Transcription factors (TFs) are DNA-binding proteins that interact with other transcriptional regulators, including chromatin remodeling/modifying proteins, to recruit or block access of RNA polymerases to the DNA template. Plant genomes devote approximately 7% of their coding sequence to TFs, which is a testament to the complexity of transcriptional regulation in these organisms. Extensive sequencing of cDNA and genomic DNA indicates that legumes encode upwards of 2,000 TFs per genome. Less than 1% of these have been characterized genetically, although TFs likely played seminal roles in legume evolution and clearly now play crucial roles in plant development and differentiation. Here we review the literature on legume TFs and describe technological developments that are paving the way for rapid and systematic characterization of TFs and the genetic regulatory networks they control.
Plants are amazing organisms. Not only are they able to build complex organic superstructures from simple inorganic molecules that ensure their growth and reproductive success, but they do this while fixed in space and subject to environmental extremes of light, temperature, water, and nutrients, and to biological challenges from competitors, pests, and pathogens. Evolution has endowed plants with a flexible developmental program that enables them to elaborate new vegetative organs and attune reproduction to prevailing environmental conditions. Plant cells can also differentiate in the short term to cope with more immediate environmental challenges. Plant development and differentiation are programmed primarily at the level of gene transcription, which is controlled by TFs and other proteins that either recruit or block access of RNA polymerases to the DNA template. TFs are usually defined as sequence-specific DNA-binding proteins that are capable of activating and/or repressing transcription. Plant genomes appear to encode many more TFs than those of animals, such as Caenorhabditis elegans and Drosophila melanogaster, which indicates that transcriptional regulation in plants is at least as complex as in animals (Riechmann et al., 2000). Arabidopsis (Arabidopsis thaliana) possesses upwards of 1,800 TF genes representing more than 7% of all protein-coding genes (Riechmann et al., 2000; Guo et al., 2005; Iida et al., 2005). Surprisingly, only one-tenth of these have been characterized genetically (Qu and Zhu, 2006) despite the enormous resources that have been devoted to Arabidopsis research over the past decade. Not surprisingly, we know far less about the role of TFs in other plant species. For instance, less than 1% of TF genes in the model legumes Lotus japonicus (or simply Lotus) and Medicago truncatula (or Medicago) have been genetically characterized. This makes review of the literature on legume TFs a relatively simple task at present, although there are signs that this situation will change rapidly over the next few years. First of all, it is already apparent that TFs play crucial roles in agriculturally important processes in legumes, such as symbiotic nitrogen fixation (SNF), so there is great incentive to learn more about this important class of regulatory proteins. Second, the genome of three legume species, Medicago, Lotus, and soybean (Glycine max), will be completed or largely so in the next 2 years, which will enable the identification of most of the TFs in these species via bioinformatics approaches. Finally, numerous tools for functional genomics have been and are being developed that will facilitate rapid and systematic functional characterization of large numbers of TFs. This review summarizes our current state of knowledge about legume TFs and considers the opportunities and challenges for continued research in this area.
THE DYNAMIC TRANSCRIPTOME
As a backdrop to our discussion of legume TFs, it is salient that recent transcriptomic studies, using arrays of cDNA or oligonucleotides to measure transcript levels, have identified thousands of legume genes that are differentially expressed during various types of plant-microbe interactions (Colebatch et al., 2002, 2004; Liu et al., 2003; Barnett et al., 2004; El-Yahyaoui et al., 2004; Kouchi et al., 2004; Lee et al., 2004; Manthey et al., 2004; Mitra et al., 2004; Moy et al., 2004; Suganuma et al., 2004; Hohnjec et al., 2005; Zou et al., 2005; Alkharouf et al., 2006; Lohar et al., 2006; Starker et al., 2006; Zabala et al., 2006), development and differentiation (Vodkin et al., 2004; Aziz et al., 2005; Firnhaber et al., 2005; Dhaubhadel et al., 2007), and in response to abiotic stress (Ainsworth et al., 2006; Buitink et al., 2006). Invariably, TFs have been found among differentially expressed genes, implicating them in the regulation of specific developmental processes or responses to the biotic and abiotic environment. Such guilt by association is a theme that is elaborated upon below.
IDENTIFICATION OF PUTATIVE TFs
Bioinformatics approaches have been instrumental in identifying putative TF genes in plants. TF families are generally defined by the types of DNA-binding domain contained by proteins in the family (Table I; Fig. 1) and putative TF genes have been identified primarily on the basis of DNA sequences within the gene that encode known DNA-binding domains (Riechmann et al., 2000; Guo et al., 2005; Iida et al., 2005). BLAST and similar searches that look for extended sequence homology between query sequences and known TFs have also been used to identify putative TFs (Iida et al., 2005). BLAST searches that utilize well-curated protein databases, such as UniProt (http://www.expasy.uniprot.org/database/knowledgebase.shtml), can also be used to support TF annotations that were made initially on the basis of the presence of a DNA-binding or other characteristic domain. We searched the current International Medicago Gene Annotation Group (IMGAG) dataset, which contains 40,568 predicted proteins, for the presence of sequences encoding DNA-binding and other TF domains to identify putative TF genes in this species (Supplemental Table S1). A subset of these TFs, obtained from an earlier release of IMGAG gene annotations, was verified by BLAST analysis, which resulted in a list of 1,084 putative TF genes (Table I). We have designed and tested gene-specific primers for each of these for use in high-throughput quantitative reverse transcription (qRT)-PCR analysis and have plans to develop this resource further to facilitate transcript analysis of all Medicago TF genes in the future (K. Kakar and M.K. Udvardi, unpublished data).
Table I.
Classification of putative TFs of Medicago into families and subfamilies
IMGAG proteins were classified as putative TFs if they contained characteristic DNA-binding or other characteristic TF domains and if annotations of matching proteins obtained by BLAST searches were consistent with such a classification. TF families previously identified in plants are presented in the first part of the table, whereas potentially novel plant TF families, which were identified by the presence of domains associated with TFs and other transcriptional regulators outside the plant kingdom, are presented in the latter part of the table. Plant-specific TF families and subfamilies are indicated in bold (according to Riechmann, 2002). D, DNA-binding domain; P, protein-protein interaction domain; NA, nucleic acid (DNA and RNA) binding domain; RD, receiver domain; LBD, ligand binding; TA, transcriptional coactivator.
