Abstract
Increasing pea (Pisum sativum) seed nutritional value and particularly seed protein content, while maintaining yield, is an important challenge for further development of this crop. Seed protein content and yield are complex and unstable traits, integrating all the processes occurring during the plant life cycle. During filling, seeds are the main sink to which assimilates are preferentially allocated at the expense of vegetative organs. Nitrogen seed demand is satisfied partly by nitrogen acquired by the roots, but also by nitrogen remobilized from vegetative organs. In this study, we evaluated the respective roles of nitrogen source capacity and sink strength in the genetic variability of seed protein content and yield. We showed in eight genotypes of diverse origins that both the maximal rate of nitrogen accumulation in the seeds and nitrogen source capacity varied among genotypes. Then, to identify the genetic factors responsible for seed protein content and yield variation, we searched for quantitative trait loci (QTL) for seed traits and for indicators of sink strength and source nitrogen capacity. We detected 261 QTL across five environments for all traits measured. Most QTL for seed and plant traits mapped in clusters, raising the possibility of common underlying processes and candidate genes. In most environments, the genes Le and Afila, which control internode length and the switch between leaflets and tendrils, respectively, determined plant nitrogen status. Depending on the environment, these genes were linked to QTL of seed protein content and yield, suggesting that source-sink adjustments depend on growing conditions.
The last two decades have seen an exponential increase in the number of plant sequences in databases and the explosion of investigations on the molecular functions and physiological roles of these genes. At the same time, new concepts, such as quantitative trait loci (QTL) mapping followed by the development of statistical tools, have emerged in quantitative genetics to identify the genes involved in the genetic variability of complex traits (Lander and Botstein, 1989). The functions of thousands of genes have been identified mainly through knockout mutant analysis (Østergaard and Yanovsky, 2004), but also through QTL identification (for review, see Paran and Zamir, 2003). These tools can now be used to address the question of phenotypic plasticity—which genes control plant functioning in which environments—and to provide some clues about which forces shaped natural variation and the strategies that should be used to breed more adapted cultivars (Paran and Zamir, 2003; Reymond et al., 2003; Koornneef et al., 2004; Mitchell-Olds and Schmitt, 2006). In this respect, Tonsor et al. (2005) proposed to analyze natural genetic variation in the model species Arabidopsis (Arabidopsis thaliana) to identify the role of genes having subtle, partially redundant, and/or environment-dependent effects on phenotypes, and to better understand gene interactions and pleiotropy. The study of natural genetic variation in crop species can provide complementary information on the genetic architecture of agronomic traits. Because of their utility for humans, whether for food, feed, or any other purpose, natural variation in crop species has been shaped both by natural and human selection and has long been studied in a range of environments, allowing for a large number of mutations to be described. Moreover, crop physiologists have acquired substantial knowledge of the crop plant cycle and functioning and the influence of the environment. In these respects, pea (Pisum sativum) is a good model crop species (Blixt, 1978; Marx, 1985; Ellis and Poyser, 2002).
Legumes have the unique property of being able to acquire nitrogen through symbiotic fixation of atmospheric dinitrogen by bacteria. Probably linked to this feature, legume seeds are often rich in protein. However, European legume crops have shown only moderate increase in yield in the last three decades (http://www.prolea.com) and great instability in performance, which has limited their development. Identifying the genes determining seed protein content and yield (Seed PC and Seed Y, respectively, in figures and tables) would allow, through marker-assisted selection, increased and stabilized seed protein content, which is an important component of seed nutritional value, while maintaining seed yield. Natural variation and QTL analysis for seed traits, including seed size, composition, and yield, have been reported in several legume crop species (Sax, 1923; Fatokun et al., 1992; Maughan et al., 2000; Hyten et al., 2004; Abbo et al., 2005; Nichols et al., 2006; Panthee et al., 2006; Upadhyaya et al., 2006). In pea, Blixt (1978) reported extensive variability for several seed traits in the 2,200 accessions of the Weibullsholm Pea gene bank. One thousand seed weight (1-seed W is seed weight in figures and tables) varied from 34 to 480 g, seed productivity varied from 0.4 to 113.2 g/plant, and seed protein content varied from 15.8% to 32.1%. QTLs for seed traits, including seed size, protein content, and yield, have been mapped (Timmerman-Vaughan et al., 1996, 2005; Tar'an et al., 2004).
Seed traits result from the integration of a series of processes occurring during the plant growth cycle and are controlled by both genotype and environment. Long-standing studies in crop physiology have provided a framework for describing plant functioning (Monteith, 1972, 1977; Pate, 1985; Ney et al., 1993; Gastal and Lemaire, 2002). In legumes, nitrogen acquisition relies both on symbiotic fixation of atmospheric nitrogen in root nodules and on soil nitrate assimilation by roots. The contribution of these two pathways to the global nitrogen nutrition of the plant varies according to the genotype and the environment. Early steps of symbiosis (plant-bacteria recognition, nodule initiation) do not seem to be major limiting factors in Europe, where soils contain inocula from diverse strains of Rhizobium leguminosarum, which form efficient nitrogen-fixing nodules with pea plants (Laguerre et al., 2003). Root growth, nodule development, and functioning are highly sensitive to variation in carbon supply from aerial parts of the plant (Voisin et al., 2003a, 2003b). In favorable conditions, symbiotic fixation and assimilation contribute 80% and 20%, respectively, of the nitrogen acquired (Salon et al., 2001), but environmental conditions—water status and structure of the soil, mineral nitrogen availability, and temperature—can modulate the respective contributions because symbiotic fixation is more susceptible than assimilation to environmental stresses (Sprent et al., 1988; Salon et al., 2001). During the seed-filling phase, assimilates are preferentially furnished to the seeds, which are the main sink, at the expense of vegetative organs and nodules. Nitrogen remobilization is then exploited to satisfy the high nitrogen demand of filling seeds. It occurs in almost all vegetative organs (Schiltz et al., 2005). In leaves, it is associated with the degradation of the photosynthetic machinery (Sinclair and de Witt, 1975, 1976; Peoples and Dalling, 1988; Schiltz et al., 2004), thus, in turn, influencing carbon fixation. Remobilized nitrogen accounts for a significant part of nitrogen accumulated in the seeds (Sinclair and de Witt, 1975; Schiltz et al., 2005).
Seed number (Seed N in figures and tables) and size are determined at different times. Seed number is determined during the phase of seed morphogenesis, where abortions can occur depending on assimilate availability (Ney et al., 1993). Seed size is determined by the rate and duration of growth. Seed growth rate is determined by the number of cotyledon cells (NcotCel; Munier-Jolain and Ney, 1998), which is fixed during embryogenesis and depends on the trophic conditions of the embryo (Weber et al., 1996; Lemontey, 1999). Thus, seed size genetic variability may be explained by cotyledon cell number variability (Munier-Jolain and Ney, 1995). The duration of seed growth is determined during the storage phase: Seed filling stops when the resources available for seed growth are exhausted or when the maximal size of the seed is reached. Seed protein content depends on the relative accumulation of starch and proteins (Lhuillier-Soundele et al., 1999). Seed protein content thus depends, on one hand, on nitrogen availability during seed filling and, on the other hand, on the embryo's intrinsic capacity to accumulate storage compounds, as exemplified by wrinkled seed peas. Pea seeds contain around 50% starch. In rugosus pea mutants, one of the five major genes controlling starch accumulation in the seed is defective (Wang and Hedley, 1985; Turner et al., 1990; Craig et al., 1998, 1999). In these mutants, the shape of the seed is wrinkled, starch accumulation is reduced, and seed protein content is elevated.
In this article, our goal was to evaluate the respective role of nitrogen source capacity and sink strength on genetic variability of seed protein content and yield in smooth-seeded peas. We assessed the roles of source capacity and sink strength in limiting seed weight and protein content in the field for eight pea genotypes of diverse origin. Then we mapped QTL for seed protein content and yield as well as QTL for variables describing the source-sink relationship in the plants in a population derived from the cross between two genotypes of contrasting plant morphology.
RESULTS
Genetic Variability of the Potential Seed Size and Protein Content of Eight Pea Genotypes
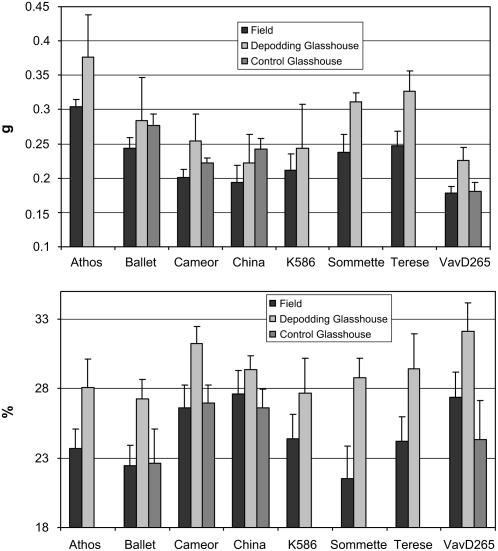
By maximizing the flux of assimilate to seeds at the beginning of seed filling (BSF) through depodding experiments in which only the pods on the second-flowering node were left to develop on the plant, we determined the potential seed size and protein content of eight genotypes (‘Ballet’, ‘Cameor’, ‘Térèse’, ‘Sommette’, ‘Athos’, VavD265, K586, and China). These potential values were compared to the performances of untreated pea plants in the field (Fig. 1). ANOVA revealed significant genotype, depodding treatment, and genotype by depodding treatment interactions (P < 0.001) for both seed weight and protein content. For most genotypes, the maximal protein content obtained after depodding was significantly higher than the field values.
