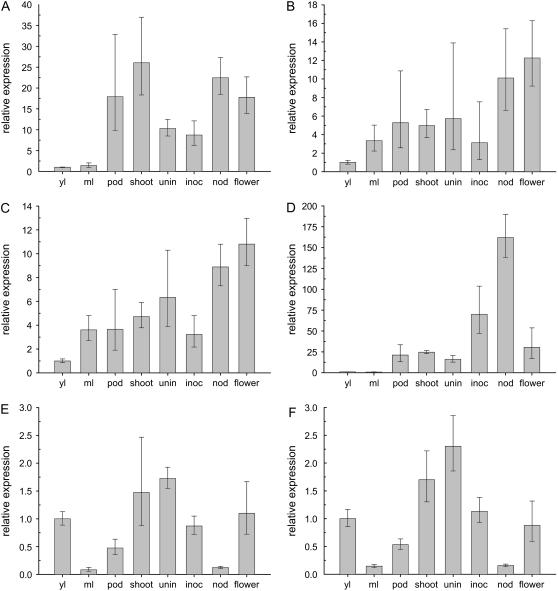
Quantitative reverse transcription (qRT)-PCR experiments were performed (Table I) to analyze the expression patterns of the different Suc synthase genes in L. japonicus. All Suc synthase genes, except LjSUS4, were expressed in all organs analyzed (Fig. 4, A–H). Calculations of the contribution of each gene to overall Suc synthase transcript levels have been made based on the assumption that L. japonicus has six isoforms of Suc synthase. LjSUS1 was the gene most highly expressed in the majority of organs examined, contributing more than 60% to the total amount of Suc synthase transcript (Fig. 4, A–E, and H). LjSUS2 and LjSUS2a were expressed at levels between 2% and 8% in all organs examined, except leaves, where they contributed up to 30% of total Suc synthase transcript (Fig. 4, A and B). LjSUS2 was more abundant than LjSUS2a in all organs analyzed. LjSUS3 transcript contributed between 9% and 29% of transcript in most organs (Fig. 4, A–E, and H), but more than 60% of the total Suc synthase transcript in inoculated roots and nodules (Fig. 4, F and G). LjSUS4 was detected only in flowers, where it contributed less than 1% of total Suc synthase transcript (Fig. 4H). LjSUS5 and LjSUS6 were expressed in all organs, but at very low levels. Only in young leaves were they expressed to an extent greater than 1% (Fig. 4A). When expression of the different genes was compared between plant organs relative to their expression in young leaves (Fig. 5), it became apparent that expression of LjSUS3 is enhanced in nodules and inoculated roots (Fig. 5D). A similar enhancement was not found for any other gene in any other organ (Fig. 5, A–C, and E and F). Hence, LjSUS3 can be regarded as a nodule-enhanced SUS gene.
Table I.
Primers used for qRT-PCR analysis
| Genes | Target Gene | Primer (Forward/Reverse) |
|---|---|---|
| Suc synthases | LjSUS1 | 5′-CGTCTCTCCTGGAGCTGATATGGA-3′ |
| 5′-GCTCCCCGTTCCTTATGCGGTC-3′ | ||
| LjSUS2 | 5′-AGGGAACTGGTCAATCTTGTCATC-3′ | |
| 5′-TAGGTTTGAAGCTTGGTCAGGGTG-3′ | ||
| LjSUS2 and LjSUS2a | 5′-CAAAGGATGATGCAAGTTAACCAGC-3′ | |
| 5′-CAAGACATTAAACAACTACTGCCCTG-3′ | ||
| LjSUS3 | 5′-TCTCACCCGGAGCTGATCAGAC-3′ | |
| 5′-GCTCTCCGTTCCTGACCCTGTTC-3′ | ||
| LjSUS4 | 5′-GACCGAGTCAAAAACATATCTGGG-3′ | |
| 5′-CGAAGAATTCGACCACCAGCTCAG-3′ | ||
| LjSUS5 | 5′-CAATGAAGAACACATAGGATATTTGG-3′ | |
| 5′-AAGGCCTCATACAATGCTGGTTG-3′ | ||
| LjSUS6 | 5′-ACGCTGAGCATATTGGATATCTAGC-3′ | |
| 5′-CACAAAAGCCTCCCTTTGAGTCAG-3′ | ||
| Actin | TC14249 | 5′-GGACAGACTCGTGAGCACGCAC-3′ |
| 5′-CAGGCTTCAAGACACCAGTTTCAAC-3′ | ||
| EF-1α | TC14056 | 5′-CTAAGGGTGAATATGATGAGTCCGGC-3′ |
| 5′-GAAAGAAGAATCACAGTCACTCCC-3′ |
Figure 4.
qRT-PCR analysis of the LjSUS1 to LjSUS6 genes in different organs of L. japonicus. A, Young leaves. B, Mature leaves. C, Pods. D, Shoots. E, Uninoculated roots. F, Inoculated roots. G, Nodules. H, Flowers. Transcript levels were normalized to levels of EF-1α or actin in the case of flowers. Values are the means ± sd from three individual plants. Note the asymmetrical distribution of the sd caused by conversion of an exponential process into a linear comparison (Livak and Schmittgen, 2001).
Figure 5.
Relative transcript levels of L. japonicus Suc synthases LjSUS1 to LjSUS6 in young leaves (yl), mature leaves (ml), pod, shoot, uninoculated (unin), and inoculated (inoc) roots, nodule (nod), and flower. Expression pattern of SUS genes was analyzed by qRT-PCR. A, LjSUS1. B, LjSUS2. C, LjSUS2 and LjSUS2a. D, LjSUS3. E, LjSUS5. F, LjSUS6. Transcript levels were expressed relative to the level in young leaves after normalization to levels of EF-1α or actin in the case of flowers. Values are means ± sd from three individual plants. Note the asymmetrical distribution of the sd caused by conversion of an exponential process into a linear comparison (Livak and Schmittgen, 2001). The profile for LjSUS4 is absent from this figure because LjSUS4 is only expressed in flowers.
Mutants Generated by TILLING Reverse Genetics Are Impaired in Suc Synthase Activity
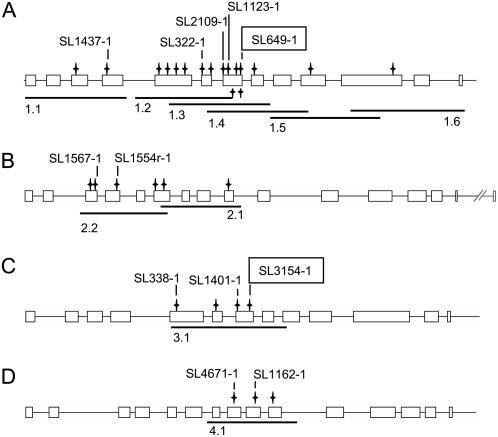
Several nonsilent mutations for LjSUS1 to LjSUS4 were identified via TILLING: 17 for LjSUS1, six for LjSUS2, four for LjSUS3, and three for LjSUS4 (Fig. 6, A–D). The large number of mutants in the LjSUS1 gene was found because the whole genomic sequence was analyzed using overlapping PCR fragments. This was done as part of a general exercise to examine the depth of allelic variation in the ethyl methanesulfonate-mutagenized population (see Supplemental Tables S1–S4 for full details). For the other genes, only part of the genomic sequence was analyzed. The mutants predicted by CODDLE (for codons optimized to discover deleterious lesions) to have potentially deleterious effects on gene function were subjected to further analysis: five alleles for LjSUS1, three for LjSUS3, and two for LjSUS2 and LjSUS4 (Fig. 6, A–D). One allele for each of LjSUS1 and LjSUS3 was found to have a premature stop codon in the middle of the sequence: at amino acid 384 for the line isolated from SL649-1 (sus1-1) and at amino acid 376 for the line isolated from SL3154-1 (sus3-1). Due to the high transcript levels of the genes encoding both of these isoforms in all organs, including nodules (Fig. 4, A–H), we focused on sus1 and sus3 mutants to elucidate the role of these Suc synthase isoforms in nodule metabolism. We also generated a double mutant, sus1-1/sus3-1.