| TF Family | No. of Genes | Characteristic Domain (InterPro No.) | Domain Function | Domain Description |
|---|---|---|---|---|
| MYB/HD like | 77 | IPR001005; IPR009057 | D | Myb, DNA binding; homeodomain like |
| MYB | 59 | IPR001005 | D | Myb, DNA binding |
| C2H2 (Zn) | 64 | IPR007087 | NA | Zn-finger, C2H2 type |
| AP2/EREBP | 55 | IPR001471 | D | Pathogenesis-related transcriptional factor and ethylene response factor |
| bHLH | 50 | IPR001092 | D | Basic helix-loop-helix dimerization region bHLH |
| HD like | 50 | IPR009057 | D | Homeodomain like |
| HD family | IPR001356 | D | Homeobox | |
| HD | 25 | |||
| HD-ZIP | 5 | IPR006712 | P | HD-ZIP protein, N terminus |
| HD-PHD-finger | 2 | IPR001965 | P | Zn-finger like, PHD-finger |
| MADS | 48 | IPR002100 | D | TF, MADS-box |
| bZIP | 42 | IPR004827 | D | Basic Leu zipper (bZIP) TF |
| PHD | 34 | IPR001965 | P | Zn-finger like, PHD-finger |
| WRKY family | IPR003657 | D | DNA-binding WRKY | |
| WRKY | 29 | |||
| LLR WRKY | 1 | IPR001611 | Leu-rich repeat | |
| ABI3/VP1 | 29 | IPR003340 | D | TF B3 |
| NAC | 29 | IPR003441 | D | No apical meristem (NAM) protein |
| C3H-type 1 (Zn) | 27 | IPR000571 | D | Zn-finger, C-x8-C-x5-C-x3-H type |
| ARF | 23 | IPR003340, IPR010525, IPR011525 | D | |
| JUMONJI | 20 | IPR003347 | D | TF jumonji, jmjC |
| GRAS | 19 | IPR005202 | P | GRAS TF |
| HMG | 15 | IPR000637 | D | HMG-I and HMG-Y, DNA binding |
| AS2 | 14 | IPR004883 | P | Lateral organ boundaries |
| C2C2 (Zn) | ||||
| Dof | 14 | IPR003851 | D | Zn-finger, Dof type |
| GATA | 7 | IPR000679 | D | Zn-finger, GATA type |
| CO like | 6 | IPR000315 | D | Zn-finger, B-box |
| YABBY | 5 | IPR006780 | D | YABBY protein |
| CCAAT-HAP3 type | 12 | IPR003958 | D | TF CBF/NF-Y/archaeal histone |
| GRF | 8 | IPR010666 | D | Zn-finger, GRF type |
| SBP | 8 | IPR004333 | D | SBP |
| EIL | 7 | IPR006957 | D | Ethylene insensitive 3 |
| LIM | 7 | IPR001781 | P | Zn-binding protein, LIM |
| SNF2 | 6 | IPR000330 | D | SNF2 family N-terminal domain |
| E2F/DP | 5 | IPR003316 | D | TF E2F/dimerization partner (TDP) |
| TCP | 5 | IPR005333 | D | TCP TF |
| FHA | 5 | IPR000253 | D | Forkhead associated |
| ARID | 4 | IPR001606 | D | AT-rich interaction region |
| HSF | 4 | IPR000232 | D | Heat shock factor (HSF)-type, DNA binding |
| AUX/IAA | 3 | IPR003311 | D | AUX/IAA protein |
| SRS | 3 | IPR006510 | D | Zn-finger, LRP1 type |
| TUB | 3 | IPR000007 | D | Tubby |
| ZIM | 3 | IPR010399 | D | ZIM |
| DDT | 3 | IPR004022 | D | DDT |
| ZF-HD | 2 | IPR006455 | D | Homeobox domain, ZF-HD class |
| MBF1 | 2 | IPR001387 | D | Helix-turn-helix type 3 |
| S1Fa like | 2 | IPR006779 | D | DNA-binding protein S1FA |
| CAMTA | 2 | IPR005559 | D | CG-1 |
| LFY | 1 | IPR002910 | D | Floricaula/leafy protein |
| NIN like | 1 | IPR003035 | D | Plant regulator RWP-RK |
| TAZ | 1 | IPR000197 | P | Zn-finger, TAZ type |
| Potentially novel plant TFs and transcriptional regulators | ||||
| CCHC (Zn) | 112 | IPR001878 | NA | Zn-finger, CCHC type |
| RR | 16 | IPR001789, IPR011006 | RD | Response regulator receiver |
| DHHC (Zn) | 14 | IPR001594 | D or P | Zn-finger, DHHC type |
| HTH | ||||
| FIS | 11 | IPR002197 | D | Helix-turn-helix, Fis type |
| AraC | 2 | IPR000005 | D | Helix-turn-helix, AraC type |
| BTB/POZ | 7 | IPR000210 | P | BTB |
| TTF-type (Zn) | 6 | IPR006580 | D | Zn-finger, TTF type |
| BD | 6 | IPR001487 | P | Bromodomain |
| λ-DB | 3 | IPR010982 | D | λ_DNA_bd |
| TrpR | 3 | IPR010921 | D | Trp repressor/replication initiator |
| TPR | 3 | IPR001440 | P | Tetratricopeptide TPR_1 |
| KRAB-box | 2 | IPR001909 | P | KRAB box |
| NRs | 2 | IPR008946 | LBD | Steroid nuclear receptor, ligand binding |
| R3H | 2 | IPR001374 | NA | Single-stranded nucleic acid binding R3H |
| YEATS | 2 | IPR005033 | TA | YEATS |
| U1-type (Zn) | 2 | IPR003604 | NA | Zn-finger, U1 type |
| A20 like | 2 | IPR002653 | P | Zn-finger, A20 type |
| Euk_TF | 1 | IPR008917 | D | Euk_TF_DNA_bd |
| NGN | 1 | IPR006645 | D | NGN |
| p53 like | 1 | IPR008967 | D | p53-like TF, DNA binding |
| SSB protein | 1 | IPR011344 | D | Single-strand binding protein |
| ssDB TR | 1 | IPR009044 | D | Single-strand DNA-binding transcriptional regulator |
| TCoAp15 | 1 | IPR003173 | D | Transcriptional coactivator p15 |
| BED-type (Zn) | 1 | IPR003656 | D | Zn-finger, BED-type predicted |
| TCoA | 1 | IPR009255 | TA | Transcriptional coactivation |
| Tc/PD | 1 | IPR001533 | TA | Transcriptional coactivator |
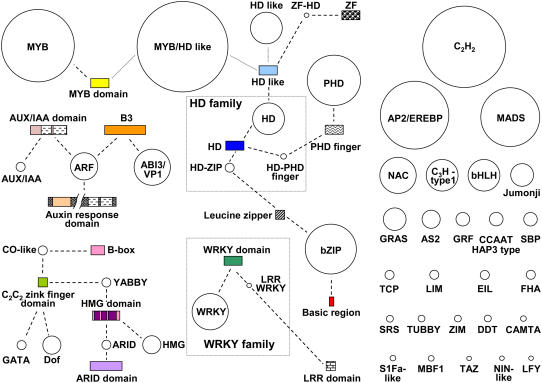
Figure 1.
Relationships and domain shuffling between Medicago TF families. TF families are represented by circles whose size is proportional to the number of members in the family. Domains are represented by rectangles, whose size is proportional to the domain length. DNA-binding domains appear in color. Protein-binding and other domains are hatched. Dashed lines indicate that a given domain is characteristic of the family or subfamily to which it is attached. Dotted lines indicate domains that define a potentially novel TF family or subfamily. Based on Figure 1 of Riechmann (2002).
FUNCTIONAL CHARACTERIZATION OF TFs
As noted above, very few legume TFs have been characterized genetically so far (Table II). An important feature of legumes that sets them apart from plants in other families is their ability to form nitrogen-fixing symbioses with soil bacteria, called rhizobia. These bacteria take up intracellular residence in specialized organs, called nodules, that develop on roots and stems specifically for the purpose of SNF. Given the importance of SNF to sustainable agriculture, it has been a major focus of legume research over the past few decades and one of the few areas of legume biology where the role of TFs has been firmly established. The first TF gene implicated in SNF was Nin, for nodule inception, which was cloned from a transposon-tagged mutant of Lotus that was unable to form nodules (Schauser et al., 1999). NIN was the founding member of a novel family of putative TFs in higher plants, now called the NIN-like family (Fig. 1), and shares homology with Chlamydomonas minus dominance proteins, which are developmental regulators in these algae. Nin-like genes are widespread in the plant kingdom. However, their predicted DNA-binding and gene regulation activities are yet to be proven formally. Classical or forward-genetics approaches have subsequently identified three other TF genes in Lotus (Nishimura et al., 2002) or Medicago (Kaló et al., 2005; Smit et al., 2005) that are essential for nodule development. NODULATION SIGNALING PATHWAY1 (NSP1) and NSP2 of Medicago are both GRAS-family proteins, putative TFs that transduce the bacterial Nod factor signal and induce expression of plant nodulin genes, which are presumably required for nodule development (Kaló et al., 2005; Smit et al., 2005). Both NSP1 and NSP2 were isolated by map-based cloning. Orthologs of NSP1 and NSP2 were subsequently isolated from Lotus by candidate gene approaches (Heckmann et al., 2006) and positional cloning (Murakami et al., 2007). The Lotus LjBzf gene encodes a bZIP TF that negatively regulates nodule development (Nishimura et al., 2002). The bzf/sym77 mutant exhibits not only faster and more prolific nodulation, but also light and gravity response defects reminiscent of the hy5 mutant of Arabidopsis. A Lotus homolog (LjBzf) of the Arabidopsis HY5 gene was subsequently cloned and found to cosegregate with the mutant phenotype. The bzf/sym77 mutant contained a single base pair mutation in a splice donor site of this gene. Finally, a wild-type version of LjBzf complemented the mutant phenotype in transgenic plants, confirming its role in the regulation of nodule development. All four of the genes mentioned above have homologs in nonlegume species, such as Arabidopsis, which suggests that they have been recruited rather than invented during evolution to fulfill roles in nodule development (Szczyglowski and Amyot, 2003). This certainly seems to be the case for LjBzf and Mszpt2-1 (see below) given the additional nonsymbiotic phenotypes of these mutants (Frugier et al., 2000; Nishimura et al., 2002). Interestingly, the nonsymbiotic phenotypes of the Lotus bzf/sym77 and Arabidopsis hy5 mutants are not identical (the latter shows enhanced lateral root initiation, whereas the former does not), reflecting evolutionary divergence in gene function in the two plant lineages quite apart from the acquisition of the novel symbiotic function in the legume lineage. This theme is reiterated below for TFs involved in floral development.