Figure 1.
Seed weight (A) and seed protein content (B) of eight genotypes measured in three different conditions: plants were grown in glasshouse and all pods, except those on the second-flowering node, were removed (depodding treatment), control plants grown in glasshouse, and control plants grown in field trials.
Variation for Seed and Plant Development and Growth-Related Characters in the ‘Térèse’ × K586 Recombinant Inbred Population
RECOMBINANT INBRED LINE1 (RIL1), a population of 139 RILs derived from a cross between ‘Térèse’ and K586, a mutant obtained from ‘Torsdag’ (Laucou et al., 1998), was assessed in four one-row trellised field trials and one 10-m2 plot trial in 2000 and 2002. Three genes controlling plant architecture segregate in this population: Le, which controls internode length (Lester et al., 1997), Afila (Af), which controls the switch between leaflets and tendrils (Gourlay et al., 2000), and Rms6, controlling the degree of basal branching (Rameau et al., 2002). A range of plant characters describing plant development and growth was measured, seed protein content and productivity traits were determined, and variables related to nitrogen source capacity and seed sink strength were calculated. The ANOVA over the five trials showed highly significant effects of genotype, environment (P < 0.0001), and genotype × environment interaction (P < 0.01) for all traits analyzed. Phenotypic plasticity observed in the RIL population was consistent with changes in the rank of mean values observed for K586 and ‘Térèse’ among different environments for various traits. ANOVA in each trial revealed a significant effect of genotype for most traits (Supplemental Table S1). However, broad-sense heritability varied among traits and trials. Whereas phenological traits, such as the date of beginning of flowering (BegFlo) and plant height (Height), had the highest h2, calculated variables (quantity of nitrogen [QN] remobilized from vegetative parts to the seeds [QNmob] and QN accumulated during seed filling [QNacc]), respectively, had the lowest. Values obtained from plants harvested in plots were less heritable than values obtained from plants harvested in rows.
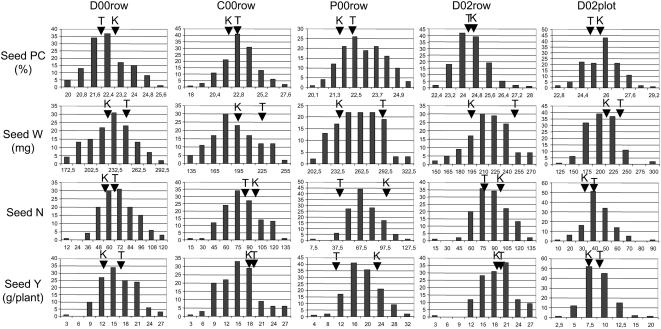
In spite of similar values being measured for ‘Térèse’ and K586 for a number of traits, high genetic variability for these traits was observed in the RIL1 population (Fig. 2; Supplemental Table S1). Notably, the mean seed protein content over the five environments was similar in ‘Térèse’ and K586 (23% and 23.4%, respectively), but varied from 20.6% to 27.3% in the RIL1 population, revealing transgressions toward higher values. Mean seed weight and straw nitrogen content (%Nstraw) varied from 215 to 249 mg and 0.96% to 1.00%, respectively, in ‘Térèse’ and K586 and varied from 154 to 291 mg and 0.68% to 1.74% in RIL1. This was even more dramatic for mean seed weight per plant (seed yield) and seed number per plant (seed number), which were also similar in ‘Térèse’ and K586 (15.1 and 16.2 g/plant and 61 and 76 seeds/plant, respectively), but varied from 2.4 to 28.8 g/plant and 11.9 to 125.7 seeds/plant in RIL1. The large range of variation and transgressive segregations observed for most traits in RIL1 (Fig. 2) suggested complex control of these traits, with positive alleles shared between the two parents of the RIL population.
Figure 2.
Frequency distribution of the seed protein content, seed weight, seed number, and seed yield of the ‘Térèse’ × K586 recombinant inbred population in the five field trials: n = 2; Dijon, France (2000) one-row trial, D00row; Dijon, France (2002) one-row trial, D02row; Dijon, France (2002) plot trial, D02plot; Chartainvilliers, France (2000) one-row trial, C00row; Premesques, France (2000) one-row trial, P00row. The arrowheads indicate the mean value of the parental lines. K, K586; T, ‘Térèse’.
The range of variation observed among the five environments was significant, but lower than the range of variation observed among RIL1 inbreds. The mean seed protein content of RIL1 varied from 22.4% to 25.7% among environments (Dijon, France [2000], row trial; Dijon, France [2002], row trial). The mean seed yield and number ranged from 8.9 to 18.8 g and 44 to 87 seeds per plant (Dijon, France [2002], plot and row trials, respectively). The mean seed weight varied from 191 to 262 mg (Premesques and Chartainvilliers trials, respectively). The %Nstraw varied from 0.9% to 1.18% (Dijon and Premesques, France [2000] trials, respectively). These variations suggested contrasting growing conditions in the different environment, reflecting contrasted climates, soils, and mode of cultivation.
Correlations between Seed Traits and Plant Development and Growth-Related Traits
In all environments, seed number was highly correlated to seed yield and to the QN accumulated in seeds and straw at harvest (Supplemental Fig. S1). It was negatively correlated to seed weight in all trials but Chartainvilliers. Seed number was positively correlated with the date of the end of flowering (EndFlo), the duration of flowering, plant height, as well as the QN per plant at BSF (QNBSF) in all one-row trials but not in the plot trial. Depending on the environment, seed weight was weakly positively correlated with seed yield, seed protein content, and date of harvest (Harvest), and weakly negatively correlated with the dates of BegFlo and EndFlo and number of basal branches (Nbranch). It was positively correlated with the mean cotyledon cell volume (VcotCel), but only weakly correlated with the NcotCel. In all environments, seed yield was highly correlated to seed number and to straw biomass at harvest (DMstraw). It was highly correlated with the QNacc and weakly correlated with the QNmob. In all environments, except the Dijon, France (2002) one-row trial, seed protein content was highly positively correlated to plant nitrogen content at BSF and %Nstraw at harvest. The Dijon, France (2002) one-row trial was the only environment in which plant biomass and nitrogen content at BSF (DMBSF and %NBSF, respectively, in figures and tables) were not significantly negatively correlated. Many correlations were modified when calculated for the subpopulations carrying the le or Le allele. For example, the correlation between plant nitrogen content and seed protein content was decreased when calculated for the subpopulations carrying the le or Le allele (Supplemental Fig. S1), suggesting a pleiotropic effect of this gene on these traits.
Mapping QTL for Seed Protein Content and Yield Components
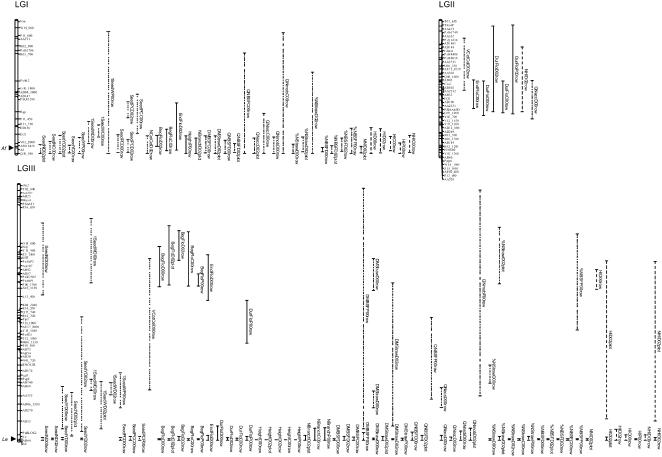
To dissect the genetic control of seed protein content and yield variability and identify the genes determining this variability, we mapped QTL for seed protein content and yield components together with QTL for variables suggested by crop physiology models (indicators of sink strength and of source capacity; Larmure and Munier-Jolain, 2004). A framework map was computed from marker data reported in Laucou et al. (1998), Loridon et al. (2005), and Aubert et al. (2006). Composite interval mapping using a log of the odds (LOD) threshold of 2.6 representing a type I risk of 10% genome wide (1,000-permutation test) identified 261 QTL across five environments for all traits measured (Fig. 3; Supplemental Table S2): 68 and 64 QTL were detected for 20 variables in one-row trials in Dijon, France (2000; D00row) and Dijon, France (2002; D02row); 45 QTL were detected for 19 variables in the plot trial of Dijon, France (2002; D02plot), and 47 and 37 QTL were detected for 18 variables in one-row trials of Premesques, France (2000; P00row) and Chartainvilliers, France (2000; C00row).
Figure 3.
CIs of QTL detected by composite interval mapping for all traits in five field trials. For each QTL, the names of the variable and of the trial are indicated. Dotted lines indicate seed trait QTL (Seed N, Seed Y, 1-seed W, Seed PC, NcotCel, and VcotCel), full lines indicate phenology and morphology trait QTLs (BegFlo, EndFlo, Harvest, Height, NBranch); dashed lines indicate source capacity QTL (DMBSF, %NBSF, DMstraw, %Nstraw; QNBSF, QNmob, and QNacc; NNI); dashed and dotted lines indicate QTL of indicators of assimilate allocation (HI and NHI). Field trials: Dijon, France (2000) one-row trial, D00row; Dijon, France (2002) one-row trial, D02row; Dijon, France (2002) plot trial, D02plot; Chartainvilliers, France (2000) one-row trial, C00row; Premesques, France (2000) one-row trial, P00row. Arrowheads indicate the Af, Le, and Rms6 loci.