Figure 6.
Positions of nonsilent mutations found via TILLING. Gene structures of LjSUS1 (A), LjSUS2 (B), LjSUS3 (C), and LjSUS4 (D) with amplicons 1.1 to 4.1 derived from primer pairs used for TILLING. Positions of nonsilent mutations are indicated together with the M2 plant number used to isolate mutants. Mutants carrying premature stop codons are framed.
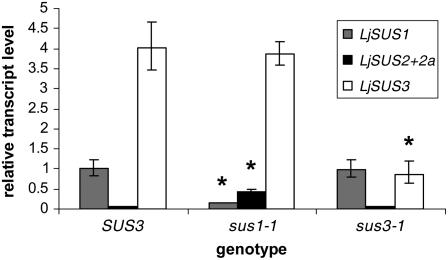
Nodules of sus1-1 had 15% of wild-type levels of LjSUS1 transcript, and nodules of sus3-1 had 23% of wild-type levels of LjSUS3 transcript (Fig. 7). Hence, the aberrant stop codons in sus1-1 and sus3-1 probably resulted in nonsense-mediated decay of the aberrant LjSUS1 and LjSUS3 transcripts, respectively. Interestingly, qRT-PCR data also showed that sus1-1 mutants had 7-fold increased LjSUS2 and LjSUS2a levels, whereas they were unchanged in sus3-1 mutants (Fig. 7).
Figure 7.
Transcript levels of LjSUS1, LjSUS2 and LjSUS2a, and LjSUS3 in inoculated roots bearing nodules from SUS3, sus1-1, and sus3-1 plants. Levels were expressed relative to LjSUS1 in the SUS3 wild type. Values are means ± sd from three individual plants. Values significantly different from wild-type levels are marked with an asterisk (P < 0.05; Student's t test).
Suc synthase activity was measured in nodule extracts of sus1 and sus3 homozygous mutants predicted to carry a deleterious mutation and in wild-type plants. A statistically significant difference in the activity between mutant and wild-type plants was observed for all sus3 mutants analyzed, but not for any of the sus1 mutants (Table II). Lines sus1-1 and sus3-1 were then backcrossed a second time (see “Materials and Methods”), and the SUS activity in nodules was compared to that in their respective wild-type plants. The sus1-1 mutant showed a 38% reduction, and the sus3-1 mutant showed a 67% reduction in Suc synthase activity compared to their respective wild-type levels (significant decrease at the 5% level; Student's t test). The activity of Suc synthase in the leaves was also compared between sus1-1 and wild type because LjSUS1 transcript made a greater contribution to leaf Suc synthase transcript (Fig. 4, A and B) than to nodule Suc synthase transcript (Fig. 4G). Activity in sus1-1 leaves was about 50% of the activity measured for the wild type (0.145 ± 0.005 compared to 0.294 ± 0.021 μmol min−1 mg−1 protein; means ± se from three individual plants). It seems likely from these results that sus1-1 and sus3-1 alleles give rise to little or no functional corresponding SUS protein.
Table II.
Suc synthase activities (assayed in cleavage direction) in nodules of sus1 and sus3 lines from the first backcross at 21 dpi; comparison to two different wild-type lines (SL649-1 as SUS1 wild type, SL338-1 as SUS3 wild type) from the segregating population
Values are means ± se from three individual plants.
| Genotype | Enzyme Activity (μmol min−1 g−1 Fresh Weight)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SUS1 | SUS3 | sus1-1 | sus1-2 | sus1-3 | sus1-4 | sus1-5 | sus3-1 | sus3-2 | sus3-3 | |
| SL line | 649-1 | 338-1 | 649-1 | 1437-1 | 322-1 | 2109-1 | 1123-1 | 3154-1 | 338-1 | 1401-1 |
| Mean | 3.46 | 3.01 | 2.65 | 2.86 | 3.27 | 2.87 | 3.02 | 0.75a | 2.28a | 2.26a |
| ±se | 0.36 | 0.09 | 0.23 | 0.07 | 0.28 | 0.30 | 0.06 | 0.11 | 0.14 | 0.09 |
Mean values are significantly different from wild-type levels at the 5% level (Student's t test).
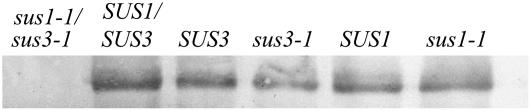
Further evidence to support the idea that the sus3-1 allele does not give rise to protein came from western-blot analysis of nodule proteins. An antiserum against PsSUS3 was used that detects all three known isoforms in pea (Barratt et al., 2001) and all isoforms in Arabidopsis, except AtSUS5 (P. Barratt, personal communication). Western blots showed that sus3-1 plants had decreased Suc synthase content in nodules (Fig. 8). No difference was observed in the content of Suc synthase protein between nodules of wild-type and sus1-1 plants; any difference in SUS1 protein content was likely to be small. This was supported by the fact that the expression of LjSUS1 in nodules is relatively low compared with LjSUS3 (Fig. 4G). The relative transcript levels and the measurements of enzyme activity (Table II) indicate that LjSUS3 makes a larger contribution to both SUS transcript and Suc synthase activity in nodules than LjSUS1.
Figure 8.
Western blot of sus1-1/sus3-1, sus3-1, sus1-1, and corresponding wild-type nodules. Equal amounts of protein (10 μg) were loaded in each lane. Blots were probed with PsSUS3 antiserum at a dilution of 1:5,000.
Primary metabolites in nodules at 57 d postinoculation (dpi) from plants grown in a mixture of perlite and vermiculite were analyzed by gas chromatography (GC)-mass spectrometry (MS). The analysis showed that sus3-1 mutants had significantly greater Suc content (Student's t test at the 5% level) than wild-type nodules (7.13 ± 0.22 and 2.20 ± 0.02 μmol g−1 dry weight, respectively; means ± se from three extractions). Furthermore, we confirmed that alkaline invertase activity was not up-regulated in both sus1-1 or sus3-1 mutants, and acid invertase activity was not measurable (data not shown). These data are consistent with the idea that the sus3-1 mutant has severely reduced capacity for Suc metabolism.
sus3-1 Mutants Show Symptoms of Nitrogen Starvation under Certain Defined Conditions
Suc synthase mutants were grown in several different media in a glasshouse. A medium that gives excellent root and nodule growth is Terragreen (a calcined attapulgite clay) mixed 1:1 with sand. This is frequently used for arbuscular mycorrhiza experiments. In this medium, as well as in compost, sus1-1 and sus3-1 mutants were not visibly different from wild-type plants. Although Terragreen has been reported to be inert in some publications (Boddington and Dodd, 1999), others (Edwards et al., 1998) and our own investigations have shown that this is not the case. It has substantial amounts of available nitrogen (equivalent to 99.5 kg N ha−1) and excessive amounts of sulfur (213 mg L−1). It also contains significant amounts of phosphate, potassium, magnesium, and normal soil amounts of iron (21 mg L−1). When plants were grown on vermiculite and fed with nitrogen-free nutrient solution, again no phenotypes for either the sus1-1 or sus3-1 mutants were observed (data not shown). In contrast to the situation in the glasshouse, when plants were grown in a mixture of perlite and vermiculite in a controlled environment room (CER), a phenotype was observed in sus3-1 mutants (Fig. 9A).