Table II.
Genetically characterized TFs in legumes
| TF Name | TF Family | Process Regulated | Species | Method | References |
|---|---|---|---|---|---|
| NIN | NIN like | Nodule development | Lotus; pea | Transposon mutagenesis | Schauser et al. (1999); Borisov et al. (2003) |
| LjBzf | bZIP | Nodule development | Lotus | Positional cloning | Nishimura et al. (2002) |
| NSP1 | GRAS | Nodule development | Medicago; Lotus | Positional cloning | Smit et al. (2005); Heckmann et al. (2006) |
| NSP2 | GRAS | Nodule development | Medicago; Lotus | Positional cloning | Kaló et al. (2005); Heckmann et al. (2006) |
| Mszpt2-1 | C2H2 (Zn) | Nodule development | Medicago | Antisense | Frugier et al. (2000) |
| LjNDX1, LjNDX2 | HD | Nodule function | Lotus | Antisense | Gronlund et al. (2003) |
| MtHAP2-1 | CCAAT binding | Nodule development | Medicago | RNAi | Combier et al. (2006) |
| UNI/LjFLO | LFY | Flower and leaf development | Pea; Lotus | Candidate gene approach | Hofer et al. (1997); Dong et al. (2005) |
| PIM, MtPIM | MADS | Floral meristem identity | Pea; Medicago | Candidate gene approach, transposon mutagenesis | Berbel et al. (2001); Taylor et al. (2002); Benlloch et al. (2006) |
| PsPi | MADS | Floral development | Pea | Complementation | Berbel et al. (2005) |
| LjCYC2 | TCP | Floral development | Lotus | Candidate gene approach | Feng et al. (2006) |
| PHANTASTICA | MYB | Compound leaf development | Pea | Candidate gene approach | Tattersall et al. (2005) |
| PvNAP | NAC | Leaf senescence | Common bean | Ectopic overexpression | Guo and Gan (2006) |
| WXP1, WXP2 | AP2 domain | Wax biosynthesis | Medicago | Ectopic overexpression | Zhang et al. (2005); Zhang et al. (2007) |
| Mszpt2-1 | Kruppel like | Salt tolerance | Medicago | Antisense | Merchan et al. (2003) |
| CAP2 | AP2 | Salt, drought tolerance, growth development | Chickpea (Cicer arietinum) | Ectopic overexpression | Shukla et al. (2006) |
| SCOF-1 | C2H2 (Zn) | Cold tolerance | Soybean | Ectopic overexpression | Kim et al. (2001) |
| Alfin1 | Zn-finger | Salt tolerance, growth development | Alfalfa | Overexpression, antisense | Winicov (2000) |
Many TF genes have been found to be expressed during nodule development and differentiation (see above) and several groups are now using the tools of reverse genetics to decipher the roles of such genes in SNF. Three TFs have been implicated in nodule development or function in this way (Table II). The first of these was Mszpt2-1, a Kruppel-like TF of the C2H2 (Zn) family that was found to be essential for differentiation of the nitrogen-fixing zone of alfalfa (Medicago sativa) nodules via an antisense RNA approach (Frugier et al., 2000). A similar approach implicated the Lotus ndx gene family in nodule function and maintenance (Gronlund et al., 2003). Most recently, RNA interference (RNAi) revealed a key role in nodule development for MtHAP2-1, a member of the CCAAT-binding family of TFs (Combier et al., 2006). Interestingly, MtHAP2-1 was found to be regulated by microRNA169, revealing an important role for microRNA in the regulation of legume development.
Whereas research to identify TFs involved in SNF has profited little from previous work in nonlegumes such as Arabidopsis, knowledge from nonlegume models has been instrumental in identifying a number of TF genes involved in common plant processes, such as flower and leaf development (Table II). In fact, the first legume TF gene to be characterized functionally was pea (Pisum sativum) FLO, which was isolated by virtue of its sequence homology to the TFs FLO and LFY of snapdragon (Antirrhinum majus) and Arabidopsis, respectively. FLO and LFY control floral development in snapdragon and Arabidopsis, and a defect in pea FLO was subsequently found to be responsible for aberrant floral and leaf development in the pea unifoliata (uni) mutant (Hofer et al., 1997). The Lotus ortholog of FLO was later identified in the same way (Dong et al., 2005). Similar approaches were used to assign functions for PIM (a MADS family TF) in pea floral meristem determination (Taylor et al., 2002) and for PHANTASTICA (a MYB TF) in pea compound leaf development (Tattersall et al., 2005). Interestingly, Lotus has a duplicate pair of PHANTASTICA-like genes that probably have divergent functions in compound leaf development (Luo et al., 2005).
The value of computational approaches in identifying genes likely to be involved in various aspects of flowering was nicely illustrated by Hecht et al. (2005), who utilized sequence information from Arabidopsis TFs to identify homologs in model legume sequence databases, which were then used to design PCR-cloning strategies to isolate homologs from pea. The majority of Arabidopsis flowering genes were represented in pea and other legume sequence databases. However, several gene families, including the MADS-box, CONSTANS, and FLOWERING LOCUS T/TERMINAL FLOWER1 families, appeared to have undergone differential expansion, whereas other genes important in Arabidopsis, including FRIGIDA and members of the FLOWERING LOCUS C clade, were conspicuously absent from legumes. Several pea and Medicago orthologs mapped to syntenic chromosomal positions, demonstrating the benefit of parallel model systems for understanding flowering phenology in crop and model legume species.
TFs of the TCP family, named after the founding members TB1, CYC, and PCF, help to establish the pattern of flower petals (Cubas, 2004), which gives legume flowers their typical bilateral symmetry. Citerne et al. (2003) sequenced a number of CYC homologs, sorted them into clades by phylogenetic analysis, and discussed the difficulties of assigning orthologs in cross-species comparisons. Thus, genetic map position, mutant phenotypes, and/or complementation of Arabidopsis mutants have been used in addition to sequence similarity to infer orthology. The squared standard mutant of Lotus is defective in the TCP gene LjCYC2, which is required to establish floral bilateral symmetry and was cloned by a candidate gene/sequence homology approach (Feng et al., 2006). Adaxial expression of two CYC genes was observed in the developing floral meristem of lupin (Lupinus albus), which also has bilaterally symmetrical flowers (Citerne et al., 2006). Interestingly, evolution of radially symmetrical flowers in Cadia, which belongs to the same subclade as Lupinus, may have resulted from an expanded domain of expression of an orthologous CYC gene in the former (Citerne et al., 2006).