Fourteen QTL for seed protein content were detected in five environments, corresponding to eight genomic regions, each region controlling this trait in one to three environments. Positive additive effects were shared equally between the two parents, with five regions showing a positive effect of the K586 allele (LGI-85, LGI-Af, LGIV-17, LGV-81, and LGVI-120 cM), and two of the ‘Térèse’ alleles (LGIII-Le and LGVII-30 cM). The QTL located on LGV at 160 and 170 cM showed opposite effects, suggesting that this region could harbor two distinct QTL despite their overlapping confidence intervals (CIs). QTL accounted for from 9% to 46% of the genetic variation. Twenty-two QTL were detected for seed weight in five environments, corresponding to nine genomic regions, each controlling this trait in one to four environments. Six regions were associated with a positive effect from ‘Térèse’, the large-seeded parent (LGIII-67, LGIII-189, LGV-165, LGVII-28, LGVII-104, and LGVII-150 cM), and three from K586 (LGI-98, LGIV-17, and LGVI-30 cM). QTL comprised from 9% to 31% of the genetic variation. Nine QTL controlled seed number in five environments, corresponding to five genomic regions, each controlling this trait in one to three environments. Four regions were associated with a positive effect from K586 (LGI-Af, LGIII-91, LGIII-238, and LGVII-97 cM) and one from ‘Térèse’ (LGV-169). QTL accounted for from 10% to 44% of the genetic variation. Finally, 11 QTL corresponding to six genomic regions were detected for seed yield, each controlling this trait in one to three environments. Four regions were associated with positive effects of K586 (LGI-Af, LGIII-Le, LGIV-96, and LGVII-87) and two of ‘Térèse’ (LGIII-207 and LGVII-105). QTL accounted for from 9% to 53% of genetic variation.
Colocalization of QTL for Seed and Plant Growth-Related Traits
Altogether, QTL detected for seed traits mapped to 16 genomic regions, which corresponded to three configurations (Fig. 3; Supplemental Table S2): (1) regions affecting many traits, including plant morphology and phenology, as well as plant biomass, nitrogen source capacity, and seed production (LGI-Af, LGIII-84, LGIII-Le, LGIV-95, LGV-Rms6, LGV-RbcS, LGVII-95, LGVII-104 cM); (2) regions harboring only seed trait QTL (LGI-Rgp, LGVI-30, LGVI-120, LGVII-150, LGIV-17 cM); and (3) regions controlling seed traits and traits related to nitrogen availability (LGIII-189, LGIII-203, LGVII-28). We also searched for QTL for VcotCel and NcotCel as indicators of sink strength. Two genomic regions were involved in the genetic variability of cotyledon cell number (LGI-Af only at D00row, LGIV-70 at D00row and D02row) and four genomic regions in the genetic variability of cotyledon cell size (LGII-25, LGIII-114 in one of the two environments, LGIV-70 and LGVII-40 in the two environments analyzed).
Loci Controlling Plant Architecture Are Associated with QTL for Seed and Plant Growth-Related Traits
Numerous QTL were located close to developmental genes Le, Af, and Rms6 (beginning, end, and duration of flowering, height and vegetative biomass, but also seed weight, number, yield, and protein content; Fig. 3). In all environments, except C00row, the af mutation was associated with a negative effect on plant nitrogen content at BSF, and in three of five environments, with a negative effect on %Nstraw at harvest and on indicators of nitrogen availability for seed filling (QNBSF, QN remobilized and accumulated during seed filling). In two of five environments, af was also associated with a negative effect on seed protein content and in three other environments with a negative effect on seed yield. As expected, the le allele was associated with reduced plant height in all environments, but also to reduced vegetative biomass at BSF and at harvest, with increased plant nitrogen content at BSF, with later flowering and with a shorter duration of flowering (except in D02plot). It was associated with an increase of the Nbranch, except in D00row and D02row, where seed number and yield were reduced. Seed protein content was increased in three environments (D00row, C00row, and P00row). The Rms6 allele was associated with a lower Nbranch and in four environments, with an earlier and longer flowering. In D00row and D02row, Rms6 was associated with a reduction in biomass at BSF and a slight increase in plant nitrogen content at BSF. In D02plot, it was associated with an increase in plant height and a decrease in seed protein content.
QTL for Seed and Plant Traits Were Supported by Independent Experiments
Despite QTL instability across environments, including for the most heritable traits (dates of flowering, seed weight), most QTL were consistent in independent environments. Some QTL instability could be due to variations in heritability. For example, fewer QTL were detected in the D02plot trial than in the D02row trial, probably linked to reduced genetic variation for several traits and particularly for dry matter accumulation in the D02plot trial (Supplemental Table S1), probably because of interplant interactions. QTL may also vary according to environmental conditions if adjustments to different conditions involve different regulatory networks, as suggested by the different patterns of trait correlation in the different environments (Supplemental Fig. S1). Looking for common QTL in different environments may reveal regulatory networks involved in the adaptation to limiting factors occurring in these environments. For example, for seed yield, QTL III-203 acted in environments D00row, D02row, and D02plot, whereas LGI-af acted in D02plot, C00row, and P00row (Table I). Finally, a limiting factor occurring in an environment may have revealed the genetic variability of susceptibility to this factor.
Table I.
Common QTL and correlation coefficients among environments, for Seed PC, 1-seed W, Seed N, and Seed Y
Indicated for each of the seed traits: above the diagonal, the R2 value (%) and the genomic region or candidate gene close to the QTL detected in common for all pairs of environments; below the diagonal, the correlation coefficients (R) and the level of significance (***, P < 0.01; **, P < 0.01; *, P < 0.05; ns, not significant) for all pairs of environments. NQTL, Number of QTLs detected in each environment for each variable.
| Traits and Trials | D00row | D02row | D02plot | C00row | P00row | NQTL |
|---|---|---|---|---|---|---|
| Seed PC | ||||||
| D00row | Af 21% RbcS 13% | VI-120 13% | Le 29% | Le 23% | 4 | |
| D02row | 0.24* | Rgp 13% | 4 | |||
| D02plot | 0.45*** | 0.40*** | 2 | |||
| C00row | 0.42*** | 0.21* | 0.25* | Le 23% | 2 | |
| P00row | 0.40*** | ns | ns | 0.25* | 2 | |
| 1-Seed W | ||||||
| D00row | III-189 20% IV-17 14% Gbsts2 9% VII-104 19% | III-189 20% IV-17 14% Gbsts2 9% | I-98 18% RbcS 9% | I-98 18% III-189 20% VII-104 19% | 7 | |
| D02row | 0.79*** | III-189 23 IV-17 14% Gbsts2 12 | III-189 23 VII-104 19% | 4 | ||
| D02plot | 0.71*** | 0.84*** | VI I-28 10% | III-189 18% | 4 | |
| C00row | 0.65*** | 0.52*** | 0.41** | I-98 11 | 4 | |
| P00row | 0.84*** | 0.74*** | 0.67** | 0.67*** | 3 | |
| Seed N | ||||||
| D00row | Le 42% | VI-87 12% | VI-87 12% | 3 | ||
| D02row | 0.64*** | 1 | ||||
| D02plot | ns | ns | Af 11% | 2 | ||
| C00row | 0.37*** | 0.21* | ns | VI-87 12% | 2 | |
| P00row | 0.43*** | 0.43*** | ns | 0.39*** | 1 | |
| Seed Y | ||||||
| D00row | III-203 10% Le 53% | III-203 12% | 3 | |||
| D02row | 0.73*** | III-203 10% | 2 | |||
| D02plot | ns | ns | Af 12% | Af 10% | 3 | |
| C00row | 0.36*** | 0.31*** | ns | Af 10% | 2 | |
| P00row | 0.44*** | 0.45*** | ns | 0.42*** | 1 |
Identification of Candidate Genes
The projection of the pea functional map (Aubert et al., 2006) onto the map of QTL detected for RIL1 in this study allowed us to identify a number of candidate genes in addition to Le, Af, and Rms6. A flowering QTL was mapped in all environments to the LGV-49-cM region, where Det (=TFL1a) is located. This gene has been shown to be involved in the regulation of flowering time and of inflorescence architecture (Foucher et al., 2003).
A seed protein content QTL was mapped in the genomic region LGI-85, which harbors the gene Rgp. This gene has a putative role in cell wall biosynthesis (Delgado et al., 1998). Another QTL for seed protein content and seed number was mapped in the LGV-160- to 170-cM genomic region where two potential candidate genes are located: the dwarfism gene Ls, involved in an early step of GA biosynthesis (Ait-Ali et al., 1997), and the gene encoding for the small Rubisco subunit (Rbcs4). A QTL for seed protein content and weight was mapped in the LGVII-30-cM genomic region, in the vicinity of an N3-like gene homologous to the Medicago truncatula nodulin MtN3 described by Gamas et al. (1996) and of a fabatin-like gene, which is expressed in filling seeds. Several QTL mapped in the LGVII-95-cM genomic region, which harbors the Ptrans (LGVII-97) and Htrans (LGVII-99) genes encoding a translocator of phosphate and a transporter of hexose, respectively. The developmental genes Cry and Stp are also located in this region (Ellis and Poyser, 2002).
A stable seed weight QTL, detected in four of five environments, mapped in the LGIII-189 genomic region in the vicinity of the candidate gene PepC (LGIII-181) encoding a phosphoenolpyruvate carboxylase. In the same region, another candidate could be the PGK1 gene encoding a phosphoglycerate kinase (LGIII-189). Other seed weight QTL were found (1) in the LGIV-17 region, which harbors the gene Elsa (LGIV-14) encoding for a Cys proteinase, a marker of monocarpic senescence in pea (Pic et al., 2002) and Sucsyn (LGIV-19), encoding a Suc synthase map; (2) in the LGVI-30 region, close to the gene Gbsts2, encoding starch synthase II; and (3) in the LGVII-150 region, near the Pip2 gene, which encodes a transmembrane protein.