Figure 9.
Phenotypes of Suc synthase mutants in a CER. A, sus1-1, sus3-1, sus1-1/sus3-1, and their corresponding wild-type plants grown on nitrogen-free medium 1 d before harvest at 57 dpi. A sus3-1 plant is shown to illustrate the most extreme phenotype observed. B, Phenotype of the sus1-1/sus3-1 double mutant and its wild type supplied with nitrogen in the medium. [See online article for color version of this figure.]
Growth parameters of wild-type plants, sus1-1, and sus3-1 mutants in the CER were recorded over a time course. Plants were inoculated with rhizobia 5 d after germination and watered twice weekly with nitrogen-free nutrient solution. Control plants were watered with Hornum solution containing nitrogen (see “Materials and Methods”). Measurements of height, the number of branches, expanded leaves, flowers, and pods, the fresh and dry weight above and below ground revealed no differences between the mutant and corresponding wild-type plants before 34 dpi (data not shown). At 47 dpi, sus1-1 mutants were the same as the corresponding wild-type plants in all parameters measured, whereas sus3-1 plants started to differ from wild-type plants in various aspects (Table III): The number of fully expanded leaves and the number of branches of sus3-1 plants were decreased and basal leaves were yellow (Fig. 9A). There was no difference between the plants in height (Table III), the numbers of flowers, or the time of flower and pod formation (data not shown). The phenotype of sus3-1 plants became more pronounced as the plants aged and an extreme example is shown in Figure 9A. The lower leaves of sus3-1 mutants had a much lower chlorophyll a/b content than those of wild-type plants, whereas sus1-1 and wild-type plants did not differ from each other in this respect (Table IV). Seed pods were harvested as they matured and seed fresh and dry weights were determined. There were no significant differences between sus1-1 and wild-type plants, whereas both the fresh and dry weight of seed was decreased in sus3-1 plants compared to wild type (Table IV). sus3-1 mutants did not show yellowing or growth retardation relative to wild-type plants when grown with the addition of nitrogen fertilizer (data not shown). These results indicated that sus3-1 plants suffered from nitrogen starvation in the absence of added nitrogen.
Table III.
Growth parameters for sus1-1, sus3-1, and corresponding wild-type plants
Values are means ± sd of biological replicates. Between three and eight replicates were measured per genotype.
| Growth Parameter | dpi | Genotype
|
|||
|---|---|---|---|---|---|
| sus1-1 | SUS1 | sus3-1 | SUS3 | ||
| No. of expanded leaves | 47 | 24.0 ± 4.5 | 23.7 ± 2.1 | 11.6a ± 1.5 | 16.9 ± 4.2 |
| 54 | 33.0 ± 9.2 | 34.0 ± 8.0 | 16.0a ± 2.0 | 27.0 ± 7.1 | |
| No. of branches | 47 | 2.5 ± 0.8 | 2.3 ± 0.6 | 1.0a ± 0.0 | 2.3 ± 0.7 |
| 54 | 3.8 ± 1.6 | 4.3 ± 2.1 | 1.6a ± 0.9 | 3.1 ± 0.8 | |
| Height (cm) | 47 | 13.4 ± 3.7 | 13.1 ± 0.4 | 13.2 ± 3.1 | 15.8 ± 3.1 |
| 54 | 17.0 ± 4.7 | 16.8 ± 0.8 | 16.1 ± 3.3 | 18.7 ± 4.4 | |
Mean values are significantly different from the corresponding wild type at the 5% level (Student's t test).
Table IV.
Growth parameters for sus1-1, sus3-1, and corresponding wild-type plants at 57 dpi
Values are means ± se of three individual plants. Seeds were collected from three to eight individuals as pods matured. FW, Fresh weight; DW, dry weight.
| Parameter Measured | Genotype
|
|||
|---|---|---|---|---|
| sus1-1 | SUS1 | sus3-1 | SUS3 | |
| Chlorophyll a content (μg mg−1 FW) | 1.02 ± 0.25 | 0.94 ± 0.17 | 0.38a ± 0.02 | 0.81 ± 0.15 |
| Chlorophyll b content (μg mg−1 FW) | 0.36 ± 0.07 | 0.39 ± 0.09 | 0.13a ± 0.02 | 0.26 ± 0.04 |
| FW above ground (g) | 1.23 ± 0.45 | 1.40 ± 0.36 | 0.69a ± 0.07 | 1.28 ± 0.10 |
| FW roots (g) | 1.23 ± 0.28 | 1.08 ± 0.33 | 0.67 ± 0.16 | 0.69 ± 0.05 |
| FW per seed (mg) | 1.17 ± 0.01 | 1.11 ± 0.04 | 1.06a ± 0.04 | 1.30 ± 0.03 |
| DW per seed (mg) | 1.01 ± 0.01 | 1.02 ± 0.04 | 0.96a ± 0.03 | 1.19 ± 0.03 |
Mean values are significantly different from the corresponding wild type at the 5% level (Student's t test).
The sus3-1 Mutant Has Functional Nodules
An acetylene reduction assay was used to analyze the capacity of nodules for nitrogen fixation. At 57 dpi, nodules of wild type, sus1-1, and sus3-1 plants grown without nitrogen fertilizer in the CER were healthy and pink and showed the same amount of acetylene reduction, regardless of whether ethylene production per nodule or ethylene production per root system was determined (Table V). There was also no difference between wild-type and mutant plants in nodule number or the number of senescent nodules (Table V). To analyze the nitrogen status of mutant plants, δ15N and carbon-to-nitrogen ratios were measured on leaf material at 57 dpi (Table V). Analysis showed that all plants were fixing nitrogen because the values were close to zero or negative (Craig et al., 1999). Much higher δ15N values are expected from plants using sources of nitrogen other than nitrogen gas. The δ15N values of sus3-1 material and corresponding wild-type plants were, however, statistically significantly different from each other. δ15N values for sus1-1 material and that from the respective wild-type plants were very similar. Carbon-to-nitrogen ratios of sus3-1 mutants were nearly twice as high as their respective wild-type plants, whereas sus1-1 mutants had very similar carbon-to-nitrogen ratios to wild-type plants (Table V).
Table V.
Nodule number, number of senescent nodules, acetylene reduction, and leaf δ15N values and carbon-to-nitrogen ratios of sus1-1, sus3-1, and corresponding wild-type plants at 57 dpi
Values are means ± se of three individual plants.
| Parameter Measured | Genotype
|
|||
|---|---|---|---|---|
| sus1-1 | SUS1 | sus3-1 | SUS3 | |
| Nodule no. | 55.3 ± 12.3 | 43.7 ± 9.4 | 37.7 ± 7.4 | 44.3 ± 10.3 |
| No. of senescent nodules | 6.7 ± 3.5 | 3.0 ± 1.0 | 8.0 ± 1.5 | 6.3 ± 1.9 |
| Acetylene reduction per root system (nmol ethylene h−1) | 504.3 ± 84.3 | 299.6 ± 93.0 | 345.5 ± 59.7 | 290.5 ± 45.5 |
| Acetylene reduction per nodule (nmol ethylene h−1) | 10.5 ± 0.3 | 7.3 ± 1.8 | 12.0 ± 1.3 | 8.7 ± 2.6 |
| δ15N (parts per thousand) | −1.3 ± 0.2 | −1.2 ± 0.1 | −1.7a ± 0.1 | −0.9 ± 0.1 |
| Carbon-to-nitrogen ratio | 5.4 ± 0.2 | 5.2 ± 0.2 | 10.2a ± 1.9 | 5.4 ± 0.1 |
Mean values are significantly different from the corresponding wild type at the 5% level (Student's t test).