Cross-species complementation studies have indicated possible roles for several legume TFs. For example, Berbel et al. (2001) rescued the Arabidopsis apetala1 (ap1) floral development mutant using a PIM cDNA clone, which they called PEAM4. Later, the same group characterized PsPI, a pea MADS-box gene homologous to the petal and stamen identity genes PISTILLATA (PI), from Arabidopsis and GLOBOSA, from snapdragon. Interestingly, constitutive expression of PsPI in Arabidopsis rescued the floral defects caused by the strong pi-1 mutant allele, despite the fact that the pea protein, PsPI, lacked a particular C-terminal motif (Berbel et al., 2005). Similarly, Guo and Gan (2006) showed that overexpression of PvNAP, a kidney bean (Phaseolus vulgaris) NAC TF homologous to Arabidopsis AtNAP, successfully complemented the leaf abscission phenotype of an atnap null mutant, indicating a possible role of NAP in bean leaf abscission.
Finally, five legume TFs have been implicated in abiotic stress tolerance (Table II). One of these, alfalfa Mszpt2-1, which was mentioned previously in the context of nodule development, was found to be induced in roots by salt treatment. Inhibition of Mszpt2-1 by antisense RNA resulted in increased sensitivity of transgenic plants to salinity (Merchan et al., 2003). Overexpression of CAP2 and Alfin1 TFs in transgenic plants conferred salt tolerance and increased growth (Winicov, 2000; Shukla et al., 2006). Constitutive overexpression of SCOF-1, a soybean protein, increased cold tolerance of transgenic Arabidopsis and tobacco (Nicotiana tabacum) plants (Kim et al., 2001). Another example of successful leveraging of knowledge from nonlegumes for legume research is provided by the Medicago WXP1 gene, a member of the AP2/ethylene-responsive element-binding protein (EREBP) family of TFs. Several members of this family have been implicated in drought tolerance in Arabidopsis and other nonlegumes. Overexpression of Medicago WXP1 in alfalfa resulted in enhanced tolerance to drought stress, which correlated with increased wax deposition in the leaf cuticle (Zhang et al., 2005).
Whereas work on deciphering the roles of legume TFs is just beginning, considerable efforts have already been made to demonstrate the functionality of such proteins in terms of their DNA-binding and transactivation abilities and subcellular localization (Table III). Approaches for isolating legume TF genes have varied widely. Homology-based methods using TF DNA from other plant families have been successfully employed to identify specific classes of legume TFs. For instance, GmEREBP1 was isolated from a soybean root cDNA library screened with a probe that was PCR amplified using degenerate primers matching the conserved EREBP-coding domain (Mazarei et al., 2002). Three soybean DRE-binding proteins were identified by sequence homology to the AP2/EREBP consensus sequence via a BLAST search of the soybean EST database (Li et al., 2005). Several groups have used degenerate primers matching conserved TF domains for PCR amplification of legume TF sequences (Chern et al., 1996a, 1996b; Heard et al., 1997; Uimari and Strommer, 1997; Zucchero et al., 2001; Tucker et al., 2002).
Table III.
Biochemical and molecular characterization of legume TFs
NL, Nuclear localization. All other abbreviations are defined in the text.
| TF Name | TF Family | Proposed Role | Species | Method | References |
|---|---|---|---|---|---|
| Ph_acut_ AY026054 | bZIP | Abiotic stress | Phaseolus acutifolius | NL | Rodriguez-Uribe and O'Connell (2006) |
| Ph_vulg_ AF350505 | bZIP | Abiotic stress | Kidney bean | NL | Rodriguez-Uribe and O'Connell (2006) |
| GmDREBa | AP2/EREBP | Abiotic stress | Soybean | Yeast one hybrid | Li et al. (2005) |
| GmDREBb | AP2/EREBP | Abiotic stress | Soybean | Yeast one hybrid | Li et al. (2005) |
| GmDREBc | AP2/EREBP | Abiotic stress | Soybean | Yeast one hybrid | Li et al. (2005) |
| SCOF-1 | C2H2 (Zn) | Abiotic stress | Soybean | NL, yeast two hybrid | Kim et al. (2001) |
| CAP2 | AP2/EREBP | Abiotic stress and development | Chickpea | EMSA, yeast one hybrid, NL | Shukla et al. (2006) |
| Alfin1 | Alfin-like/PHD-finger | Abiotic stress and development | Alfalfa | EMSA | Bastola et al. (1998) |
| PLATZ1 | PLATZ (Zn) | Cell division | Pea | EMSA | Nagano et al. (2001) |
| GmHZ1 | HD-ZIP | Defense | Soybean | EMSA, NL | Wang et al. (2005) |
| SGBF-1 | GBF | Development | Soybean | EMSA | Hong et al. (1995) |
| SGBF-2 | GBF | Development | Soybean | EMSA | Hong et al. (1995) |
| STF1 | bZIP with RING Zn-finger motif | Development | Soybean | EMSA | Cheong et al. (1998) |
| STF2 | bZIP with RING Zn-finger motif | Development | Soybean | EMSA | Cheong et al. (1998) |
| STGA1 | TGA-type bZIP | Development | Soybean | EMSA | Cheong et al. (1994) |
| PvTGA1.1 | TGA-type bZIP | Leaf abscission | Kidney bean | EMSA | Tucker et al. (2002) |
| PvTGA2.1 | TGA-type bZIP | Leaf abscission | Kidney bean | EMSA | Tucker et al. (2002) |
| PvTGA2.2 | TGA-type bZIP | Leaf abscission | Kidney bean | EMSA | Tucker et al. (2002) |
| Myb26 | MYB | Flower development | Pea | EMSA | Uimari and Strommer (1997) |
| ngl9 | MADS box | Nodule and flower development | Alfalfa | EMSA | Zucchero et al. (2001) |
| nmhc5 | MADS box | Nodule development | Alfalfa | EMSA | Heard et al. (1997) |
| GBP | GAGA-binding protein | Nodule function | Soybean | EMSA, yeast one hybrid | Sangwan and O'Brian (2002) |
| G/HBF-1 | bZIP | Pathogen defense response | Soybean | EMSA | Dröge-Laser et al. (1997) |
| KAP-2 | H-box binding | Phenylpropanoid biosynthesis | Kidney bean and Medicago | EMSA, in vitro transcription assay | Lindsay et al. (2002) |
| GmHdl56 | HD-ZIP | Phosphate responses | Soybean | EMSA, DNase-I footprinting | Tang et al. (2001) |
| GmHdl57 | HD-ZIP | Phosphate responses | Soybean | EMSA | Tang et al. (2001) |
| PvALF | ABI3 like | Seed development | Kidney bean | EMSA, transient expression assay, transactivation in planta | Bobb et al. (1997); Nag et al. (2005) |
| ROM1 | bZIP | Seed development | Kidney bean | Transient expression assay, EMSA, DNase-I footprinting | Chern et al. (1996b) |
| ROM2 | bZIP | Seed development | Kidney bean | Transient expression assay, EMSA, DNase-I footprinting | Chern et al. (1996a) |
| TGA1a | TGA-type bZIP | Seed development | Pea | EMSA, DNase-I footprinting, methyl interference assay | de Pater et al. (1994) |
| GmGT-2 | Trihelix | Light responses | Soybean | EMSA | O'Grady et al. (2001) |
| PCF1 | HMG | Photosynthesis | Pea | Filter-binding assay, DNase-I footprinting | Pwee et al. (1994); Webster et al. (1997) |
| VR-EIL1/2 | EIL | Ethylene-signaling pathway | Vigna radiata | EMSA, NL, transient expression, transactivation in yeast | Lee and Kim (2003) |
| GmEREBP1 | AP2/EREBP | Wounding and pathogen response | Soybean | EMSA | Mazarei et al. (2002) |
Other TFs have been identified based on their ability to interact with known gene cis-elements. The use of cDNA expression libraries has been valuable in this regard. Two HD-ZIP proteins (GmHDL56/57) were identified using a 160-bp fragment of the VspB promoter (Tang et al., 2001). Similarly, G/HBF-1, SGBF-1/2, and GmGT-2 were isolated based on their ability to bind specific promoter elements (Hong et al., 1995; Dröge-Laser et al., 1997; O'Grady et al., 2001). Yeast (Saccharomyces cerevisiae) one-hybrid screens have been effective in the isolation of legume proteins that bind specific cis-elements. Sangwan and O'Brian (2002) constructed a soybean nodule cDNA library in a vector containing a GAL4-activation domain to produce GAL4 fusion proteins. Introduction of this library into a yeast His-auxotroph engineered to have a HIS3 gene preceded by a (GA)27/(CT)27 dinucleotide repeat sequence resulted in the identification of a His prototroph containing a GBP-GAL4 fusion. Rarely have legume TF protein-protein interactions been identified, presumably because most of the research in this area has focused on protein-DNA interactions. Kim et al. (2001) identified a C2H2-type Zn-finger protein, SCOF-1, which failed to exhibit DNA-binding activity with several candidate cis-elements, but was subsequently found to enhance the abscisic acid response element-dependent gene expression mediated by SGBF-1.