Effect of Le and Af on Seed and Plant Traits in Isogenic and Near-Isogenic Backgrounds
To test for the effects of Le and Af in an isogenic background, isogenic and near-isogenic lines (NILs) were sown in 2003 and 2005 in three-block, replicated-design one-row trials at the Institut National de la Recherche Agronomique (INRA; Dijon, France). The mutant allele le (whether le-1 or le-3) was associated with significantly lower aerial vegetative plant biomass at BSF, QNBSF, QNmob, and QNacc during seed filling, and with higher plant nitrogen content at BSF and increased Nbranch (Table II). Seed number and yield were reduced, but seed protein content was unchanged. Effects varied according to the background genotype, suggesting epistatic interactions. The mutant allele af led to lower plant nitrogen content at BSF and to lower seed weight. No significant effect was observed on seed number, yield, or protein content (Table III).
Table II.
Effect of different alleles of Le on plant morphology, plant source capacity, and seed traits, in isogenic lines and NILs in trellised field trials in Dijon, France (2003 and 2005)
Significant effect of Le (F test; *, P < 0.05; **, P < 0.01; ***, P < 0.00).
| Traits | ‘205’
|
‘Térèse’
|
‘Torsdag’
|
||||
|---|---|---|---|---|---|---|---|
| Le | le-1 | Le | le-1 | Le | le-3 | ||
| Height*** | Mean | 121.00 | 69.00 | 149.17 | 74.17 | 137.50 | 67.50 |
| sd | 13.42 | 6.52 | 15.94 | 11.14 | 11.73 | 7.58 | |
| Nbranch*** | Mean | 2.24 | 2.36 | 1.57 | 2.23 | 1.70 | 1.76 |
| sd | 0.39 | 0.25 | 0.39 | 0.32 | 0.14 | 0.30 | |
| DMBSF*** | Mean | 20.12 | 15.69 | 20.80 | 19.12 | 26.26 | 15.60 |
| sd | 5.08 | 2.66 | 2.72 | 4.41 | 8.85 | 3.52 | |
| %NBSF*** | Mean | 3.60 | 4.03 | 3.09 | 3.48 | 3.73 | 4.38 |
| sd | 0.31 | 0.38 | 0.10 | 0.15 | 0.53 | 0.50 | |
| DMStraw*** | Mean | 18.11 | 12.16 | 16.15 | 14.34 | 22.19 | 13.75 |
| sd | 5.52 | 1.75 | 2.10 | 3.07 | 3.16 | 1.39 | |
| %Nstraw*** | Mean | 1.05 | 1.32 | 0.78 | 0.93 | 0.80 | 1.21 |
| sd | 0.13 | 0.17 | 0.10 | 0.06 | 0.08 | 0.17 | |
| Seed N*** | Mean | 142.60 | 138.96 | 97.30 | 88.97 | 139.40 | 81.00 |
| sd | 29.13 | 19.23 | 7.32 | 18.06 | 10.28 | 13.15 | |
| 1-Seed W | Mean | 210.20 | 156.31 | 246.30 | 249.60 | 232.23 | 242.00 |
| sd | 60.82 | 7.19 | 15.62 | 10.39 | 11.01 | 13.96 | |
| Seed PC | Mean | 23.77 | 24.49 | 26.39 | 25.73 | 26.73 | 27.40 |
| sd | 2.22 | 2.31 | 1.89 | 0.87 | 1.02 | 1.34 | |
| Seed Y*** | Mean | 29.14 | 21.73 | 23.90 | 22.29 | 32.42 | 19.54 |
| sd | 5.93 | 3.09 | 1.57 | 5.20 | 3.24 | 3.03 | |
| QNBSF** | Mean | 71.44 | 63.11 | 64.38 | 66.26 | 94.20 | 67.12 |
| sd | 13.57 | 11.26 | 8.68 | 14.89 | 19.68 | 11.40 | |
| Qnmob** | Mean | 52.49 | 46.86 | 51.88 | 52.87 | 76.52 | 50.64 |
| sd | 9.32 | 14.08 | 8.01 | 13.60 | 17.29 | 9.76 | |
| Qnacc* | Mean | 57.44 | 38.93 | 49.14 | 38.96 | 62.04 | 35.25 |
| sd | 23.33 | 8.71 | 10.96 | 21.37 | 20.48 | 21.31 | |
Table III.
Effect of different alleles of Af on plant morphology, plant source capacity, and seed traits in NILs of ‘Térèse’
*, Significant F test (P < 0.01).
| Traits | ‘Térèse-Af’ | ‘Térèse-af’ |
|---|---|---|
| Height | 75.00 | 74.17 |
| Nbranch | 2.34 | 2.23 |
| DMBSF | 18.47 | 19.12 |
| %NBSF* | 3.9 | 3.48 |
| DM Straw | 13.08 | 14.34 |
| %Nstraw | 1.00 | 0.93 |
| Seed N | 79.20 | 88.97 |
| 1-Seed W* | 269.1 | 249.6 |
| Seed PC | 26.1 | 25.7 |
| Seed Y | 21.4 | 22.3 |
| QNBSF | 71.7 | 66.3 |
| QNmob | 58.7 | 52.9 |
| QNacc | 30.4 | 39.0 |
DISCUSSION
Improving the level and stability of seed protein content while maintaining seed yield is an important challenge for pea breeding. In this study, we carried out an integrated analysis of source-sink relationships to dissect the genetic control of seed protein content and yield, two complex traits controlled by numerous small-effect genes and submitted to environmental variations (Matthews and Arthur, 1985; Karjalainen and Kortet, 1987; Atta et al., 2004).
Source Capacity Is a Major Limiting Factor of Seed Protein Content and Weight in Pea
A model predicting pea seed protein content (Larmure and Munier-Jolain, 2004) indicated that genetic variability for this trait can originate from (1) variability in the availability of nitrogen for seed filling, whether remobilized from vegetative parts at BSF and/or assimilated during seed filling; and (2) variability in the maximal rate of nitrogen accumulation in the seed arising from variations in the rate of accumulation of nitrogenous storage compounds. By maximizing the flux of assimilates from the mother plant to the seeds through depodding experiments, we estimated the potential seed weight and seed protein content in eight genotypes of various origins. We identified significant genetic variability both for the maximal rate of nitrogen accumulation in the seeds and for the availability of nitrogen assimilates. In particular, a contrasting response to variations in assimilate availability was observed among ‘Térèse’ and K586, the two parents of our mapping population, the seed protein content of ‘Térèse’ being more limited than K586. For most genotypes, seed protein content was lower in control plants than in depodded plants, indicating that nitrogen assimilate availability is often limiting, even in a nitrogen-fixing species.
Colocations between QTL Controlling Seed Traits and Plant Characteristics Provide Clues to the Identification of Candidate Genes
To identify the genetic factors responsible for seed protein content and yield variation, we have searched for QTL for seed traits and for indicators of sink strength (NcotCel) and source nitrogen capacity (QNBSF and QNacc, nitrogen nutrition index [NNI]). Very few studies identified concomitantly QTL for seed traits and for plant traits potentially associated with source capacity. In grain legumes, some studies searched QTL for seed traits and plant developmental traits, such as flowering, maturity, leaf area, height, node numbers (Mansur et al., 1993; Chung et al., 2003; Zhang et al., 2004; Timmerman-Vaughan, et al., 2005), but, to our knowledge, none searched QTL for seed traits and plant traits potentially associated with nitrogen source capacity. In cereals, a few studies reported clusters of QTL for plant nitrogen utilization efficiency, nitrogen remobilization, and grain yield (Tuberosa et al. 2002; Coque and Gallais, 2006; Laperche et al., 2006).
In this study, we detected 261 QTL across five environments for all traits measured (Fig. 3; Supplemental Table S2). Most QTL were consistent in independent environments and some QTL may correspond to QTL reported for other pea mapping populations. A seed protein QTL located on LGVI between the markers G4_2000 and B7_1750 and a seed yield QTL located on LGVII near marker G12_650 identified in pea by Tar'an et al. (2004) could correspond to the QTL on LGVI-110 cM and on LGVII between 87 or 105 cM in our study. Other authors described a syntenic seed weight QTL in legumes located on LGIII near marker M27 (Fatokun et al., 1992; Timmerman-Vaughan et al., 1996, 2004), which probably corresponds to our seed weight QTL on LGIII-189 cM.