Growth of the sus3-1/sus1-1 Double Mutant Is Severely Impaired in the Absence of Nitrogen
Plants homozygous for the sus3-1 allele may be able to rely on LjSUS1 to provide sufficient carbon for fixation because they were able to grow normally under certain circumstances. We generated a double mutant, sus1-1/sus3-1, therefore, to shed further light on the importance of LjSUS1 and LjSUS3 in Suc metabolism in the nodule. Nodules of the double mutant showed a much more severe reduction in Suc synthase activity relative to wild-type plants than did those of sus3-1 mutants. Nodules from double mutants grown in a CER without added nitrogen had 94% lower activity than wild-type plants grown in the same condition (0.28 ± 0.06 compared with 4.63 ± 0.13 μmol min−1 g−1, means ± se for three individual plants), and Suc synthase protein was undetectable on western blots (Fig. 8). Under these growth conditions, double mutants started to differ visibly from the corresponding wild-type plants earlier than sus3-1 mutants, at 38 dpi. Leaf number was much more strongly reduced than that of sus3-1 mutants, and height was also reduced, whereas it was not affected in sus3-1 mutants (data not shown; Table III). At 47 dpi, double mutants had yellow leaves and at 57 dpi they had a significantly reduced shoot weight, but did not differ in root weight from wild-type plants (double-mutant shoot weight was 0.73 ± 0.14 g and wild-type weight was 1.54 ± 0.22 g; double-mutant root weight was 0.61 ± 0.21 g and wild-type weight was 0.86 ± 0.15 g; all values are means ± se of three replicates). When plants were fed with nitrogen-containing nutrient solution, their appearance was very similar to the wild type (Fig. 9B) and there were no differences with respect to chlorophyll content or shoot and root weight (data not shown).
DISCUSSION
We have shown that Suc synthase is encoded by a small family of at least six genes in L. japonicus (Fig. 1). This is the same number of genes reported for Arabidopsis (Baud et al., 2004) and rice (Huang et al., 1996; Harada et al., 2005) and extends the understanding of the gene family in legumes, where several isoforms have been reported in Medicago truncatula (Hohnjec et al., 1999) and pea (Barratt et al., 2001). As in Arabidopsis, the encoded SUS proteins appear to fall into three distinct groups of isoforms, each group containing two proteins (Fig. 3). In L. japonicus, these pairs of SUS proteins are LjSUS1 and LjSUS3, LjSUS2 and LjSUS4, and LjSUS5 and LjSUS6. The equivalent protein pairs in Arabidopsis are AtSUS1 and AtSUS4, AtSUS2 and AtSUS3, and AtSUS5 and AtSUS6, respectively. The similarities between the amino acid sequences of the isoforms belonging to the different groups between species suggest that the genes encoding members of the different groups diverged a relatively long time ago, at least before the separation of monocots and dicots. Conservation of sequence differences over such long periods of time usually reflects functional differences between members of the different subclades. The divergence between the genes encoding the pairs of proteins belonging to each subclade appears to have been more recent. Interestingly, LjSUS3, the gene most highly expressed in nodules of L. japonicus, is closely related to pea SUS1 (PsSUS1), which is known to have an important function in carbon metabolism in nodules (Craig et al., 1999). Unlike other species studied so far, a subisoform of LjSUS2, called LjSUS2a, was predicted as a result of alternative splicing of the LjSUS2 transcript (Fig. 2). This leads to the formation of two different mRNAs. LjSUS2a was present at very low levels in all organs analyzed and was always lower than LjSUS2 (Fig. 4, A–H), suggesting that the LjSUS2a transcript might be less stable.
Our expression analysis showed that the genes encoding the different isoforms of Suc synthase are differentially expressed in L. japonicus (Fig. 4, A–H), as has been reported for pea, M. truncatula, and Arabidopsis (Craig et al., 1999; Barratt et al., 2001; Hohnjec et al., 2003; Baud et al., 2004). LjSUS1 was the most highly expressed gene in all organs apart from inoculated roots and nodules (Fig. 4, F and G). LjSUS2 was expressed at low levels in the organs analyzed apart from leaves (Fig. 4, A and B). Its expression was induced in sus1-1 nodules (Fig. 7), which indicates that the LjSUS2 isoform might be able to compensate for the loss of LjSUS1 activity. LjSUS3 is a nodule-enhanced gene that is 8-fold more highly expressed in nodules than in uninoculated roots (Fig. 5D). High levels of transcript of this gene in nodules were also reported by Flemetakis et al. (2006). LjSUS4 was found only in flowers, at very low levels, and does not contribute significantly to the overall LjSUS mRNA content. LjSUS5 and LjSUS6 were found in all organs analyzed, which is in agreement with data from AtSUS5 and AtSUS6 (Baud et al., 2004).
Phylogenetic analysis of Suc synthases (Fig. 3) showed a clear legume cluster of Suc synthase isoforms as well as a monocot (rice SUS5 and SUS6) and a dicot (LjSUS5 and LjSUS6 and AtSUS5 and AtSUS6) cluster in the “New Group” (Komatsu et al., 2002). The first three Suc synthase isoforms of M. truncatula (MtSucS1, MtSucS2, and MtSucS3) are closely related to the three pea Suc synthase isoforms and share a similar expression pattern with them (Hohnjec et al., 1999). There are, however, some distinct differences in expression patterns between the L. japonicus genes and those in other legumes. LjSUS1 and PsSUS3 have very different expression patterns, although they are closely related (Fig. 3). PsSUS3 was hardly detectable in the organs analyzed apart from flowers and young testas (Barratt et al., 2001), whereas LjSUS1 was the most highly expressed gene in all organs, except nodules and inoculated roots (Fig. 4, A–H). Both LjSUS3 and PsSUS1 are nodule-enhanced SUS genes, but LjSUS3 is expressed in all other organs analyzed (Fig. 4), whereas PsSUS1 mRNA was not detected in mature leaves (Barratt et al., 2001). Predictions about expression patterns, therefore, cannot be drawn from phylogeny alone. This holds true for the stress-induced expression of other Suc synthase isoforms (Harada et al., 2005).
To analyze the role of Suc synthase in the nodule, we concentrated on mutations in genes encoding isoforms 1 and 3 because their expression was high in this organ (Fig. 4G). The greatest effects on Suc synthase activity were observed in mutants bearing premature stop codons in the SUS genes (Fig. 6). sus1-1 and sus3-1 were good candidates for being null alleles for the production of the respective SUS enzymes. Line sus3-1 showed a 67% reduction in Suc synthase activity in its nodules, as well as a decrease in the Suc synthase protein content on western blots (Fig. 8). This reduction was sufficient to affect the Suc content of the nodule: Suc levels were elevated in sus3-1 nodules. These results are consistent with more than a 60% contribution of LjSUS3 to total Suc synthase transcript levels in this organ (Fig. 4G). The sus1-1 allele showed a 38% decrease in Suc synthase activity in nodules, consistent with the estimate of LjSUS1 transcript in nodules being about 34% of the total SUS transcript (Fig. 4G). In leaves, the enzyme activity in sus1-1 plants dropped to 50% of wild-type levels, in accordance with the 60% contribution of LjSUS1 to Suc synthase transcript in this organ (Fig. 4, A and B).