Approaches to demonstrate DNA binding of legume TFs include electrophoretic mobility shift assays (EMSAs), hybridization of labeled DNA to TFs on filters, DNase-I footprinting, and yeast one-hybrid assays (Table III). In one interesting example, Bastola et al. (1998) identified cDNA encoding Alfin1, a protein with a putative Zn-binding domain, by differential screening of salt-tolerant alfalfa cells. The DNA-binding specificity of Alfin1 was determined by binding of purified protein to random oligonucleotides in an EMSA followed by PCR amplification to identify the preferential target sites. In most instances, however, DNA-binding specificity has been tested only on a few select cis-element sequences that have been identified by work in other plant families. Such biased approaches are likely to miss important TF-DNA interactions. Alternative, nonbiased approaches are now available that should solve this problem (see below).
Further evidence of TF activity has occasionally been provided using transactivation assays. Some groups have demonstrated in vivo transactivation in cell culture and transient transformation systems, including particle bombardment of bean cotyledons (Chern et al., 1996a, 1996b; Bobb et al., 1997), polyethylene glycol-mediated transfection of Arabidopsis protoplasts (Kim et al., 2001), and in vitro transcription activation in rice (Oryza sativa) cell extracts (Lindsay et al., 2002).
One verification of a protein's role as a TF is its localization to the nucleus. For legume TFs, this has been done with immunohistochemical localization (Rodriguez-Uribe and O'Connell, 2006) or using GFP or GUS fusion proteins (Kim et al., 2001; Kaló et al., 2005; Smit et al., 2005; Wang et al., 2005; Shukla et al., 2006). In the case of NSP2, a regulator of legume-rhizobium symbiosis, nuclear relocalization was detected following application of purified Nod factors, suggesting that posttranslational modification is required to activate this protein (Kaló et al., 2005).
So far, there has been a major disconnect between TFs that have been ascribed a biological role based on genetic data and TFs that have been characterized at the biochemical and/or molecular levels. Clearly, to understand better the function of genetically characterized TFs, we need to identify the genes and network of genes that they control. On the other hand, for TFs that have been characterized in terms of their DNA-binding ability, it is now important that biological function be established via forward or reverse genetics. Furthermore, despite the knowledge that TFs often work as part of a team or complex of proteins to recruit or block recruitment of RNA polymerase to the DNA (Lee and Young, 2000), virtually nothing is known about the proteins that interact with legume TFs to ensure their biological activity. Tissue and organ development and differentiation and plant responses to specific environmental challenges require the concerted activity of networks of TFs, which orchestrate global changes in transcription. The details of these networks remain unknown in legumes. Addressing these open questions in legume TF biology in an efficient manner will require a coordinated effort on the part of the scientific community. The final section of this review offers a roadmap for this enterprise.
A ROADMAP FOR FUTURE RESEARCH ON LEGUME TFs
Within the next 2 or 3 years, complete or near-complete genomic sequence for the euchromatic regions of three legumes, Medicago, Lotus, and soybean, will be available. This will greatly facilitate systematic approaches to TF functional analysis. Bioinformatics approaches will rapidly identify putative TFs among the new genomic sequences, as described above, which will provide grist for the functional analysis mill. Whereas forward genetics will gather momentum as genomic sequencing results in more complete and better integrated physical and genetic maps of chromosomes, which will facilitate map-based cloning of additional TFs involved in legume development and differentiation, reverse-genetics approaches are likely to play a more significant role in the functional characterization of TFs in the future. Certainly, reverse genetics offers a more systematic way to characterize all putative TF genes.
Some of the tools for systematic reverse-genetics analysis of TF function in legumes, such as plant transformation protocols for RNAi and overexpression (Thykjaer et al., 1997; Chabaud et al., 2003; Ott et al., 2005; Zhang et al., 2005) and ethyl methanesulfonate mutant populations for TILLING (Perry et al., 2003), are already in place. Others, such as transposon-insertion (Tadege et al., 2005) and fast neutron bombardment deletion mutant populations (Wang et al., 2006) are being developed, and a mutant from a small Tnt1 insertion population has already been described (Benlloch et al., 2006). Viral-induced gene silencing is another promising tool for high-throughput reverse genetics that has proven successful in pea (Constantin et al., 2004).
In view of the TF content of Arabidopsis and rice, we expect that each of the three model legumes mentioned above will possess at least 2,000 TF genes. It will be an impossible task for any one group to characterize this number of genes, at least at the genetic level. A coordinated international effort would help to make the process of TF gene function discovery most efficient. One way to give direction to such an enterprise would be to determine first the developmental and environmental expression profiles of each TF in the context of the whole transcriptome. This would serve several purposes. First, it would reveal any organ/developmental specificity. Second, it would reveal any environmental stress specificity, which would constrain hypotheses about possible roles of each TF. Third, by setting TF gene expression profiles into the broader, whole-genome context of transcription, correlations between individual TFs and groups of other genes would be revealed, which would help to refine hypotheses about possible TF function, especially if correlated sets of genes are predicted to be involved in one or just a few biological processes. Many of the tools required for such transcriptome analyses are now available for Medicago, Lotus, and soybean, including Affymetrix GeneChips containing probe sets for the majority of genes in these three models. In addition, we are currently developing gene-specific primers for all Medicago TFs for qRT-PCR to complement data obtained using the corresponding Affymetrix GeneChip (K. Kakar and M.K. Udvardi, unpublished data), and a similar resource is being developed for soybean (G. Stacey, personal communication). Transcript quantification by qRT-PCR is more sensitive than by DNA array hybridization methods (Czechowski et al., 2004) so the resources being developed for qRT-PCR profiling in Medicago and soybean will provide a more comprehensive picture of TF expression patterns in this species. Hierarchical cluster analysis (Yu et al., 2005) and Pearson correlation (Zar, 1999; Persson et al., 2005) are two ways to identify genes that are coordinately regulated, which will not only provide clues about the possible function of TF genes (see above), but also identify possible downstream target genes of specific TFs.
There have been few attempts to confirm the physical interaction between a genetically characterized legume TF and a target gene, although possible target genes have been identified by transcriptome analysis of TF mutants (e.g. Kaló et al., 2005; Smit et al., 2005). Methods have been developed to identify TF target genes in a nonbiased, high-throughput manner. Perhaps the most powerful of these is chromatin immunoprecipitation (ChIP) followed by DNA array hybridization (called ChIP-chip) to identify DNA fragments covalently bound to immunoprecipitated DNA-binding proteins (Thibaud-Nissen et al., 2006). Specific immunoprecipitation can be facilitated by in planta expression of the TF of interest as a fusion protein with a short, nonplant peptide epitope at one end, which enables the use of commercially available monoclonal antibodies directed toward the epitope to precipitate the fusion protein and any covalently linked proteins and DNA. This approach has been used, for example, to identify genomic DNA bound by the Arabidopsis FLC TF (Helliwell et al., 2006). Affinity purification can also be used to identify associated proteins in DNA-binding complexes (Wood et al., 2006). A prerequisite for ChIP-chip is an array containing probes for promoter DNA. Arrays designed to detect gene transcripts, such as the Affymetrix GeneChips for Medicago, Lotus, and soybean, are not suitable for such applications. However, tiling arrays, which contain oligonucleotide probes covering the entire genome (coding and noncoding) with short or no gaps between probed sequences are well suited to ChIP-chip (Thibaud-Nissen et al., 2006; Zhang et al., 2006). Although no tiling array exists for a legume yet, we have plans to develop a Medicago tiling array upon completion of the genome sequence in 2008.