Most QTL for seed and plant traits mapped in clusters. These clusters of QTL can correspond to a single gene having pleiotropic effects on different traits. We hypothesize that different types of clusters may correspond to different underlying processes and genes. Alternatively, clusters of QTL can correspond to closely linked genes. Clusters of QTL for seed traits only may correspond to genes specifically involved in seed development and metabolism. These genes may control the processes determining sink strength and the rate of assimilate accumulation in seeds. An example of such a QTL was identified in tomato (Lycopersicon esculentum; Fridman et al., 2000). In this study, the genomic region LGI-85 harbored a seed protein content QTL and the gene Rgp. This gene is thought to play a role in cell wall biosynthesis (Delgado et al., 1998) and, interestingly, was found to be expressed in M. truncatula filling seeds (Gallardo et al., 2003). Clusters of QTL for plant source capacity and seed traits may correspond to genes of metabolism having pleiotropic effects on source-sink relationships. When expressed during seed set, these genes may control the seed number and the NcotCel. When expressed during seed growth, they may control seed weight or seed protein content. In maize (Zea mays), two cytosolic Gln synthase isoforms (gln1-3 and gln1-4) expressed in the leaves were found to control seed number and seed weight, respectively (Martin et al., 2006). In this study, a seed weight QTL detected in four of five environments mapped in the vicinity of the candidate gene PepC (LGIII-181). In the same region, another candidate is the PGK1 gene encoding a phosphoglycerate kinase (LGIII-189). Clusters of QTL for plant morphology, phenology, growth, nitrogen status, and seed traits may correspond to developmental genes controlling vegetative development and/or flowering and having pleiotropic effects on plant morphology, source capacity, and, eventually, on seed protein content and productivity. By modifying plant morphology and phenology, these genes may modify the size and/or structure of the different plant organs and thus assimilate partitioning among organs. QTL regions encompassing the development genes Le and Af lie in this category. Similarly, in Arabidopsis, the gene erecta controls a range of phenotypic traits (Koornneef et al., 2004). In rice (Oryza sativa), a gene involved in erect leaf phenotype is associated with higher grain yield (Sakamoto et al., 2006). In wheat (Triticum aestivum), the dwarfism gene Rht-B1 is associated with numerous QTL, including root development and grain yield QTL (Laperche et al., 2006).
Genes Determining Plant Architecture Are Linked to QTL of Seed Protein Content and Yield
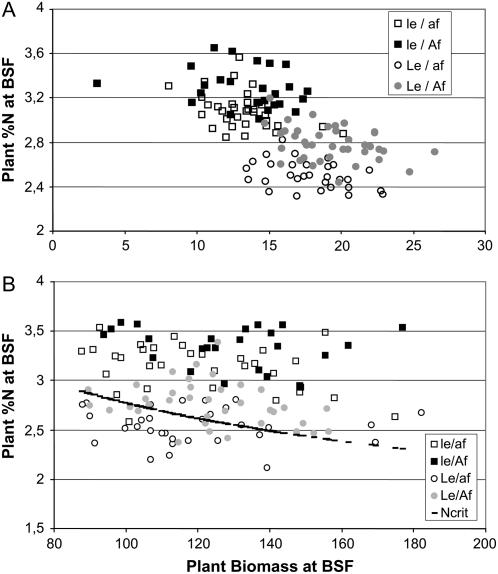
Both the le and af mutations were introduced into older varieties to produce current field pea varieties adapted to mechanical harvesting. Shorter internodes and tendrils replacing the leaflets answer a major concern in the pea crop, its standing ability at harvest under normal field conditions. In this study, the af mutation was associated with a negative effect on plant nitrogen content in four of five environments and the le mutation was associated with increased plant nitrogen content in all environments. The relationship between plant nitrogen content and biomass accumulation serves as the basis for the diagnosis of crop nitrogen status (Gastal and Lemaire, 2002). In the mapping population RIL1 (Fig. 4), this relationship differentiated lines according to le and af, indicating that Le/af lines may encounter suboptimal nitrogen status as compared to their le/Af counterparts. Consistently, both Le and af were associated with reduced NNI and Kof et al. (2006) observed a reduction of root weight in af NILs. These results suggest that genes such as af or le, which affect the development of aerial vegetative organs (in le mutants, GA concentrations were not affected in developing seeds nor in roots, but only in growing shoots; Swain et al., 1995; Yaxley et al., 2001), may induce root modifications. Other examples of cross talk between roots and shoots have been reported in legumes (e.g. the shoot control of the hypernodulating phenotype of sym29 and Har1 mutants; Krusell et al., 2002).
Figure 4.
Relationship between plant nitrogen content and plant biomass at BSF in the RIL1 population. Different allelic combinations at Le and Af genes are indicated. A, Dijon, France (2000) one-row field trial. Plant biomass is expressed in grams/plant. B, Dijon, France (2002) plot trial. Plant biomass is expressed in g/0.15 m2 square. In this trial, Ncrit was calculated.
Limiting nitrogen availability during seed set may reduce seed number, whereas reduced nitrogen availability per filling seed may reduce seed protein content (Larmure and Munier-Jolain, 2004). Consistently, reduced plant nitrogen content in both Le and af genotypes was associated with reduced seed protein content in some environments. This suggests that seed protein content could be regulated by plant nitrogen content as an indicator of plant nitrogen status. Plant nitrogen content represents the pool of remobilizable nitrogen. Schiltz et al. (2005) showed that remobilized nitrogen is a unique pool, distributed to seeds independently from their position on the plant. However, the effect of le and af on seed traits varied among environments. The af mutation was associated with a negative effect on seed number and seed yield in three environments, whereas seed protein content was reduced in the other two environments. And we could not detect any effect of af on seed protein content, number, and yield in NILs, but detected a reduction in seed weight. Earlier reports gave conflicting results on the effect of the af mutation on seed protein content (Kielpinski and Blixt, 1982; Matthews and Arthur, 1985; Sagan et al., 1993). Different factors may be responsible for these discrepancies: (1) different environmental conditions (e.g. in the 2003 and 2005 experiments on af NILs, the mean temperature rose above 25°C for several days during seed development); (2) epistatic interactions in the RIL1 population (significant interaction effects were found between af and markers from the LGVII-Ptrans region for plant nitrogen content at BSF in three of five environments; P < 0.005; data not shown); (3) af may not be responsible for seed protein content variability, but may be closely linked to the effective gene; or (4) several genes with compensatory effects may lie in the region introgressed in NILs. Deeper analysis of source-sink relationships in af/normal isogenic lines would be useful. Once af is identified at the molecular level, it will be possible to screen TILLING pea resources to find isogenic mutants for this gene and confirm its effect on seed traits. The le mutation was associated in RIL1 with later flowering and shorter duration of flowering. In two environments, seed number and seed yield were reduced. In the other environments, the reduction in height was compensated for by an increased NBranch and no effect was detected on seed number and seed yield in these environments. In le isogenic and NILs, seed number and yield were reduced, but seed protein content was unchanged. Consistently, Blixt (1978) mentioned for trellised plants a decreasing effect of le on seed yield through an effect on pod number and Swain et al. (1995) reported a slight maternal effect of le-3 on seed number. A way forward in pea breeding could be to try to compensate for the limitations induced by le and af (reduced seed protein content for af and seed number of le).
CONCLUSION
In this study, we found that seed protein content and seed weight may be limited both by sink strength and by source capacity. In our mapping population, source capacity was the major source of genetic variation for seed protein content and productivity. The developmental genes Le and af were linked to the most stable and significant seed protein content and yield QTL. However, QTL regions can cover several closely linked genes and further study will be necessary to unravel the molecular basis of the detected seed trait QTL. Novel tasks should be undertaken: (1) increase the power of detection of QTL, for example, by using multiparent populations (Blanc et al., 2006); (2) study the effect of candidate genes in isogenic backgrounds when possible; (3) identify the genetic determinants of nitrogen source capacity and seed sink strength by mapping QTL for traits associated with root nitrogen assimilation efficiency and seed capacity to synthesize storage proteins; and (4) characterize limiting environmental factors and address the molecular basis of seed trait phenotypic plasticity.
MATERIALS AND METHODS
Plant Material
Eight genotypes ( ‘Ballet’, ‘Cameor’, ‘Térèse’, ‘Sommette’, ‘Athos’, VavD265, K586, and China) were used to determine, by depodding experiments, the seed protein content and weight potential. These genotypes were described in Baranger et al. (2004). The mapping population, RIL1, comprised 139 RILs derived from a cross between ‘Térèse’ and K586, a ramified mutant obtained from ‘Torsdag’ (Laucou et al., 1998). Three genes controlling plant architecture segregate in this population: Le controls internode length, Af controls the switch between leaflets and tendrils, and Rms6 controls the degree of basal branching. To confirm the effect of Le and Af on seed nitrogen content and productivity, isogenic and NILs obtained for these genes were tested in field trials. Two pairs of isogenic lines for Le were kindly provided by J. Ross (205+ and 205−, and ‘Torsdag’ and NGB5839, University of Tasmania). NILs for Le and Af (‘Térèse’, ‘Térèse-Af’, ‘Térèse-Le’) were kindly provided by C. Rameau (INRA-Versailles).
Glasshouse Depodding Experiments
Three experiments were conducted successively in glasshouses in March 2001, March 2004, and June 2004 in Dijon, France. The eight genotypes (‘Ballet’, ‘Cameor’, ‘Térèse’, ‘Sommette’, ‘Athos’, VavD265, K586, and China) were sown in 2001. Only four of the eight genotypes were sown in 2004 (‘Ballet’, ‘Cameor’, VavD265, and China). Plants were grown in 5-L pots containing a sterile mix (1:1 [v/v]) of atapulgite and expanse clay. The temperature and minimal daylength were controlled (22°C/16°C, 16-h photoperiod). Five days a week, plants were watered with a nutritive solution at 4.5 mEq of nitrogen and with deionized water otherwise. The flowers of the three first-flowering nodes were tagged on the day of pollination. From 12 d after pollination (at BSF) to maturity, pods from the first-, third-, and subsequent flowering nodes were removed. Only the pods of the second-flowering node were left to develop until the end of desiccation. Then, seed weight was measured and seed nitrogen content was determined according to the Dumas method. In 2004, we also kept control plants on which all flowering nodes were left to develop and only seeds from the second-flowering node were harvested and analyzed for their seed size and seed nitrogen content.