It was difficult to observe an aberrant phenotype of sus mutants. Signs of nitrogen starvation in sus3-1 mutants were found when plants were grown without nitrogen in CERs, but were not observed for plants grown without nitrogen under glasshouse conditions. sus3-1 mutants grown in the CER started to differ from the wild-type and sus1-1 plants relatively late in growth, after 47 dpi (Table III). After this stage, sus3-1 mutants showed obvious signs of nitrogen starvation: fewer expanded leaves, fewer branches (Table III), yellow basal leaves with reduced chlorophyll content (Fig. 9A; Table IV), reduced fresh weight above ground, and decreased seed fresh and dry weight compared with wild-type plants (Table IV). There was no difference between sus3-1 and wild-type plants with respect to height (Table III), numbers of flowers, or the time of flower and pod formation (data not shown). The glasshouse and the CER differed in temperature, light quality, and intensity, as well as humidity and water saturation of the growth medium. One or several of these differing environmental conditions might explain why nitrogen starvation of sus3-1 mutants is only observed in the CER. We are investigating which factors might be responsible for the induction of nitrogen starvation. sus1-1 mutants did not show signs of nitrogen starvation, which is in agreement with the lower contribution of LjSUS1 to Suc synthase transcript in the nodule (Fig. 4G). Overall, our results showed that LjSUS3 was necessary for normal growth in the CER; LjSUS1 alone could not maintain an adequate level of nitrogen fixation. However, in the glasshouse, LjSUS1 alone could maintain normal growth and LjSUS3 was dispensable.
We also examined the morphology of the wild-type and sus nodules at the peak of LjSUS3 expression (Flemetakis et al., 2006), but found no sign of early senescence at that stage (data not shown). We also counted the number of functioning and senescent nodules of wild-type and sus mutants and again found no difference between them (Table V). Determinate nodules, like the ones from L. japonicus, are continuously generated as older ones go through programmed senescence (Szczyglowski et al., 1998), which could explain why early senescence was not found. In contrast, rug4 mutants of pea show an increased number of both healthy and senescing nodules (Craig et al., 1999). The increase in nodule number of rug4 plants may be related to the indeterminate nature of pea nodules. Acetylene reduction assays indicated that nodules of sus1-1 and also sus3-1 mutants had the capacity to fix nitrogen and were not quantitatively different in this respect from nodules of wild-type plants (Table V). This was true both for the acetylene reduction per nodule and per root system because nodule numbers were the same between mutant and wild-type plants (Table V). δ15N measurements on leaf material confirmed that nodules of both sus1-1 and sus3-1 mutants were fixing nitrogen. There was a significant difference, however, between the δ15N values for wild-type and sus1-1 mutants and those for sus3-1 mutants (Table V). Thus, sus3-1 mutants were capable of fixing nitrogen, but not to the same extent as wild-type plants. This conclusion was supported by the decrease in carbon-to-nitrogen ratios of sus3-1 plants (Table V).
Results from sus1-1/sus3-1 mutants grown in the CER showed that the aberrant phenotype in these plants was stronger than in sus3-1 mutants (Fig. 9A). These plants lacked 94% of Suc synthase enzyme activity in the nodule and developed an aberrant phenotype at 38 dpi. In contrast to sus3-1 mutants, the sus1-1/sus3-1 double mutants showed significantly reduced height compared with their corresponding wild-type plants at 38 dpi. Plants supplied with nitrogen did not show any signs of nitrogen starvation (Fig. 9B). The more severe phenotype of double mutants confirmed that LjSUS1 contributes to the ability of L. japonicus plants to assimilate nitrogen. Hence, analysis of sus1-1 and sus3-1 lines showed that both the nodule-enhanced isoform LjSUS3 and LjSUS1 can contribute to nitrogen fixation in the L. japonicus nodule. Interestingly, the biomass of the double mutant in the presence of nitrogen was very similar to the wild type, indicating that only the lack of nitrogen was responsible for the difference in growth. Hence, lack of these two isoforms was not significantly impairing the carbon supply to the rest of the plant.
Taken as a whole, our data indicate that invertases cannot metabolize the available Suc to compensate for the loss of Suc synthase activity. A role for nodule-enhanced alkaline/neutral invertase, LjINV1, in nodule function has been proposed by Flemetakis et al. (2006). Their developmental study showed a slight (2-fold) increase in LjINV transcript levels in nodules up to 10 dpi and increased LjSUS3 transcript level from 14 to 21 dpi. This observation and our data indicate that there may be a role for LjINV at early stages of nodule development and this would be in agreement with previous findings by Morell and Copeland (1984), who showed that alkaline invertase was the predominant enzyme cleaving Suc at early stages of nodule development.
Prior to our work, the only Suc synthase mutant available in legumes was the rug4 mutant of pea, which lacks the major nodule isoform PsSUS1. Comparison of the rug4 phenotype with that of the L. japonicus mutants allows us to draw the following conclusions, which may well be applicable to legumes generally, namely: both pea and L. japonicus require Suc synthase activity in their root nodules for normal levels of nitrogen fixation; activity of invertases cannot compensate for the role played by Suc synthase in Suc metabolism in either species; and a reduction of total Suc synthase activity in nodules to levels of around 10% or less of those in wild-type plants induces a severe nitrogen starvation phenotype. The relative importance of different isoforms of SUS in contributing to Suc synthase activity in nodules differs between pea and L. japonicus; LjSUS3 is less dominant (68%) than the equivalent isoform (PsSUS1) in pea (89%). Nodules of the sus3-1 mutant do not show premature senescence like their counterparts in PsSUS1 (rug4) mutants. Furthermore, the growth of the double sus1-1/sus3-1 mutant is considerably more impaired than that of either single mutant. Hence, both the nodule-enhanced isoform LjSUS3, as well as LjSUS1, are important for nitrogen fixation in L. japonicus. It will be interesting to discover whether pea is exceptional in the high level of dependence on a single isoform of Suc synthase for carbon utilization by nodules or whether other legumes also display this phenomenon.
We have shown that TILLING in L. japonicus is a powerful tool for systematically dissecting the contribution of each single isoform of Suc synthase to nodule metabolism. Whether L. japonicus nodules rely solely on Suc synthase for their supply of ATP, carbon precursors, and reductant or whether the nodule-enhanced alkaline/neutral invertase has a role early in nodule induction, remains to be established. We are currently examining TILLING mutants to determine the role of invertases in the nodule.
MATERIALS AND METHODS
Plant Growth Conditions
Plants for RNA extracts were grown in the glasshouse on a Terragreen:sand mixture (1:1; Oil-Dri Ltd.). Plants for enzyme assays, western blots, and GC-MS analysis were grown in the glasshouse on perlite:vermiculite (1:1). Seeds for growth experiments were scarified, sterilized with 10% (v/v) bleach (containing 1% available chlorine), and incubated in sterile water overnight at 4°C. They were germinated on water agar for 5 d before transfer to the respective growth medium: sterilized perlite:vermiculite mixture (1:1) or Terragreen:sand (1:1). Seedlings were immediately inoculated with Mesorhizobium loti strain Tono. Plants on perlite:vermiculite were fed twice a week either with nitrogen-free nutrient solution (Broughton and Dilworth, 1971) or Hornum solution, containing 5 mm NH4NO3, 3 mm KNO3 (Handberg and Stougaard, 1992). Elemental analysis of Terragreen was performed by NRM Laboratories Ltd. Plants for growth experiments were either grown in the glasshouse or in a CER as stated in the text. Plants in the glasshouse were grown under a 16/8-h photoperiod using supplemental lighting (high-pressure sodium; minimum 300 μmol s−1 m−2) at a 25°C day/20°C night temperature. Plants grown in a CER were at 25°C with 16/8-h photoperiods (maximum light level at 140 μmol s−1 m−2).