The preceding paragraphs may give the impression that the road is mostly clear for rapid progress in TF function discovery in model legumes. However, it is likely that there will be bumps, potholes, and unexpected turns in the road ahead. For instance, some TFs appear to job share with one or more close relatives, so that loss of function of one gene may go unnoticed in a mutant plant (Riechmann and Ratcliffe, 2000). Functional redundancy between some TFs makes it difficult, if not impossible, to isolate mutations in these genes via forward genetics. However, reverse-genetics approaches that utilize phylogenetic and transcriptomic information to identify potentially redundant genes prior to the creation of double or higher order mutants should overcome this problem (e.g. Liljegren et al., 2000; Zhang et al., 2003). Obviously, the availability of well-curated mutant populations (see above) will be essential for this endeavor. An alternative approach to overcome functional redundancy is to create dominant-negative mutants by fusing TFs to known repressor domains (Markel et al., 2002). Yet another approach is to overexpress the TF of interest in transgenic plants and to monitor the effects of this on the expression of other genes and on the phenotype (biochemical, physiological, developmental, or otherwise) of the altered plants. Ideally, TF overexpression should be confined to the same cell, tissue, and organ types as the endogenous gene and preferably under the control of an inducible promoter. There is mounting evidence that diversification of TF function in plants often results from changes in nontranscribed sequences that alter the expression domain of the gene, rather than from changes in the coding sequence that alter the DNA- or protein-binding properties of TFs. Thus, TF mutant phenotypes have been suppressed by ectopic expression of related TFs that are not normally expressed in the same tissue/organ as the mutated TF (Riechmann, 2002). Overexpression of TFs can also interfere with processes totally unrelated to the normal function of the protein (Riechmann and Ratcliffe, 2000). Clearly, care must be taken in designing and interpreting TF overexpression experiments.
TFs interact physically with other proteins, in addition to the RNA polymerase complex itself, to effect changes in gene transcription (Lee and Young, 2000). The specific makeup of these complexes is unknown for the majority of plant genes, although this knowledge is a prerequisite to understanding the combinatorial control of transcription (Singh, 1998). Approaches such as yeast two-hybrid screening (e.g. Zhang et al., 1999) and ChIP followed by proteomic analysis of the resulting protein complexes (Helliwell et al., 2006; Wood et al., 2006) will be useful in this context. Finally, the network of genes regulated by a single TF and its partners is just a small part of a larger genetic regulatory network that ensures coordinated expression of genes involved in many different cellular processes during plant development and differentiation. Deciphering these global genetic networks will require integration of many of the genomic, functional genomic, and bioinformatic approaches described above.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Putative TFs among IMGAG-annotated proteins.
This work was supported by the Samuel Roberts Noble Foundation, the Max Planck Society, the European Union FP6 Program, and U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service-National Research Initiative.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Michael K. Udvardi (mudvardi@noble.org).
The online version of this article contains Web-only data.
References
- Ainsworth EA, Rogers A, Vodkin LO, Walter A, Schurr U (2006) The effects of elevated CO2 concentration on soybean gene expression: an analysis of growing and mature leaves. Plant Physiol 142 135–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkharouf NW, Klink VP, Chouikha IB, Beard HS, MacDonald MH, Meyer S, Knap HT, Khan R, Matthews BF (2006) Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224 838–852 [DOI] [PubMed] [Google Scholar]
- Aziz N, Paiva NL, May GD, Dixon RA (2005) Transcriptome analysis of alfalfa glandular trichomes. Planta 221 28–38 [DOI] [PubMed] [Google Scholar]
- Barnett MJ, Tolman CJ, Fisher RF, Long SR (2004) A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc Natl Acad Sci USA 101 16636–16641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastola DR, Pethe VV, Winicov I (1998) Alfin1, a novel zinc-finger protein in alfalfa roots that binds to promoter elements in the salt-inducible MsPRP2 gene. Plant Mol Biol 38 1123–1135 [DOI] [PubMed] [Google Scholar]
- Benlloch R, d'Erfurth I, Ferrandiz C, Cosson V, Beltran JP, Canas LA, Kondorosi A, Madueno F, Ratet P (2006) Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol 142 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbel A, Navarro C, Ferrandiz C, Canas LA, Beltran J-P, Madueno F (2005) Functional conservation of PISTILLATA activity in a pea homolog lacking the PI motif. Plant Physiol 139 174–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbel A, Navarro C, Ferrandiz C, Canas LA, Madueno F, Beltran JP (2001) Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J 25 441–451 [DOI] [PubMed] [Google Scholar]
- Bobb AJ, Eiben HG, Bustos MM (1997) Conserved RY-repeats mediate transactivation of seed-specific promoters by the developmental regulator PvALF. Nucleic Acids Res 25 641–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N, et al (2003) The sym35 gene required for root nodule development in pea is an ortholog of nin from Lotus japonicus. Plant Physiol 131 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitink J, Leger JJ, Guisle I, Vu BL, Wuilleme S, Lamirault G, Le Bars A, Le Meur N, Becker A, Kuester H, et al (2006) Transcriptome profiling uncovers metabolic and regulatory processes occurring during the transition from desiccation-sensitive to desiccation-tolerant stages in Medicago truncatula seeds. Plant J 47 735–750 [DOI] [PubMed] [Google Scholar]
- Chabaud M, de Carvalho-Niebel F, Barker DG (2003) Efficient transformation of Medicago truncatula cv Jemalong using the hypervirulent Agrobacterium tumefaciens strain AGL1. Plant Cell Rep 22 46–51 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Park JM, Yoo CM, Bahk JD, Cho MJ, Hong JC (1994) Isolation and characterization of STGA1, a member of the TGA1 family of bZIP transcription factors from soybean. Mol Cells 4 405–412 [Google Scholar]
- Cheong YH, Yoo CM, Park JM, Ryu GR, Goekjian VH, Nagao RT, Key JL, Cho MJ, Hong JC (1998) STF1 is a novel TGACG binding factor with a zinc finger motif and a bZIP domain which heterodimerizes with GBF proteins. Plant J 15 199–209 [DOI] [PubMed] [Google Scholar]
- Chern MS, Bobb AJ, Bustos MM (1996. a) The regulator of MAT2 (ROM2) protein binds to early maturation promoters and represses PvALF-activated transcription. Plant Cell 8 305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M-S, Eiben HG, Bustos MM (1996. b) The developmentally regulated bZIP factor ROM1 modulates transcription from lectin and storage protein genes in bean embryos. Plant J 10 135–148 [DOI] [PubMed] [Google Scholar]
- Citerne HL, Luo D, Pennington RT, Coen E, Cronk QCB (2003) A phylogenomic investigation of CYCLOIDEA-like TCP genes in the Leguminosae. Plant Physiol 131 1042–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citerne HL, Pennington RT, Cronk QC (2006) An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proc Natl Acad Sci USA 103 12017–12020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39 487–512 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Kloska S, Trevaskis B, Freund S, Altmann T, Udvardi MK (2002) Novel aspects of symbiotic nitrogen fixation uncovered by transcript profiling with cDNA arrays. Mol Plant Microbe Interact 15 411–420 [DOI] [PubMed] [Google Scholar]
- Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernie T, Ott T, Gamas P, Crespi M, et al (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev 20 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin GD, Krath BN, MacFarlane SA, Nicolaisen M, Johansen IE, Lund OS (2004) Virus-induced gene silencing as a tool for functional genomics in a legume species. Plant J 40 622–631 [DOI] [PubMed] [Google Scholar]