Field Trials
Field data obtained from 1998 to 2005 for the eight genotypes (‘Ballet’, ‘Cameor’, ‘Térèse’, ‘Sommette’, ‘Athos’, VavD265, K586, and China) were gathered to compare the seed protein content and size obtained on depodded plants to the values obtained in the field (Supplemental Table S3). The mapping population RIL1 was sown in three field trials in 2000, in a two-block replicated design (March 7, 2000 at INRA-Dijon, Domaine d'Epoisses 21, France; March 17, 2000 at Nickerson, Chartainvilliers 28, France, and at Serasem, Presmesques 59, France). In these trials, a plot consisted of a row of 40 plants grown on trellises. To test for the effect of lodging and interplant competition on the QTL detected in these first trials, the population was sown again in March 5, 2002, at INRA, Dijon, Domaine d'Epoisses 21, France, in a two-block one-row trial, and in a two-block plot trial, where plots were 8.5 × 1.2 m2, sown with 90 seeds m−2. These 10-m2 plots were separated by a barley (Hordeum vulgare) plot to avoid competition between morphologically different pea (Pisum sativum) genotypes. To test for the effect of Le and Af in isogenic background, ‘Térèse(le-1/af)’, ‘Térèse(le-1/Af)’, ‘Térèse(Le/af)’, ‘Torsdag(Le/Af)’, ‘Torsdag(le-3/Af)’, ‘205(Le/Af)’, ‘205(le-1/Af)’ isogenic, and NILs were sown on February 26, 2003 and March 17, 2005 in three-block replicated one-row trials at INRA-Dijon, Domaine d'Epoisses 21, France. Different types of measurements describing plant development and growth were done and parameters related to nitrogen availability were calculated. Phenology and morphology traits were scored along the plant life cycle (BegFlo, EndFlo, Harvest, Height, Nbranch). In one-row trials, samples of 10 plants were harvested at BSF and after seeds had ripened (Harvest). In the plot trial, plants located in squares of 60 × 25 cm2 delineated at emergence in the middle of the plots, were harvested at BSF and at harvest. At BSF, plant dry weight (DMBSF) was measured after 48 h at 80°C and plant nitrogen content (%NBSF) was estimated by near-infrared spectroscopy. At harvest, seed productivity and its components (Seed N, Seed Y, and 1-seed W) were measured as well as DNStraw. Seed and %Nstraw were estimated by near-infrared spectroscopy. Seed protein content was calculated from seed nitrogen content.
Protein and Nitrogen Content Determination
Seed samples were ground at 0.2 mm with a ZM100 Retsch grinder. Plants harvested at BSF and straws harvested at plant maturity were ground at 1.0 mm with a SM100 Retsch grinder. For near-infrared predictions of nitrogen content, we used a NIRS 6500 (Foss) apparatus equipped with an autosampler module and a set of 48 small ring cups. The calibrations for seeds, plants at BSF, and straws were developed with Math Treatment: PLS SNV Detrend 1.4.4.1. The chemical reference method for nitrogen content used in our laboratory is the Kjeldahl determination norm (NF V07-350). Each year, new equations are developed and validated by comparing the predicted values with new reference measurements. For seeds, a calibration was developed based on protein content values obtained between 1998 and 2002 for 490 pea seed flowers, ranging from 16.4% to 35%. The characteristics of the calibration (Pg0002b.eqa) built on these 490 reference values were standard error of calibration (SEC) = 0.343, standard error of cross-validation (SECV) = 0.411, R2 = 0.989, slope = 1.001. This equation was validated with 87 samples harvested in 2000 (R2 = 0.963) and with 16 samples harvested in 2002 (R2 = 0.992). For plants at BSF, a calibration was developed based on a 3-year sample (1999, 2000, 2002) database, including protein content values for 138 pea plant samples harvested at BSF, which ranged from 8.20% to 29.5%. The characteristics of the calibration (Pfv02c.eqa) built on these 138 reference values were: SEC = 0.495, SECV = 0.651, R2 = 0.988, slope = 0.992. This equation was validated with 114 samples (R2 = 0.980). For straws, a calibration (2002d.eqa) was developed based on a 3-year sample (1999, 2000, 2002) database, including protein content values for 156 pea straw samples, ranging 4.1% to 15.2%. The characteristics of the calibration built on these 156 reference values were: SEC = 0.46, SECV = 0.584, R2 = 0.965, slope = 0.983. This equation was validated with 70 samples (R2 = 0.957). Seed protein content was estimated as 6.25 × seed nitrogen content. Parameters linked to nitrogen availability were calculated: QN BSF = dry biomass per plant × nitrogen content of the plants; QNStraws = dry straw biomass per plant × nitrogen content of straws; QN seed weight per plant × nitrogen content of seeds, QNacc = QNStraws + QN seeds − QNBSF, and QNmob = QNBSF − QNStraws, with the hypothesis that seeds are the only sink for nitrogen after BSF. The critical nitrogen concentration (Ncrit) and the NNI were calculated for the Dijon, France (2002) plot trial as described in Gastal and Lemaire (2002). The harvest index (HI; seed biomass/total biomass at harvest, per plant) and nitrogen HI (NHI; seed nitrogen biomass/total nitrogen biomass, per plant) were also calculated.
Cell Volume and Cell Number
Cotyledon cell number, which relates to seed sink strength, and cell volume were measured using the Coulter method for 66 randomly chosen RILs on seeds from the Dijon, France (2000 and 2002) row trials (five seeds/line in 2000 and three seeds/line in 2002). Seeds were soaked in distilled water for 4 h. Then, the seed coat and the embryonic axis were removed. Cotyledons were cut in small pieces (about 1 mm3) using a razor blade. Cotyledon pieces were fixed in 3 volumes glacial ethanol/1 volume glacial acetic acid at 4°C overnight, then rinsed three times in 3 mL distilled water for 5 min, immersed in a 1 n HCl solution for 45 min at 60°C, and rinsed again three times in 3 mL distilled water for 5 min. Then, cotyledon pieces were immersed in 3 mL enzymatic solution digested in a pectinase solution (1% pectinase [w/v]; ICN Biomedicals), sorbitol 0.2 m (Kalys), 0.2 m NaHCO2 in glacial acetic acid, pH 5.3 (Sigma-Aldrich), and slowly agitated at 37°C for 2 h. Then, tubes were kept on ice. Cells were filtrated on a 250-μL nylon mesh, rinsed using 50 to 100 mL distilled water, left to deposit in pellet, and homogenized in 20 mL distilled water. A 1-mL aliquot was analyzed using a Coulter Multisizer II (Coulter Electronics Ltd), which measured the exact volume of cells, classified cell population, and calculated the mean cell volume of the sample. Then, cell number was calculated as described in Lemontey et al. (2000).
Statistical Analysis
ANOVAs were performed on the data obtained from depodding glasshouse experiments and field trials on the eight genotypes (‘Ballet’, ‘Cameor’, ‘Térèse’, ‘Sommette’, ‘Athos’, VavD265, K586, and China) to determine the level of significance of the treatment, genotype, and treatment by genotype effects.
ANOVAs were performed for each trial on RIL1 to test for genotype and block effects using the SAS GLM procedure (SAS Institute, 2000) and broad sense heritabilities (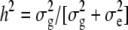 ) were estimated for each trial. ANOVA was also performed on the whole dataset, including data for the five trials, to test for the genotype, environment, and genotype × environment effects. Genotype mean values per trial were obtained using the lsmeans command of the SAS GLM procedure. Values obtained from squares in the plot trial were divided by the number of plants/square to be compared with values obtained per plant in the row trials. Genotypic correlations between all traits were calculated in each trial on the whole RIL population, using the SAS CORR procedure. Correlations between environments were also calculated for seed traits.
) were estimated for each trial. ANOVA was also performed on the whole dataset, including data for the five trials, to test for the genotype, environment, and genotype × environment effects. Genotype mean values per trial were obtained using the lsmeans command of the SAS GLM procedure. Values obtained from squares in the plot trial were divided by the number of plants/square to be compared with values obtained per plant in the row trials. Genotypic correlations between all traits were calculated in each trial on the whole RIL population, using the SAS CORR procedure. Correlations between environments were also calculated for seed traits.
For QTL analysis, a framework genetic map was built for RIL1, using map data described in Loridon et al. (2005) and Aubert et al. (2006). This framework map was tested using the ripple command of MAPMAKER (Lander and Botstein, 1989). The framework map spanned 1,113 cM and included 249 markers distributed over the seven linkage groups of the pea map. QTL analysis was performed using MCQTL software (Jourjon et al., 2005) by the iterative QTL mapping method (iQTLm), which is an iterative QTL detection method using genetics cofactors. Cofactor selection and QTL detection were performed using F tests. F thresholds were determined for several traits showing various h2, by 1,000 permutation tests, for a global genome-wide type I risk of 20% for cofactor selection, and 10% for QTL detection. A mean F value over the traits was used. Cofactors were searched by forward regression, using a threshold of F = 9 (equivalent to LOD = 1.95). Then, QTL were searched by iQTLm using a threshold of F = 12.3 (equivalent to LOD = 2.6). Two-LOD support CI corresponding to a conservative 95% CI in a RIL population (van Ooijen, 1992) were determined. For each trait, a global R2, individual R2, and allelic effect at each QTL were estimated. For some genomic regions, epistatic interactions were searched by two-way ANOVA: Markers of selected regions were taken as one main factor; all other markers, one at a time, were considered as main effect, and interaction of the two markers were tested. Finally, the pea candidate gene reference map (Aubert et al., 2006) was projected onto the RIL1 QTL map to search for colocations, using BioMercator (Arcade et al., 2004). The QTL map was drawn using MapChart, version 2.1 (Voorrips, 2002).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Correlations between each seed trait and all the other traits.