Gene Sequence Assembly
LjSUS1
A full-length consensus coding sequence for LjSUS1 was obtained by sequencing and aligning EST clones MSQL074d09_f, TM0013.1, MSQL002f04_f, LP844-11-b4, MWL056a11_r, LjNEST21d10r, MWL061h08_r, MSQ014c12_f, MWL018h01_r, MWL068h11_r, MWM135e07_r, LJA13372, MWM128c01_r, MSQL066f11_f, MWM128c01_f, MSQL047c12_f, MSQL087e01_f, MWM141d03_r, and MWL053g06_r. This coding sequence was used to search for a corresponding genomic sequence. The complete sequence was identified at EMBL, accession AP004481, clone LjT14007, TM0013, chromosome 6. Coding and genomic sequence aligned 100%.
LjSUS2
A partial coding sequence for LjSUS2 was obtained by sequencing and aligning EST clones GNLf004e06, LjNEST17c12rc, MWM120b01_r, LjNEST12e10r, MWM072e06_r, MWM237h11_r, MWL013h03_r, MWL080e04_r, LjNEST13d7rc, and LjNEST7B10r. This partial sequence was used to screen a TAC library. Genomic sequence corresponding to SUS2 partial coding sequence was identified in clone LJT30M08, TM1573 contig 10, chromosome 1. A predicted full-length coding sequence was obtained from the genomic sequence by using the Genscan program (available at http://genes.mit.edu/GENSCAN.html). An additional EST clone, BP064260, was sequenced, which had an additional splice site at the 3′ end. This resulted in five additional amino acids before the stop codon, a diverse 3′ UTR, and an additional intron of 5,389 bases.
LjSUS3
A partial coding sequence for LjSUS3 was obtained by sequencing and aligning EST clones MWL036f03_r, MWL079e09_r, MWM199e09_r, and MWL039g10_r. This partial sequence was used to screen a bacterial artificial chromosome library. A genomic sequence corresponding to this partial sequence was identified on clone LjB09G07, BM1684 contig 11. A predicted full-length coding sequence was obtained from the genomic sequence using Genscan.
LjSUS4
TAC clone LjT36C12, TM0773, was identified as containing a genomic sequence that corresponded to a SUS gene. The coding sequence was predicted using Genscan and sequence alignments confirmed it to be a fourth SUS gene, LjSUS4.
LjSUS5
The Lotus japonicus genome sequence database (Kazusa) was searched using Arabidopsis (Arabidopsis thaliana) sequence At5g37180. A candidate sequence, Ljwgs_025455.1, was found in the whole-genome shotgun assembly, which showed good homology with the Arabidopsis AtSUS5 gene. A TAC clone, LjT08I01, TM2173, contained sequence corresponding to Ljwgs_025455.1. Annotated gene sequence and alignments showed that this was a partial sequence only. There are no ESTs corresponding to the 3′ end of this gene; therefore, the partial sequence was used in gene sequence analysis.
LjSUS6
The L. japonicus genome sequence database was searched using Arabidopsis sequence At1g73370. A candidate sequence, Ljwgs_017710.1, was found in the whole-genome shotgun assembly, which showed good homology with the Arabidopsis AtSUS6 gene. A TAC clone, LjT38B12, TM2118, was identified that contained the whole SUS6 gene.
Sequences for LjSUS1 to LjSUS6 have been deposited in GenBank with the following accession numbers: AP004481, AP009335, AP009336, AP007582, AP009338, and AP009337, respectively.
Gene Structure Prediction and in Silico Protein Analysis
Suc synthase gene structures from start to stop codons were predicted using the CODDLE program (http://www.proweb.org./coddle). The ClustalW program (http://www.ebi.ac.uk/clustalw) was used for determining the percent identity between Suc synthase genes.
Phylogenetic Analysis
Protein sequences for 43 Suc synthases were aligned using the ClustalW program. The ends of the alignment (at amino acid 877) were trimmed and seven insertions specific to particular species were removed before performing phylogenetic analysis with Phylip programs (version 3.65). A distance matrix method employing the Dayhoff PAM matrix model was used to compare the sequences. The tree was built using the neighbor-joining clustering method and midpoint rooted as described in the Phylip documentation. Bootstrap values were calculated by analyzing datasets 1,000 times to indicate the confidence of each tree clade.
qRT-PCR
Plant material was harvested and ground in liquid nitrogen. Total RNA was extracted with CONCERT reagent (Invitrogen) and purified using the RNeasy kit (Qiagen) with on-column DNase I treatment (Qiagen) as described by the manufacturer. Genomic DNA-specific primers were used as controls to confirm the absence of genomic DNA in the RNA extract. RT was performed with 5 μg of RNA using SuperScript reverse transcriptase (Invitrogen) and oligo(dT)15 primers (Promega) according to the manufacturer's instructions.
All samples were measured in technical triplicates on three biological triplicates, consisting of three single plants harvested and extracted individually. Two controls lacking cDNA were included on each 96-well microplate (MJ Research). Master mixes were prepared; each sample contained SYBR Green JumpStart Taq ReadyMix (Sigma), 10 ng cDNA, and gene-specific primers (200 nm). Actin (for flowers) and elongation factor-1α (EF-1α; for all other tissues) were used as internal standards. The specificity of primers designed for qRT-PCR (Table I) was confirmed by sequencing after qRT-PCR reaction and by running products on agarose gels. Two different primer pairs were used to quantify the expression of LjSUS2 and LjSUS2a: One primer pair was designed to specifically amplify the longer transcript, LjSUS2, whereas the other primer pair amplifies both transcripts LjSUS2 and LjSUS2a. Standard curve experiments are as described elsewhere (Livak and Schmittgen, 2001; see “Statistical Analysis”) and proved that primers designed for all isoforms bind with the same efficiency to the DNA template. qRT-PCR was performed on the following organs: young and mature leaves, green pods, stems, uninoculated roots, roots inoculated with M. loti and bearing nodules, nodules, and flowers. Plants were 42 dpi and only young, pink nodules were picked for nodule analysis.
PCR reactions were performed on an Opticon machine (MJ Research). The PCR program started at 94°C for 2 min, and this was followed by 40 cycles of incubation at 94°C (15 s), 60°C (30 s), 72°C (1 min), and 76°C (1 s). Melting curves were recorded from 65°C to 95°C, reading every 0.5°C. For quantification of gene expression, the method described by Livak and Schmittgen (2001; see “Statistical Analysis”) was used.
TILLING
TILLING was carried out to isolate mutants for genes LjSUS1 to LjSUS4 using the general TILLING population and the preselected nodule mutant population described previously (Perry et al., 2003). Primers were designed using the CODDLE program to identify the region that had the maximum likelihood of being affected by ethyl methanesulfonate-induced mutation to produce a deleterious allele. Additional primer sets to screen other regions for the different genes were as follows: six primer sets, each amplifying a product of 1 kb to cover the entire SUS1 gene; SUS2, two primer sets each amplifying a product of 1.2 kb; SUS3 and SUS4, one primer set to screen 1.2 kb. Gene-specific primers were directly labeled with the fluorescent dyes 6-carboxyfluorescein and 4,7,2′,7′-tetrachloro-6-carboxyfluorescein for primers specific for SUS1, SUS2, and SUS3 genes for analysis with an ABI 377 sequencer as previously described (Perry et al., 2003), and IRD 700 and IRD 800 labels for SUS4 primers for analysis with LI-COR sequencer as previously described (Colbert et al., 2001).