- Cubas P (2004) Floral zygomorphy, the recurring evolution of a successful trait. Bioessays 26 1175–1184 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38 366–379 [DOI] [PubMed] [Google Scholar]
- de Pater S, Katagiri F, Kijne J, Chua N-H (1994) bZIP proteins bind to a palindromic sequence without an ACGT core located in a seed-specific element of the pea lectin promoter. Plant J 6 133–140 [DOI] [PubMed] [Google Scholar]
- Dhaubhadel S, Gijzen M, Moy P, Farhangkhoee M (2007) Transcriptome analysis reveals a critical role of CHS7 and CHS8 genes for isoflavonoid synthesis in soybean seeds. Plant Physiol 143 326–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong ZC, Zhao Z, Liu CW, Luo JH, Yang J, Huang WH, Hu XH, Wang TL, Luo D (2005) Floral patterning in Lotus japonicus. Plant Physiol 137 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge-Laser W, Kaiser A, Lindsay WP, Halkier BA, Loake GJ, Doerner P, Dixon RA, Lamb C (1997) Rapid stimulation of a soybean protein-serine kinase that phosphorylates a novel bZIP DNA-binding protein, G/HBF-1, during the induction of early transcription-dependent defenses. EMBO J 16 726–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Yahyaoui F, Küster H, Amor BB, Hohnjec N, Pühler A, Becker A, Gouzy J, Vernié T, Gough C, Niebel A, et al (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136 3159–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Zhao Z, Tian Z, Xu S, Luo Y, Cai Z, Wang Y, Yang J, Wang Z, Weng L, et al (2006) Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA 103 4801–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firnhaber C, Puhler A, Kuster H (2005) EST sequencing and time course microarray hybridizations identify more than 700 Medicago truncatula genes with developmental expression regulation in flowers and pods. Planta 222 269–283 [DOI] [PubMed] [Google Scholar]
- Frugier F, Poirier S, Satiat-Jeunemaitre B, Kondorosi A, Crespi M (2000) A kruppel-like zinc finger protein is involved in nitrogen-fixing root nodule organogenesis. Genes Dev 14 475–482 [PMC free article] [PubMed] [Google Scholar]
- Gronlund M, Gustafsen C, Roussis A, Jensen D, Nielsen LP, Marcker KA, Jensen EO (2003) The Lotus japonicus ndx gene family is involved in nodule function and maintenance. Plant Mol Biol 52 303–316 [DOI] [PubMed] [Google Scholar]
- Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J (2005) DATF: a database of Arabidopsis transcription factors. Bioinformatics 21 2568–2569 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46 601–612 [DOI] [PubMed] [Google Scholar]
- Heard J, Caspi M, Dunn K (1997) Evolutionary diversity of symbiotically induced nodule MADS box genes: characterization of nmhC5, a member of a novel subfamily. Mol Plant Microbe Interact 10 665–676 [DOI] [PubMed] [Google Scholar]
- Hecht V, Foucher F, Ferrandiz C, Macknight R, Navarro C, Morin J, Vardy ME, Ellis N, Beltran JP, Rameau C, et al (2005) Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol 137 1420–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann AB, Lombardo F, Miwa H, Perry JA, Bunnewell S, Parniske M, Wang TL, Downie JA (2006) Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol 142 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46 183–192 [DOI] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michael A, Ellis N (1997) Unifoliata regulates leaf and flower morphogenesis in pea. Curr Biol 7 581–587 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Vieweg ME, Puhler A, Becker A, Kuster H (2005) Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol 137 1283–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JC, Cheong YH, Nagao RT, Bahk JD, Key JL, Cho MJ (1995) Isolation of two soybean G-box binding factors which interact with a G-box sequence of an auxin-responsive gene. Plant J 8 199–211 [DOI] [PubMed] [Google Scholar]
- Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K (2005) RARTF: database and tools for complete sets of Arabidopsis transcription factors. DNA Res 12 247–256 [DOI] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kim JC, Lee SH, Cheong YH, Yoo C-M, Lee SI, Chun HJ, Yun D-J, Hong JC, Lee SY, Lim CO, et al (2001) A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J 25 247–259 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Shimomura K, Hata S, Hirota A, Wu GJ, Kumagai H, Tajima S, Suganuma N, Suzuki A, Aoki T, et al (2004) Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res 11 263–274 [DOI] [PubMed] [Google Scholar]
- Lee H, Hur CG, Oh CJ, Kim HB, Park SY, An CS (2004) Analysis of the root nodule-enhanced transcriptome in soybean. Mol Cells 18 53–62 [PubMed] [Google Scholar]
- Lee JH, Kim WT (2003) Molecular and biochemical characterization of VR-EILs encoding mung bean ETHYLENE INSENSITIVE3-LIKE proteins. Plant Physiol 132 1475–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA (2000) Transcription of eukaryotic protein-coding genes. Annu Rev Genet 34 77–137 [DOI] [PubMed] [Google Scholar]
- Li X-P, Tian A-G, Luo G-Z, Gong Z-Z, Zhang J-S, Chen S-Y (2005) Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor Appl Genet 110 1355–1362 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman J, Yanofsky M (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404 766–770 [DOI] [PubMed] [Google Scholar]
- Lindsay WP, McAlister FM, Zhu Q, He X-Z, Drage-Laser W, Hedrick S, Doerner P, Lamb C, Dixon RA (2002) KAP-2, a protein that binds to the H-box in a bean chalcone synthase promoter, is a novel plant transcription factor with sequence identity to the large subunit of human Ku autoantigen. Plant Mol Biol 49 503–514 [DOI] [PubMed] [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohar DP, Sharopova N, Endre G, Penuela S, Samac D, Town C, Silverstein KA, VandenBosch KA (2006) Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol 140 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JH, Yan J, Weng L, Yang J, Zhao Z, Chen JH, Hu XH, Luo D (2005) Different expression patterns of duplicated PHANTASTICA-like genes in Lotus japonicus suggest their divergent functions during compound leaf development. Cell Res 15 665–677 [DOI] [PubMed] [Google Scholar]
- Manthey K, Krajinski F, Hohnjec N, Firnhaber C, Puhler A, Perlick AM, Kuster H (2004) Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Mol Plant Microbe Interact 17 1063–1077 [DOI] [PubMed] [Google Scholar]
- Markel H, Chandler J, Werr W (2002) Translational fusions with the engrailed repressor domain efficiently convert plant transcription factors into dominant-negative functions. Nucleic Acids Res 30 4709–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazarei M, Puthoff DP, Hart JK, Rodermel SR, Baum TJ (2002) Identification and characterization of a soybean ethylene-responsive element-binding protein gene whose mRNA expression changes during soybean cyst nematode infection. Mol Plant Microbe Interact 15 577–586 [DOI] [PubMed] [Google Scholar]
- Merchan F, Breda C, Hormaeche JP, Sousa C, Kondorosi A, Aguilar OM, Megias M, Crespi M (2003) A kruppel-like transcription factor gene is involved in salt stress responses in Medicago spp. Plant Soil 257 1–9 [Google Scholar]
- Mitra RM, Shaw SL, Long SR (2004) Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 101 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy P, Qutob D, Chapman BP, Atkinson I, Gijzen M (2004) Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol Plant Microbe Interact 17 1051–1062 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Miwa H, Imaizumi-Anraku H, Kouchi H, Downie JA, Kawaguchi M, Kawasaki S (2007) Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res 13 255–265 [DOI] [PubMed] [Google Scholar]
- Nag R, Maity MK, Dasgupta M (2005) Dual DNA binding property of ABA insensitive 3 like factors targeted to promoters responsive to ABA and auxin. Plant Mol Biol 59 821–838 [DOI] [PubMed] [Google Scholar]