Supplemental Table S1. Mean parental and recombinant inbred values, sd, heritability, and significance of genotype effect in the five field trials for different types of measurements describing plant development and growth.
Supplemental Table S2. QTL detected for all traits in five field environments by genomic regions.
Supplemental Table S3. Description of experiments used to assess potential and field value for one-seed weight and seed protein content in eight genotypes.
Supplementary Material
Acknowledgments
We are very grateful to our colleagues involved in glasshouse experiments (F. Jacquin and P. Mathey), in field experiments (H. Houtin, C. Rond, P Mangin, and N. Blanc) and biochemical analysis at INRA-Dijon (B. Roy and J. Gonthier), and in field experiments at Nickerson (D. Corre) and Serasem (H. Havegeer and E. Margalé). We would also like to thank S. Schnee for help in entering QTL data in the Genoplante database and R. Thompson, K. Gallardo, and G. Aubert for their helpful suggestions on the manuscript. Many thanks to J. Ross for providing Le isogenic lines, C. Rameau for providing us with the RIL1 population and ‘Térèse’ NILs, and to M. Bouchez, B. Ngom, J. Marcel, and S. Jasson for their help during QTL analysis at INRA-Toulouse.
This work was supported by the French national programs Genoplante GOP-PeaC and GOP-PeaC2.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Judith Burstin (burstin@epoisses.inra.fr).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abbo S, Molina C, Jngmann R, Grusak MA, Berkovitch Z, Kahl G, Winter P, Reifen R (2005) Quantitative trait loci governing carotenoid concentration and weight in seeds of chickpea (Cicer arietinum L.). Theor Appl Genet 111 185–195 [DOI] [PubMed] [Google Scholar]
- Ait-Ali T, Swain SM, Reid JB, Sun T, Kamiya Y (1997) The LS locus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J 11 443–454 [DOI] [PubMed] [Google Scholar]
- Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J (2004) BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20 2324–2326 [DOI] [PubMed] [Google Scholar]
- Atta S, Maltese S, Cousin R (2004) Protein content and dry weight of seeds from various pea genotypes. Agronomie 24 257–266 [Google Scholar]
- Aubert G, Morin J, Jacquin F, Loridon K, Quillet MC, Petit A, Rameau C, Lejeune-Hénaut I, Huguet T, Burstin J (2006) Functional mapping in pea, as an aid to the candidate gene approach and for investigating the synteny with the model species Medicago truncatula. Theor Appl Genet 112 1024–1041 [DOI] [PubMed] [Google Scholar]
- Baranger A, Aubert G, Arnau G, Lainé AL, Deniot G, Potier J, Weinachter C, Lejeune-Hénaut I, Lallemand J, Burstin J (2004) Genetic diversity within Pisum sativum using protein-and PCR-based markers. Theor Appl Genet 108 1309–1321 [DOI] [PubMed] [Google Scholar]
- Blanc G, Charcosset A, Mangin B, Gallais A, Moreau L (2006) Connected populations for detecting quantitative trait loci and testing for epistasis: an application in maize. Theor Appl Genet 113 206–224 [DOI] [PubMed] [Google Scholar]
- Blixt S (1978) Problems relating to pea breeding. Agri Hortique Genetica 36 56–87 [Google Scholar]
- Chung J, Babka HL, Graef GL, Staswick PE, Lee DJ, Cregan PB, Shoemaker RC, Specht JE (2003) The seed protein, oil, and yield QTL on soybean linkage group I. Crop Sci 43 1053–1067 [Google Scholar]
- Coque M, Gallais A (2006) Genomics regions involved in response to grain yield selection at high and low nitrogen fertilization in maize. Theor Appl Genet 112 1205–1220 [DOI] [PubMed] [Google Scholar]
- Craig J, Barratt P, Tatge H, Déjardin A, Handley L, Gardner CD, Barber L, Wand TL, Hedley C, Martin C, et al (1999) Mutations at the rug4 locus alter carbon and nitrogen metabolism of pea plants through an effect on sucrose synthase. Plant J 17 353–362 [Google Scholar]
- Craig J, Lloyd JR, Tomlinson K, Barber L, Edwards A, Wang TL, Martin C, Hedley CL, Smith AM (1998) Mutations in the gene encoding starch synthase II profoundly alter amylopectin structure in pea embryos. Plant Cell 10 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado IJ, Wang Z, de Rocher A, Keegstra K, Raikhel NV (1998) Cloning and characterization of AtRGP1: a reversibly autoglycosylated Arabidopsis protein implicated in cell wall biosynthesis. Plant Physiol 116 1339–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis THN, Poyser SJ (2002) An integrated and comparative view of pea genetic and cytogenetic maps. New Phytol 153 17–25 [Google Scholar]
- Fatokun CA, Menancio-Hautea DI, Danesh D, Young ND (1992) Evidence for orthologous seed weight genes in cowpea and mung bean based on RFLP mapping. Genetics 132 841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucher F, Morin J, Courtiade J, Cadioux S, Ellis N, Banfield MJ, Rameau C (2003) DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell 15 2742–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman E, Pleban T, Zamir D (2000) A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA 97 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Le Signor C, Vanderkerckhove J, Thompson R, Burstin J (2003) Proteomics of Medicago truncatula (line J5) seed development establishes the timeframe of metabolic processes related to reserve accumulation. Plant Physiol 133 664–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P, de Carvalho Niebel F, Lescure N, Cullimore JV (1996) Use of subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol Plant Microbe Interact 9 233–242 [DOI] [PubMed] [Google Scholar]
- Gastal F, Lemaire G (2002) N uptake and distribution in crops: an agronomical and ecophysiological perspective. J Exp Bot 53 789–799 [DOI] [PubMed] [Google Scholar]
- Gourlay CW, Hofer JMI, Ellis THN (2000) Pea compound leaf architecture is regulated by interactions among the genes UNIFOLIATA, COCHLEATA, AFILA, and TENDRIL-LESS. Plant Cell 12 1279–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten DL, Pantalone VR, Sams CE, Saxton AM, Landau-Ellis D, Stefaniak TR, Schmidt ME (2004) Seed quality QTL in a prominent soybean population. Theor Appl Genet 109 552–561 [DOI] [PubMed] [Google Scholar]
- Jourjon MF, Jasson S, Marcel J, Ngom B, Mangin B (2005) MCQTL: multi-allelic QTL mapping in multi-cross design. Bioinformatics 21 128–130 [DOI] [PubMed] [Google Scholar]
- Karjalainen R, Kortet S (1987) Environmental and genetic variation in protein content of peas under northern conditions and breeding implications. J Agric Sci Finl 59 1–9 [Google Scholar]
- Kielpinski M, Blixt S (1982) The evaluation of the afila character with regard to its utility in new cultivars of dry pea. Agri Hortique Genetica 40 51–74 [Google Scholar]
- Kof EM, Vinogradova IA, Oorzhak AS, Kalibernaya ZV (2006) The rates of shoot and root growth in intact plants of pea mutants in leaf morphology. Russ J Plant Physiol 53 116–125 [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55 141–172 [DOI] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn D, et al (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420 422–426 [DOI] [PubMed] [Google Scholar]
- Laguerre G, Louvrier P, Allard MR, Amarger N (2003) Compatibility of rhizobial genotypes within natural populations of Rhizobium leguminosarum biovar viciae for nodulation of host legumes. Appl Environ Microbiol 69 2276–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121 185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laperche A, Devienne-Barret F, Maury O, Le Gouis J, Ney B (2006) A simplified conceptual model of carbon/nitrogen functioning for QTL analysis of winter wheat adaptation to nitrogen deficiency. Theor Appl Genet 113 1131–1146 [DOI] [PubMed] [Google Scholar]
- Larmure A, Munier-Jolain NG (2004) A crop model component simulating N partitioning during seed filling in pea. Field Crops Res 85 135–148 [Google Scholar]
- Laucou V, Haurogné K, Ellis N, Rameau C (1998) Genetic mapping in pea: 1-RAPD-based genetic linkage map of Pisum sativum. Theor Appl Genet 97 905–915 [Google Scholar]
- Lemontey C (1999) Influence du génotype maternel sur les divisions cellulaires dans l'embryon: conséquences pour le potentiel de croissance de la graine de pois. PhD thesis. L'Institut National Agronomique Paris, Grignon, France
- Lemontey C, Mousset-Declas C, Munier-Jolain N, Boutin JP (2000) Maternal genotype influences pea seed size by controlling both mitotic activity during early embryogenesis and final endoreduplication level/cotyledon cell size in mature seed. J Exp Bot 51 167–175 [DOI] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB (1997) Mendel's stem length gene (Le) encodes a gibberellin 3 β-hydroxylase. Plant Cell 9 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhuillier-Soundele A, Munier-Jolain NG, Ney B (1999) Influence of nitrogen availability on seed nitrogen accumulation in pea. Crop Sci 39 1741–1748 [Google Scholar]
- Loridon K, McPhee K, Morin J, Dubreuil P, Pilet-Nayel ML, Aubert G, Rameau C, Baranger A, Coyne C, Lejeune-Hénaut I, et al (2005) Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor Appl Genet 111 1022–1031 [DOI] [PubMed] [Google Scholar]