Genomic DNA isolated from the forward screen plant population and general TILLING population plants were quantified and diluted to 5 ng μL−1. Normalized DNA was pooled 4-fold and 1 μL pooled DNA was used in a 10-μL PCR reaction using gene-specific labeled primers and a touchdown PCR program as described (Colbert et al., 2001), but without the addition of unlabeled primers. All primer sets were used to screen the nodulation-defective population. In addition, 2,304 individuals for SUS1 and SUS3 and 3,804 individuals for SUS2 and SUS4 were screened from the general population. PCR products were digested with Cel1, filtered though a Sephadex G50 fine column, concentrated, and loaded as described (Colbert et al., 2001). Samples were run on either ABI 377 or LI-COR 4300 sequencer depending on the labeled primers used.
Individuals from a pool containing a positive sample were rescreened to identify the individual plant containing the mutation. The individual was then sequenced and the effect of the mutation predicted using the PARSESNP program as described in Taylor and Greene (2003). Mutations that resulted in a premature stop codon and those with a Position-Specific Scoring Matrix difference score >10 were selected for further analysis. M3 seed was sown and homozygous mutants were selected from segregates in the case of heterozygous mutations at M2, which was the case for the majority of selected lines. Homozygous mutants of L. japonicus accession Gifu B-129 direct from TILLING or from segregating F2 populations were backcrossed to accession Miyakojima MG20.
Suc Synthase Assays
Cleavage Direction
Between five and 10 nodules were harvested with fine forceps into ice-cold Eppendorf tubes and extracted immediately in the tube in ice-cold 50 mm HEPES (pH 7.5), 5 mm MgCl2, 1 mm EDTA, 2 mm dithiothreitol (DTT), and 0.1% (w/v) polyvinylpolypyrrolidone. Samples were centrifuged at 15,800g for 10 min at 4°C, and the supernatant was used for assays. Assays were performed at 30°C and contained 50 mm HEPES (pH 7.0), 2 mm MgCl2, 1 mm EDTA, 200 mm Suc, 2 mm DTT, and 2 mm UDP. Assays were started by addition of extract and stopped by boiling for 2 min. The assay was confirmed to be linear with time and extract amount. Two sets of control assays were performed: one stopped immediately after addition of the extract, and the other lacking UDP. Fru produced during the incubation was assayed according to Stitt et al. (1989).
Synthesis Direction
Leaf samples of 200 to 400 mg from three individual plants were harvested, ground in liquid nitrogen, and extracted in an ice-cold mortar containing 30 mg polyvinylpolypyrrolidone per sample, 50 mm HEPES (pH 7.5), 5 mm MgCl2, 1 mm EDTA, 2 mm DTT, and protease inhibitor cocktail (Sigma). Samples were centrifuged at 15,800g for 10 min at 4°C and the supernatant was used for assays. Assays were at 25°C for 30 min and performed according to Bieniawska et al. (2007).
Alkaline and Acid Invertase Activity
Nodules were harvested and extracted as described for the Suc synthase assay in the cleavage direction. Alkaline and acid invertase activities were assayed as described by Hill et al. (2003). Enzyme assays were stable over time and amount of extract added.
Acetylene Reduction Assay
One root system per sample was placed in a blood tube (Vacutainer; Becton-Dickinson) and sealed with a rubber lid. Acetylene (0.5 mL) was injected and samples were incubated for 2 h at room temperature. The amount of ethylene produced was quantified by comparison with an ethylene standard. A gas chromatograph equipped with a hydrogen flame ionization detector was used. The column was 1.5 m long and 3.25 mm in diameter and packed with Porapak N.
Western-Blot Analysis
Nodule extracts were prepared as described for the Suc synthase assay (cleavage direction). PAGE and western blotting were performed as described by Barratt et al. (2001). A ProSieve prestained color marker (Cambrex) was used as a reference, and 10 μg of extracted protein were loaded per lane. Recombinant SUS3 from pea (Pisum sativum), as described by Barratt et al. (2001), was used as the primary antigen.
δ15N Analysis and Carbon-to-Nitrogen Ratio
Gas isotope analysis was carried out using a ThermoFinnigan (GmbH) system with a Conflo III interface. Approximately 10 mg of freeze-dried leaf material was weighed into tin capsules, which were sealed and placed inside a zero-blank autosampler attached to a Costech Elemental Analyzer ECS4010 (Costech International). Samples were analyzed according to Vallet et al. (1991).
Chlorophyll Measurement
Approximately 100 mg of lower leaves were harvested and ground in liquid nitrogen. After addition of 10 mL of 80% (v/v) Tris-buffered acetone (pH 8.0), samples were centrifuged for 5 min at 1,500g. The absorbance at 663.6 nm and 646.6 nm of the supernatant was measured and chlorophyll content calculated following the equations by Porra (2002).
Seed Weight
Exactly 100 dry, mature seeds were counted and the fresh weight determined. For seed dry weight, mature seeds were dried at 80°C until seed weight was stable.
GC-MS Analysis
Nodules were harvested, immediately frozen in liquid nitrogen, and freeze dried for 24 h. Approximately 14 mg of sample were ground in liquid nitrogen and extracted with 70% (v/v) ethanol according to a protocol by Kadlec (2001), with slight modifications. Phenyl-α-d-glucoside (0.5 g/L) was added as an internal standard. Derivatization and GC-MS analysis was performed as described by Desbrosses et al. (2005).
Statistical Analysis
All samples were measured as a minimum of biological triplicates. Student's t test was used to analyze the statistical significance of the difference between mutant and wild-type data. For qRT-PCR data, the method of Livak and Schmittgen (2001) was adopted. Briefly, it was confirmed that the amplification efficiencies of the target and the reference were equal and that the ΔΔCt method for the relative quantification of target could be used. Data were analyzed using the 2−ΔΔCt method. This method leads to error bars that are asymmetrically distributed above and below the average value because an exponential process is converted into a linear comparison of amounts.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. L. japonicus SUS1 alleles.
Supplemental Table S2. L. japonicus SUS2 alleles.
Supplemental Table S3. L. japonicus SUS3 alleles.
Supplemental Table S4. L. japonicus SUS4 alleles.