- Nagano Y, Furuhashi H, Inaba T, Sasaki Y (2001) A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences. Nucleic Acids Res 29 4097–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura R, Ohmori M, Fujita H, Kawaguchi M (2002) A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proc Natl Acad Sci USA 99 15206–15210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Grady K, Goekjian VH, Nairn RT, Nagao RT, Key JL (2001) The transcript abundance of GmGT-2, a new member of the GT-2 family of transcription factors from soybean, is down-regulated by light in a phytochrome-dependent manner. Plant Mol Biol 47 367–378 [DOI] [PubMed] [Google Scholar]
- Ott T, van Dongen JT, Günther C, Krusell L, Desbrosses G, Vigeolas H, Bock V, Czechowski T, Geigenberger P, Udvardi MK (2005) Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol 15 531–535 [DOI] [PubMed] [Google Scholar]
- Perry JA, Wang TL, Welham TJ, Gardner S, Pike JM, Yoshida S, Parniske M (2003) A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol 131 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pwee KH, Webster CI, Gray JC (1994) HMG protein binding to an A/T-rich positive regulatory region of the pea plastocyanin gene promoter. Plant Mol Biol 26 1907–1920 [DOI] [PubMed] [Google Scholar]
- Qu L-J, Zhu Y-X (2006) Transcription factor families in Arabidopsis: major progress and outstanding issues for future research. Curr Opin Plant Biol 9 544–549 [DOI] [PubMed] [Google Scholar]
- Riechmann JL (2002) Transcriptional regulation: a genomic overview. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. The American Society of Plant Biologists, Rockville, MD [DOI] [PMC free article] [PubMed]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3 423–434 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Uribe L, O'Connell MA (2006) A root-specific bZIP transcription factor is responsive to water deficit stress in tepary bean (Phaseolus acutifolius) and common bean (P. vulgaris). J Exp Bot 57 1391–1398 [DOI] [PubMed] [Google Scholar]
- Sangwan I, O'Brian MR (2002) Identification of a soybean protein that interacts with GAGA element dinucleotide repeat DNA. Plant Physiol 129 1788–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195 [DOI] [PubMed] [Google Scholar]
- Shukla RK, Raha S, Tripathi V, Chattopadhyay D (2006) Expression of CAP2, an APETALA2-family transcription factor from chickpea, enhances growth and tolerance to dehydration and salt stress in transgenic tobacco. Plant Physiol 142 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KB (1998) Transcriptional regulation in plants: the importance of combinatorial control. Plant Physiol 118 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debelle F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791 [DOI] [PubMed] [Google Scholar]
- Starker CG, Parra-Colmenares AL, Smith L, Mitra RM, Long SR (2006) Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol 140 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma N, Yamamoto A, Itou A, Hakoyama T, Banba M, Hata S, Kawaguchi M, Kouchi H (2004) cDNA macroarray analysis of gene expression in ineffective nodules induced on the Lotus japonicus sen1 mutant. Mol Plant Microbe Interact 17 1223–1233 [DOI] [PubMed] [Google Scholar]
- Szczyglowski K, Amyot L (2003) Symbiosis, inventiveness by recruitment? Plant Physiol 131 935–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Ratet P, Mysore KS (2005) Insertional mutagenesis: a Swiss army knife for functional genomics of Medicago truncatula. Trends Plant Sci 10 229–235 [DOI] [PubMed] [Google Scholar]
- Tang Z, Sadka A, Morishige DT, Mullet JE (2001) Homeodomain leucine zipper proteins bind to the phosphate response domain of the soybean Vspb tripartite promoter. Plant Physiol 125 797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall AD, Turner L, Knox MR, Ambrose MJ, Ellis THN, Hofer JMI (2005) The mutant crispa reveals multiple roles for PHANTASTICA in pea compound leaf development. Plant Cell 17 1046–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SA, Hofer JMI, Murfet IC, Sollinger JD, Singer SR, Knox MR, Ellis THN (2002) Proliferating inflorescence meristem, a MADS-box gene that regulates floral meristem identity in pea. Plant Physiol 129 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud-Nissen F, Wu H, Richmond T, Redman JC, Johnson C, Green R, Arias J, Town CD (2006) Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J 47 152–162 [DOI] [PubMed] [Google Scholar]
- Thykjaer T, Finnemann J, Schauser L, Christensen L, Poulsen C, Stougaard J (1997) Gene targeting approaches using positive-negative selection and large flanking regions. Plant Mol Biol 35 523–530 [DOI] [PubMed] [Google Scholar]
- Tucker ML, Whitelaw CA, Lyssenko NN, Nath P (2002) Functional analysis of regulatory elements in the gene promoter for an abscission-specific cellulase from bean and isolation, expression, and binding affinity of three TGA-type basic leucine zipper transcription factors. Plant Physiol 130 1487–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uimari A, Strommer J (1997) Myb26: a MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J 12 1273–1284 [DOI] [PubMed] [Google Scholar]
- Vodkin LO, Khanna A, Shealy R, Clough SJ, Gonzalez DO, Philip R, Zabala G, Thibaud-Nissen F, Sidarous M, Stromvik MV, et al (2004) Microarrays for global expression constructed with a low redundancy set of 27,500 sequenced cDNAs representing an array of developmental stages and physiological conditions of the soybean plant. BMC Genomics 5 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HL, Li G, Chen R (2006) Fast neutron bombardment (FNB) mutagenesis for forward and reverse genetic studies in plants. In J Teixeira da Silva, ed, Floriculture, Ornamental and Plant Biotechnology: Advances and Special Issues, Vol 1, Ed 1. Global Science Books, Isleworth, UK, pp 629–639
- Wang Y-H, Li Y-D, Luo G-Z, Tian A-G, Wang H-W, Zhang J-S, Chen S-Y (2005) Cloning and characterization of an HDZip I gene GmHZ1 from soybean. Planta 221 831–843 [DOI] [PubMed] [Google Scholar]
- Webster CI, Packman LC, Pwee KH, Gray JC (1997) High mobility group proteins HMG-1 and HMG-I/Y bind to a positive regulatory region of the pea plastocyanin gene promoter. Plant J 11 703–715 [DOI] [PubMed] [Google Scholar]
- Winicov I (2000) Alfin1 transcription factor overexpression enhances plant root growth under normal and saline conditions and improves salt tolerance in alfalfa. Planta 210 416–422 [DOI] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Tang Y, Li F, Frank MB, Huang H, Dozmorov I, Zhu Y, Centola M, Cao W (2005) Protection against hydrogen peroxide-induced cell death in cultured human retinal pigment epithelial cells by 17beta-estradiol: a differential gene expression profile. Mech Ageing Dev 126 1135–1145 [DOI] [PubMed] [Google Scholar]
- Zabala G, Zou J, Tuteja J, Gonzalez DO, Clough SJ, Vodkin LO (2006) Transcriptome changes in the phenylpropanoid pathway of Glycine max in response to Pseudomonas syringae infection. BMC Plant Biol 6 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar JH (1999) Biostatistical Analysis, Ed 4. Prentice-Hall, Englewood Cliff, NJ
- Zhang J-Y, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, Wang Z-Y (2005) Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J 42 689–707 [DOI] [PubMed] [Google Scholar]
- Zhang J-Y, Broeckling CD, Sumner LW, Wang Z-Y (2007) Heterologous expression of two Medicago truncatula AP2 domain transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol Biol (in press) [DOI] [PubMed]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Fan W, Kinkema M, Li X, Dong X (1999) Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci USA 96 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tessaro MJ, Lassner M, Li X (2003) Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Rodriguez-Zas S, Aldea M, Li M, Zhu J, Gonzalez DO, Vodkin LO, DeLucia E, Clough SJ (2005) Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Mol Plant Microbe Interact 18 1161–1174 [DOI] [PubMed] [Google Scholar]
- Zucchero JC, Caspi M, Dunn K (2001) ngl9: a third MADS box gene expressed in alfalfa root nodules. Mol Plant Microbe Interact 14 1463–1467 [DOI] [PubMed] [Google Scholar]