- Mansur LM, Lark KG, Kross H, Oliveira A (1993) Interval mapping of quantitative trait loci for reproductive, morphological, and seed traits of soybean (Glycine max L.). Theor Appl Genet 86 907–913 [DOI] [PubMed] [Google Scholar]
- Martin A, Lee J, Kichey T, Gerentes D, Zivy M, Tatout C, Dubois F, Balliau T, Valot B, Davanture M, et al (2006) Two glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell 18 3252–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx GA (1985) The pea genome: a source of immense variation. In PD Hebblethwaite, MC Heath, TCK Dawkins, eds, The Pea Crop: A Basis for Improvement. Butterworths, London
- Matthews P, Arthur E (1985) Genetic and environmental components of variation in protein content of peas. In PD Hebblethwaite, MC Heath, TCK Dawkins, eds, The Pea Crop: A Basis for Improvement. Butterworths, London
- Maughan PJ, Saghai-Maroof MA, Buss GR (2000) Identification of quantitative trait loci controlling sucrose content in soybean (Glycine max). Mol Breed 6 105–111 [Google Scholar]
- Mitchell-Olds T, Schmitt J (2006) Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature 441 947–952 [DOI] [PubMed] [Google Scholar]
- Monteith JL (1972) Solar radiation and productivity in tropical ecosystems. J Appl Ecol 9 747–766 [Google Scholar]
- Monteith JL (1977) Climate and the efficiency of crop production in Britain. Philos Trans R Soc Lond B Biol Sci 281 277–294 [Google Scholar]
- Munier-Jolain N, Ney B (1998) Seed growth rate in grain legumes. II. Seed growth rate depends on cotyledon cell number. J Exp Bot 49 1971–1976 [Google Scholar]
- Munier-Jolain NG, Ney B (1995) Variability of seed growth rate of various pea cultivars (Pisum sativum L.) in relation to their seed cell number in: improving production and utilisation of grain legumes. Proceedings of the Second European Conference on Grain Legumes, Copenhagen, pp 32–34
- Ney B, Duthion C, Fontaine E (1993) Timing of reproductive abortions in relation to cell division, water content, and growth of pea seeds. Crop Sci 33 267–270 [Google Scholar]
- Nichols DM, Glover KD, Carlson SR, Specht JE, Diers DW (2006) Fine mapping of a seed protein QTL on soybean linkage group and its correlated effects on agronomic data. Crop Sci 46 834–839 [Google Scholar]
- Østergaard L, Yanovsky MF (2004) Establishing gene function by mutagenesis in Arabidopsis thaliana. Plant J 39 682–696 [DOI] [PubMed] [Google Scholar]
- Panthee DR, Pantalone VR, Sams CE, Saxton AM, West DR, Orf JH, Killam AS (2006) Quantitative trait loci controlling sulfur containing amino acids, methionine and cysteine, in soybean seeds. Theor Appl Genet 112 546–553 [DOI] [PubMed] [Google Scholar]
- Paran I, Zamir D (2003) Quantitative traits in plants: beyond the QTL. Trends Genet 19 303–306 [DOI] [PubMed] [Google Scholar]
- Pate JS (1985) Physiology of pea—a comparison with other legumes in terms of economy of carbon and nitrogen in whole-plant and organ functioning. In PD Hebblethwaite, MC Heath, TCK Dawkins, eds, The Pea Crop: A Basis for Improvement. Butterworths, London
- Peoples MB, Dalling MS (1988) The interplay between proteolysis and amino acid metabolism during senescence and nitrogen reallocation. In LD Nooden, AC Leopold, eds, Senescence and Aging in Plants. Academic Press, New York, pp 182–217
- Pic E, de la Serve BT, Tardieu F, Turc O (2002) Leaf senescence induced by mild water deficit follows the same sequence of macroscopic, biochemical, and molecular events as monocarpic senescence in pea. Plant Physiol 128 236–246 [PMC free article] [PubMed] [Google Scholar]
- Rameau C, Murfet IC, Laucou V, Floyd RS, Morris SE, Beveridge CA (2002) Pea Rms6 mutants exhibit increased basal branching. Physiol Plant 115 458–467 [DOI] [PubMed] [Google Scholar]
- Reymond M, Muller B, Leonardi A, Charcosset A, Tardieu F (2003) Combining quantitative trait loci analysis and an ecophysiological model to analyze the genetic variability of the responses of maize leaf growth to temperature and water deficit. Plant Physiol 131 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan M, Ney B, Duc G (1993) Plant symbiotic mutants as a tool to analyse nitrogen nutrition and yield relationship in field-grown peas (Pisum sativum L.). Plant Soil 153 33–45 [Google Scholar]
- Sakamoto T, Morinka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al (2006) Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotechnol 24 105–109 [DOI] [PubMed] [Google Scholar]
- Salon C, Munier-Jolain NG, Duc G, Voisin AS, Grandgirard D, Larmure A, Emery RJN, Ney B (2001) Grain legume seed filling in relation to nitrogen acquisition: a review and prospects with particular reference to pea. Agronomie 21 539–552 [Google Scholar]
- SAS Institute (2000) SAS/STAT User's Guide. SAS Institute, Cary, NC
- Sax K (1923) The association of size differences with seed-coat pattern and pigmentation in Phaseolus vulgaris. Genetics 8 552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiltz S, Gallardo K, Huart M, Negroni L, Sommerer N, Burstin J (2004) Proteome reference maps of vegetative tissues in Pisum sativum: an investigation of nitrogen mobilization from leaves during seed filling. Plant Physiol 135 2241–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiltz S, Munier-Jolain N, Jeudy C, Burstin J, Salon C (2005) Dynamics of exogenous nitrogen partitioning and nitrogen remobilization from vegetative organs in pea revealed by N-15 in vivo labeling throughout seed filling. Plant Physiol 137 1463–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair TR, de Witt CT (1975) Photosynthate and nitrogen requirements for seed production by various crops. Science 189 565–567 [DOI] [PubMed] [Google Scholar]
- Sinclair TR, de Witt CT (1976) Analysis of the carbon and nitrogen limitations of soybean yield. Agron J 68 319–324 [Google Scholar]
- Sprent JI, Stephens JH, Rupela OP (1988) Environmental effects on nitrogen fixation. In RJ Sumerfield, ed, World Crops: Cool Season Food Legumes. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 801–810
- Swain SM, Ross JJ, Reid JB, Kamiya Y (1995) Gibberellins and pea seed development: expression of the lhi, ls, and le5839 mutations. Planta 195 426–433 [Google Scholar]
- Tar'an B, Warkentin T, Somers DJ, Miranda D, Vandenberg A, Blade S, Bing D (2004) Identification of quantitative trait loci for grain yield, seed protein concentration and maturity in field pea (Pisum sativum L.). Euphytica 136 297–306 [Google Scholar]
- Timmerman-Vaughan GM, McCallum JA, Frew TJ, Weeden NF, Russel AC (1996) Linkage mapping of quantitative trait loci controlling seed weight in pea (Pisum sativum L.). Theor Appl Genet 93 431–439 [DOI] [PubMed] [Google Scholar]
- Timmerman-Vaughan GM, Mills A, Whitfield C, Frew TJ, Butler R, Murray S, Lakeman M, McCallum JA, Russel AC, Wilson D (2005) Linkage mapping of QTL for seed yield, yield components, and developmental traits in pea. Crop Sci 45 1336–1344 [Google Scholar]
- Tonsor SJ, Alonso-Blanco C, Koornneef M (2005) Gene function beyond the single trait: natural variation, gene effects, and evolutionary ecology in Arabidopsis thaliana. Plant Cell Environ 28 2–20 [Google Scholar]
- Tuberosa R, Sanguinetti MC, Landi P, Giuliani MM, Salvi S, Conti S (2002) Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the filed at two water regimes. Plant Mol Biol 48 697–712 [DOI] [PubMed] [Google Scholar]
- Turner SR, Barrat DHP, Casey R (1990) The effect of different alleles at the r locus on the synthesis of seed storage proteins in Pisum sativum. Plant Mol Biol 14 793–803 [DOI] [PubMed] [Google Scholar]
- Upadhyaya HD, Kumar S, Gowda CLL, Singh S (2006) Two major genes for seed size in chickpea (Cicer arietimum L.). Euphytica 147 311–315 [Google Scholar]
- van Ooijen JW (1992) Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet 84 803–811 [DOI] [PubMed] [Google Scholar]
- Voisin AS, Salon C, Jeudy C, Warembourg FR (2003. a) Root and nodule growth in Pisum sativum L. in relation to photosynthesis: analysis using 13C labelling. Ann Bot (Lond) 92 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin AS, Salon C, Jeudy C, Warembourg FR (2003. b) Symbiotic N2 fixation in relation to C economy of Pisum sativum L as function of plant phenology. J Exp Bot 54 2733–2744 [DOI] [PubMed] [Google Scholar]
- Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93 77–78 [DOI] [PubMed] [Google Scholar]
- Wang TL, Hedley CL (1985) Genetic and developmental analysis of the seed. In R Casey, DR Davies, eds, Peas: Genetics, Molecular Biology and Biotechnology. CAB International, London
- Weber H, Borisjuk L, Wobus U (1996) Controlling seed development and seed size in Vicia faba: a role for seed coat-associated invertases and carbohydrate state. Plant J 10 823–834 [Google Scholar]
- Yaxley JR, Ross JJ, Sherriff LJ, Reid JB (2001) Gibberellin biosynthesis mutations and root development in pea. Plant Physiol 125 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WK, Wang YJ, Luo GZ, Zhang JS, He CY, Wu XL, Gai JY, Chen SY (2004) QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theor Appl Genet 108 1131–1139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.