Supplementary Material
Acknowledgments
We thank Dr. Paul Barratt for the kind gift of antibodies against pea Suc synthase, Alan Jones for GC-MS analysis, and Paul Bailey for assistance with phylogeny. We also thank Jillian Perry for help with the TILLING platform and Andrea Pitzschke for providing EST sequences for some of the LjSUS genes. We are grateful to Professors Cathie Martin and Alison Smith and to Dr. Claire Domoney for helpful comments on the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Trevor L. Wang (trevor.wang@bbsrc.ac.uk).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Avigad G (1982) Sucrose and other disaccharides. In FA Loewus, W Tanner, eds, Encyclopedia of Plant Physiology, Vol 13A. Springer-Verlag, Berlin, pp 217–347
- Barratt DHP, Barber L, Kruger NJ, Smith AM, Wang TL, Martin C (2001) Multiple, distinct isoforms of sucrose synthase in pea. Plant Physiol 127 655–664 [PMC free article] [PubMed] [Google Scholar]
- Baud S, Vaultier MN, Rochat C (2004) Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot 55 397–409 [DOI] [PubMed] [Google Scholar]
- Bieniawska Z, Barratt DHP, Garlick AP, Thole V, Kruger NJ, Martin C, Zrenner R, Smith AM (2007) Analysis of the sucrose synthase gene family in Arabidopsis. Plant J 49 810–828 [DOI] [PubMed] [Google Scholar]
- Boddington CL, Dodd JC (1999) Evidence that differences in phosphate metabolism in mycorrhizas formed by species of Glomus and Gigaspora might be related to their life-cycle strategies. New Phytol 142 531–538 [Google Scholar]
- Broughton WJ, Dilworth MJ (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey PS, Taliercio EW, Carlson SJ, Ruan YL (1998) Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol Gen Genet 259 88–96 [DOI] [PubMed] [Google Scholar]
- Colbert T, Till BJ, Tompa R, Reynolds S, Steine MN, Yeung AT, McCallum CM, Comai L, Henikoff S (2001) High-throughput screening for induced point mutations. Plant Physiol 126 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J, Barratt P, Tatge H, Dejardin A, Handley L, Gardner CD, Barber L, Wang T, Hedley C, Martin C, et al (1999) Mutations at the rug4 locus alter the carbon and nitrogen metabolism of pea plants through an effect on sucrose synthase. Plant J 17 353–362 [Google Scholar]
- Desbrosses GG, Kopka J, Udvardi MK (2005) Lotus japonicus metabolic profiling: development of gas chromatography-mass spectrometry resources for the study of plant-microbe interactions. Plant Physiol 137 1302–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan KA, Hardin SC, Huber SC (2006) The three maize sucrose synthase isoforms differ in distribution, localization, and phosphorylation. Plant Cell Physiol 47 959–971 [DOI] [PubMed] [Google Scholar]
- Edwards J, ap Rees T (1986) Sucrose partitioning in developing embryos of round and wrinkled varieties of Pisum sativum. Phytochemistry 25 2027–2032 [Google Scholar]
- Edwards SG, Young JPW, Fitter AH (1998) Interactions between Pseudomonas fluorescence biocontrol agents and Glomus mosseae, an arbuscular mycorrhizal fungus, within the rhizosphere. FEMS Microbiol Lett 166 297–303 [Google Scholar]
- Flemetakis E, Efrose RC, Ott T, Stedel C, Aivalakis G, Udvardi MK, Katinakis P (2006) Spatial and temporal organization of sucrose metabolism in Lotus japonicus nitrogen-fixing nodules suggests a role for the elusive alkaline/neutral invertase. Plant Mol Biol 62 53–69 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Stitt M (1993) Sucrose synthase catalyzes a readily reversible reaction in vivo in developing potato tubers and other plant tissues. Planta 189 329–339 [DOI] [PubMed] [Google Scholar]
- Gordon AJ (1995) Sucrose metabolism to support N2 fixation in legume root nodules. In IA Tikhonovich, NA Povorov, VI Romanov, WE Newton, eds, Nitrogen Fixation: Fundamentals and Applications. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 533–538
- Gordon AJ, Minchin FR, James CL, Komina O (1999) Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol 120 867–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti L, Perata P, Alpi A (1995) Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiol 108 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handberg K, Stougaard J (1992) Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J 2 487–496 [Google Scholar]
- Harada T, Satoh S, Yoshioka T, Ishizawa K (2005) Expression of sucrose synthase genes involved in enhanced elongation of pondweed (Potamogeton distinctus) turions under anoxia. Ann Bot (Lond) 96 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill LM, Morley-Smith ER, Rawsthorne S (2003) Metabolism of sugars in the endosperm of developing seeds of oilseed rape. Plant Physiol 131 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec N, Becker JD, Puhler A, Perlick AM, Kuster H (1999) Genomic organization and expression properties of the MtSucS1 gene, which encodes a nodule-enhanced sucrose synthase in the model legume Medicago truncatula. Mol Gen Genet 261 514–522 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Perlick AM, Puhler A, Kuster H (2003) The Medicago truncatula sucrose synthase gene MtSucS1 is activated both in the infected region of root nodules and in the cortex of roots colonized by arbuscular mycorrhizal fungi. Mol Plant Microbe Interact 16 903–915 [DOI] [PubMed] [Google Scholar]
- Huang JW, Chen JT, Yu WP, Shyur LF, Wang AY, Sung HY, Lee PD, Su JC (1996) Complete structures of three rice sucrose synthase isogenes and differential regulation of their expressions. Biosci Biotechnol Biochem 60 233–239 [DOI] [PubMed] [Google Scholar]
- Kadlec P (2001) Chemical analysis of the carbohydrates. In CL Hedley, ed, Carbohydrates in Grain Legume Seeds. CABI Publishing, Norwich, UK, pp 31–59
- Komatsu A, Moriguchi T, Koyama K, Omura M, Akihama T (2002) Analysis of sucrose synthase genes in citrus suggests different roles and phylogenetic relationships. J Exp Bot 53 61–71 [PubMed] [Google Scholar]
- Koonjul PK, Minhas JS, Nunes C, Sheoran IS, Saini HS (2005) Selective transcriptional down-regulation of anther invertases precedes the failure of pollen development in water-stressed wheat. J Exp Bot 56 179–190 [DOI] [PubMed] [Google Scholar]
-
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the
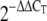 method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar] - Martin T, Frommer WB, Salanoubat M, Willmitzer L (1993) Expression of an Arabidopsis sucrose synthase gene indicates a role in metabolization of sucrose both during phloem loading and in sink organs. Plant J 4 367–377 [DOI] [PubMed] [Google Scholar]
- Morell M, Copeland L (1984) Enzymes of sucrose breakdown in soybean nodules—alkaline invertase. Plant Physiol 74 1030–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SN, van Dongen JT, Alfred SC, Mamun EA, Zhao XC, Saini HS, Fernandes SF, Blanchard CL, Sutton BG, Geigenberger P, et al (2005) Cold-induced repression of the rice anther-specific cell wall invertase gene OsINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ 28 1534–1551 [Google Scholar]
- Perry JA, Wang TL, Welham TJ, Gardner S, Pike JM, Yoshida S, Parniske M (2003) A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol 131 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73 149–156 [DOI] [PubMed] [Google Scholar]
- Rolletschek H, Borisjuk L, Koschorreck M, Wobus U, Weber H (2002) Legume embryos develop in a hypoxic environment. J Exp Bot 53 1099–1107 [DOI] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT (2003) Suppression of sucrose synthase gene expression represses cotton fiber cell initiation, elongation, and seed development. Plant Cell 15 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva LI, Keurentjes JJB, Bentsink L, Vonk J, van der Plas LHW, Koornneef M, Vreugdenhil D (2006) Vacuolar invertase regulates elongation of Arabidopsis thaliana roots as revealed by QTL and mutant analysis. Proc Natl Acad Sci USA 103 2994–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Lilley RM, Gerhardt R, Heldt HW (1989) Metabolite levels in specific cells and subcellular compartments of plant leaves. Methods Enzymol 174 518–552 [Google Scholar]
- Subbaiah CC, Sachs MM (2001) Altered patterns of sucrose synthase phosphorylation and localization precede callose induction and root tip death in anoxic maize seedlings. Plant Physiol 125 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski K, Shaw RS, Wopereis J, Copeland S, Hamburger D, Kasiborski B, Dazzo FB, de Bruijn FJ (1998) Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol Plant Microbe Interact 11 684–697 [Google Scholar]
- Taylor NE, Greene EA (2003) PARSESNP: a tool for the analysis of nucleotide polymorphisms. Nucleic Acids Res 31 3808–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Day DA (1997) Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol 48 493–523 [DOI] [PubMed] [Google Scholar]
- Vallet C, Arendt M, Mabon F, Naulet N, Martin GJ (1991) Combination of mass spectrometry and site-specific NMR isotope analyses in the characterisation of amino acids. J Sci Food Agric 56: 167–185
- Vance CP, Heichel GH (1991) Carbon in N2 fixation—limitation or exquisite adaptation. Annu Rev Plant Physiol Plant Mol Biol 42 373–392 [Google Scholar]
- Zrenner R, Salanoubat M, Willmitzer L, Sonnewald U (1995) Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J 7 97–107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.